Abstract
Glucose metabolism in mitochondria through oxidative phosphorylation (OXPHOS) for generation of adenosine triphosphate (ATP) is vital for cell function. However, reactive oxygen species (ROS), a by-product from OXPHOS, is a major source of endogenously produced toxic stressors on the genome. In fact, ATP could be efficiently produced in a high throughput manner without ROS generation in cytosol through glycolysis, which could be a unique and critical metabolic pathway to prevent spontaneous mutation during DNA replication. Therefore glycolysis is dominant in robust proliferating cells. Indeed, aerobic glycolysis, or the Warburg effect, in normal proliferating cells is an example of homeostasis of redox status by transiently shifting metabolic flux from OXPHOS to glycolysis to avoid ROS generation during DNA synthesis and protect genome integrity. The process of maintaining redox homeostasis is driven by genome wide transcriptional clustering with mitochondrial retrograde signaling and coupled with the glucose metabolic pathway and cell division cycle. On the contrary, the Warburg effect in cancer cells is the results of the alteration of redox status from a reprogramed glucose metabolic pathway caused by the dysfunctional OXPHOS. Mutations in mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) disrupt mitochondrial structural integrity, leading to reduced OXPHOS capacity, sustained glycolysis and excessive ROS leak, all of which are responsible for tumor initiation, progression and metastasis. A “plumbing model” is used to illustrate how redox status could be regulated through glucose metabolic pathway and provide a new insight into the understanding of the Warburg effect in both normal and cancer cells.
Keywords: Mitochondrial bioredox, glucose metabolic pathway, the Warburg effect, mitochondrial dynamics, glycolysis
Introduction
Increased aerobic glycolysis in cancer cells, known as the Warburg effect [1], is a hallmark of cancer and used as a diagnostic tool for solid tumors. However, the mechanism of the Warburg effect in cancer cells is still unclear because of its complexity. It is hypothesized that glycolysis is dominant in cancer cells as the result of mitochondrial damage or reduced OXPHOS capacity [2,3], but the functional mitochondria and upregulation of OXPHOS are revealed in breast cancer tissue [3] and ROS generation is shown to be positively associated with tumor metastasis [4]. Not surprisingly, the concept of ‘reverse Warburg effect’ has emerged owing to the fact that dominant glycolysis is found in adjacent stromal cells, but not in tumor cells [5].
Accumulating evidence across species indicates that the Warburg effect could be a universal phenomenon in normal robust proliferating cells, such as those of budding yeast, Drosophila, mammalian early embryonic cells, stem cells and primary spermatocytes, and even in virus infected cells. Thus, the understanding of the Warburg effect common in normal proliferating cells could be a breakthrough point helping to elucidate the Warburg effect in cancer cells. An early study using mitogen-activated rat thymocyte showed that resting cells use OXPHOS to produce 88% of ATP while proliferating cells use glycolysis to generate about the same amount of ATP [2] at the expense of massive glucose consumption. But ROS generated from OXPHOS in the resting cells was nearly abolished in proliferating cells by the use of glycolysis. It was suggested that adopting aerobic glycolysis in those cells could be a protective strategy against reactive oxygen species [2]. The budding yeast study showed that dominant glycolysis during DNA synthesis could reduce spontaneous mutation rate by coupling DNA replication with glycolysis in cell cycle [6] and revealed that although spontaneous mutations in mutants are avoidable under glycolysis, a high spontaneous mutation rate would result from those same mutants under glycolysis/OXPHOS oscillation. Therefore the authors suggested that glycolysis during DNA replication could protect genome integrity [6]. In addition, cyclinD1 expression during cell cycle G1/S transition correlated with low mitochondrial activity with reduced OXPHOS capacity through either inhibiting pyruvate dehydrogenase in fragmented mitochondria in vitro [7], or repressing nuclear respiratory factor 1 (NRF-1) in vivo [8], indicating an intrinsic strategy coupled with cell cycle to quench ROS during DNA replication in S phase.
The aerobic environment could generate mutations and inhibit growth of E.coli with a mutant gene in scavenging H2O2, but the same mutant strain could grow well under anaerobic condition [9]. Therefore, mitochondrion has been adopted by eukaryotic cells through symbiosis to serve not only as a ‘powerhouse’ for generating energy, but also a safety place for handling ambient oxygen during energy production. Genome wide analysis for the profiling of oxidative-stress-response genes in budding yeast revealed that 211 genes associated with mitochondrial respiratory function were overrepresented upon H2O2 assault, but very few of those genes were involved in the response to the assault by other oxidative stressors [10], indicating that cells are equipped with a specific defense system to recognize and respond to H2O2 as an endogenous oxidative stressor. Thus the mitochondrion in eukaryotic cell is more likely as a ‘nuclear power plant’, which is designed and constructed in such a sophisticated way that the ATP energy could be generated safely through OXPHOS, but ROS leak must be reduced to the minimal to avoid its genotoxic effect [11]. Mitochondrial respiratory system, an electron transport chain (ETC) on the inner membrane, is made up of five complexes (I-V) from multi protein subunits, which are directed by 13 mtDNA and 79 nDNA structural genes and 37 nDNA assembly factor genes [12]. Those subunits are physically contacted and arranged within a distance of ≤ 14 Å, which is a conserved distance from evolution for highly efficient electron transfer, called the ‘tunneling effect’ [13] to reduce ROS leakage. Thus the ETC complex I, as the most sophisticated complex involving 7 mtDNA- and 48 nDNA-encoded proteins in its constitution, is the most ROS leaking site [14]. Its dysfunction can lead to many chronic diseases such as neurodegenerative disorders, ageing and cancer.
Mitochondrial proton electrochemical gradient between mitochondrial matrix and inter membrane space across the inner membrane could create a membrane potential (∆Ψm), which is used for ATP synthesis from ADP and inorganic phosphate (iP) [3], or heat generation from brown adipose tissue (BAT) via uncoupling proteins [15]. As the electron carriers, mitochondrial ETC complexes, especially the complex I and III, are the major sources of endogenous ROS generation, depending on ∆Ψm, the NADH/NAD+ ratio, CoQH2/CoQ ratio and the local O2 concentration [16] and the integrity of ETC’s infrastructure as well [14]. Under non-stressful conditions, ROS emission level will be kept to a minimal by both intact mitochondrial structure and ROS scavenging system despite high respiration rate [17]. Excess mitochondrial ROS could be generated by damages either in respiratory function or structural integrity caused by mutations as seen in cancer cells [3]. For example, a nonsense mutation of G44A in NADH dehydrogenase (ubiquinone) Fe-S protein (NDUFS) gene of complex I is associated with increased oxidative stress in fibroblasts from patients with mutations [18]. This probably occurs as a result of the alteration of protein structure leading to the loss the ‘tunneling effect’ [13]. Aerobic organisms have a wide array of ROS scavenging mechanisms to reduce genome toxicity of ROS, including constitutive and inducible antioxidant components. Manganese superoxide dismutase (MnSOD) within mitochondrial matrix mainly catalyzes the formation of O2 and H2O2 from superoxide (O2 -•) generated from complex I, as only H2O2 could be diffused through membranes from mitochondria into cytosol, while Cu/Zn SOD in intermembrane space and glutathione peroxidase (GPx) in cytosol are responsible for catalyzing H2O2 into water [15]. Sublethal H2O2 could act as an independent signal to regulate cell function, such as the modulation of cell cycle progression in Drosophila developmental models [18-20], but H2O2 could be converted to hydroxyl radical (•HO), a highly oxidative reactive molecule via ‘Fenton chemistry’ with Fe2+, which could attack all of cellular components including DNA, proteins and lipids. The ROS-induced DNA damage includes single and double-stranded DNA breaks (DSB), base and sugar modifications, and DNA-protein crosslinks [21,22].
Studies on the correlation of mitochondrial respiratory capacity with its morphology and dynamics including fusion/fission and cytosol localization revealed that some mitochondrial morphologies, such as orthodox form and fission (fragmentation), were associated with low respiration capacity and linked to many human diseases, including cancer [23,24]. A pioneer study on mitochondrial ultrastructure coupling with respiration capacity revealed that three forms of mitochondria as condensed, orthodox and intermediate coexisted with different respiration capacity [25,26]. The condensed form of mitochondrion has high respiration capacity while the orthodox form has low respiration rate and the intermediate form has the capacity between condensed and orthodox forms. Interestingly, those forms with different respiration rate could be switched back and forth quickly under different metabolic conditions [27,28]. Mitochondria with different respiration capacity could represent different redox states as well. For example, the condensed form with high OXPHOS capacity could be termed as ‘oxidative state’ with more ROS generation, and the orthodox form with low respiration capacity as ‘reductive state’ with less ROS generation while the intermediate form could be a ‘neutral state’. As mitochondrion is the major source of endogenous ROS with potential genomic toxicity, reductive state created by glycolysis without ROS (the Warburg effect) and its synchronization with DNA replication could be vital to prevent spontaneous mutation [6], which is the top priority and far more important than the ‘economy’ of glucose catabolism for ATP production. In other words, the economic ATP production with ROS generation during respiration is exchanged to a less economic glycolysis with no ROS generation as a tradeoff for creating a transient reductive milieu during DNA replication to protect genome integrity [2,6]. Moreover, glycolysis could generate ATP as almost equally efficient as OXPHOS in a high throughput manner [29]. The correlation of metabolic flux with cell redox status has been recognized and led to the question “how alteration in glucose metabolism affects cytosolic and mitochondrial redox potential and ATP generation” [27]. Although alteration in mitochondrial energy production, termed mitochondrial bioenergetics, has a role in tumorigenesis as currently recognized [3], the homeostasis of redox status in normal proliferating cells coupled with glucose metabolism, the alteration of redox status derived from ‘reprogrammed energetics’ by mutations in cancer cells and its importance in the origin of cancer need to be elucidated. Instead of mitochondrial bioenergetics, we introduce a new concept of mitochondrial ‘bioredox’, a dynamics in regulation of cellular redox potential through glucose metabolic pathway via 1) glucose metabolic ‘flux rate’ (OXPHOS in mitochondria) and 2) glucose metabolic ‘flux route’ (glycolysis in cytosol and/or respiration in mitochondria) in cell survival and function. Glycolytic flux could be boosted by reducing OXPHOS rate [2,6], or inhibited by ROS through inactivating pyruvate kinase M2 (PKM2), an essential regulator of aerobic glycolysis in early embryo and cancer cells, by way of oxidizing the cysteine 358 of the protein [25]. On the other hand, OXPHOS flux could be boosted by hyperfusion of fragmented mitochondria with low respiration capacity through mitochondrial dynamics in normal cells [26], or inhibited by metabolic intermediates of Krebs cycle through mitochondria-cytosol shuttle systems such as citrate-α-ketoglutarate (α-KG) shuttle for reductive NADPH generation, or malate (MAL)-oxaloacetate (OOA)-aspartate (ASP) shuttle for impermeable NADH transport in energy rejuvenation [3,30]. Both shuttle systems involved in redox regulation could be interconnected through antiporters and both could regulate glucose metabolism, as high glucose could saturate MAL-OOA-ASP shuttle, decrease NAD+ level in cytosol and increase glycolysis with lactate production as a compensation process for less NAD+ in cytosol [31]. Meanwhile the alteration of citrate-α-KG shuttle system by mutation in isocitrate dehydrogenase (IDH) in cancer cells could further convert α-KG to 2-hydroxyglutarate (2HG) as an ‘oncometabolite’ with decreased NADPH [3]. The conversion of 2HG from α-KG may also reduce the efficacy of citrate-α-KG/MAL-ASP shuttles via antiporters, resulting in the shift to glycolysis in cytosol [3]. Calorie restriction therefore could keep those shuttle systems working efficiently partly through increased activity of malate dehydrogenase (MDH) in mitochondria for the regeneration of OOA from MAL, which in turn maintains glucose flux on the route of OXPHOS with high efficiency [32].
Homeostasis of redox status derived from glucose metabolism is vital to reduce oxidative stress and maintain genome fidelity during DNA replication
The budding yeast shares a high degree of conservation of cellular and molecular pathways with human cells and several features with cancer cells [33,34]. Therefore it has been a valuable tool not only for the study of the mechanism of human diseases, including cancer [35] but also for the discovery of cancer therapeutic agents [36]. The most important contributions to cancer research from budding yeast study are the pioneer discovery of cell division cycle genes (CDC-genes) in the regulation of cell cycle in early 1970s [37] and the study of the coupling between the yeast metabolic cycle and cell division cycle [38]. Yeast cells without mitochondrial DNA (rho0) would have low genome instability only under fast dividing condition, but would have high genome instability under non-dividing condition [39], supporting the notion that glycolysis without ROS generation in fast dividing cells is protective for the genome stability. Glycolysis is therefore preferred for robust proliferation when cells are growing in high glucose medium owing to the Crabtree effect [40]. But under limited nutrient condition such as growing in phosphate limited medium, the majority of the yeast cells would stop ‘buddying’ with only G1 phase coupling with oxygen consumption oscillation, but no cell division occurred [41]. Interestingly, after brief starvation, followed by growing in low glucose medium, a specific scenario created in laboratory to study cell cycle and metabolism, the buddying yeast would resume cell division cycle which is synchronized with oxygen consumption oscillation [6]. It was revealed that low or deceased oxygen consumption phase called reductive building (RB) was coupled with DNA replication and associated with dominant glycolytic metabolism while G2/M phases proceeded during oxygen consumption phase including reductive recharge phase (RC) and oxidative phase (OX) [6], indicating that glycolysis is synchronized with DNA replication only during cell cycle. Yeast mutants with aberrations in cell cycle, especially with fast growth rate, could increase the spontaneous mutation with oxygen consumption oscillation [6]. But strikingly, no extra spontaneous mutation was found for the same mutants if growing solely under glycolysis. This suggested that glycolytic pathway could protect genome integrity [6].
Interestingly, the spontaneous mutation rate is negatively correlated with the cell-cycle length (cell growth rate) [6]. For example, mutant strains with similar growth rate to the wild type (WT) would have a similar spontaneous mutation rate. But the shorter the cycle length (faster growth) is, the higher the mutation rate will be with oxygen consumption oscillation. It was shown that the mutant with the shortest cell cycle harboring mutation of Sic1, a cyclin-dependent kinase (CDK) inhibitor, would have the highest mutation rate [6]. This phenomenon could be explained by another yeast study showing that the DNA replication would synchronize more closely with RB phase with glycolysis in cells with slow growth rate (longer cell cycle length) but the dynamic of DNA replication shifted right away into high oxygen consumption phase in cells with fast growth rate [41]. We speculate that the faster the growth rate is, the further the shift away from reductive environment (glycolysis) to high oxidative stress environment (OXPHOS) will be during DNA synthesis, leading to higher spontaneous mutation rate. The fastest growing cells would suffer the most oxygen assault during their DNA replication and result in the highest mutation rate, which is the case in Chen’s observation [6]. Moreover, the fast-growing cells have less cells arrested in G1/G0 with more cells entering dividing cycle under oxidative stress [42]. These cells ignore the genomic toxicity of ambient oxygen [9] and therefore are more vulnerable to mutagenesis. In other words, slowing down cell growth rate could have protective effect against spontaneous mutagenesis because DNA replication would be synchronized more closely with reductive environment. Genome-wide transcription oscillation analysis revealed that cells with slow growth rate would have 67% less gene transcripts during high oxygen consumption phase [42], owing to low oxidative stress in cells with slow growth rate [43]. On the other hand, in fast growing cells, a total of 494 (only 25%) gene transcripts in S phase showed up compared to that in slow growing cells. Those 25% gene products might represent the ‘core’ or ‘short pulse’ gene products necessary for DNA replication and cell division [41,42]. However, the remaining 1485 (75%) gene transcripts in S phase in slow growing cells could be critical in protecting their genome integrity as one gene transcript called “DNA-dependent DNA replication” [42] could be vital for genome fidelity in daughter cells.
It has been generally accepted that the Warburg effect is one of the hallmarks in the development of the early mammalian embryo [44], especially at pre-implantation stage [45], in self –renewing stem cells [46], and even in non-mammalian and human tumor virus infection [47-50]. In early embryo development, redox status is tightly and precisely controlled by changes in OXPHOS capacity through the regulation of mtDNA replication, mitochondrial biogenesis, mitochondrial dynamics and cytosol localization in different stages of embryo proliferation and differentiation [51]. For example, following fertilization and up to implantation when cell division begins, the total number of mitochondria within each blastomere decreases owing to dilution, with little mitochondrial biogenesis occurring at this stage [51]. During the oocyte maturation, each cell has about 75,000-100,000 mitochondria, which form clusters located around the nucleus. These mitochondria have shown an elevated capacity of OXPHOS based on the high levels of mitochondrial transcription factor A (mtTFA) and mtDNA polymerase subunit gamma expression for mitochondrial biogenesis. Starting from pre-implantation, the mitochondria decrease in numbers after each cell division to about half in the inner cell mass and to only 20 per cell in the embryonic stem cells at the blastocyst stage. Indeed, even new mitochondrial generation is blocked by low expression of mtTFA during the two-cell stage, then the mitochondrial biogenesis increases in the eight-cell stage, reaching a maximum at the morula stage [51]. Strikingly, mitochondria start to gain regenerating capacity first at the trophectoderm, then throughout the entire embryo [51]. This sequence indicates that OXPHOS is prioritized for cells involved in the functioning of placenta formation in the trophectoderm for transporting nutrition, while low number of mitochondria in the inner cell mass is maintained for cell mass production during organ generation.
The mtDNA replication is also controlled at the transcriptional level during mouse embryo development. It was shown that even when transcriptional factors were abundant, mtDNA replication did not occur until the blastocyst stage [52], suggesting that a strategy of reducing oxidative stress is restrictedly applied to this stage. It has also been reported that glycolysis is dominant in less-formed cells or stemness cells to boost the formation of invadopodia as seen in metastatic cells with lamellipodia/pseudopodia features [53] or in earliest primordial germ cells (PGCs) with amoeboid appearance [54]. In other words, glycolysis or reductive state is associated with functionless and formless of cells [1], such as early embryo, stem cells and cancer cells. A study using cultured human osteosarcoma cells (143B) showed that the mitochondrial shape and size were synchronized with the cell cycle and oscillated between reticular and fragmented forms during cell cycle [55]. Using fluorescence in situ hybridization (FISH) imaging of mtDNA combined with mitochondrial staining, it showed that 60-75% of cells with fragmented mitochondria were in S phase and that reticular mitochondria increased during mitosis and in G1 phase [55], confirming the findings in buddying yeast study [6]. Interestingly, the whole copy set of 16 kb of mtDNA that encodes the 13 polypeptides for the OXPHOS system is in the form of nucleoids and segregated into each fragmented mitochondrion during the fission process possibly with one copy of nucleoid for every fragment [55]. This suggests that the mitochondrial mass in reticular shape could be made up of fragmented mitochondria that still have potentially all the functional components for OXPHOS, which could be the foundation of mitochondrial dynamics in fusion/fission to modulate cellular metabolism and stress [56]. Mitochondrial bioredox was also coupled with spermatogenesis [57] in which mitochondria in spermatogonial cells undergoing DNA replication adopted the orthodox form with reduced OXPHOS/ROS generation. But beyond this stage, mitochondria changed to condensed form with high OXPHOS capacity to meet the needs in different stages of meiosis [57].
Mitochondrial bioredox is governed by genome-wide transcriptional expression clustering and mitochondrial retrograde signaling
Mitochondrial dynamics, especially mitochondrial fusion/fission and the distribution in cytosol could be important in regulating redox status and energy production of cells [24]. The alteration of mitochondrial dynamics could be the etiology of many human diseases [58]. Genetic studies have revealed that mitochondrial fusion process is governed by three guanosine triphosphate hydrolase enzyme (GTPase) which include two dynamin-like proteins mitofusin-1 (MFN1) and mitofusin-2 (MFN2), and another optic atrophy protein 1 (OPA1) located at different cellular compartments [58]. OPA1 is located in the inner membrane, whereas MFN1 and MFN2 are both located in the outer membrane for mitochondrial fusion. Other proteins, including fission 1 protein (FIS1) located in the outer membrane as a receptor and dynamin related protein 1 (DRP1) located in cytosol, are involved in mitochondrial fission [58]. Disruption of mitochondrial fusion process is associated with neurodegenerative diseases such as Parkinson’s, Alzheimer’s and Huntington’s diseases [59], probably by the alteration in both bioredox and bioenergetics of cells. Indeed, a study of virus infection in neuron revealed that virus could high-jack the moving ability of mitochondria along microtubule and down-regulate OXPHOS capacity with increased fragmented mitochondria [59] to create a reductive milieu for virus replication. Mitochondrial fusion could generate a functioning mitochondrial population that could survive from autophagy during starvation [60]. Disruption of mitochondrial fusion/fission processes, either by overexpression of MFN2 or knockdown of DRP1, would increase OXPHOS capacity to prevent cell-cycle progression in lung cancer cells [61], probably due to change in oxidative state and increase in ROS leak, whereas fragmented mitochondria, either genetically induced or obesity-related, were associated with dysfunctional mitochondria as seen in insulin resistance and aging [62,63]. These dysfunctional mitochondria are probably in excessive reductive state. Moreover, decreased ATP synthesis, either by inhibiting ETC complexes activity via oligomycin in vitro [60] or by altering protein crystal structure via site-directed mutation on cytochrome bc1 complex [64], could delay S phase entry of cells through excessive ROS leak, which in turn disturbs homeostasis of redox status during cell division cycle.
The mitochondrial redox proteins with known crystal structure, such as cytochrome bc1, cytochrome C oxidase and cytochrome oxidase, were collected for the determination of electron transfer efficiency. Interestingly, the study revealed that as close as 14 Å or less apart in distance, these redox centers could have very high free energy optimized for tunneling rates in the 1013-1017 per second range, called ‘tunneling effect’ [13]. This magic distance was conserved from evolution as this distance was applied to all of redox centers collected with known crystal structure at that time, indicating that the compacted or more closed redox centers could be vital for highly efficient electron transfer to minimize ROS leakage. It could explain the finding that dark electron dots precipitated in matrix and along cristae of inner membrane in condensed form could represent the compactness/interaction of enzymes of Kreb’s cycle in matrix and ETC complexes on cristae to prevent ROS leakage during high capacity of OXPHOS [13,27,28]. Indeed, genetic study of the cytochrome bc1 showed that changing the length of the flexible linker region in the iron-sulfur redox region by adding one or two alanine residues could reduce this ubiquinol-cytochrome C reductase activity by 50% and 90% respectively [64]. This result revealed the correlation of mitochondrial ROS leak with the integrity of mitochondrial structure and function and supported the notion that any findings in excessive ROS generation could be a sign of damage of mitochondrial structure and/or function especially in cancer tissue or cancer cell lines with functional mitochondria or with high OXPHOS [4,65]. The tunneling effect could also explain the fact that most of intracellular ROS comes from complex I [66] because it is composed of 55 different subunits with unusual and unique coupling mechanism via long range conformational changes [65], which could be a potential ‘heel of Achilles’ and source of ROS leak.
What is the driver of mitochondrial bioredox coupling with cell metabolism and function, especially during DNA replication? A yeast study showed that a genome-wide oscillation in transcriptional clustering occurred at three nearly equally spaced intervals within the cell cycle and correlated with oxygen consumption cycle/metabolic cycle [67]. Using microarray analysis on a total of the 5329 genes expressed during cell cycle in buddying yeast, it was found that almost half of the genes were expressed under high oxygen consumption phases with 2250 (42%) in RC and 650 (12%) in OX phase, whereas 2429 (46%) genes were expressed in RB phase. Moreover, genes expressed in OX phase were highly dynamic with higher amplitude than genes expressed in RC and RB phases [67]. Those results support the notion that most of those genes “with constant low expression level throughout cell division cycle” could represent G1 cycling in quiescent cells [41], while genes in OX phases could represent ‘short pulse’ [42] along with concentrated gene expression, such as cyclins [7,8], amino acid synthesis and RNA metabolism for DNA synthesis and cell division.
Based on these observations, we hypothesized that gene expression in cell cycle could be stratified into two parts: ‘G1/G0 cycling’ as background and ‘S/G2/M’ as pulse. The background of G0/G1 cycling was in quiescent cells and regulated by the majority of gene transcription (88%). Amazingly S phase pulse triggered cell division on the top of G1/G0 background by only those 12% of the genes which could be involved in cell division signaling, such as nutrition, redox status, oncogenes, and the process of “concentrating the expression of biosynthetic genes and active metabolism into short pulses” [41]. This is demonstrated by a positive correlation of over nutrition and cancer incidence in human society [50].
Using a 39S mitochondrial ribosomal protein L10 (MRPL10) as a probe to identify genes clustering with metabolism in cell cycle, it was found that 73 clustered genes for nuclear-encoded mitochondrial ribosomes were highly expressed shortly after cells ceased respiration in RB phase and reached the peak expression at the end of DNA synthesis, indicating that ‘grouping’ of those genes could be vital during DNA synthesis to quench ROS generation and protect DNA integrity [68]. Interestingly gene transcripts in RC phase, prior to OX phase, were correlated with autophagy, peroxisome, and ubiquitin/proteasome, suggesting that ‘clean-up’ could be the major tasks in the preparation for the transition from redox state dominant with glycolysis to oxidative state of OXPHOS [68]. Proteasome-mediated proteolysis for defined target proteins is an important regulatory mechanism to control various pathways in eukaryotic cell, such as in DNA repair mechanism [69]. A mutation in the yeast proteasome gene RPN11/MPR1 resulted in cell-cycle arrest, overproduction of nDNA and mtDNA, and fragmentation of the mitochondria [70], indicating that the proteasome plays a vital role in transition process during cell cycle. Indeed, prolonged mitochondrial fusion with over oxidative stress in mutant could result in chromosome misalignment during mitosis [26]. A similar study of Dacapo, a cyclin-dependent kinase inhibitor, in Drosophila embryogenesis, revealed that timely ‘turn-on’ and ‘turn-off’ of this protein expression were critical for normal cell division, as the loss-of-function mutation resulted in its prolonged expression causing inappropriate cell-cycle entry and lethality [71]. Moreover, proteasome might play a role in DNA damage repair (DDR) as it was enriched at the sites of DNA damage as a part of DDR system [69]. Therefore proteasome inhibitors are widely used as targeted therapy in treatment of cancer [72].
Strikingly, the Warburg effect was also observed in virus infection in host cells [47-50] as virus DNA replication was found to be synchronized with reduced H2O2 level, increased glycolysis and glucose consumption, increased lactate production and enzyme activity of glucose-6-phosphate dehydrogenase (G6PDH) and PPP [47]. Human Hepatitis C virus (HCV) infection could increase mitochondrial fission and mitophagy to reduce HCV- induced host cell apoptosis [48] and create a reductive environment for virus replication. Another example is that high risk human papillomavirus protein E2, HPV-18 E2 and 16 E2, could localize to the mitochondrial inner membrane and modify the cristae morphology, leading to the increase of glycolysis and stabilization of hypoxia-inducible factor (HIF) [49]. Taken together, these results indicate that avoiding ROS attack during DNA replication could be a conserved universal strategy from evolution by all living organisms, including virus, through adoption of glycolysis for DNA replication to protect their genome integrity.
Unbalanced mitochondrial bioredox along with the alteration of glucose metabolism owing to mtDNA and nDNA mutation is responsible for the Warburg effect in cancer cells
The Warburg effect in cancer cells could be partially explained by the facts that cancer cells share many common properties in gene expression with normal embryonic cells especially at very early stage [1], such as the amoeboid primordial germ stage in the yolk sack [54] or pre-implantation stage [73]. The embryonic isoform of pyruvate kinase (PKM2), which catalyzes the last step of glycolysis to yield pyruvate, is highly expressed in tumors, yet is absent from adult tissues except in adipocytes [74]. Dominant glycolysis with reduced mitochondrial respiration has been widely observed in patients with different kinds of solid tumors [30,75]. Using a trans-mitochondrial approach, the mitochondria were switched among different tumor cell lines while the cytosol of the cells was kept intact (cybrid) [76]. Aggressive osteosarcoma cell line (143B TK-p0 cells) cybrid harboring mitochondria from benign breast epithelial cells (MCF10A) showed reduced ability in metastasis, reduced ROS generation, increased complex I expression, and reduced cell proliferation and tumorigesis in nude mice, while the cybrid with mitochondria from moderately metastatic cells showed lower capacity in tumorigenesis in nude mice [76], suggesting a role of mitochondria in tumorigenesis. This role of mitochondria was probably the result of mitochondrial retrograde signaling. In another study, the ability of tumorigenesis of a prostate cancer cell line PC3, harboring a mutation in cytochrome oxidase subunit I (COI) with reduced OXPHOS and increased ROS leak, could be neutralized by harboring normal mitochondria in nude mouse model [77], indicating the intact structure and function of mitochondria are crucial in preventing tumorigenesis.
Interestingly, mitochondria from highly metastatic tumor cells could not transform non-transformed mouse NIH3T3 cells into tumor cells, even with defect on OXPHOS capacity due to a mutation at complex I [76], implying that the interaction of mitochondria and cytosol compartments played a role in tumor initiation. Microarray analysis in a cybrid of highly metastatic breast cell line harboring non-cancerous mitochondria revealed that normal mitochondria could decrease the expression of oncogenes such as RAS, HER2, SRC, etc and increase the expression of tumor suppressor genes such as RB1, PTEN and VHL [78], again demonstrating the importance of intact mitochondrial in cancer prevention. Importantly, the ROS generating capacity but not excessive glycolysis, could be responsible for tumor metastasis, as only enhanced glycolysis without ROS generation in hybrid harboring mitochondria without mtDNA (mt∆) cannot trigger metastasis [79], supporting the hypothesis that alteration of mitochondrial bioredox with excessive ROS could be the driver for tumorigenesis and metastasis.
ROS plays a vital role in antimicrobial host defense as well as signal transduction in inflammation process [80]. During acute inflammation process, it is critical to stop microorganisms’ invasion via excessive ROS generated from mitochondria of damaged cells and peroxisome from neutrophils. This strategy is realized through metabolic alterations by increasing OXPHOS, fatty acid oxidation and UCPs [80]. However chronic inflammation process with sustained ROS leak could be tumorigenic and responsible for many types of cancers [81]. Indeed, H2O2 pulse could induce DNA replication even under oxidative stress environment in yeast study [6]. Meanwhile excessive and sustained ROS could act as a mutagen for initiating cancer by creating ‘gain-of-function’ mutations in oncogenes for proliferation and metastasis [82] or ‘loss-of-function’ mutations in tumor suppressor genes, because lack of mutation variation, called genostasis, could prevent cells from adapting to stress [80], due to less available choices. It is highly possible that excessive ROS from damaged mitochondria could play a critical role in cancer initiation and evolution [81] by generating a vast diversity of mutations, some of which are oncogenic mutations such as mutations in oncogenes KRAS and PI3K [81,83]. Tumor cells could be nourished by glycolysis for survival, recurrence and metastasis.
Mutation of mtDNA and nDNA are responsible for the alteration of mitochondrial redox potential [31] and reprogramming of bioenergetics [4]. How does mutation be generated in the first place even with intact mitochondria with normal function? The key could be in the synchronization of DNA replication with reductive state which is coupled with glucose metabolism, cell growth rate and cell cycling [41-43]. Budding yeast study showed that glucose concentration in the medium would influence the growth rate, as 40% of glucose reduction could slow down cell growth rate from 2 hour to 4 hours [43]. The slower the growth rate (longer the cell cycle length), the more closed the synchronization of DNA replication with reductive environment [43]. And more genes were expressed during DNA synthesis to improve genome fidelity [42]. On the other hand, the faster the growth rate (shorter the cell cycle), the farther away the DNA synthesis from reductive phase [43]. As a result, less genes were expressed in DNA synthesis and more cells were entering cell cycle under high oxidative stress [42]. All of these scenarios lead to increased vulnerability of spontaneous mutations [6], ultimately causing mutations critical in cell cycle control, such as TP53 mutation in cancer [84]. The effect of calorie restriction on tumor prevention was demonstrated in mouse liver regeneration model in vivo [85]. It showed that under 40% less calorie intake, newly generated liver cells showed a shorter time for both DNA synthesis (accelerated or fast pass though S phases) and low protein expression of p21, cyclin A and E2F1, compared with ad libitum-fed mice [86]. Clinical study showed that low E2F1 expression could favor positive outcome of breast cancer patients [87], while the fast pass-through S phase could be beneficial in the prevention of spontaneous mutation, which was demonstrated in yeast study [7,42]. The quick pass of S phase under calorie restriction could represent a new mechanism among others to reduce spontaneous mutation rate, including s phase closely synchronized with reductive milieu [42], higher level of fidelity of DNA polymerases and lower level of mitochondrial membrane potential for less ROS leakage [88]. Moreover, highly efficient generation of ATP via glycolysis makes it possible for DNA replication to complete faster and for the time of exposure in unavoidable oxidative stress to shorten during S phase to prevent spontaneous mutation. In the above studies, no data were collected in mutation rate in newly generated liver tissue under calorie restriction or with ad libitum diet mice. Meanwhile, high glucose level could saturate MAL-OOA-ASP shuttle to shun glucose metabolic flux rout from OXPHOS to glycolysis as the Warburg effect [89].
Mechanisms involved in the homeostasis of mitochondrial bioredox which couples with glucose metabolic pathway and cell function
It has been revealed that the condensed form of mitochondria with high respiration rate is associated with a phenomenon of ‘contraction of matrix’ which reduces the volume of matrix by 50% compared to the orthodox form [27,28]. How can the volume of mitochondrial matrix regulate mitochondrial respiration capacity or mitochondrial bioredox? Studies of mitochondrial membrane ion channels revealed that four potassium channels are located in the inner membrane [85,90]. Among those potassium channels, an ATP-sensitive potassium channel (mitoKATP) could be regulated by ATP status, similar to the potassium channel on the pancreatic cell membrane for insulin secretion [4,85,91]. MitoKATP could be regulated by redox states as well. Oxidative agents such as H2O2 and O2 - could activate this channel [90,92], while molecules such as high level of NADPH and ATP could close the channel to stop K+ influx from entering matrix from inner membrane space [93], leading to the retention of K+ within the intermembrane space, which in turn results in increased osmotic pressure and swelling of intermembrane space and cristae [90]. At the same time, swelling of the intermmebrane space causes the matrix being squeezed or contracted to about 50% of its original volume as seen in condensed form [27,28] while low ATP and high ROS could active/open mitoKATP and let K+ influx into matrix via ionic concentration gradient to restore the volume of matrix to its original through osmotic pressure by water as seen in orthodox form [93]. This phenomenon points to the possibility of switching from condensed form to orthodox form within minutes [25,26] just through potassium flux-induced osmotic pressure and water. It was shown in one study that the orthodox form with low respiration capacity and low ∆Ψm could act as cardioprotective preconditioning in heart ischemia-reperfusion injury model to prevent ROS-induced damage [94]. In cancer cells, low mitoKATP expression was associated with low respiration and high ∆Ψm leading to the resistance of apoptosis [95,96]. Dichloroacetate (DCA), an inhibitor of mitochondrial pyruvate dehydrogenase kinase (PDK), an inhibitor of mitochondrial pyruvate dehydrogenase kinase (PDK), which phosphorylates and inhibits the activity of pyruvate dehydrogenase complex to prevent pyruvate entry into mitochondria for OXPHOS, could increase respiration, ROS generation and expression of Kv1.5 of potassium channel in plasma membrane and decrease proliferation and induce apoptosis in cancer cells, but not in normal cells [96]. MitoKATP channel opener, diazoxide and pinacidil at the pharmacological dosage, could slightly increase K+ ion flux into matrix with reduced membrane potential and respiration, leading to a significant increase in matrix volume which is “sufficient to reverse matrix contraction caused by oxidative phosphorylation” [97].
How could OXPHOS capacity be regulated simply by squeezing the volume of mitochondrial matrix? It revealed that condensed form of mitochondria with high respiration feature would accompany a phenomenon of high electron precipitate in matrix where the enzymes of Krebs cycle locate, and in the cristae with ETC complexes. SIRT3, a NAD+-dependent deacetylase in mitochondria, is an enzyme to remove acetyl molecule from acetylated proteins. And its activation could reduce ROS leakage [92], via interaction with ETC complex I subunits [95]. The effect of SIRT3 on reducing ROS from mitochondria could be demonstrated by its effect on improving protein-protein interaction [98,99], probably through increasing compactness among enzymes in matrix via ‘spatial factor’ which was revealed from the enzyme activity in bacterial carbon fixation [13,100]. Increased interaction among ETC complexes on cristae via ‘spatial factor’ could prevent ROS leakage to trigger ‘tunneling effect’ [14].
The process of DNA replication is vulnerable for genome integrity itself, in term of spontaneous mutagenesis, owing to exogenous and endogenous ROS assault [9,10,78]. Therefore balanced redox status through glucose metabolic pathway could be crucial for genome integrity in daughter cells [6]. A pioneer isotope labeling study on fertilized rabbit ovum from 1-day to 6-day stage (before the blastocyst stage and after) revealed that glucose was metabolized mainly through pentose phosphate pathway (PPP) prior the blastocyst stage, whereas glycolytic and respiration were established after the blastocyst stage. This study also revealed that no respiration could be detected in 6-day blastocysts [101], indicating that reductive milieu was critical to ensure the fidelity of DNA replication, especially within the first few copies of DNA during the very early stage of embryo development [1]. The 13C-metabolic flux analysis was used for probing intracellular metabolic fluxes [102] and showed in Chinese hamster ovary (CHO) cells that in non-growing cells, more than 97% of consumed glucose was first diverted through PPP with a high NADPH production as redox equivalents. Then the metabolic flux continued into pyruvate entering Krebs cycle with little or no lactate production [103]. On the other hand, in the dividing cells the glucose flux was diverted mainly toward glycolysis for pyruvate and lactate production, but little was diverted to PPP and OXPHOS [2,103]. Similar results were obtained from a study in human glioblastoma cells showing that PPP was upregulated with acute oxygenation but down-regulated by glycolysis while glycolysis itself was upregulated with hypoxia [104]. It seems that the homeostasis of redox status is maintained and coupled intrinsically by pairing OXPHOS to generate ROS as oxidative stress with PPP to produce NADPH as reductive equivalent. This pairing has been selected from evolution to protect cells from oxidative damage.
It was revealed that cancer cells, probably normal cells as well, cannot survive without glucose [105], therefore, the glucose metabolic pathway could be a cellular metabolic ‘magic machine’, consisting of PPP and glycolysis in cytosol and respiration in mitochondria, and coupling with redox status and cell function. Glucose molecule could be one of few critical native players of the ‘game’. When a glucose molecule enters this machine, it is not simply for energy generation at the end, but it could generate different ‘side effects’ before entering OXPHOS at the end in mitochondria. Glucose molecule might ‘stop’ or trap at glycolysis (2 ATP/glucose) by avoiding ROS generation during DNA replication in S phase, or ‘enter’ into mitochondria through OXPHOS for thorough ATP generation (36 ATP/glucose) for cellular differentiation and functioning where energy is much needed. In other words, energy generation in the form of ATP at the end is the consequence but the ‘side effect’, such as creating reductive state via glycolysis is the driver. Maslow’s hierarchy of needs [106] represent the order, by which, survival located in the bottom is the first need to meet in order to fulfill other physiological needs. Any bio-organism will take whatever it takes to survive, then thrive, which is an excellent example of homeostasis of bioredox status adopted during cell cycle to protect genome integrity.
Based on the facts that iron-sulfur (Fe-S) clusters has been used as redox centers for anaerobic metabolism in anaerobic time [107] and extended into ETC of mitochondria in aerobic world [108], it is possible that the sequence of glucose metabolic pathway from PPP and glycolysis in cytosol to OXPHOS in mitochondrion could be another example of “ontogeny repeating phylogeny” at the cellular metabolic level. This metabolic recapitulation with DNA synthesis, either for cell duplication or virus replication in host cells, must be hooked intrinsically by evolution, programmed into the genome in all of living organisms and will repeat again and again whenever a life needs to renew.
Closing remarks
The mitochondrial bioredox derived from glucose metabolic pathway and coupled with cell function is shown in a plumbing model, which would illustrate our understanding of the Warburg effect in normal quiescent and proliferating cells as well as in cancer cells shown in Figure 1A-C.
Figure 1.
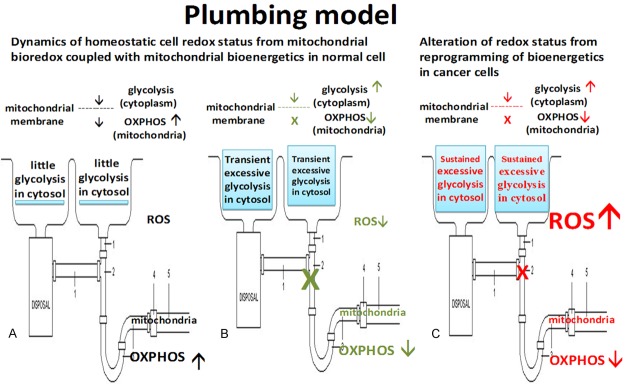
A plumbing model illustrate the understanding of the Warburg effect in (A) normal quiescent cells; (B) normal proliferating cells and (C) cancer Cells. Glucose metabolic transiently shifting from (A) and (B) represents the homeostasis of mitochondrial bioredox coupling with bioenergetics in normal proliferating cells. However, cancer cells have excessive ROS as well as sustained glycolysis and reduced OXPHOS, owing to the alteration of mitochondrial bioredox derived from reprogramming of bioenergetics from mutation.
Figure 1A represents the oxidative state of mitochondrial bioredox derived from glucose metabolic pathway in normal quiescent cells, or healthy life style such as low calorie intake and aerobic exercise, which is featured with high efficiency of OXPHOS with low ROS leak and little glycolysis in cytosol [109]. While Figure 1B could represent the reductive state of mitochondrial bioredox either in normal proliferating cells during DNA replication, which is featured by reduced OXPHOS, but transient increase in glycolysis with little ROS to protect genome integrity, as the Warburg effect in normal cells [2,7]. The transition between Figure 1A and 1B represents the homeostasis of mitochondrial bioredox regulating cell redox status, which is governed by genome-wide gene transcription and retrograde mitochondrial signaling coupled with cell function. However Figure 1C alone could represent the Warburg effect in cancer cells or unhealthy life style with over nutrition or lack of aerobic exercise. It is featured by excessive ROS leak plus reduced OXPHOS and sustained increase in glycolysis, an alteration of mitochondrial bioredox derived from reprogrammed bioenergetics from mutation of mtDNA and nDNA [1,3]. Importantly, this model shows that OXPHOS of mitochondrion as an evolutionary youngster at the end of glucose metabolic pathway, could regulate upstream of glycolytic flux effectively through a ‘reverse bottle-neck effect’, or ‘funnel effect’. The transition between OXPHOS and glycolysis could be a dynamic and transient event coupled mainly with DNA synthesis in S phase, but not G2/M phases to avoid ROS and protect genome integrity. However, the alteration of mitochondrial bioredox and bioenergetics make cancer cells trapped into Figure 1B, which could be rescued by simply reviving the OXPHOS capacity [96].
In summary, our hypothesis in understanding the Warburg effect could be high-lighted as the following: 1) Glycolysis as ancient metabolic pathway has three unique features that could be beneficial to protect genome integrity during DNA replication by a) by providing fast ATP generation in a simple and efficient manner as “ATP production by glycolysis can be more rapid than OXPHOS ” to support DNA replication [29,110]; b) by creating reductive milieu without ROS to reduce spontaneous mutation rate [2,7]; c) by providing NADPH and macromolecules for DNA synthesis with PPP [111], all of which could be critical to protect genome integrity; 2) Avoiding ROS generation during DNA replication is the top priority in all living organisms to protect genome integrity which is conserved from evolution for all living organisms; 3) Dynamic homeostasis of cellular redox status during cell division in healthy cells is governed by genome wide transcription clustering and mitochondrial retrograde signaling through mitochondrial bioredox coupled with bioenergetics and cell cycle; 4) The Warburg effect in cancer cells is the alteration of mitochondrial bioredox derived from reprogramming of mitochondrial bioenergetics by mutations [3], featured by excessive ROS, sustained with increased glycolysis and reduced OXPHOS, leading to tumorigenesis, progression and metastasis. Therefore, our hypothesis emphasizes on the importance of mitochondrial bioredox coupling with bioenergetics for DNA replication and provides a new insight into the Warburg effect in proliferating normal cells as well as in cancer origin, treatment and prevention.
Acknowledgements
This work was supported by SPORE grant from National Cancer Institute (NCI) to Dr. Gordon Mills. This work was supported in part by grants from the National Science Foundation of China (81071631, 81472729), the Key Foundation of Tianjin Health Bureau (2013KR14) and the foundation of committee on science and technology of Tianjin (13JCYBJC42700).
Disclosure of conflict of interest
None.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Brand KA, Hermfisse U. Aerobic glycolysis by proliferating cells: a protective strategy against reactive oxygen species. FASEB J. 1997;11:388–395. doi: 10.1096/fasebj.11.5.9141507. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG, Broda P, Minetti C, Lisanti MP, Sotgia F. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle. 2011;10:4047–4064. doi: 10.4161/cc.10.23.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro MC, Wang C, Fortina P, Addya S, Pestell RG, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 7.Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, Jiao X, Li A, Zhang X, Lu Y, Wang C, Byers S, Nicholson R, Link T, Shemluck M, Yang J, Fricke ST, Novikoff PM, Papanikolaou A, Arnold A, Albanese C, Pestell R. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26:5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, Pestell RG. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci U S A. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci U S A. 2004;101:6564–6569. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung HJ, Ma W, Starost MF, Lago CU, Lim PK, Sack MN, Kang JG, Wang PY, Hwang PM. Ambient oxygen promotes tumorigenesis. PLoS One. 2011;6:e19785. doi: 10.1371/journal.pone.0019785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koopman WJ, Distelmaier F, Smeitink JA, Willems PH. OXPHOS mutations and neurodegeneration. EMBO J. 2013;32:9–29. doi: 10.1038/emboj.2012.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 14.Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stier A, Bize P, Habold C, Bouillaud F, Massemin S, Criscuolo F. Mitochondrial uncoupling prevents cold-induced oxidative stress: a case study using UCP1 knockout mice. J Exp Biol. 2014;217:624–630. doi: 10.1242/jeb.092700. [DOI] [PubMed] [Google Scholar]
- 16.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortassa S, O’Rourke B, Aon MA. Redox-optimized ROS balance and the relationship between mitochondrial respiration and ROS. Biochim Biophys Acta. 2014;1837:287–295. doi: 10.1016/j.bbabio.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iuso A, Scacco S, Piccoli C, Bellomo F, Petruzzella V, Trentadue R, Minuto M, Ripoli M, Capitanio N, Zeviani M, Papa S. Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J Biol Chem. 2006;281:10374–10380. doi: 10.1074/jbc.M513387200. [DOI] [PubMed] [Google Scholar]
- 19.Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- 20.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 21.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 22.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 23.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 24.Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta. 2012;1817:1833–1838. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 25.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackenbrock CR. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J Cell Biol. 1966;30:269–297. doi: 10.1083/jcb.30.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hackenbrock CR. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J Cell Biol. 1968;37:345–369. doi: 10.1083/jcb.37.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara KY, Mori H. An efficient method for quantitative determination of cellular ATP synthetic activity. J Biomol Screen. 2006;11:310–317. doi: 10.1177/1087057105285112. [DOI] [PubMed] [Google Scholar]
- 30.Raimundo N, Baysal BE, Shadel GS. Revisiting the TCA cycle: signaling to tumor formation. Trends Mol Med. 2011;17:641–649. doi: 10.1016/j.molmed.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011;14:443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagopian K, Soo Hoo R, Lopez-Dominguez JA, Ramsey JJ. Calorie restriction influences key metabolic enzyme activities and markers of oxidative damage in distinct mouse liver mitochondrial sub-populations. Life Sci. 2013;93:941–948. doi: 10.1016/j.lfs.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Ruiz R, Uribe-Carvajal S, Devin A, Rigoulet M. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim Biophys Acta. 2009;1796:252–265. doi: 10.1016/j.bbcan.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Gi aever G, Prokisch H, Oefner PJ, Davis RW. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 35.Pereira C, Coutinho I, Soares J, Bessa C, Leao M, Saraiva L. New insights into cancer-related proteins provided by the yeast model. FEBS J. 2012;279:697–712. doi: 10.1111/j.1742-4658.2012.08477.x. [DOI] [PubMed] [Google Scholar]
- 36.Matuo R, Sousa FG, Soares DG, Bonatto D, Saffi J, Escargueil AE, Larsen AK, Henriques JA. Saccharomyces cerevisiae as a model system to study the response to anticancer agents. Cancer Chemother Pharmacol. 2012;70:491–502. doi: 10.1007/s00280-012-1937-4. [DOI] [PubMed] [Google Scholar]
- 37.Hartwell LH, Culotti J, Reid B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci U S A. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh AK, Chance B, Pye EK. Metabolic coupling and synchronization of NADH oscillations in yeast cell populations. Arch Biochem Biophys. 1971;145:319–331. doi: 10.1016/0003-9861(71)90042-7. [DOI] [PubMed] [Google Scholar]
- 39.Dirick L, Bendris W, Loubiere V, Gostan T, Gueydon E, Schwob E. Metabolic and environmental conditions determine nuclear genomic instability in budding yeast lacking mitochondrial DNA. G3 (Bethesda) 2014;4:411–423. doi: 10.1534/g3.113.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz-Ruiz R, Rigoulet M, Devin A. The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim Biophys Acta. 2011;1807:568–576. doi: 10.1016/j.bbabio.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Slavov N, Macinskas J, Caudy A, Botstein D. Metabolic cycling without cell division cycling in respiring yeast. Proc Natl Acad Sci U S A. 2011;108:19090–19095. doi: 10.1073/pnas.1116998108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin SL, Marcus IM, Klevecz RR, Li CM. Dynamics of oscillatory phenotypes in Saccharomyces cerevisiae reveal a network of genome-wide transcriptional oscillators. FEBS J. 2012;279:1119–1130. doi: 10.1111/j.1742-4658.2012.08508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slavov N, Botstein D. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol Biol Cell. 2011;22:1997–2009. doi: 10.1091/mbc.E11-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krisher RL, Prather RS. A role for the Warburg effect in preimplantation embryo development: metabolic modification to support rapid cell proliferation. Mol Reprod Dev. 2012;79:311–320. doi: 10.1002/mrd.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen PJ, Fear JM. Cheating death at the dawn of life: developmental control of apoptotic repression in the preimplantation embryo. Biochem Biophys Res Commun. 2011;413:155–158. doi: 10.1016/j.bbrc.2011.08.064. [DOI] [PubMed] [Google Scholar]
- 46.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen IT, Aoki T, Huang YT, Hirono I, Chen TC, Huang JY, Chang GD, Lo CF, Wang HC. White spot syndrome virus induces metabolic changes resembling the warburg effect in shrimp hemocytes in the early stage of infection. J Virol. 2011;85:12919–12928. doi: 10.1128/JVI.05385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A. 2014;111:6413–6418. doi: 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai D, Tan CL, Gunaratne J, Quek LS, Nei W, Thierry F, Bellanger S. Localization of HPV-18 E2 at mitochondrial membranes induces ROS release and modulates host cell metabolism. PLoS One. 2013;8:e75625. doi: 10.1371/journal.pone.0075625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nykky J, Vuento M, Gilbert L. Role of mitochondria in parvovirus pathology. PLoS One. 2014;9:e86124. doi: 10.1371/journal.pone.0086124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.St John JC, Facucho-Oliveira J, Jiang Y, Kelly R, Salah R. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum Reprod Update. 2010;16:488–509. doi: 10.1093/humupd/dmq002. [DOI] [PubMed] [Google Scholar]
- 52.Thundathil J, Filion F, Smith LC. Molecular control of mitochondrial function in preimplantation mouse embryos. Mol Reprod Dev. 2005;71:405–413. doi: 10.1002/mrd.20260. [DOI] [PubMed] [Google Scholar]
- 53.Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jansen RP, Burton GJ. Mitochondrial dysfunction in reproduction. Mitochondrion. 2004;4:577–600. doi: 10.1016/j.mito.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 55.Margineantu DH, Gregory Cox W, Sundell L, Sherwood SW, Beechem JM, Capaldi RA. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- 56.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meinhardt A, Wilhelm B, Seitz J. Expression of mitochondrial marker proteins during spermatogenesis. Hum Reprod Update. 1999;5:108–119. doi: 10.1093/humupd/5.2.108. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DuBoff B, Feany M, Gotz J. Why size matters - balancing mitochondrial dynamics in Alzheimer’s disease. Trends Neurosci. 2013;36:325–335. doi: 10.1016/j.tins.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Peralta S, Moraes CT. Mitochondrial alterations during carcinogenesis: a review of metabolic transformation and targets for anticancer treatments. Adv Cancer Res. 2013;119:127–160. doi: 10.1016/B978-0-12-407190-2.00004-6. [DOI] [PubMed] [Google Scholar]
- 61.Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jheng HF, Tsai PJ, Guo SM, Kuo LH, Chang CS, Su IJ, Chang CR, Tsai YS. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol Cell Biol. 2012;32:309–319. doi: 10.1128/MCB.05603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo AY, Joseph AM, Dutta D, Hwang JC, Aris JP, Leeuwenburgh C. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci. 2010;123:2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nett JH, Hunte C, Trumpower BL. Changes to the length of the flexible linker region of the Rieske protein impair the interaction of ubiquinol with the cytochrome bc1 complex. Eur J Biochem. 2000;267:5777–5782. doi: 10.1046/j.1432-1327.2000.01650.x. [DOI] [PubMed] [Google Scholar]
- 65.Sazanov LA, Baradaran R, Efremov RG, Berrisford JM, Minhas G. A long road towards the structure of respiratory complex I, a giant molecular proton pump. Biochem Soc Trans. 2013;41:1265–1271. doi: 10.1042/BST20130193. [DOI] [PubMed] [Google Scholar]
- 66.Grivennikova VG, Vinogradov AD. Generation of superoxide by the mitochondrial Complex I. Biochim Biophys Acta. 2006;1757:553–561. doi: 10.1016/j.bbabio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 67.Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci U S A. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 69.Stone HR, Morris JR. DNA damage emergency: cellular garbage disposal to the rescue? Oncogene. 2014;33:805–813. doi: 10.1038/onc.2013.60. [DOI] [PubMed] [Google Scholar]
- 70.Rinaldi T, Ricci C, Porro D, Bolotin-Fukuhara M, Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol Biol Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Nooij JC, Letendre MA, Hariharan IK. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- 72.Bringhen S, Gay F, Donato F, Troia R, Mina R, Palumbo A. Current Phase II investigational proteasome inhibitors for the treatment of multiple myeloma. Expert Opin Investig Drugs. 2014:1–17. doi: 10.1517/13543784.2014.920821. [DOI] [PubMed] [Google Scholar]
- 73.Monk M, Hitchins M, Hawes S. Differential expression of the embryo/cancer gene ECSA(DPPA2), the cancer/testis gene BORIS and the pluripotency structural gene OCT4, in human preimplantation development. Mol Hum Reprod. 2008;14:347–355. doi: 10.1093/molehr/gan025. [DOI] [PubMed] [Google Scholar]
- 74.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 75.Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaria G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 76.Kaipparettu BA, Ma Y, Park JH, Lee TL, Zhang Y, Yotnda P, Creighton CJ, Chan WY, Wong LJ. Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PLoS One. 2013;8:e61747. doi: 10.1371/journal.pone.0061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ishikawa K, Hashizume O, Koshikawa N, Fukuda S, Nakada K, Takenaga K, Hayashi J. Enhanced glycolysis induced by mtDNA mutations does not regulate metastasis. FEBS Lett. 2008;582:3525–3530. doi: 10.1016/j.febslet.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 80.Bell G. Evolutionary rescue and the limits of adaptation. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120080. doi: 10.1098/rstb.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murphy MP. Modulating mitochondrial intracellular location as a redox signal. Sci Signal. 2012;5:pe39. doi: 10.1126/scisignal.2003386. [DOI] [PubMed] [Google Scholar]
- 82.Ostrow SL, Barshir R, DeGregori J, Yeger-Lotem E, Hershberg R. Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 2014;10:e1004239. doi: 10.1371/journal.pgen.1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bykov VJ, Wiman KG. Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett. 2014;588:2622–2627. doi: 10.1016/j.febslet.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 85.Szewczyk A, Jarmuszkiewicz W, Kunz WS. Mitochondrial potassium channels. IUBMB Life. 2009;61:134–143. doi: 10.1002/iub.155. [DOI] [PubMed] [Google Scholar]
- 86.Cuenca AG, Cress WD, Good RA, Marikar Y, Engelman RW. Calorie restriction influences cell cycle protein expression and DNA synthesis during liver regeneration. Exp Biol Med (Maywood) 2001;226:1061–1067. doi: 10.1177/153537020122601114. [DOI] [PubMed] [Google Scholar]
- 87.Vuaroqueaux V, Urban P, Labuhn M, Delorenzi M, Wirapati P, Benz CC, Flury R, Dieterich H, Spyratos F, Eppenberger U, Eppenberger-Castori S. Low E2F1 transcript levels are a strong determinant of favorable breast cancer outcome. Breast Cancer Res. 2007;9:R33. doi: 10.1186/bcr1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delaney JR, Murakami C, Chou A, Carr D, Schleit J, Sutphin GL, An EH, Castanza AS, Fletcher M, Goswami S, Higgins S, Holmberg M, Hui J, Jelic M, Jeong KS, Kim JR, Klum S, Liao E, Lin MS, Lo W, Miller H, Moller R, Peng ZJ, Pollard T, Pradeep P, Pruett D, Rai D, Ros V, Schuster A, Singh M, Spector BL, Wende HV, Wang AM, Wasko BM, Olsen B, Kaeberlein M. Dietary restriction and mitochondrial function link replicative and chronological aging in Saccharomyces cerevisiae. Exp Gerontol. 2013;48:1006–1013. doi: 10.1016/j.exger.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 90.Laus MN, Soccio M, Trono D, Liberatore MT, Pastore D. Activation of the plant mitochondrial potassium channel by free fatty acids and acyl-CoA esters: a possible defence mechanism in the response to hyperosmotic stress. J Exp Bot. 2011;62:141–154. doi: 10.1093/jxb/erq256. [DOI] [PubMed] [Google Scholar]
- 91.Wang C, Geng B, Cui Q, Guan Y, Yang J. Intracellular and extracellular adenosine triphosphate in regulation of insulin secretion from pancreatic beta cells (beta) J Diabetes. 2014;6:113–119. doi: 10.1111/1753-0407.12098. [DOI] [PubMed] [Google Scholar]
- 92.Bause AS, Matsui MS, Haigis MC. The protein deacetylase SIRT3 prevents oxidative stress-induced keratinocyte differentiation. J Biol Chem. 2013;288:36484–36491. doi: 10.1074/jbc.M113.472324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leanza L, Zoratti M, Gulbins E, Szabo I. Mitochondrial ion channels as oncological targets. Oncogene. 2014;33:5569–5581. doi: 10.1038/onc.2013.578. [DOI] [PubMed] [Google Scholar]
- 94.Queliconi BB, Wojtovich AP, Nadtochiy SM, Kowaltowski AJ, Brookes PS. Redox regulation of the mitochondrial K(ATP) channel in cardioprotection. Biochim Biophys Acta. 2011;1813:1309–1315. doi: 10.1016/j.bbamcr.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Harry G, Hashimoto K, Porter CJ, Andrade MA, Thebaud B, Michelakis ED. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 97.Kowaltowski AJ, Seetharaman S, Paucek P, Garlid KD. Bioenergetic consequences of opening the ATP-sensitive K(+) channel of heart mitochondria. Am J Physiol Heart Circ Physiol. 2001;280:H649–657. doi: 10.1152/ajpheart.2001.280.2.H649. [DOI] [PubMed] [Google Scholar]
- 98.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park SH, Ozden O, Jiang H, Cha YI, Pennington JD, Aykin-Burns N, Spitz DR, Gius D, Kim HS. Sirt3, Mitochondrial ROS, Ageing, and Carcinogenesis. Int J Mol Sci. 2011;12:6226–6239. doi: 10.3390/ijms12096226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 101.Fridhandler L. Pathways of glucose metabolism in fertilized rabbit ova at various pre-implantation stages. Exp Cell Res. 1961;22:303–316. doi: 10.1016/0014-4827(61)90109-4. [DOI] [PubMed] [Google Scholar]
- 102.Yang TH. 13C-based metabolic flux analysis: fundamentals and practice. Methods Mol Biol. 2013;985:297–334. doi: 10.1007/978-1-62703-299-5_15. [DOI] [PubMed] [Google Scholar]
- 103.Sengupta N, Rose ST, Morgan JA. Metabolic flux analysis of CHO cell metabolism in the late non-growth phase. Biotechnol Bioeng. 2011;108:82–92. doi: 10.1002/bit.22890. [DOI] [PubMed] [Google Scholar]
- 104.Kathagen A, Schulte A, Balcke G, Phillips HS, Martens T, Matschke J, Gunther HS, Soriano R, Modrusan Z, Sandmann T, Kuhl C, Tissier A, Holz M, Krawinkel LA, Glatzel M, Westphal M, Lamszus K. Hypoxia and oxygenation induce a metabolic switch between pentose phosphate pathway and glycolysis in glioma stem-like cells. Acta Neuropathol. 2013;126:763–780. doi: 10.1007/s00401-013-1173-y. [DOI] [PubMed] [Google Scholar]
- 105.Polet F, Feron O. Endothelial cell metabolism and tumour angiogenesis: glucose and glutamine as essential fuels and lactate as the driving force. J Intern Med. 2013;273:156–165. doi: 10.1111/joim.12016. [DOI] [PubMed] [Google Scholar]
- 106.Coutts R. A pilot study for the analysis of dream reports using Maslow’s need categories: an extension to the emotional selection hypothesis. Psychol Rep. 2010;107:659–673. doi: 10.2466/09.PR0.107.5.659-673. [DOI] [PubMed] [Google Scholar]
- 107.Kornas A, Kuzniak E, Slesak I, Miszalski Z. The key role of the redox status in regulation of metabolism in photosynthesizing organisms. Acta Biochim Pol. 2010;57:143–151. [PubMed] [Google Scholar]
- 108.Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Hoek P, Van Dijken JP, Pronk JT. Effect of specific growth rate on fermentative capacity of baker’s yeast. Appl Environ Microbiol. 1998;64:4226–4233. doi: 10.1128/aem.64.11.4226-4233.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 111.Du W, Jiang P, Mancuso A, Stonestrom A, Brewer MD, Minn AJ, Mak TW, Wu M, Yang X. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol. 2013;15:991–1000. doi: 10.1038/ncb2789. [DOI] [PMC free article] [PubMed] [Google Scholar]


