Highlight
The lack of the jasmonic acid precursor 12-oxophytodienoic acid in rice led to increased salt tolerance, which is correlated with an increased ROS-scavenging activity in stress conditions.
Key words: ALLENE OXIDE CYCLASE (AOC), jasmonate, Oryza sativa, oxidative stress, 12-oxophytodienoic acid (12-OPDA), reactive oxygen species (ROS), salinity.
Abstract
Salinity stress represents a global constraint for rice, the most important staple food worldwide. Therefore the role of the central stress signal jasmonate for the salt response was analysed in rice comparing the responses to salt stress for two jasmonic acid (JA) biosynthesis rice mutants (cpm2 and hebiba) impaired in the function of ALLENE OXIDE CYCLASE (AOC) and their wild type. The aoc mutants were less sensitive to salt stress. Interestingly, both mutants accumulated smaller amounts of Na+ ions in their leaves, and showed better scavenging of reactive oxygen species (ROS) under salt stress. Leaves of the wild type and JA mutants accumulated similar levels of abscisic acid (ABA) under stress conditions, and the levels of JA and its amino acid conjugate, JA–isoleucine (JA-Ile), showed only subtle alterations in the wild type. In contrast, the wild type responded to salt stress by strong induction of the JA precursor 12-oxophytodienoic acid (OPDA), which was not observed in the mutants. Transcript levels of representative salinity-induced genes were induced less in the JA mutants. The absence of 12-OPDA in the mutants correlated not only with a generally increased ROS-scavenging activity, but also with the higher activity of specific enzymes in the antioxidative pathway, such as glutathione S-transferase, and fewer symptoms of damage as, for example, indicated by lower levels of malondialdehyde. The data are interpreted in a model where the absence of OPDA enhanced the antioxidative power in mutant leaves.
Introduction
Soil salinization is a global threat that causes a huge reduction of agricultural yield worldwide. More than 20% of all irrigated land on earth is affected by salinization. Rice (Oryza sativa), as one of the world’s most important cereal crops, provides the primary source of food and calories for about half of mankind (Khush, 2005). Rice as a so-called glycophyte is very sensitive to salinity stress especially at the seedling stage, with height, root length, emergence of new roots, and dry matter affected significantly by salinity (Pearson et al., 1966; Akbar and Yabuno, 1974). Salinity stress is generally defined as the presence of excessive amounts of soluble salt that hinder or negatively affect the functions needed for normal plant growth and development. Salt stress is comprised of two harmful effects: osmotic stress leading to reduced water uptake, and ionic stress caused by the toxicity of specific ions (mainly Na+ and Cl–). Ionic stress leads to unrestrained overproduction of ROS (reactive oxygen species), such as superoxide radicals (O2·), hydrogen peroxide (H2O2), and hydroxyl radicals (OH·–). These reactive molecules accumulate to toxic levels and trigger oxidative damage in the cells and organelles by destroying membranes, proteins, enzymes, and nucleic acids (Abogadallah, 2010; Sharma et al., 2012; Ismail et al., 2014a, b ).
The adaptive response of salt-stressed plants is controlled by chemical signals that will compensate adjustment of growth and development in response to such unfavourable conditions. It should be noted that some of these signals play a dual role—if controlled in space and time, they can act as signals triggering adaptation, if developing unconstrained, they accompany stress-related damage (for a review, see Ismail et al., 2014b ). Central players among these stress signals are jasmonic acid (JA), its biologically active precursor 12-oxophytodenoic acid (OPDA), and its derivatives such as methyl jasmonate (MeJA) or the amino acid-conjugated jasmonate, JA–isoleucine (JA-Ile), in the following collectively termed as jasmonates (JAs). JAs have been reported to accumulate in response to salinity stress (tomato, Pedranzani et al., 2003; rice, Moons et al., 1997). Whether this accumulation is a signal triggering adaptation or just a by-product or consequence of adaptation is not very clear. However, the fact that a salt-tolerant cultivar of rice shows higher endogenous JA contents as compared with a salt-sensitive cultivar, as well as the observation that exogenous MeJA can reduce the uptake of sodium in this salt-tolerant cultivar (Kang et al., 2005), indicates a function for JAs in salt adaptation. Overexpression of a wheat AOC (ALLENE OXIDE CYCLASE) gene in wheat and Arabidopsis resulted in an improved salt tolerance of these species (Zhao et al., 2014). However, it is not possible to draw a general connection between high levels of JA and adaptation; during a comparison of two grapevine cell lines differing in their salinity tolerance, the accumulation of JA and JA-Ile was more pronounced in the sensitive Vitis riparia rather than in the salt-tolerant Vitis rupestris (Ismail et al., 2014a ). These discrepancies underscore that it is not the presence or absence of JAs that decides the salinity response, but rather the right timing and control (for a review, see Ismail et al., 2014b ). The complexity in the relationship between JAs and salinity adaptation is further accentuated by the recent finding that the precursor OPDA (but not JA itself) was significantly induced in drought-stressed Arabidopsis leaves (Savchenko et al., 2014). Moreover, in rice roots, JA biosynthesis was reported to be strongly induced by drought stress, but only marginally by salt stress, indicating that the two components of salinity stress might differ in their transduction events (Takeuchi et al., 2011).
The complexity extends into the signalling triggered by JAs, because this signalling has been found to interact with the signalling triggered by other plant hormones known to be involved in the adaptation to salt stress such as abscisic acid (ABA). These interactions, often referred to as ‘hormonal cross-talk’, need more investigations, especially in crops of economic importance. For instance, MYC2, a transcription factor conveying the stimulation of transcription by JA, also acts downstream of ABA, providing a mechanism for co-regulation of the respective target genes (Kazan and Manners, 2012). The addition of exogenous MeJA caused an elevation in the endogenous level of ABA in rice (Seo et al., 2001), indicating that ABA synthesis is modulated by JA signalling. In Arabidopsis thaliana, JA induces expression of genes encoding ABA receptor proteins, thereby contributing to a maintenance of the balance between growth and defence (for a review, see Wasternack and Hause, 2013). However, the interactions can also be antagonistic, as found for the regulation of salt-stress-related gene expression by JAs and ABA in rice and Arabidopsis (Moons et al., 1997; Anderson et al., 2004). In addition to MYC2, JA and ABA signalling interact at the level of the JASMONATE ZIM-DOMAIN (JAZ) gene family, acting as repressors for JA signalling (Staswick, 2008). These two, only partially elucidated mechanisms provide a tight regulatory interaction between JA- and ABA-mediated signalling during the response to salt stress (for a review, see Kumar et al., 2013).
As one of the most salt-sensitive glycophytes, rice has very limited strategies to deal effectively with salt stress and therefore does not grow well on saline soil (Galvan-Ampudia and Testerink, 2011). Therefore, lowering the amount of sodium loaded into the transpiration stream is vital to circumvent damage of photosynthetic tissues. Salinity and osmotic stress induce, either directly or indirectly via hormonal regulation, stomatal closure, which reduces evaporation and overall water transport (Horie et al., 2012). At micromolar concentrations, ABA causes closure of stomata, but the effect of ABA on the stomatal aperture is dependent on the intercellular CO2 concentration and on the presence of the signal substance nitric oxide (NO) (Heldt and Heldt, 2005). Due to the close interaction between JA and ABA signalling, it is to be expected that JAs also play a role in stomatal aperture. However, it should be noted that due to its negative impact on photosynthesis, stomatal closure can only provide short-term protection due to reduced transpiration, but cannot be a mechanism for long-term adaptation to salinity stress.
To show the functionality of JAs for adaptation, in previous studies exogenous MeJA was added (Kang et al., 2005) and/or fluctuations in the levels of endogenous JAs were measured (Moons et al., 1997; Pedranzani et al., 2007). However, these fluctuations could be by-products of adaptation rather than their cause, and exogenous MeJA is present constitutively and therefore is not a good equivalent for the tightly controlled, transient accumulation of JAs that is characteristic for successful adaptation. To address the functionality of JAs for adaptation, it is necessary to create a condition in which the accumulation of endogenous JAs is blocked. This became possible for the rice model by mutants impaired in JA biosynthesis (Riemann et al., 2003). The mutant hebiba is deficient in JA, and this phenotype could be attributed to the locus for AOC, a synthetic enzyme driving the formation of the JA precursor OPDA (Riemann et al., 2013). A second mutant, coleoptile photomorphogenesis 2 (cpm2), shows a similar phenotype and could be shown to be allelic to hebiba. Using these two mutants of JA biosynthesis in comparison with their wild-type background, the role of JA for salinity adaptation was analysed in an integrative approach considering the response of growth, ion uptake, and redistribution, ROS, antioxidants, accumulation of phytohormones, and the expression patterns of specific response genes. A model was arrived at where the absence of OPDA accumulation in the mutants enables a better capability to ward off oxidative stress.
Materials and methods
Plant materials, growth, and stress conditions
In this study, Oryza sativa L. ssp. japonica cv. Nihonmasari was used as the wild type. The two mutant lines cpm2 and hebiba were generated in the same cultivar (Riemann et al., 2013). The caryopses were dehusked and surface sterilized by incubating the seeds in 70% ethanol for 1min then washed briefly twice with double-distilled water. Subsequently, the seeds were incubated in a sodium hypochlorite solution containing ~5% of active chlorine for 30min followed by five washing steps in sterilized double-distilled water. The seeds were sown on 0.5% phytoagar medium (Duchefa, The Netherlands) and incubated for 10–12 d in a culture room (at 25 °C, continuous light of 120 μmol m–2s–1). After 10–12 d, well-grown seedlings were transferred to custom-made sterilized floating racks and moved to a glass container containing double-distilled water as control or a solution containing NaCl to cause salt stress. The shoots of control and stressed plants were harvested, frozen in liquid nitrogen, and then stored in –80 °C to be used for enzymatic or non-enzymatic antioxidants, or for gene expression analysis.
Analysis of root elongation
Root elongation was evaluated as the mean of the seminal root length of seedlings raised in darkness (25 °C, 7 d). The seeds were surface sterilized as described above, and sown on 0.5% phytoagar medium with different concentrations of NaCl (0, 7.8, 15.6, 31.3, 62.3, 125, and 250mM). The seedlings were scanned and the root length was measured using Image J (http://imagej.nih.gov/ij/, last accessed 22 January 2015).
Measurement of sodium ion content
Dry cells of each biological replicate were transferred into digestion tubes (Gerhardt, UK), supplemented with 5ml of concentrated nitric acid (HNO3), and then incubated for at least 24h at room temperature with vortexing at 6h and 24h. Samples were placed on a water bath at 100 °C for 2h. After cooling, the final volume of each sample was adjusted to 10ml with distilled water and vortexed. Contents of sodium ions were measured by flame atomic absorption spectrometry (AAnalyst200, Perkin Elmer) in an air acetylene flame (Institute of Applied Geosciences, Aquatic Geochemistry, Karlsruhe Institute of Technology). Blank samples were prepared by adding 5ml of concentrated nitric acid to an empty digestion vessel and processed as described above.
Determination of lipid peroxidation and aqueous peroxide levels
Lipid peroxidation of shoots was estimated by the level of malondialdehyde (MDA) using the thiobarbituric acid (TBA) method as described by Heath and Packer (1968). Briefly, 500mg of rice shoots were homogenized using a mortar and pestle in 1ml of 0.1% trichloroacetic acid (TCA, w/v). The homogenate was then centrifuged at 10 000 g for 20min, and 0.5ml of the supernatant was added to 1ml of 0.5% TBA in 20% TCA. This mixture was then heated in a boiling water bath for 1h. The reaction was stopped by transferring the tubes to an ice bath for 10min, and then the tubes were centrifuged for 10min at 10 000 g. The absorbance of the supernatant was recorded at 532nm and 600nm. The value of the non-specific absorption at 600nm was subtracted. The amount of MDA–TBA complex (red pigment) was calculated from the extinction coefficient 155mM–1 cm–1.
Steady-state levels of H2O2 in the shoots of control and salt-stressed rice seedlings were measured using the FOX-1 method (ferrous oxidation with Xylenol Orange) (Wolf, 1994) with some modifications. Leaves (70mg fresh weight) were perfectly ground in 5ml of 5% TCA containing 100 μg of active charcoal. The mixture was filtered using No. 1 filter paper (Whatman), and then a measured volume of the filtrate was incubated with FOX-1 reagent (100 μM Xylenol Orange, 250 μM ammonium sulphate, 100mM sorbitol, and 25mM H2SO4) for 30min. The absorbance was recorded at 560nm. The values of aqueous peroxide were referred to as micromoles H2O2 using a standard curve.
Estimation of soluble proline
Accumulation of free proline was monitored according to Bates et al. (1973). Briefly, 200mg of fresh tissue of leaves were homogenized using a mortar and pestle containing a small amount of quartz sand. The homogenate was filtered through filter paper No. 1 (Whatman). The filtrate was centrifuged (10 000 g, 10min) at room temperature. A 1ml aliquot of the supernatant was treated with 2ml of reaction buffer (1ml of glacial acetic acid and 1ml of ninhydrin reagent), and mixed well. The reaction was boiled in a water bath for 1h, and then cooled to room temperature gradually. The absorbance was recorded at 520nm. Soluble proline content was expressed as μmol proline per g fresh weight according to a standard curve.
Free radical-scavenging activity (DPPH-scavenging activity)
In order to measure the antioxidant ability of the rice shoot extract, 2, 2-diphenyl-1-picrylhydrazyl (DPPH), a stable free radical, was used according to Goffman and Bergman (2004). A crude extract (100 μl) of rice leaves at different concentrations (37–370 μg extract ml–1 reaction) was added to 900 μl of freshly prepared DPPH methanolic solution (80 ppm). The reduction in the absorbance (515nm) of the methanolic solution of DPPH was monitored. The percentage DPPH inhibition was calculated using the following formula: (Abcont–Absample)/Abcont×100, with the Abcont absorbance value at 515nm. The IC50 value for each sample was calculated to determine the amount in micrograms of extract sufficient to scavenge 50% or half of the DPPH radical substance. Butylated hydroxyanisole (BHA), as a very efficient antioxidant, was used as an internal control.
Antioxidant enzyme activity measurement
For estimating the activity of catalase (CAT), peroxidase (POD), glutathione reductase (GR), glutathione S-transferase (GST), and superoxide dismutase (SOD), leaves of the treated seedlings were homogenized in 1ml of ice cold extraction buffer according to Venisse et al. (2001). For ascorbate peroxidase (APX), the same procedure was followed but using the extraction buffer of Nakano and Asada (1981). SOD (EC 1.5.1.1) activity was assayed by monitoring the inhibition of the photochemical reduction of nitroblue tetrazolium at 560nm (Beauchamp and Fridovich, 1971). Total protein content was estimated according to Bradford et al. (1976).
RNA extraction and quantitative real-time PCR
Total RNA was isolated from the shoots of control and salinity-stressed plants (100mM NaCl, 24h and 72h) using the InnuPrep plant RNA kit (Analytika Jena RNA kit) according to the manufacturer’s instructions. cDNA synthesis was performed with a Dynamo cDNA synthesis kit (Finnzymes, Finland) using total RNA as a template. Real-time PCR was done with a SYBR green dye protocol using an Opticon 2 system (Biorad, USA). The primer sequences for the genes of interest are listed in Supplementary Table S1 available at JXB online.
Endogenous level of ABA, OPDA, JA, and JA-Ile
OPDA, JA, JA-Ile, and ABA were quantified simultaneously using a standardized ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS)-based method according to Balcke et al. (2012) using [2H5]OPDA, [2H6]JA, [2H2]JA-ile, and [2H6]ABA as internal standards.
Chlorophyll content
Total chlorophyll, chlorophyll a, and chlorophyll b contents were determined based on the method of Arnon (1949). A 100mg aliquot of leaves was homogenized with acetone. The extract was filtered through Whatman No. 1. filter paper and washed 2–3 times with 80% acetone. The final volume of the extract was made up to 25ml. Chlorophyll contents were calculated based on the absorbance measured at 645, 652, and 663nm, respectively.
Preparation of plant extracts for the determination of specific ROS-scavenging activities
Rice leaves were collected and washed gently several times with Millipore water. The washed leaves were ground in 5ml of warm Millipore water (80 °C). Subsequently the homogenate was mixed with 50ml of Millipore water, stirred for 3h, and filtered through filter paper No. 2 (Whatman). This process was repeated once and the combined filtrate was freeze-dried. The yellowish solid crude extracts were kept at –20 °C to be used for testing scavenging activities of O2·, H2O2, and OH·– as described below.
Superoxide anion-scavenging activity
Measurement of superoxide-scavenging activity was done based on the method described by Zhishen et al. (1999) with slight modifications. All solutions were prepared in 0.05M phosphate buffer (pH 7.8). The reaction was carried out in light for 40min in a total volume of 3ml. Crude extracts were dissolved in phosphate buffer to obtain aqueous extracts in the final concentrations of 25, 50, 100, 150, and 250 μg per reaction, respectively. Absorbance was measured at 560nm. Low absorbance of the reaction mixture indicates increased O2·-scavenging activity. The percentage inhibition of O2· generation was calculated by using the following formula:
A0 is the absorbance of the control with water instead of aqueous extract, while A1 is the absorbance of sample or standard, respectively. The IC50 value is the concentration of sample in micrograms of aqueous extract per reaction required for scavenging 50% of O2· in the reaction mixture. It was calculated based on a regression curve.
Hydrogen peroxide-scavenging assay
H2O2-scavenging activity of the extract was determined by the method of Ruch et al. (1989). Different amounts (60, 120, 240, 300, and 420 μg) of the extract were dissolved in 3.4ml of 0.1M phosphate buffer (pH 7.4), and mixed with 600 μl of a 43mM solution of H2O2. The absorbance value of the reaction mixture was recorded at 230nm and butylhydroxytoluol (BHT) was used as a standard. The percentage scavenging of H2O2 was calculated as follows
The IC50 value is the concentration of sample in micrograms of aqueous extract per reaction required for scavenging of 50% of H2O2 in the reaction mixture, and it was calculated based on a regression curve.
Hydroxyl radical-scavenging assay
OH·-scavenging activity was measured according to the method of Kunchandy and Rao (1990). The reaction mixture contained 0.8ml of phosphate buffer solution (50mM, pH 7.4), 0.2ml of a sample of different concentrations (25, 50, 100, 150, and 250 μg of aqueous extract per reaction mixture), 0.2ml of EDTA (1.04mM), 0.2ml of FeCl3 (1mM), and 0.2ml of 2-deoxyribose (60mM). The mixtures were kept in a water bath at 37 °C and the reaction was started by adding 0.2ml of ascorbic acid (2mM) and 0.2ml of H2O2 (10mM). After incubation at 37 °C for 1h, 2ml of cold TBA (10 mgml–1) was added to the reaction mixture followed by 2ml of HCl (25%). The mixture was heated at 100 °C for 15min and then cooled down with water. The absorbance of the solution was measured at 532nm. The scavenging percentage was calculated according to the following formula:
where A0 is the absorbance of the control without a sample. A1 is the absorbance after adding the sample and deoxyribose, and A2 is the absorbance of the sample without deoxyribose. The IC50 value is the concentration of sample in micrograms of total aqueous extract per reaction required for the scavenging of 50% of OH· in the reaction mixture. It is calculated based on the regression curve.
Results
JA-deficient mutants are less sensitive to salinity stress
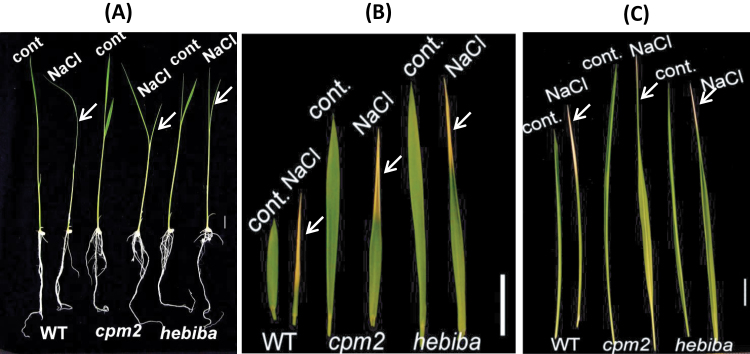
Rice seedlings (age 10 d) were subjected to salt stress either at a very high concentration for short-term experiments (285mM NaCl for 24h), or at a high concentration for long-term experiments (100mM for 72h, subsequently referred to as mid-term exposure). For both stress treatments, leaves of hebiba and cpm2 appeared to be less sensitive with respect to wilting and necrosis (Fig. 1). In the short-term exposure at high concentrations, the wild-type leaves were entirely rolled as a response to salt stress, whereas the mutant leaves remained unfolded (Fig. 1A). Moreover, the second leaf of the wild type was almost entirely necrotic after mid-term salt exposure, whereas the second leaves of both mutants showed necrosis only at the tip (Fig. 1B). For the same treatment, the third leaf of the wild type was entirely rolled and almost half of the leaf was necrotic, starting from the tip (Fig. 1C). In contrast, the third leaves of both hebiba and cpm2 were only marginally necrotic, at the tip, and approximately two-thirds of the leaf remained unfolded. In addition, they were also able to maintain higher chlorophyll levels under salt stress (Supplementary Fig. S1 at JXB online). Hence, these mutants, which are deficient in the phytohormone JA, displayed a clearly reduced salt sensitivity in their phenotype. It is noteworthy that mutants develop longer leaves due to deficiency in JA, which has been described for hebiba previously (Riemann et al., 2003).
Fig. 1.
Effects of salt stress on the phenotype of the wild type (WT) and JA biosynthesis mutants. (A) Ten-day-old rice seedlings were subjected to salt stress at a high concentration (285mM NaCl) for 24h. (B) The second leaf of 10-day-old rice seedlings which were subjected to a 100mM NaCl solution for 3 d. (C) The third leaf of 10-day-old rice seedlings which were subjected to a 100mM NaCl solution for 3 d. Arrows indicate the salt-treated leaves. Scale bar=10mm.
Effect of salt stress on root elongation
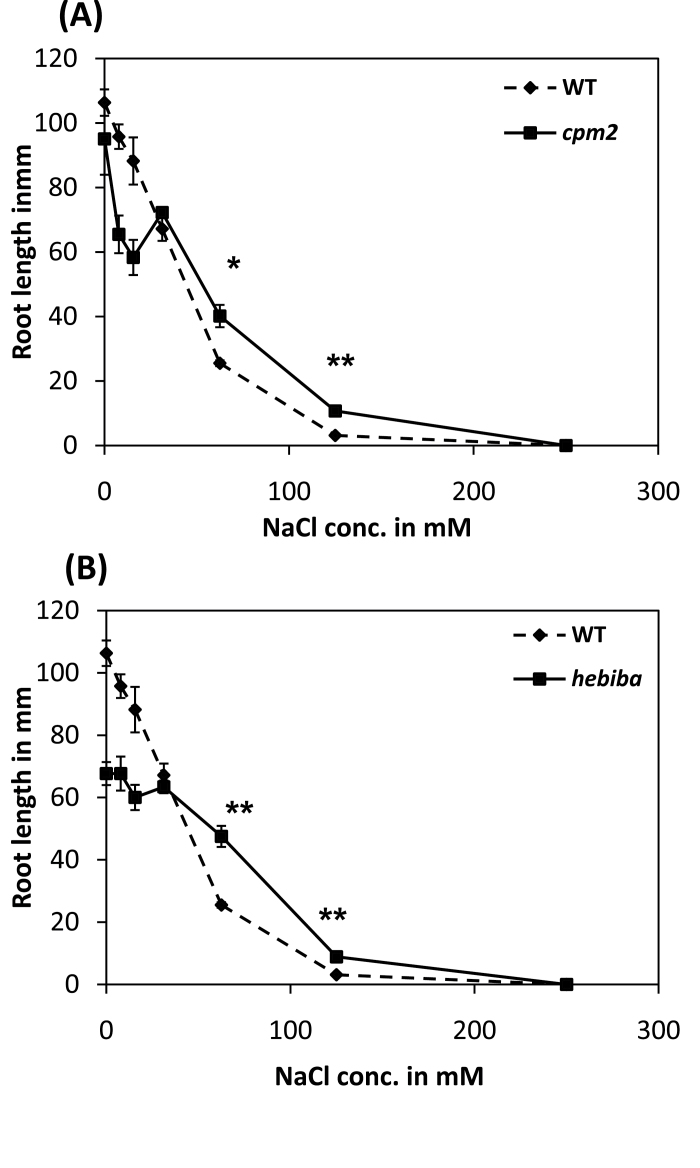
In order to compare the effect of salinity on growth in the wild type and mutants, the length of seminal roots was examined after 7 d in complete darkness when caryopses were germinated on medium containing various amounts of NaCl. Roots of hebiba and cpm2 were shorter under these conditions when grown in the absence of supplemented NaCl or at concentrations <12mM (Fig. 2), indicating a function for AOC in seminal root elongation. When salt concentrations were raised to 62.5mM and 125mM, both wild-type and mutant roots decreased in length. However, this decline was less pronounced in the mutants, such that, under these conditions, mutant roots were significantly longer than those of the wild type (Fig. 2). Neither the wild type nor the JA biosynthesis mutants was able to germinate on medium containing 250mM NaCl.
Fig. 2.
Root length of the wild type (WT) and JA biosynthesis mutants grown in medium containing different concentrations of NaCl. (A) Comparison of the root length of etiolated WT and cpm2 seedlings grown on medium containing 0–250mM NaCl. (B) Comparison of the root length of etiolated WT and hebiba seedlings grown on medium containing 0–250mM NaCl. n=70–100 seedlings. *Significant difference at P<0.05; **significant difference at P<0.01 in a Student’s t-test.
JA biosynthesis mutants accumulate less Na+ ions in shoots, but not in roots
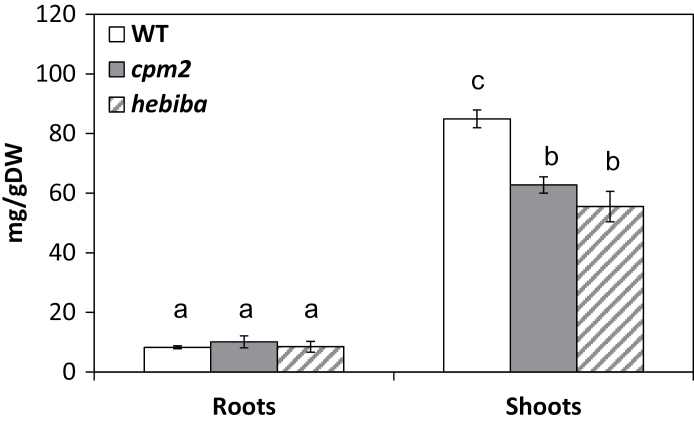
The degree of stress will depend on the uptake and translocation of NaCl. Therefore, the content of sodium ions was examined in roots and shoots of seedlings grown for 3 d on 100mM NaCl. The shoots of the mutants accumulated significantly (by 25–30%) less sodium ions compared with the wild type. However, there was no significant difference between the mutants and the wild type in sodium contents in the roots (Fig. 3). Hence, the weaker stress symptoms observed in mutant leaves correlate with a lower level of sodium ions in the tissue.
Fig. 3.
Content of sodium ions in roots and shoots of the wild type (WT) and JA biosynthesis mutants. Ten-day-old rice WT (white bars), cpm2 (grey bars), and hebiba (striped bars) seedlings were stressed in aqueous NaCl (100mM) solution for 3 d in roots and in shoots. Values represent the mean of at least three independent experiments ±SE. Significant differences amongst different treatments or genotypes are indicated by different letters, according to Tukey’s Honestly Significant Difference (HSD) test (P<0.05).
Enhanced antioxidant activity in JA-deficient mutants
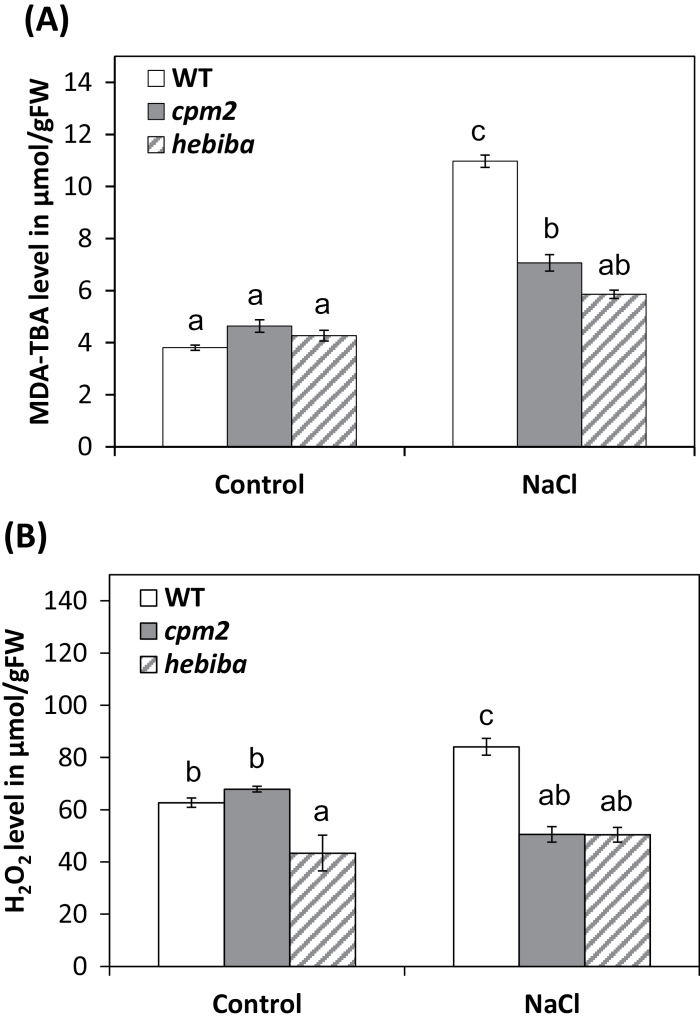
Oxidative damage is a manifestation of stress susceptibility. Therefore oxidative events and the antioxidative system of wild-type and JA biosynthesis mutant plants were analysed under salinity stress. First, the level of lipid peroxidation, as a readout for oxidative degradation of the membrane, was estimated by measuring the product MDA. Generally, the level of MDA was increased in both the wild type and JA mutants in response to salt stress. Nevertheless, the salinity-induced level of MDA in wild-type shoots was ~50% higher compared with cpm2 and almost twice that observed in hebiba (Fig. 4A). To understand the reduced lipid peroxidation in the mutants, the levels of H2O2 in the shoot were determined using Xylenol Orange (Fig. 4B). In the unchallenged controls, the content of H2O2 was similar for the wild type and cpm2, whereas hebiba displayed significantly lower (30%) values. In response to salt stress, the wild type accumulated ~ 60% more H2O2 compared with the mutants. Thus, the pattern for H2O2 paralleled the level of lipid peroxidation reported by MDA.
Fig. 4.
Lipid peroxidation and hydrogen peroxide levels in the wild type (WT) and JA biosynthesis mutants under salinity stress. (A) The level of malondialdehyde (MDA) was estimated in the WT and JA biosynthesis mutants in shoots of control and salt-stressed seedlings. (B) Levels of aqueous peroxide in the WT and JA biosynthesis mutants in shoots of control and salt-stressed seedlings. Values represent the mean of at least three independent experiments ±SE. Results for the WT, cpm2, and hebiba are indicated by white, grey, and striped bars, respectively. Significant differences amongst different treatments or genotypes are indicated by different letters, according to Tukey’s Honestly Significant Difference (HSD) test (P<0.05).
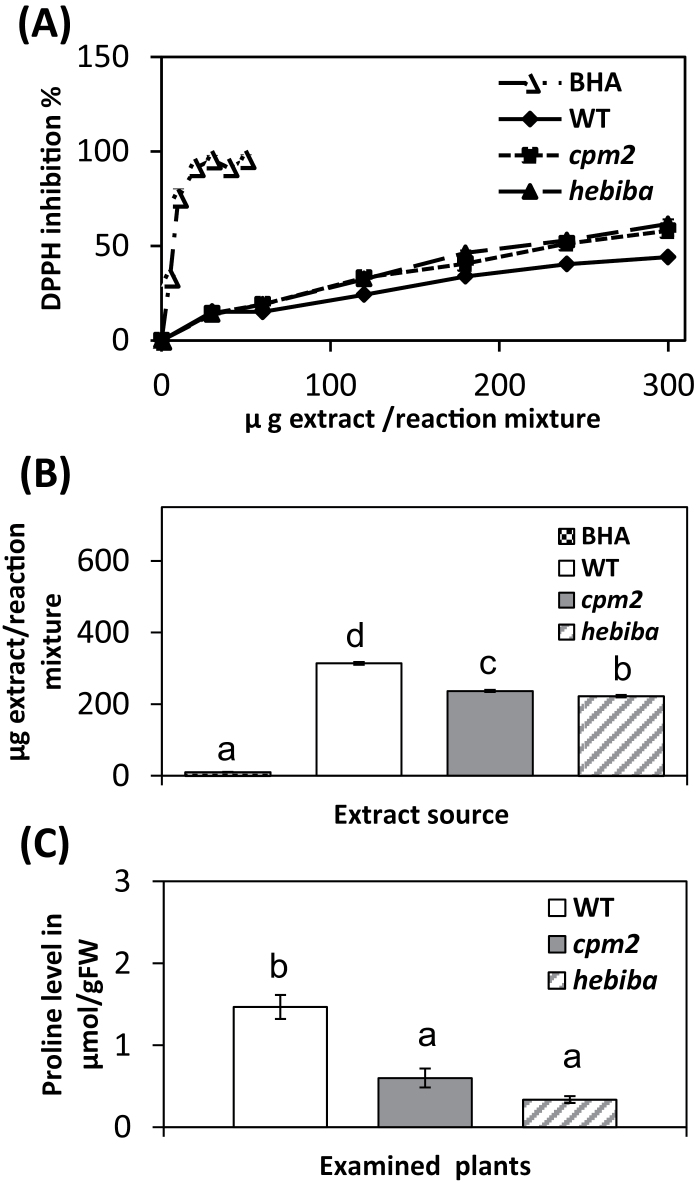
In order to test the ability of the plants to detoxify the ROS, the total antioxidative ability was estimated, by quantifying scavenging of the stable radical DPPH by methanolic leaf extracts. The methanolic extract of all three genotypes succeeded in scavenging the stable radical DPPH in a dose-dependent manner, however, with different efficiency (Fig. 5A). To quantify these differences, the concentrations required to reach 50% inhibition were determined and these IC50 values were plotted as a measure of scavenging activity (Fig. 5B). Compared with the wild type, leaf extracts from the mutants were more efficient, with IC50 values being reduced by about a third, whereby hebiba was most efficient. As a positive control, the powerful scavenger BHA was used. The very low IC50 value of 8.23 μg ml–1 confirms that the assay system was working properly. Thus, the reduced accumulation of MDA in the JA biosynthesis mutants correlates with an elevated activity of antioxidants. To understand this improved ROS detoxification in more detail, additional assays were used that can discriminate between the different species of active oxygen, namely O2·–, H2O2, and OH·–, testing extracts from both control and salt-stressed plants. As shown in Supplementary Fig. S2 at JXB online, aqueous leaf extracts from both cpm2 and hebiba scavenged O2· better than the wild type, with IC50 values that were 30% (cpm2) to 40% (hebiba) lower than in the wild type. In contrast, there was no difference between the wild type and JA biosynthesis mutants in the detoxification of H2O2 and OH· (Supplementary Figs S3, S4). In addition to scavenging superoxide, plants under salt stress accumulate soluble proline as a protection against ion-dependent protein degradation. The level of soluble proline was therefore estimated. The control samples of both the wild type and the mutants did not contain a detectable amount of soluble proline (Fig. 5C). In all the genotypes tested, exposure to salinity led to a considerable increase in proline content. However, the estimated soluble proline amount in the shoots of the mutants was significantly lower than in those of the wild type (by 30% in cpm2, and even by 60% in hebiba).
Fig. 5.
Free radical-scavenging ability and level of soluble proline in the wild type (WT) and JA biosynthesis mutants. (A) DPPH free radical-scavenging activity of standard butylated hydroxyanisole (BHA) and methanolic extract of WT and JA biosynthesis mutants under salinity stress of 100mM NaCl for 3 d. (B) IC50 values were calculated from (A) depending on regression analysis. (C) Soluble proline in the WT, cpm2, and hebiba under salt stress. Proline was not detectable in control samples of the same genotypes. Values represent the mean of at least three independent experiments ±SE. Results for the WT, cpm2, and hebiba are indicated by white, grey, and striped bars, respectively. The dotted symbols in (A) and (B) represent the measurement for BHA. Significant differences amongst different treatments or genotypes are indicated by different letters, according to Tukey’s Honestly Significant Difference (HSD) test (P<0.05).
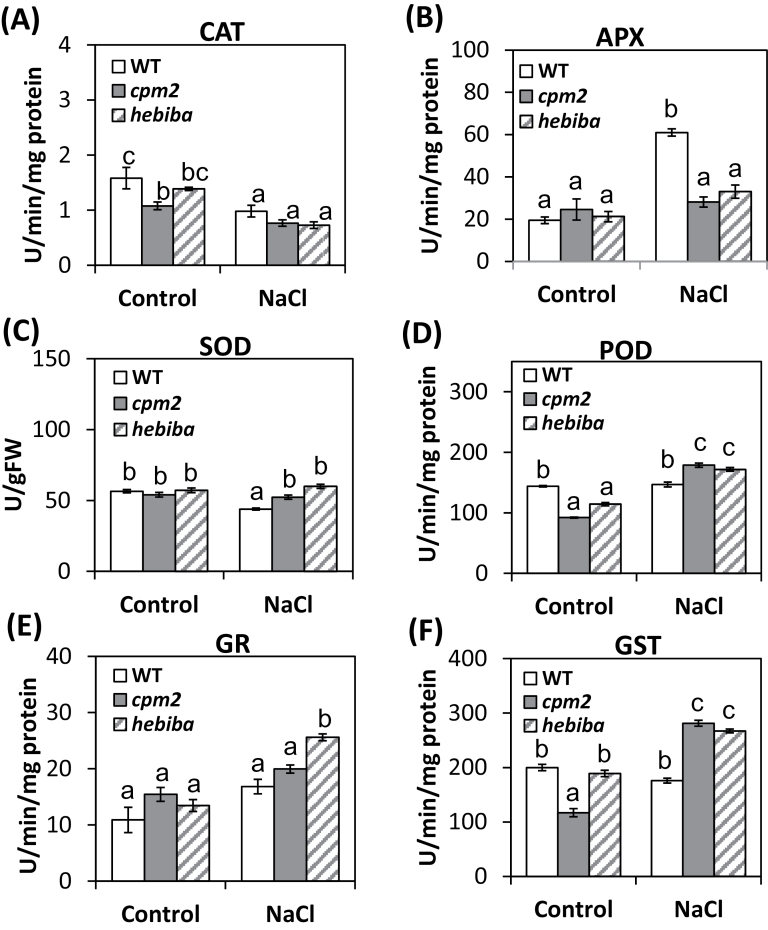
Effect of salinity on the activity of antioxidative enzymes
To understand in more detail by what mechanisms the mutants achieve their improved antioxidant homeostasis, the activities of the enzymatic scavengers CAT, APX, SOD, POD, GR, and GST were determined. Activities were estimated in the shoots of the wild type and JA biosynthesis mutants following treatment with 100mM NaCl over 72h. The activity of CAT was diminished in the shoots of the wild type, cpm2, and hebiba after salt stress (Fig. 6A), and the level of activity under salt stress was not significantly different between the wild type and mutants. In contrast, APX activity was increased in response to salinity stress by almost 3-fold in the wild type but was not significantly increased in the mutants (Fig. 6B). The activity of SOD decreased slightly under salt stress in the wild type, while it remained stable in the mutants (Fig. 6C).
Fig. 6.
Profiles of antioxidative enzymes in both the wild type (WT) and JA biosynthesis mutants under salinity stress. Activity of (A) catalase (CAT), (B) ascorbate peroxidase (APX), (C) superoxide dismutase (SOD), (D) peroxidase (POD), (E) glutathione reductase (GR), and (F) glutathione S-transferase (GST). Values represent the mean of at least three independent experiments ±SE. Results for the WT, cpm2, and hebiba are indicated by white, grey, and striped bars, respectively. Significant differences amongst different treatments or genotypes are indicated by different letters, according to Tukey’s Honestly Significant Difference (HSD) test (P<0.05).
The pattern was different for POD (Fig. 6D), GR (Fig. 6E), and GST (Fig. 6F). Here, the activities of these enzymes were elevated in the mutants in response to salt stress, whereas the wild type did not show significant changes. In summary, it was found that the antioxidative enzyme machinery in cpm2 and hebiba is altered compared with the wild type, whereby the activities of APX and SOD in response to salt were down-regulated, while the activities of POD and GST were up-regulated in the mutants. Mild differences in the activity of several enzymes were also detected in the untreated control plants, indicating that JA affects the antioxidative activity not only in response to stress but also during normal development of plants.
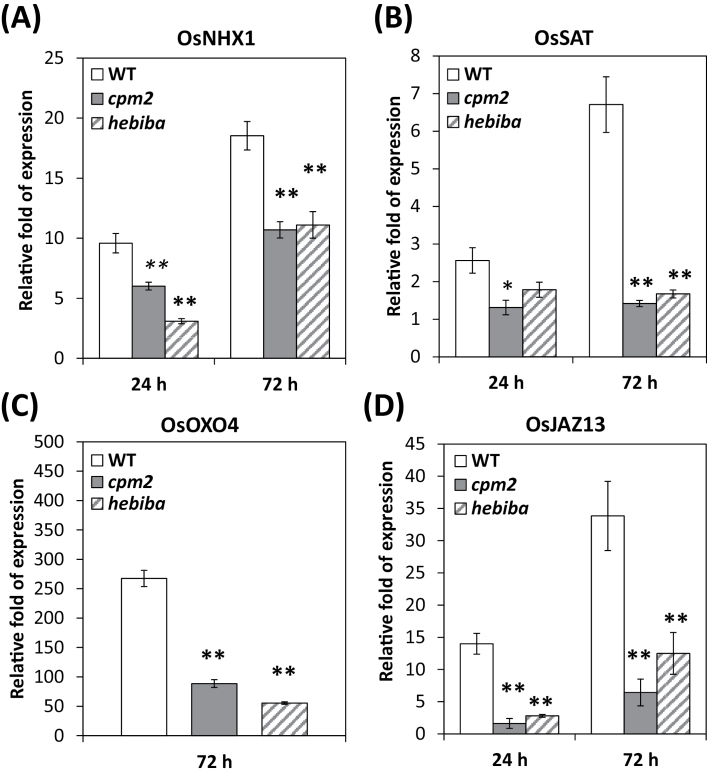
JA deficiency alters gene expression in response to salt stress
In order to assess the adaptation of rice plants to salinity, the expression of salt-stress-related genes was examined after 24h and 72h of growth on saline medium. In both the mutants and the wild type, the relative expression of the OsNHX1 gene (encoding a vacuolar Na+/H+ vacuolar antiporter) was induced by salinity (Fig. 7). However, levels were reduced by 40% in the mutants as compared with the wild type at the two time points investigated. A similarly different pattern was also detected for a gene encoding a protein similar to the enzyme serine acetyltransferase important for oxidative stress responses in Arabidopsis (Dominguez-Solis et al., 2008), referred to as OsSAT (AK287779). In contrast to OsNHX1, this gene was only slightly induced after 24h, but showed an almost 7-fold increase in its transcripts at 72h. However, this gene was not induced in cpm2 and hebiba. Expression of OsOXO.4 (encoding oxalic acid oxidase, catalysing the production of H2O2 and CO2 from oxalic acid) was strongly induced under salinity stress (Fig. 7), but again its induction in the mutants was only 30% (cpm2) or even 20% (hebiba) of that observed in the wild type.
Fig. 7.
Alterations in transcript accumulations of stress-related genes in response to salinity. Plants were treated with 100mM NaCl for 24h and 72h as indicated. Values represent the mean of at least three independent experiments ±SE. Results for the wild type (WT), cpm2, and hebiba are indicated by white, grey, and striped bars, respectively. *Significant difference at P<0.05; **significant difference at P<0.01 in a Student’s t-testt.
In order to investigate the role of JA signalling in modulating the salinity stress response, for the transcript abundance of OsJAZ13 (encoding JAZ protein 13) was determined as a marker of JA signalling. The expression of OsJAZ13 was induced by salt stress. However, the relative change of transcripts was found to be significantly smaller in the mutants at both time points examined (Fig. 7). Nevertheless, the JA biosynthesis mutants showed some induction of this gene, although this induction was only marginal as compared with the wild type.
While OsOXO.4 and OsJAZ13 transcripts accumulated strongly under salinity stress, other typical JA-responsive genes did not show such clear relative changes. The relative change of the transcripts of OsJAR1 and OsJAZ8, two JA-dependent genes which are inducible by wounding, was weak and occurred late (Supplementary Fig. S5 at JXB online), suggesting that an only a subset of JA-dependent genes responds to salt stress.
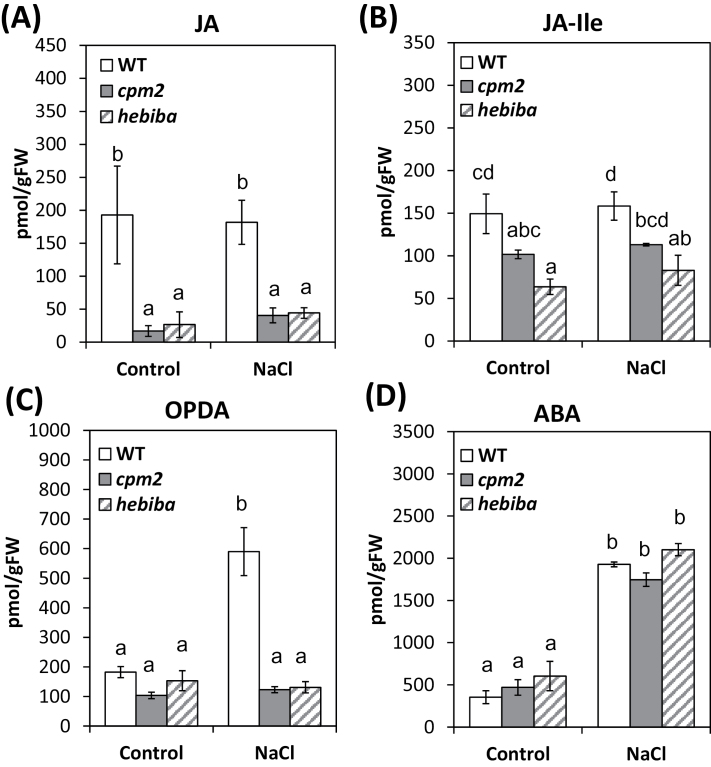
OPDA accumulates in response to salt stress
Levels of OPDA, JA, JA-Ile, and ABA in response to salt stress were compared in the wild type, cpm2, and hebiba. Since the early response was of interest, hormonal levels were compared 6h after challenging the plants with NaCl. As expected, the mutants produced less JA and JA-Ile compared with the wild type (Fig. 8A, B), but no significant increase in JA or JA-Ile levels in response to salt stress was observed, either in the wild type or in either of the mutants. However, in the wild type, OPDA levels increased ~3-fold in response to NaCl, while in the mutants OPDA levels remained very low even after salt stress (Fig. 8C).
Fig. 8.
The level of jasmonates in shoots of the wild type (WT) and JA biosynthesis mutants under control conditions and 6h of salinity stress. Levels of (A) jasmonic acid (JA), (B) JA–isoleucine (JA-Ile), (C) 12-cis-oxophytodienoic acid (OPDA), and (D) abscisic acid (ABA). Values represent the mean of at least three independent experiments ±SE. Results for the WT, cpm2, and hebiba are indicated by white, grey, and striped bars, respectively. Means followed by different letters among treatments are significantly different, according to ANOVA single-factor test with respect to Tukey’s Honestly Significant Difference (HSD) test (P<0.05).
Because the absence of JAs in the mutants may affect ABA biosynthesis, ABA levels were also examined in the same samples. This hormone was clearly induced (~4-fold) by NaCl treatment, but no significant differences in the levels were observed between the different genotypes.
Discussion
The function of JA in salt stress is under debate. On the one hand, exogenous application of MeJA leads to the reduction of sodium uptake in salt-sensitive, but not in salt-tolerant rice cultivars (Kang et al., 2005). On the other hand, exogenous application of MeJA promoted the translocation of sodium ions from root to shoot in maize, but at the same time reduced the overall amount of sodium ions in both roots and shoots (Shahzad, 2011). Part of the discrepancies in the literature might be caused by the fact that the constitutive presence of exogenous MeJA is not equivalent to the tightly controlled and mostly transient accumulation of endogenous JAs observed in the context of stress adaptation (for a review, see Ismail et al., 2014b ). To address the function of endogenous jasmonates, the strategy of analysing the salinity responses of jasmonate synthesis rice mutants was therefore employed. Surprisingly, these mutants performed significantly better than the wild type if challenged by salt stress. They showed fewer salt damage symptoms with respect to wilting of the second and third leaf (Fig. 1), produced longer roots under medium and high concentrations of NaCl (Fig. 2), and maintained a higher content of chlorophyll under salt stress (Supplementary Fig. S1 at JXB online). A detailed analysis of the mutant response revealed three potential phenomena that correlated with this improved salt tolerance: (i) a reduced translocation of sodium into the shoots; (ii) an elevated induction of antioxidants; and (iii) a lack of salt induction of the JA precursor OPDA. In the following, the potential role of these phenomena in salt tolerance or adaptation is discussed.
Jasmonate synthesis mutants translocate less sodium into the shoots
The mutants accumulated smaller amounts of sodium ions in the leaves (Fig. 3). However, the concentration of sodium in the roots was generally much lower than that observed in the leaves, and here the mutants and wild type displayed the same values. The low concentration of sodium in the roots is consistent with the assumption that these concentrations reflect the steady-state levels resulting from uptake, translocation into the leaves, and extrusion (for a review, see Munns and Tester, 2008), whereas the leaves act as sodium sinks. However, it should further be kept in mind that the sodium measurements cannot discriminate between intracellular and apoplastic sodium in the root. The fact that the dose–response curve of root growth as regards sodium is shifted to higher concentrations in both cpm2 and hebiba (Fig. 2) indicates that the effective concentration inside the cells of the root might well be lower in the mutants. It should be noted in this context that the 30% reduction of sodium accumulation in the leaves matched closely the shift of the dose–response curves measured for root growth. Moreover, for the cpm2 mutant, the somewhat weaker reduction of sodium in the leaves is mirrored by a somewhat weaker shift of the dose–response curve for root growth as compared with hebiba. These correlations are consistent with a working model, where the reduced translocation of sodium into the symplasmic transport pathway (Erickson, 1986) accounts for the higher salt tolerance of the mutants. The present data are consistent with the published record on salt-tolerant varieties of rice. For instance, one of the most important parameters that distinguishes the rice salt-tolerant cultivars, such as Pokkali, from salt-sensitive cultivars, such as IR29, is their ability to take up less sodium ions into the leaves when exposed to salt stress (Golldack et al., 2003; Kader and Lindberg, 2005). When, in the mutants, less sodium is translocated to the leaves, but the steady-state level of sodium in the roots does not change, this means either that uptake of sodium through the non-selective cation channels (Ismail et al., 2014a ) is reduced, and/or that the extrusion of sodium through the SOS1 exporter is promoted (for a review, see Munns and Tester, 2008). Irrespective of the underlying mechanism, the elevated salt tolerance of the JA biosynthesis mutants correlates with a reduced translocation of sodium ions into the photosynthetic tissues.
Jasmonate synthesis mutants show elevated induction of antioxidants
ROS accompany many abiotic and biotic stress responses and, although often just considered as a manifestation of stress-related cellular damage, they play a second role as important signals triggering stress adaptation (for a review, see Dat et al., 2000). To use a cellular event as a signal requires that the cell can control both its generation and its elimination. The wild type and mutants were therefore probed for indications of oxidative damage (Pang and Wang, 2008), and for indications of active control of ROS by salt-inducible antioxidants. This set of experiments supports two conclusions: (i) JA biosynthesis mutants display fewer indications of oxidative stress upon challenge by salinity and (ii) JA biosynthesis mutants induce antioxidants more readily upon challenge by salinity stress.
As a first readout for oxidative stress, MDA, an important intermediate in ROS scavenging widely used as an indicator of the extent of oxidation damage under stress (Borsani et al., 2001; Apel and Hirt, 2004), was measured. The MDA level was estimated under salinity, and it was found that its level was significantly lower in the leaves of mutants compared with the wild type exactly parallel with the reduced translocation of sodium into the leaves (compare Figs 4A and 3). This parallelism is to be expected—when less salt is transferred into the leaves this should evoke less oxidative damage in the leaves. A similar result was reported by Vaidyanathan et al. (2003), where the MDA level was lower in Pokkali (a salt-tolerant rice cultivar) compared with Pusa Basmati (a salt-sensitive rice cultivar).
As a second readout for oxidative stress, the level of H2O2 was measured (Fig. 4B). H2O2 is not a free radical (Halliwell et al., 2000), and is therefore comparatively innocuous. However, if converted non-enzymatically, the extremely noxious hydroxyl radical anion (OH·) will be generated which reacts strongly and rapidly (in <1 μs) with proteins, lipids, and DNA, causing cell membrane damage (Ishada et al., 1999). Accumulation of H2O2 in the context of salinity damage has been documented for rice and soybean (Mandhania et al., 2006; Weisany et al., 2012). Similarly to MDA, the mutants do not show elevated levels of H2O2 under salinity, consistent with the reduced translocation of sodium into the leaves (compare Figs 4B and 3). In accordance with the observations described, two genes induced by oxidative stress (OsOXO.4 and OsSAT) were not induced as strongly in the JA biosynthesis mutants as in the wild type (Fig. 7).
As a third readout for oxidative stress, soluble proline was measured. Soluble proline is considered as an osmoprotectant, probably associated with osmotic regulation and membrane stability under stress (Maggio et al., 2000). Although its role under salt stress is not well established, there are many reports, for example supporting a function for proline in osmotic adjustment under salt stress in rice and wheat (Lutts et al., 1996; Poustini et al., 2007). On the other hand, accumulation of proline seems to accompany salinity-induced damage in rice (Garcia et al., 1997; Hoai et al., 2003). In fact, similarly to MDA and H2O2, increased levels of soluble proline were observed under salt stress. However, these increases were significantly lower in the mutants. Again this can be explained by the lower translocation of sodium ions into the mutant leaves. Proline accumulation is therefore seen as an indicator for salinity damage rather than as an indicator for stress tolerance.
Plants are equipped with an array of enzymatic and non-enzymatic antioxidative molecules to alleviate cellular damage caused by ROS (Foyer and Noctor, 2000). Whereas the reduced salinity damage in mutant leaves can be understood mostly in terms of reduced sodium translocation into the transpiration stream, there might also be differences in the capacity or inducibility of the antioxidative system. Among the enzymatic antioxidants, the activity of APX was strongly induced in the wild type, but not in the mutants. This enzyme catalyses the reduction of H2O2 to water and oxygen using ascorbate as an electron donor (Asada, 1999) and, since the mutants accumulate less H2O2 under salt stress (Fig. 4B), their need to rely on APX is less pronounced. Again, this can be explained in terms of reduced sodium translocation and would classify the induction of APX activity as a direct response to salinity-related damage.
However, not all antioxidant responses fell into this pattern. Some events showed an inverse pattern: for instance, the non-enzymatic antioxidative activity measured by the DPPH assay was higher for leaf extracts from challenged JA biosynthesis mutants as compared with the wild type. Moreover, the activities of POD, GR, and especially GST were induced by salt in the mutants, but not in the wild type. When a phenomenon is induced more strongly, although sodium levels are lower, this means that the respective event cannot be a mere manifestation of salinity-induced damage, but probably is adaptive in nature. Peroxidases comprise a group of specific enzymes such as NADPH-peroxidase, NADP-peroxidase, fatty-acid peroxidase, and others, and a group of non-specific enzymes from different sources catalyse the dehydrogenation of antioxidants such as phenolics, aromatic amines, and others in order to break down and/or produce H2O2 and ROS (Malik and Singh, 1980; Donald and Cipollini, 1997). The salt-induced higher POD activity in the mutants might be linked to a better scavenging activity for H2O2 which could be one reason for the lower H2O2 level in the salt-stressed mutants (Fig. 6D). GR and GST are well known to confer general protection against various stress conditions by detoxifying the cellular products of oxidative stress (Marrs, 1996; Yousof et al., 2012). The stronger induction of non-enzymatic antioxidants, as well as the enzymatic activities of POD, GR, and GST in the mutants is observed on the basis of a significantly reduced translocation of sodium, and therefore on the basis of a significantly reduced level of sodium-induced damage. This stronger induction must therefore be caused by elevated sensitivity of sodium-triggered signalling and is evidence for an improved stress adaptation of the JA biosynthesis mutants.
The overlooked damage signal: OPDA
The most striking observation of the plant hormone analysis was a strong increase of OPDA in the wild type by salt stress (Fig. 8). It is known that OPDA, an intermediate of JA biosynthesis, can by itself induce a specific set of genes which are not regulated by JA, and therefore OPDA can be regarded as a signalling molecule in its own right (Taki et al., 2005). OPDA is also discussed as one of the highly reactive electrophile species (RES) responsible for signalling in chloroplasts (for a review, see Farmer and Müller, 2013). Upon binding to its receptor cyclophilin 20–3, OPDA can induce retrograde signalling (Park et al., 2013). When the receptor-–hormone complex interacts with serine acetyltransferase, which stabilizes the formation of cysteine synthase, the redox homeostasis in plastids is altered. This signalling pathway is not active in the mutants, and this may be advantageous for the adaptation of rice to salt stress. In future, once suitable mutants in rice are identified, it will be interesting to compare aoc mutants and mutants of OPDA REDUCTASE, which are able to synthesize OPDA, but not other JAs, in order to compare their sensitivity to salt.
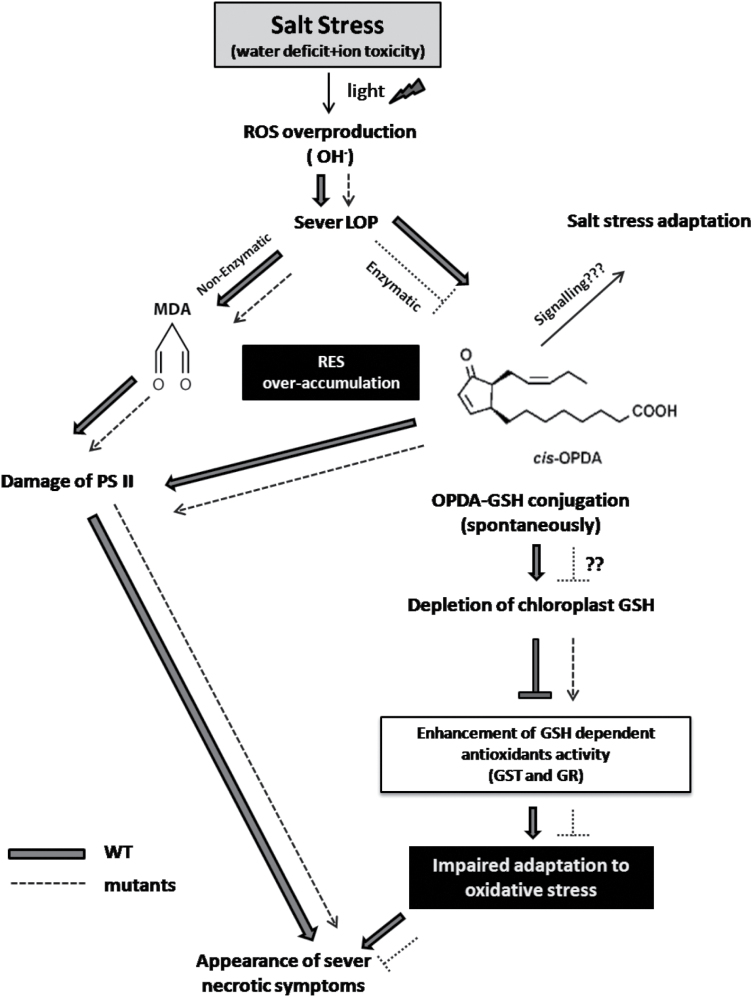
In the proposed model (Fig. 9), the accumulation of more sodium ions in the wild-type leaves in concert with the NaCl-triggered water deficit stress led to overproduction of ROS, especially OH· radicals. This will cause vigorous lipid peroxidation (LOP) (Ishada et al., 1999; Mittler et al., 2004; Halliwell, 2006). LOP in turn will cause, through enzymatic (eLOP) and non-enzymatic (nLOP) routes, the accumulation of RES, such as MDA and OPDA (Farmer and Müller, 2013). Since eLOP is blocked in the JA biosynthesis mutants due to inactivation of AOC (Riemann et al., 2013), OPDA levels will be much higher in the wild type than in cpm2 and hebiba (Fig. 8C). As a result, the wild type will be depleted of free GSH (a nucleophilic thiol-containing antioxidant) as result of stronger spontaneous OPDA–GSH conjugation (Davione et al., 2006). Additionally, the higher abundance of OPDA and MDA is expected to impair the electron transport system in photosystem II (PSII; Alméras et al., 2003). The model predicts higher levels of free GSH, which should result in a higher activity of GSH-dependent antioxidant enzymes such as GST and GR (Chen et al., 2012). Consistent with this prediction from this model, both mutants, in contrast to the wild type, showed elevated activities of GR and GST under salinity stress (Fig. 6E, F).
Fig. 9.
A model representing the effects of salt stress on the wild type and jasmonate biosynthesis mutants under salt stress. The observed phenotype of increased salt sensitivity in the wild type could be linked to the accumulation of higher amounts of MDA and OPDA as a result of higher lipid peroxidation due to ROS accumulation. Both MDA and OPDA are thought to destroy PSII and induce GSH depletion, resulting in disarmed antioxidative ability in the wild type. The mutants showed less accumulation of OPDA and MDA under salt stress conditions and less necrotic symptoms.
As one of the most salt-sensitive glycophytes, rice has very limited effective strategies for dealing with salt stress and does not grow well on saline soil (Galvan-Ampudia and Testerink, 2011). Therefore, lowering the amount of uploaded sodium into the transpiration stream is vital for adaptation under salt stress in rice seedlings. Since sodium contents in the roots were similar in roots and differed in the shoots (Fig. 3), it can be concluded that in the JA biosynthesis mutants less sodium is transferred from the roots to the shoots compared with the wild type. To explain this reduced transfer there are basically three mechanisms: (i) stimulation of stomatal closure resulting in reduction of the transpiration stream; (ii) stimulation of sodium extrusion in the roots by the SOS1 exporter; and (iii) reduction of sodium influx from the apoplastic transport route in the root cortex into the endodermal cells by reduced activity of non-selective cation channels (NSCCs). Future studies, combining molecular and cell biological approaches in a time-resolved manner, will be required to identify the underlying mechanism responsible for the reduced sodium translocation.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The total chlorophyll contents of both WT and JA biosynthesis mutants under control and salt stress conditions.
Figure S2. Superoxide-scavenging assay (SOSA) of both wild-type and JA biosynthesis mutants under salinity stress.
Figure S3. Hydrogen peroxide-scavenging assay (HPSA) of both wild-type and JA biosynthesis mutants under salinity stress.
Figure S4. Hydroxyl radical-scavenging activity (HRSA) of both wild-type and JA biosynthesis mutants under salinity stress.
Figure S5. Alterations in transcript accumulations of stress-related genes in response to salinity.
Table S1. The sequences of forward and reverse primers for the genes of interest and the two genes used for normalization.
Acknowledgements
This work was supported by a fellowship (German-Egyptian Research Long Term Scholarship, GERLS) funded by the Deutsche Akademische Austauschdienst (DAAD), Germany, and the Ministry of Higher Education and Scientific Research, Egypt. We acknowledge Gesine Preuss (KIT) for excellent sodium content analysis by atomic absorption spectroscopy, and Gerd Balcke (IPB Halle) for help with LC-MS measurements. The authors declare to have no conflict of interest.
References
- Abogadallah GM. 2010. Antioxidants defense under salt stress. Plant Signaling and Behavior 5, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar M, Yabuno Y. 1974. Breeding of saline-resistant varieties of rice. II. Comparative performance of some rice varieties to salinity during early developing stages. Japanese Journal of Breeding 25, 1761–1781. [Google Scholar]
- Alméras E, Stolz S, Vollenweider S, Reymond P, Mène-Saffrané L, Farmer EE. 2003. Reactive electrophile species activate defense gene expression in Arabidopsis. The Plant Journal 34, 205–216. [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. 2004. Antagonistic interaction between abscisic acid and jasmonate–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis . The Plant Cell 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. 2004. Reactive oxygen species metabolism oxidative stress, and signals transduction. Annual Review of Plant Biology 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Arnon DI. 1949. Copper enzyme in isolated chloroplast polyphenoloxidase in Beta vulgaris L. Plant Physiology 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. 1999. The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annual Review of Plant Physiology 50, 601–639. [DOI] [PubMed] [Google Scholar]
- Balcke G, Handrick V, Bergau N, Fichtner M, Henning A, Stellmach H, Tissier A, Hause B, Frolov A. 2012. An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 8, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates LS, Waldren BP, Teare ID. 1973. Rapid determination of free proline of water-stress studies. Plant and Soil 39, 205–207. [Google Scholar]
- Beauchamp C, Fridovich I. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Analytical Biochemistry 44, 276–287. [DOI] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA. 2001. Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedling. Plant Physiology 126, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 278–254. [DOI] [PubMed] [Google Scholar]
- Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP. 2012. Drought and salt stress tolerance of an Arabidopsis glutathione-S-transferase U17 knockout mutants are attributed to the combined effect of glutathione and abscisic acid. Plant Physiology 158, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, Van Breusegem F. 2000. Dual action of the active oxygen species during plant stress responses. Cellular and Molecular Life Sciences 57, 779–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davione C, Falletti O, Douki T, Lacazio G, Ennar N, Montillet JL, Triantaphylidés C. 2006. Adduct of oxylipin electrophiles to glutathione reflect a 13 specificity of the downstream lipoxygenase pathway in the tobacco hypersensitive response. Plant Physiology 140, 1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Solis JR, He Z, Lima A, Ting J, Buchanan BB, Luan S. 2008. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts Proceedings of the National Academy of Sciences, USA 105, 16386–16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald F, Cipollini JR. 1997. The induction of soluble peroxidase activity in bean leaves and wind induced mechanical perturbation. American Journal of Botany 85, 1589–1591. [PubMed] [Google Scholar]
- Erickson RO. 1986. Symplastic growth and symplasmic transport. Plant Physiology 82, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Müller MJ. 2013. ROS mediated lipid peroxidation and RES-activated signalling. Annual Review of Plant Biology 64, 429–450. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. 2000. Oxygen processing in photosynthesis: regulation and signaling. New Phytologist 146, 359–388. [Google Scholar]
- Galvan-Ampudia CS, Testerink C. 2011. Salt stress signals shape the plant root. Current Opinion in Plant Biology 14, 296–302. [DOI] [PubMed] [Google Scholar]
- Garcia AB, de Almeida JE, Iger S, Gerats T, van Montagu M, Caplan AB. 1997. Effect of osmoprotectants upon NaCl stress in rice. Plant Physiology 115, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman FD, Bergman CJ. 2004. Rice kernel phenolic content and its relationship with antiradical efficiency. Journal of the Science of Food and Agriculture 84, 1235–1240. [Google Scholar]
- Golldack D, Quigley F, Michalowski CB, Kamasani UR, Bohnert HJ. 2003. Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Molecular Biology 51, 71–81. [DOI] [PubMed] [Google Scholar]
- Halliwell B. 2006. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiology 141, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Clement M, Long L. 2000. Hydrogen peroxide in the human body. FEBS Letters 486, 10–13. [DOI] [PubMed] [Google Scholar]
- Heath R, Packer L. 1968. Photoperoxidation in isolated chloroplasts. I. Kinetics and stochiometry of fatty acid peroxidation. Archives of Biochemistry and Biophysics 56, 189–198. [DOI] [PubMed] [Google Scholar]
- Heldt HW, Heldt F. 2005. Plant biochemistry. 3rd edn. Elsevier Acedemic Press. [Google Scholar]
- Hoai NTT, Shin IS, Kobayashi Kenji U. 2003. Accumulation of some nitrogen compounds in response to salt stress and their relationship with salt tolerance in rice (Oryza sativa L.) seedlings. Plant Growth Regulation 41, 159–164. [Google Scholar]
- Horie T, Karahara I, Katsuhara M. 2012. Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice 5, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishada H, Makino A, Mae T. 1999. Fragmentation of the large subunit if ribulose-1,5-bisphosphate carboxylase by reactive oxygen species occurs near Gly-329. Journal of Biological Chemistry 274, 5222–5226. [DOI] [PubMed] [Google Scholar]
- Ismail A, Seo M, Takebayashi Y, Kamiya Y, Eiche E, Nick P. 2014. a Salt adaptation requires efficient fine-tuning of jasmonate signaling. Protoplasma 251, 881–898. [DOI] [PubMed] [Google Scholar]
- Ismail A, Takeda S, Nick P. 2014. b Life and death under salt stress: same players, different timing? Journal of Experimental Botany 65, 2963–2979. [DOI] [PubMed] [Google Scholar]
- Kader MA, Lindberg S. 2005. Uptake of sodium in protoplast of salt-sensitive and salt-tolerant cultivars of rice, Oryza sativa L., determined by the fluorescent dye SBFI. Journal of Experimental Botany 56, 3149–3158. [DOI] [PubMed] [Google Scholar]
- Kang DJ, Seo YJ, Lee JD, Ishii R, Kim KU, Shin DH, Park SK, Jang SW, Lee IJ. 2005. Jasmonic acid differentially affect growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice culture. Journal of Agronomy and Crop Science 191, 273–282. [Google Scholar]
- Kazan K, Manners JM. 2012. JAZ repressor and the orchestration of phytohormones cross talk. Trends in Plant Science 17, 22–31. [DOI] [PubMed] [Google Scholar]
- Khush GS. 2005. What it will take to feed 5.0 billion rice consumers in 2030. Plant Molecular Biology 59, 1–6. [DOI] [PubMed] [Google Scholar]
- Kumar K, Kumar M, Kim SR, Ryu H, Cho YG. 2013. Insights into genomics of salt stress responses in rice. Rice 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunchandy E, Rao MNA. 1990. Oxygen radical scavenging activity of curcumin. International Journal of Pharmaceutics 58, 237–240. [Google Scholar]
- Lutts S, Kinet JM, Bouharmont J. 1996. Effects of salt stress on growth, mineral nutrition and proline accumulation in relation to osmotic adjustment in rice cultivars differing in salt resistance. Plant Growth Regulation 19, 207–218. [Google Scholar]
- Maggio A, Reddy MP, Joly RJ. 2000. Leaf gas exchange and solute accumulation in the halophyte Salvadora persica grown at moderate salt. Environmental and Experimental Botany 44, 31–38. [DOI] [PubMed] [Google Scholar]
- Malik CP, Singh MB. 1980. Plant enzymology and histoenzymology . New Delhi: Kalyani Publishers, 53. [Google Scholar]
- Mandhania S, Madan S, Sawhney V. 2006. Antioxidants defense mechanism under salt stress in wheat seedlings. Biologia Plantarum 227, 227–231. [Google Scholar]
- Marrs KA. 1996. The functions and regulation of glutathione S-transferases in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 127–158. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Moons A, Prinsen E, Bauw G, Montagu MV. 1997. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transgenic transcripts in rice roots. The Plant Cell 9, 2243–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Physiology and Plant Molecular Biology 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Asada K. 1981. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplast. Plant and Cell Physiology 22, 867–880. [Google Scholar]
- Pang CH, Wang BS. 2008. Oxidative stress and salt tolerance in plants. In: Luttge U, Beyschlag W, Murata J, eds. Progress in botany . Berlin: Springer, 231–245. [Google Scholar]
- Park SW, Li W, Viehhauser A, et al. 2013. Cyclophilin 20–3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proceedings of the National Academy of Sciences, USA 110, 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson GA, Ayers SD, Eberhand DL. 1966. Relative salt tolerance of rice during germination and early seedling development. Soil Science 102, 151–156. [Google Scholar]
- Pedranzani H, Racagni G, Alemano S, Miersch O, Ramirez I, Pena-Cortes H, Taleisnik E, Machado-Domenech E, Abdala G. 2003. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regulation 41, 149–158. [Google Scholar]
- Pedranzani H, Sierra-de-Grado R, Vigliocco A, Miersch O, Abdala G. 2007. Cold and water stresses produce changes in endogenous jasmonates in two populations of Pinus pinaster Ait. Plant Growth Regulation 52, 111–116. [Google Scholar]
- Poustini K, Siosemardeh A, Ranjbar M. 2007. Proline accumulation as a response to salt stress in 30 wheat (Triticum aestivum L.) cultivars differing in salt tolerance. Genetic Resources and Crop Evolution 54, 925–934. [Google Scholar]
- Riemann M, Haga K, Shimizu T, et al. 2013. Identification of rice allene oxide cyclase mutants and the function of Jasmonate for defense against Magnaporthe oryza . The Plant Journal 74, 226–238. [DOI] [PubMed] [Google Scholar]
- Riemann M, Müller A, Korte A, Furuya M, Weiler EW, Nick P. 2003. Impaired induction of the jasmonate pathway in the rice mutant hebiba . Plant Physiology 133, 1820–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch RJ, Cheng SJ, Klaunig JE. 1989. Prevention of cytotoxicity and inhibition of intracellular communication by antioxidant catechins isolated from Chinese green tea. Carcinogenesis 10, 1003–1008. [DOI] [PubMed] [Google Scholar]
- Savchenko T, Kolla VA, Wang CQ, Nasafi Z, Hicks DR, Phadungchob B, Chehab WE, Brandizzi F, Froehlich J, Dehesh K. 2014. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiology 164, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Hwang I, Lee J.S, Choi YD. 2001. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proceedings of the National Academy of Sciences, USA 98, 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad AN. 2011. The role of jasmonic acid (JA) and abscisic acid (ABA) in salt resistance of maize (Zea mays L.). Dissertation University of Giessen, Germany. [Google Scholar]
- Sharma P, Jha AB, Dubey RS, Pessarakli M. 2012. Reactive oxygen species, oxidative damage, and antioxidant defense mechanisms in plants under stressful conditions. Journal of Botany 2012, 1–26. [Google Scholar]
- Staswick PE. 2008. JAZing up jasmonate signaling. Trends in Plant Science 13, 66–71. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Gyohda A, Tominaga M, et al. 2011. RSOsPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant and Cell Physiology 52, 1686–1696. [DOI] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 2005. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiology 139, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan H, Sivakumar P, Chakrabarty R, Thomas G. 2003. Scavenging of reactive species in NaCl-stressed rice (Oryza sativa L.): differential response in salt-tolerant and sensitive varieties. Plant Science 105, 1411–1418. [Google Scholar]
- Venisse JS, Gullner G, Brisset MN. 2001. Evidences for the involvement of an oxidative stress in the initiation of infection of pear by Erwinia amylovora. Plant Physiology 125, 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisany W, Sohrabi Y, Heidari G, Siosemardeh A, Ghassemi-Golezani K. 2012. Changes in antioxidant enzymes activity and plant performance by salinity stress and zinc application in soybean (Glycine max L.). Plant Omics Journal 5, 60–67. [Google Scholar]
- Wolf SP. 1994. Ferrous ion oxidation in the presence of ferric ion indicator xylenol orange for measurement of hydrogen peroxidase. Methods in Enzymology 233, 182–189. [Google Scholar]
- Yousof PY, Hakeem KUR, Chandana R, Ahmad P. 2012. Role of glutathione reductase in plant abiotic stress. In: Ahmad P, Prasad MNV, eds. Abiotic stress responses in plants: metabolism, productivity and sustainability . Berlin: Springer, 149–158. [Google Scholar]
- Zhao Y, Dong W, Zhang N, Ai X, Wang M, Huang Z, Xiao L, Xia G. 2014. A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiology 164, 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhishen J, Mengcheng T, Jianming W. 1999. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chemistry 64, 555–559. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.