Highlight
Here, a function for SlARF9, of the tomato ARF gene family, is defined and new insight provided into the mechanism by which auxin controls cell division and early fruit development.
Key words: Auxin, AUXIN RESPONSE FACTOR 9 (ARF9), cell division, fruit development, fruit size, tomato (Solanum lycopersicum L.).
Abstract
The transformation of the ovary into a fruit after successful completion of pollination and fertilization has been associated with many changes at transcriptomic level. These changes are part of a dynamic and complex regulatory network that is controlled by phytohormones, with a major role for auxin. One of the auxin-related genes differentially expressed upon fruit set and early fruit development in tomato is Solanum lycopersicum AUXIN RESPONSE FACTOR 9 (SlARF9). Here, the functional analysis of this ARF is described. SlARF9 expression was found to be auxin-responsive and SlARF9 mRNA levels were high in the ovules, placenta, and pericarp of pollinated ovaries, but also in other plant tissues with high cell division activity, such as the axillary meristems and root meristems. Transgenic plants with increased SlARF9 mRNA levels formed fruits that were smaller than wild-type fruits because of reduced cell division activity, whereas transgenic lines in which SlARF9 mRNA levels were reduced showed the opposite phenotype. The expression analysis, together with the phenotype of the transgenic lines, suggests that, in tomato, ARF9 negatively controls cell division during early fruit development.
Introduction
The development of the closed carpel is thought to be one of the features that contributed to the evolutionary success of the angiosperms (Scutt et al., 2006). The carpel is the female reproductive organ that differentiates into stigma, style, and the ovary, the latter of which encloses the ovules. After successful completion of pollination and fertilization, the ovary develops into a fruit, with the ovary wall becoming the pericarp and the ovules developing into seeds. The fruit creates a protected environment for the seeds to mature and may mediate dispersal of the mature seeds (Gillaspy et al., 1993). It is widely assumed that reproductive development occurs at the expense of vegetative growth (Snow and Whigham, 1989). Accordingly, the wild progenitors of many fruit crop species produce smaller fruits compared to the domesticated species (Tanksley, 2004; Doebley et al., 2006). A good example is the tomato, with wild relatives such as Solanum pimpinellifolium bearing small fruit, and cultivated tomato species (Solanum lycopersicum L.) producing large fruit with a more than 100-fold increase in weight compared to wild species (Grandillo et al., 1999; Tanksley, 2004). The tomato has been extensively used as a plant model species to study fruit development. Although research has mainly focussed on the later stages of fruit growth, processes occurring during fruit set and early stages of fruit development also have implications on the traits of the mature fruit, such as fruit size and shape (Paran and van der Knaap, 2007).
During flower development, cells at the floral meristems proliferate and differentiate to form the floral organs. The rate, duration, and direction of cell divisions in the developing ovary may already substantially impact final fruit size and shape (Bohner and Bangerth, 1988; van der Knaap et al., 2014). When the ovary has reached its mature size, cell division activity stops. After a few days, the flower may abscise or, upon successful completion of pollination and fertilization, set fruit by resuming further cell division (Gillaspy et al., 1993). This period continues for 10–14 days, and largely determines the final number of cells in the fruit (Bohner and Bangerth, 1988). In the next stage of development, fruit growth essentially depends on cell expansion, with cells increasing up to a 100-fold in volume (Tanksley, 2004). After this 6–7-week period the fruit has reached its final size and will start to ripen (Mapelli et al., 1978; Bünger-Kibler and Bangerth, 1982; Gillaspy et al., 1993).
Auxin plays an important role in tomato fruit set and fruit development. The auxin concentration in the ovary rapidly increases within 2 days after pollination (Mariotti et al., 2011). The application of auxin on unpollinated ovaries leads to the formation of fruits without the need for pollination and fertilization (Gustafson, 1936; Bünger-Kibler and Bangerth, 1982). Similarly, affecting auxin synthesis or responsiveness by the ovary-specific expression of the iaaM or rolB genes from Agrobacterium spp. (Ficcadenti et al., 1999; Carmi et al., 2003) or the overexpression of the auxin receptor TRANSPORT INHIBITOR RESPONSE 1 (TIR1) (Ren et al., 2011) resulted in the formation of seedless tomato fruits. Down-regulation of transcription factors involved in the regulation of auxin-mediated gene expression, like Aux/indole-3-acetic acid (IAA) 9 and AUXIN RESPONSE FACTOR 7 (ARF7) also results in fruit development without the need for pollination and fertilization (Wang et al., 2005a ; de Jong et al., 2009). Transgenic lines in which ARF7 transcript levels were reduced produced fruits with a thick pericarp, due to an increase in cell expansion, and formed a pointy tip at the blossom end of the fruit, demonstrating the effects of cell division and expansion that occur early in development on both size and shape of the mature fruit.
The regulatory role of auxin during the early stages of tomato fruit development is also demonstrated by the rapid increase in expression of auxin-related genes after pollination and fertilization. Previously, cDNA-amplified fragment length polymorphism–based transcript profiling (cDNA-AFLP) has been used to identify genes that are differentially expressed during tomato fruit set (Vriezen et al., 2008). One of the genes induced by pollination appeared to be the putative tomato orthologue of the Arabidopsis ARF9 gene, Solanum lycopersicum ARF9 (SlARF9). Here, a functional analysis of this member of the tomato ARF gene family is described. The phenotypes of transgenic plants with either increased or reduced transcript levels of SlARF9 indicate that SlARF9 negatively controls cell division during early fruit growth.
Materials and methods
Plant materials and growth conditions
Tomato plants (S. lycopersicum L. ‘Moneymaker’) were grown as described in de Jong et al. (2009). For expression analysis of SlARF9 in ovaries, flowers were emasculated 3 days before anthesis. Hand pollination or hormone treatments were carried out at the stage of anthesis. SlARF9 expression under the influence of auxin was analysed in ovaries of flowers treated with 2 µL of 1mM 4-Cl-IAA (Sigma-Aldrich, http://www.sigmaaldrich.com) in 2% ethanol. The treatment was repeated 6h after the first application. Control flowers were collected at the stage of anthesis. For analysis of SlARF9 and SlARF9B expression in the transgenic lines, pericarp tissue was collected from ovaries and fruit that were formed by the second generation (T2) of the SlARF9 overexpression (SlARF9-OE) lines, and the first generation (T1) of SlARF9-RNAi lines. The pericarp of fruits 3–4mm in diameter from the same lines was collected for the transcript profiling analysis. All collected tissues were frozen in liquid N2 and stored at −80°C until RNA extraction.
RNA isolation, cDNA synthesis, and real-time quantitative PCR
Total RNA was extracted from the frozen tomato plant tissues with the TRIzol Reagent (Invitrogen, http://www.invitrogen.com), using a standard protocol from Invitrogen (Chomczynski and Mackey, 1995). Equal amounts of RNA were treated with RNase-free DNase I (Fermentas, http://www.fermentas.com), and used as a template for cDNA synthesis (iScriptTM cDNA synthesis Kit, Bio-Rad, http://bio-rad.com). For real-time quantitative PCR the same conditions were used as described by de Jong et al. (2009). The primers were as follows: SlARF9 (Solyc08g082630), forward 5ʹ-CGTAGGCGTCAACAAATACTTAGAGG-3ʹ, reverse 5ʹ-TCCACTGTGAAGAAAGATCATCAATTCC-3ʹ; SlARF9B (Solyc08g008380), forward 5ʹ-TTGCGTCCTCACAATTCGGAAAGC-3ʹ, reverse 5ʹ-CCAGAGCACCCTTCAGCAGAGC-3ʹ. As reference genes SlActin2/7 (forward 5ʹ-GGACTCTGGTGATGGTGTTAG-3ʹ, reverse 5ʹ-CCGTTCAGCAGTAGTGGTG-3ʹ, based on SGN-U579547), and SlUbi7 (forward 5ʹ-CCCTGGCTGATTACAACATTC-3ʹ, reverse 5ʹ-TGGTGTCAGTGGGTTCAATG-3ʹ, based on SGN- U576276) were used. Each real-time quantitative PCR experiment was based on two technical and two biological repeats, except for the verification of the SlARF9 expression pattern as obtained from the cDNA-AFLP analysis (Vriezen et al., 2008) for which only one biological series was used.
Isolation of the SlARF9 promoter sequence
Genomic DNA was isolated from leaf tissue to generate a SnaI (Fermentas) GenomeWalker tomato library (GenomeWalker Universal Kit, BD Biosciences, http://www.bdbiosciences.com). The use of gene-specific primer 5ʹ-TTCTTCAGCCAGGAAATGACTATTGATAACTCG-3ʹ and nested primer 5ʹ-GGAGAATTCATATTCGGCTGAGAC-3ʹ resulted in the isolation of a 3kb fragment corresponding to the SlARF9 promoter. The Erase-a-Base system (Promega, http://www.promega.com) was used to generate subclones containing progressive unidirectional deletions of this fragment. Subsequently, these subclones were sequenced and aligned using ClustalW (http://www.ebi.ac.uk/clustalW).
Plant transformation
To generate fruit-specific SlARF9 overexpression lines, the coding sequence of SlARF9 (forward 5ʹ-CACCATGGCAACTATAAATGGGTGGTG-3ʹ, reverse 5ʹ- TTAACTGTCTGCGCGAGACAGGG-3ʹ) was cloned into the pENTR/D-TOPO entry vector (Invitrogen). This clone was recombined with the pARC983 binary vector, in which the cauliflower mosaic virus (CaMV) 35S promoter was replaced for the ovary- and young fruit-specific TPRP-F1 promoter (Czerednik et al. 2012). For the generation of the SlARF9-RNAi lines, a fragment of the SlARF9 mid-region (amino acids 367–506, forward 5ʹ-AAAAAGCAGGCTGTCCCACCAACCGCAGAGAAGAAC-3ʹ; reverse 5ʹ-AGAAAAGCTGGGTGCTGTAGTCGTGCCTCAGTAGTGC-3ʹ) was cloned into the pDONR221 entry vector (Invitrogen), which was subsequently recombined with the binary vector pK7GWIWG2(I) (Karimi et al., 2002) in both sense and antisense orientation under the transcriptional regulation of the CaMV 35S promoter and terminator. To generate the pSlARF9::GUS (β-glucuronidase) lines, the promoter fragment of SlARF9 (2200bp, forward 5ʹ-CACCTTTTCAAAGAGGTGTGACATTTTCAATAAC-3ʹ; reverse 5ʹ-CAACCTTCAATTCCAAAAACTAAAGAACACCC-3ʹ) was cloned into the pENTR/D-TOPO entry vector. This entry clone was recombined with the destination vector pKGWFS7 (Karimi et al., 2002).
The transgenic tomato plants were generated by Agrobacterium tumefaciens–mediated transformation, as described in de Jong et al. (2009). Although grown on kanamycin-containing medium, possible escapes were detected by PCR with primers specific for the kanamycin-resistance gene (forward 5ʹ-GACTGGGCACAACAGACAATCG-3ʹ, reverse 5ʹ-GCTCAGAAGAACTCGTCAAGAAGG-3ʹ) on genomic DNA. Subsequently, lines were tested for tetraploidy, as only diploid lines were used for further analysis.
Histochemical analysis of GUS activity
Tissues of first-generation adult plants (T1) and 15-day-old seedlings (T2) of the pSlARF9::GUS lines were submerged in GUS-staining buffer containing 0.1% Triton X-100, 0.5mM Fe2+CN, 0.5mM Fe3+CN, 10mM EDTA, 1mg mL-1 X-Gluc, 0.1mg mL-1 in 50mM phosphate buffer, pH 7.0. After incubation at 37ºC, the tissues were cleared with 70% ethanol and viewed under a stereomicroscope (Leica MZFL III, Leica Microsystems, http://leica-microsystems.com). For detailed analysis of lateral roots and ovules by light microscopy, the GUS-stained tissues were embedded in Technovit 7100 (Heraeus Kulzer, http://www.heraeus-kulzer.com). The embedded tissues were sliced into sections of 5 μm. The sections of the lateral roots were counterstained with 0.5% safranine, and subsequently partly de-stained with 70% ethanol. The sections were viewed under a Leitz Orthoplan microscope (Leica Microsystems). Images were made with a Leica digital camera (model DFC 420C; Leica Microsystems).
Quantification of cell area and number of cell layers
Pericarp tissues of fruits 7–8mm in diameter were fixed in a 2% glutaraldehyde in 0.1M phosphate buffer, pH 7.2, overnight at 4ºC. Subsequently, the tissues were dehydrated in an ethanol series and embedded in Spurr resin. Sections of 1 μm were stained with a toluidine blue solution (0.1% in 1% borax). Pericarp tissue of mature fruits at the breaker stage were fixed in formalin–acetic acid–alcohol solution (3.7–4.1% formaldehyde solution, 5% acetic acid, and 50% ethanol), dehydrated in an ethanol series, and subsequently embedded in Technovit. Sections of 5 μm were stained with a toluidine blue solution. The sections were viewed under a Leitz Orthoplan microscope (Leica Microsystems), and micrographs were made with a Leica digital camera (model DFC 420C; Leica Microsystems). These micrographs were used for further analysis.
For analysis of the 7–8mm fruits, square sections of 0.16mm2 were delimited and positioned approximately 0.1mm from the inner pericarp, including the epidermal layer. For analysis of the mature fruits, sections of 9mm2 were delimited and positioned approximately 1mm from the inner pericarp. Then the total number of cells inside these squares was counted. Cells that were positioned with two-thirds or more of their size in the sections were included. For estimation of the number of cell layers within the pericarp, a line was drawn across the pericarp sections. The number of cells along this line, including cell layers of the epidermis and the three distinct layers of the pericarp (exocarp, mesocarp, and endocarp) were scored (Supplementary Fig. S1). In total, one region per fruit and 4–15 fruits per line were analysed, deriving from 5–12 (7–8mm fruits) or 5–15 (mature fruit) plants per line.
Statistics
For statistical analysis of quantitative PCR data, log2-transformed, reference-gene-corrected Ct values were used (Rieu and Powers, 2009). ANOVA was applied, with a Tukey’s post-hoc test being invoked when multiple comparisons of tomato lines were made for a given (target) gene.
For the statistical analysis of the different phenotypic traits, fruits of 7–8mm and at breaker stage were collected from multiple plants of the different transgenic and wild-type lines arranged in four complete statistical blocks. Because the number of fruit collected for each transgenic line differed per block, the imbalance precluded use of ANOVA and so the method of residual maximum likelihood was used to fit a linear mixed model to the data, taking into account the blocks as a random effect. Where necessary, the method also took into account the nesting of fruit within clusters and clusters within plants as random effects, so that comparison of transgenic lines was based on the correct degrees of freedom for underlying residual (plant-to-plant) variation. A log (to base e) transformation was used for fruit weight to account for some heterogeneity of variance across the lines. Means for the lines were compared using standard error of the difference (SED), invoking the least significant difference values at the 5% or 1% level of significance. The GenStat (14th edition, VSN International Ltd, Hemel Hempstead, UK) was used for this analysis.
Microarray data analysis
The transcript profiling analysis was done using pericarp tissue from fruits 3–4mm in diameter, with each sample containing the pericarp of two fruits. For each separate line, tissues were collected from two to three plants, resulting in a total of six samples for SlARF9-OE, seven samples for SlARF9-RNAi, and two samples for wild type. Total RNA was extracted as described for real-time quantitative PCR. To synthesize cDNA, 100ng of total RNA was used with the Ambion WT expression kit (Applied Biosystems/Life Technologies, Nieuwerkerk aan den IJssel, The Netherlands). Subsequently, the cDNA was labelled with biotin, using the Affymetrix GeneChip WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA, USA), and hybridized to the Affymetrix EUTOM3 tomato exon arrays. The microarray signals were determined using MadMax microarray analysis software (hhtp://madmax.bioinformatics.nl). Data were deposited in the NCBI GEO repository, accession number GSE63637. Further analysis was performed using GeneMaths XT microarray data analysis software (Applied Maths, http://www.applied-maths.com/genemaths-xt). Prior to analysis, the data was normalized using 2log transformation, followed by mean subtraction scaling. Student’s t-test and principal component analysis (PCA) analyses were performed in GeneMaths XT and PAST3 (http://folk.uio.no/ohammer/past/). The Pearson correlation coefficients were calculated using the corresponding function of Microsoft Office Excel 2010. Pairwise comparison between the transcriptomes of wild-type, SlARF9-OE, and SlARF9-RNAi fruits (t-test, P < 0.05) resulted in a list of 753 differentially expressed genes (Supplementary Table S1).
All genes represented on the array, with a maximum expression value of >10 across the samples, were classified according to the MapMan functional categories (Thimm et al., 2004), which have been assigned based on the ITAG2.3 genome annotation (http: //mapman.gabipd.org). The distribution of the functional categories was evaluated with gene set enrichment analysis (GSEA) (Subramanian et al., 2005). The natural scale intensity data were used as the input for the GSEA (April 2014), and the genes were ranked based on the signal-to-noise metric. Gene sets with <10 and >500 members were ignored, leaving out 843 of the 1209 MapMan categories. With these settings, four gene sets with a false discovery rate (FDR) <25% were found to be up-regulated in the SlARF9-RNAi lines. Leading edge analysis was used to extract the genes that accounted for the gene set’s enrichment signal.
Results
Expression of SlARF9
The transcriptome of ovaries before and after fruit induction has previously been analysed to study the role of phytohormones in tomato fruit set (Vriezen et al., 2008). One of the characterized transcripts specifically modulated after pollination encoded an ARF protein homologous to Arabidopsis thaliana ARF9 (GenBank Accession No. BT013639), and was therefore designated SlARF9 (ITAG2.3 Solyc08g082630) (Zouine et al., 2014). The derived protein sequence of SlARF9 contains 658 amino acids, and comprises the N-terminal B3-derived DNA binding domain (amino acids 74–236) and two C-terminal homo- and heterodimerization domains, III and IV (CTD, amino acids 566–602 and 609–651, respectively) that are typically present in ARFs (Supplementary Fig. S2) (Guilfoyle and Hagen, 2007). The middle region (MR) of ARF proteins, located between the DNA binding domain and CTD, functions as a transcriptional activation or repression domain depending on its amino acid composition (Ulmasov et al., 1999a ; Tiwari et al., 2003). The MR of SlARF9 is enriched with serine, which represents 11.6% of amino acid residues, suggesting that this ARF may act as a transcriptional repressor.
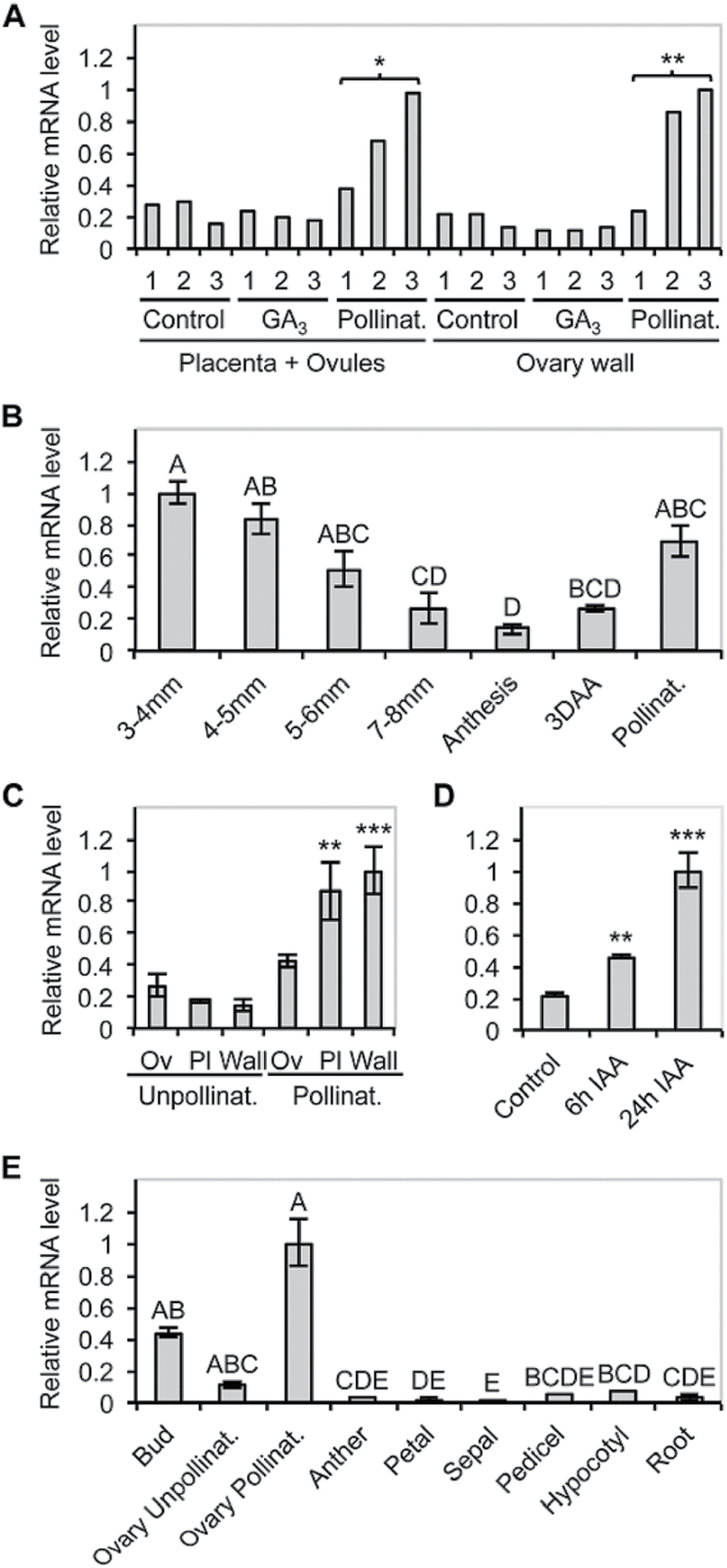
The transcript profiling described in Vriezen et al. (2008) showed that SlARF9 expression increased within 48h after pollination, but not after treatment with gibberellin (GA). Furthermore, SlARF9 was shown to be expressed in the placental and ovular tissues as well as the ovary wall. In the current study, these patterns were verified by real-time quantitative PCR (Fig. 1A). Expression analysis in ovaries collected at various stages of flower development showed that the SlARF9 transcript was also highly abundant in the early stages of flower development. The transcript levels were reduced during the later stages of flower development, reaching the lowest level of expression at anthesis and remaining low unless successful pollination and fertilization occurred (Fig. 1B). These processes increased SlARF9 expression mainly in the placental tissue and the ovary wall (Fig. 1C).
Fig. 1.
SlARF9 mRNA levels during tomato fruit set. (A) Verification of the SlARF9 expression pattern as obtained from the cDNA-AFLP analysis (Vriezen et al., 2008) by real-time quantitative PCR on placenta together with ovular tissue and the ovary wall, at 1, 2 and 3 days after treatment. Total RNA was isolated from emasculated flowers (Control), emasculated flowers treated with gibberellic acid (GA3), and emasculated flowers after hand pollination (Pollinat.) (pools of 3–5 ovaries per sample). *significantly different from control treatment, P < 0.05; **P < 0.01. (B) Relative mRNA levels of SlARF9 in tomato ovaries collected throughout different stages of flower development, at anthesis, from unpollinated flowers 3 days after anthesis (3DAA), and flowers 3 days after hand pollination (Pollinat.). Standard errors are indicated (n = two pools of 3–5 flowers). Capital characters above bars indicate homogenous categories (Tukey) with differences at P < 0.05. (C) Relative mRNA levels of SlARF9 in unpollinated tomato ovaries at anthesis and ovaries collected 3 days after hand pollination, dissected into ovule (Ov), placenta (Pl), and ovary wall tissue (Wall) samples. Standard errors are indicated (n = two pools of 10 ovaries). **significantly different from the unpollinated control, P < 0.01; ***P < 0.001. (D) Relative mRNA levels of SlARF9 in tomato ovaries of emasculated flowers collected 6 or 24h after auxin treatment (IAA). Untreated ovaries were used as a control. Standard errors are indicated (n = two pools of 3–5 ovaries). **significantly different from the control, P < 0.01; ***P<0.001. (E) Relative mRNA levels of SlARF9 in young flower buds, unpollinated ovaries, and various other floral organs collected from flowers at the stage of emasculation, pollinated ovaries (3 DAP), and in the hypocotyl and root of 10-day-old seedlings. Standard errors are indicated (n = 2). Capital characters above bars indicate homogenous categories (Tukey) with differences at P < 0.05.
Although GA-treatment of unpollinated mature ovaries had no effect on SlARF9 expression, treatment with IAA resulted in an increase of SlARF9 transcript levels (Fig. 1D). In silico analysis of the 1.5kb promoter sequence for the presence of auxin-related cis-acting regulatory elements using PlantCARE (Lescot et al., 2002) and PLACE (Higo et al., 1999) software resulted in the identification of two degenerated auxin response elements (AuxREs; Supplementary Table S2). These elements are typically found in the promoter sequences of auxin response genes and are bound by the ARF transcription factors (Ulmasov et al., 1999b ). Furthermore, the SlARF9 promoter sequence contains several NTBBF1ARROLB-elements. These elements were first identified in the promoter sequence of rolB, one of the oncogenes present in the T-DNA sequence of Agrobacterium rhizogenes, and are involved in the auxin-inducible expression of the rolB-gene in plants (Baumann et al., 1999). Both AuxREs and NTBBF1ARROLB-elements are also present in the promoter regions of SlIAA2 and SlIAA14. These are transcriptional repressors that regulate the expression of auxin-responsive genes. However, many Aux/IAA genes are auxin-inducible themselves (Reed, 2001). Similar to SlARF9, the expression of SlIAA2 and SlIAA14 was found to be up-regulated in ovaries by pollination (Vriezen et al., 2008) and treatment with auxin (Supplementary Fig. S3).
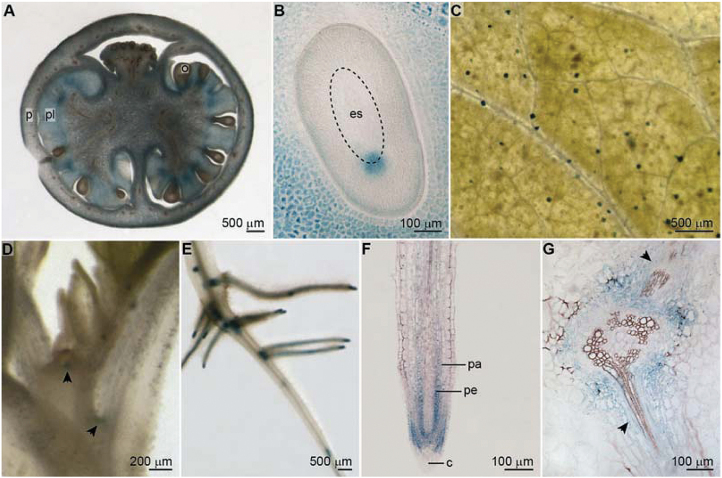
To investigate the expression of SlARF9 in more detail, an SlARF9 promoter-GUS fusion was constructed using the 2200bp 5ʹ-end flanking sequence of SlARF9 and the GUS-coding sequence of the uidA gene. Subsequently, this pSlARF9::GUS construct was introduced into tomato by Agrobacterium-mediated gene transfer. In seven out of the fourteen independent lines that were generated, uidA-expression was observed in tomato fruits of 5–6mm in diameter, corresponding to approximately 8 days after pollination (DAP). The GUS staining was visible in the pericarp, the outer cell layers of the placenta that develop into a gel-like substance, and in the ovules (Fig. 2A). Microscopic analysis of cross-sections through the ovules showed that the GUS staining was located at the micropylar end of the embryo sac, corresponding to the location of the suspensor or wall ingrowths that develop quickly around the base of the suspensor (Fig. 2B) (Briggs, 1995). The levels of SlARF9 transcript were low in plant tissues other than the ovary (Fig. 1E). Nevertheless, GUS staining could also be seen in the glandular hairs at the surface of leaf and stem (Fig. 2C) and in the axillary shoot apical meristem (Fig. 2D). Furthermore, GUS staining was observed in the primary root tips, early lateral root primordia, and outgrowing lateral roots (Fig. 2E). Here, the staining was located in the meristematic zone of the root tips, the pericycle, and in a few cell layers of the parenchyma (Fig. 2F,G). These findings indicate that although SlARF9 is predominantly expressed in the fruit, SlARF9 may also function in other tissues, mostly those in which many cell divisions occur.
Fig. 2.
Histochemical GUS staining of pSlARF9::GUS tomato lines. (A) Tomato fruit, 5–6mm in diameter, corresponding to approximately 6 DAP. The GUS staining is visible in the ovules (o), placenta (pl), and pericarp (p). (B) Cross section of an ovule from a 5–6mm tomato fruit. The GUS staining is localized at the micropylar end of the embryo sac (es), which is encircled. (C) Glandular hairs and trichomes on the leaf surface. Only the glandular hairs showed GUS activity. (D) The apex of a 15-day-old seedling. GUS is expressed at the axillary meristems, at the base of the leaves (arrows). (E) Primary and lateral roots of a 15-day-old seedling. (F) Longitudinal section of a lateral root tip of a 15-day-old seedling. The GUS staining is located in the meristematic zone, but not in the columella (c). The pericycle (pe) and a few cell layers of parenchyma (pa) were also stained. (G) Longitudinal section through two emerging lateral roots (arrows).
Overexpression and silencing of SlARF9 have opposite effects on fruit size
To explore the physiological role of SlARF9 in tomato fruit set and development, transgenic tomato lines were generated in which the gene was overexpressed or silenced. For the production of the SlARF9 overexpression lines (SlARF9-OE), the coding sequence of SlARF9 was ligated to the TPRP-F1 promoter, which is specific for the ovary and young fruit (Carmi et al., 2003). From the 11 independent transgenic lines that were generated, the two SlARF9-OE lines with the highest expression, i.e. lines 4 and 5, were selected for further analysis. Transgenic tomato lines in which the SlARF9 gene was silenced were generated by an RNAi approach, using a 420bp fragment based on the MR of SlARF9 (amino acids 367–506, Supplementary Fig. S2). This fragment was cloned into an RNAi binary vector, under the transcriptional regulation of the CaMV 35S promoter, and transferred to tomato by Agrobacterium-mediated transformation. In four out of the twelve generated transgenic lines the SlARF9 transcript levels were reduced. These SlARF9-RNAi lines (numbers -1, -6, -9 and -12) were used for further analysis.
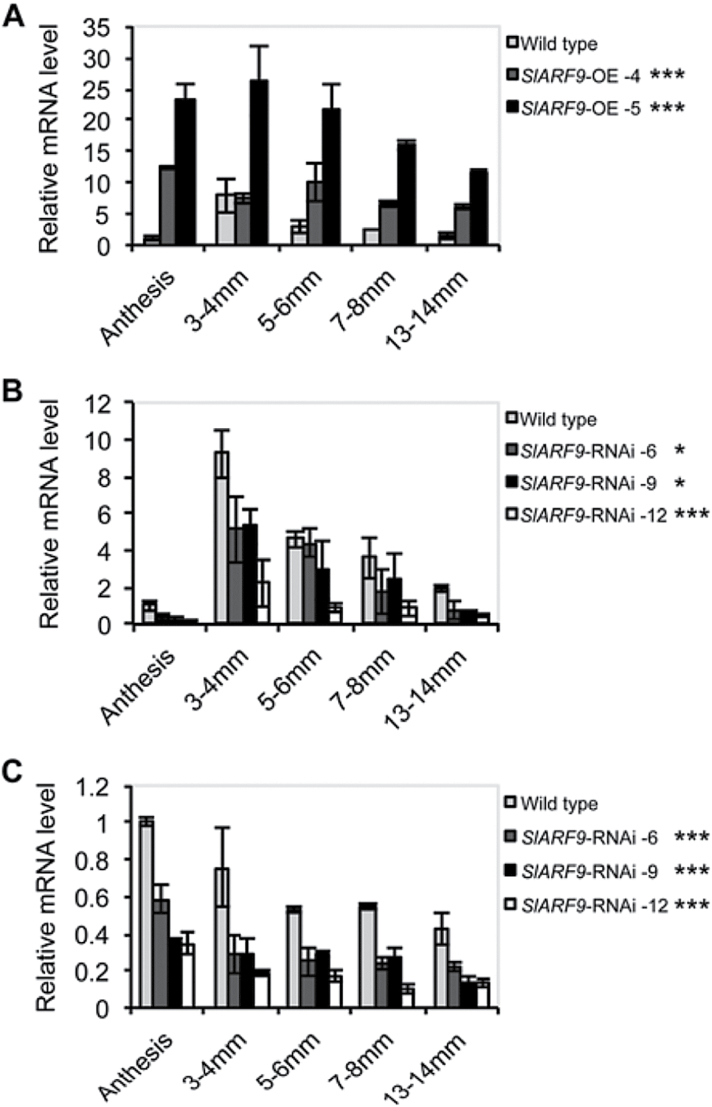
Expression analysis of SlARF9 during several early stages of fruit development showed that in wild type the relative mRNA level of SlARF9 rapidly increased after pollination and fertilization, and was highest in fruits of 3–4mm in diameter, corresponding to 6 DAP. In subsequent stages the transcript levels decreased again (Fig. 3A,B; Supplementary Table S3). In the SlARF9-OE lines, SlARF9 transcript levels were already high at anthesis independently of pollination, and remained high for a longer period of time than in wild-type fruits (Fig. 3A). In the SlARF9-RNAi lines the expression pattern of SlARF9 was similar to that in wild type, but the overall transcript level was reduced by 40–70% and most prominently in the 3–4mm fruit (Fig. 3B). Recently, Zouine et al. (2014) identified 22 putative functional ARF genes in the tomato genome. The authors’ phylogenetic analysis showed that SlARF9 clusters with another ARF gene located on chromosome 8, referred to as SlARF9B (ITAG2.3 Solyc08g008380). Although closely related to SlARF9, the expression of SlARF9B in the ovary was not affected by pollination (Fig. 3C). The specificity of the 420bp fragment of SlARF9 used to generate the SlARF9-RNAi lines was tested by genomic DNA Southern blot analysis, which resulted in a single strong hybridization signal (Supplementary Fig. S4). However, analysis of SlARF9B expression in ovaries and young fruits collected from the SlARF9-RNAi lines showed that the transcript level of SlARF9B was decreased compared to that in wild type (Fig. 3C). The SlARF9 fragment used for generation of the RNAi lines contained three stretches of 21–23 nucleotides highly similar to the sequence of SlARF9B, only containing one or two mismatches. It is possible that these fragments brought about the degradation of the SlARF9B mRNA.
Fig. 3.
SlARF9 and SlARF9B mRNA levels in developing wild-type and transgenic fruits. Relative mRNA levels of SlARF9 in ovaries and fruits collected from (A) wild-type and SlARF9-OE lines, and (B) SlARF9-RNAi lines. (C) Relative mRNA levels of SlARF9B in wild-type and SlARF9-RNAi ovaries and fruits. Standard errors are indicated (n = two pools of 3–5 ovaries). Statistical analysis of SlARF9 expression in the wild type (a,b) is presented in Table S2. *significantly different from the wild type, P < 0.05; *** P < 0.001.
Although the RNAi construct was under regulation of the constitutive 35S promoter, no obvious vegetative phenotypes were observed. By contrast, both SlARF9-OE and SlARF9-RNAi lines showed a clear and opposite phenotype in the fruit. Overexpression of SlARF9 resulted in a reduction in the final size and weight of fruits, as determined at breaker stage (Table 1). SlARF9 silencing, on the other hand, resulted in bigger and heavier fruit as compared to the wild type (Table 1, Supplementary Fig. S1D).
Table 1.
Analysis of fruit size and weight of mature wild-type and transgenic fruits, as determined at breaker stage
| Line | Width (cm) | P (line) | P (type) | Height (cm) | P (line) | P (type) | Weight (g) | P (line) | P (type) |
|---|---|---|---|---|---|---|---|---|---|
| Wild type | 5.685 | 5.220 | 84.775 (4.440) |
||||||
| SlARF9-OE-4 | 5.332 | *** | ** | 3.718 | *** | *** | 64.457 (4.166)a |
*** | *** |
| SlARF9-OE-5 | 5.482 | ns | 3.944 | *** | 73.553 (4.298) |
* | |||
| SlARF9-RNAi-1 | 6.095 | *** | *** | 6.005 | *** | *** | 101.799 (4.623) |
*** | *** |
| SlARF9-RNAi-6 | 6.357 | *** | 6.118 | *** | 107.878 (4.681) |
*** | |||
| SlARF9-RNAi-9 | 6.184 | *** | 6.044 | *** | 108.636 (4.688) |
*** | |||
| SlARF9-RNAi-12 | 5.962 | ns | 96.641 (4.571) |
ns | |||||
| Maximum SED Degrees of freedom |
0.1787 121 |
0.1835 62 |
0.0893 131 |
To determine the width and weight of wild-type and transgenic fruit, fruits were collected from plants grown in one to four complete statistical blocks with 5–143 replicates per line per block. The height of the fruit was determined in two of the blocks with 18–143 replicates per genotype per block. The level of significance compared to wild-type fruits is indicated for each individual line and per type of transgenic line. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. a The means on the log (to base e) scale, are used for statistical comparisons.
Modulation of SlARF9 expression level affects cell expansion and division at early stage
To understand the cause of the SlARF9-dependent changes in fruit size, cell size was determined by examining the number of cells per surface unit, and number of cell layers in the pericarp of breaker stage fruits. The pericarp of the fruits of the SlARF9- silenced lines, which bore bigger fruits, had smaller average cell size (greater cells/mm2), but more cell layers (Table 2, Supplementary Fig. S1). In the SlARF9-OE lines, which bore smaller fruits, the opposite was seen because the pericarp in the fruits contained fewer cell layers. Cell size was not significantly (P < 0.05, least significant difference) increased in these lines compared to wild type.
Table 2.
Quantification of cell number per surface unit and number of cell layers in the pericarp of mature wild-type and transgenic fruits, collected at breaker stage
| Line | Cells/mm2 | P (line) | P (type) | Cell layers | P (line) | P (type) |
|---|---|---|---|---|---|---|
| Wild type | 6.192 | 27.53 | ||||
| SlARF9-OE-4 | 5.676 | ns | ns | 24.39 | * | * |
| SlARF9-OE-5 | 5.395 | ns | 23.96 | * | ||
| SlARF9-RNAi-1 | 7.391 | ns | ** | 36.03 | *** | *** |
| SlARF9-RNAi-6 | 7.307 | ns | 32.85 | *** | ||
| SlARF9-RNAi-9 | 7.241 | ns | 34.40 | *** | ||
| SlARF9-RNAi-12 | 9.755 | *** | 34.13 | *** | ||
| Maximum SED | 1.1514 | 2.034 | ||||
| Degrees of freedom | 49 | 50 |
To determine the cell number and number of cell layers in the pericarp, fruits were collected from plants grown in three complete statistical blocks with 2–7 replicates per genotype per block. The level of significance compared to wild-type fruits is indicated for each individual line and per type of transgenic line. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Several TILLING mutants with mutations in the SlARF9 gene were also identified and analysed, but none of them had larger fruits than the corresponding wild type. One of the lines, slarf9-1, carried a C-to-T mutation translating into a histidine 179-to-tyrosine substitution in the middle of the B3 DNA binding domain. BLAST analysis showed that this amino acid is highly conserved among tomato ARFs and completely conserved among ARF9-related proteins up to the monocots. Although fruit weight of this mutant was normal (Table 3), cytological analysis showed it had significantly (P < 0.05, least significant difference) more cell layers in the pericarp at breaker stage than the wild type (Table 3).
Table 3.
Quantification of fruit weight and number of cell layers in the pericarp of mature wild-type and slarf9-1 fruits, collected at breaker stage
| Line | Weight (g) | P | Cell layers | P |
|---|---|---|---|---|
| Wild type | 51.05 | 26.50 | ||
| slarf9-1 | 55.14 | ns | 30.17 | * |
*significantly different from wild type, P < 0.05; ns, not significant.
To test whether the SlARF9-modulated transgenic lines already differentiated from wild type in the earlier stages of fruit development, histological cross-sections of 7–8mm fruits were also analysed. In the pericarp of SlARF9-OE fruits, cells were on average bigger than in wild-type fruits, but they still contained a normal number of cell layers. In the SlARF9-RNAi lines, cells were smaller and the number of cell layers was already increased (Table 4, Supplementary Fig. S5).
Table 4.
Quantification of cell number per surface unit and number of cell layers in the pericarp of wild-type and transgenic fruits, 7–8mm in diameter
| Line | Cells/mm2 | P (line) | P (type) | Cell layers | P (line) | P (type) |
|---|---|---|---|---|---|---|
| Wild type | 779.2 | 25.77 | ||||
| SlARF9-OE-4 | 585.1 | * | ** | 26.41 | ns | ns |
| SlARF9-OE-5 | 582.0 | * | 25.13 | ns | ||
| SlARF9-RNAi-1 | 1164.7 | *** | *** | 30.93 | * | *** |
| SlARF9-RNAi-6 | 1296.3 | *** | 31.87 | *** | ||
| SlARF9-RNAi-9 | 1077.0 | ** | 30.66 | ** | ||
| SlARF9-RNAi-12 | 1441.1 | *** | 29.97 | * | ||
| Maximum SED | 136.30 | 2.517 | ||||
| Degrees of freedom | 47 | 46 |
To determine the cell number and number of cell layers in the pericarp, fruits were collected from plants grown in two complete blocks with 2–6 replicates per genotype per block. The level of significance compared to wild–type fruits is indicated for each individual line and per type of transgenic line. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Transcriptional analysis of early fruit development in SlARF9-OE and SlARF9-RNAi lines
To identify possible transcriptomic changes associated with the fruit developmental phenotypes observed in the transgenic lines, the gene expression profiles of 3–4mm fruits from wild-type and transgenic plants were analysed. At this stage, the SlARF9 transcript reached its maximum level in wild-type fruits (Fig. 3), but no phenotypic differences were observed between wild-type and transgenic fruits. The transcript profiling analysis was carried out using Affymetrix EUTOM3 tomato exon arrays.
The microarray analysis showed a 2.6-fold difference in SlARF9 expression between wild-type and SlARF9-OE fruits (P < 0.001, Student’s t-test), and a 1.6-fold difference between wild-type and SlARF9-RNAi lines (P < 0.05, Student’s t-test), closely corresponding to the results obtained by real-time quantitative PCR analysis (Fig. 3). PCA of the normalized microarray data indicated that, overall, the transcriptomes of the different types were somewhat similar, but especially those of wild-type and SlARF9-OE fruit (Supplementary Fig. S6A). Comparable results were obtained when applying the PCA to only those genes with significantly different expression levels between any two types based on a per gene ANOVA test, as the first principal component (PC1) did not separate wild-type from OE plants (Supplementary Fig. S6B). Possibly, the SlARF9-dependent transcriptional responses were already nearly saturated in 3–4mm wild-type fruits.
Because the SlARF9-OE and SlARF9-RNAi fruits had opposite phenotypes, pairwise comparison between the transcriptomes of these lines was done. Unexpectedly, the analysis showed that only SlARF9 expression levels remained significantly different between the two types after correcting for multiple comparisons (FDR = 0.03); adding the next most significant group of 14 genes raised the FDR to 0.50. Therefore, instead of looking at individual genes, GSEA was applied to the whole dataset (Subramanian et al., 2005). Using MapMan functional gene categories as gene sets (Thimm et al., 2004), three biological processes were found to be significantly overrepresented in the SlARF9-RNAi fruits (FDR < 0.25). These were brassinosteroid (BR) biosynthesis and degradation (bins 17.3.1 and 17.3.1.2), polyamine metabolism (22.1), and cell death (31.5; Supplementary Table S4).
Discussion
After successful completion of pollination and fertilization, molecular, biochemical, and structural changes transform the ovary into a fruit. These changes are likely to be preceded by changes at the transcriptomic level, giving rise to a dynamic and complex regulatory network that includes signalling by phytohormones such as auxin, GA, ethylene, and abscisic acid (Lemaire-Chamley et al., 2005; Vriezen et al., 2008; Molesini et al., 2009; Mounet et al., 2012; Pascual et al., 2009; Wang et al., 2009).
SlARF9 as a regulator of cell division during early tomato fruit development
In tomato, 22 putative functional ARF genes have been identified (Zouine et al., 2014). Many of these genes show dramatic changes in expression during fruit set and throughout the different stages of fruit development, suggesting that the family of ARF-transcription factors plays an important role in the control of tomato fruit growth (Wang et al., 2009; Kumar et al., 2011; Wu et al., 2011; Zouine et al., 2014). SlARF9 transcript levels increase within 48h after pollination and decrease again in the following days (Fig. 1A and Fig. 3) (Vriezen et al., 2008; Wu et al., 2011). This initial increase in SlARF9 expression was not observed in parthenocarpic fruit formed after GA application (Vriezen et al., 2008), but could be induced in unpollinated ovaries by treating with auxin (Fig. 1D). Mariotti et al. (2011) analysed the IAA content in ovaries before and after pollination, and found a 5-fold increase in free IAA levels in pollinated ovaries collected 2 days post anthesis compared to ovaries collected at anthesis. These levels declined 5 days post anthesis. This pattern is very similar to the expression of SlARF9 in pollinated ovaries. Furthermore, use of the auxin-inducible DR5 promoter coupled to a fluorescent reporter gene showed that before fertilization the auxin response was mainly localized in the embryo sac and the integuments, whilst 6 days post anthesis the auxin activity was mainly observed in the funiculus and outer layer of the placental cells surrounding the seeds (Pattison and Catala, 2012). These placental cells were also stained in the 5–6mm fruit from the pSlARF9::GUS lines (Fig. 2A,B). It is well established that ARFs regulate gene expression in response to auxin, but the considerable overlap between auxin distribution and SlARF9 expression indicates that the transcriptional regulation of SlARF9 itself also depends on the auxin dynamics during tomato fruit set and development.
Normally, during the first 10–14 days of development, tomato fruit growth mainly depends on cell division (Mapelli et al., 1978; Bünger-Kibler and Bangerth, 1982; Gillaspy et al., 1993), but in fruits induced by the auxin IAA the period of cell division was shorter, only lasting 10 days, although cell division took place at a higher rate as compared to that in seeded control fruits. Nevertheless, these IAA-induced fruits remained smaller than control fruits because cell expansion was strongly impaired (Bünger-Kibler and Bangerth, 1982). Treatments with synthetic auxins stimulated cell division for an extended period, resulting in the formation of fruits with a higher number of pericarp cells (Bünger-Kibler and Bangerth, 1982; Serrani et al., 2007). These findings suggest that during the early stages of tomato fruit development, cell division activity is tightly regulated by auxin. SlARF9 might be part of this regulatory mechanism, because decreased SlARF9 transcript levels resulted in the formation of bigger fruits due to extra cell divisions in the pericarp, whereas increased SlARF9 transcript levels led to the formation of smaller fruits as compared to wild type. These opposing phenotypes indicate that SlARF9 acts as a repressor of cell division during fruit growth. Although there was silencing of SlARF9B too, the fact that this gene is not affected by pollination, together with the phenotype of the slarf9-1 TILLING line, indicates that SlARF9 has a major role in the process. In the fruit, SlARF9 showed highest expression in the cell division phase (Fig. 3). The induction of SlARF9 during this phase could create a negative feedback loop in the signal transduction pathway of auxin that promotes cell proliferation, allowing the fine-tuning of cell division activity during early fruit development.
To date, one member of the Arabidopsis ARF gene family, AtARF2 has been identified as a repressor of cell division, since the ovules of the megaintegumenta (mnt)/arf2 mutant were increased in volume due to extra anticlinal cell divisions in the integuments, which continued for a longer period than in wild-type ovules. The expression of genes that promote cell division was not increased in young dividing tissues, but prolonged during maturation (Schruff et al., 2006). In a similar way, the period of cell division might have been prolonged in the pericarp of the SlARF9-RNAi fruits, whilst in the SlARF9-OE fruits this period may have been reduced with pericarp cells starting to expand at an earlier time point than in developing wild-type fruits. To verify this hypothesis, more detailed analysis on the timing of cell division and expansion will be necessary.
SlARF9 as a transcriptional regulator
ARF family members that contain a serine-rich MR, like SlARF9, are putative transcriptional repressors (Tiwari et al., 2003), which was recently confirmed by Zouine et al. (2014). However, the mechanism by which ARF repressors regulate the expression of auxin-dependent genes is still unclear. Several studies suggest that their interaction with Aux/IAAs or with activating ARFs is very weak (Tiwari et al., 2003; Hardtke et al., 2004; Shen et al., 2010). However, Rademacher et al. (2012) have been able to show that AtARF9 could interact with Aux/IAA10 in protoplasts. Alternatively, the ARF repressors may compete with the ARF activators for the AuxRE binding sites in the promoters of auxin response genes, thus inhibiting the expression of these genes independently of Aux/IAAs and providing an alternative mechanism of gene regulation (Guilfoyle and Hagen, 2007).
Despite SlARF9 being a transcriptional repressor and the obvious phenotype of the transgenic lines, no statistically significant differences in expression of single genes were found when comparing the transcriptomes of 3–4mm fruit collected from the SlARF9-OE and SlARF9-RNAi plants. This fruit size was selected as it is the developmental stage at which SlARF9 is most highly expressed in wild-type fruits. It is, however, conceivable that it is not the stage with the highest ARF9 protein level, because many ARFs are subject to post-transcriptional regulation (Mallory et al., 2005; Wang et al., 2005b ; Williams et al., 2005; Wu et al., 2006; Nogueira et al., 2007; Finet et al., 2013; Zouine et al., 2014). Alternatively, it is possible that very subtle changes at the transcriptomic level already have considerable effects on early fruit growth.
GSEA is a powerful tool to help identify such subtle changes by focussing on groups of genes that are involved in similar biological pathways or processes instead of looking at the differential expression of individual genes (Thimm et al., 2004). GSEA of the microarray data from SlARF9-OE versus SlARF9-RNAi fruits resulted in the identification of four functional gene categories that were overrepresented in the SlARF9-RNAi fruits. This included sets of genes involved in synthesis of BRs and polyamine (PA), compounds that have previously been associated with fruit development.
Pollinated ovaries from the tomato cultivar ‘Micro-Tom’, which has several mutations including one in the BR biosynthesis gene DWARF (D/CYP85A1), develop normally (Martí et al., 2006). Also plants with a null mutation in the tomato D gene, extreme dwarf (d x), produce fruits similar to wild type (Nomura et al., 2005). Interestingly, however, high levels of brassinolide could still be detected in the fruits of this mutant, suggesting that brassinolide synthesis in the fruits might be independent of D/CYP85A1. Therefore, with a lack of a fruit phenotype in these mutants, BR cannot be dismissed as a regulator of fruit set and early fruit development. Several studies have demonstrated that the signalling pathways of BR and auxin interact at the level of transcriptional regulation (Nakamura et al., 2003; Goda et al., 2004; Nemhauser et al., 2004). Even direct interactions have been observed between ARFs and BR signalling components (Vert et al., 2008; Je and Goh, 2010; Je et al., 2010; Oh et al., 2014), but ARF9 has not been implicated so far.
The second biological process overrepresented in the SlARF9-RNAi fruits was PA synthesis. High levels of free PAs have been detected at anthesis and during the cell division stage of fruit growth in pollinated fruit, as well as in auxin- and GA-induced fruit (Egea-Cortines et al., 1993; Alabadi et al., 1996). PA levels are altered in the parthenocarpic transgenic IAA9-antisense line (Wang et al., 2009) and have been associated with parthenocarpic fruit development in the pat-2 mutant (Fos et al., 2003). Furthermore, application of PAs to unpollinated tomato ovaries could induce partial parthenocarpy (Fos et al., 2003), whilst application of α-difluoromethylornithine, an inhibitor of the PA biosynthesis enzyme ornithine decarboxylase, to pollinated flowers resulted in a reduction in fruit fresh weight, possibly due to a reduction in cell division (Cohen et al., 1982; Teitel et al., 1985). However, more work will be required to understand the mechanisms by which PAs control fruit growth.
Further studies, including one on direct targets of SlARF9, should reveal if and how these pathways mediate SlARF9 function in early fruit development.
Modifying fruit size
The final number of cells in the pericarp is mostly determined during the cell division phase of tomato fruit development, and is an important factor in determining the size and weight of the mature fruit (Bohner and Bangerth, 1988). So far, quantitative trait loci studies have identified a number of loci for tomato fruit size and weight that have been selected during domestication (reviewed in Grandillo et al., 1999; Paran and van der Knaap, 2007), but in only a few of these studies have the causative genes been cloned. One of them is FW3.2/KLUH, which encodes a P450 enzyme of the CYP78A subfamily. A single nucleotide polymorphism in the promoter of the gene has been associated with increased cell division and fruit weight (Chakrabarti et al., 2013). Another is FW2.2/CNR (CELL NUMBER REGULATOR), the first fruit weight gene identified by quantitative trait loci analysis (Frary et al., 2000). FW2.2 controls cell division in the early stages of tomato fruit development, possibly by interacting with a CKII kinase, which plays an important role in the signalling cascade that modulates the cell cycle (Cong and Tanksley, 2006; Liu et al., 2003). Polymorphisms in members of the FW2.2 gene family have been associated with increased fruit size during domestication across species, for example in eggplant (Doganlar et al., 2002), pepper (Chaim et al., 2001; van der Knaap and Tanksley, 2003), and even in un-related species such as avocado (Dahan et al., 2010), maize (Guo et al., 2010), and sweet and sour cherry (De Franceschi et al., 2013). Thus, further elucidation of the SlARF9 signalling pathway may not just provide more insight into the regulatory mechanism by which auxin controls cell division during the early stages of fruit development in tomato, but may also be of interest to improve agronomic yield of this and other major crop species.
Supplementary Data
Fig. S1. Wild type and transgenic fruits at breaker stage.
Fig. S2. Alignment of the predicted amino acid sequences of SlARF9 and AtARF9.
Fig. S3. Auxin-induced expression of SlIAA2 and SlIAA14.
Fig. S4. Southern blot analysis, verifying the specificity of the SlARF9 DNA fragment used to generate the SlARF9-RNAi lines.
Fig. S5. Microscopic analysis of the pericarp during early fruit development of wild-type and transgenic fruits.
Fig. S6. Principal component analysis of the normalized microarray data.
Table S1. Transcriptomic changes due to modulations in SlARF9 expression identified by microarray analysis.
Table S2. Auxin-related cis-acting regulatory elements.
Table S3. SlARF9 expression during tomato fruit set.
Table S4. Leading edge subsets from the GSEA comparing the transcriptomes of the SlARF9-OE and SlARF9-RNAi lines.
Acknowledgements
We would like to thank Frank Schepers (Bayer CropScience Vegetable Seeds, The Netherlands) for the generation of the transgenic SlARF9-RNAi lines. We are also grateful to Anouk ten Elzen for technical assistance and Dr Elisabeth Pierson (Radboud University Nijmegen, The Netherlands) for help with microscopy.
Glossary
Abbreviations:
- AuxRE
auxin response elements
- BR
brassinosteroid
- CaMV
cauliflower mosaic virus
- c-DNA-AFLP
cDNA-amplified fragment length polymorphism–based transcript profiling
- CTD
C-terminal homo- and heterodimerization domains
- DAP
days after pollination
- FDR
false discovery rate
- GA
gibberellin
- GSEA
genome set enrichment analysis
- GUS
β-glucuronidase
- IAA
indole-3-acetic acid
- kb
kilobase
- MR
middle region
- OE
overexpression line
- PA
polyamine
- PCA
principal component analysis
- SED
standard error of the difference.
References
- Alabadi D, Aguero MS, Perez-Amador MA, Carbonell J. 1996. Arginase, arginine decarboxylase, ornithine decarboxylase, and polyamines in tomato ovaries (changes in unpollinated ovaries and parthenocarpic fruits induced by auxin or gibberellin). Plant Physiology 112, 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G. 1999. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. The Plant Cell 11, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohner J, Bangerth F. 1988. Effects of fruit-set sequence and defoliation on cell number, cell size and hormone levels of tomato fruits (Lycopersicon esculentum Mill) within a truss. Plant Growth Regulation 7, 141–155. [Google Scholar]
- Briggs CL. 1995. The initiation, development and removal of embryo sac wall ingrowths in the developing seeds of Solanum nigrum L. An ultrastructural study. Annals of Botany 76, 429–439. [Google Scholar]
- Bünger-Kibler S, Bangerth F. 1982. Relationship between cell number, cell size and fruit size of seeded fruits of tomato (Lycopersicon esculentum Mill), and those induced parthenocarpically by the application of plant-growth regulators. Plant Growth Regulation 1, 143–154. [Google Scholar]
- Carmi N, Salts Y, Dedicova B, Shabtai S, Barg R. 2003. Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217, 726–735. [DOI] [PubMed] [Google Scholar]
- Chaim AB, Paran I, Grube RC, Jahn M, van Wijk R, Peleman J. 2001. QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theoretical and Applied Genetics 102, 1016–1028. [Google Scholar]
- Chakrabarti M, Zhang N, Sauvage C, et al. 2013. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proceedings of the National Academy of Sciences of the United States of America 110, 17125–17130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Mackey K. 1995. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. BioTechniques 19, 942–945. [PubMed] [Google Scholar]
- Cohen E, Arad SM, Heimer YM, Mizrahi Y. 1982. Participation of ornithine decarboxylase in early stages of tomato fruit development. Plant Physiology 70, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong B, Tanksley SD. 2006. FW2.2 and cell cycle control in developing tomato fruit: a possible example of gene co-option in the evolution of a novel organ. Plant Molecular Biology 62, 867–880. [DOI] [PubMed] [Google Scholar]
- Czerednik A, Busscher M, Bielen BAM, Wolters-Arts M, de Maagd RA, Angenent GC. 2012. Regulation of tomato fruit pericarp development by an interplay between CDKB and CDKA1 cell cycle genes. Journal of Experimental Botany 63, 2605–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan Y, Rosenfeld R, Zadiranov V, Irihimovitch V. 2010. A proposed conserved role for an avocado FW2.2-like gene as a negative regulator of fruit cell division. Planta 232, 663–676. [DOI] [PubMed] [Google Scholar]
- De Franceschi P, Stegmeir T, Cabrera A, et al. 2013. Cell number regulator genes in Prunus provide candidate genes for the control of fruit size in sweet and sour cherry. Molecular Breeding 32, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. 2009. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. The Plant Journal 57, 160–170. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. 2006. The molecular genetics of crop domestication. Cell 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD. 2002. Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161, 1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M, Coben E, Arad S, Bagni N, Mizrahi Y. 1993. Polyamine levels in pollinated and auxin-induced fruit of tomato (Lycopersicon esculentum) during development. Physiologia Plantarum 87, 14–20. [Google Scholar]
- Ficcadenti N, Sestili S, Pandolfini T, Cirillo C, Rotino G, Spena A. 1999. Genetic engineering of parthenocarpic fruit development in tomato. Molecular Breeding 5, 463–470. [Google Scholar]
- Finet C, Berne-Dedieu A, Scutt CP, Marletaz F. 2013. Evolution of the ARF gene family in land plants: old domains, new tricks. Molecular Biology and Evolution 30, 45–56. [DOI] [PubMed] [Google Scholar]
- Fos M, Proano K, Alabadi D, Nuez F, Carbonell J, Garcia-Martinez JL. 2003. Polyamine metabolism is altered in unpollinated parthenocarpic pat-2 tomato ovaries. Plant Physiology 131, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. 2000. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. [DOI] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. 1993. Fruits: a developmental perspective. The Plant Cell 5, 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. 2004. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiology 134, 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandillo S, Ku HM, Tanksley SD. 1999. Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theoretical and Applied Genetics 99, 978–987. [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Current Opinion in Plant Biology 10, 453–460. [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Dieter JA, Zou J, Spielbauer D, Duncan KE, Howard RJ, Hou Z, Simmons CR. 2010. Cell Number Regulator1 affects plant and organ size in maize: implications for crop yield enhancement and heterosis. The Plant Cell 22, 1057–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson FG. 1936. Inducement of fruit development by growth-promoting chemicals. Proceedings of the National Academy of Sciences of the United States of America 22, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C, Ckurshumova W, Vidaurre D, Singh S, Stamatiou G, Tiwari S, Hagen G, Guilfoyle T, Berleth T. 2004. Overlapping and non–redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis–acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je BI, Goh C-H. 2010. Brassinosteroid homeostasis via coordinate regulation of signaling and synthetic pathways. Plant Signaling and Behavior 5, 1440–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je BI, Piao HL, Park SJ, et al. 2010. RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. The Plant Cell 22, 1777–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kumar R, Tyagi AK, Sharma AK. 2011. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Molecular Genetics and Genomics 285, 245–260. [DOI] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C. 2005. Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiology 139, 750–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cong B, Tanksley SD. 2003. Generation and analysis of an artificial gene dosage series in tomato to study the mechanisms by which the cloned quantitative trait locus fw2.2 controls fruit size. Plant Physiology 132, 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. 2005. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. The Plant Cell 17, 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli S, Frova C, Torti G, Soressi GP. 1978. Relationship between set, development and activities of growth regulators in tomato fruits. Plant and Cell Physiology 19, 1281–1288. [Google Scholar]
- Mariotti L, Picciarelli P, Lombardi L, Ceccarelli N. 2011. Fruit-set and early fruit growth in tomato are associated with increases in indoleacetic acid, cytokinin, and bioactive gibberellin contents. Journal of Plant Growth Regulation 30, 405–415. [Google Scholar]
- Martí E, Gisbert C, Bishop GJ, Dixon MS, García-Martínez JL. 2006. Genetic and physiological characterization of tomato cv. Micro-Tom. Journal of Experimental Botany 57, 2037–2047. [DOI] [PubMed] [Google Scholar]
- Molesini B, Rotino GL, Spena A, Pandolfini T. 2009. Expression profile analysis of early fruit development in iaaM-parthenocarpic tomato plants. BMC Research Notes 2, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounet F, Moing A, Kowalczyk M, et al. 2012. Down-regulation of a single auxin efflux transport protein in tomato induces precocious fruit development. Journal of Experimental Botany 63, 4901–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S. 2003. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiology 133, 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biology 2, E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC. 2007. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes and Development 21, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S. 2005. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. Journal of Biological Chemistry 280, 17873–17879. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. 2014. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife , e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I, van der Knaap E. 2007. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. Journal of Experimental Botany 58, 3841–3852. [DOI] [PubMed] [Google Scholar]
- Pascual L, Blanca JM, Canizares J, Nuez F. 2009. Transcriptomic analysis of tomato carpel development reveals alterations in ethylene and gibberellin synthesis during pat3/pat4 parthenocarpic fruit set. BMC Plant Biology 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Catala C. 2012. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. The Plant Journal 70, 585–598. [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, et al. 2012. Different auxin response machineries control distinct cell fates in the early plant embryo. Developmental Cell 22, 211–222. [DOI] [PubMed] [Google Scholar]
- Reed JW. 2001. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends in Plant Science 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Ren Z, Li Z, Miao Q, Yang Y, Deng W, Hao Y. 2011. The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. Journal of Experimental Botany 62, 2815–2826. [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ. 2009. Real-time quantitative RT-PCR: design, calculations, and statistics. The Plant Cell 21, 1031–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. 2006. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. [DOI] [PubMed] [Google Scholar]
- Scutt CP, Vinauger-Douard M, Fourquin C, Finet C, Dumas C. 2006. An evolutionary perspective on the regulation of carpel development. Journal of Experimental Botany 57, 2143–2152. [DOI] [PubMed] [Google Scholar]
- Serrani J, Fos M, Atarés A, García-Martínez J. 2007. Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv Micro-Tom of tomato. Journal of Plant Growth Regulation 26, 211–221. [Google Scholar]
- Shen C, Wang S, Bai Y, Wu Y, Zhang S, Chen M, Guilfoyle TJ, Wu P, Qi Y. 2010. Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). Journal of Experimental Botany 61, 3971–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow AA, Whigham DF. 1989. Costs of flower and fruit production in Tipularia discolor (Orchidaceae). Ecology 70, 1286–1293. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD. 2004. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. The Plant Cell 16, S181–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitel DC, Cohen E, Arad S, Birnbaum E, Mizrahi Y. 1985. The possible involvement of polyamines in the development of tomato fruits in vitro. Plant Growth Regulation 3, 309–317. [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Hagen G, Guilfoyle T. 2003. The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999. a . Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences of the United States of America 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999. b . Dimerization and DNA binding of auxin response factors. The Plant Journal 19, 309–319. [DOI] [PubMed] [Google Scholar]
- van der Knaap E, Chakrabarti M, Chu YH, et al. 2014. What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Frontiers in Plant Science 5, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Tanksley SD. 2003. The making of a bell pepper-shaped tomato fruit: identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theoretical and Applied Genetics 107, 139–147. [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. 2008. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proceedings of the National Academy of Sciences of the United States of America 105, 9829–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. 2008. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. The New Phytologist 177, 60–76. [DOI] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech J, Bouzayen M. 2005. a . The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. The Plant Cell 17, 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latche A, Pech JC, Fernie AR, Bouzayen M. 2009. Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. The Plant Cell 21, 1428–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. 2005. b . Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. The Plant Cell 17, 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC. 2005. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proceedings of the National Academy of Sciences of the United States of America 102, 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, Yu X, Lu G. 2011. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum . Plant Cell Reports 30, 2059–2073. [DOI] [PubMed] [Google Scholar]
- Wu M-F, Tian Q, Reed JW. 2006. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. [DOI] [PubMed] [Google Scholar]
- Zouine M, Fu Y, Chateigner-Boutin AL, Mila I, Frasse P, Wang H, Audran C, Roustan JP, Bouzayen M. 2014. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLOS One 9, e84203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.