Abstract
Steroid conjugates, which often occur as metabolites, are challenging to characterize. One application is female-mouse urine, where steroid conjugates serve as important ligands for the pheromone-sensing neurons. Although the two with the highest abundance in mouse urine were previously characterized with mass spectrometry (MS) and NMR to be sulfated steroids, many more exist but remain structurally unresolved. Given that their physical and chemical properties are similar, they are likely to have a sulfated steroid ring structure. Because these compounds occur in trace amounts in mouse urine and elsewhere, their characterization by NMR will be difficult. Thus, MS methods become the primary approach for determining structure. Here, we show that a combination of MS tools is effective for determining the structures of sulfated steroids. Using 4-pregnene analogs, we explored high-resolving power MS (HR-MS) to determine chemical formulae; HD exchange MS (HDX-MS) to determine number of active, exchangeable hydrogens (e.g., OH groups); methoxyamine hydrochloride (MOX) derivatization MS, or reactive desorption electrospray ionization with hydroxylamine to determine the number of carbonyl groups; and tandem MS (MSn), high-resolution tandem MS (HRMS/MS), and GC-MS to obtain structural details of the steroid ring. From the fragmentation studies, we deduced three major fragmentation rules for this class of sulfated steroids. We also show that a combined MS approach is effective for determining structure of steroid metabolites, with important implications for targeted metabolomics in general and for the study of mouse social communication in particular.
Introduction
Determining the structure of steroid conjugates is a major challenge in metabolomics. A particular need is to understand animal communication. Mammals use their sense of smell to identify and learn the status of other animals. Natural cues such as urine contain metabolites informative about reproductive status, social dominance, and levels of stress [1]. In most cases, however, the identity of the signaling compounds remains unknown. A recent study [2] used the responses of olfactory neurons in the mouse vomeronasal organ, specializing in the detection of “social odors” or “pheromones” produced by other animals, to assay the activity of purified urine fractions. The structures of two active compounds were determined [3], by using a combination of mass spectrometry (MS) and nuclear magnetic resonance (NMR), were shown to be sulfated steroids of the glucocorticoid family. Because steroids are among the main regulators of mammalian physiology [4], their metabolites are attractive candidates for conveying many aspects of an animal’s status. Indeed, several lines of evidence suggest that urine contains other active, but unknown, sulfated steroids. For the urine from female mice of the BALB/c strain, sulfated compounds account for an estimated 80% of the total activation of these sensory neurons [2].
Unfortunately, like many metabolites, many of the unknown cues in natural stimuli are present in much lower abundance than the two compounds identified by Hsu et al. [3], and consequently present a greater challenge for structural analysis. MS is particularly apt for obtaining structural information from small sample quantities, but we felt that a single MS method would not be adequate. Thus, the purpose of the research reported here is to explore the effectives of various combinations of MS methods to determine steroid-sulfate structure.
For many years, steroidal hormones have been analyzed by gas chromatography-mass spectrometry (GC-MS) since Sweeley and Horning’s [5] first success in 1960. More recently, steroids were also analyzed by using HPLC coupled with tandem mass spectrometry (LC-MS/MS) [6–12]. GC has the advantages over LC of higher chromatographic resolution, affording specificity for minor structural differences, as well as a vast EI online database for structural identification of analytes. GC-MS, however, requires some knowledge of the analytes’ structure, because GC’s reliance on volatility often necessitates functional-group-specific derivatization. In contrast, LC-MS/MS requires less initial knowledge of the structure of the sample, as derivatization is not a requirement for ionization. This eases the handling requirement for the sample and reduces possible losses of material during sample preparation.
Here, we describe a combination of methods, taking advantage of both GC-MS and LC-MS/MS, that constitute a MS-based “tool box” for studying structures of sulfated steroids. Although each tool is not new, we wished to explore whether a combination would offer an effective approach to steroid identification. To demonstrate, we focused on a set of six commercial or custom-synthesized 4-pregnene steroids, a key class of compounds because they are central in steroid biosynthetic pathways in the mouse [13]. The 4-pregnenes sulfates were chosen because they are structurally similar to previously identified sulfated steroids found in mouse urine and active for mouse communication. We also anticipate that more unknown sulfated steroids belonging to this category exist in mouse urine. The combination we selected include different MS-based approaches including 1) high mass-resolving-power MS (HR-MS) to determine chemical formula; 2) HD exchange MS (HDX-MS) to determine number of active hydrogens (e.g., –OH groups); 3) methoxyamine hydrochloride (MOX) derivatization MS, or online derivatization with hydroxylamine by reactive desorption electrospray ionization, to determine the number of carbonyl groups; and 4) tandem MS (MSn), high-resolution tandem MS (HRMS/MS), and 5) GC-MS to probe details of the steroid-ring structure. From the fragmentation studies, we can draw some generalized fragmentation rules for sulfated-steroid CID MS/MS. This combination of MS-based methods provides an opportunity to “footprint” steroid sulfates metabolites, providing a foundation for sequel studies of mouse social communication.
Experimental
Materials
Sulfated steroids, including 4-pregnen-11β, 21-diol-3, 20-dione 21-sulfate (termed SS425, standing for sulfated steroid 425, the number of which is the molecular mass of [M - H]−), 4- pregnen-11β, 17, 21-triol-3, 20-dione 21-sulfate (SS441) and 4- pregnen-17, 21-diol-3, 11, 20-trione 21-sulfate (SS439), and steroids, including 5β-pregnan-11β, 21-diol-3, 20-dione, 5α-pregnan-3α, 11β, 21-triol-20-oneand 4- pregnen-11β, 20β, 21-triol-3-one were purchased from Steraloids Inc (Newport, RI), and dissolved in methanol to 5 mM as stock solution. Strata-X 33u polymeric reversed phase 60 mg/3 m for solid phase extraction (SPE) was purchased from Phenomenex (Torrance, CA). All other chemicals and reagents were purchased from Sigma Aldrich (St. Louis, MO).
Sulfation of steroids
Sulfation methods to synthesize sulfated steroids from steroids were modified from that of Kornel et al. [14] to suit for micro-synthesis. Acidified pyridine was prepared by adding 0.3 mL chlorosulfonic acid dropwise to 5 mL dry pyridine while cooling on an ice bath. To sulfate steroids 5β-pregnan-11β, 21-diol-3, 20-dione, 5α-pregnan-3α, 11β, 21-triol-20-one and 4- pregnen-11β, 20β, 21-triol-3-one, 5 μL of 5 mM stock steroid solution was dried by using a SpeedVac at room temperature, and the residue redissolved in 80 μL pyridine. Acidified pyridine (30 μL) was then added, and the mixture was shaken for 5 min, as optimized, following which saturated NaHCO3 solution was used to neutralize the reaction solution, which was then dried in a SpeedVac at room temperature. The residue was redissolved in 3 mL H2O and desalted with SPE columns, and the final product was eluted with 1 mL methanol. Sulfation of 5β-pregnan-11β, 21-diol-3, 20-dione and 4- pregnen-11β, 20β, 21-triol-3-one occurred at C21, as indicated by LC-MS/MS results (data not shown), leading to single products named SS427 (standing for sulfated steroid 425, the number of which is the molecular mass of [M - H]−) and iSS427 (standing for isomer SS427), respectively; sulfation of 5α-pregnan-3α, 11β, 21-triol-20-one happened at either C3 or C21, referred to as 3-SS429 and 21-SS429 respectively, which were separated with a custom-packed C18 column before infusion into ESI source for MS/MS analysis.
Mass Spectrometry
Electrospray ionization Mass Spectrometry (ESI-MS)
Sample stock solutions were diluted to 1 μM and infused to an ESI source at 3 μL/min. Both low-mass-resolving power and high-mass-resolving power ESI-MS were conducted in the negative-ion mode on a LTQ Orbitrap Velos and a LTQ Orbitrap from Thermo Scientific (West Palm Beach, FL), equipped with the Xcalibur operating system. Spray voltage in the negative ion mode was set to 3.5 kV; capillary voltage and temperature was at −32 V and 275 °C; and the tube lens was −222 V. Tandem MS (MSn) experiments were carried out after low energy CID by using a relative collision energy ranging from 30 to 40% of the maximum and high energy HCD with 150 eV collision energy.
Reactive Desorption Electrospray ionization (reactive DESI)
All DESI experiments were carried out by using an electrosonic spray ionization (ESSI) source to generate charged microdroplets, and a silica capillary to introduce the sample [15, 16]. The ESSI sprayer was aimed to the outlet of the sample introduction capillary at an angle of 30°–45°, and the horizontal distance between the two was ~0.5 mm. The solvent for ESSI consisted of methanol/water in DESI and methanol/water/5% hydroxylamine/0.05% acetic acid in reactive DESI, and was injected at 10 μL/min, comparing to 4 μL/min of the sample flow rate. The control DESI experiments were done in the negative-ion mode, whereas the reactive DESI experiments were done in the positive-ion mode. The spray voltage was set at 5 kV for positive ions and −5 kV for negative ions; the nebulizing gas (N2) pressure for ESSI was 80 psi. MS detection was on a Thermo Finnigan LCQ DecaXP plus. Capillary temperature and voltage was set at 250 °C and 21 V, and the tube lens was 10 V for positive ions; they were set to 250 °C, −47 V, and −50 V, respectively, for negative ions.
Gas Chromatography- Mass Spectrometry (GC-MS)
The sulfate group of sulfated steroids was removed by solvolysis as described in [17], under conditions for micro-synthesis. Acidified ethyl acetate was prepared by shaking 1 M sulfuric acid in water and ethyl acetate (V:V= 1:2) vigorously for 2–3 min and then discarding the water phase. The dried sample was dissolved in acidified ethyl acetate to a concentration of 1 μM to give a total of 5 mL and incubated for ~ 6 h at 38 °C, followed by neutralization with ammonium hydroxide. The reaction time was optimized for both SS425 and SS441 to 6 h. The desulfated sample was then dried, and redissolved in 15 μL pyridine, to which 15 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% TMS was added and incubated at 70 °C for 1 h for SS425 and 2 h for SS441. Derivatized samples were then loaded into an Agilent (Santa Clara, CA) 7693 autosampler, and 1 μL injections were analyzed on an Agilent 7890A gas chromatograph system interfaced to an Agilent 7200 accurate-mass Q-TOF mass spectrometer. The samples were injected with a 10:1 split. The GC column was a DB-5MS (12 m, 0.25 mm i.d., 0.33 um film coating, P.J. Cobert St. Louis, MO). A linear temperature gradient was used: the initial temperature of 80° was held for 2 min and increased to 280 at 15°/min. The temperature was held at 280° for 5 min. The samples were ionized by electron ionization (EI), and the source temperature, electron energy, and emission current were set at 280 °C, 70 eV and 300 uA, respectively. The injector and transfer line temperatures were also 280°C.
Hydrogen Deuterium Exchange (HDX) and methoxyamine hydrochloride (MOX) derivatization
HDX of sulfated steroids was initiated by dissolving a dried sample in D2O, to give a sample concentration of ~ 2 μM, at neutral pH. The same volume of acetonitrile was added prior to conducting ESI MS and MS/MS. MOX derivatization involved 18 h incubation of dried sample (20 μL × 1 uM) in 100 μL MOX reagent (2% in pyridine) at 60 °C. The product solution was then dried under nitrogen and re-dissolved in 20 μL MeOH and infused to the ESI source for analysis.
Results and Discussion
Electrospray ionization (ESI-MS), HDX, and MOX derivatization
Given that these sulfated steroids contain a negative charge on the sulfate group, all ESI mass spectra and product-ion spectra (MSn) spectra were obtained in the negative-ion mode to give an [M - H]− molecular ion. High-resolving-power mass measurement gave mass accuracy within 5 ppm for both 32S and 34S isotopic peaks, based on the molecular ion formulae (Table 1, Figure S1a, b), as expected.
Table 1.
Outcomes of the application of HRMS, HDX, MOX derivatization, reactive DESI, and HCD MS/MS of SS425 and SS441
| Sulfated steroids | Structure | HR/MS | HDX MS (# of –OH) | MOX deriv. (# of C=O) | Reactive DESI (# of C=O) | HCD MS/MS (-SO4−?) | |
|---|---|---|---|---|---|---|---|
| 32S (error) | 34S (error) | ||||||
| SS425 |
|
425.1641 (3 ppm) | 427.1670 (3 ppm) | 426 (1) | 483 (2) | 491/492/493 (2) | 97 (yes) |
| SS441 |
|
441.1584 (1 ppm) | 443.1549 (3 ppm) | 443 (2) | 499 (2) | 507/508/509 (2) | 97 (yes) |
Upon HDX, the molecular ion profile cleanly shifted 1 Da for SS425, indicating the existence of one exchangeable proton in the compound (Figure S1c). For SS441, the shift is 2 Da (Figure S1d), indicating two exchangeable protons. For an unknown, its accurate mass would reveal the number of oxygens; that number, minus those in the sulfate group, indicates the maximum number of exchangeable sites, provided there are no nitrogens in the molecule. Under neutral conditions, hydrogens alpha to a carbonyl group exchange very slowly and will not be “counted” here.
MOX derivatization converts the carbonyl group to an oxime (C=NOCH3), leading to an increase of 29 Da in mass for every carbonyl of the compound. Thus, HDX and MOX derivatization afford a count of the number of ketone or aldehyde groups (Table 1 indicates two carbonyl groups in both SS425 and SS441).
Reactive Desorption Electrospray ionization (reactive DESI)
As an alternative to MOX derivatization in counting carbonyl groups in sulfated steroids, we followed Huang et al. [18] in performing reactive DESI on steroids in the positive-ion mode, utilizing hydroxylamine and ambient-pressure ionization to derivatize steroidal carbonyl groups. Hydroxylamine was used instead of methoxyamine because it is more volatile than the latter and is compatible with on-line derivatization and detection. Unlike traditional DESI, we introduced the sulfated steroid samples in a capillary with a constant flow instead of depositing them on a plate. In this process, protonated hydroxylamine ions are formed during electrospray and carried in microdroplets toward the substrate where they react. The proposed product ions include a positively charged tetrahedral intermediate (hydroxylamine adduct, with a mass increase of 34 Da) and an oxime formed by loss of water from the tetrahedral intermediate (Scheme 2 in literature [18]). Because the reaction with hydroxylamine ions give a more readily protonated group in lieu of the origianal carbonyl group, the derivatization adds a positive charge to the molecule, counterbalanced in part by the negative charge of the highly acidic sulfate group. Provided there is more than one carbonyl group in the compound, we expect the products to be zwitterions both in solution and in a mass spectrometer ion source operating in the positive-ion mode,
As shown in Table 1 and Figure S2, product ions from reactive DESI on SS425 and SS441 are of m/z 493 and 509, respectively, both with mass increases of 68 Da, indicating the presence of two carbonyl groups in each compound. Unlike Huang et al.’s study [18], the oxime product wasn’t observed in either reaction, and the tetrahedral intermediate product was accompanied by an oxidation reaction that resulted in products with 1 or 2 Da decrease in mass, such as those at m/z 491/492 and m/z 507/508 in reactive DESI of SS425 and SS441, respectively. These differences likely result from the different instrumentation and DESI setup (we used ESSI instead of DESI). Although the mechanisms for the formation of the oxidized species of the tetrahedral intermediates is not a focus of this study, a possible explanation is that the ESI capillary (in this case, DESI capillary) is an electrolytic half-cell where redox reactions occur under the conditions of DESI [19].
HCD MS/MS of ESI produced steroid sulfates
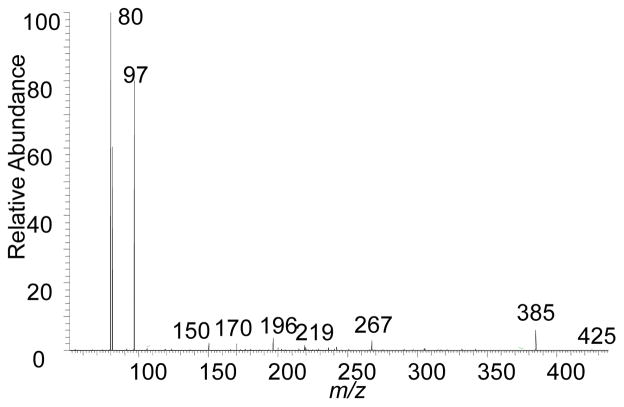
Verifying the presence of a sulfate group to account for at least one sulfur atom in the compounds is straightforward. With HCD MS2, the product-ion spectra of SS425 and SS441 are dominated by ions of m/z 97, HOSO3−, confirming that one sulfur atom is part of a sulfate group. One example is the HCD product-ion spectrum (MS2) of SS425 (Figure 1). Aside from the ion of m/z 97, the ion of m/z 80 is SO3−, offering more confirmation for the presence of a sulfate group. The formation of HSO4− and SO3− is expected and was reported previously in product-ion spectra of many steroidal sulfates [20–23]. The proton-transfer mechanism as proposed in [24], occurs with the elimination of a ketene and a ketone. However, other vinyl or phenyl sulfates, such as estron-3-yl 3-sulfate, do not undergo bisulfate elimination to form HSO4− in mass spectrometric fragmentation [22,23, 25], because that reaction would form a prohibitively high-energy benzyne. We term the formation of the product ion HOSO3− of m/z 97 upon HCD MS/MS as the “97” rule, which indicates the presence of a sulfate as a signature product ion. Although HCD gives highly obvious evidence for a sulfate substituent via the abundant product ions of m/z 80 and 97, the other product ions, requiring higher dynamic range, provide incomplete structural details of the sulfated steroid. Therefore, we utilized an alternative tandem MS method with CID fragmentation in an ion trap, which prevented observation of the m/z 80 and 97 ions and provided higher dynamic range for product ions arising from ring cleavages.
Figure 1.
HCD fragmentation of the negatively charged molecular ion of m/z 425 of SS425.
CID MSn of ESI-produced steroid sulfates
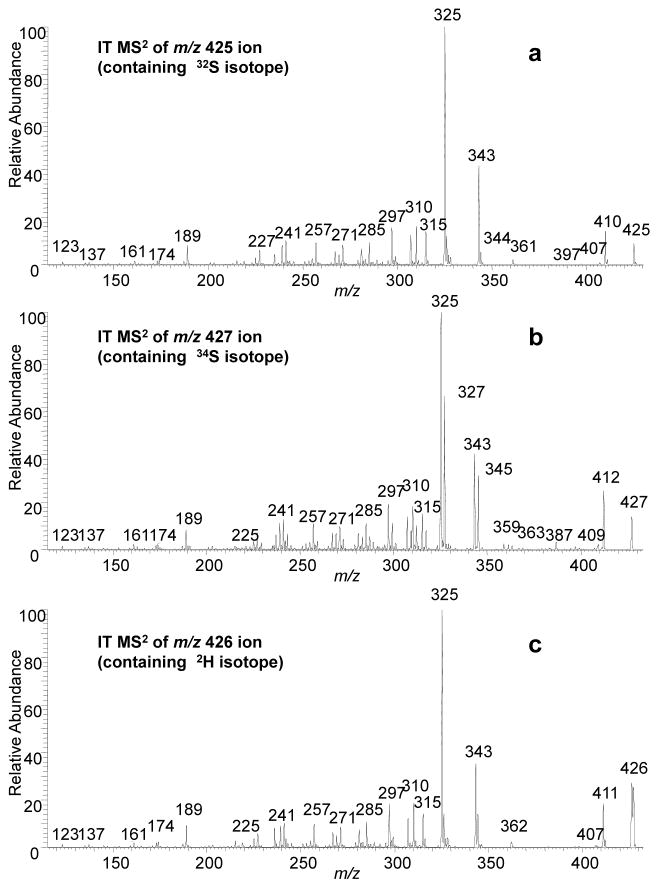
Today’s MS/MS technology allows the low-energy, negative-ion CID spectra of metabolites such as SS425 and SS441 (presented in Figures 2a and 3a) to be obtained with accurate m/z measurement (Table 2). We were able to obtain product-ion (MS2) spectra (Figure 2b and 3b) from precursor ions of m/z 427 for SS425 and 443 for SS441, which are a mixture of isotopologs containing approximately 53% 34S. These spectra also confirm the presence/absence of sulfate group. For example, the single ion of m/z 410 in Figure 2a must contain the sulfate group, consistent with the calculated elemental composition and the shift of 2 Da to m/z 412 in Figure 2b. More significantly, the product ions of m/z 325 and 343 must be formed by losses including the sulfate groups as the m/z 325 ion now appears as both m/z 325 (losses of H234SO3 + H2O) and 327 ions (losses of H232SO3 + H2O); the latter retaining two 13C atoms. This is consistent with accurate mass measurements for ions of m/z 343 and 325 (Table 2).
Figure 2.
Product ion spectra of the [M - H]− ions of SS425 at (a) m/z 425 containing 32S isotopolog (b) m/z 427 containing 34S isotopolog (c) m/z 426 after H/DX containing 2H isotopolog.
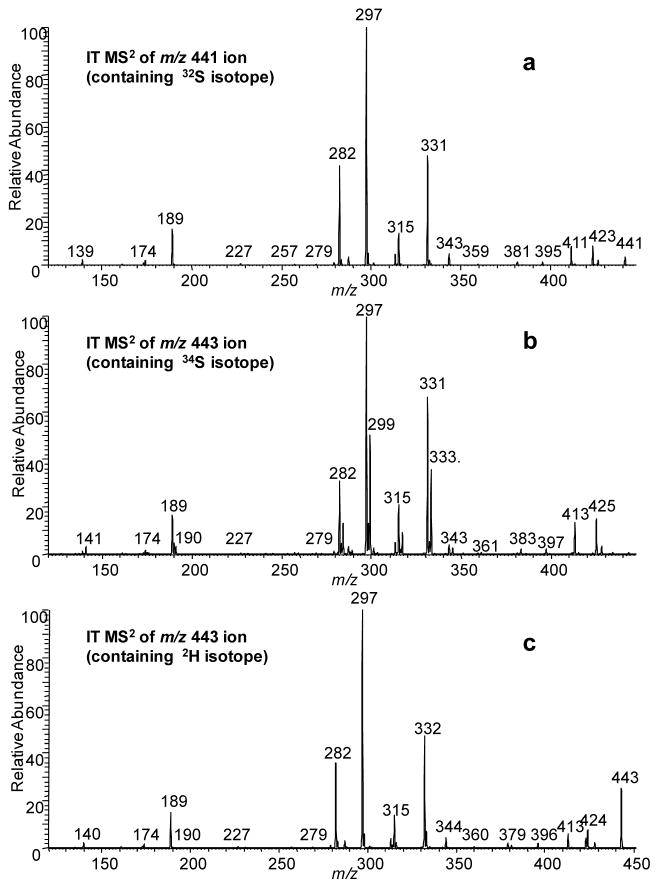
Figure 3.
Product ion spectra of the [M - H]− ions of SS441 at (a) m/z 441 containing 32S isotopolog (b) m/z 443 containing 34S isotopolog (c) m/z 443 after H/DX containing 2H isotopolog.
Table 2.
Calculated fragment-ion elemental compositions based on accurate mass detection of CID fragmentations with HRMS2 for SS425 and SS441.
| a. SS425 | b. SS441 | ||||
|---|---|---|---|---|---|
|
| |||||
| m/z | Calc. mass | Comp. | m/z | Calc. mass | Comp. |
| 410.1374 | 410.1394 | C20H26O7S | 426.1354 | 426.1354 | C20H26O8S |
| 343.1890 | 349.1904 | C21H27O4 | 423.1479 | 423.1483 | C21H27O7S |
| 325.1785 | 325.1798 | C21H25O3 | 411.1479 | 411.1483 | C20H27O7S |
| 315.1943 | 315.1955 | C20H27O3 | 343.1909 | 343.1915 | C21H27O4 |
| 310.1552 | 310.1563 | C20H22O3 | 331.1909 | 331.1915 | C20H27O4 |
| 297.1839 | 297.1849 | C20H25O2 | 315.1596 | 315.1602 | C19H23O4 |
| 285.1475 | 285.1485 | C18H21O3 | 313.1803 | 313.1809 | C20H25O3 |
| 271.1684 | 271.1693 | C18H23O2 | 297.1487 | 297.1496 | C19H21O3 |
| 257.1529 | 257.1536 | C17H21O2 | 282.1256 | 282.1261 | C18H18O3 |
| 241.1582 | 241.1587 | C17H21O | 189.0921 | 189.0921 | C12H13O2 |
| 189.0911 | 189.0910 | C12H13O2 | 138.9710 | 138.9709 | C2H3O5S |
To assign with more confidence the compositions of the product ions, MS2 was also conducted on samples where the -OH group has become deuterated (Fig 2c and 3c). By comparing product ions in Figure 2a and 2c, or Figure 3a and 3c, any mass shift indicates, in the absence of HD scrambling, the number of –OH groups retained in the related product ion. Noteworthy is the lack of any significant shifts of the m/z 325 and 343 ions (Figure 2c), indicating that the loss of H2SO3 and H2O now occur as those of HDSO3 and H2O. The results suggest that the loss of H2SO3 involves the H transfer from –OH on C11 to the sulfate group while transferring the charge from the sulfate group to the oxygen on C11.
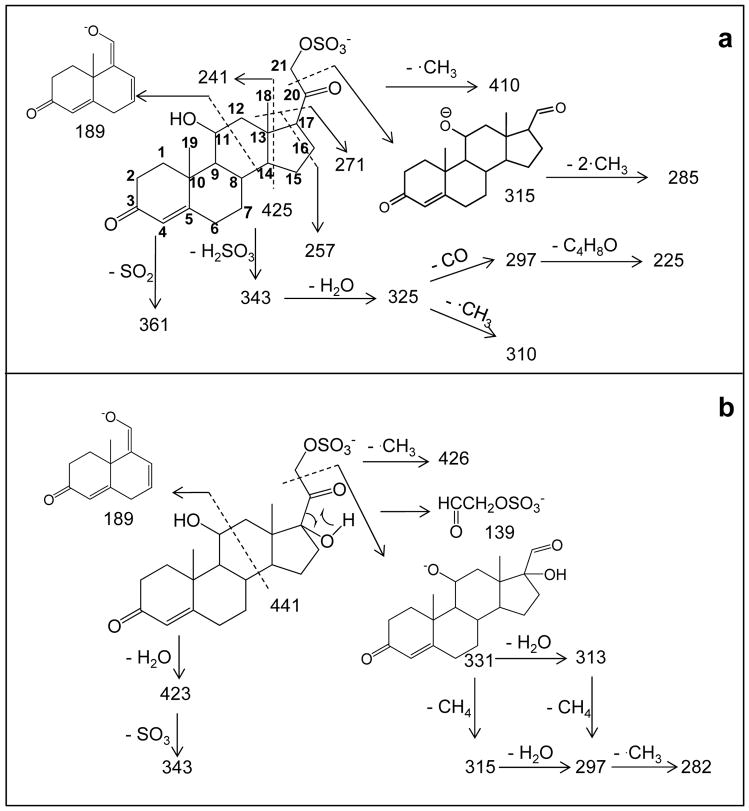
This combination of isotopic labeling and accurate m/z measurement in the product-ion spectra permits a fragmentation pathway scheme to be proposed. Those schemes (Scheme 1a and 1b, and reiterated in a simpler form in Figures 4a and 4e) are informative of the steroid-ring structure (the carbons are numerically labeled according to convention on structure of SS425 in Scheme 1a for convenience). Additional support and complementary information to characterize the steroid ring were found in a MS3 experiment (Figures S3 – S8) and are reiterated in Figures 4b, c, d, f, g, and h. The fragmentation pathways proposed here for either MS2 or MS3 are not validated by molecular orbital theory or isotope labeling. Instead, the ion structures are proposed according to their accurate masses to be consistent with the fragmentation. The purpose is to show correlations in the fragmentation patterns of compounds from the same class of sulfated steroids and to provide insights on how CID fragmentation can be applied to track functional groups and their changes on the ring. One outcome is that the ring topology of steroid sulfates of this type can be characterized by the rich fragmentation seen in both MS2 and MS3 experiments, as illustrated in Figure 4. Collisional activation, however, does not always produce all possible fragmentations of the ring, in which case, the specific functional group location cannot be made. For example, the location of C17-OH in SS441 cannot be established because there’s no cleavage for C16-C17 bond; in which case if SS441 were an unknown compound, the –OH group can only be vaguely assigned to C16 or C17.
Scheme 1.
Fragmentation pathway for (a) SS425 and (b) SS441 with CID MS/MS. Ion structures are proposed to be consistent with fragmentation and accurate masses of product ions from HRMS/MS.
Figure 4.
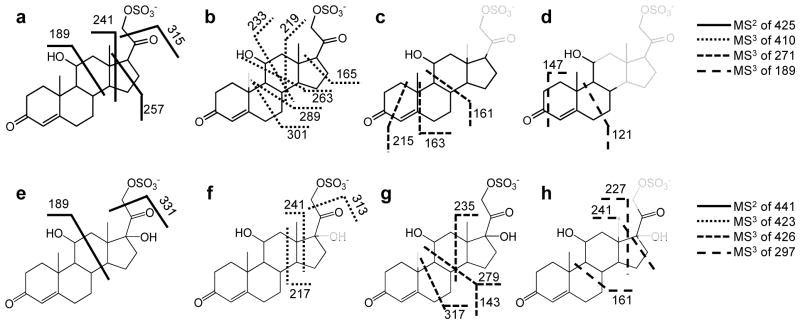
Fragmentation sites of (a) molecular ion and (b–d) various product ions of SS425. Fragmentation sites of (e) molecular ion and (f–h) various product ions of SS441. Those parts of the structures drawn in fainter lines represent the regions lost as neutrals in the MS/MS fragmentation.
Functional Group Location based on CID MS/MS
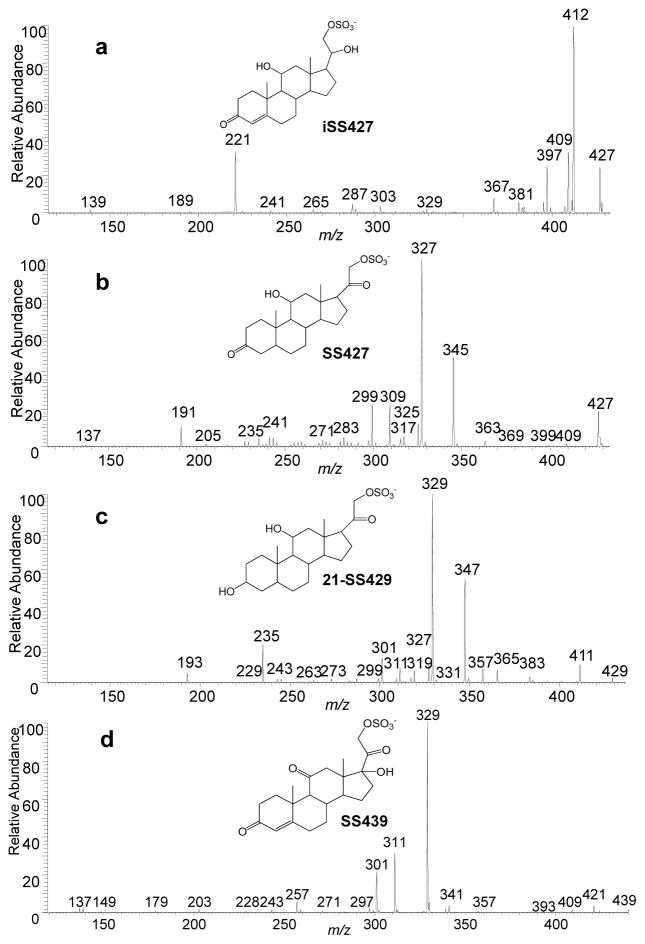
One goal of interpreting tandem MS and accurate m/z data is to locate a functional group on the steroid ring. The fragment ions of m/z 189 found for both SS425 and SS441 are depicted to contain steroid rings A and B, as well as C11. The formation of ions of m/z 189 is not simply due to the two homolytic cleavages of C11-C12 and C8-C14 bonds (scheme 1 and 2), but also must involve a proton transfer from C11-OH to the weak SO3− base, possibly followed by loss of the H2 and C9H14O5S. Although the reasoning to assign structure is similar to that used by Hsu et al. [3], and also supported by chemical composition from accurate mass and the lack of a sulfate group by 34S-MS2, we want to emphasize that we are not conducting a mechanism study but rather to use pathways to study substitutions on C11. To determine whether there is a correlation between the presence of a hydroxyl group on C11 and the occurrence of this product ion of m/z 189 in MS2, four more sulfated steroids, namely, iSS427, SS427, 21-SS429 and SS439, were prepared, and their product-ion spectra were studied to search for similar cleavages. As expected, three of the four compounds, all of which possess a hydroxyl group at C11, gave product ions of m/z 189, 191 and 193 (Figure 5a, b, and c), respectively, as a result of the cleavage of the C11-C12 and C8-C14 bonds. By contrast, the product-ion spectrum of SS439 contains no such product ion, theoretically of m/z 187, owing to this cleavage, because C11 is now a carbonyl group (Figure 5d). Thus, we summarize this correlation between the presence of C11-OH and the product ion of m/z 189/191/193 as the “C11-OH” rule, indicating that only when there’s a hydroxyl group on C11 will this fragmentation happen in sulfated steroids. On the other hand, when interpreting product-ion spectra of unknown sulfated steroids, MS3 should be conducted to fragment the m/z 189/191/193 ion, if present, to confirm its structure before invoking this rule and making conclusions. Although there are many other carbon sites on the steroid ring to accommodate a hydroxyl group and likely other fragmentation rules for steroids with hydroxyl group at those sites, C11, based on biological relevance, is an important position on the steroid ring structure because it is often hydroxylated by 11β-hydroxylase during steroid hormone synthesis in mouse, and sometimes followed by further oxidation [26]. These differences are readily detected by MS2.
Scheme 2.
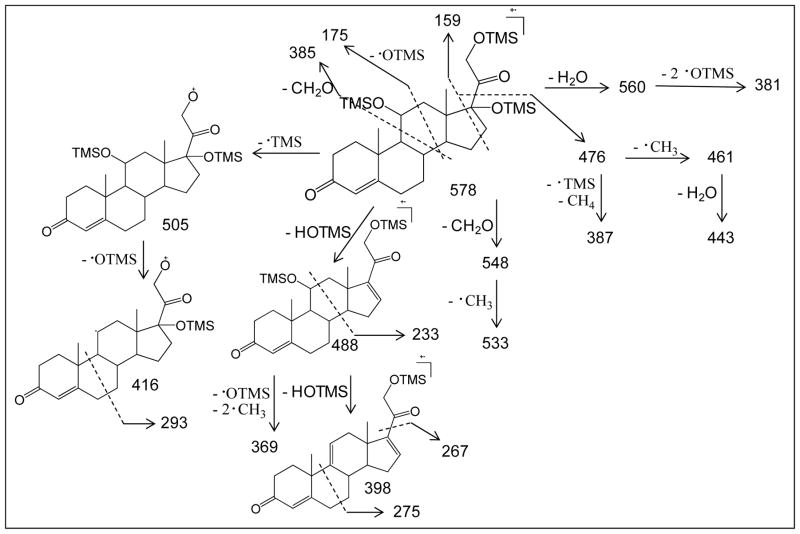
EI fragmentation pathway of derivatized product from SS441. Ion structures are proposed to be consistent with accurate masses and fragmentation outcomes.
Fig. 5.
CID fragmentation in the negative ion mode of (a) iSS427, (b) SS427, (c) 21-SS429, and (d) SS439.
Another important functional group, located at C20, is similarly deciphered by tandem MS. A derivative of many cholesterol steroids formed in steroid hormone synthesis involves reduction of cholesterol side-chain length and the hydroxylation of C21 [13] by two important steroid enzymes, cholesterol 20–22 desmolase and 21-hydroxylase. C20 retains a -OH or =O after C20-C22 cleavage with 20–22 desmolase action [27], and hydroxylated C21 could be then sulfated by a sulfatase to form sulfated steroids. As a result, sulfated steroids occurring biologically often differ at C20 as either a C=O or a C-OH. Again, this difference can be revealed by a product ion formed by the cleavage of bond C20-C21 and loss of a neutral molecule CH2SO4 to give a product [M - H+- 110]−, which only occurs when C20 is C=O. For example, as in Figure 2a, the loss of 110 from the precursor ion of m/z 425 leads to a product ion of m/z 315 (relative abundance of more than 20%). Accurate mass of the m/z 315 ion from HRMS2 (Table 2) and 34S-MS2 (Figure 2b) indicates this ion forms by the loss of CH2SO4, allowing us to write an ion structure (Scheme 1a). The same fragmentation occurs for SS441. More support for this fragmentation chemistry comes from the product-ion spectra of iSS427, SS427, 21-SS429 and SS349, for which the loss of 110 occurs for the latter three (Figure 5b, c, d), all of which have an C20 oxidized to C=O. In contrast, when C20 retains a hydroxyl group, this cleavage is inhibited. As shown in Figure 5a, no obvious product ion at m/z 317 is present. Thus, we summarize this as the “−110” rule, which addresses the correlation between C20 =O and fragmentation C20-C21 upon CID.
Gas Chromatography Mass Spectrometry (GC-MS)
GC combined with EI-MS is a powerful approach for the identification of unknown compounds, steroids being one important class of them. GC is particularly advantageous because it provides effective separation, which is needed for the analysis of complex biological mixtures in metabolomics. Further, EI mass spectra has always been informative in structure studies, but it is even more so with today’s high-resolving power MS instruments that give accurate m/z for all fragments at speeds sufficient for GC separation. Exhaustive studies were reported by others to characterize the EI mass spectra of steroids and delineate fragmentation [28]. Furthermore, online databases of EI mass spectra have expanded tremendously during the past three decades. For example, Wiley Registry 9th edition contains approximately 662,000 EI mass spectra of 592,000 compounds; and Wiley also published a database “Mass Spectra of Physiologically Active Substances: Including drugs, steroid hormones, and endocrine disruptors” and contains 4182 mass spectra and chemical structures of steroids. Thus, structural studies of unknown steroids can again take advantage of GC-MS, and, thus, we included it in our MS approach for unknown sulfated steroids.
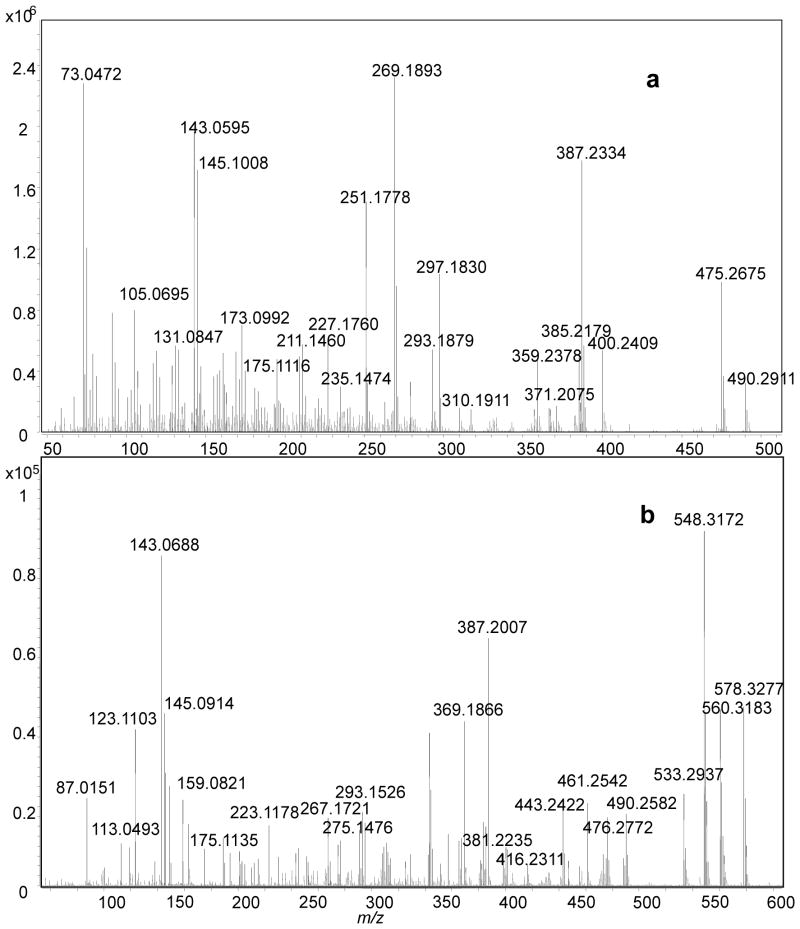
To utilize GC-MS, we had to remove chemically the sulfate group from the two standards, SS425 and SS441, and we then derivatized the products for GC/MS analysis. Solvolytic removal of the sulfate group was successful for nanomoles of samples, and the product yields were greater than 50% for both SS425 and SS441 (as determined by LC/MS). Removing the sulfate group on sulfated steroids is important for GC/MS analysis because the resulting samples are considerably more volatile. It is noteworthy, however, that solvolysis may convolute quantitative analysis for complex biological fluids for which a requirement may be to compare free versus sulfated steroids. For that purpose, the combination of LC/MS and GC/MS for may be more suitable for quantitative analysis. After BSTFA/TMS derivatization of the desulfated steroids, we analyzed 0.1 nmol of sample. This low requirement of sample is compatible with studying low-abundance, urinary sulfated steroids, an application of this methodology. Accurate-mass mass spectra for SS425 and SS441 (Fig 6a and 6b) were searched in NIST MS database. A match of corticosterone bis(trimethylsilyl) ether was found for SS425, with a high probability score of 93%. However, no match was found for SS441. Although this structure may appear in other databases, we turned to the EI fragmentation pathway of SS441, based on accurate masses of fragments, to extract signature fragmentations that assist in identification. As before, the ion structures in this scheme are not established but only proposed to be consistent with accurate masses and fragmentation outcomes, and to organize our conclusions on how EI fragmentation might report on the structural details of a steroid ring. Scheme 2 shows that under EI fragmentation, cleavages in the ring, mostly near the functional groups, reveal more detailed information about the ring structure. Therefore, the rich fragmentation from EI-MS complements that from ESI-MSn, and, taken together, allows one or several structure candidates to be proposed for an unknown.
Fig. 6.
GC-MS spectrum for derivatized product from (a) SS425 and (b) SS441.
Conclusions
A difficulty in assigning the structures of steroid sulfate metabolites as constituents in natural stimuli is their low abundance, precluding the application of most structural tools. To deal with this problem, we implemented a combined approach including HRMS to give the chemical formula, and HDX, MOX derivatization and reactive DESI to count various functional groups. The combination also includes MSn and GC-MS to resolve the arrangement of the functional groups and ring structure. Three major fragmentation rules were deduced in this study, the “97” rule, the “C11-OH” rule and the “−110” rule, collectively addressing the three metabolic important carbon sites for this family of compounds from the mouse. Proper chemical manipulation and derivatization permit the analysis of sulfated steroids by GC-MS to take advantage of its highly effective separation and the availability of online database searching to confirm structural assignments, taking advantage of the complementary and rich EI fragmentation. This MS-based combined approach consumes approximately 0.1 nmol of sample for ESI-MS with direct infusion when analyzed in the negative-ion mode and 1 nmol was required for simple chemical modification and derivatization for GC-MS. An outcome is that the ring topology of steroid sulfates of this type can often be well characterized, except when an important fragmentation is missing and only vague assignments can be made. In addition, although the fragmentation “rules” will likely change for other classes of steroids, this approach should be general for rare steroid metabolites, affording one or several putative structure that can be synthesized and its MS properties compared with those of the unknown.
Another important application may be premarin, a pharmaceutical from urine of female pregnant horses, which is comprised of a large number of steroid sulfates, many still resisting identification.
Supplementary Material
Acknowledgments
We thank Dr. Hao Chen for providing us with electrosonic spray ionization and helping set up the DESI experiments, Jan Crowley for helping with the GC-MS experiments, and Fong-Fu Hsu for assistance in accurate-mass, product-ion spectra. This research was supported by the National Institute of General Medical Sciences (8 P41 GM103422-35 to MLG) and the National Institute on Deafness and Other Communication Disorders (R01 DC005964 to TEH), both of the National Institutes of Health.
References
- 1.Ferrero DM, Liberles SD. The secret codes of mammalian scents. Wiley Interdiscip Rev Syst Biol Med. 2010;2:23–33. doi: 10.1002/wsbm.39. [DOI] [PubMed] [Google Scholar]
- 2.Nodari F, Hsu FF, Fu XY, Holekamp TF, Kao LF, Turk J, Holy TE. Sulfated steroids as natural ligands of mouse pheromone-sensing neurons. J Neurosci. 2008;28:6407–6418. doi: 10.1523/JNEUROSCI.1425-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu FF, Nodari F, Kao LF, Fu XY, Holekamp TF, Turk J, Holy TE. Structural characterization of sulfated steroids that activate mouse pheromone-sensing neurons. Biochemistry-Us. 2008;47:14009–14019. doi: 10.1021/bi801392j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkenstein E, Tillmann HC, Christ M, Feuring M, Wehling M. Multiple actions of steroid hormones - a focus on rapid, nongenomic effects. Pharmacological Reviews. 2000;52:513–555. [PubMed] [Google Scholar]
- 5.Sweeley CC, Horning EC. Microanalytical separation of steroids by gas chromatography. Nature. 1960;187:144–145. doi: 10.1038/187144a0. [DOI] [PubMed] [Google Scholar]
- 6.Stanczyk FZ, Clarke NJ. Advantages and challenges of mass spectrometry assays for steroid hormones. J Steroid Biochem Mol Biol. 2010;121:491–495. doi: 10.1016/j.jsbmb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Kang S, Park S, Kim MJ, Oh SM, Chung KH, Lee S. A sensitive and selective lc-ms/ms analysis coupled with an online sample enrichment technique for h295r steroidogenesis assay and its application in the investigation of the effect of sildenafil on steroidogenesis. Analytical and bioanalytical chemistry. 2013;405:9489–9496. doi: 10.1007/s00216-013-7380-5. [DOI] [PubMed] [Google Scholar]
- 8.Fayad PB, Prevost M, Sauve S. On-line solid-phase extraction coupled to liquid chromatography tandem mass spectrometry optimized for the analysis of steroid hormones in urban wastewaters. Talanta. 2013;115:349–360. doi: 10.1016/j.talanta.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 9.Keevil BG. Novel liquid chromatography tandem mass spectrometry (lc-ms/ms) methods for measuring steroids. Best practice & research Clinical endocrinology & metabolism. 2013;27:663–674. doi: 10.1016/j.beem.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Galuska CE, Hartmann MF, Sanchez-Guijo A, Bakhaus K, Geyer J, Schuler G, Zimmer KP, Wudy SA. Profiling intact steroid sulfates and unconjugated steroids in biological fluids by liquid chromatography-tandem mass spectrometry (lc-ms-ms) The Analyst. 2013;138:3792–3801. doi: 10.1039/c3an36817c. [DOI] [PubMed] [Google Scholar]
- 11.Couchman L, Vincent RP, Ghataore L, Moniz CF, Taylor NF. Challenges and benefits of endogenous steroid analysis by lc-ms/ms. Bioanalysis. 2011;3:2549–2572. doi: 10.4155/bio.11.254. [DOI] [PubMed] [Google Scholar]
- 12.Soldin SJ, Soldin OP. Steroid hormone analysis by tandem mass spectrometry. Clinical chemistry. 2009;55:1061–1066. doi: 10.1373/clinchem.2007.100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne AH, Hales DB. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev. 2004;25:947–970. doi: 10.1210/er.2003-0030. [DOI] [PubMed] [Google Scholar]
- 14.Kornel L, Kleber JW, Connie JW. Studies on steroid conjugates: Ii. Chemical synthesis and characterization of sodium cortisol-21-sulfate and sodium tetrahydrocortisol-3,21-disulfate. Steroids. 1964;4:67–75. [Google Scholar]
- 15.Takats Z, Wiseman JM, Gologan B, Cooks RG. Electrosonic spray ionization. A gentle technique for generating folded proteins and protein complexes in the gas phase and for studying ion-molecule reactions at atmospheric pressure. Anal Chem. 2004;76:4050–4058. doi: 10.1021/ac049848m. [DOI] [PubMed] [Google Scholar]
- 16.Miao Z, Chen H. Direct analysis of liquid samples by desorption electrospray ionization-mass spectrometry (desi-ms) Journal of the American Society for Mass Spectrometry. 2009;20:10–19. doi: 10.1016/j.jasms.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 17.Burstein S, Lieberman S. Hydrolysis of ketosteroid hydrogen sulfates by solvolysis procedures. J Biol Chem. 1958;233:331–335. [PubMed] [Google Scholar]
- 18.Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Rapid screening of anabolic steroids in urine by reactive desorption electrospray ionization. Anal Chem. 2007;79:8327–8332. doi: 10.1021/ac0711079. [DOI] [PubMed] [Google Scholar]
- 19.Van Berkel GJ. The electrolytic nature of electrospray. In: Cole RB, editor. Electrospray Ionization Mass Spectrometry. 2. Wiley; New York, NY, USA: 1997. pp. 65–105. [Google Scholar]
- 20.Chatman K, Hollenbeck T, Hagey L, Vallee M, Purdy R, Weiss F, Siuzdak G. Nanoelectrospray mass spectrometry and precursor ion monitoring for quantitative steroid analysis and attomole sensitivity. Analytical Chemistry. 1999;71:2358–2363. doi: 10.1021/ac9806411. [DOI] [PubMed] [Google Scholar]
- 21.Gaskell SJ. Evaluation of the quantitative-analysis of a steroid sulfate using fast atom bombardment and tandem mass-spectrometry. Biomedical and Environmental Mass Spectrometry. 1988;15:99–104. doi: 10.1002/bms.1200150207. [DOI] [PubMed] [Google Scholar]
- 22.Weidolf LO, Lee ED, Henion JD. Determination of boldenone sulfoconjugate and related steroid sulfates in equine urine by high-performance liquid chromatography/tandem mass spectrometry. Biomed Environ Mass Spectrom. 1988;15:283–289. doi: 10.1002/bms.1200150508. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Henion J. Quantitative and qualitative determination of estrogen sulfates in human urine by liquid chromatography/tandem mass spectrometry using 96-well technology. Anal Chem. 1999;71:3955–3964. doi: 10.1021/ac990162h. [DOI] [PubMed] [Google Scholar]
- 24.Attygalle AB, Garcia-Rubio S, Ta J, Meinwald J. Collisionally-induced dissociation mass spectra of organic sulfate anions. Journal of the Chemical Society-Perkin Transactions. 2001;2:498–506. [Google Scholar]
- 25.Rudewicz P, Straub KM. Rapid structure elucidation of catecholamine conjugates with tandem mass-spectrometry. Analytical Chemistry. 1986;58:2928–2934. doi: 10.1021/ac00127a008. [DOI] [PubMed] [Google Scholar]
- 26.Domalik LJ, Chaplin DD, Kirkman MS, Wu RC, Liu WW, Howard TA, Seldin MF, Parker KL. Different isozymes of mouse 11 beta-hydroxylase produce mineralocorticoids and glucocorticoids. Molecular endocrinology. 1991;5:1853–1861. doi: 10.1210/mend-5-12-1853. [DOI] [PubMed] [Google Scholar]
- 27.Burstein S, Gut M. Intermediates in the conversion of cholesterol to pregnenolone: Kinetics and mechanism. Steroids. 1976;28:115–131. doi: 10.1016/0039-128x(76)90131-8. [DOI] [PubMed] [Google Scholar]
- 28.Gaskell SJ. Analysis of steroids by mass spectrometry. Methods Biochem Anal. 1983;29:385–434. doi: 10.1002/9780470110492.ch7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.