Abstract
Background:
One of the major clinical problems regarding Pseudomonas aeruginosa is attributed to metallo-beta-lactamases (MBL). This group of enzymes is a subset of beta lactamases which belong to group B of Ambler classification and cause hydrolysis of carbapenems. Based on epidemiological studies conducted worldwide, it is proved that prevalence of genes coding MBLs in P. aeruginosa species are different in various geographic zones and even in various hospitals. Therefore, according to the clinical importance of organisms generating MBLs, it is necessary to identify and control these bacteria in hospitals for therapeutic purposes.
Objectives:
The current study aimed to investigate the Metallo-beta-Lactamase VIM-1, SPM-1, and IMP-1 genes among clinical P. aeruginosa species isolated in Zahedan, Iran.
Materials and Methods:
The current study investigated the presence of MBL through phenotypic and genotypic methods and also the pattern of antibiotic resistance in P. aeruginosa species isolated in hospitals. The Minimum Inhibitory Concentration (MIC) against imipeneme was measured for 191 P. aeruginosa species isolated from Zahedan hospitals after identification through biochemical methods and determination of the antibiotic resistance pattern. Strains with MIC > 4 µg/mL were studied by phenotypic and genotypic methods.
Results:
The rate of resistance against imipeneme was 5.7% and after carrying out the phenotypic experiments, nine species were identified as of MBL producer. Seven species were confirmed by Polymerase Chain Reaction (PCR) method. Gene VIM-1 was the predominant gene among the positive (antibiotic resistant) species.
Conclusions:
The study results showed that MBL genes were present in some of the species isolated from Zahedan hospitals. Regarding the importance of MBL producer bacteria in hospitals, quick identification and evaluation of these clinical species can be considered as an important and basic step for treatment and control of pseudomonad infections.
Keywords: Metallo-beta-lactamase, Pseudomonas aeruginosa, Imipeneme, Antibiotic Resistance
1. Background
Pseudomonas aeruginosa is one of the leading antibiotic-resistant bacterium, which plays an important role in the inception of nosocomial infections worldwide (1). In most areas of the world, resistance of P. aeruginosa to β-Lactam compounds is increasing (2). Recently, by carbapenems, infection due to resistant species is controlled better. In recent decades, P. aeruginosa is reported to be resistant against these compounds (3, 4). Although different reasons such as generation of chromosomal Amp-c, loss of purine D2 or efflux pumps can be considered for this resistance, the most important reason is generation of carbapeneme hydrolyzing enzymes (5).
Metallo-beta-lactamase-generative P. aeruginosa species were reported for the first time in Japan in 1991. However, these species are reported worldwide as well as Iran, in which isolation of multidrug-resistant P. aeruginosa species has reached a critical point (6). Metallo-beta-lactamases (MBLs) such as VIM, IMP, and SPM, which belong to group B of Ambler classification, cause hydrolysis of a variety of beta lactams except monobactam (aztreonam) (7). Among beta lactamases, MBLs are unique in requiring the presence of zinc ion in the active site of the enzyme. Their activity is inhibited by chelating agents such as Ethylene Diamine Tetra Acetic Acid (EDTA), Sodium Mercapto Acetic Acid (SMA), and Dipicolinc Acid, but beta lactams inhibitors such as clavulanic acid, sulbactam, and tazobactam have no effects on them (8-10).
Based on epidemiological studies conducted worldwide, it is proved that the prevalence of genes coding MBL in P. aeruginosa species is different in various geographic zones and even in various hospitals (11-13). Therefore, according to the clinical importance of organisms generating MBL, it is necessary to identify and control them in hospitals for therapeutic purposes (13).
2. Objectives
One of the major clinical problems related to P. aeruginosa is attributed to MBL. Given that there was no information about MBL genes in Zahedan hospitals, the current study was designed to investigate Metallo-beta-Lactamase VIM-1, SPM-1, and IMP-1 genes among clinical P. aeruginosa species isolated in Zahedan, Iran by phenotypic (E test) and genotypic (Polymerase Chain Reaction) methods.
3. Materials and Methods
3.1. Bacterial Isolates
Samples included 191 species of P. aeruginosa isolated from Zahedan hospitals (Emam Ali, Boali, Nabi Akram, and Tamin-e-Ejtemaei) from October 2012 to March 2013. The isolates were identified using microbiological and biochemical methods.
3.2. Antibiotic Resistance Investigation
Measurement of antibiotic resistance was conducted by disk diffusion method for the following antibiotics: ceftizoxime (ZOX), gentamicin (GM), ciprofloxacin (CP), ceftazidime (CAZ), piperacillin (PLP), tazobactam and piperacillin (TZP), cefteriaxon (CRO), cefotaxime (CTX), and imipenem (IPM) (Mastdiscs, Britain) according to the standards of Clinical and Laboratory Standards Institute (CLSI).
3.3. Susceptibility Testing
Minimum Inhibitory Concentration (MIC) of imipenem resistant isolates was measured by E-test, according to CLSI protocol. Pseudomonas aeruginosa ATCC 27853 was used as a control reference strain. In order to detect MBL generative isolates, E-test tapes were used (AB Biodisk, Solna, Sweden) according to the manufacturer’s instructions. These tapes have two parts. In one part (IP), there is a gradient of different imipenem ratios (4 - 256 µg/mL) and in the other one (IPI), a gradient of different imipenem ratios plus a constant concentration of EDTA (1 - 64 µg/mL) (10, 14). To interpret E-test tapes, it should be considered that if MIC of IP/IPI ratio is equal or greater than eight, (MBL) enzyme is generated (10).
3.4. Investigation of bla-VIM-1, bla-IMP-1, and bla-SPM-1 Genes by PCR
Imipenem resistant isolates were used to investigate bla-VIM-1, bla-IMP-1, and bla-SPM-1 genes by PCR. For DNA extraction, boiling method was applied. First, three to five colonies were extracted from fresh culture medium and then suspension was prepared using 200 mL of distilled water boiled at 100˚C for 10 minutes. Next, it was centrifuged at 12000 rpm for 10 minutes. The supernatant containing DNA was transferred to new Eppendorf tubes for PCR in order to amplify MBL genes, bla-VIM-1, bla-IMP-1, and bla-SPM-1. The following primers (Sinaclone, Iran) were used to amplify MBL genes (15):
VIM-1:
Forward: 5′-AGTGGTGAGTATCCGACAG-3′
Reverse: 5′-ATGAAAGTGCGTGGAGAC-3′
IMP-1:
Forward: 5′- ACCGCAGCAGAGTCTTTGCC-3′
Reverse: 5′-ACAACCAGTTTTGCCTTACC-3′
SPM-1:
Forward: 5′- GCGTTTTGTTTGTTGCTC -3′
Reverse: 5′-TTGGGGATGTGAGACTAC-3′
To perform PCR, 1 µL of the extracted DNA was added to PCR Master Mix with the final volume of 25 µL containing 1.5 Mm of MgCl2, 0.24 Mm of dNTPs, 5 pm of each primer, and one unit of Taq polymerase enzyme. PCR was controlled by P. aeruginosa species carrying out SPM-1, VIM-1, and IPM-1 genes. In order to control the molecular weight, 100 base pair DNA ladder was applied and electrophoresed on 1% agarose gel and then stained using ethidium bromide. Result was observed by transilluminator under ultraviolet (UV) light.
Amplification reactions were carried out in a 25 µL volume using one unit of Taq polymerase enzyme (Sinaclone, Iran), 20 ng of template DNA, 1.5 Mm of MgCl2 , 0.24 Mm of dNTPs and 5 pm of each primer in the reaction buffer recommended by the enzyme manufacturer. PCR was performed using Taq polymerase kit (Sinaclone, Iran). The products were amplified with 30 cycles in the following program: denaturation at 95°C for 5 minutes, denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds. Pseudomonas aeruginosa isolate generating SPM-1, VIM-1, and IPM-1was used as positive control. PCR product was electrophoresed on 1% agarose gel.
4. Results
Among the 191 isolated species of P. aeruginosa, the maximum and minimum number of isolated species belonged to ICU (43) and CCU (one), respectively. Furthermore, the species were isolated from different clinical samples; most of them (71 species) belonged to mid-stream urine and the least (one specie) to biopsy samples.
4.1. Antibiotic Resistance Pattern
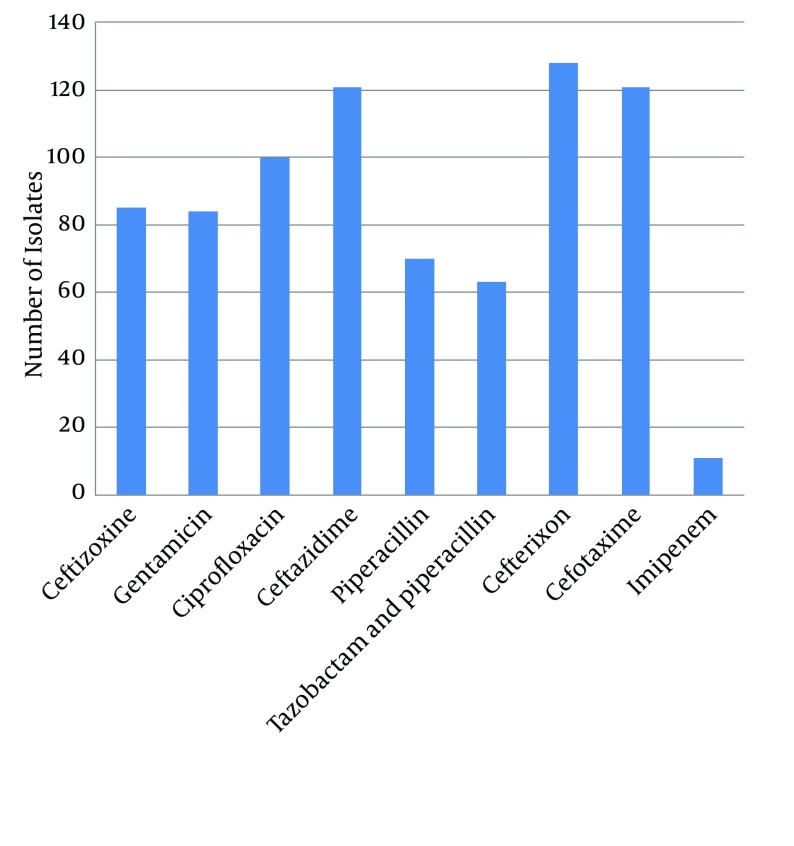
The results of antibiotic resistance pattern, obtained by Kirby-Bauer method on 191 P. aeruginosa species isolated from different areas of Zahedan hospitals for piperacillin, cefotaxime, ceftizoxime, ceftazidime, piperacillin, imipenem, tazobactam, piperacillin, and ciprofloxacin are shown in Figure 1.
Figure 1. Antibiotic Resistance Pattern of the Isolated Pseudomonas aeruginosa Species.
4.2. Metallo-bata-Lactamase Producers Detection
The imipenem-nonsusceptible isolates were evaluated by E-test to detect MBLs. Eleven out of 191 isolates were resistant against imipenem. For these resistant species, E-test was applied and nine out of eleven resistant isolates were identified as MBL producers.
4.3. PCR Detection of Metallo-beta-Lactamase Genes
PCR was conducted on nine isolates to investigate the frequency of IPM-1, SPM-1, and VIM-1 genes. In seven isolates, only VIM-1 gene was found, while none of the genes were found in two isolates (Figure 2). Among the isolates containingVIM-1 gene, three samples were isolated from lesions, three from mid-stream urine, and one from endotracheal tube (ETT). Totally, four isolates belonged to ICU and other three isolates to CCU. These seven isolates were resistant against all the studied antibiotics, but they were sensitive to polymyxin B and colistin.
Figure 2. PCR Products Gel Electrophoresis for VIM-1 Gene (261 bp).
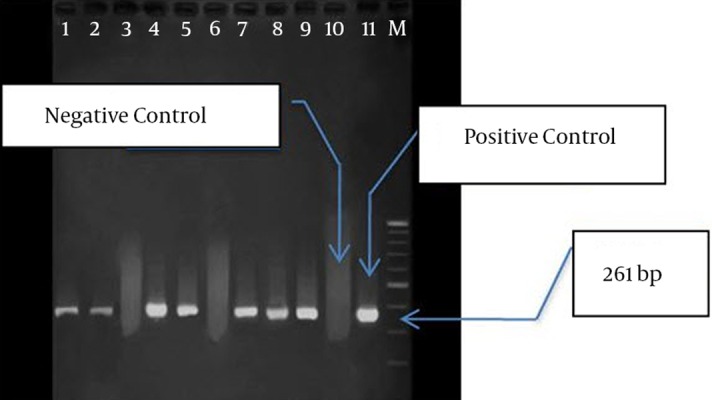
Lanes: M: 100 bp Marker, 11: Positive Control, 1 to 9: PCR Product for VIM-1 Gene,10: Negative Control.
5. Discussion
It is a great challenge to treat infections caused by P. aeruginosa, due to its resistance against drugs. Different antibiotics including beta lactames, aminoglycosides and quinolones are applied to treat the infections caused by P. aeruginosa. Antibiotic resistance pattern of these bacteria (P. aeruginosa) is changing quickly. However, studies showed that the resistance of P. aeruginosa against antibiotics fluctuates, especially regarding imipenem (16). Carbapenems and beta lactam antibiotics are using considerably to treat infections due to multidrug-resistant P. aeruginosa since they are resistant against most of the beta lactamases and have great membrane permeability (17). However, in last decades, warnings about an increase in P. aeruginosa resistance against antimicrobial agents are reported. These organisms have various self-protection ways against antibiotics.
One of the most important ways for resistance against imipeneme is generation of MBL enzymes (18, 19). VIM, IMP, and SPM from MBL family are quite prevalent in P. aeruginosa (20). Since various reports indicate the increased resistance of P. aeruginosa isolates against antibiotics (especially imipeneme), proper use of these antibiotics and time-consuming identification of isolates generating MBL should be considered. It leads to successful treatment and prevents propagation of resistant genes, which can be seriously harmful for societies (21). There are different methods to identify MBL-generative species. The present study used E-test tapes. These tapes contain EDTA, which is a MB inhibitor (22, 23).
In the current study, seven out of nine isolates, which were positive in E-test (phenotypic method), contained VIM-1 gene and two isolates were negative in both phenotypic and genotypic (PCR) methods, indicating the presence of other factors such as lack of opr D, which results in membrane permeability change, efflux pumps or chromosomal Ampc beta lactamase. Two isolates, positive in phenotypic method, were reported negative in PCR reaction indicating that other genes are involved in this process. Among the isolates containing VIM-1 gene, four species (57%) were isolated from the patients hospitalized in ICU for approximately a long time and had undergone antibiotic therapy. It indicates the presence and propagation of the above mentioned gene under such circumstances. In the current investigation, IMP-1 and SMP-1 genes were not observed in any of the strains resistant to imipenem. The study by Shahcheraghi et al. on 243 P. aeruginosa species isolated from Imam Khomeini Hospital in Tehran, showed that 15 isolates contained VIM-1 gene (24). Also, Sepehriseresht et al. reported that 94 out of 483 species of P. aeruginosa isolated from the patient lesions were resistant and intermediate resistant species and carried vim1, vim2, ipm1, and ipm2 genes by PCR (25).
The results of the above mentioned researches are nearly consistent with those of the present study indicating that VIM-1 is the predominant MBL in Iran. Another research showed that 34 out of 111 (43%) species isolated carried VIM gene (6, 26) and their results are not consistent with those of the current study. It can be said that the prevalence of MBL in the species isolated from that hospital was more than expected. Since Imam Musa Kazem Hospital is a burn center, a great amount of imipenem is prescribed there and resistance against this antibiotic has reached 94.9%. Therefore, it is not unexpected to find genes driving drug resistance, especially MBL ones located more on dynamic elements (plasmide or transpozone) are propagated more in this center.
The current study showed that fortunately the prevalence of MBL in Zahedan is less than other cities. However it should be noted that according to the studies in most areas of the world, the prevalence is incremental and if preventive measures are not taken, there will be major clinical problems. Since most of integrones containing VIM-1 gene coding aminoglycosides destructing enzyme, the importance of the problem is intensified (27). Therefore diagnostic methods should be routinely used to determine these species in clinical laboratories; also synthesis of more effective antimicrobial compounds with new effecting mechanisms should be noted.
Acknowledgments
The current study was supported by Tropical Diseases Research Center, Zahedan University of Medical Sciences, Zahedan, Iran. Authors wish to thank the manager of the center and his colleague for useful discussions and critical reading of the manuscript.
Footnotes
Authors’ Contributions:Study concept and design: Shahram Shahrekizahedani, Farshid Kafilzadeh and Zakaria Bameri; acquisition of data: Mehdi Ghamgosha; analysis and interpretation of data: Shahram Shahreki zahedani, Gholamreza Farnoosh, and Ramezan Ali Taheri; drafting of the manuscript: Mehdi Ghamgosha; critical revision of the manuscript for important intellectual content: Gholamreza Farnoosh, and Ramezan Ali Taheri; statistical analysis: Zakaria Bameri.
Funding/Support:This study was financially supported by grant from Zahedan University of Medical Sciences, Zahedan, Iran.
References
- 1.Gailiene G, Pavilonis A, Kareiviene V. The peculiarities of Pseudomonas aeruginosa resistance to antibiotics and prevalence of serogroups. Medicina (Kaunas). 2007;43(1):36–42. [PubMed] [Google Scholar]
- 2.Takigawa K, Fujita J, Negayama K, Xu G, Yamagishi Y, Miyawaki H, et al. Comparing antimicrobial activity against resistant Pseudomonas aeruginosa using an index for the absence of cross-resistance. J Antimicrob Chemother. 1995;35(3):425–7. doi: 10.1093/jac/35.3.425. [DOI] [PubMed] [Google Scholar]
- 3.Lagatolla C, Edalucci E, Dolzani L, Riccio ML, De Luca F, Medessi E, et al. Molecular evolution of metallo-beta-lactamase-producing Pseudomonas aeruginosa in a nosocomial setting of high-level endemicity. J Clin Microbiol. 2006;44(7):2348–53. doi: 10.1128/JCM.00258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaskulski MR, Medeiros BC, Borges JV, Zalewsky R, Fonseca ME, Marinowic DR, et al. Assessment of extended-spectrum beta-lactamase, KPC carbapenemase and porin resistance mechanisms in clinical samples of Klebsiella pneumoniae and Enterobacter spp. Int J Antimicrob Agents. 2013;42(1):76–9. doi: 10.1016/j.ijantimicag.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Gupta AK, Chauhan DS, Srivastava K, Das R, Batra S, Mittal M, et al. Estimation of efflux mediated multi-drug resistance and its correlation with expression levels of two major efflux pumps in mycobacteria. J Commun Dis. 2006;38(3):246–54. [PubMed] [Google Scholar]
- 6.Sarhangi M, Motamedifar M, Sarvari J. Dissemination of Pseudomonas aeruginosa Producing blaIMP1, blaVIM2, blaSIM1, blaSPM1 in Shiraz, Iran. Jundishapur J Microbiol. 2013;6(7) [Google Scholar]
- 7.Hall BG, Salipante SJ, Barlow M. The metallo-beta-lactamases fall into two distinct phylogenetic groups. J Mol Evol. 2003;57(3):249–54. doi: 10.1007/s00239-003-2471-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, et al. Novel acquired metallo-beta-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005;49(11):4485–91. doi: 10.1128/AAC.49.11.4485-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan C, Beceiro A, Gutierrez O, Alberti S, Garau M, Perez JL, et al. Characterization of the new metallo-beta-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain. Antimicrob Agents Chemother. 2008;52(10):3589–96. doi: 10.1128/AAC.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou F, Ji B, Zhang H, Jiang H, Yang Z, Li J, et al. Synergistic effect of thymol and carvacrol combined with chelators and organic acids against Salmonella Typhimurium. J Food Prot. 2007;70(7):1704–9. doi: 10.4315/0362-028x-70.7.1704. [DOI] [PubMed] [Google Scholar]
- 11.Fiett J, Baraniak A, Mrowka A, Fleischer M, Drulis-Kawa Z, Naumiuk L, et al. Molecular epidemiology of acquired-metallo-beta-lactamase-producing bacteria in Poland. Antimicrob Agents Chemother. 2006;50(3):880–6. doi: 10.1128/AAC.50.3.880-886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouda S, Ohara M, Onodera M, Fujiue Y, Sasaki M, Kohara T, et al. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J Antimicrob Chemother. 2009;64(1):46–51. doi: 10.1093/jac/dkp142. [DOI] [PubMed] [Google Scholar]
- 13.Upadhyay S, Sen MR, Bhattacharjee A. Presence of different beta-lactamase classes among clinical isolates of Pseudomonas aeruginosa expressing AmpC beta-lactamase enzyme. J Infect Dev Ctries. 2010;4(4):239–42. doi: 10.3855/jidc.497. [DOI] [PubMed] [Google Scholar]
- 14.Lim YM, Shin KS, Kim J. Distinct antimicrobial resistance patterns and antimicrobial resistance-harboring genes according to genomic species of Acinetobacter isolates. J Clin Microbiol. 2007;45(3):902–5. doi: 10.1128/JCM.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, et al. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41(12):5407–13. doi: 10.1128/JCM.41.12.5407-5413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patzer JA, Dzierzanowska D. Increase of imipenem resistance among Pseudomonas aeruginosa isolates from a Polish paediatric hospital (1993-2002). Int J Antimicrob Agents. 2007;29(2):153–8. doi: 10.1016/j.ijantimicag.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 17.Sunagawa M, Kanazawa K, Nouda H. [Antipseudomonal activity of carbapenem antibiotics]. Jpn J Antibiot. 2000;53(7):479–511. [PubMed] [Google Scholar]
- 18.Hocquet D, Plesiat P, Dehecq B, Mariotte P, Talon D, Bertrand X, et al. Nationwide investigation of extended-spectrum beta-lactamases, metallo-beta-lactamases, and extended-spectrum oxacillinases produced by ceftazidime-resistant Pseudomonas aeruginosa strains in France. Antimicrob Agents Chemother. 2010;54(8):3512–5. doi: 10.1128/AAC.01646-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozyurt M, Haznedaroglu T, Sahiner F, Oncul O, Ceylan S, Ardic N, et al. [Antimicrobial resistance profiles of community-acquired uropathogenic Escherichia coli isolates during 2004-2006 in a training hospital in Istanbul]. Mikrobiyol Bul. 2008;42(2):231–43. [PubMed] [Google Scholar]
- 20.Lee K, Yong D, Yum JH, Lim YS, Bolmstrom A, Qwarnstrom A, et al. Evaluation of Etest MBL for detection of blaIMP-1 and blaVIM-2 allele-positive clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2005;43(2):942–4. doi: 10.1128/JCM.43.2.942-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niitsuma K, Saitoh M, Kojimabara M, Kashiwabara N, Aoki T, Tomizawa M, et al. [Antimicrobial susceptibility of Pseudomonas aeruginosa isolated in Fukushima Prefecture]. Jpn J Antibiot. 2001;54(2):79–87. [PubMed] [Google Scholar]
- 22.Vahdani M, Azimi L, Asghari B, Bazmi F, Rastegar Lari A. Phenotypic screening of extended-spectrum ss-lactamase and metallo-ss-lactamase in multidrug-resistant Pseudomonas aeruginosa from infected burns. Ann Burns Fire Disasters. 2012;25(2):78–81. [PMC free article] [PubMed] [Google Scholar]
- 23.Gales AC, Jones RN, Turnidge J, Rennie R, Ramphal R. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001;32 Suppl 2:S146–55. doi: 10.1086/320186. [DOI] [PubMed] [Google Scholar]
- 24.Shahcheraghi F, Nikbin VS, Feizabadi MM. Identification and genetic characterization of metallo-beta-lactamase-producing strains of Pseudomonas aeruginosa in Tehran, Iran. New Microbiol. 2010;33(3):243–8. [PubMed] [Google Scholar]
- 25.Sepehriseresht S, Boroumand MA, Pourgholi L, Sotoudeh Anvari M, Habibi E, Sattarzadeh Tabrizi M. Detection of vim- and ipm-type metallo-beta-lactamases in Pseudomonas aeruginosa clinical isolates. Arch Iran Med. 2012;15(11):670–3. [PubMed] [Google Scholar]
- 26.Golshani Z, Ahadi AM, Sharifzadeh A. Occurrence of Ambler Class B Metallo-β-Lactamase Gene in Imipenem-Resistant Pseudomonas aeruginosa Strains Isolated from Clinical Samples. Zahedan J Res Med Sci. 2014;16(2):6–9. [Google Scholar]
- 27.Lee K, Park AJ, Kim MY, Lee HJ, Cho JH, Kang JO, et al. Metallo-beta-lactamase-producing Pseudomonas spp. in Korea: high prevalence of isolates with VIM-2 type and emergence of isolates with IMP-1 type. Yonsei Med J. 2009;50(3):335–9. doi: 10.3349/ymj.2009.50.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]