Abstract
Implant-associated infections can have severe effects on the longevity of implant devices and they also represent a major cause of implant failures. Treating these infections associated with implants by antibiotics is not always an effective strategy due to poor penetration rates of antibiotics into biofilms. Additionally, emerging antibiotic resistance poses serious concerns. There is an urge to develop effective antibacterial surfaces that prevent bacterial adhesion and proliferation. A novel class of bacterial therapeutic agents, known as antimicrobial peptides (AMP’s), are receiving increasing attention as an unconventional option to treat septic infection, partly due to their capacity to stimulate innate immune responses and for the difficulty of microorganisms to develop resistance towards them. While host- and bacterial- cells compete in determining the ultimate fate of the implant, functionalization of implant surfaces with antimicrobial peptides can shift the balance and prevent implant infections. In the present study, we developed a novel chimeric peptide to functionalize the implant material surface. The chimeric peptide simultaneously presents two functionalities, with one domain binding to a titanium alloy implant surface through a titanium-binding domain while the other domain displays an antimicrobial property. This approach gains strength through control over the bio-material interfaces, a property built upon molecular recognition and self-assembly through a titanium alloy binding domain in the chimeric peptide. The efficiency of chimeric peptide both in-solution and absorbed onto titanium alloy surface was evaluated in vitro against three common human host infectious bacteria, S. mutans, S. epidermidis, and E. coli. In biological interactions such as occurs on implants, it is the surface and the interface that dictate the ultimate outcome. Controlling the implant surface by creating an interface composed chimeric peptides may therefore open up new possibilities to cover the implant site and tailor it to a desirable bioactivity.
Keywords: Antimicrobial coatings, Bio-material interfaces, Biological surface functionalization, Biointerface, Bioactivity, Infection-free implants
1. INTRODUCTION
Titanium and its alloys have been extensively used in orthopedic and dental implants, mainly due to their unique combination of excellent mechanical properties, corrosion resistance, biocompatibility and osseointegration [1-5]. However, the risk of failure of these implants, which can lead to suboptimal clinical outcomes, still poses a significant threat to patients and post-surgical challenges to their clinicians [6, 7]. Although recent enhancements in the design of prosthetic devices and the advancements in surgical procedures have reduced the number of complications leading to failure, implant associated bacterial infections is still a serious challenge and a major cause of post-surgical morbidity and mortality [8].
Implant materials provides an ideal surface to the growth of common pathogens such as Staphylococcus aureus, Staphylococcus epidermidis, and Pseudomonas aeruginosa, which could be acquired shortly after surgery or at later date. Failure to adequately combat these bacterial infections at the implant-tissue interface often results in complex revision procedures, with an economic burden to the health-care system, and in most cases the removal of the implant with the re-instrumentation at a later date is the only remedy. Moreover, formation by these pathogens of complex biofilm structures on the implant surface or its periphery can make the problem more difficult to address. Bacterial biofilm formed on the implant creates a barrier that decreases penetration of antimicrobial agents reducing the susceptibility of the biofilms members to drug delivery [6, 9, 10]. Even with substantial interest and efforts to improve local delivery of systemic antibiotics to implant surfaces, challenges regarding diluted drug levels at the target site and the potential toxicity of conventional antibiotics still need to be addressed [11, 12]. Furthermore, the potential development and spread of antibiotic-resistant pathogens such as the methicillin-resistant Staphylococcus aureus (MRSA) remain a challenge to hospital acquired infections [10, 13].
One of the successful survival strategies of bacteria is their ability to adhere to virtually any surfaces through their various types of adhesins. Free floating bacteria can activate the biofilm related phenotype after their attachment to implant surfaces. Initial phase of their attachment is relatively less stable compared to later stage where bacteria start expressing biofilm specific genes. The window of opportunity relies on designing the surfaces prior to bacterial attachment moving into almost an irreversible phase, where the biofilm formation starts. Development of implant surfaces that would prevent bacterial adhesion becomes critical. Several surface coating and functionalization strategies have been reported to overcome implant failure associated with infections. In an attempt to render the implant surface non-adhesive and/or to introduce antimicrobial surfaces, the use of polyethylene glycol (PEG) and its derivatives [14, 15], coatings of albumin [16], covalent attachment of conventional antibiotics [17-20], chlorhexidine [21], silver, nitrogen oxide [22, 23] and quaternary ammonia compounds [19, 24] have been used. While the activation of implant surfaces by these agents have been shown to reduce bacterial adhesion, existing covalent coupling strategies often require complex chemistry to execute, with the unwieldy requirement of specific functional groups on the surface with extensive optimization steps. Moreover, the limited capacity of this chemical derivatization used for modification of different implant materials makes them far from providing a comprehensive solution [25-27]. Additionally, the slow release of derivatized antimicrobial agents from preloaded implant devices raises concern about a possible link to increased incidences of bacterial resistance and cytotoxicity [28].
Bioactivation of implant surfaces with more biocompatible and nontoxic biomolecules such as antimicrobial peptides (AMP’s) would be a feasible approach to overcome infection derived implant failure without evoking either toxicity or antibiotic resistance. These short, cationic AMP’s are evolutionary conserved constituents of the innate immune defense systems of many organisms, including insects, plants, animals and humans [29-31]. AMP’s are believed to specifically target and disrupt the integrity of negatively charged cell membrane of microorganisms. Although there is no consensus in their sequence and structure, AMP’s usually have an amphipathic structure which serves as efficient ionic recognition between the cationic residues of peptide and the phospholipids of the bacterial membrane [32, 33]. Furthermore, in contrast to conventional antibiotics, it is extremely difficult for microorganisms to develop resistance against these peptides because of their highly sophisticated reaction mechanisms and considerably rapid rate action [29, 34]. More importantly, AMP’s have broad-spectrum antimicrobial activity against gram-positive and gram-negative bacteria, fungi and viruses. AMP’s can work synergistically with conventional antibiotics and facilitate antibiotic penetration to the infection site, thus enabling more aggressive biofilm treatment [29]. It has been also demonstrated that the sequence and/or resulting structure of natural AMP’s can be utilized as templates for the design of synthetic variants with enhanced antimicrobial activities [35-37]. By retaining their localized effect through tethering and assembly of AMP’s as an antimicrobial coating to implant surfaces agents it could greatly increase effectiveness while reducing potential cytotoxic consequences and collateral damage through vascular re-distribution [38].
Unlike other approaches utilizing covalent linkages to tether AMP’s on implant surface, we have created bifunctional chimeric peptides composed of implant surface binding peptide and an AMP agent. These bifunctional chimeric peptides rely on the properties of the solid-binding peptides [39] that preferentially bind to the titanium surface, a common implant surface, while freely exposing the AMP motif to combat invading bacteria. This interface on the implant surface is constructed by combining combinatorially selected solid-binding peptides with AMP sequences in different combinations through an intervening flexible linker [40-44]. These bifunctional chimeric peptides were characterized in terms of their binding properties to titanium surface and their antimicrobial efficacy either in-solution or immobilized on surface. Keeping in mind the importance of assessing any therapeutic target against a range of problematic bacteria due to varying responses, we chose three different types of bacteria, Streptococcus mutans, Staphylococcus epidermidis, and Escherichia coli to test the efficacy of our bifunctional chimeric peptides against. S. mutans is a gram-positive, biofilm-forming bacterium commonly found in oral implant infections [45]. S. epidermidis is a gram-positive, biofilm-forming bacterium commonly found in orthopedic implant infections, making up 32% of clinical isolates from orthopedic implant infections. E. coli is a gram-negative, slime-producing bacterium occasionally found in orthopedic implant infections [46, 47].
Here we provide details on an alternative method of implant-surface functionalization that does not require complex procedures or the covalent modification of the implant surface [47-49]. The principles laid out in the paper can be applied to other identified AMP sequences, and expanded to a wide-range of biomaterials other than titanium by deploying different solid binding sequences that binds to other biomaterials [40, 49, 50]. In addition, structure-function relationships found in this paper can be applied for control of functionality over designing chimeric peptides by following simple rules [51].
2. MATERIALS and METHODS
2.1 Target Implant Material Characterization and Preparation
Surface properties of titanium grade V powder (Sigma-Aldrich, St Louis, MO, USA) and titanium grade V implant (Vetimplants, St. Augustine, FL, USA) were determined by scanning electron microscopy (SEM). Elemental composition of the substrate was analyzed by collecting energy-dispersive X-ray spectroscopy (EDS) spectra for 100 seconds at 9 keV using a LaB6 filament. Titanium grade V implant (Vetimplants, St. Augustine, FL, USA) was cut into approximately 1cm × 1cm squares and sharp edges were removed by hand polishing with a 600-grit finish silicon carbide metallurgical paper. Before experimental use, titanium grade V powder and implant pieces were cleaned by sonicating sequentially in a 1:1 acetone/methanol mixture, then isopropyl alcohol, and finally de-ionized water. Then, substrates were sterilized for 15 minutes under UV light.
2.2 Selection of Titanium Binding and Antimicrobial Binding Peptides
Titanium binding peptides (TiBP) were selected by cell surface display [50] and phage display method. Briefly, for cell surface display approach, the FliTrx bacterial cell surface display system (Invitrogen, Carlsbad, CA, USA) was used to select peptide sequences against titanium substrates [52-54]. After four-rounds of successful biopanning enrichment, the DNA sequences for each of the 60 isolated clones were analyzed. Binding properties for each peptide were characterized by quantitative fluorescent microscopy, employing a Nikon Eclipse TE-2000U fluorescent microscope (Nikon Inc., Melville, NY, USA) equipped with a Hamamatsu ORCA-ER cooled CCD camera (Hamamatsu Corp., Bridgewater, NJ, USA), imaged using a FITC filter (exciter 460–500 nm, dichroic 505 nm, emitter 510–560 nm) and MetaMorph imaging system (Universal Imaging, West Chester, PA, USA). Finally, the binding affinity for each peptide was calculated by determining in triplicate samples, the average number of adherent bacterial cells expressing the peptide identified on the titanium surface. Consequently to this analysis, the titanium binding peptides (TiBP) were grouped as strong, moderate or weak binders, according to their binding capacity.
In phage display method, the Ph.D.-12 phage display peptide library kit (New England Biolabs, Ipswich, MA, USA) containing 1.2 × 109 different randomized peptide sequences was used as previously described [55]. The peptide library is incubated with titanium grade V powder in potassium carbonate (PC) buffer containing 0.1% Tween 20 detergent (Merck, Whitehouse Station, NJ, USA) and then the unbound phages are removed by washing the surface with PC buffer containing 0.1% detergent (Tween 20 and Tween 80). The bound phages are eluted specifically from the surface using elution buffer; and the eluted pool is amplified in Escherichia coli ER2738. Amplified phages are purified and subsequently used for additional panning rounds. After each round, the phages are grown on solid media, and single clones are selected by picking single-phage plaques that constitute a clone for each selected peptide. Genomic DNA’s of single-phage clones are then isolated and their nucleotide sequence determined. Finally, individual clones were characterized by quantitative fluorescent microscopy employing a Nikon Eclipse TE-2000U, as described above.
Computationally-designed and characterized short AMP sequences were chosen by data mining from the literature [44, 56]. The molecular weight (MW), isoelectronic point (pI), charge and grand average of hydropathy (GRAVY) value parameters for each peptide were calculated using ExPASy Proteomics Server.
2.3 Peptide Synthesis
An automated solid-phase peptide synthesizer (CS336X, CS-Bio Inc., Menlo Park, CA, USA) was utilized to synthesize peptides through Fmoc-chemistry. In this approach, modified amino acids with the N-terminus and amino acid side chains protected by Fmoc-group and an appropriate protecting group, respectively, were used. In the reaction vessel, the Wang resin (Novabiochem, West Chester, PA, USA), pre-loaded with Fmoc protected first amino acid, was treated with 20% piperidine in DMF to remove the Fmoc group and monitored by UV-absorbance at 301 nm. The incoming amino acid, separately activated with HBTU (Sigma-Aldrich, St Louis, MO, USA) in dimethylformamide (DMF), was transferred into the vessel and incubated with the resin for 45 min. After washing the resin with DMF, this protocol was applied for addition of each of the next amino acids.
Following synthesis, the resulting resin-bound peptides were cleaved and the side-chain de-protected using reagent-K (TFA/thioanisole/H2O/phenol/ethanedithiol (87.5:5:5:2.5)) and precipitated by cold ether. Crude peptides were purified by RP-HPLC with up to >98% purity obtained (Gemini 10u C18 110A column). The sequence of the peptides was confirmed by mass spectroscopy (MS) using a MALDI-TOF mass spectrometry with reflectron (RETOF-MS) on an Autoflex II (Bruker Daltonics, Billerica, MA, USA). Stock solutions of each peptide at 4 mM were made in sterile-, de-ionized-water by dissolving the peptides. Subsequent dilutions were accomplished with sterile 1X PBS.
2.4 Binding Characterization of Peptides onto the Titanium Alloy Implant Surfaces
Fluorescent microscopy characterization procedure was applied to investigate the binding affinities for both the titanium binding peptide as was the AMP conjugated bifunctional chimeric peptides. In this assay, the biotinylated peptide was incubated with pre-cleaned substrates for 3 hours at room temperature. Following, substrates were washed three times with 1X phosphate-buffered saline (PBS) and bound peptides were labelled with streptavidin-Alexa Fluor 488 (Molecular Probes, Eugene, OR, USA) by incubation for 15 minutes in the dark. Substrates were washed with de-ionized water three times and bound peptides were visualized on the substrate surface by fluorescent microscope. All measurements were carried out by triplicate independent experiments.
2.5 Bacterial Maintenance and Culturing
Three bacteria species - Escherichia coli American Type Culture Collection (ATCC) 2592, Streptococcus mutans ATCC 25175, and Staphylococcus epidermidis ATCC 29886 were used in the present study. All of them were cultured according to ATCC protocol using the following media: Trypticase Soy Broth (TSB) for E. coli, Brain Heart Infusion (BHI) Broth for S. mutans, and Nutrient Broth (NB) for S. epidermidis. For all three bacterial species, the bacterial pellet obtained from ATCC was rehydrated in 0.5 mL of the above-specified media, and several drops of the suspension were immediately streaked on the relevant solid media. The agar-plate was then incubated aerobically at 37°C for 24 hours, except in the case of S. mutans which was incubated in the presence of 5% CO2 supplemented atmosphere. S. mutans overnight cultures were made by aseptically transferring a single-colony forming unit into 10 mL of BHI, followed by aerobic incubation at 37°C in the presence of 5% CO2 for 16 hours under static conditions. Overnight cultures of S. epidermidis and E. coli were made by aseptically transferring a single-colony forming unit into 10 mL of NB or TSB (respectively), followed by aerobic incubation at 37°C with constant agitation (200 rpm) for 16 hours.
2.6 In-Solution Antimicrobial Activity of Chimeric Peptides
The in-solution antimicrobial activity of the chimeric peptide was analyzed against S. mutans, S. epidermidis, and E. coli spectrophotometrically. For each bacteria species, solutions of selected antimicrobial peptides were added in specified media to reach pre-determined final concentrations and inoculated with the bacteria to a final concentration of 107 cells/mL. Bacterial growth at 37°C was monitored over the course of 24 hours by optical density measurements at 600 nm on a Tecan Safire spectrophotometer. Each experiment contained control samples consisted solely of 107cells/mL of bacteria in the specified media.
2.7 Bacterial Adhesion and Quantification on Chimeric Peptide Coated Implant Surfaces
Pre-cleaned titanium substrates were incubated at 37°C under constant agitation (200 rpm) with chimeric peptide solution and removed after 3 hours. An aliquot of 1 mL of sterile 1X PBS was then added to each well, agitated by pipetting three times and removed from the well. A second 1mL aliquot of sterile 1X PBS was added to each well, agitated as before, and removed from the well. Using sterile forceps, each titanium substrate was moved to a clean well that was free of any peptides.
To proceed with bacterial adhesion experiments, overnight cultures for each bacterium were prepared as described above. Bacteria from the overnight cultures were used to inoculate fresh media to a final concentration of 107 cells/ml. Cultures were incubated until they reached the mid-log phase as determined by optical density measurement at 600 nm, collected by centrifugation at 2000 xg for 5 minutes. The supernatant decanted and the bacterial pellet was re-suspended in 500 μL of specified media. This suspension was transferred to a 2 mL centrifuge tube and centrifuged at room temperature at 2000 xg for 3 minutes. The supernatant was carefully removed and the bacterial pellet was re-suspended in sterile 1X PBS to a final concentration of 108 cells/mL. An aliquot of 1mL of the 108 cells/mL cell suspension was added to each well containing a chimeric peptide-modified titanium substrate, and incubated for 2 hours. For S. mutans experiments, incubation was carried out at 37°C in the presence of 5% CO2 under static conditions; for S. epidermidis and E. coli experiments, incubation was carried out aerobically at 37°C under constant agitation (200 rpm). After 2 hours incubation the bacterial suspension was removed by aspiration and the surfaces were washed two times with 1mL of 1X PBS. Bacterial cells adhered to the titanium substrates were fixed with 500 μL of 2% glutaraldehyde for 30 minutes, followed by dehydration in a series of increasing gradient of water: alcohol baths, consisting of 50% ethanol for 10 minutes, 70% ethanol for 10 minutes, 90% ethanol for 10 minutes, and a final 1 mL of 100% ethanol. Detection of the bacterial cells was carried out by addition of 500 μL of 5 μM SYTO9 green fluorescent nucleic acid stain (Invitrogen, Carlsbad, CA, USA) added to each well containing a substrate, protected from light, and incubated for 20 minutes. Substrates were washed 3 times with 1mL of 1X PBS, and each aliquot was agitated by re-pipetting twice. After washing, the substrates were secured onto a clean microscope slide and viewed with a Nikon Eclipse TE2000-U fluorescent microscope. Images were obtained from five random sites of implant surfaces and analyzed for percent surface coverage using MetaMorph (Version 6.r6) software.
2.8 Chimeric Peptide Structure Determination and Structure-Function Analysis
The structure of the peptides was investigated by the fragment insertion method using the Robetta server, followed by energy minimization routing using PyRosetta software [57-59]. Two hundred decoys were energy minimized for each sequence. The lowest energy structure was taken as the best estimate of the structure in-solution for the molecular descriptor analysis. All decoys were used for the secondary structure analysis of the length of alpha helix, the length of right-handed alpha-helices and the length of left-handed alpha helices. DSSP [60] was used to calculate the secondary structure based upon three-dimensional atomic coordinates for a peptide structure. Rules were induced by the MLEM2 algorithm modified learning from experience module version 2 [61]. Only the rules that label all cases in the data more accurately than a rule without conditions are given in the results.
3. RESULTS and DISCUSIONS
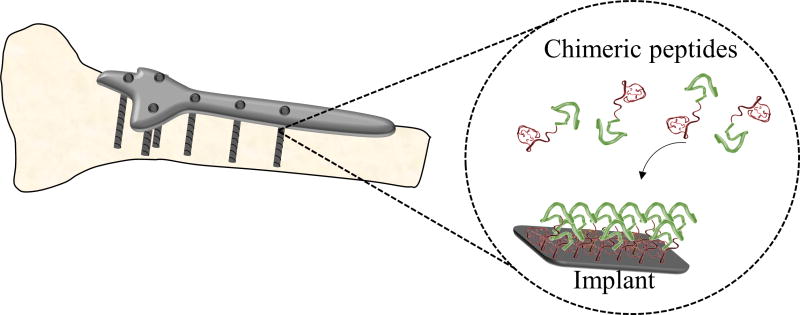
In this study, we demonstrated the use of chimeric peptides as antimicrobial coating agents on titanium grade V implants (Figure 1). Bifunctional chimeric peptides, comprising a combinatorially-selected titanium binding and computationally-designed antimicrobial domains were constructed. Surface-binding characterizations of these peptides were investigated using fluorescent microscopy. Antimicrobial activity of these bifunctional peptides were demonstrated against different pathogens common to implant infections; S. mutans, S. epidermidis, and E. coli. The potential molecular property rules were elaborated for the related sequence- activity relationships of the peptides following their folding patterns in the secondary structures.
Figure 1.
Schematics of biological self-assembly of chimeric antimicrobial peptide coating of titanium implant surface.
3.1 Selection and Characterization of Solid Binding Peptides
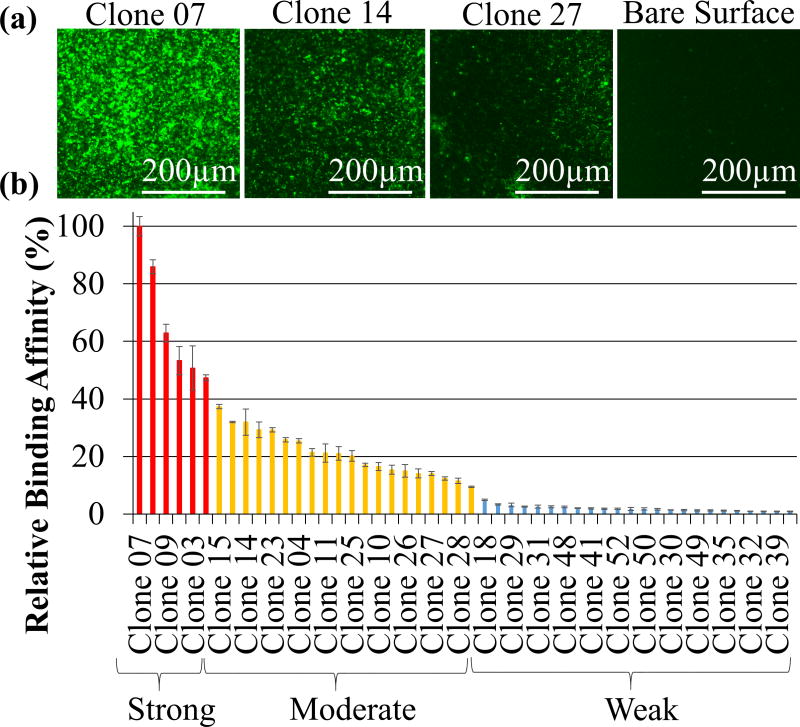
The phage display technique [55] was applied on titanium grade V powder to select peptides that could serve as potential molecular linkers to tether antimicrobial peptides on implant material surfaces. Throughout the selection process, four successive rounds of biopanning were performed, resulting in 50 unique clones, which were subjected to DNA nucleotide sequence determination and analysis. Fluorescent microscopy technique is a semi-quantitative binding assay that was applied to investigate the affinity level among each of the titanium binding peptides expressed by individual clones. For this assay, each clone was incubated with titanium grade V powder and then visualized using an anti-M13 specific antibody and fluorophore labeled secondary antibody. To evaluate the specific surface affinity of individual clones, the bound phage clones expressing titanium binding sequences were visualized as uniformly distributed bright green rods on a dark background of the implant material, as opposed to wild type M13 phage, which fail to bind. Based on these results, all the identified peptides were successfully categorized as strong, moderate and weak binders (Figure 2).
Figure 2.
Phage bound titanium binding peptides (TiBP’s) selected by phage display: (a) examples of FM images of TiBP’s with different binding affinities; (b) categorization of the titanium binding phage clones based on relative binding affinity analysis via FM.
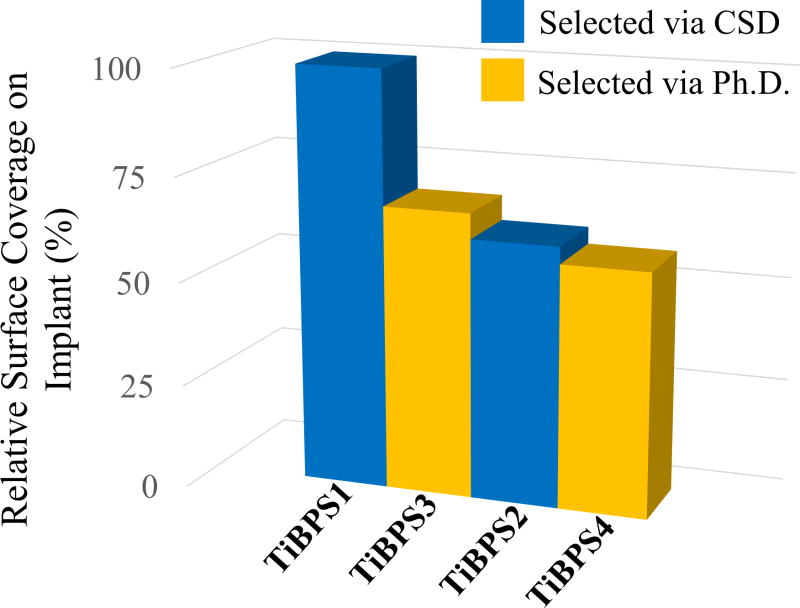
To eliminate the internal bias resulting from the amino acid distribution for each phage-display and cell-surface display library, the two strongest peptides selected via phage display were further characterized and compared with titanium binding peptides that are previously selected by cell surface display and characterized [50]. Each identified peptide was synthesized with biotin and incubated with titanium implants. After removing the unbound peptides, surface coverage of each peptide were visualized by using a fluorophore probes attached via interaction of biotin with streptavidin. Surface coverage ratios as well as the predicted MW, pI, net charge and G.R.A.V.Y. values of synthesized titanium binding peptides are depicted in Figure 3 and Table 1.
Figure 3.
Relative binding affinities of titanium binding peptides (TiBP’s) selected by cell surface display (CSD) and phage display (Ph.D.). Phage bound TiBPS3 and TiBPS4 are represented in previous figure as Clone 7 and Clone 22, respectively.
Table 1.
The physicochemical properties of the selected titanium binding peptides (TiBP), MW, pI, net charge and the hydropathy values.
| Peptide Name | Sequence | MW (kDa) | pI | Charge | G.R.A.V.Y score |
|---|---|---|---|---|---|
| TiBPS1 | RPRENRGRERGL | 1495.6 | 11.82 | +3 | -2.633 |
| TiBPS2 | SRPNGYGGSESS | 1197.1 | 5.72 | 0 | -1.567 |
| TiBPS3 | HAYKQPVLSTPF | 1387.6 | 8.60 | +1 | -0.333 |
3.2 Selection and Characterization of Antimicrobial Peptides
Bacteria growth curves in the presence of antimicrobial peptides with two-fold increment concentrations ranging from 1 μg/mL (0.66 μM) to 512 μg/mL (337.92 μM) were analyzed for a 24 h period to determine the minimum inhibition concentration (MIC) value for the each of the bacterial strains common for oral and orthopedic implant infections, i.e., S. mutans, S. epidermidis, and E. coli. As shown in Table 2, AMP1 and AMP2 are both effective against three of these organisms yet each revealed with different MIC values. The MIC values of AMP1 against E. coli, S. epidermidis, and S. mutans were determined as 9.45 μM, 4.72 μM, and 37.81 μM, respectively. For AMP2, these MIC values against E. coli, S. epidermidis, and S. mutans were found as 21.08 μM, 0.66 μM, and 10.54 μM, respectively. These concentrations indicate that AMP1 is more effective against E. coli than AMP2. Whereas, AMP2 prevented S. mutans and S. epidermidis growth with much lower concentrations than AMP1.
Table 2.
Minimum inhibitory concentration (MIC) values of AMP-1 and AMP-2 against E. coli, S. epidermidis and S. mutans.
| Peptide | Sequence | E. coli (μg/ml) | S. epidermidis (μg/ml) | S. mutans (μg/ml) |
|---|---|---|---|---|
| AMP1 | LKLLKKLLKLLKKL | 16 (9.45 μM) | 8 (4.72 μM) | 64 (37.81 μM) |
| AMP2 | KWKRWWWWR | 32 (21.08 μM) | 1 (0.66 μM) | 16 (10.54 μM) |
3.3. Construction and Characterization of Bifunctional Chimeric Peptides
Bifunctional chimeric peptides having both titanium binding affinity and antimicrobial activity were constructed by coupling the titanium binding peptide (TiBP) domain with the antimicrobial peptides, i.e. either AMP1 or AMP2, in different combinations. In this design, TiBP’s were inserted to the C’-terminal ends of the AMP’s with a structurally flexible triple glycine (Gly-Gly-Gly) linker sequence to enable the surface display and thus preserve the functionalities of both the titanium binding peptide and the AMP’s. The amino acid sequences and theoretical parameters, such as MW and pI, for each of the bifunctional peptide were listed in Table 3. Successful design of any bifunctional chimeric peptide requires that the multifunctional activities embedded in the final construct are confirmed. Therefore, the efficiency of the resulting bifunctional chimeric peptide was investigated with respect to titanium binding affinity as well as its antimicrobial activity. To investigate the antimicrobial activity, bifunctional chimeric peptides were tested against strains of E. coli, S. epidermidis, and S. mutans and the resulting MIC values were calculated by spectrophotometrically monitoring bacterial growth in the presence of these bifunctional chimeric peptides. Concentration for each peptide was chosen such that the lowest test concentration was set to the predetermined MIC value of each AMP to be ensure that the same number of AMP molecules were present in the solution. The dynamic range was determined using two fold increment to reach the highest peptide concentrations tested.
Table 3.
MW, pI, net charge and the hydropathy of designed chimeric peptides.
| Peptide Name | Sequence | MW (kDa) | pI | Charge | G.R.A.V.Y score |
|---|---|---|---|---|---|
| TiBPS1-AMP1 | RPRENRGRERGLGGGLKLLKKLLKLLKKL | 3341.1 | 11.85 | +9 | -0.890 |
| TiBPS2-AMP1 | SRPNGYGGSESSGGGLKLLKKLLKLLKKL | 3042.6 | 10.93 | +6 | -0.448 |
| TiBPS1-AMP2 | RPRENRGRERGLGGGKWKRWWWWR | 3166.6 | 12.13 | +7 | -2.254 |
| TiBPS3-AMP2 | HAYKQPVLSTPFGGGKWKRWWWWR | 3058.5 | 11.17 | +5 | -1.104 |
As shown in Table 4, among five different bifunctional chimeric peptides, those that harboring AMP1 as an antimicrobial counterpart, i.e. TiBPS1-AMP1 and TiBPS2-AMP1, showed the most effective antibacterial activity against E. coli, at concentrations of 9.58 μM and 21 μM, respectively. Furthermore, compared to the TiBPS2-AMP1, TiBPS1-AMP1 was revealed to be two times more effective in its antimicrobial activity. The attenuation in the antimicrobial efficiency of AMP1 depended on the titanium binding peptide to which it is coupled. This outcome can be attributed to differences in the amino acid composition and the sequences of these bifunctional chimeric peptides. In the case of S. epidermidis, it was revealed that the TiBPS1-AMP2 is the most effective peptide in-solution, being able to prevent bacterial growth at as low as 2.52 μM concentration. In addition, TiBPS1-AMP1, TiBPS2-AMP1 and TiBPS3-AMP2 also showed considerable antimicrobial activity, with MIC values of 4.78 μM, 5.23 μM and 5.23 μM, respectively. Interestingly, in contrast to two fold reduction in the antimicrobial efficiency of TiBPS2-AMP1 compared to TiBPS1-AMP1 against E. coli, these bifunctional peptides did not show the same trend against S. epidermidis. This can be attributed to the complex interactions between the bacterial cell membrane and the antimicrobial peptides during targeting and penetration into the bacterial cell membrane. It also implies that overall antimicrobial activity depends not only on amino acid sequence, but also the membrane structure and composition of the targeted microorganism. Also, the hydrophobicity of the AMP, the presence of positively charged residues, amphipatic nature of the peptide, and secondary structure are some of known factors that can effect both the antimicrobial activity and antimicrobial selectivity to specific organisms.
Table 4.
Minimum inhibitory concentration (MIC) values of chimeric peptides against E. coli, S. epidermidis and S. mutans.
| Peptide Name | Sequence | E. coli (μg/ml) | S. epidermidis (μg/ml) | S. mutans (μg/ml) |
|---|---|---|---|---|
| TiBPS1-AMP1 | RPRENRGRERGLGGGLKLLKKLLKLLKKL | 32 (9.58 μM) | 16 (4.78 μM) | 512 (153.25 μM) |
| TiBPS2-AMP1 | SRPNGYGGSESSGGGLKLLKKLLKLLKKL | 64 (21 μM) | 16 (5.23 μM) | 1024 (336.5 μM) |
| TiBPS1-AMP2 | RPRENRGRERGLGGGKWKRWWWWR | 256 (80.8 μM) | 8 (2.52 μM) | 256 (80.8 μM) |
| TiBPS3-AMP2 | HAYKQPVLSTPFGGGKWKRWWWWR | 512 (167.4 μM) | 16 (5.23 μM) | 256 (83.7 μM) |
In the case of S. mutans both TiBPS1-AMP2 and TiBPS3-AMP2 showed highest antimicrobial activity with MIC of 80.8 μM while for the TiBPS1-AMP1 it was 153.25 μM which again suggests the complex mechanism of antimicrobial action of peptides.
3.4. Bacterial Adhesion on Peptide Functionalized Implant Surfaces
Following the determination of the MIC values of each peptide against three different bacterium in-solution, antimicrobial efficacy of these peptides were further characterized on the titanium implant surfaces against S. mutans, S. epidermidis and E. coli.
With this aim, 100 μM of each bifunctional chimeric peptide was incubated for 4 hours at 37°C with constant agitation with the sterile titanium implant. The excess peptide was removed by washing surface 3 times with PBS buffer. Surfaces were incubated with bacteria culture at 108 cells/mL for 2 hours. After incubation, the cells were fixed and labeled with SYTO 9 dye, which penetrates through the bacterial membranes and stains the cells green. The bacterial binding and antimicrobial efficacy of peptide functionalized titanium implant surfaces were analyzed by visualizing under fluorescent microscopy.
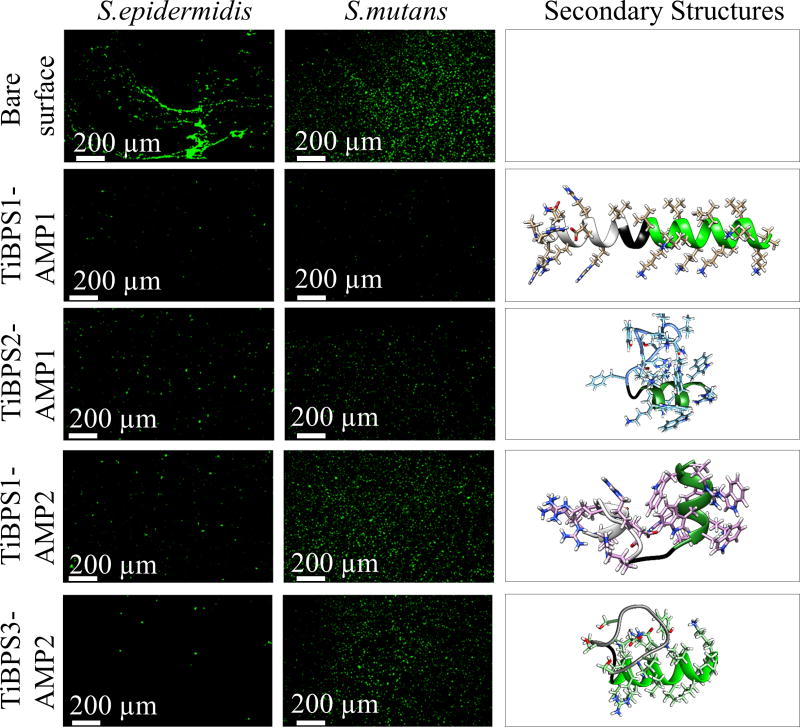
As shown in Figure 4, both the S. mutans and S. epidermidis adhesion on titanium implants coated with bifunctional chimeric peptides was significantly reduced compared to bare titanium implant surface. Moreover, in contrast to in-solution antimicrobial activities discussed above, surfaces coated with TiBPS1-AMP1 and TiBPS2-AMP1 showed similar antimicrobial activities against S. mutans and S. epidermidis. Compared to AMP2 containing bifunctional chimeric peptides, those harboring AMP1 showed better surface antimicrobial activity against S. mutans. These results demonstrates that with 30-fold reduction in bacterial adhesion compared to bare surface, the TiBPS1-AMP1 is the most efficient bifunctional chimeric peptide to be utilized as titanium implant surface functionalization against S. mutans. On the other hand, against S. epidermidis acquired infections, TiBPS1-AMP1 and TiBPS3-AMP2 would be the better choice for surface functionalization of titanium implants.
Figure 4.
Bacterial adhesion on peptide modified titanium implant surfaces against Streptococcus mutans (middle column), Staphylococcus epidermidis (left column) and their predicted secondary structures (right column).
3.5. Peptide Molecular Property Rules
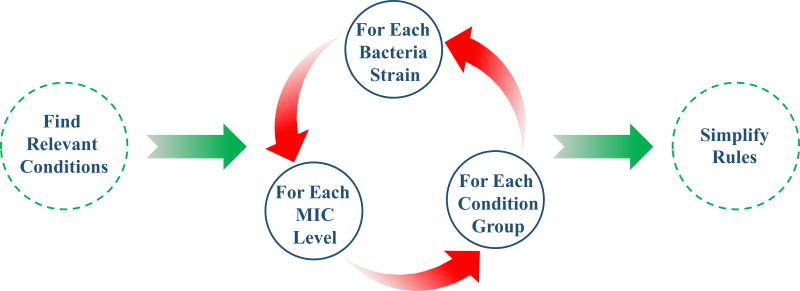
AMP-sequences tested, AMP1 and AMP2, have complex sequence-activity relationships with bacterial cell membranes that are sensitive to the change between chimeric and single forms. To decipher the sequence-activity relationships of these variants, patterns from multiple sources were considered in deriving two rule sets. Figure 5 provides a diagram of the derivation of each of the two rule sets. The first rule set is based on molecular properties, such as isoelectric point, overall charge, average hydropathy and molecular weight. The second rule set is based on folding patterns of secondary structure in terms of alpha helix length. The peptide AMP efficiency results were divided first by bacteria strain. Then, they were subdivided by level of efficiency, such as low, medium and high.
Figure 5.
The algorithm flowchart showing inducing rules to describe the antimicrobial functionality of peptides.
Rules for the most relevant and specific patterns were derived by the rough set theory approach. The rules are simplified if removing a part of a rule does not affect its specificity. A rule is removed if other remaining rules cover the results that it also covers.
For the first rule set, the molecular properties that were the most relevant and specific according to rough set theory to each efficiency level are given by the rules in Table 5. The first two columns of the table describe which molecular properties are relevant and specific. The next two columns give the divisions of the data by bacterial strain and by the MIC level. The last column describes how relevant the rule is by stating the number of applicable results to which the rules applies and it describes how specific the rule is by stating the number of those results to which the rule correctly applies.
Table 5.
Rules set induced from MW, pI, net charge and the hydropathy for minimum inhibitory concentration (MIC) by molarity.
| Property | Value Interval | Pathogen | MIC Interval | Correct Cases/Applicable Cases (7 Total Cases) |
|---|---|---|---|---|
| pI | 9.65 –12.13 | E. coli | 9.45 μM –21 μM | 3/3 |
| charge | 5.5 –9.0 | E. coli | 9.45 μM –21 μM | 3/3 |
| GRAVY score | -2.177 –0.5 | E. coli | 9.45 μM –21 μM | 3/3 |
| Molecular Weight | 1279.5–3341.1 | E. coli | 9.45 μM –21 μM | 3/3 |
|
| ||||
| pI | 10.965 –12.13 | E. coli | 167.4 μM –167.6 μM | 2/2 |
| Charge | 0 – 5.5 | E. coli | 167.4 μM –167.6 μM | 2/2 |
| Molecular Weight | 3050.55–3341.1 | E. coli | 167.4 μM –167.6 μM | 2/2 |
|
| ||||
| Charge | 3.5 –9.0 | S. epidermidis | 4.72 μM -5.23 μM | 5/5 |
| GRAVY score | -1.8375 –0.5 | S. epidermidis | 4.72 μM -5.23 μM | 5/5 |
| Molecular Weight | 1593.95–3341.1 | S. epidermidis | 4.72 μM -5.23 μM | 5/5 |
The first rule set uses the molecular properties as conditions. These rules are certain rules, meaning all of the results that apply are classified correctly. The first two rules imply that to have the lowest observed MIC value of under 21 μM for E. coli, the charge of the peptide should be +6 or greater. When the charge is less positive, the MIC level is near 167 μM is expected. The molecular weight range of the first rule indicates that either AMP’s in the chimeric or singular form can be effective against E. coli at the low MIC value of under 21 μM. The observed constraints for an MIC level for S. epidermidis near 5 μM are a positive charge from +4 to +9 with an average hydropathy score of the amino acid residues between -1.84 and 0.5. Negative hydropathy scores relate to hydrophilic amino acids such as arginine. Again, the range of molecular weight shows that the results include an AMP in chimeric form and in an AMP singular form with an MIC efficiency of near 5 μM.
3.6. Peptide Structure Function Rules
Peptide structure tendencies may influence the antimicrobial functionality. We performed structural analysis to investigate if there is a trend for their secondary structure tendencies. Figure 4 shows the antimicrobial functionality for four of the bifunctional chimeric sequences against S. epidermidis and S. mutans and their lowest predicted energy structures. At first glance, comparing the first two peptides with the last two peptides seems to indicate that the longer the alpha helix, the stronger the antimicrobial functionality against these strains.
Here, in order to obtain the lowest energy structure of these chimeric peptides we performed a further analysis among the hundreds of decoys that are generated to obtain the lowest energy structure also uncovered trends using the peptide structure tendencies, in addition to the lowest energy structures.
The second rule set (Table 6) was generated to identify predicted secondary structural features that may be responsible for the antimicrobial functionality. Right-handed alpha helices were commonly predicted for the peptides, but no beta-sheet formations were predicted. Left-handed alpha helices are uncommon for systems that use L-amino acids. The side chains of L-amino acids would point toward the crowded axis of the helix instead of away. D-amino acids may form left-handed alpha helices, and for the same reason D-amino acids generally do not form right-handed helices [62].
Table 6.
Rules set induced from lengths of alpha-helices and the chirality of the member residues for minimum inhibitory concentration (MIC) level by molarity.
| Alpha Helix Property | Pathogen | MIC Interval | Correct Cases/Applicable Cases (1400 Total Cases) |
|---|---|---|---|
| 5-a.a.-helix 4-a.a.-right-handed helix |
E. coli | 9.45 μM –21 μM | 164/179 |
| 5-a.a.-helix 4-a.a.-right-handed helix |
S. epidermidis | 4.72 μM -5.23 μM | 164/179 |
| 5-a.a.-helix 4-a.a.-right-handed helix |
S. mutans | 10.54 μM –37.81 μM | 151/179 |
| 8-a.a.-helix 6 or 8-a.a.-right-handed helix |
S. mutans | 336.5 μM | 8/11 |
The peptide structure prediction scheme does not explicitly constrain the predicted structures to any particular secondary structure type. The backbone angles were selected from similar fragments in the Protein Databank returned from the Robetta server. A specific conformation of a 5 amino-acid alpha helix in which 4 of the amino acids are predicted to turn toward a right-handed axis seems to be a common predicted secondary structure feature in peptides with MIC values on the order of 10 μM across all three strains. The other identified predicted secondary structural feature is an 8 amino-acid long alpha-helix associated with an MIC on the order of 100 μM in S. mutans. The results from the E. coli and S. epidermidis were the same because the same sequences were effective against both strains.
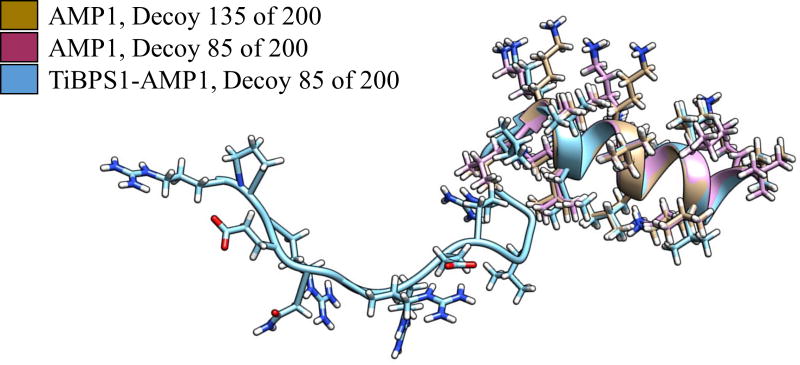
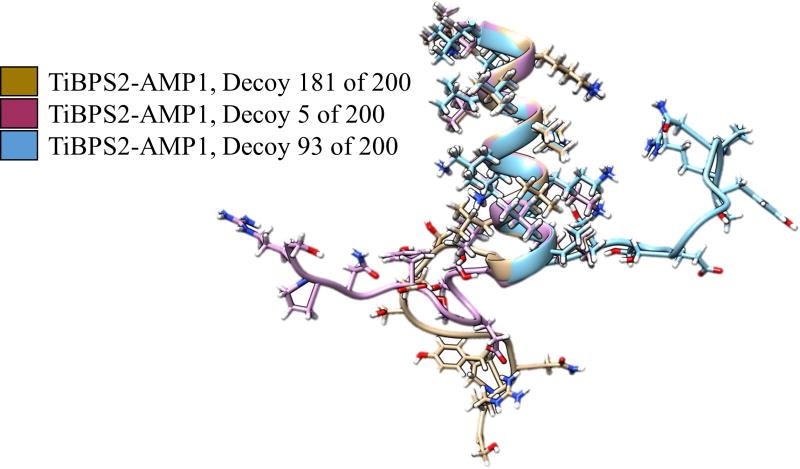
Among the predicted structures that the rule induction algorithm identified are many cases of the AMP unit showing the same folded structure in the bifunctional chimeric peptide and the AMP-only sequence were observed. Figure 6 shows decoys of a bifunctional chimeric sequence and two AMP-only sequences that have the same predicted folded structure for the AMP unit. Figure 7 shows three bifunctional chimeric sequence decoys with varied TiBP structure and conserved AMP structure. The AMP-secondary structure patterns may be sequence-order specific. If the AMP structure was at the N-terminus, instead of the C-terminus, the TiBP may be more disruptive to its secondary structure patterns. Our results provide an initial scheme to develop design rules for chimeric peptides where their secondary structure predictions could be linked to their observed antimicrobial properties. Combining secondary structure analysis with the experimental evaluations may provide an iterative path for effective design of engineered peptides to utilize their biological tasks while expanding the functional repertoire with materials selective biological self –assembly property.
Figure 6.
Structural alignment showing the conserved AMP1 structure in bifunctional chimeric peptides and AMP-only peptides.
Figure 7.
Structural alignment showing the flexibility of TiBPS2 compared to the secondary structure pattern seen AMP1 in decoys of TiBPS2-AMP1.
4. CONCLUSIONS
Here we describe a peptide-based implant surface functionalization approach to prevent implant failure due to bacterial infection. Bifunctional chimeric peptides having a specific surface recognition and binding ability, as well as an antimicrobial activity were designed. The implant surface was coated with these chimeric peptides and inoculated with a standard bolus of bacteria culture to test for bacterial adhesion and/or growth on implant surface. The in-solution activity tests revealed that the functionality of the antimicrobial peptide is conserved when combined in the bifunctional chimeric peptide. The bacterial adhesion studies demonstrated that chimeric peptides coatings provided antimicrobial property for the titanium implants. Collectively, the use of solid binding peptides as molecular recognition units for creating an interface on the implant surface that can be combined with antimicrobial peptides may enable better control of the tissue-implant interface and thereby leads to modalities that prevent infection and subsequent implant failure. Structure-function relationships used here recognize features putatively leading to the antimicrobial functionality based on individual pathogen data. The analysis of the peptide secondary structures provides guidance for future de novo antimicrobial peptide design specific for diverse surfaces.
By offering single-step and bio-friendly alternative to the conventional chemical and physical immobilization methods, without the requirement of undesired surface activation processes, solid binding peptides provides new approaches towards merging biological tasks into self-assembly pathways. These short peptides can be the key components to achieve integrated materials-tissue interfaces coupling large repertoire of the biological tasks to the specific sites.
Acknowledgments
Authors gratefully acknowledge the financial support from National Institute of Health (NIH) - Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), Musculoskeletal Tissue Engineering Section 7R21AR062249-03 and University of Kansas New Faculty General Research Fund (NFGRF) as well as National Institute of Dental and Craniofacial Research grant DE13045.
Contributor Information
Deniz T. Yucesoy, GEMSEC, Genetically Engineered Materials Science and Engineering Center, Department of Materials Science and Engineering, University of Washington, Seattle, WA 98195, USA.
Marketa Hnilova, GEMSEC, Genetically Engineered Materials Science and Engineering Center, Department of Materials Science and Engineering, University of Washington, Seattle, WA 98195, USA.
Kyle Boone, Bioengineering Program and Bioengineering Research Center, University of Kansas, Lawrence, KS-66045.
Paul M. Arnold, Department of Neurosurgery, Spinal Cord Injury Center, School of Medicine, University of Kansas, Kansas City, KS 66160, USA.
Malcolm L. Snead, Ostrow School of Dentistry of USC, Center for Craniofacial Molecular Biology, University of Southern California, Los Angeles, CA 90089, USA.
Candan Tamerler, Department of Mechanical Engineering and Bioengineering Research Center, University of Kansas, Lawrence, KS-66045 ctamerler@ku.edu, + 7858642984.
References
- 1.Bauer S, Schmuki P, von der Mark K, Park J. Prog Mater Sci. 2013;58(3):261–326. [Google Scholar]
- 2.Puleo D, Nanci A. Biomaterials. 1999;20(23):2311–2321. doi: 10.1016/s0142-9612(99)00160-x. [DOI] [PubMed] [Google Scholar]
- 3.Geetha M, Singh A, Asokamani R, Gogia A. Prog Mater Sci. 2009;54(3):397–425. [Google Scholar]
- 4.Le Guéhennec L, Soueidan A, Layrolle P, Amouriq Y. Dent Mater. 2007;23(7):844–854. doi: 10.1016/j.dental.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 5.Pennekamp PH, Gessmann J, Diedrich O, Burian B, Wimmer MA, Frauchiger VM, Kraft CN. J Orthop Res. 2006;24(3):531–540. doi: 10.1002/jor.20066. [DOI] [PubMed] [Google Scholar]
- 6.Costerton J, Stewart PS, Greenberg E. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein RA, Darouiche RO. Clin Infect Dis. 2001;33(9):1567–1572. doi: 10.1086/323130. [DOI] [PubMed] [Google Scholar]
- 8.Uçkay I, Hoffmeyer P, Lew D, Pittet D. J Hosp Infect. 2013;84(1):5–12. doi: 10.1016/j.jhin.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Antoci V, Jr, Adams CS, Parvizi J, Davidson HM, Composto RJ, Freeman TA, Wickstrom E, Ducheyne P, Jungkind D, Shapiro IM. Biomaterials. 2008;29(35):4684–4690. doi: 10.1016/j.biomaterials.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazemzadeh-Narbat M, Kindrachuk J, Duan K, Jenssen H, Hancock RE, Wang R. Biomaterials. 2010;31(36):9519–9526. doi: 10.1016/j.biomaterials.2010.08.035. [DOI] [PubMed] [Google Scholar]
- 11.Rathbone CR, Cross JD, Brown KV, Murray CK, Wenke JC. J Orthop Res. 2011;29(7):1070–1074. doi: 10.1002/jor.21343. [DOI] [PubMed] [Google Scholar]
- 12.Hetrick EM, Schoenfisch MH. Chem Soc Rev. 2006;35(9):780–789. doi: 10.1039/b515219b. [DOI] [PubMed] [Google Scholar]
- 13.Campoccia D, Montanaro L, Speziale P, Arciola CR. Biomaterials. 2010;31(25):6363–6377. doi: 10.1016/j.biomaterials.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Harbers GM, Emoto K, Greef C, Metzger SW, Woodward HN, Mascali JJ, Grainger DW, Lochhead MJ. Chem Mater. 2007;19(18):4405–4414. doi: 10.1021/cm070509u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimotoyodome A, Koudate T, Kobayashi H, Nakamura J, Tokimitsu I, Hase T, Inoue T, Matsukubo T, Takaesu Y. Antimicrob Agents Chemother. 2007;51(10):3634–3641. doi: 10.1128/AAC.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An Y, Stuart G, McDowell S, McDaniel S, Kang Q, Friedman R. J Orthop Res. 1996;14(5):846–849. doi: 10.1002/jor.1100140526. [DOI] [PubMed] [Google Scholar]
- 17.Jose B, Antoci V, Jr, Zeiger AR, Wickstrom E, Hickok NJ. Chem Biol. 2005;12(9):1041–1048. doi: 10.1016/j.chembiol.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Price J, Tencer A, Arm D, Bohach G. J Biomed Mater Res. 1996;30(3):281–286. doi: 10.1002/(SICI)1097-4636(199603)30:3<281::AID-JBM2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 19.Russell A, Tattawasart U, Maillard J-Y, Furr J. Antimicrob Agents Chemother. 1998;42(8):2151–2151. doi: 10.1128/aac.42.8.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wininger DA, Fass RJ. Antimicrob Agents Chemother. 1996;40(12):2675. doi: 10.1128/aac.40.12.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris L, Mead L, Müller-Oberländer E, Richards R. J Biomed Mater Res A. 2006;78(1):50–58. doi: 10.1002/jbm.a.30611. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee I, Pangule RC, Kane RS. Adv Mater. 2011;23(6):690–718. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, Chu PK, Zhang Y, Wu Z. J Biomed Mater Res B Appl Biomater. 2009;91(1):470–480. doi: 10.1002/jbm.b.31463. [DOI] [PubMed] [Google Scholar]
- 24.Gottenbos B, van der Mei HC, Klatter F, Nieuwenhuis P, Busscher HJ. Biomaterials. 2002;23(6):1417–1423. doi: 10.1016/s0142-9612(01)00263-0. [DOI] [PubMed] [Google Scholar]
- 25.Mrksich M, Whitesides GM. Annu Rev Biophys. 1996;25(1):55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 26.Mohorčič M, Jerman I, Zorko M, Butinar L, Orel B, Jerala R, Friedrich J. J Mater Sci Mater Med. 2010;21(10):2775–2782. doi: 10.1007/s10856-010-4136-z. [DOI] [PubMed] [Google Scholar]
- 27.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Chem Rev. 2005;105(4):1103–1170. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 28.Harris LG, Richards RG. Injury. 2006;37(2):S3–S14. doi: 10.1016/j.injury.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Brogden KA. Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 30.Giuliani A, Pirri G, Nicoletto SF. Cent Eur J Biol. 2007;2(1):1–33. [Google Scholar]
- 31.Zasloff M. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 32.Reddy K, Yedery R, Aranha C. Int J Antimicrob Agents. 2004;24(6):536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Pasupuleti M, Schmidtchen A, Malmsten M. Crit Rev Biotechnol. 2012;32(2):143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 34.Jenssen H, Hamill P, Hancock RE. Clin Microbiol Rev. 2006;19(3):491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingham AB, Moore RJ. Biotechnol Appl Biochem. 2007;47(1):1–9. doi: 10.1042/BA20060207. [DOI] [PubMed] [Google Scholar]
- 36.Hilpert K, Elliott MR, Volkmer-Engert R, Henklein P, Donini O, Zhou Q, Winkler DF, Hancock RE. Chem Biol. 2006;13(10):1101–1107. doi: 10.1016/j.chembiol.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 37.Fjell CD, Jenssen H, Hilpert K, Cheung WA, Pante N, Hancock RE, Cherkasov A. J Med Chem. 2009;52(7):2006–2015. doi: 10.1021/jm8015365. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Z, Vasil AI, Hale JD, Hancock RE, Vasil ML, Hodges RS. J Pept Sci. 2008;90(3):369–383. doi: 10.1002/bip.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarikaya M, Tamerler C, Jen AK-Y, Schulten K, Baneyx F. Nat Mater. 2003;2(9):577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 40.Campoccia D, Montanaro L, Arciola CR. Biomaterials. 2013;34(34):8533–8554. doi: 10.1016/j.biomaterials.2013.07.089. [DOI] [PubMed] [Google Scholar]
- 41.Kwakman PH, te Velde AA, Vandenbroucke-Grauls CM, Van Deventer SJ, Zaat SA. Antimicrob Agents Chemother. 2006;50(12):3977–3983. doi: 10.1128/AAC.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etienne O, Picart C, Taddei C, Haikel Y, Dimarcq J, Schaaf P, Voegel J, Ogier J, Egles C. Antimicrob Agents Chemother. 2004;48(10):3662–3669. doi: 10.1128/AAC.48.10.3662-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Appendini P, Hotchkiss J. J Appl Polym Sci. 2001;81(3):609–616. [Google Scholar]
- 44.Zhou Y, Snead ML, Tamerler C. Nanomedicine: Nanotechnology, Biology and Medicine. 2015;11(2):431–434. doi: 10.1016/j.nano.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Socransky SS, Haffajee AD. Periodontol 2000. 2002;28(1):12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 46.Campoccia D, Montanaro L, Arciola CR. Biomaterials. 2006;27(11):2331–2339. doi: 10.1016/j.biomaterials.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 47.Costa F, Carvalho IF, Montelaro RC, Gomes P, Martins MCL. Acta Biomater. 2011;7(4):1431–1440. doi: 10.1016/j.actbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Meyers SR, Khoo X, Huang X, Walsh EB, Grinstaff MW, Kenan DJ. Biomaterials. 2009;30(3):277–286. doi: 10.1016/j.biomaterials.2008.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshinari M, Kato T, Matsuzaka K, Hayakawa T, Shiba K. Biofouling. 2010;26(1):103–110. doi: 10.1080/08927010903216572. [DOI] [PubMed] [Google Scholar]
- 50.Yazici H, Fong H, Wilson B, Oren E, Amos F, Zhang H, Evans J, Snead M, Sarikaya M, Tamerler C. Acta Biomater. 2013;9(2):5341–5352. doi: 10.1016/j.actbio.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oren EE, Tamerler C, Sahin D, Hnilova M, Seker UOS, Sarikaya M, Samudrala R. Bioinformatics. 2007;23(21):2816–2822. doi: 10.1093/bioinformatics/btm436. [DOI] [PubMed] [Google Scholar]
- 52.Lu Z, Murray KS, Van Cleave V, LaVallie ER, Stahl ML, McCoy JM. Nat Biotechnol. 1995;13(4):366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 53.Tamerler C, Khatayevich D, Gungormus M, Kacar T, Oren EE, Hnilova M, Sarikaya M. J Pept Sci. 2010;94(1):78–94. doi: 10.1002/bip.21368. [DOI] [PubMed] [Google Scholar]
- 54.Tamerler C, Sarikaya M. Acta Biomater. 2007;3(3):289–299. doi: 10.1016/j.actbio.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Seker UOS, Wilson B, Dincer S, Kim IW, Oren EE, Evans JS, Tamerler C, Sarikaya M. Langmuir. 2007;23(15):7895–7900. doi: 10.1021/la700446g. [DOI] [PubMed] [Google Scholar]
- 56.Fjell CD, Jenssen H, Cheung WA, Hancock RE, Cherkasov A. Chem Biol Drug Des. 2011;77(1):48–56. doi: 10.1111/j.1747-0285.2010.01044.x. [DOI] [PubMed] [Google Scholar]
- 57.Chaudhury S, Lyskov S, Gray JJ. Bioinformatics. 2010;26(5):689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simons KT, Kooperberg C, Huang E, Baker D. J Mol Biol. 1997;268(1):209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 59.Bradley P, Misura KM, Baker D. Science. 2005;309(5742):1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 60.Kabsch W, Sander C. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 61.Grzymala-Busse JW, Rzasa W. Fund Inform. 2010;100(1):99–116. [Google Scholar]
- 62.Shepherd NE, Hoang HN, Abbenante G, Fairlie DP. J Am Chem Soc. 2009;131(43):15877–15886. doi: 10.1021/ja9065283. [DOI] [PubMed] [Google Scholar]