Significance
KRAS (Kirsten rat sarcoma viral oncogene homolog) is frequently mutated in pancreatic, colon, and lung tumors, which predicts poor clinical outcome, whereas germ-line mutations are associated with developmental disorders, including Noonan syndrome. Although K-RAS is an attractive anticancer target, no clinically successful inhibitors are available. Most disease-associated mutations elevate the activated GTP-bound form of KRAS; however, some remain unexplained. KRAS signals from cellular membranes; however, our studies revealed that its association with the membrane surface sequesters its binding site for effector proteins, hampering signaling. Some disease-associated KRAS mutations disrupt this autoinhibition, identifying a new gain-of-function mechanism and explaining how certain Noonan syndrome mutations activate K-RAS signaling. Importantly, these findings open new avenues for therapeutic strategies to target oncogenic K-RAS through stabilizing autoinhibitory interactions with the membrane.
Keywords: KRAS, nuclear magnetic resonance, lipid bilayer nanodisc, oncogenic mutation, Noonan syndrome
Abstract
K-RAS4B (Kirsten rat sarcoma viral oncogene homolog 4B) is a prenylated, membrane-associated GTPase protein that is a critical switch for the propagation of growth factor signaling pathways to diverse effector proteins, including rapidly accelerated fibrosarcoma (RAF) kinases and RAS-related protein guanine nucleotide dissociation stimulator (RALGDS) proteins. Gain-of-function KRAS mutations occur frequently in human cancers and predict poor clinical outcome, whereas germ-line mutations are associated with developmental syndromes. However, it is not known how these mutations affect K-RAS association with biological membranes or whether this impacts signal transduction. Here, we used solution NMR studies of K-RAS4B tethered to nanodiscs to investigate lipid bilayer-anchored K-RAS4B and its interactions with effector protein RAS-binding domains (RBDs). Unexpectedly, we found that the effector-binding region of activated K-RAS4B is occluded by interaction with the membrane in one of the NMR-observable, and thus highly populated, conformational states. Binding of the RAF isoform ARAF and RALGDS RBDs induced marked reorientation of K-RAS4B from the occluded state to RBD-specific effector-bound states. Importantly, we found that two Noonan syndrome-associated mutations, K5N and D153V, which do not affect the GTPase cycle, relieve the occluded orientation by directly altering the electrostatics of two membrane interaction surfaces. Similarly, the most frequent KRAS oncogenic mutation G12D also drives K-RAS4B toward an exposed configuration. Further, the D153V and G12D mutations increase the rate of association of ARAF-RBD with lipid bilayer-tethered K-RAS4B. We revealed a mechanism of K-RAS4B autoinhibition by membrane sequestration of its effector-binding site, which can be disrupted by disease-associated mutations. Stabilizing the autoinhibitory interactions between K-RAS4B and the membrane could be an attractive target for anticancer drug discovery.
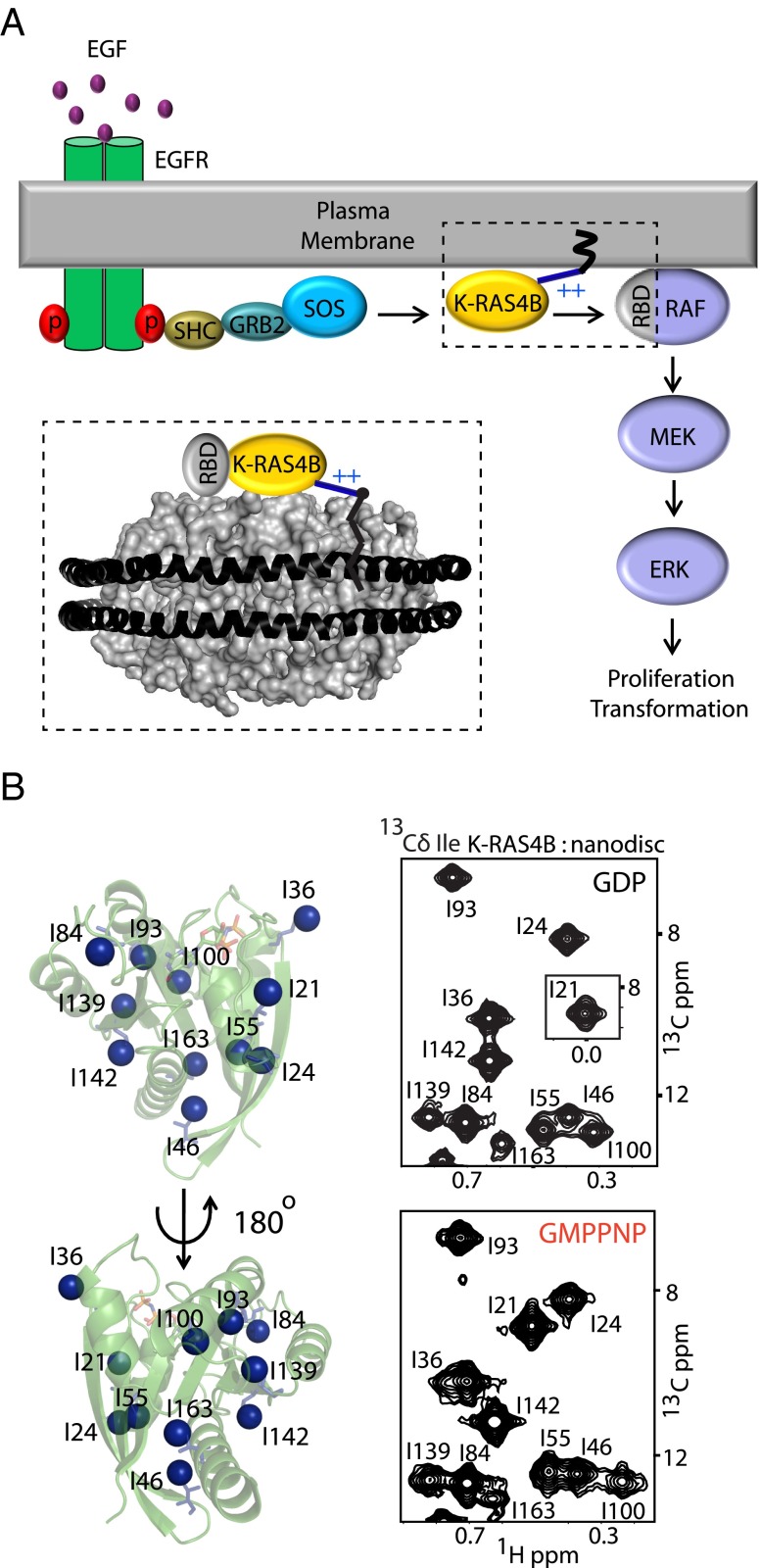
The K-RAS4B (Kirsten rat sarcoma viral oncogene homolog 4B) protein product of the KRAS gene undergoes posttranslational farnesylation and C-terminal processing, which, in conjunction with a poly-basic hypervariable region (HVR), targets K-RAS4B to anionic lipid rafts on the intracellular side of the plasma membrane (Fig. 1A) (1). This localization is essential for K-RAS4B function and enhances signaling fidelity (2). Although the significance of membrane tethering of K-RAS4B is well appreciated, a high-resolution map of how K-RAS4B interacts with the membrane is lacking. Because membrane-anchored RAS presents a major challenge to crystallization, current structural insights into the behavior of membrane-anchored RAS have come from a variety of lower-resolution techniques including in vivo FRET-based studies (3), fluorescence and infrared spectroscopic studies (4–6), and in silico models (3). These pioneering studies suggested that the K-RAS4B GTPase domain adopts certain preferred orientations on the anionic membrane and that these orientations could be influenced by the bound nucleotide. Here we present high-resolution NMR-derived models of the dynamic interactions between K-RAS4B and the lipid bilayer and how they can be impacted by disease-associated mutations or interactions with effector proteins.
Fig. 1.
K-RAS4B signaling at the plasma membrane and the nanodisc lipid bilayer model for NMR studies. (A) Schematic illustration of K-RAS4B signaling on plasma membrane. (Inset) Schematic of a K-RAS4B:nanodisc complex. (B) Distribution of eleven isoleucine Cδ throughout the K-RAS4B GTPase-domain. (Right) 1H-13C HMQC spectra of nanodisc-conjugated K-RAS4B in the GDP- (Upper) and GMPPNP- (Lower) bound forms.
Results
K-RAS4B Activation State Determines Population of Two Major Orientations on Lipid Bilayer.
Recent developments in solution NMR technology have made it possible to study integral membrane proteins and those anchored to membranes (7). To overcome the inherent challenges associated with solution NMR studies of membrane-associated proteins, we used selective 13C-labeling of the Cδ methyl groups (8) of the 11 isoleucine residues in K-RAS4B, which produced excellent quality (signal/noise > 100) 1H-13C heteronuclear multiple quantum coherence (HMQC) spectra with 11 well-resolved peaks, in solution (Fig. S1A) and tethered to nanodiscs (Fig. 1B). Tethering was achieved by conjugation of the C-terminal farnesylation site (Cys185) of a K-RAS4B variant (C118S) lacking a surface-exposed Cys (Fig. S2A) to a thiol-reactive maleimide-functionalized lipid {1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidomethyl)cyclohexane-carboxamide] (PE-MCC)} (9).
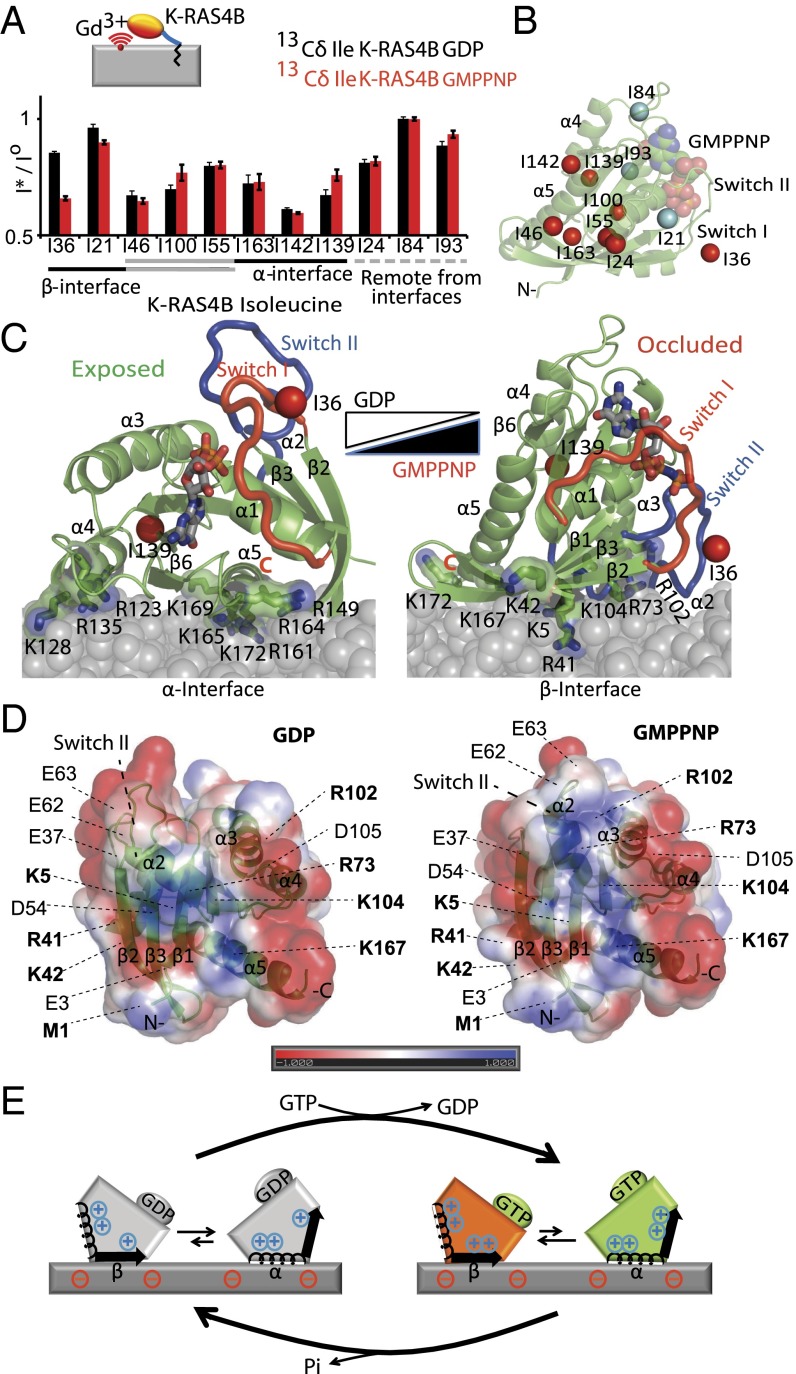
All 11 isoleucines are located <4 Å from the protein surface and are well distributed throughout the protein to provide excellent probes for molecular interactions. To unambiguously identify regions of the GTPase domain interacting with or in close proximity to the lipid bilayer, we prepared nanodiscs incorporating lipids conjugated to the paramagnetic ion gadolinium (Gd3+) and measured broadening of the K-RAS4B Ile Cδ resonances induced by paramagnetic relaxation enhancement (PRE) (10). Mapping the PRE-broadened Ile probes on the structure of K-RAS4B (Fig. 2 A and B and Table S1) revealed that not all of the observed PRE-derived constraints can be simultaneously satisfied by a single interface, indicative of equilibrium between at least two orientations with distinct sites of membrane association.
Fig. 2.
Activated K-RAS4B adopts an occluded orientation on anionic membranes. (A) PRE-induced broadening of K-RAS4B H(13C) resonances by Gd3+-conjugated lipid incorporated into nanodiscs. PRE effects detected for each isoleucine residue of K-RAS4B presented as ratios of H(13C) resonance intensities of K-RAS4B conjugated to nanodiscs in the presence (I*) to those in the absence (Io) of Gd3+. Residues are grouped according to their location with respect to the α and β membrane interfaces defined below. Error bars based on spectral noise. (B) Map of the PRE effect on K-RAS4B Ile-Cδ. Residues broadened more than 20% are highlighted in red and the remainder in cyan. (C) PRE-driven models of K-RAS4B-nanodisc complexes (see Materials and Methods for details). (D) Surface electrostatics of K-RAS4B-GDP (Left; PDB ID code 4LPK) and K-RAS4B-GMPPNP (Right; modeled from PDB ID code 3GFT). K-RAS4B is oriented to view the occluded interface from the membrane. Blue, positive; red, negative. Surface exposed positive (bold) and negative charged residues are indicated. Increased exposure of R102 and reorientation of E63 contribute to a more positively charged occluded interface in the GMPPNP-bound form. (E) Schematic illustration of K-RAS4B reorientation on GTPase cycling. The α- and β-interfaces are shown with a helix and an arrow, respectively.
A comparison of the PRE profiles of the active and inactive forms of K-RAS4B reveals that Ile36 and Ile139 are the residues most sensitive to the activation state (Fig. 2A). Ile36 is located at the C terminus of switch I in close proximity to switch II, and Ile139 is on the opposite side of the GTPase domain in proximity to helix-α4, suggesting that nucleotide loading shifts the equilibrium of membrane association of these two surfaces. To construct structural models of membrane-bound K-RAS4B and identify preferred orientations of the active and inactive forms, we carried out high ambiguity-driven biomolecular docking (HADDOCK) simulations (11) (Table S2) using distance restraints derived from the PRE experiments (10) (Fig. 2 A and B). The ensemble of models computed by HADDOCK for activated and inactive K-RAS4B each clearly exhibit two distinct clusters of orientations (Fig. 2C and Fig. S2B), consistent with orientational flexibility of the K-RAS4B GTPase domain (6); however, the relative population of each cluster was dependent on the activation state (Table S2). In the cluster that predominates in the GDP-bound form, most of the K-RAS4B helices are oriented parallel to the membrane, and a surface comprised of the N terminus of β1 (N-β1), α4, β6, α5, and the loop connecting β2 and β3 interfaces with the membrane (Fig. 2 C and D). This surface, located opposite to switch I, is therefore designated the α-interface. By contrast, the helices are oriented semiperpendicular to the membrane in the major cluster for activated K-RAS4B-GMPPNP (a nonhydrolyzable analog of GTP), and the membrane interface is formed by structural elements surrounding C terminus of switch I, including β-strands 1–3 of the GTPase domain β-sheet core, α2, and the C-terminal part of α3 (the β-interface). These results are consistent with previous infrared spectroscopy data, which showed that the helical trajectories of K-RAS4B bound to GDP, but not GMPPNP, are mainly parallel with the bilayer plane, although the membrane interfaces were not determined (4, 5).
The structural basis for this change in orientation likely involves the nucleotide-dependent conformational change in switch II, whereby GMPPNP binding both increases the surface-exposed positive charge and decreases the negative charge on the β-interface (Fig. 2D), electrostatically stabilizing the semiperpendicular K-RAS4B orientation (Table S2). Intriguingly, the nucleotide-dependent orientation preference of K-RAS4B (Fig. 2 B and C) differs strikingly from that of H-RAS [by molecular dynamics (MD)- and FRET-based studies] (12, 13) and Rheb (by NMR) (10), where the activated and inactive forms favor helix-parallel and -perpendicular orientations, respectively. Consistent with this finding, previous MD simulation studies of K-RAS4B on neutral membrane bilayers did not recapitulate the H-RAS–specific orientations (3). Importantly, the RAS isoforms H-RAS, N-RAS, and even the splice variant K-RAS4A have C-terminal HVR sequences and lipidation motifs distinct from those of K-RAS4B (1), as well as isoform-specific substitutions in the C-terminal lobe of the GTPase domain (14), which likely lead to different orientational equilibria of the GTPase domain.
Interaction with RAS-Binding Domains Requires K-RAS4B Reorientation to Expose Effector-Binding Site.
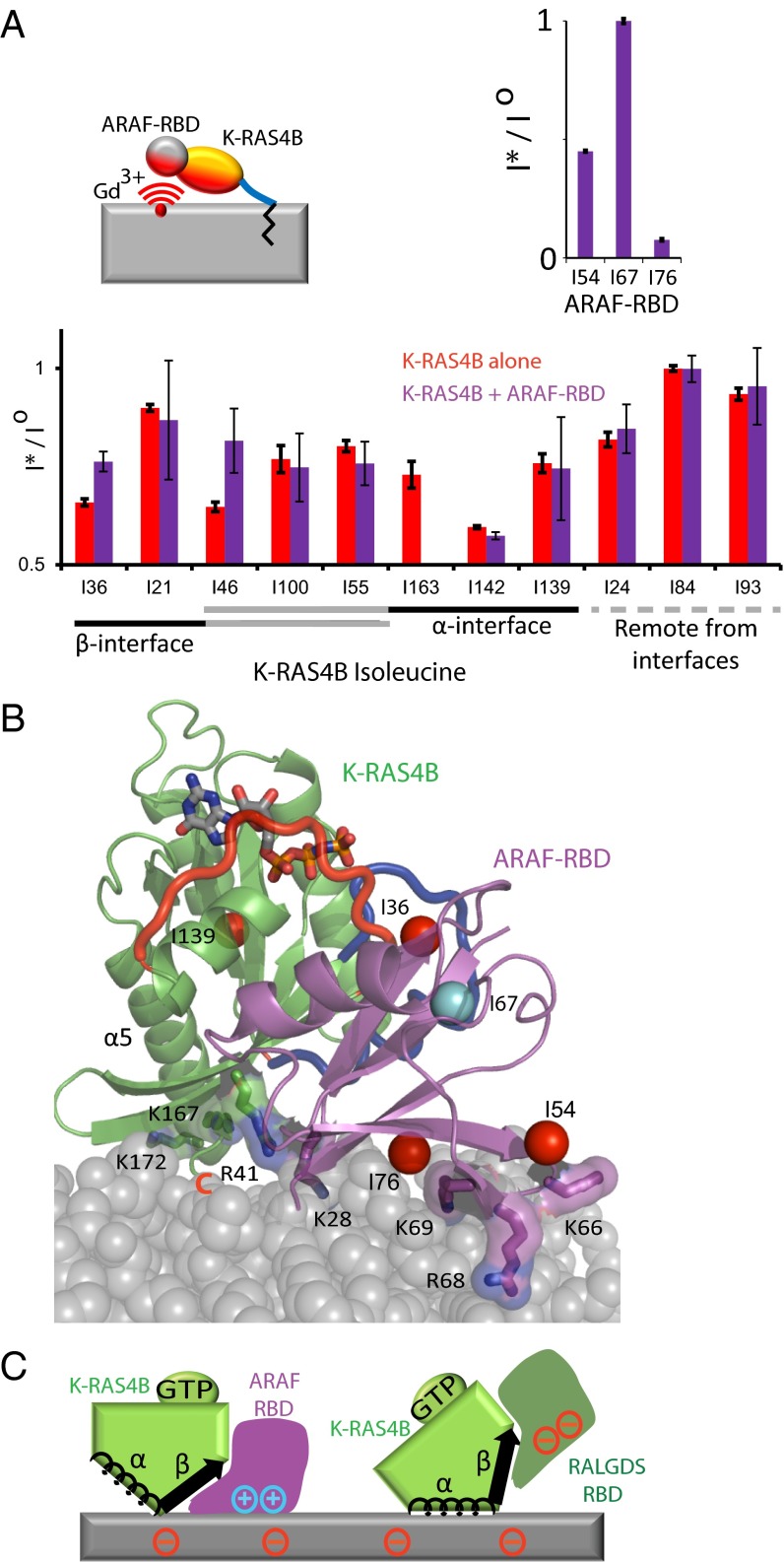
To our surprise, the preferred orientation of activated K-RAS4B sequesters the C terminus of switch I (i.e., the effector-binding loop) at the membrane surface (Fig. S3) and appears incompatible with binding of effector proteins. Thus, we examined how the orientation of activated K-RAS4B on the membrane is affected by interaction with the RAS-binding domain (RBD) of the rapidly accelerated fibrosarcoma (RAF) isoform ARAF. Like the RBD of CRAF that is preferentially recruited to K-RAS4B nanoclusters in cells (15), ARAF-RBD contains a positively charged patch adjacent to the RAS-binding site (Fig. S4A) and contains three isoleucine residues well distributed to provide PRE-based restraints. We reconstituted nanodisc-tethered complexes of isotopically labeled K-RAS4B with ARAF-RBD (total molecular weight, ∼200 kDa), performed NMR measurements (Fig. S1B) to deduce PRE distance restraints from the lipid bilayer to both K-RAS4B and ARAF-RBD (Tables S1 and S3), and derived HADDOCK models of the membrane-associated complex. Within this complex, the GTPase domain was found in a new semiexposed orientation intermediate between the exposed and occluded orientations (Table S2), which places the cationic surface of the ARAF-RBD in contact with the anionic membrane surface (Fig. 3 A–C and Fig. S4B) and reduces the PRE effect on Ile36, reflecting global reorientation away from the occluded configuration, as well as Ile46, which suggests the β-interface rotates away from the membrane on complex formation. Interestingly, the RBD of RAS-related protein (RAL) guanine nucleotide dissociation stimulator (GDS) possesses an anionic surface and shifts the orientational equilibrium of K-RAS4B away from the occluded orientation toward a fully exposed orientation, as evidenced by decreased PRE on Ile36 and Ile100, increased PRE on Ile139, Ile142, and Ile163, and only minor PRE perturbations of the RALGDS-RBD resonances, whereas reduced PRE on Ile24 and Ile46 indicate that K-RAS4B adopts an altered exposed orientation in complex with RALGDS (Fig. S5). These observations strongly argue that K-RAS4B interactions with different RBDs can lead to stabilization of distinct GTPase domain/membrane orientations that vary depending on the nature of the effector (Fig. 3C). K-RAS4B complexes with other RBDs remain to be investigated, including the RBD of PI3 kinase, which has been challenging to express in a soluble form. The probability of forming productive complexes with various effector proteins would be expected to depend on the dynamic conformational equilibrium of K-RAS4B on the membrane and correlate inversely with the transient population of the occluded orientation. Based on the intimate engagement of membranes by K-RAS4B in multiple orientations, we postulate that K-RAS4B signaling output might be altered by mutations that remodel the membrane orientational equilibrium.
Fig. 3.
Occluded K-RAS4B orientation is incompatible with RBD binding. (A) Effect of interaction with ARAF-RBD on the PRE profile of nanodisc-tethered K-RAS4B. (Upper) PRE profile of ARAF-RBD Ile-Cδ resonances in the presence of Gd3+-containing (I*) vs. Gd3+-free (Io) nanodisc-tethered K-RAS4B-GMPPNP. (Lower) PRE profile of nanodisc-tethered K-RAS4B-GMPPNP (red) vs. nanodisc-tethered K-RAS4B-GMPPNP in complex with ARAF-RBD (magenta). Residues are grouped according to their location with respect to the α and β membrane interfaces. (B) PRE-based model of nanodisc-tethered K-RAS4B in complex with ARAF-RBD. The K-RAS4B:ARAF-RBD complex was modeled on the basis of homology to H-RAS:C-RAFRBD (PDB ID code 4G0N) (37), and this complex was docked to the bilayer surface based on PRE-derived constraints using HADDOCK. The lowest HADDOCK-scored structure within the major cluster is shown. (C) Schematic illustration of membrane orientations of K-RAS4B complexed with RBDs of ARAF vs. RALGDS.
Disease-Associated Mutations Relieve Effector-Binding Site Occlusion.
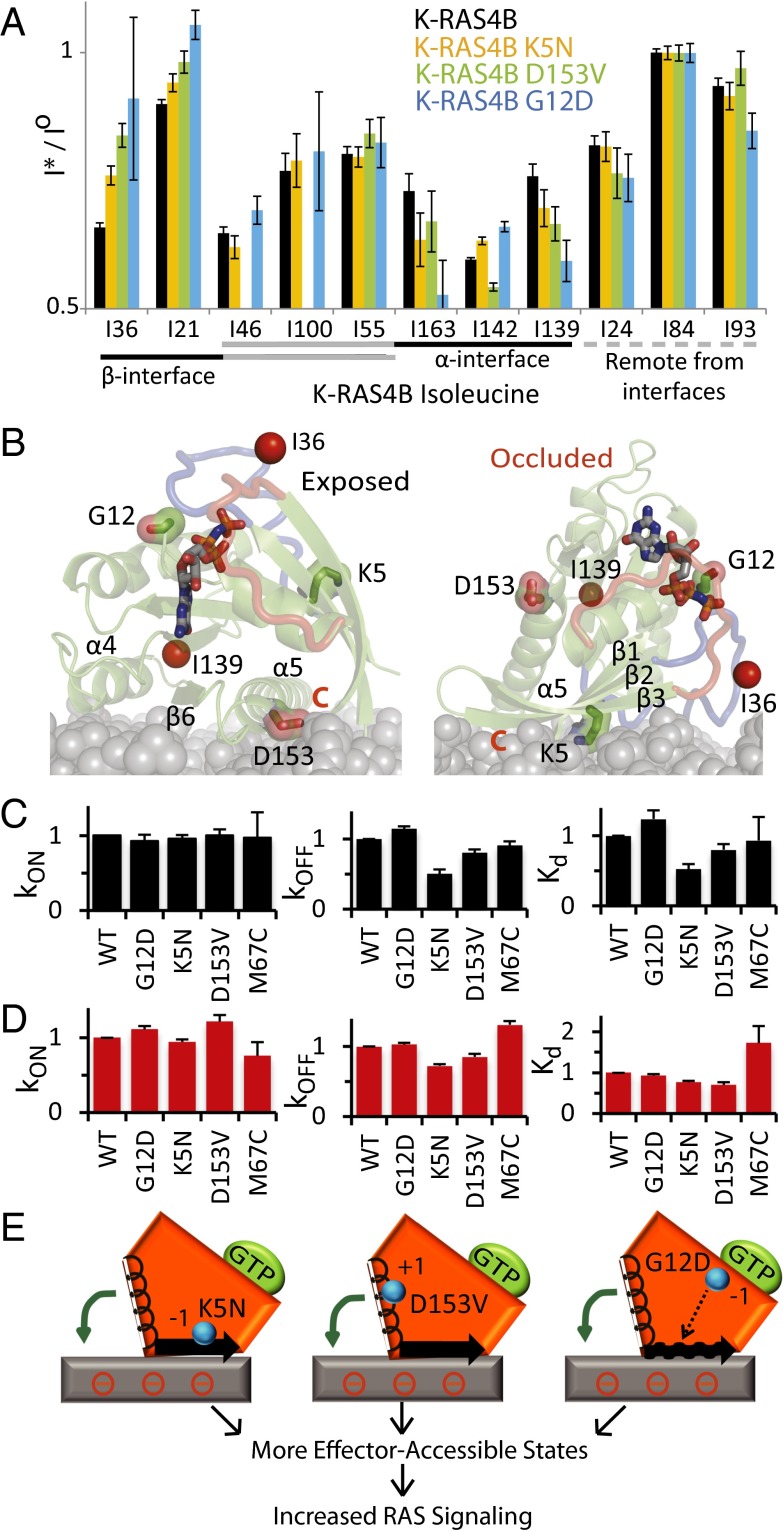
We then sought to address whether oncogenic mutations of KRAS affect the membrane orientation. Generally, oncogenic K-RAS mutants, such as G12D, are known to increase GTP loading, thus leading to hyperactivation of RAS signaling pathways (16). Here we unveiled that the K-RAS4B G12D mutation markedly releases the effector-occluded configuration, as evidenced by reduced PRE broadening of Ile21/Ile36 and increased broadening of Ile139/Ile163 (Fig. 4A), suggesting that oncogenic mutations could alter the normal function of RAS proteins through multiple actions. Gly12 is not spatially proximal to the membrane in either orientation (Fig. 4B); however, the G12D substitution was previously shown to increase the population of a nucleotide-free–like conformation of RAS-GTP called state 1, which increases the entropy (i.e., mobility) of the switch I/II and helix α3 region (17, 18). The enhanced dynamics in the nucleotide-binding region (19) is most likely responsible for destabilizing the β-interface. To further investigate whether the state 1 population influences the membrane orientation, we measured PRE broadening of the state 1-selective K-RAS4B mutant V29G (20). The V29G substitution is not proximal to the membrane; nevertheless, the V29G mutant recapitulated the G12D reorientation toward the exposed state (Fig. S6), indicating that the state 1/2 equilibrium plays a role in determining K-RAS4B membrane orientation, apparently through allosteric modulation of the β-interface (19).
Fig. 4.
K-RAS4B oncogenic and Noonan syndrome mutations relieve the occluded orientation and alter RBD binding kinetics on lipid bilayer. (A) PRE profiles of WT K-RAS4B vs. K5N, D153V, and G12D mutants. 1H-13C HMQC spectra are shown in Fig. S1. Error bars based on spectral noise. Residues are grouped according to their location with respect to the α and β membrane interfaces. (B) Location of mutations and Ile36/Ile139 probes on exposed and occluded K-RAS4B models. (C and D) Relative association rates (kON), dissociation rates (kOFF), and dissociation constants (Kd) for (C) free K-RAS4B and (D) nanodisc-tethered K-RAS4B binding to ARAF-RBD. Binding kinetics of K-RAS4B (analyte) to immobilized ARAF-RBD (ligand) was measured by biolayer interferometry (Fig. S7). Each kON and kOFF rate was determined using three concentrations of K-RAS4B. Error bars represent SD. Kd values were determined from kON and kOFF, and SD was propagated accordingly. (E) Schematic illustration of K-RAS4B reorientation induced by mutations. The mutation locations are shown by cyan spheres and changes in net charge are depicted by ±1. The wavy arrow represents the increased dynamics of the β-interface associated with the state 1 population of the G12D mutant. The curved green arrow represents the reorientation.
Our model further predicts that K-RAS4B signaling could be modulated by mutations that directly alter the membrane interaction surfaces. Two germ-line KRAS mutations identified in Noonan and cardio-facio-cutaneous (CFC) syndrome patients, K5N (21) and D153V (22–24), were shown to induce phosphorylation of MEK1/2 more strongly than WT KRAS; however, the mechanism of their activation has remained elusive. Unlike oncogenic K-RAS mutations such as G12D, G13D, and Q61H, neither of these germ-line mutations appreciably altered intrinsic GTPase activity or nucleotide exchange or sensitivity to GAPs or guanine nucleotide exchange factors (GEFs) in solution; thus, they have been classified as mutants whose phenotypes are not explained by their biochemical properties (25, 26). Intriguingly, we find that these mutations change the electrostatic surface of the two main membrane interfaces of K-RAS4B (Fig. 4B). By reducing the positive charge on the occluded β-interface and reducing the negative charge on the α-interface of the exposed orientation, respectively, our models predict that both the K5N and D153V mutations would shift the orientational equilibrium of K-RAS4B away from the occluded orientation (Fig. 4 B and E), therefore unleashing the membrane-dependent autoinhibition of K-RAS4B signaling. Indeed, PRE experiments demonstrated that both mutations reduced the PRE effect on Ile36 and increased that on Ile139 (Fig. 4A), indicating that mutation-induced surface electrostatic changes can cause reorientation of K-RAS4B on anionic membranes. To determine whether the increased population of the binding-competent exposed state enhances interaction of these K-RAS4B mutants with effector proteins, we used biolayer interferometry (BLI; ForteBio Octet RED96) to directly measure the binding kinetics of K-RAS4B to immobilized ARAF-RBD. We postulate that the degree of exposure of the effector binding site will influence the association rate (kON) of the K-RAS4B–effector interaction. Remarkably, the two mutations that most strongly destabilize the occluded orientation, G12D and D153V lead to measurable increases in the association rates (10% and 20%, respectively) of nanodisc-tethered K-RAS4B with ARAF-RBD, whereas the mutations had no significant effect on the association rate of free K-RAS4B (Fig. 4 C and D and Fig. S7). The K5N mutation, which has a more subtle effect on orientation, did not have a detectable effect on the association rates of free or nanodisc-tethered K-RAS4B, but decreased the dissociation rate in a lipid bilayer-independent manner (Fig. 4 C and D). Overall, these disease-associated mutations enhanced the RBD interaction of lipid bilayer-tethered K-RAS4B by 10–25% (Fig. 4D), suggesting that perturbation of the conformational equilibrium contributes to their increased downstream signaling and is a particularly important factor in the case of D153V. To further delineate the effect of K-RAS4B membrane orientation on RBD binding, we rationally designed a Cys mutation that, on conjugation to PE-MCC, would favor the occluded orientation: A M67C substitution introduced on the edge of the occluded interface to avoid spoiling the effector-binding site had no effect on free K-RAS4B binding to immobilized ARAF-RBD. Interestingly, this Cys mutant reduced the affinity of nanodisc-conjugated K-RAS4B twofold (Fig. 4D), presumably by stabilizing the autoinhibited occluded conformation through a covalent bond between Cys67 and PE-MCC.
Discussion
The present data provide, to our knowledge, the first evidence for release of membrane-dependent autoinhibition as a previously unidentified mechanism by which K-RAS4B mutations can activate signaling (Fig. 4E). The extent to which orientational effects contribute to K-RAS4B G12D signaling in the cell relative to effects on the GTPase cycle remains to be investigated; however, the K5N and D153V Noonan/CFC syndrome mutations suggest that perturbing membrane orientation to unleash autoinhibition may alone be sufficient to produce a disease phenotype. Importantly, KRAS K5N and D153V mutations have also been detected in lung, stomach, and renal cancers (27, 28), whereas germ-line and somatic mutations encoding K5E were recently identified in CFC syndrome (29) and chronic myelomonocytic leukemia, respectively (27, 30).
The oncogenic and RASopathy-associated K-RAS4B mutations examined here relieve a membrane-associated orientation that occludes effector binding, which may define a new mechanism by which KRAS mutations can enhance signaling (Fig. 4E). The K-RAS4B orientational equilibrium may further be modulated by (i) protein–protein interactions such as Ca2+/calmodulin binding to the HVR (1), (ii) posttranslational modifications such as PKC phosphorylation of Ser181 in the HVR (1) and acetylation or monoubiquitinylation of K104 (1) in the β-interface, or (iii) alterations of the membrane lipid composition. Further studies are needed to elucidate how the intimate communication between RAS and membrane may be dynamically regulated by these regulatory factors. Interestingly switch II is exposed in the major conformation of the inactive GDP-bound form of K-RAS4B, suggesting it is accessible to activation by RAS GEFs.
Finally, K-RAS4B, a well-validated oncogenic driver for many cancer types, is a challenging drug target (16). Although some lead compounds have been identified (31), there are still no clinically effective RAS inhibitors available. The propensity of K-RAS4B to associate with the membrane in a manner that occludes its effector-binding site may reveal a novel and unexplored therapeutic target at the protein–membrane interface. Our model suggests that pharmacological modulation of the orientational equilibrium may be exploited to sequester activated K-RAS4B mutants.
Materials and Methods
Preparation of proteins and nanodisc-tethered protein complexes, NMR measurements, and BLI assays are fully described in SI Materials and Methods.
NMR- and PRE-Guided Molecular Docking Simulations.
All docking simulations were performed using HADDOCK 2.0 (11, 32) as described previously (10). Derivation of the full-length K-RAS4B and nanodisc models are described in SI Materials and Methods. The K-RAS4B: nanodisc [MSP1D1, 80% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 20% 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS)] complex was then subjected to energy minimization in CNS before HADDOCK simulations. HADDOCK ambiguous restraints were generated from the PRE measurements of K-RAS4B tethered to Gd3+-containing nanodiscs. Because the lateral diffusion rate of DOPC at room temperature (∼8.2 × 103 nm2/ms) (33) is high relative to T2 relaxation times of macromolecules (e.g., nanodisc-tethered Rheb ∼30 ms), an assumption was made that the paramagnetic Gd3+ ion conjugated to 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid (gadolinium salt; PE-DTPA) uniformly sampled all positions on the nanodisc surface (78 nm2) during the time course of the measurement. Thus, ambiguous restraints were generated between PRE-affected isoleucine residues on K-RAS4B and any lipid headgroup atom on the nanodisc surface. Isoleucine residues exhibiting peak broadening of >20% were defined as active, and the adjacent N- and C-terminal residues were defined as passive residues. For simulation of K-RAS4B-GDP, isoleucines 24, 46, 55, 100, 139, 142, and 163 were defined as active and residues 23, 25, 45, 47, 54, 56, 99, 101, 138, 140, 141, 143, 162, and 164 were defined as passive. For simulation of K-RAS4B-GMPPNP, isoleucines 24, 36, 46, 55, 100, 139, 142, and 163 were identified as active residues and 23, 25, 35, 37, 45, 47, 54, 56, 99, 101, 138, 140, 141, 143, 162, and 164 as passive residues. Surface electrostatics of K-RAS4B were generated by the Poisson-Boltzmann Solver (34–36). The K-RAS4B:ARAF–RBD complex was modeled by assembling the K-RAS4B model described above with the NMR structure of ARAF-RBD [Protein Data Bank (PDB) ID code 1WXM], guided by the H-RAS:C-RAFRBD crystal structure (PDB ID code 4G0N) (37). In simulations of the complex, isoleucines 24, 36, 46, 55, 100, 139, 142, and 163 of K-RAS4B and isoleucines 54 and 76 of ARAF-RBD were identified as active residues, and the flanking residues were selected as passive. To normalize the contribution of both molecules to the K-RAS4B:ARAF-RBD complex models, ARAF-RBD restraints were weighted more heavily to compensate for fewer Ile probes. Ambiguous distance restraints were assigned a range of 2–5 Å. Although the paramagnetic effect of Gd3+ can extend to a 20-Å radius (38), each lipid headgroup on the membrane surface is only partially (2.5%) occupied by Gd3+-conjugated lipid; thus, a reduced upper limit of 5 Å was estimated. An upper limit of 2 Å was selected for isoleucines broadened >50%, as well as K-RAS4B Cys185. Additionally, the Lys175 at the beginning of the polybasic region was identified as an active residue. The CNS topology and the parameter files for GDP, GMPPNP, DOPC, and DOPS were generated using the HIC-UP server (39) and modified according to the data reported for these small molecules in the Automated Topology Builder (ATB) and Repository (40). The docking protocol was composed of a 3,000 rigid-body docking stage, where the top 200 ranked structures based on the HADDOCK scores were refined using semiflexible simulated annealing, followed by water refinement. The docking protocol was executed with default HADDOCK script parameters with an additional Powell energy minimization step of the lipid headgroups before the semiflexible refinement stage.
Cluster Analysis of HADDOCK Models.
Pairwise backbone RMSD values were tabulated for the K-RAS4B G-domain (residues 4–171) after alignment of the nanodiscs within the same plane and a translational and 360° rotational (in 5° increments) RMSD minimization search that were confined only to movements within the 2D plane of the membrane surface, as described previously (10). The K-RAS4B:nanodisc position in each solution was kept constant during pairwise RMSD calculation. Cluster analysis was performed on the calculated pairwise RMSD values using a previously described algorithm (41), setting an RMSD cutoff of 7.5 Å (11, 32) and cluster size cutoff of 20 structures. The HADDOCK scores of all 200 structures were plotted against RMSD relative to the global mean structure, defined as the solution with the lowest average pairwise RMSD to all other 199 solutions. All structural manipulations and measurements in 3D space were performed using CNS (42).
Supplementary Material
Acknowledgments
We thank Sharon Campbell (University of North Carolina at Chapel Hill) for providing the H-RAS resonance assignments, Bob Berno (McMaster University) for 31P NMR data collection, and Trevor Moraes and Anastassia Pogoutse (University of Toronto) for the use of BLI instrumentation. This work was supported by Cancer Research Society (Canada) Grant 14014, the Princess Margaret Cancer Foundation, and Canadian Cancer Society Grant 703209. NMR spectrometers were funded by the Canada Foundation for Innovation. L.E.K., B.G.N., and M.I. hold Canada Research Chairs.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.C.P. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes: 2MSC, 2MSD, and 2MSE), and in the Biological Magnetic Resonance Bank, www.bmrb.wisc.edu (BMRB entry ID codes: 25114, 25115, and 25116).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419895112/-/DCSupplemental.
References
- 1.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: Post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian T, et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9(8):905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 3.Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc Natl Acad Sci USA. 2010;107(3):1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor S, et al. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur Biophys J. 2012;41(10):801–813. doi: 10.1007/s00249-012-0841-5. [DOI] [PubMed] [Google Scholar]
- 5.Weise K, et al. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J Am Chem Soc. 2011;133(4):880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- 6.Werkmuller A, Triola G, Waldmann H, Winter R. Rotational and translational dynamics of ras proteins upon binding to model membrane systems. Chemphyschem. 2013;14(16):3698–3705. doi: 10.1002/cphc.201300617. [DOI] [PubMed] [Google Scholar]
- 7.Maslennikov I, Choe S. Advances in NMR structures of integral membrane proteins. Curr Opin Struct Biol. 2013;23(4):555–562. doi: 10.1016/j.sbi.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1(2):749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 9.Schulz TA, et al. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187(6):889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazhab-Jafari MT, et al. Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc. J Am Chem Soc. 2013;135(9):3367–3370. doi: 10.1021/ja312508w. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 12.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem. 2007;50(4):674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 13.Abankwa D, et al. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27(5):727–735. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker JA, Mattos C. The Ras-membrane interface: Isoform-specific differences in the catalytic domain. Molec Cancer Res. 2015;13(4):595–603. doi: 10.1158/1541-7786.MCR-14-0535. [DOI] [PubMed] [Google Scholar]
- 15.Plowman SJ, Ariotti N, Goodall A, Parton RG, Hancock JF. Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Mol Cell Biol. 2008;28(13):4377–4385. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araki M, et al. Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers. J Biol Chem. 2011;286(45):39644–39653. doi: 10.1074/jbc.M111.227074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geyer M, et al. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996;35(32):10308–10320. doi: 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]
- 19.Grant BJ, Gorfe AA, McCammon JA. Ras conformational switching: Simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLOS Comput Biol. 2009;5(3):e1000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spoerner M, Wittinghofer A, Kalbitzer HR. Perturbation of the conformational equilibria in Ras by selective mutations as studied by 31P NMR spectroscopy. FEBS Lett. 2004;578(3):305–310. doi: 10.1016/j.febslet.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Zenker M, et al. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2007;44(2):131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubbert S, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38(3):331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 23.Niihori T, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38(3):294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 24.Carta C, et al. Germline missense mutations affecting KRAS Isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79(1):129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gremer L, et al. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum Mutat. 2011;32(1):33–43. doi: 10.1002/humu.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubbert S, et al. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007;27(22):7765–7770. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abaan OD, et al. The exomes of the NCI-60 panel: A genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73(14):4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, et al. BRAF and KRAS mutations in stomach cancer. Oncogene. 2003;22(44):6942–6945. doi: 10.1038/sj.onc.1206749. [DOI] [PubMed] [Google Scholar]
- 29.Nava C, et al. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: Genotype-phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44(12):763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 31.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 32.de Vries SJ, et al. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69(4):726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 33.Filippov A, Orädd G, Lindblom G. Influence of cholesterol and water content on phospholipid lateral diffusion in bilayers. Langmuir. 2003;19(16):6397–6400. [Google Scholar]
- 34.Jo S, Vargyas M, Vasko-Szedlar J, Roux B, Im W. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 2008;36(web server issue):W270–W275. doi: 10.1093/nar/gkn314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 36.Im W, Beglov D, Roux B. Continuum solvation model: Computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comput Phys Commun. 1998;111(1-3):59–75. [Google Scholar]
- 37.Fetics SK, et al. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure. 2015;23(3):505–516. doi: 10.1016/j.str.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otting G. Protein NMR using paramagnetic ions. Annu Rev Biophys. 2010;39:387–405. doi: 10.1146/annurev.biophys.093008.131321. [DOI] [PubMed] [Google Scholar]
- 39.Kleywegt GJ, Henrick K, Dodson EJ, van Aalten DM. Pound-wise but penny-foolish: How well do micromolecules fare in macromolecular refinement? Structure. 2003;11(9):1051–1059. doi: 10.1016/s0969-2126(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 40.Malde AK, et al. An automated force field topology builder (ATB) and repository: Version 1.0. J Chem Theory Comput. 2011;7(12):4026–4037. doi: 10.1021/ct200196m. [DOI] [PubMed] [Google Scholar]
- 41.Daura X, et al. Peptide folding: When simulation meets experiment. Angew Chem Int Ed. 1999;38(1-2):236–240. [Google Scholar]
- 42.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.