Significance
The relationship between proteins unfolded under physiological conditions and those unfolded by chemical denaturation remains controversial. Specifically, although FRET studies suggest that unfolded proteins invariably contract with falling denaturant levels, scattering studies argue that they do not. Here, we explore the origins of this discrepancy using PEG as a negative control. Scattering indicates that, as expected, the polymer’s dimensions are denaturant-independent. The dye-labeled polymer, nevertheless, exhibits denaturant-dependent changes in measured transfer efficiency similar to those seen for unfolded proteins. This similarity raises questions regarding the interpretation of such changes as being indicative of hydrophobic or hydrogen bond-driven collapse.
Keywords: protein folding, SAXS, two state, statistical coil, Flory scaling
Abstract
Small-angle scattering studies generally indicate that the dimensions of unfolded single-domain proteins are independent (to within experimental uncertainty of a few percent) of denaturant concentration. In contrast, single-molecule FRET (smFRET) studies invariably suggest that protein unfolded states contract significantly as the denaturant concentration falls from high (∼6 M) to low (∼1 M). Here, we explore this discrepancy by using PEG to perform a hitherto absent negative control. This uncharged, highly hydrophilic polymer has been shown by multiple independent techniques to behave as a random coil in water, suggesting that it is unlikely to expand further on the addition of denaturant. Consistent with this observation, small-angle neutron scattering indicates that the dimensions of PEG are not significantly altered by the presence of either guanidine hydrochloride or urea. smFRET measurements on a PEG construct modified with the most commonly used FRET dye pair, however, produce denaturant-dependent changes in transfer efficiency similar to those seen for a number of unfolded proteins. Given the vastly different chemistries of PEG and unfolded proteins and the significant evidence that dye-free PEG is well-described as a denaturant-independent random coil, this similarity raises questions regarding the interpretation of smFRET data in terms of the hydrogen bond- or hydrophobically driven contraction of the unfolded state at low denaturant.
Recent years have seen a significant controversy arise regarding the behavior of the unfolded states of single-domain proteins in response to changing levels of chemical denaturant. That is, although small-angle X-ray scattering (SAXS) and single-molecule FRET (smFRET) results are all consistent with the argument that, at high levels of chemical denaturant (∼6 M or above), unfolded proteins adopt a swollen, self-avoiding ensemble well-approximated as an excluded volume random coil (1–3), the two approaches produce seemingly discrepant results for the dimensions of the unfolded states populated at lower (∼1 M) denaturant (4). For example, time-resolved SAXS studies of the unfolded state transiently populated when protein L is rapidly shifted from high guanidine hydrochloride (GuHCl) to low denaturant conditions produce no experimentally significant evidence for the compaction of this single-domain protein before folding (4, 5) [e.g., estimated radii of gyration of 25.1 ± 0.3 Å for the unfolded state at equilibrium in 7 M GuHCl and 24.9 ± 1.1 Å for the transient unfolded state populated at 0.67 M GuHCl; confidence intervals are SEs (4)]. In contrast, multiple smFRET studies conducted at equilibrium suggest that the unfolded state of dye-labeled protein L contracts by 20–40% over this same range of denaturant concentrations (6, 7) (Fig. 1). Moreover, this discrepancy seems to be nearly universal among single-domain proteins: whereas the results of at least a half-dozen SAXS studies on chemically unmodified, single-domain proteins fail to detect any experimentally significant evidence (at experimental precision of, typically, a few percent) of contraction (8–15), all of the more than one-dozen smFRET studies of dye-labeled, single-domain proteins reported to date have been interpreted in terms of unfolded states that contract as the denaturant concentration is lowered to ∼1 M (2, 6, 7, 16–26).
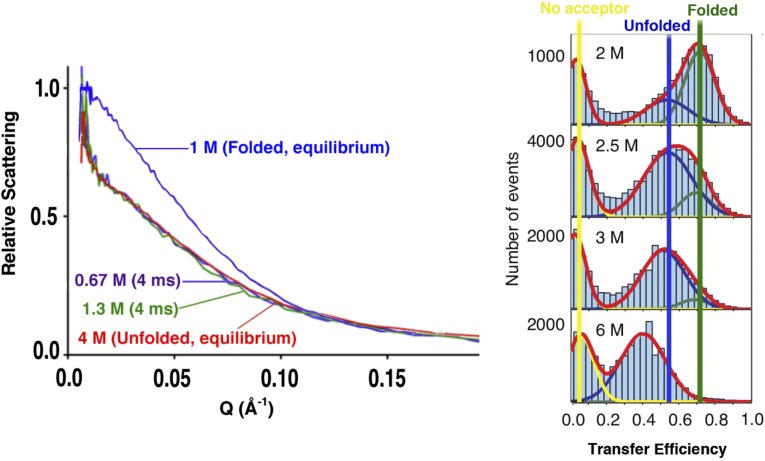
Fig. 1.
Although it is widely accepted that chemically denatured proteins adopt an ensemble of conformations well-described as random coil (2), significant debate remains regarding the nature of the unfolded states populated at low denaturant concentrations (4). Specifically, time-resolved SAXS measurements collected within milliseconds of transfer from high to low denaturant suggest that chemically denatured single-domain proteins do not undergo any significant conformational change before folding itself (5, 12–14). (Left) Shown, for example, are SAXS profiles collected on protein L at equilibrium at 1 M (native conditions), at equilibrium at 4 M GuHCl (unfolding conditions), and transiently before folding on transfer to denaturant concentrations as low as 0.67 M; the near-superimposability of the scaled scattering data for the various unfolded states suggests that the unfolded chain does not change dimensions (to within experimental uncertainty of a few percent) before folding (4). (Right) The efficiency of smFRET observed across dye-labeled unfolded proteins, in contrast, invariably decreases significantly as the level of denaturant rises (6, 7, 16, 19, 20, 23), an observation that has universally been interpreted in terms of an unfolded state that expands significantly at higher denaturant concentrations. Shown are equilibrium smFRET data collected on dye-labeled protein L as a function of GuHCl concentration (6); the large shift in the placement of the peak corresponding to unfolded molecules (indicated by the blue bar) suggests that the chain contracts significantly when transferred from high to low denaturant. Fig. 1 (Left) reproduced with permission from ref. 4. Fig. 1 (Right) reproduced with permission from ref. 6.
The discord between the views arising from SAXS and FRET presumably occurs because of some as yet unrecognized systematic or interpretational error associated with converting scattering profiles and/or observed transfer efficiencies into unfolded-state dimensions. Its exact origins, however, remain elusive. For example, although denaturant-dependent background scattering from the buffer must be subtracted to calculate molecular dimensions, native-state dimensions predicted from SAXS data are typically within experimental uncertainty of those calculated from crystal structures (27) and independent of denaturant concentration until the population of unfolded protein becomes significant (9). Similarly, although smFRET, by comparison, relies on several denaturant-dependent assumptions and approximations, no attempt to explain the discrepancy between smFRET and scattering as arising because of these assumptions and approximations has yet proven successful (4). Studies of rigid constructs, such as polyproline (23, 28–32), for example, indicate that the denaturant dependence of the index of refraction, the quantum yield, and the spectral shift of the dyes are all too small to account for the observed changes in transfer efficiency. Denaturant-dependent viscosity effects on the rate of conformational averaging have, likewise, been shown to be too small to account for the seeming inconsistency between SAXS- and FRET-based observations (33). Again, the source of this experimentally robust discrepancy remains stubbornly unclear (a more detailed discussion of this issue is in ref. 4).
Key to the proper interpretation of any experiment is the availability of the appropriate controls. Because the pertinent SAXS experiments fail to produce evidence of unfolded-state contraction, the most relevant controls for these studies are positive controls establishing the technique’s ability to detect unfolded-state compaction, if it were to occur, of the magnitude seen by smFRET. Fortunately, these controls are already in hand: SAXS has repeatedly been shown to distinguish (at confidence intervals of many σ) between the dimensions of chemically unfolded, disulfide-free proteins, which generally coincide closely with expected random coil dimensions (2), and the 20–30% more compact (but still flexible and unfolded) states seen when the same chemically denatured protein is cross-linked through disulfide bonds (11, 12, 34, 35). Conversely, because smFRET studies of unfolded states are universally interpreted in terms of significant collapse, the most critical controls for smFRET would be negative controls (that is, the demonstration of the denaturant independence of smFRET across an unfolded polymer, the dimensions of which are known to be denaturant-independent). Here, for the first time to our knowledge, we report the results of just such a negative control. Specifically, we have performed smFRET and scattering studies of a flexible polymer known to adopt a conformation well-approximated as an expanded, excluded volume random walk (1–3) (herein referred to as a random coil) in aqueous solution across a wide range of conditions.
Results and Discussion
As our negative control, we have used PEG [also known as poly(ethylene oxide) or polyoxyethylene], an uncharged but extremely hydrophilic polymer [e.g., 3 kDa PEG is soluble in water to greater than 55% (wt/wt) (36)] that, according to extensive prior literature, is well-approximated as a swollen random coil in aqueous solution, even in the absence of denaturant. Evidence for the random coil nature of aqueous PEG is provided by studies monitoring many different dimensionally dependent properties. These sources of evidence include gel filtration (37), light scattering (38, 39), and sedimentation (40) studies of its hydrodynamic radius as a function of its molecular weight, studies of the viscosity of PEG solutions as a function of molecular weight (41), small-angle neutron scattering (SANS) (42) and simulations-based (43) studies of the polymer’s radius of gyration as a function of molecular weight, and single-molecule pulling studies of PEG’s extension as a function of force (44). In short, all of these physically distinct methods of monitoring the behavior of PEG produce dimensional scaling behaviors within experimental uncertainty of those expected for an expanded, excluded-volume random coil (43).
Given PEG’s great hydrophilicity (reducing or eliminating hydrophobic interactions) and its inability to donate hydrogen bonds (precluding intramolecular hydrogen bonding), it is, perhaps, not surprising that the polymer adopts a swollen random coil conformation in the absence of denaturant. For the same reasons, we would also expect the dimensions of this random coil to be denaturant-independent. To confirm this expectation, we have measured the radius of gyration of dye-free PEG in the presence and absence of the denaturants GuHCl and urea using SANS. (The use of SAXS is precluded by the sensitivity of this polymer to ionizing radiation. The relatively large mass of material required for SANS studies, in turn, precludes use of dye-labeled PEG in these studies.) Specifically, we have characterized 3 and 5 kDa PEG with polydispersity indices (the ratio of the mass-averaged molecular mass to the number-averaged molecular mass) of 1.02 for both. These constructs are, thus, composed of 68 ± 9 and 114 ± 15 ethylene glycol monomers, respectively, corresponding to contour lengths equivalent to those of unfolded ∼65- and ∼108-residue proteins, respectively.
SANS data suggest that dimensions of PEG are, to within experimental uncertainty, independent of denaturant concentration (Fig. 2). To illustrate these observations, we first fit our scattering data to the Debye approximation (45):
| [1] |
where I(Q) is the scattering intensity at the scattering vector Q, I0 is the scattering intensity at Q = 0, Rg is the radius of gyration, and Iic is the background incoherent scattering [which is assumed to be constant at all Q (46)]. This relationship, which explicitly models scattering by a Gaussian chain using only three fitted parameters, fits our 3 kDa PEG scattering data well except at the very lowest values of Q, where the errors are largest (because of the small number of neutrons at these energies), providing additional evidence for the random coil nature of this highly soluble polymer (Fig. 2 and Figs. S1 and S2). Moreover, the error-weighted best-fit radii of gyration produced by these fits are, to within experimental uncertainty, independent of denaturant concentration (Fig. 2, Lower Right), suggesting that the dimensions of this random coil are unchanged by the addition of urea or GuHCl. We observe similarly denaturant-independent, random coil-like scattering for 5 kDa PEG in both denaturants (Figs. S3 and S4). We have also fit our data to the perhaps better-known Guinier approximation, which unlike the Debye approximation, makes no a priori assumptions regarding the shape of the scatterer. Over the narrower range of Q for which the Guinier approximation is valid, we, once again, see no significant evidence for denaturant-driven changes in the molecular dimensions of PEG, finding that the fitted Rg values are nearly all within reasonable confidence intervals of the average value (Figs. S5–S7).
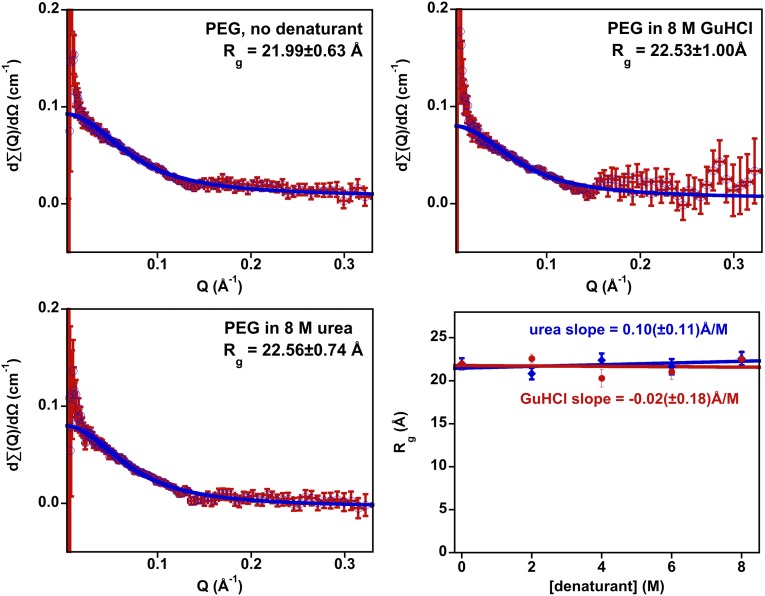
Fig. 2.
(Upper Left) Background-subtracted neutron scattering data collected on 3 kDa PEG in the absence of denaturant are well-fitted by the Debye approximation describing the scattering by a Gaussian chain, providing additional evidence that the aqueous polymer adopts a conformation well-modeled as a random coil. (Shown in blue is the error-weighted fit to scattering intensity using Eq. 1.) Background-subtracted scattering data collected in (Upper Right) 8 M GuHCl and (Lower Left) 8 M urea are, likewise, well-fitted by the Debye approximation, producing radii of gyration within experimental uncertainty of the value seen in the absence of denaturant. (Lower Right) Data collected over a range of GuHCl (Fig. S1) and urea (Fig. S2) concentrations provide additional evidence that the dimensions of the PEG random coil are denaturant-independent. A linear fit of Rg vs. denaturant concentration, for example, produces best-fit slopes of ≤0.1 Å/M for both denaturants. This slope corresponds to less than one-half of 1% change in dimensions per molar change in denaturant, a value within experimental uncertainty of zero. The error bars shown are estimated 95% confidence intervals. The poor signal-to-noise ratios seen at low Q are a consequence of the TOF measurement technique used in these studies. The magnitudes of the deviations observed at low Q are independent of denaturant concentration, suggesting that they do not reflect a denaturant-dependent change in PEG conformation. Figs. S1 and S2 show data collected at intermediate denaturant concentrations.
The apparent denaturant independence of the dimensions of dye-free PEG renders the molecule a good candidate for the flexible but noncollapsing polymer that we seek as a negative control. Moreover, the polymer’s extreme hydrophilicity and inability to form intramolecular hydrogen bonds suggest that, even if the dimensions of PEG were proven denaturant-dependent (counter to both our chemistry-informed expectations and the above scattering results), comparison of its behavior with that of unfolded proteins might provide insights into the origins of the denaturant dependence of smFRET. Thus motivated, we have measured the denaturant dependence of smFRET across PEG constructs terminally modified with FRET-reporting dyes. To ensure ready comparison with prior work, we have used Alexa-488 and -594 as our FRET reporters; this dye pair has been used in the large majority of previous smFRET studies of protein folding (6, 7, 17, 21–24).
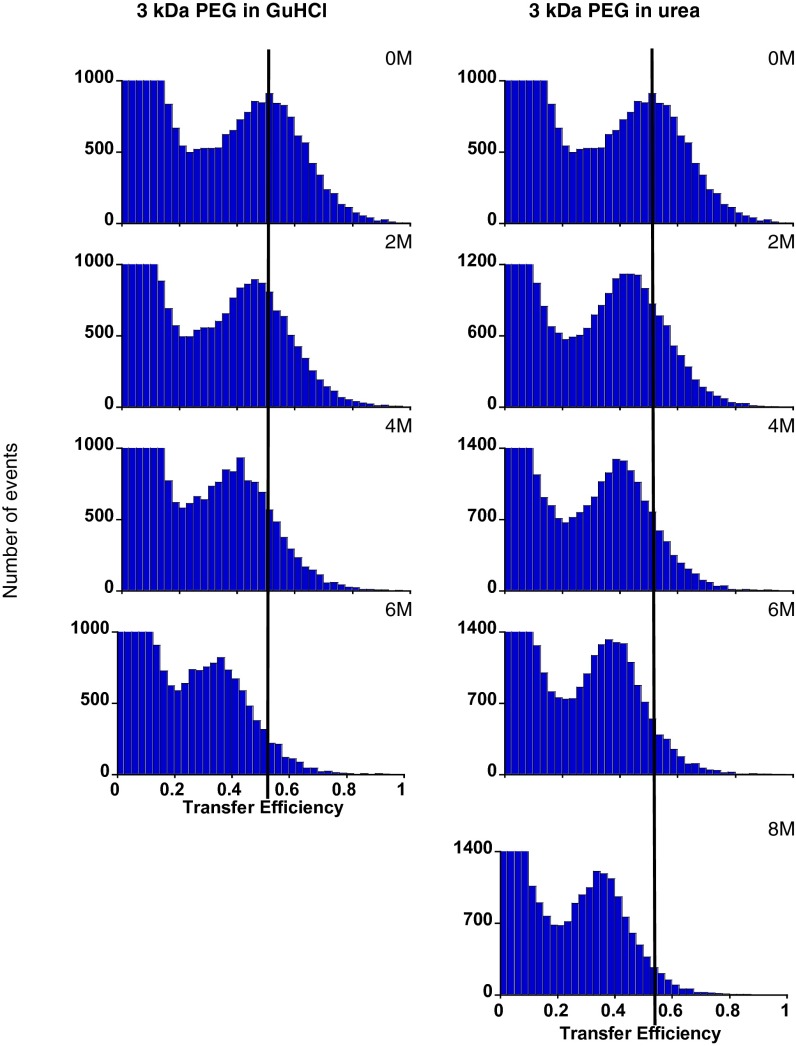
In contrast to the apparently random coil behavior and denaturant-independent dimensions of dye-free PEG, the efficiency of FRET across the dye-labeled polymer decreases significantly with increasing denaturant concentration. Specifically, in the absence of denaturant, we observe two peaks in histograms of observed transfer efficiency for 3 kDa PEG: one near zero arising because of molecules lacking a functional acceptor dye and one at ∼48% efficiency corresponding to properly dual-labeled molecules (Fig. 3). The observed transfer efficiency across the population of properly labeled molecules drops significantly with rising denaturant concentration, falling to ∼35% in 6 M GuHCl or 8 M urea. Changes of similar magnitude are also seen for 5 kDa PEG (Fig. S8), albeit with lower overall transfer efficiency because of this construct’s larger end-to-end separation.
Fig. 3.
In contrast to the apparent denaturant independence of the dimensions and random coil behavior of dye-free PEG, the observed efficiency of energy transfer across the dye-labeled polymer is denaturant-dependent. Shown is transfer across 3 kDa PEG labeled at the termini with Alexa-488 and -594, the most commonly used smFRET dye pair, as a function of (Left) GuHCl or (Right) urea. The peaks at near-zero transfer efficiency arise because of molecules lacking a functional acceptor dye. smFRET data collected on 5 kDa PEG exhibit similar denaturant dependence (Fig. S8).
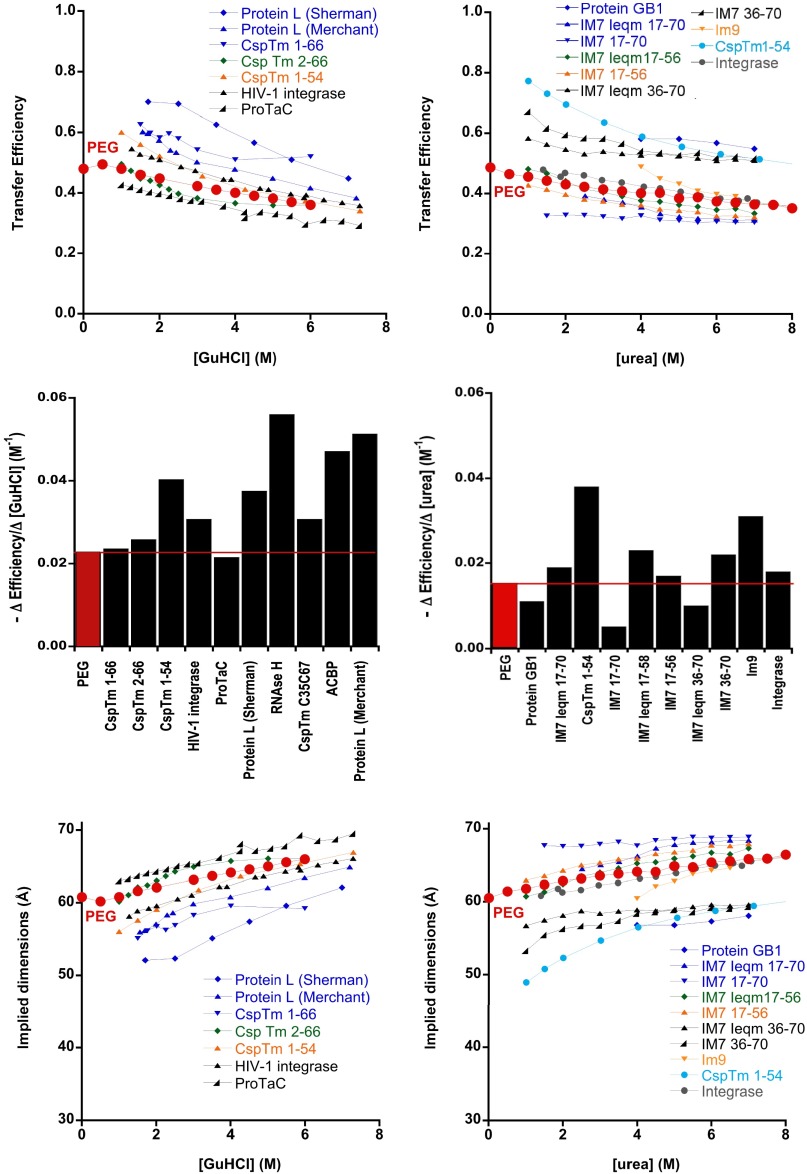
The absolute (i.e., raw and uninterpreted) denaturant-dependent changes in FRET that we observe for dye-labeled PEG are quite similar to those seen for a number of chemically denatured, dye-labeled proteins (Fig. 4, Top and Middle). Under the standard assumptions used in the interpretation of smFRET data from proteins (namely, that both FRET efficiency and the FRET measurement are themselves independent of denaturant, suppositions that we believe are now suspect), the observed changes in FRET would correspond to 10% expansion of the labeled PEG at high denaturant (i.e., a 30% decrease in monomer density). Also, although this change may seem small, such expansion is outside of the 95% confidence intervals (one-tailed t test) for the largest possible expansions, consistent with our four SANS datasets (Fig. 2 and Figs. S3 and S4). Moreover, the denaturant-driven changes in dimensions implied by the observed FRET changes are, likewise, quite similar to those seen for a number of dye-labeled proteins over the range down to ∼1 M denaturant (Fig. 4, Bottom).
Fig. 4.
(Top and Middle) The denaturant dependence of energy transfer across dye-labeled PEG falls within the range of the denaturant dependencies seen for transfer across unfolded proteins modified with the same dye pair. (Bottom) The expansions in interdye distance that these changes in transfer efficiency would imply (were alterations in molecular dimensions the true source of these changes) are, likewise, similar to the denaturant-driven expansions reported for unfolded, dye-labeled proteins. Given the argument that the dimensions of dye-free PEG are likely independent of denaturant concentration, however, and given the vast chemical differences between PEG and polypeptides, these similarities raise questions regarding the extent to which changes in FRET across unstructured polymers can be interpreted as changes in their molecular dimensions. All of the proteins shown here were, like our PEG constructs, labeled with Alexa-488 and -594. Protein data were taken from CspTm 1–66 (25); CspTm 2–66 (23); Csp 1–54, HIV-1 integrase, and ProtaC (22); Protein GB1 (18); a number of IM7 variants (26); Im9 (24); and protein L (6, 7). Middle represents the absolute values of the slopes obtained from linear least squares fits of the data in Top over the range of 1–8 M denaturant. The dimensions displayed in Bottom assume a Förster radius of 60 Å as appropriate for this dye pair.
We close by admitting that we cannot prove the negative. That is, our results do not prove that the dimensions of all unfolded states remain fixed over all denaturant concentrations. Indeed, many larger (generally >120 residues) proteins have been shown to undergo transient contraction by SAXS (ref. 11 and references therein), energy transfer across even a single-domain protein sometimes changes quite dramatically (i.e., much more dramatically than the changes seen for dye-labeled PEG) at denaturant concentrations lower than those explored by SAXS (22), and even at denaturant concentrations above 1 M, some single-domain proteins exhibit larger changes in FRET than those that we observe for dye-labeled PEG (7). Here, however, we have replicated the more subtle changes in transfer efficiency observed for many unfolded single-domain proteins at denaturant concentrations above 1 M using (dye-labeled) PEG, a polymer that, in the absence of dye modification, (i) is extremely hydrophilic and incapable of forming intramolecular hydrogen bonds and (ii) forms an expanded conformation in the absence of denaturant, the properties of which are consistent with random coil behavior and the dimensions of which, by scattering, appear invariant on the addition of denaturant. Also, although we are not yet able to provide an explanation as to the origins of this effect (see the Introduction and the discussion in ref. 4), our results would, nevertheless, seem to raise important questions regarding the near-universal interpretation of such behavior in terms of hydrophobicity- or hydrogen bond-driven contraction of protein unfolded states.
Materials and Methods
Sample Preparation.
Sodium monophosphate (reagent grade; Sigma), Tween-20 (reagent grade; Sigma), D2O (99.6% purity; Sigma), and unlabeled 3 and 5 kDa α-methoxy-ω-hydroxy–terminated or end-labeled PEG (Rapp-Polymere) were used as received. Urea (ultrapure; MP Biomed) and GuHCl (Thermo Fisher) were either used as received (smFRET) or per-deuterated by three cycles of dissolution in D2O and lyophilization. The requisite dye-labeled PEG constructs were synthesized starting with N-hydrosuccinamide– and aldehyde-terminated 3 kDa PEG (Rapp-Polymere), which was then sequentially modified using Alexa-488 hydrazide and Alexa-594 maleimide and HPLC-purified at a commercial synthesis house (Biosearch Technologies). For the SANS studies, unmodified α-methoxy-ω-hydroxy–terminated PEG constructs were used. All PEG constructs had a polydispersity index of 1.02.
Single-Molecule Spectroscopy.
Dye-labeled PEG was dissolved at 15 pM in 50 mM sodium phosphate (pH 7), 0.001% (vol/vol) Tween-20, and the relevant concentration of denaturant. Single-molecule fluorescence was then observed using a custom-built confocal optical system (47, 48). A 488-nm continuous-wave laser beam (Coherent Sapphire 488–25) was attenuated to 100 μW using absorptive neutral density filters, spatially filtered, collimated, and focused 15 μm into the sample by a 60×, 1.2 N.A. water immersion objective (Olympus UPLSAPO 60XW). Resulting donor and acceptor fluorescence was collected by the same objective, separated from scattered laser light by a dichroic mirror (Chroma Z488/633RPC), and focused by a tube lens (Olympus LU074700) through a 100-μm confocal pinhole (Thorlabs P100S). After the pinhole, the beam was long pass-filtered (Omega 493AELP) to further reduce scattered laser light. Donor and acceptor signals were separated using a second dichroic mirror (Omega 560DCLP), and band pass-filtered (Chroma D525/50m and D630/60m for the donor and the acceptor, respectively) before being focused by 10× objectives (Newport M-10X) onto avalanche photodiode detectors (PerkinElmer SPCM-AQR-16). The arrival time of each detected photon was recorded by a time digitizer with 100 ps resolution (Ortec 9353).
For each sample, data were collected continuously for ∼30 min. Signal bursts from fluorescent dye labels were identified using Wald–Yang sequential analysis (49, 50). Background and burst threshold levels were determined by examining binned data for each sample. Bursts with 10 or fewer counts were discarded, and processing of the remaining data was carried out as described by Pfeil (50) to reduce the effect of photobleaching and remove quantization artifacts from the measured transfer efficiencies, which were then plotted.
SANS Measurements.
SANS measurements were conducted using dye-free PEG at 3 mM in phosphate-buffered D2O at the appropriate concentration of per-deuterated denaturant. All measurements were carried out with the time-of-flight Low Q Diffractometer (LQD) at the Los Alamos Neutron Science Center at the Los Alamos National Laboratory (51). The details of this instrument along with the data acquisition and reduction techniques that we used are described in detail elsewhere (52). Briefly, this instrument uses neutrons produced in pulses by spallation caused by the deposition of 800 MeV protons on a tungsten target followed by moderation by a liquid hydrogen moderator (20 K). Neutrons over an incident wavelength range λ = 2–16 Å encoded by the TOF at the 64 × 64-cm position-sensitive proportional counter provide a momentum transfer (Q) domain of 0.0035–0.35 Å−1 [Q = (4π/λ) sinθ, where θ is one-half of the scattering angle]. The TOF scattering data were reduced to intensity as a function of Q and placed on an absolute scale of differential scattering cross-section per unit volume using standard techniques corrected for (i) solution, cell, and instrument background scattering; (ii) detector nonlinearity; and (iii) sample transmission. Finally, the error-weighted data were fit to scattering intensity using Eq. 1. Secondary standard samples measured at the National Institute of Standards and Technology Center for Neutron Research were used to calibrate the data to an absolute scale (centimeters−1).
Supplementary Material
Acknowledgments
This work benefited from the use of the Low Q Diffractometer, LQD, at the Manuel Lujan Jr. Neutron Scattering Center at the Los Alamos National Laboratory. Los Alamos National Security LLC operates the Los Alamos National Laboratory under Department of Energy Contract DE-AC52-06NA25396. This work was supported by NIH Grants GM055694 (to T.R.S.) and GM093263 (to K.W.P.). H.M.W. was supported by NIH Grant GM097463 and a Whitaker Fellowship, and E.A.L. was partially supported by National Science Foundation Grant DMR-0960331.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418673112/-/DCSupplemental.
References
- 1.McCarney ER, et al. Site-specific dimensions across a highly denatured protein; a single molecule study. J Mol Biol. 2005;352(3):672–682. doi: 10.1016/j.jmb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Kohn JE, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci USA. 2004;101(34):12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann H, et al. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109(40):16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo TY, et al. Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J Mol Biol. 2012;418(3-4):226–236. doi: 10.1016/j.jmb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plaxco KW, Millett IS, Segel DJ, Doniach S, Baker D. Chain collapse can occur concomitantly with the rate-limiting step in protein folding. Nat Struct Biol. 1999;6(6):554–556. doi: 10.1038/9329. [DOI] [PubMed] [Google Scholar]
- 6.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc Natl Acad Sci USA. 2007;104(5):1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman E, Haran G. Coil-globule transition in the denatured state of a small protein. Proc Natl Acad Sci USA. 2006;103(31):11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia P, Serrano L, Durand D, Rico M, Bruix M. NMR and SAXS characterization of the denatured state of the chemotactic protein CheY: Implications for protein folding initiation. Protein Sci. 2001;10(6):1100–1112. doi: 10.1110/ps.52701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segel DJ, Fink AL, Hodgson KO, Doniach S. Protein denaturation: A small-angle X-ray scattering study of the ensemble of unfolded states of cytochrome c. Biochemistry. 1998;37(36):12443–12451. doi: 10.1021/bi980535t. [DOI] [PubMed] [Google Scholar]
- 10.Semisotnov GV, et al. Protein globularization during folding. A study by synchrotron small-angle X-ray scattering. J Mol Biol. 1996;262(4):559–574. doi: 10.1006/jmbi.1996.0535. [DOI] [PubMed] [Google Scholar]
- 11.Kimura T, et al. Specifically collapsed intermediate in the early stage of the folding of ribonuclease A. J Mol Biol. 2005;350(2):349–362. doi: 10.1016/j.jmb.2005.04.074. [DOI] [PubMed] [Google Scholar]
- 12.Jacob J, Dothager RS, Thiyagarajan P, Sosnick TR. Fully reduced ribonuclease A does not expand at high denaturant concentration or temperature. J Mol Biol. 2007;367(3):609–615. doi: 10.1016/j.jmb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Jacob J, Krantz B, Dothager RS, Thiyagarajan P, Sosnick TR. Early collapse is not an obligate step in protein folding. J Mol Biol. 2004;338(2):369–382. doi: 10.1016/j.jmb.2004.02.065. [DOI] [PubMed] [Google Scholar]
- 14.Konuma T, et al. Time-resolved small-angle X-ray scattering study of the folding dynamics of barnase. J Mol Biol. 2011;405(5):1284–1294. doi: 10.1016/j.jmb.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 15.Kathuria SV, et al. Microsecond barrier-limited chain collapse observed by time-resolved FRET and SAXS. J Mol Biol. 2014;426(9):1980–1994. doi: 10.1016/j.jmb.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deniz AA, et al. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc Natl Acad Sci USA. 2000;97(10):5179–5184. doi: 10.1073/pnas.090104997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann A, et al. Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc Natl Acad Sci USA. 2007;104(1):105–110. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang F, Sato S, Sharpe TD, Ying L, Fersht AR. Distinguishing between cooperative and unimodal downhill protein folding. Proc Natl Acad Sci USA. 2007;104(1):123–127. doi: 10.1073/pnas.0609717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuzmenkina EV, Heyes CD, Nienhaus GU. Single-molecule FRET study of denaturant induced unfolding of RNase H. J Mol Biol. 2006;357(1):313–324. doi: 10.1016/j.jmb.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 20.Laurence TA, Kong X, Jäger M, Weiss S. Probing structural heterogeneities and fluctuations of nucleic acids and denatured proteins. Proc Natl Acad Sci USA. 2005;102(48):17348–17353. doi: 10.1073/pnas.0508584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci USA. 2007;104(8):2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller-Späth S, et al. From the cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107(33):14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419(6908):743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 24.Tezuka-Kawakami T, Gell C, Brockwell DJ, Radford SE, Smith DA. Urea-induced unfolding of the immunity protein Im9 monitored by spFRET. Biophys J. 2006;91(5):L42–L44. doi: 10.1529/biophysj.106.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipman EA, Schuler B, Bakajin O, Eaton WA. Single-molecule measurement of protein folding kinetics. Science. 2003;301(5637):1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 26.Pugh SD, Gell C, Smith DA, Radford SE, Brockwell DJ. Single-molecule studies of the Im7 folding landscape. J Mol Biol. 2010;398(1):132–145. doi: 10.1016/j.jmb.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svergun D, Barberato C, Koch MHJ. CRYSOL – a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28(6):768–773. [Google Scholar]
- 28.Best RB, et al. Effect of flexibility and cis residues in single-molecule FRET studies of polyproline. Proc Natl Acad Sci USA. 2007;104(48):18964–18969. doi: 10.1073/pnas.0709567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein IH, Schüller V, Böhm P, Tinnefeld P, Liedl T. Single-molecule FRET ruler based on rigid DNA origami blocks. ChemPhysChem. 2011;12(3):689–695. doi: 10.1002/cphc.201000781. [DOI] [PubMed] [Google Scholar]
- 30.Sahoo H, Roccatano D, Hennig A, Nau WM. A 10-A spectroscopic ruler applied to short polyprolines. J Am Chem Soc. 2007;129(31):9762–9772. doi: 10.1021/ja072178s. [DOI] [PubMed] [Google Scholar]
- 31.Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. Proc Natl Acad Sci USA. 2005;102(8):2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoefling M, et al. Structural heterogeneity and quantitative FRET efficiency distributions of polyprolines through a hybrid atomistic simulation and Monte Carlo approach. PLoS ONE. 2011;6(5):e19791. doi: 10.1371/journal.pone.0019791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makarov DE, Plaxco KW. Measuring distances within unfolded biopolymers using fluorescence resonance energy transfer: The effect of polymer chain dynamics on the observed fluorescence resonance energy transfer efficiency. J Chem Phys. 2009;131(8):085105. doi: 10.1063/1.3212602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshino M, Hagihara Y, Hamada D, Kataoka M, Goto Y. Trifluoroethanol-induced conformational transition of hen egg-white lysozyme studied by small-angle X-ray scattering. FEBS Lett. 1997;416(1):72–76. doi: 10.1016/s0014-5793(97)01172-1. [DOI] [PubMed] [Google Scholar]
- 35.Arai M, Kataoka M, Kuwajima K, Matthews CR, Iwakura M. Effects of the difference in the unfolded-state ensemble on the folding of Escherichia coli dihydrofolate reductase. J Mol Biol. 2003;329(4):779–791. doi: 10.1016/s0022-2836(03)00511-4. [DOI] [PubMed] [Google Scholar]
- 36.Samuel Z, Milton HJ. 1997. Introduction to Chemistry and Biological Applications of Poly(Ethylene Glycol). Poly(Ethylene Glycol), ACS Symposium Series (American Chemical Society). Available at dx.doi.org/10.1021/bk-1997-0680.ch001. Accessed July 1, 2014.
- 37.Kuga S. Pore size distribution analysis of gel substances by size exclusion chromatography. J Chromatogr A. 1981;206(3):449–461. [Google Scholar]
- 38.Devanand K, Selser JC. Asymptotic behavior and long-range interactions in aqueous solutions of poly(ethylene oxide) Macromolecules. 1991;24(22):5943–5947. [Google Scholar]
- 39.Devanand K, Selser JC. Polyethylene oxide does not necessarily aggregate in water. Nature. 1990;343(6260):739–741. [Google Scholar]
- 40.Luo Z, Zhang G. Scaling for sedimentation and diffusion of poly(ethylene glycol) in water. J Phys Chem B. 2009;113(37):12462–12465. doi: 10.1021/jp906468n. [DOI] [PubMed] [Google Scholar]
- 41.Kawaguchi S, et al. Aqueous solution properties of oligo- and poly(ethylene oxide) by static light scattering and intrinsic viscosity. Polymer (Guildf) 1997;38(12):2885–2891. [Google Scholar]
- 42.Thiyagarajan P, Chaiko DJ, Hjelm RP. A neutron scattering study of poly(ethylene glycol) in electrolyte solutions. Macromolecules. 1995;28(23):7730–7736. [Google Scholar]
- 43.Lee H, Venable RM, Mackerell AD, Jr, Pastor RW. Molecular dynamics studies of polyethylene oxide and polyethylene glycol: Hydrodynamic radius and shape anisotropy. Biophys J. 2008;95(4):1590–1599. doi: 10.1529/biophysj.108.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dittmore A, McIntosh DB, Halliday S, Saleh OA. Single-molecule elasticity measurements of the onset of excluded volume in poly(ethylene glycol) Phys Rev Lett. 2011;107(14):148301. doi: 10.1103/PhysRevLett.107.148301. [DOI] [PubMed] [Google Scholar]
- 45.Debye P. Molecular-weight determination by light scattering. J Phys Colloid Chem. 1947;51(1):18–32. doi: 10.1021/j150451a002. [DOI] [PubMed] [Google Scholar]
- 46.Grillo I. Small-angle neutron scattering and applications in soft condensed matter. In: Borsali R, Pecora R, editors. Soft Matter Characterization. Springer; New York: 2008. pp. 723–782. [Google Scholar]
- 47.Pfeil SH, Wickersham CE, Hoffmann A, Lipman EA. A microfluidic mixing system for single-molecule measurements. Rev Sci Instrum. 2009;80(5):055105. doi: 10.1063/1.3125643. [DOI] [PubMed] [Google Scholar]
- 48.Wickersham CE, et al. Tracking a molecular motor with a nanoscale optical encoder. Nano Lett. 2010;10(3):1022–1027. doi: 10.1021/nl904192m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, Yang H. Photon-by-photon determination of emission bursts from diffusing single chromophores. J Phys Chem B. 2005;109(46):21930–21937. doi: 10.1021/jp0546047. [DOI] [PubMed] [Google Scholar]
- 50.Pfeil SH. 2009. Microfluidic mixing for non-equilibrium single-molecule optical spectroscopy. PhD thesis (Univ of California, Santa Barbara, CA)
- 51.Hjelm RP, Muhrer G, Nelson RO, Russell GJ, Kupcho KM. The LQD upgrade: Challenges and opportunities of brighter neutron sources. In: Mank G, Conrad H, editors. Proceedings of the International Collaboration for Advanced Neutron Sources XVI. Forschungszentrum Jülich GmbH; Jülich, Germany: 2003. pp. 177–183. [Google Scholar]
- 52.Hjelm RP. The resolution of TOF low- Q diffractometers: Instrumental, data acquisition and reduction factors. J Appl Crystallogr. 1988;21(6):618–628. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.