Neuropathic pain is a major, intractable clinical problem and its pathophysiology is not well understood. Although recent gene expression profiling studies have enabled the identification of novel targets 1–4, classical study designs provide somewhat unclear results owing to differential expression of hundreds of genes across sham and nerve injured groups that are difficult to validate, particularly with respect to specificity of pain modulation 5. To circumvent this, we employed two outbred lines of rats 6, which are genetically similar except for being genetically segregated due to selective breeding for differences in neuropathic pain hypersensitivity 7. We identified a serine protease inhibitor, SerpinA3N, to be upregulated in the dorsal root ganglia (DRG) in a profiling screen, which was further validated for its mouse homolog. Mice lacking SerpinA3N developed more neuropathic mechanical allodynia than wild-type mice (WT) and exogenous delivery of SerpinA3N attenuated mechanical allodynia in WT mice. Adoptive experiments showed that T-lymphocytes infiltrating the DRG after nerve injury release the leukocyte elastase (LE), which was inhibited by SerpinA3N derived from DRG neurons. Genetic loss of LE or exogenous application of a LE inhibitor (Sivelastat) in WT mice attenuated neuropathic mechanical allodynia. Overall, we reveal a novel role for a member of the serpin superfamily and a leukocyte elastase in the modulation of neuropathic allodynia, which hold clinical relevance.
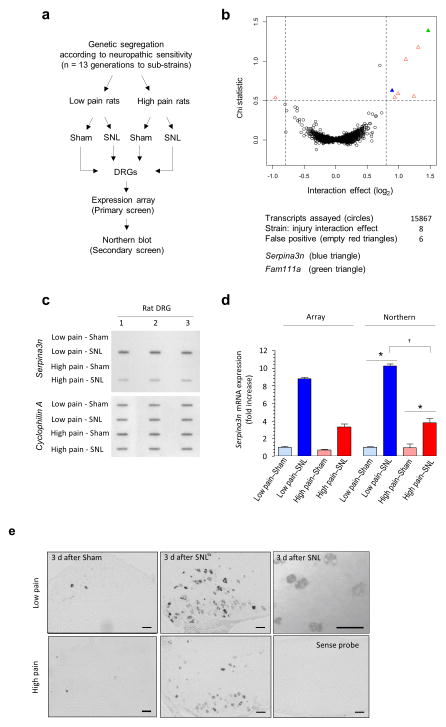
A differential pain phenotype in high and low pain sensitivity rats is apparent 3 days post injury7. To identify genes that are associated with the induction of neuropathic pain and are differentially expressed, microarray mRNA expression profiling was performed on L4–L5 DRGs 3 days after spinal nerve ligation (SNL) versus sham surgery from rats with high versus low pain-like sensitivity after nerve injury 6 (Fig. 1a). Although a very large number of genes were differentially regulated between the sham and neuropathic groups (Fig. 1b; the original screen results are depicted in Supplementary Table 1), only eight genes were found to be potentially differentially regulated between neuropathic animals with high and low sensitivity within the primary array screen (top left and right corners of the volcano plot in Fig. 1b, Supplementary Table 2). We next rescreened all eight initial ‘gene hits’ on biologically-distinct RNA samples via slot-northern blotting with criterion of injury-induced differential regulation between the high and low pain sensitivity mice (Fig. 1c and Supplementary Fig. 1). Only two transcripts encoding the rat ‘Serine Protease Inhibitor 3′ (Serpina3n, formerly known as Spin2c) (Fig. 1c) and the ‘Family with sequence similarity 111, member A′ (Fam111a) (Supplementary Fig. 1) were differentially expressed. Nerve injury induced Serpina3n expression in low pain-sensitivity to a greater extent than in rats with high neuropathic pain sensitivity (Figs. 1c,d). This was also validated via mRNA in situ hybridization in the injured L4 DRG, which revealed a primarily neuronal expression of Serpina3n (Fig. 1e).
Figure 1.
Serpina3n emerges as a key gene linked to behavioral traits associated with neuropathic pain in rats. (a) Schematic diagram of the microarray analyses on L4–L5 DRGs from rat strains demonstrating low neuropathic pain or high neuropathic pain behavior following spinal nerve ligation. (b) Chi Statistical test versus interaction size plot showing the regulated transcripts detected 3 days post-SNL. Filled blue and green triangles represent Serpina3n and Fam111a, respectively. Empty red triangles represent false positives genes. (c) Northern slot blot analysis of Serpina3n mRNA in low neuropathic and high neuropathic rats at 3 days post-SNL (normalized to Cyclophilin A), n = 3 (d) Quantitative estimation of Serpina3n mRNA regulation analyzed via Affymetrix expression array and Northern blots (n = 3). For northern data: Two-way ANOVA (P = 2.9e-6); *P < 0.05 as compared to sham, †P < 0.05 between Low pain and High pain rats post-SNL, post-hoc Tukey’s test. (e) In situ hybridization (ISH) using digoxigenin-labeled antisense riboprobes for Serpina3n expression on L4 DRG from Low pain and High pain rats, 3 days post-SNL and sham-operated rats; image on the bottom right panel represents sense control probe. Scale bars: 100 μm. Error bars: standard error of the mean.
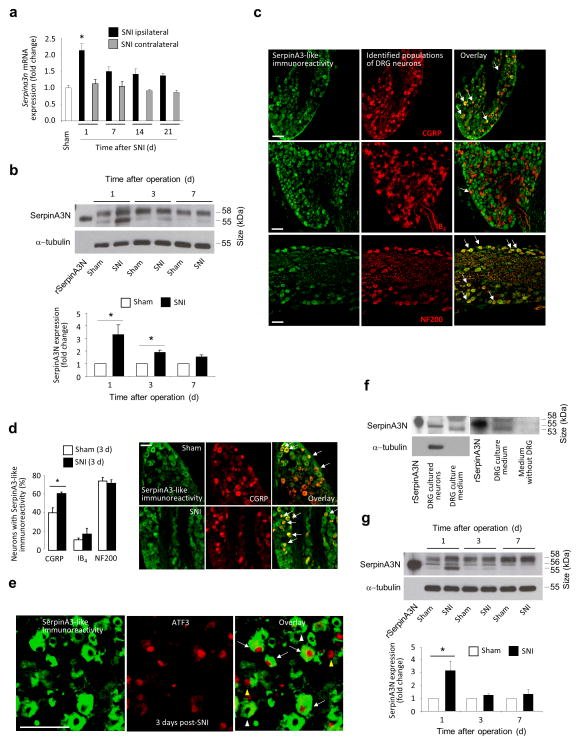
Upregulation of Serpina3n expression also occurred in mice and in the spared nerve injury (SNI) model (Fig. 2). Quantitative Real-Time PCR (qPCR) analysis demonstrated an upregulation of the expression of mouse Serpina3n mRNA in ipsilateral L3–L4 DRGs at day 1 post-SNI as compared to sham-treatment (Fig. 2a), but not in contralateral lumbar DRGs (Fig. 2a) or ipsilateral thoracic DRGs post-SNI (Supplementary Fig. 2a). In comparison with 9 other related Serpina3 genes in the mouse DRG, Serpina3n is the most abundantly expressed serpin isoform, followed by Serpina3g, whereas Serpina3h, Serpina3k and Serpina3f show very low abundance (Supplementary Fig. 2b). After SNI, Serpina3g and Serpina3k were upregulated at days 1 and 3 post-nerve injury (Supplementary Fig. 2c). Expression of SerpinA3N protein was increased at days 1 and 3, and declined to sham levels at day 7 post-SNI as compared to sham-treated mice (Fig. 2b; Supplementary Note 1).
Figure 2.
Serpina3n is upregulated in mouse lumbar DRGs following spared nerve injury, a model of neuropathic pain. (a) Real time quantitative PCR (qPCR) analysis of Serpina3n expression in L3–L5 DRGs post-SNI, Cyclophilin A serving as reference gene (n = 4 mice per time point; *P < 0.05 compared to sham DRG; 1-way ANOVA, Tukey’s post-hoc test). (b) Western blot analysis and quantification of SerpinA3N expression (55 kDa band) in L3–L4 DRGs post-SNI normalized to α-tubulin expression (n = 3–5 mice/ time point,1 way ANOVA, Tukey’s post-hoc test). (c) SerpinA3-like immunoreactivity in peptidergic (CGRP) and non-peptidergic (IB4-binding) nociceptors and large diameter neurons (NF200) in L3–L4 DRG sections from naïve mice (arrows indicate areas of colocalization). (d) Quantitative analysis of SerpinA3-like immunoreactivity in L3–L4 DRGs 3 days post-SNI as compared to sham (n = at least 3 DRG sections/mouse, 3 mice/treatment group; *P < 0.05 compared to sham; two-tailed unpaired T-test) and examples of upregulation in the CGRP-expressing population. (e) SerpinA3 and ATF3 staining in L3–L4 DRGs at day 1 post-SNI showing SerpinA3+/ATF3+ (white arrows), Serpina3+/ATF3− (white arrowheads) and SerpinA3−/ATF3+ (yellow arrowheads) neurons. (f) Western blot analysis of SerpinA3N expression in lysates and medium from cultured DRG neurons. (g) Western blot analysis and quantification of SerpinA3N expression in lumbar spinal cord post-SNI (n = 3–5 mice/time point, *P < 0.05 compared to sham, 1-way ANOVA, Tukey’s post-hoc test). Scale bars represent 100 μm in (c), (d) and (e). Error bars: standard error of mean.
Immunohistochemistry on mouse L3–L4 DRGs with an antibody raised against SerpinA3N and co-staining for identification of DRG neuronal subpopulations revealed SerpinA3-like staining (please see Supplementary Note 1) in neurofilament 200 (NF200)-positive large myelinated Aβ fiber neurons and in a large fraction of calcitonin gene-related protein (CGRP)-positive peptidergic nociceptive neurons, and it was less abundant in isolectin B4 (IB4)-positive nonpeptidergic nociceptor neurons (Figs. 2c, d and Supplementary Fig. 3a). The percentage of CGRP-positive neurons with SerpinA3-like immunoreactivity increased in the DRG after SNI, as compared to sham controls (Fig. 2d). This was not due to a change in distribution pattern of CGRP in DRG neurons post-injury as the total percentage of neurons with SerpinA3-like immunoreactivity increased at day 3 post-SNI (Supplementary Fig. 3b). This was further supported by cell size versus frequency histograms showing that neurons with a diameter of 10–20 μm exhibit increased SerpinA3-like immunoreactivity 3 days post-SNI as compared to sham-treatment (Supplementary Fig. 3c). Using ATF-3 as a marker of injured neurons 8, nerve injury-induced upregulation of Serpina3n mRNA (Supplementary Fig. 3d) and SerpinA3-like immunoreactivity (Fig. 2e) is observed in injured neurons (ATF-3-positive) as well as in neurons that are spared from injury on day 1 or 3 post-SNI (Supplementary Figs. 3d,e).
SerpinA3N, like other serpins, is a secreted protein and contains a predicted 20-amino acid N-terminal signal peptide 9,10. Western blot analysis of cell lysates and culture media from serum-starved DRG cell cultures enriched for sensory neurons revealed SerpinA3N in the superfusate and lysate (Fig. 2f, left panel; Supplementary Note 1), but not in fresh medium without contact with DRG cultures (Fig. 2f, right panel), indicating secretion from DRG neurons. Direct depolarization and activation of cultured DRG neurons did not enhance release of SerpinA3N (Supplementary Fig. 4a), suggesting that other events downstream of nerve injury may regulate secretion.
Expression of SerpinA3N in the spinal dorsal horn was increased at 1 day post-SNI as compared to sham controls (Fig. 2g. Supplementary Note 1). Serpina3n mRNA expression was upregulated in the spinal cord to a lesser extent at day 1 post-SNI (Supplementary Fig. 4b), and both Serpina3n mRNA and protein expression were higher in the DRG than in the spinal cord at day 1 post-SNI (Supplementary Fig. 4b,c, Supplementary Note 1).
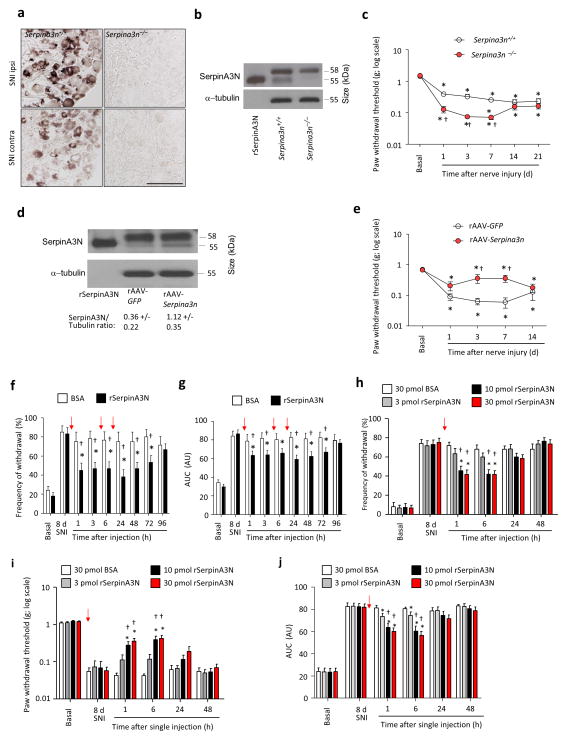
To address the functional effect of Serpina3n upregulation in nociceptive modulation, we generated null mutant mice (Serpina3n−/−) by deleting exon 2 from the mouse Serpina3n gene (Supplementary Figs. 5a, b). In situ mRNA hybridization analyses on DRGs confirmed that basal expression and nerve injury-induced upregulation of the Serpina3n transcript was not evident in Serpina3n−/− mice (Fig. 3a). SerpinA3N protein expression was not detectable in DRG lysates from Serpina3n−/− mice 1 day post-SNI selectively (Fig. 3b). In basal conditions, Serpina3n−/− mice were undistinguishable from WT littermates with respect to nociceptive behavior elicited by von Frey mechanical stimulation, plantar application of acetone (cold stimulus), noxious heat or plantar pinprick stimulation (Supplementary Fig. 6a–d). However, mechanical allodynia was considerably increased in Serpina3n−/− mice as compared to WT littermates for up to 7 days post-SNI, but not later (Fig. 3c). In contrast, nociceptive responses to acetone and pinprick following nerve-injury were comparable between Serpina3n−/− mice and WT littermates (Supplementary Figs. 6e,f). None of the mice from either genotype developed significant and reliable hypersensitivity to heat in the Hargreaves plantar test or the Hot plate test post-SNI (Supplementary Figs. 6g,h). However, responses to a brush stimulus applied to the plantar surface were stronger in Serpina3n−/− mice post-SNI than in WT littermates (Supplementary Fig. 6i), suggesting that upregulation of SerpinA3N during the early phase post-injury acts as an endogenous brake specifically against the transition from acute to chronic neuropathic mechanical hypersensitivity. To address this hypothesis further, we overexpressed Serpina3n cDNA in L3–L4 DRGs of adult WT mice by intraganglionic injection of recombinant adeno-associated virions (rAAV) carrying Serpina3n cDNA or GFP cDNA as a control (example and quantification is shown in Fig. 3d). Overexpression of SerpinA3N in the DRG did not alter basal nociceptive sensitivity, but decreased mechanical allodynia at day 3 and 7 post-SNI, but not thereafter, as compared to GFP-expressing mice (Fig. 3e).
Figure 3.
SerpinA3N attenuates mechanical allodynia following nerve injury. (a, b) Serpina3n mRNA ISH (a) and Western blot analysis (b) of SerpinA3N (55 kDa band) in L3–L4 DRGs from Serpina3n null mice (Serpina3n−/−) 1 day post-SNI. Scale bar: 100 μm. (c) Paw withdrawal thresholds to plantar von Frey application in Serpina3n−/− and WT mice (n = 16 mice/group). (d, e) Western blot analysis of SerpinA3N expression in L3–L4 DRGs (d) and mechanical thresholds (e) in mice receiving intraganglionic injection of adeno-associated virions (AAV) expressing Serpina3n or GFP (n = 8 mice/group). (f, g) Paw withdrawal responses (f) and area under the curve (AUC; g) of mechanical responses (0.04 g to 2 g; expressed in arbitrary units [AU]) in WT mice following intrathecal injection of 10 pmol rSerpinA3N or BSA (control) 3 times (red arrows) separated by 6 h intervals at 8 days post-SNI (n = 12 mice per group). (h–j) Impact of single injections of 3 – 30 pmol rSerpinA3N or BSA on mechanical allodynia, represented as responses to 0.16 g (h), thresholds (i) or AUC (0.04 g to 2 g; j) at day 8 post-SNI (n = 10–12 mice/dose or vehicle). In (c) and (e), *P < 0.05 compared to basal values and †P < 0.05 compared to the corresponding control. In (f–j), *P < 0.05 compared to day 8 post-SNI; †P < 0.05 compared to vehicle. In all panels: 2-way ANOVA of repeated measures, Tukey’s post-hoc test. Error bars: standard error of mean.
To determine whether exogenous SerpinA3N would reduce established neuropathic tactile allodynia, at 8 days post-SNI, we administered rSerpinA3N (10 pmoles) or PBS containing 10 pmoles of bovine serum albumin (BSA) as vehicle intrathecally to WT mice with mechanical allodynia (Figs. 3f,g). rSerpinA3N substantially reduced mechanical allodynia within 1 h following the first injection, which was extended until 72 h upon employing the dosing scheme shown in Figs. 3f,g. The protective effects of rSerpinA3N were dose-dependent (Figs. 3h–j). A single i.t. injection of SerpinA3N (10 pmol) at day 18 post-SNI, when chronic hypersensitivity is established, did not decrease mechanical hypersensitivity as compared to BSA-containing vehicle (Supplementary Fig. 6j). Anti-allodynic effects of intrathecal rSerpinA3N during early stages post-SNI were neither accompanied by any damage to spinal neurons, microglia and astrocytes (Supplementary Fig. 7) nor did rSerpinA3N influence any parameters of nerve injury-related neuroinflammation in the spinal cord or the DRG at 2–7 days post-SNI (Supplementary Fig. 8; see Supplementary Note 2 for details).
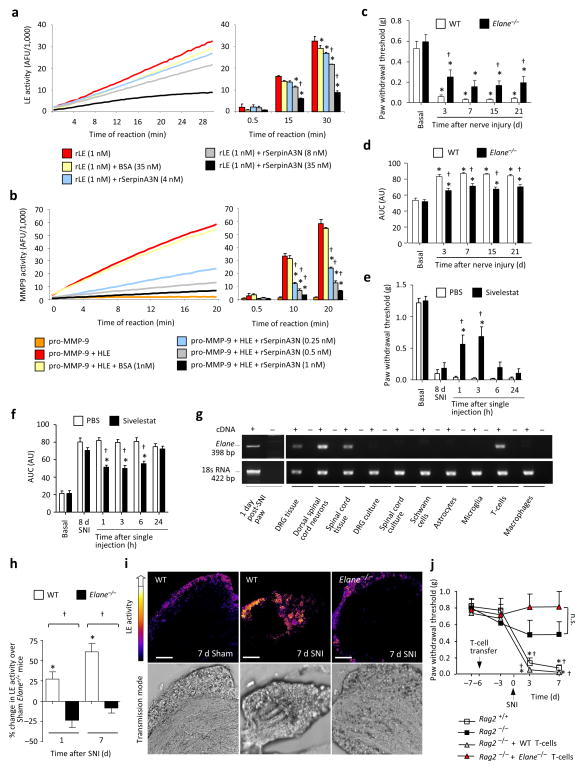
Serpins exert their function by binding and inhibiting specific serine proteases and several serine proteases can be substrates for SerpinA3N in vitro 9–11. This includes the human leukocyte elastase (HLE) 10, a serine protease that has been associated with airway inflammation 12,13, but not the nervous system to date. In an in vitro fluorometric assay, mouse rSerpinA3N dose-dependently inhibited the catalytic activity of mouse recombinant leukocyte elastase (rLE), in comparison with BSA (35 nM) (Fig. 4a). Other serine proteases such as Thrombin and Matrix Metalloproteinase 9 (MMP9), that are associated with nociception 14,15, were not affected by SerpinA3N (Supplementary Figs. 9a,b). However, consistent with studies showing a role for LE in activating MMP-9 16,17, we observed that in the presence of HLE, rSerpinA3N indirectly inhibited MMP-9 activation (Fig. 4b).
Figure 4.
Leukocyte elastase (LE) is a substrate for SerpinA3N and promotes neuropathic allodynia. (a, b) LE activity (a) or MMP9 activity (b), as assessed by in vitro fluorometric assays. AFU: arbitrary fluorescence units; n = 3, *P < 0.05 compared to protease alone, †P < 0.05 compared to BSA control. (c, d, e, f) Paw withdrawal thresholds (c, e) and AUC of responses to von Frey filaments (d, f) in LE null (Elane−/−) and WT mice (c, d) or in WT mice following injection of the LE inhibitor Sivelestat, or vehicle 8 days post-SNI (e, f); n = 8–9/group; *P < 0.05 compared to basal (c, d) or vehicle (e, f), †P < 0.05 between genotypes (c, d) or compared to vehicle (e, f). (g) Elane mRNA expression in different cell types in vitro or tissues from WT mice. (h, i) Quantification (h) and staining (i) of LE activity in L4 DRGs of Elane−/− and WT mice; n = 4–5 experiments, 4 mice/data point; *P < 0.05 compared to sham, †P < 0.05 between genotypes, 1-way ANOVA, post-hoc Tukey’s test. Scale bar: 100 μm. (j) Mechanical allodynia in Rag2−/− and WT mice following adoptive transfer of WT- or LE-deficient T cells 6 d prior to SNI; n = 10 mice/group; *P < 0.05 compared to basal values, †P < 0.05 compared to Rag2−/− mice without T-cell transfer. Unless otherwise indicated, 2-way ANOVA of repeated measures, Tukey’s post-hoc test. Error bars: standard error of mean.
SNI-induced mechanical allodynia was reduced in mice deficient in the gene encoding LE (Elane−/− mice) as compared to WT littermates, up to 21 days after nerve injury (Figs. 4c,d). Inhibiton of LE using Sivelestat 18, administered 8 days post-SNI via a single, low-dose i.t. injection (200 pmol) rapidly and considerably reduced mechanical allodynia (Figs. 4e,f). Sivelastat blocked LE activity in vitro at doses which are comparable with its effects on mechanical allodynia in vivo (Supplementary Fig. 9c). These results suggest LE is functionally involved in the induction as well as the maintenance of long-term tactile hypersensitivity and that its pharmacological blockade is effective when peak neuropathic pain is established.
LE is produced in blood leukocytes, mainly neutrophils 13. Although Elane mRNA is expressed in mouse-derived L3–L4 DRGs or spinal cord tissue, Elane mRNA was not detectable in neuronal cultures derived from the DRG or the spinal cord. Elane mRNA was also not expressed in many non-neuronal cells that have been associated with neuropathic pain, including astrocytes, microglia, macrophages or Schwann cells 19,20, but was found in T-lymphocytes (Fig. 4g). T-cells are present in the mouse DRG in vivo under naïve conditions and increase in frequency following nerve injury (Supplementary Figs. 10a–f), consistent with previous reports 21,22. Although Gr-1+ neutrophils are found in the vicinity of the DRG and increase in frequency after nerve injury, they remain primarily localized to the borders rather than invading the parenchyma of the DRG in large numbers (Supplementary Fig. 10g). LE activity 23 was increased in L3–L4 DRGs 1 and 7 days post-SNI as compared to sham-treated mice (Figs. 4h,i).
To address whether T-cells function as a source of LE in neuropathic pain, T-cells derived from WT or Elane−/− mice were adoptively transferred into Rag2−/− mice. Mechanical allodynia was reduced post-SNI in Rag2−/− mice (Fig. 4j), consistent with previous reports on Rag1−/− mice which similarly lack T-cells 24. Adoptive transfer of WT T-cells in Rag2−/− mice restored mechanical allodynia (Fig. 4j), whereas Rag2−/− recipients of LE-deficient T-cells did not develop mechanical hypersensitivity following SNI (Fig. 4j). These differences were not due to differential efficiency of adoptive transfer of T-cells derived from Elane−/− or WT mice (Supplementary Fig. 11).
SerpinA3N is a part of a large superfamily preserved and differentially amplified across species 25,26, whose members exert a myriad of functions, ranging from coagulation to chromatin condensation to control of immune cell function 11, with specificity being imparted by distinct expression patterns and substrate recognition. Our data suggest that the SerpinA3 cluster expressed and induced in peripheral sensory neurons may be a key endogenous regulator ofthe acute to chronic switch in pain hypersensitivity. This anti-allodynic activity of SerpinA3N is neither linked to modulating glial reactions post-injury nor does SerpinA3N prevent immune cells from infiltrating the DRG, but rather unfolds its actions a step further down by acting on proteases derived from immune cells in the DRG. Indeed, LE represents a novel effector for nociceptive modulation in the DRG, particularly via T-lymphocytes, underscoring their contributions in mechanical allodynia post-injury.
Unlike the close functional links observed between SerpinA3N, LE and resident as well as infiltrating T-cells over induction of mechanical allodynia (i.e. first 7 days post-SNI), we observed a divergence between the involvement of SerpinA3N and LE in mechanisms associated with the maintenance of chronic neuropathic hypersensitivity (Supplementary Fig. 12). Progressively increasing T-cell cell infiltration observed in the DRG, which is consistent with sustained involvement of LE, may result in inadequate blockade by SerpinA3N. Furthermore, LE may elicit its sustained actions via other locations and mechanisms and could be under the control of additional endogenous inhibitors. For example, in airway, colonic and cardiovascular injury, Elafin and secretory leukocyte peptidase inhibitor have been shown to act as endogenous inhibitors and key regulators of LE activity 27,28. Because relief of established neuropathic hypersensitivity was afforded by the intrathecal delivery of a low dose of Sivelestat, a drug which is currently awaiting FDA approval and is already in clinical usage in some countries for the therapy of airway inflammation, there may be opportunities for a therapeutic approach based on LE blockade.
Online Methods
Genetically-modified mice
We generated conditional Serpina3n knockout mice using a BAC recombination strategy. The129Sv BAC clone (bMQ-264h22) containing Serpina3n was obtained from BioScience. Primers were designed to retrieve a 10.3 kB region containing Serpina3n, insert an upstream LoxP site within intron 1, and insert a downstream FRT-Neo-FRT-LoxP cassette within intron 2. Exon 2 was chosen for deletion because it contains the translation start site, encodes the signal sequence for secretion, and accounts for over 50% of the Serpina3n coding sequence. The final targeting vector, with a LoxP flanked region of 1.1kB and homology arms of 4.7 kB and 4.5kB, was linearized with AATII, electroporated into W4/129S6derived ES cells, and selected with G418. DNA from G418-resistant ES cells was analyzed by long range PCR and Southern blot. Validated ES cells were injected into blastocysts and implanted into C57BL/6 pseudo-pregnant female mice. Male chimeric animals were bred for germline transmission with (1) germline FLPe-expressing females (Gt(ROSA)26Sortm1(FLP1)Dym) to delete the FRT-flanked Neo cassette and generate heterozygote conditional knockouts (Serpina3nfl/+) and (2) germline Cre-expressing females (Tg(EIIa-cre) C5379Lmgd) to generate heterozygote null mice (Serpina3n+/−). F1 heterozygotes were inter-bred, yielding WT, heterozygous, and homozygous mice in the expected 1:2:1 ratio. In subsequent generations, the EIIa-cre allele was bred out of the null line. For all behavioral and histology experiments, mixed background Serpina3n−/− and WT littermate controls were used. Serpina3n−/− mice are viable and fertile. Mice were genotyped by PCR using the following primers: F.1 (mutant forward) 5′ TCTGGATTCAGGCCACGGTTT3′; F.2 (WT forward) 5′ AGGACATTGATGGTGCTGGT3′; R.1 (common reverse) 5′ CCTACAGGGATGGATATTTCC 3′. The WT and mutant alleles were detected as fragments of 459 bp and 518 bp, respectively.
Mice lacking the leucocyte elastase (referred to here as Elane−/− mice) (B6.129X1-Elanetm1Sds/J)29 were bought commercially (Jackson Laboratories) and genotyped by PCR on mouse genomic DNA using the primers oIMR7064 (WT forward), oIMR7065 (common) and oIMR8162 (mutant reverse). Homozygote mice were identified by a 310 bp fragment corresponding to the mutant Elane gene. WT mice were identified by a 230 bp fragment corresponding to the WT allele. Heterozygote animals were identified by the presence of both fragments. Primer sequences are available on the corresponding website of the source (Jackson Laboratories).
High Neuropathic Pain and Low Neuropathic Pain sub-strain production
High neuropathic and low neuropathic rats were derived from outbred Wistar-based Sabra strain rats by genetic selection for high vs. low autotomy behavior in the neuroma model of neuropathic pain. The animals used for the present study were from the 45th (high pain) and the 42nd (Low pain) generations of selection. The selection process, including quantification of pain phenotype, has been originally described by Devor and Raber (1990)6. High pain and Low pain rats show contrasting pain phenotypes in both the neuroma and the spinal nerve ligation (SNL) models of neuropathic pain7.
Microarray analysis and secondary screen
In adult High pain and Low pain rats, induction of peripheral nerve injury, sample collection, DRG total RNA extraction, and microarray hybridization were performed as previously described30. RAE230A arrays (Affymetrix) were used for biologically-independent triplicate samples. Background correction and quantile-quantile data normalization were followed by probe set intensity computation, all using the RMA algorithm within R as implemented in the Bioconductor package31–33. For each probe, a linear model with one-way effects for sub-strain (High pain or Low pain) and injury (sham or SNL), and sub-strain by injury interaction was fit using iteratively reweighted least squares. For each probe, the chi statistic34, comparing the residual sum of squares of the full model to the model without interaction effect, was calculated. Graphical analysis of outliers in a plot of chi statistic versus interaction effect magnitude allowed identification of probes whose regulation potentially differed between sub-strains. Each of the eight outlying genes, within the top left and top right sections of Fig. 1b were subject to a secondary screen on biologically independent triplicate DRG total RNA samples, distinct from those used in the array analysis. Of the eight genes, two showed reliable differential regulation between the two sub-strains in this secondary screen. These were Serpina3n (Fig. 1c) and Fam111a (Supplementary Fig. 1).
Spared Nerve Injury
In the ‘Spared Nerve Injury’ (SNI) model for neuropathic pain35, mice were anesthetized under 2% isofluran and the fur of the lateral part of the left tight was removed. The skin on the lateral surface of the thigh was incised and a section was made directly through the biceps femoris muscle exposing the sciatic nerve and its three terminal branches: sural, common peroneal and tibial nerves. The common peroneal and tibial nerve were tightly ligated with 5.0 silk and sectioned distal to the ligation, removing 2–4 mm of the distal nerve stump. These two nerves were subsequently cut and the sural nerve was left intact. Muscle and skin were closed in two layers. The mice were housed under standard conditions in cages for 3 days before the isolation of tissues or perfusion of animals.
In some experiments, Minocycline (Sigma), was injected i.p. into mice at a concentration of 30 mg/kg (dissolved in H20) 16 and 0.5 h before SNI operations and then twice daily for the duration of the experiment.
Generation of recombinant adeno-associated virions (rAAV) expressing SerpinA3N
In order to overexpress SerpinA3N unilaterally in L3 and L4 DRG of mice, the mouse Serpina3n cDNA sequence was amplified from RNA isolated from DRG of C57Bl/6 mice using AccuPrime™ Pfx SuperMix (Invitrogen). The DNA fragment corresponding to Serpina3n was gel-purified and TOPO-cloned using the Zero Blunt® TOPO® PCR Cloning Kit (Invitrogen). The TOPO-cloned Serpina3n was transformed into competent bacteria and minipreps of single colonies from this transformation reaction were prepared. After sequencing the clones, one single colony containing the right Serpina3n open reading frame (ORF) sequence was excised from the TOPO vector using appropriate digestion enzymes and then cloned into the pAM-MCS-QERK-stop vector.
Chimeric AAV2/8 virions (a mixture of serotypes 2 and 8) carrying the Serpina3n ORF as described above or GFP as a control were produced according to the protocol described in Luo et al 201336.
Intraganglionic (DRG) injections
We have previously described intraperenchymal injections into mouse DRG in vivo36. Mice were deeply anesthetized with sleep mix (Dormicum, Domitar and Fentanyl). The back hair was removed using a rodent trimmer and the eyes were covered with eye ointment (Bepanthen) to prevent them from drying out. Mice were fixed firmly in a stereotactic frame and placed on a heating plate to maintain the body temperature constant throughout the surgery. The iliac crest and last rib were identified through palpation of the dorsal surface of mice and the area between them was sterilized by wiping the skin with 70% ethanol. L3 and L4 lumbar vertebral columns were identified from the last rib and iliac crest and a small incision directly above the iliac crest and on top of L3 and L4 level was made using a sterile scalpel. The lateral processes of the vertebral bones were exposed by moving the corresponding muscles right at the inter-vertebral junctions. A small portion at the edge of the lateral process right on the top of the DRG was removed in order to expose a small portion of the DRG and a small glass pipette (tip diameter: 25 μm) filled with viral solution containing 0.01% Fast Green was inserted in the middle of the DRG. The quality of injection was determined from a gradual increase in the coloration of the DRG and any leakage was detected by visualization. After the injection, the exposed DRG was covered by saline soaked gelatin foam and muscles were moved back to their original position.
Intrathecal delivery of drugs
To enable intrathecal delivery of pharmacological agents at the level of lumbar spinal segments, mice were deeply anaesthetized with an intraperitoneal injection of 0.65 μl/g body weight of sleep mix [0.23 μg/μl Sedator (Eurovet International), 3.08 μg/μl Dormicum (Roche), 0.01 μg/μl Fentanyl-Janssen (Janssen-Cilag)] and a polyethylene catheter (Biomedical Instruments) was stereotactically inserted through an opening in the cisterna magna into the lumbar subarachnoid space at the L3–L4 segments. The tip of the catheter was located near the lumbar enlargement of the spinal cord. The volume of dead space of the i.t. catheter was 10 μl. Mice were allowed to recover for 3 days after surgery and animals showing any motor abnormalities were excluded from further experiments. The correct placement of the catheter was verified at the end of the experiment by i.t injection of 5 μl 1% Evans blue and performing a laminectomy. Drugs and recombinant proteins were administered intrathecally in the indicated dosage using a microinjection syringe (Hamilton) in a volume of 5 μl separated from an 8 μl volume of saline through a 1 μl air bubble. For every experiment, mice were randomly allocated to experimental treatments (vehicle or test substance, e.g. recombinant proteins or drugs) by a scientist independent from the experimentor.
Reverse transcription PCR
Total RNA was extracted from mouse L3–L4 DRGs, lumbar L3–L4 spinal cord segments and ipsilateral paw skin isolated from mice at 8 days post-SNI using the phenol/chloroform extraction method; similar methods were employed for extracting RNA from cultured DRG neurons and cultured embryonic spinal cord neurons. Total RNA from acutely isolated mouse dorsal spinal cord neurons and T-cells and from primary cell cultures of Schwann cells, astrocytes, microglia and macrophages were purchased from 3H Biomedical, Uppsala, Sweden. The RNA was reverse-transcribed using RevertAid™ M-MuLV Reverse Transcriptase (Fermentas), random hexamer and Oligo(dT) primers (Roche and Invitrogen, respectively) according to standard protocols. PCR reactions were then performed on cDNA from the aforementioned cell types and tissues using the following primers:
Elane (forward): 5′-GGC CCT TGG CAG ACT ATC CAG C-3′,
Elane (reverse): 5′-ACC TGC ACG TTG GCG TTA ATG G-3′,
18s RNA (forward): 5′-AGT TAT GGT TCC TTT GGT CGC TC-3′,
18s RNA (reverse): 5′-GTT ATT TTT CGT CAC TAC CTC CC-3′.
Paw tissue from WT mice 1 day post-SNI served as a positive control, 18s RNA served as a housekeeping gene and reverse-transcription reactions performed without (–) cDNA served as a negative control.
Real-Time quantitative PCR
qPCR reactions were performed on cDNA samples using theTaqMan® Gene Expression Assay (Invitrogen) including predesigned primers for Serpina3n, GAPDH and Cyclophilin A (Invitrogen). The reactions were performed using a LightCycler® 96Real Time PCR System (Roche) and the data analyzed using the corresponding software.
Gene expression analysis of Serpina3 isoforms was performed on DRG cDNA using the iQ™ SYBR® Green Supermixsystem (Bio-Rad Laboratories). Primers were directed against the non-conserved 3′ region of the transcripts, which encodes the protease interaction domain that defines target specificity. To ensure primer specificity, 10 of the 11 proposed functional Serpina3 transcripts were cloned from mouse tissue and each primer pair was tested for its ability to amplify only the appropriate Serpina3 clone. The following primer sequences were used:
| Gene | Forward primer 5′-3′ | Reverse primer 5′-3′ | Size (bp) |
|---|---|---|---|
| Serpina3a | TCATCACAATAGCCCGATAT | CCTTACCCATAACGCTAAT | 126 |
| Serpina3b | GCAATCACAATAGTCGGATAC | ATAACCTTGCCCATTACAT | 131 |
| Serpina3c | CTTAGTAGAAGAACCAGTC | AGAGAGTAGTCTGAACAT | 79 |
| Serpina3f | CTCCAATGTTGTCAAGGTGTA | CCACATTTAGGAAGTTCTCAT | 148 |
| Serpina3g | GTGTCGGATGTTGTGCAGTT | CACCATTTGGGAAGTTCAT | 147 |
| Serpina3h | GCAGGATGCAGCAGGTC | AGGATGTGCTCCAGGCTGTT | 139 |
| Serpina3j | AATATGACTTCCTGTCCACT | GAATCTGAGTGTAAAACACAG | 85 |
| Serpina3k | TATTGGTGGCATTCGTAAG | CTTTGGCCATAAAGAGGATAC | 107 |
| Serpina3m | ACAAACTATGACTGTGC | AAGAGGGTAGTCTGAACAC | 75 |
| Serpina3n | CCCTGAGGAAGTGGAAGAAT | CCTGATGCCCAGCTTTGAAA | 119 |
Northern Blotting was performed according to the protocol described by Costigan et al. 200237.
In situ hybridization
For rat DRG, in situ hybridization was performed on 10 μm cryosections of the L4 dorsal root ganglia using digoxigenin-labeled antisense riboprobes, as previously described30,38.
In situ hybridization was performed on mouse DRG sections as follows. DRGs L4–L5 were harvested from WT and Serpina3n−/− mice 1 day post-SNI, cryosectioned (10 μm), mounted on Superfrost Plus slides (VWR) and frozen at −80°C until use. A digoxigenin-labeled anti-sense cRNA probe against the floxed exon of Serpina3n was generated by a T7 (Roche) in vitro transcription reaction using a Serpina3n cDNA (5′:ACTGCAGAACACAGAAGATGGCCT3′:TCACCAGCACCATCAATGTCCTTTT). In situ hybridization was performed as previously described39. Following ON hybridization, slides were incubated with Alkaline phosphatase conjugated anti-digoxigenin antibody (Roche, 1:200) for 1 h at RT. After several washes in PBST, slides were incubated in NBT/BCIP chromogenic substrate (Roche) according to manufacturer’s specifications for 5 h. Slides were coverslipped and brightfield images were captured on a Nikon 80i Upright Microscope.
Antibodies
For Western blot analysis, we used a goat polyclonal anti-SerpinA3N antiserum (1:500; R&D Systems, Cat. # AF4709, no references available) and a mouse monoclonal anti-α-tubulin antibody (1:2000; Sigma, Cat. # T9026)40. For immunohistochemistry, we used a goat polyclonal anti-SerpinA3N antiserum (1:200; R&D Systems, Cat. # AF4709), a rabbit polyclonal anti-CGRP antiserum (1:2000; ImmunoStar, Cat. # 24112)41, a rabbit polyclonal anti-NF200 antiserum (1:500; Sigma, Cat. # N4142)42, a rat monoclonal anti-CD3 (clone 17A2) antibody (1:100; BD Biosciences, Cat. # 555273)43, a rabbit polyclonal anti-Glial Fibrillary Acid Protein (GFAP) antibody (1:500; Millipore, Cat. # AB5804)44, biotinylated Isolectin B4 (1:100; Vector laboratories, Cat. # B-1205)41, a biotinylated rat anti-Gr-1 antibody (clone RB6-8C5) (1:500; BD Biosciences, Cat. # 553125)45, a rabbit polyclonal Iba-1 antiserum (1:500; Wako, Cat. # 019-19741)41, a rabbit-anti-ATF3 polyclonal antiserum (1:200; Santa Cruz Biotechnology, Cat. # sc-188)46, an anti-β-Tubulin Isotype III (clone SDL.3D10) monoclonal antibody (1:800; Sigma, Cat. # T5076)47, an anti-NeuN (clone A60) monoclonal antibody (1:200; Millipore, Cat. # MAB377)48 and a donkey anti-rabbit-Cy3 antiserum (1:300; Jackson ImmunoResearch, Cat. # 711-165-152)49 For FACS analysis, the following antibodies were used: FITC Rat Anti-Mouse CD4 (Clone RM4-5) (1:100; BD Biosciences, Cat. # 553046)50, Alexa Fluor® 647 anti-mouse CD8a (1:100; Biolegend, Cat. # 100727)51, PE anti-mouse/human CD45R/B220 (1:200; Biolegend, Cat. # 103207)52, PE/Cy7 anti-mouse Ly-6G/Ly-6C (Gr-1) (1:2000; Biolegend, Cat. # 108416)53 and Pacific Blue™ anti-mouse/human CD11b (1:100; Biolegend, Cat. # 101224)52.
Tissue preparation
For immunohistochemistry, animals were perfused transcardially with PBS followed by 4% PFA. Spinal cord segments at lumbar level L3–L4 and the L3 and L4 DRGs were then removed. Paw skin punches where taken out using a biopsy punch. Tissues were post-fixed for up to 16 h in 4% PFA at 4 °C. Spinal cords, DRGs and paw skin punches were stored in 0.5% PFA at 4 °C for up to 2 weeks and incubated in 30% sucrose solution ON at 4 °C for cryostat sectioning (25 μm, 16 μm and 20 μm corresponding to the thickness per section, respectively).
DAB immunohistochemistry
The immunohistochemical staining procedure started with the incubation in 1% hydrogen peroxidase (in PBS:Methanol 1:1) followed by 3 times of 15 min washing with PBST and 30 min incubation in 7% normal horse serum in 0.02% PBST (blocking solution). Sections were incubated ON with the desired primary antibody in blocking solution at 4 °C. The next day, sections were washed 3 times for 15 min with 0.02% PBST, followed by incubation with the secondary biotinylated antibody for 30 min (Vectastain Elite ABC Kit, Vector laboratories). After three 15 min washing steps with 0.02% PBST and incubation for 30 min in an avidin-biotin complex solution (Vectastain Elite ABC Kit), sections were washed two times for 15 min in 0.02% PBST and twice for 15 min in PBS before stained with 3,3′-diaminobenzidine tetra hydrochloride (DAB) solution (DAB substrate Kit for Peroxidase, Vector laboratories, Burlingame, USA). Staining reactions were terminated upon visual inspection by adding and washing with water. Sections were finally air-dried and mounted with Mowiol. Bright field colored images were captured with an automated Leica DM4000 B microscope coupled to a MBF CX9000 camera (MBF Bioscience) and displayed with PictureFrame™ software.
Immunofluorescence
Tissue sections were permeabilized for 15 min in 0.5% PBST (Triton X100 in PBS) followed by incubation for 15 min in ice cold 50 mM Glycine in PBS and 2 washings for 15 min in PBS. Afterward the slides were blocked for 1 h in 10% NHS in 0.5% PBST and incubated ON at 4 °C with the appropriate primary antibody in 1% NHS in 1% PBST. The next day, sections were washed 3 times for 15 min in PBS. Alexa Fluor® 488- or Alexa Fluor® 594-conjugated secondary antibodies (Life Technologies) were diluted 1:1000 in 1% NHS in 0.1% PBST and incubated on the sections for 1 h in the dark. Sections were then washed 3 times for 10 min with PBS and incubated for 10 min in 10 mM Tris-HCl pH 8.0 in the dark before mounting with Mowiol. For CD3 staining, we followed a previously described protocol43. For immunostaining of ATF3 on ISH sections for Serpina3n, at completion of ISH the slides were washed in H20 and blocked in blocking buffer (1% BSA, 0.1% triton) for 1 h. Slides were then incubated with rabbit-anti-ATF3 antibody ON at 4 °C. Slides were afterward washed in PBST and incubated in anti-rabbit-Cy3 antiserum for 1 h at RT. After images were captured, the ISH signal was pseudocolored green and overlaid on ATF3 staining. Fluorescence images were obtained using a Laser scanning spectral confocal microscope (Leica TCS SP2 AOBS, Bensheim, Germany) and Leica Confocal Software (v2.61) or a Nikon A1 confocal laser microscope system and the corresponding software. For GFP- and TRITC-fluorescence, a sequential Scan mode was used to rule out bleed-through between channels.
Quantitative analysis of stained cells
For images derived from co-immunostainings between SerpinA3N and NF200, IB4, CGRP or ATF3, the number of double-positive cells was determined using Fiji (ImageJ) software. The cell size of SerpinA3-like immunoreactive cells corresponded to Feret’s diameter, which is defined as “the longest distance between any two points along the selection boundary”. All images from the same experiment were processed similarly. Afterward, every positive cell within a tissue section was labeled with a number and those cells exhibiting positive immunoreactivity for two antibodies were considered double-positive. Quantification of Iba-1 immunoreactivity in spinal cord sections were done using Fiji software and corresponded to the average cell number of 3 representative ROI (regions of interest) of 1002 microns from each section. For these images, a deconvolution step was included to reduce background noise. The number of CD3-positive cells within a tissue section was estimated by counting them directly from the microscope. The final value corresponded to the average number of CD3-positive cells within every section from the same experimental group.
Western blotting
DRGs and spinal cord segments at the lumbar level L3–L4 were mechanically homogenized and lysed in ice cold RIPA buffer with complete, Mini, EDTA-free Protease inhibitor tablets (Roche) for 1 h at 4 °C. Lysates were then centrifuged for 20 min at 13.000 rpm at 4 °C to remove insoluble material. Protein concentration was then determined using the BCA method (BCA Protein Assay Kit, Thermo Scientific). The samples were separated on 10% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane. The membranes were blocked in 5% milk in TBST, incubated ON with the primary antibody in the same solution and washed 3 times for 15 min in TBST. The membranes were incubated with the secondary antibody for 1 h and washed again as before. For the α-tubulin loading control, the membranes were incubated 30 min with striping buffer (200 mM Glycine, 0.1% SDS, 1% Tween-20, pH 2.2) at RT, blocked, and incubated with the primary and secondary antibodies as described above. Detection of immunoreactive bands was performed using the enhanced chemiluminescence detection system (ECL, GE Healthcare). The intensity of immunoreactive bands was quantified using ImageJ software.
Culture of DRG neurons
5–8 weeks old female and male C57BL/6 mice were anesthetized with CO2 and killed by decapitation. DRGs were quickly collected in cold PBS and transferred to F12 medium without serum containing 1% penicillin/streptomycin. After removing axons and meningeal tissue, DRGs from each animal (30–40) were digested for 30 min at 37 °C in 1 ml of an enzymatic solution (0.25 mg/ml Trypsin [6000BAEE units/mg, Sigma], 1 mg/ml collagenase [300 units/mg, Sigma] and 0.2 mg/ml DNAse I [2.580 Kunitz units/mg solid, Sigma] in F12 medium without serum) using a thermomixer (max. velocity 14000 rpm). After mechanical dissociation with a glass pipette pre-coated with Sigmacote® solution (Sigma), the digestion was stopped by adding 120 μl of 10X Trypsin inhibitor (2.5 mg/ml, 10000 BAEE units/mg protein, Sigma) and 100 μl FBS. Cells were mixed well and centrifuged at 1000 rpm for 3 min. The supernatant was removed and the cells were carefully resuspended in 2 ml F12 medium containing 10% FBS. The cell suspension (2 ml, max. 2 mice) was carefully loaded in a tube containing a discontinuous Percoll gradient (1.5 ml 35% Percoll [Percoll: 90% Percoll (Amersham) + 10% 10x PBS] in F12 medium plus 2.5 ml 25% Percoll in DMEM medium) to allow separation of neurons from non-neuronal cells. Following centrifugation at 1000 g for 15 min, the upper part of the supernatant was removed and the lower 1–1.5 ml portion was left intact. The pellet was resuspended and mixed with F12 medium up to 13.5 ml and additional 1.5 ml 10% FBS were added and mixed with the solution. After 10 min centrifugation at 500 g, the supernatant was removed and cells were resuspended in 6 ml F12 medium plus 10% FBS. The cell suspension was mixed carefully and centrifuged 7–8 min at 500 g. The cells were resuspended in 400 μl F12 medium plus 10% FBS and plated on a Petri dish (35 mm diameter). After short time, 1.6 ml medium were added to the cells and they were incubated ON at 37 °C with 5% CO2. The next day the medium was changed with F12 medium plus 10% FBS containing a mixture of growth factors (BDNF [10 ng/ml], NGF [10 ng/ml], GDNF [5 ng/ml], NT3 [5 ng/ml], Sigma) and the mitotic inhibitor AraC (5 μM, Sigma). Cells were incubated at 37 °C for 4–7 days until treatment.
To study release of SerpinA3N from cultured DRG neurons, all DRGs from one 5-weeks old WT mouse were collected, split into 3 wells of 12-well plates and resuspended in 1 ml F12 medium plus 10% FBS per well. 4 days later, 750 μl medium were removed from each well and 500 μl serum-depleted medium containing KCl or vehicle alone were added into each well and incubated at the desired concentrations and incubation times. Cell supernatants were analyzed by Western blotting. For quantification, intensity values of bands corresponding to SerpinA3N in the supernatants were normalized to tubulin expression in the corresponding cell lysates to ensure equal quantities of cellular input.
Embryonic spinal cord cultures
E16 embryos were removed and placed in a 10 cm Petri dish containing 0.33 M glucose/PBS. After removing of the dura, spinal cords were collected in 0.33 M glucose/PBS and cut in small pieces. The dissociated tissue (2–3 spinal cords) was transferred to 1 ml of enzyme solution (the same used for DRG cultures) and digested for 15 min at 37 °C. The digestion was stopped with Trypsin inhibitor (the same used for DRG cultures), mixed gently and centrifuged for 10 min at 1000 rpm. The supernatant was removed and the cells resuspended in 0.5–1 ml complete medium (Neurobasal medium A, 2 mM Glutamax, 1x B27 supplement and 1x penicillin/streptomycin). Cells were counted using a Neubauer chamber, diluted to 60.000 cells/ml and plated on a 12-well plate (1 ml cells/well). The day after, half of the medium was changed with new medium containing 5-Fluorouracil and Uridin (15 μg/ml and 35 μg/ml respectively, Sigma). Cells were kept in culture at least 3 weeks changing half of the medium every 3–4 days.
Adoptive transfer experiments
For purification of splenic T-cells, Macs cell sorting was performed using Dynabeads (untouched mouse T-cells, Invitrogen) and neutrophils were depleted using biotinylated rat anti-Gr-1 antibody (clone RB6-8C5) as described in details previously54.
Mice were injected i.v. with 200 μl of purified T-cells in sterile PBS at a concentration of 1×107/200 μl, 6 days prior to SNI. Surface staining was performed via flow cytometry according to standard procedures. Data acquisition was carried out with an 8-color flow cytometer (Canto II, BD Biosciences) and analyzed with FlowJo (Treestar).
Fluorometric measurements of protease activity
The proteolytic activity of the desired proteases was measured through the fluorescence released after protease-induced cleavage of specific fluorogenic substrates. With the exception of the dose-response assay of sivelestat effect over rLE activity (which was performed in an Infinite® 200 PRO multimode reader from Tecan), all fluorescence measurements were performed using Fluoroskan Ascent (Thermo Electron LED GmbH) and expressed as AFU (arbitrary fluorescence units). For all experiments, the following parameters were used: kinetic-mode, 20 ms integration time, 1 s lag time, 30 s interval time. Ascent Software 2.4.2 (Thermo Scientific) was used to analyze the measurements. All reactions were done in a 96-well plate format in triplicate using a final reaction volume of 100 μl. BSA was used as negative control for rSerpinA3N activity. In all experiments, the concentration of BSA was equal to the highest concentration of rSerpinA3N tested. The tubes containing the fluorogenic substrates were always covered with aluminium foil prior to the measurements.
The activity of mouse recombinant LE (R&D Systems) was measured using the fluorogenic substrate MeOSucAAPV-AMC (Bachem) as described by the manufacturer. Pre-mixes containing a fixed amount of rLE and increasing concentrations of rSerpinA3N or Sivelestat (Sigma) were prepared in assay buffer (50 mM Tris, 1 M NaCl, 0.05% (w/v) Brij-35, pH 7.5). The solutions containing the proteins were mixed with equal volumes of substrate in assay buffer reaching a final substrate concentration of 100 μM. The samples were placed in the multi-well plate and the reading was measured immediately with the fluorescence reader at excitation and emission wavelengths of 380 nm and 460 nm (respectively) for 30 min.
The activity of purified active mouse alpha thrombin (Cell Systems Biotechnologie) was measured by its ability to cleave the fluorogenic peptide substrate benzoyl-FVR-AMC (Merck). Pre-mixes containing a fixed amount of thrombin and increasing concentrations of mouse rSerpinA3N (R&D Systems) in assay buffer (50 mM Tris-HCl, 300 mM NaCl, pH 8.2) were prepared in different tubes and incubated at RT for 10 min. The solutions containing the proteins were mixed with equal volumes of substrate in assay buffer reaching a final substrate concentration of 50 μM. The samples were placed in the multi-well plate and the reading was measured immediately with the fluorescence reader at excitation and emission wavelengths of 380 nm and 460 nm (respectively) for 20 min.
The activity of MMP-9 was measured using the FRET substrate Mca-PLGL-Dap(Dnp)-AR-NH2 (MoBiTec). To test SerpinA3N effect on MMP-9 activity, mouse recombinant pro-MMP-9 (10 ng/μl, MoBiTec) was first activated in 1 mM APMA, followed by incubation at 37 °C for 2 h. Pre-mixes containing a fixed amount of active MMP9 and increasing concentrations of rSerpinA3N were prepared in pre-warmed assay buffer (50 mM Tris, 10 mM CaCl2, 150 mM NaCl, 0.05 % Brij-35, pH7.5) and incubated at 37 °C for 10 min. For the HLE-induced MMP-9 activity assay, samples containing a fixed amount of HLE and increasing concentrations of SerpinA3N were prepared in assay buffer. For MMP9 activation, every sample was mixed with pro-MMP-9 (10 ng/μl) in pre-warmed assay buffer and incubated for 2 h at 37 °C. For both assays, the solutions containing the proteins were mixed with equal volumes of substrate in assay buffer reaching a final substrate concentration of 10 μM. The final mixtures were added to the plate and the reading was performed at excitation and emission wavelengths of 320 nm and 400 nm (respectively) for 20 min.
Measurement of LE activity in ex vivo DRG explants
The activity of LE in DRG was measured using the LE ratiometric probe NEmo-2, a ratiometric FRET-based monitoring LE reporter that detects the activity of membrane-associated LE, but not soluble LE55. Briefly, L4 DRGs from operated mice were isolated and incubated immediately with 50 μM NEmo-2 dissolved in F12 medium for 60 min at 37 °C protected from light. DRGs were shortly washed with PBS, fixed for 48 h in 4 % PFA and postfixed for 16 h in 0.5 % PFA followed by ON incubation in 30% sucrose for cryosectioning. DRGs were cut in 40 μm sections and images from every section were taken using a Leica SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany) equipped with a 20×1.4 dry objective with the following settings: The donor coumarin 343 of NEmo-2 was excited with a 405 nm diode laser and emission was sampled between 470–510 nm. Sensitized emission of the acceptor TAMRA was sampled between 570–610 nm. The pinhole was set to 300 μm. For the quantification of LE activity in ex-vivo explants of DRG, images were processed with ImageJ 1.38r software (http://rsb.info.nih.gov/ij/) using background subtraction, exclusion of saturated pixels, smoothing with a median filter, thresholding and calculation of donor/acceptor (D/A) ratio images from which the activity was quantified – these were uniformly applied over all images by an observer unaware of the identity of the groups. Because the probe accumulated unspecifically along the outer rim of all sections, this region (corresponding to 20 pixels thickness) was eliminated from each section and the region corresponding to the somata of DRG neurons was selected as region of interest. About 8 slices were analyzed per DRG and FRET intensity was averaged per DRG. The representative images shown in the manuscript were created by transformation to the LUT range. Values in Fig. 4h were calculated as % change over values derived from sham-treated WT mice and negative values in Elane−/− mice are indicative of basal LE activity in WT sham-treated mice. The experimenter was blinded to the identity of the mice from which the DRGs were derived for measurement of LE activity.
Nociceptive tests
All animal procedures were approved by the local governing bodies (Regierungspräsidium Karlsruhe, Germany, local ethics committees at Harvard University, USA and University of Jerusalem, Israel). For all experiments involving behavior, operations and delivery of drugs, 8–10 weeks old female and male (equal numbers) mice of the C57BL/6 strain were used.
All experimenters were blinded to the identity of the mice they were analyzing.
Application of mechanical force using von Frey filaments (static punctate mechanical stimuli)
Before testing, mice were habituated in a small plastic (7.5 × 7.5 × 15 cm) cage for 1 h. Mechanical sensitivity was determined with a graded series of nine von Frey filaments that produced a bending force of 0.02, 0.04, 0.07, 0.16, 0.4, 0.6, 1, 1.4 or 2 g. The stimuli were applied within the sural nerve territory (lateral part of the hindpaw). Each filament was tested 10 times in increasing order starting with the filament producing the lowest force. Von Frey filaments were applied at least 3 s after the mice had returned to their initial resting state. For baseline mechanical sensitivity test all filaments were applied and the number of withdrawals was recorded. For tactile allodynia: the minimal force filament for which animals presented either a brisk paw withdrawal and/or an escape attempt in response to at least 5 of the 10 stimulations determined the mechanical response threshold. Mechanical thresholds were defined as the minimum pressure required for eliciting 60% or 40% of withdrawal responses out of 5 stimulations and measured in grams (force application). To represent responsivity to all filament strengths, stimulus intensity (force)-response frequency curves were constructed per time point tested and the integral (area under the curve) thereof was represented per time point. Responses to plantar pinprick stimulation and plantar acetone were recorded as duration of the time spent by the animals licking, flinching or biting their paws.
Contact heat pain (Hot plate test)
Mice were placed on a metallic plate heated to a set temperature (30, 49, 52 or 55 °C) within an acrylic container (Bioseb, France), and the latency for flinching, licking one of the hindpaws, or jumping was measured. Mice were sequentially tested for 30, 52, 55 and 49 °C. One temperature was tested per day.
Radiant heat pain (Hargreaves test)
Before testing, mice were placed on an elevated glass surface and habituated in their individual cage for 30 min. Then a radiant heat source (beam intensity set to cause baseline latencies in C57Bl6/j mice ~15 s) targeted at an individual paw and the latency to paw withdrawal measured. The radiant heat apparatus used was from Ugo Basile (Italy).
Brush test (dynamic mechanical stimuli)
Mice were habituated in a small plastic (7.5 × 7.5 × 15 cm) cage for 1 h and then three successive gentle touch stimuli were applied with a paint brush of round head diameter of 2 mm (Princeton brush Co.) onto the sural territory of the paw. Each stimulation was applied from the middle of the foot to its distal part and lasted approximately 1 s. The total time spent flinching or licking the paw was measured with a stopwatch.
Statistics
For all measurements, data was calculated and presented as mean ± standard error of the mean (S.E.M.). Sample size was chosen to ensure adequate power to detect effect sizes using G-POWER program.
Two-tailed unpaired T-test, one-way Analysis of Variance (ANOVA) followed by post-hoc Tukey’s test or two-way ANOVA for random measures or repeated measures followed by Tukey’s post-hoc test were used to determine statistical significance. P ≤ 0.05 was considered significant. For all statistical analyses, the appropriate statistical tests were chosen, the data met the assumptions of the test and the variance between the statistically compared groups was similar. SigmaPlot or ORIGIN were employed for statistical analyses.
Supplementary Material
Acknowledgments
The authors are grateful to R. LeFaucheur for secretarial assistance and to D. Baumgartl-Ahlert for technical assistance. We acknowledge support from the Interdisciplinary Neurobehavioral Core, Heideiberg for the behavioral experiments performed here. We are grateful to M. Meister for his help in the analysis of FACS data. This work was supported by an ERC Advanced Investigator Grant (PAINPLASTICITY; project no. 294293) to R. Kuner, the Deutsche Forschungsgemeinschaft (DFG MA 2081/4-1) to M. Mall., the Deutsche Forschungsgemeinschaft (SFB 938) to B. Arnold., from the US National Institute of Health (5 R01 NS038253 and 2R37NS039518) to C. Woolf and R01 NS074430 to M. Costigan, a grant from the Israel Science Foundation to M. Devor. M. Mall and R. Kuner are members of the Molecular Medicine Partnership Unit, Heidelberg. R. Kuner is a principal investigator in the Excellence Cluster ‘CellNetworks’ of Heidelberg University. L. Vicuña was partially supported by a PhD fellowship from CellNetworks and by the Hartmut-Hoffmann Berling International Graduate School for Cellular & Molecular Biology. M. Simonetti was partially supported by a post-doctoral fellowship from CellNetworks. A. Latremoliere was partially supported by the RO1DE022912 grant.
Footnotes
Author contributions:
L.V., D.E.S., A.L., K.K.B., M.S., D.H., S.P., P.R., R. S. G., C.N., S.G. and M.C. performed experiments and analyzed data; R. K., M.C., C.J.W., S.D.L., M.D., B.A., M.A.M. designed and supervised experiments; R.K., L.V. and M.C. primarily wrote the manuscript.
Competing Interests: The authors have patent applications pending based on the above-described data.
References
- 1.Costigan M, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133:2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nissenbaum J, et al. Susceptibility to chronic pain following nerve injury is genetically affected by CACNG2. Genome Res. 2010;20:1180–1190. doi: 10.1101/gr.104976.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorge RE, et al. Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nat Med. 2012;18:595–599. doi: 10.1038/nm.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tegeder I, et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med. 2006;12:1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- 5.Persson AK, et al. Correlational analysis for identifying genes whose regulation contributes to chronic neuropathic pain. Mol Pain. 2009;5:7. doi: 10.1186/1744-8069-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devor M, Raber P. Heritability of symptoms in an experimental model of neuropathic pain. Pain. 1990;42:51–67. doi: 10.1016/0304-3959(90)91092-W. [DOI] [PubMed] [Google Scholar]
- 7.Ziv-Sefer S, Raber P, Barbash S, Devor M. Unity vs. diversity of neuropathic pain mechanisms: Allodynia and hyperalgesia in rats selected for heritable predisposition to spontaneous pain. Pain. 2009;146:148–157. doi: 10.1016/j.pain.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath AJ, et al. The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins. J Biol Chem. 2005;280:43168–43178. doi: 10.1074/jbc.M505598200. [DOI] [PubMed] [Google Scholar]
- 10.Sipione S, et al. Identification of a novel human granzyme B inhibitor secreted by cultured sertoli cells. J Immunol. 2006;177:5051–5058. doi: 10.4049/jimmunol.177.8.5051. [DOI] [PubMed] [Google Scholar]
- 11.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 12.Lee WL, Downey GP. Leukocyte elastase: physiological functions and role in acute lung injury. Am J Respir Crit Care Med. 2001;164:896–904. doi: 10.1164/ajrccm.164.5.2103040. [DOI] [PubMed] [Google Scholar]
- 13.Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 14.Garcia PS, Gulati A, Levy JH. The role of thrombin and protease-activated receptors in pain mechanisms. Thrombosis and haemostasis. 2010;103:1145–1151. doi: 10.1160/TH09-12-0848. [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki Y, et al. Distinct roles of matrix metalloproteases in the early- and late-phase development of neuropathic pain. Nat Med. 2008;14:331–336. doi: 10.1038/nm1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferry G, et al. Activation of MMP-9 by neutrophil elastase in an in vivo model of acute lung injury. FEBS Lett. 1997;402:111–115. doi: 10.1016/s0014-5793(96)01508-6. [DOI] [PubMed] [Google Scholar]
- 17.Jackson PL, et al. Human neutrophil elastase-mediated cleavage sites of MMP-9 and TIMP-1: implications to cystic fibrosis proteolytic dysfunction. Mol Med. 2010;16:159–166. doi: 10.2119/molmed.2009.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawabata K, et al. ONO-5046, a novel inhibitor of human neutrophil elastase. Biochem Biophys Res Commun. 1991;177:814–820. doi: 10.1016/0006-291x(91)91862-7. [DOI] [PubMed] [Google Scholar]
- 19.McMahon SB, Malcangio M. Current challenges in glia-pain biology. Neuron. 2009;64:46–54. doi: 10.1016/j.neuron.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 20.Scholz J, Woolf CJ. The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci. 2007;10:1361–1368. doi: 10.1038/nn1992. [DOI] [PubMed] [Google Scholar]
- 21.Hu P, Bembrick AL, Keay KA, McLachlan EM. Immune cell involvement in dorsal root ganglia and spinal cord after chronic constriction or transection of the rat sciatic nerve. Brain Behav Immun. 2007;21:599–616. doi: 10.1016/j.bbi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Kim CF, Moalem-Taylor G. Detailed characterization of neuro-immune responses following neuropathic injury in mice. Brain Res. 2011;1405:95–108. doi: 10.1016/j.brainres.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 23.Gehrig S, Mall MA, Schultz C. Spatially resolved monitoring of neutrophil elastase activity with ratiometric fluorescent reporters. Angew Chem Int Ed Engl. 2012;51:6258–6261. doi: 10.1002/anie.201109226. [DOI] [PubMed] [Google Scholar]
- 24.Costigan M, et al. T-cell infiltration and signaling in the adult dorsal spinal cord is a major contributor to neuropathic pain-like hypersensitivity. J Neurosci. 2009;29:14415–14422. doi: 10.1523/JNEUROSCI.4569-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10:1845–1864. doi: 10.1101/gr.gr-1478r. [DOI] [PubMed] [Google Scholar]
- 26.Irving JA, et al. Serpins in prokaryotes. Mol Biol Evol. 2002;19:1881–1890. doi: 10.1093/oxfordjournals.molbev.a004012. [DOI] [PubMed] [Google Scholar]
- 27.Alam SR, Newby DE, Henriksen PA. Role of the endogenous elastase inhibitor, elafin, in cardiovascular injury: from epithelium to endothelium. Biochemical pharmacology. 2012;83:695–704. doi: 10.1016/j.bcp.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Reardon C, et al. Thymic stromal lymphopoetin-induced expression of the endogenous inhibitory enzyme SLPI mediates recovery from colonic inflammation. Immunity. 2011;35:223–235. doi: 10.1016/j.immuni.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.