Abstract
Objective:
To quantify genetic overlap between migraine and ischemic stroke (IS) with respect to common genetic variation.
Methods:
We applied 4 different approaches to large-scale meta-analyses of genome-wide data on migraine (23,285 cases and 95,425 controls) and IS (12,389 cases and 62,004 controls). First, we queried known genome-wide significant loci for both disorders, looking for potential overlap of signals. We then analyzed the overall shared genetic load using polygenic scores and estimated the genetic correlation between disease subtypes using data derived from these models. We further interrogated genomic regions of shared risk using analysis of covariance patterns between the 2 phenotypes using cross-phenotype spatial mapping.
Results:
We found substantial genetic overlap between migraine and IS using all 4 approaches. Migraine without aura (MO) showed much stronger overlap with IS and its subtypes than migraine with aura (MA). The strongest overlap existed between MO and large artery stroke (LAS; p = 6.4 × 10−28 for the LAS polygenic score in MO) and between MO and cardioembolic stroke (CE; p = 2.7 × 10−20 for the CE score in MO).
Conclusions:
Our findings indicate shared genetic susceptibility to migraine and IS, with a particularly strong overlap between MO and both LAS and CE pointing towards shared mechanisms. Our observations on MA are consistent with a limited role of common genetic variants in this subtype.
Migraine is a primary headache disorder characterized by recurrent attacks of severe, often throbbing, headache associated with autonomic dysfunction. Although the majority of patients have migraine without aura (MO), one third have headaches preceded by transient neurologic disturbances (migraine with aura [MA]).1 Ischemic stroke (IS) is etiologically heterogeneous and a leading cause of premature death and disability.2
Results of epidemiologic studies show increased risk of IS in migraine patients.3 A large meta-analysis of case-control and observational cohort studies reported an increased risk of IS for patients with MO and MA,4 whereas more recent meta-analyses reported the association to be restricted to MA.3,5,6 Pathophysiology linking these neurovascular disorders remains poorly understood; suggested mechanisms include cortical spreading depression,7 endothelial dysfunction,8 enhanced platelet activation,9 and vasoconstriction.10
Recent genome-wide association studies (GWAS) identified common genetic variants associated with migraine11 and its subtypes MO12 and MA.13 Similarly, GWAS results point to variants associated with IS subtypes such as large artery atherosclerotic14,15 and cardioembolic.16 We combined GWAS from the International Headache Genetics Consortium (IHGC)11 and METASTROKE15 to assess shared genetic bases for migraine and IS. We first examined known genome-wide risk loci in the respective phenotypes. Using 2 methodologies, we then evaluated shared genetic risk for migraine with IS: (1) analysis of shared polygenic risk with subsequent estimation of genetic correlation between phenotypes and (2) detailed investigation of overlapping regions.
METHODS
Standard protocol approvals, registrations, and patient consents.
Ethics statement.
For all study cohorts, all participants gave informed consent and local research ethics boards approved all protocols.11,15
Cohorts.
Investigators of the IHGC study, a meta-analysis of GWAS data, enrolled 23,285 patients with migraine and 95,425 population-based or headache-free controls from 29 studies.11 When possible, researchers considered subclassifications of migraine with (MA: 5,118 cases vs 74,239 controls) or without aura (MO: 7,107 cases vs 69,427 controls). The METASTROKE study consists of combined data from 15 GWAS of IS (12,389 cases vs 62,004 controls).15 We used TOAST criteria17 to classify IS as large artery stroke (LAS) (2,167 cases/49,159 controls from 11 studies), cardioembolic stroke (CE) (2,365 cases/56,140 controls from 13 studies), and small vessel disease (SVD) (1,894 cases/51,976 controls from 12 studies) (tables e-1 and e-2 on the Neurology® Web site at Neurology.org).11,15 We removed overlapping controls between the migraine and stroke samples from deCODE, WTCCC2 (B58C and KORA), and the Rotterdam studies from the stroke datasets for polygenic score analyses, cross-phenotype spatial mapping (CPSM), and correlation analyses to avoid inflation of statistics.
Genome-wide association studies.
Both the IHGC migraine11 and METASTROKE15 studies consisted of independently performed genome-wide single nucleotide polymorphism (SNP) genotyping using standard technologies and imputation to HapMap release 21 or 22 CEU phased genotype18 or 1000 Genome reference panels. Investigators contributed summary statistical data from association analyses using frequentist additive models for meta-analysis after application of appropriate quality control measures (e-Methods). Subtle differences in allele frequencies between migraine and stroke could lead to deviation from the expected test statistic. Thus, as a final quality control step, we meta-analyzed results from the IHGC study and the METASTROKE study and constructed quantile-quantile plots (figure e-1).
Statistical analysis.
For analysis of previously discovered risk loci for IS or migraine, we extracted relevant loci from the literature and the 2 described consortia.11,15 We examined all SNPs within a window of ±50 kb surrounding the original reported risk SNP (coordinates from human genome build hg18) and reported the most significant p values of all genotyped or imputed SNPs within this window. We applied Bonferroni correction for association, integrating all tested SNPs for IS risk loci (650 tested SNPs), migraine risk loci (1,175 tested SNPs), and MO risk loci (213 tested SNPs) with resulting p value thresholds of 7.69E-5, 4.25E-5, and 2.30E-4, respectively.
Polygenic scores reveal combined effects of multiple nonsignificant variants derived from a derivation sample and tested in an independent replication sample. We derived polygenic scores for multiple p value cutoffs (0.5, 0.25, 0.1, 0.05, 0.01, 0.001, and 0.0001) in derivation samples. Further, we performed testing on summary statistics using the grs.summary function of the gtx package for R, a technique previously used in multiple studies, which estimates the polygenic component with high reliability.19 We use the term replication to describe analyses across phenotypes.
Use of linkage disequilibrium (LD) pruned data (r2 > 0.25) ensured approximate independence of SNPs. We retained the SNP with the lowest p value in an independent region and calculated the proportion of variance explained in the testing set by the polygenic scores using Nagelkerke's pseudo R2. Outcome measures include the p value of the association of the polygenic score in the testing dataset and the variance explained.
CPSM identifies genomic windows exhibiting similar association patterns across 2 phenotypes using a signal processing approach. CPSM allows analysis of pleiotropy across multiple diseases. Peak heights serve as an intuitive measure for description of shared risk loci in different phenotypes. This method corrects for background noise in the p value distribution and is thus superior to comparisons of single p values. We computed Pearson covariance between p values from 2 traits across a 60-kb sliding window. In each iteration, the window slides to the next SNP; thus, we obtained a covariance coefficient for each SNP in the analysis. We then detected signal peaks across the genome in the covariance trace and calculated the signal sn for a given SNP with index n, position bn (base pairs), and association p values p1,n, p2,n for 2 phenotypes as follows:
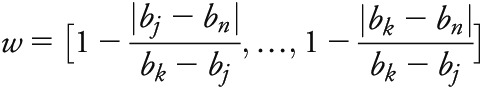 |
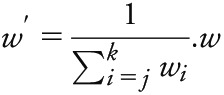 |
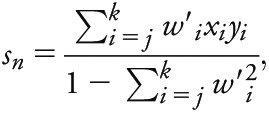 |
where each bi Є bj,…bk is the position of SNPi within the window of SNPsj,…,k containing SNPn. For a given window size d (base pairs), the window of SNPsj,…,k is defined such that j is the smallest SNP index where 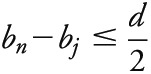 and k is the largest SNP index where
and k is the largest SNP index where 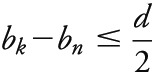 .
.
After constructing the CPSM signal for all SNPs, we corrected for strong associations identified in only one phenotype by permuting the association p values for one phenotype 1,000 times while holding the other phenotype constant, and then recalculating CPSM signals. From the total set of 2,000 permutation signals (1,000 per phenotype), we subtracted the upper 0.95 quantile at each SNP as the background signal threshold from the observed covariance as a correction. We then identified regions of shared association as peaks above the 99.95 (approximately corresponding to a height of 1.5) percentile of the covariance signal. We highlighted regions with a height >2.5 (corresponding to approximately 99.985 percentile) and with a height >5 (corresponding to approximately 99.998 percentile). CPSM only provides a signal when the effect in 2 traits is the same, implying shared causality in the discovered regions.
Utilizing a recently developed framework for polygenic analyses and based on the number of SNPs, the dataset sample sizes, and estimates of disease prevalence and pseudo-heritability, we estimated the power to detect an association indexing on a given degree of genetic correlation between the 2 phenotypes. We used the same framework, including p values from polygenic analysis, to estimate the overall degree of genetic correlation between each of the IS and migraine phenotypes, a posteriori to the polygenic analysis. We estimated genetic correlation in both the forward direction (using results from polygenic analysis of IS and subtypes as a discovery sample and migraine and subtypes as a replication sample) and the reverse direction (using results from the polygenic analysis from migraine and subtypes as a discovery sample and stroke and subtypes as a replication sample) to evaluate consistency of results using the estimateCorrFromP method. An implementation of the procedure was downloaded from http://sites.google.com/site/fdudbridge. This method approximates SNP correlation from cross-disorder applications of polygenic scores and can be compared to GREML-SNP genetic correlation. All analyses used R statistical software (http://www.R-project.org). Using stroke prevalence data from the British Heart Foundation for IS20 (1.7% in the United Kingdom) and the proportional incidence of IS events from all stroke events in the OXVASC study21 (59%), we estimated the prevalence of IS (∼1%). We then used the proportion of IS subtypes (CE, LAS, or SVD) from a meta-analysis of population-based incidence studies22 to estimate the prevalence of each subtype. We estimated stroke heritability on a liability scale.23 Although we acknowledge that migraine prevalence may vary across countries, we estimated migraine prevalence to be 17% for all migraine, 11% for MO, and 5% for MA based on published data.1,24 Migraine heritability estimates vary in the literature, with MA being highest. We chose heritability measures of 0.65 for MA25 and 0.61 for MO26 and a more conservative measure of 0.57 for all migraine.
RESULTS
Information on clinical subtypes was available for 12,225 (52.5%) of the migraine and for 6,426 (51.9%) of the IS patients (tables e-1 and e-2). We identified 38,338 potentially overlapping controls and excluded them from analyses where necessary. QQ plots revealed no inflation of test statistics (lambda inflation factors below 1.05 in all analyses of migraine subtypes vs all IS; figure e-1 and e-Methods).
All migraine.
We first evaluated risk loci identified in previous GWAS on IS or its subtypes,15 in all migraine11 and vice versa. Although we identified several variants reaching nominal association (p < 0.05), when controlling for all tested SNPs, none of the tested variants surpassed Bonferroni-corrected p value thresholds (tables e-3 and e-4).
Polygenic scores.
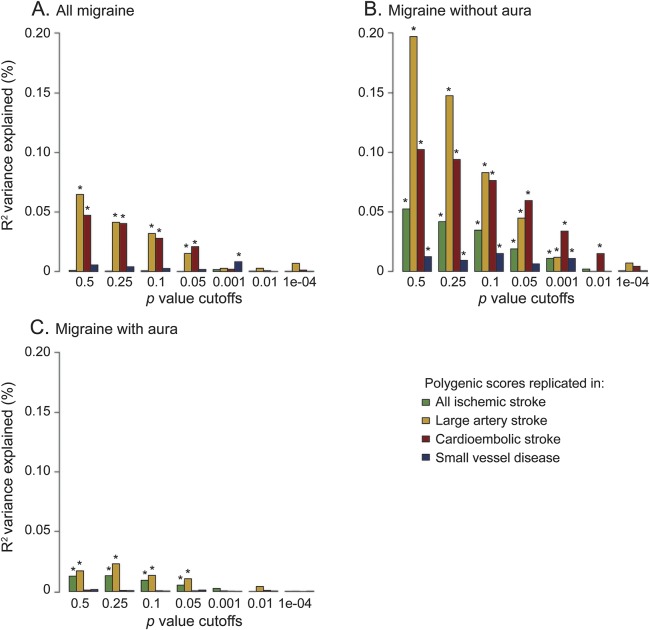
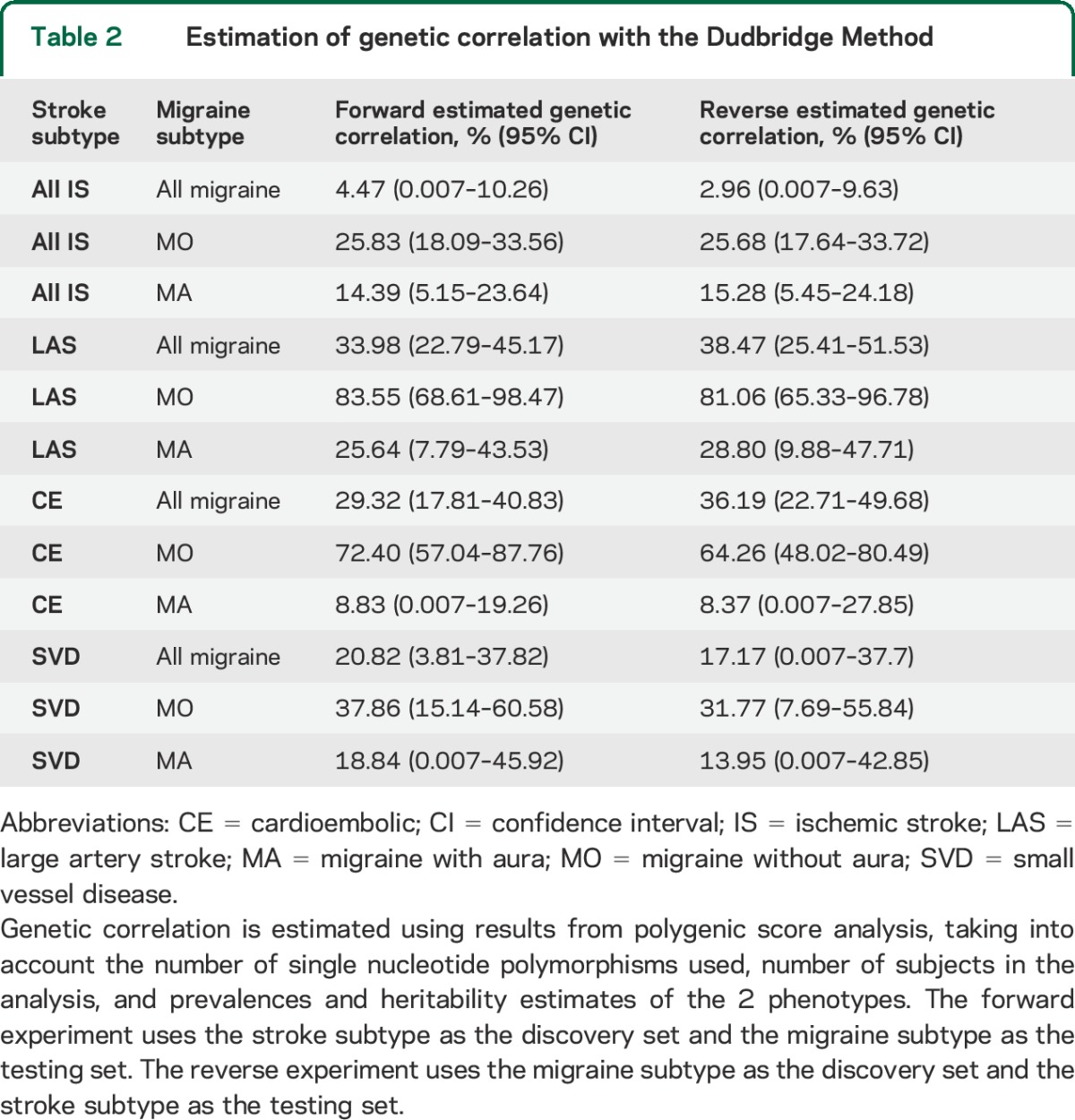
Scores derived from LAS, CE, and SVD each showed significant associations with all migraine (figure 1, tables 1 and e-5) with replication p values ranging from 2.7E-9 for LAS to 0.017 for SVD. Explained variance ranged from 0.005% to 0.03% (figure 1, table 1). Conversely, polygenic scores derived from all migraine significantly associated with LAS, CE, and SVD (figure 2, tables 1 and e-5) with replication p values between 8E-9 (replication in LAS) and 0.03 (replication in SVD) and an explained variance between 0.008% and 0.065% (figure 2, tables 1 and e-5). Calculated estimates of genetic correlation between all migraine and IS ranged from approximately 3% for correlation with all IS to 38% for correlation with LAS (table 2).
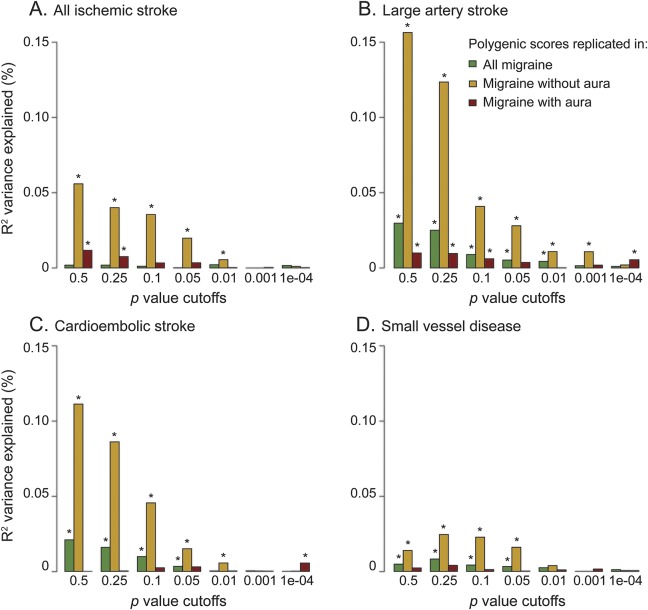
Figure 1. Results from polygenic score analysis using ischemic stroke as a discovery phenotype.
(A) All ischemic stroke. (B) Large artery stroke. (C) Cardioembolic stroke. (D) Small vessel disease. Migraine was used as a replication phenotype. The x-axis describes the p value cutoffs used in the polygenic score; the y-axis describes the pseudo-R2 variance explained by the score. Asterisks on top of a bar designate p values < 0.05. Raw values can be found in table e-5.
Table 1.
Polygenic score results
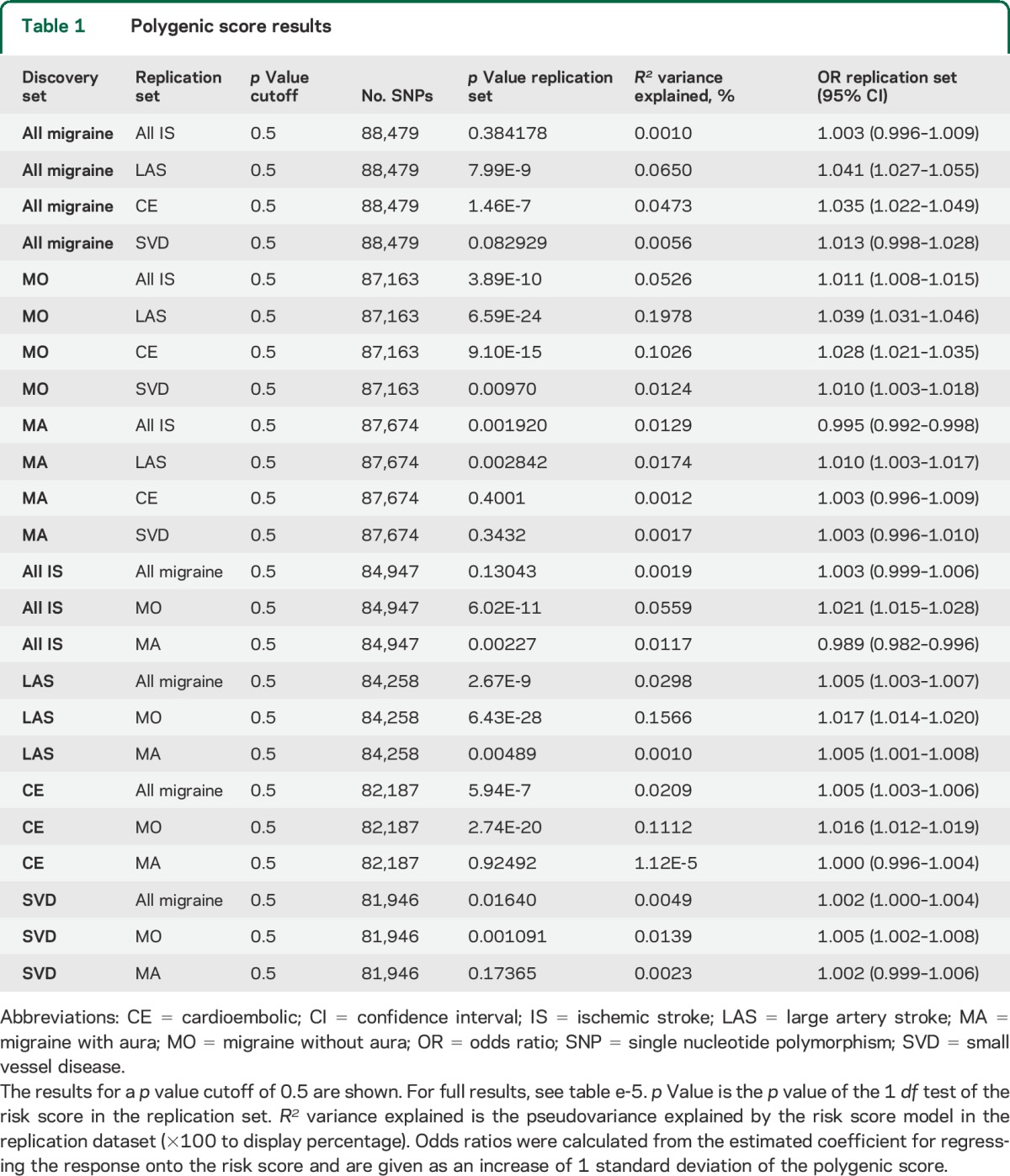
Figure 2. Results from polygenic score analysis using migraine as a discovery phenotype.
(A) All migraine. (B) Migraine without aura. (C) Migraine with aura. Stroke was used as a replication phenotype. The x-axis describes the p value cutoffs used in the polygenic score; the y-axis describes the pseudo-R2 variance explained by the score. Asterisks on top of a bar designate p values < 0.05. Raw values can be found in table e-5.
Table 2.
Estimation of genetic correlation with the Dudbridge Method
CPSM analysis.
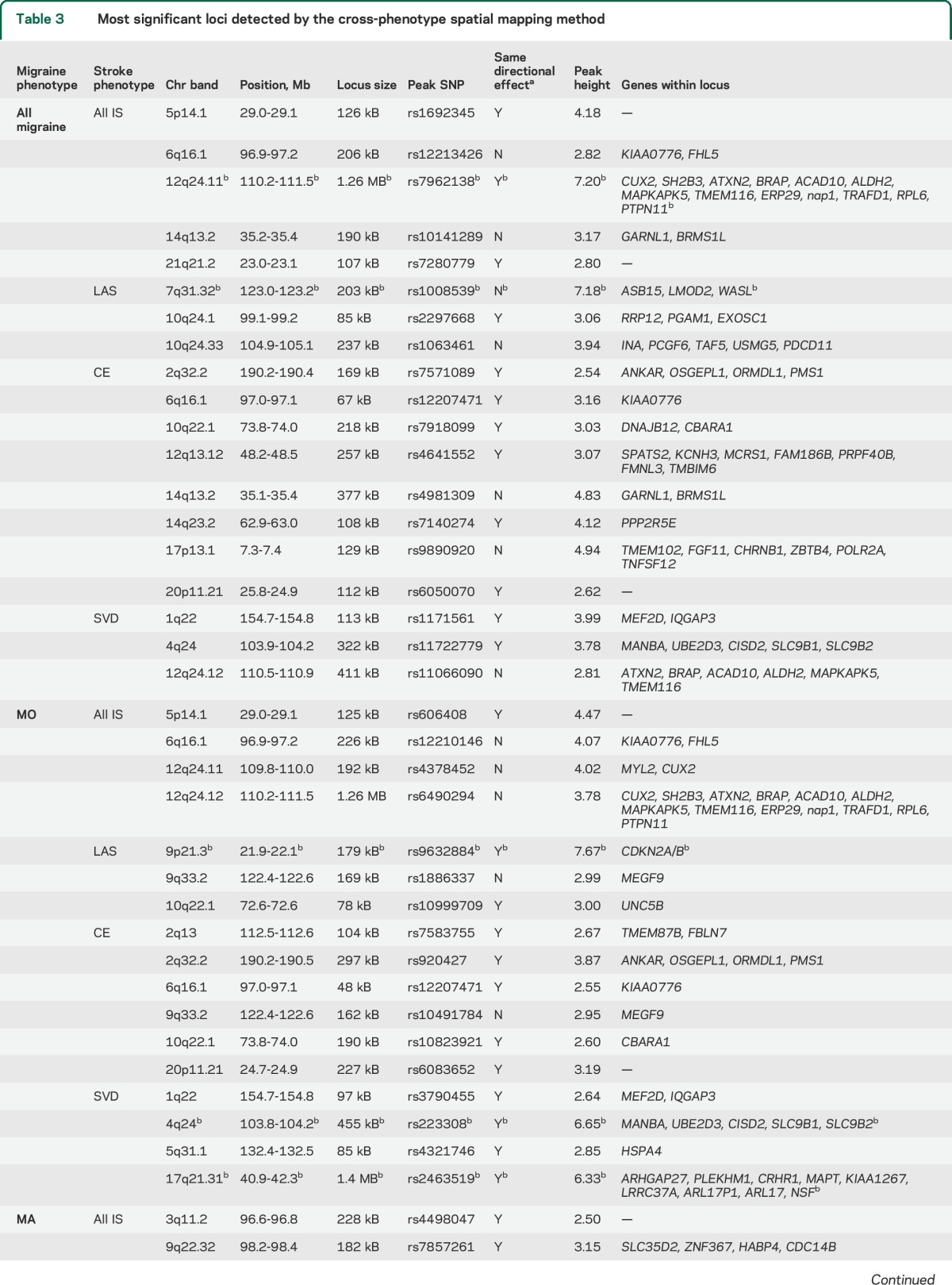
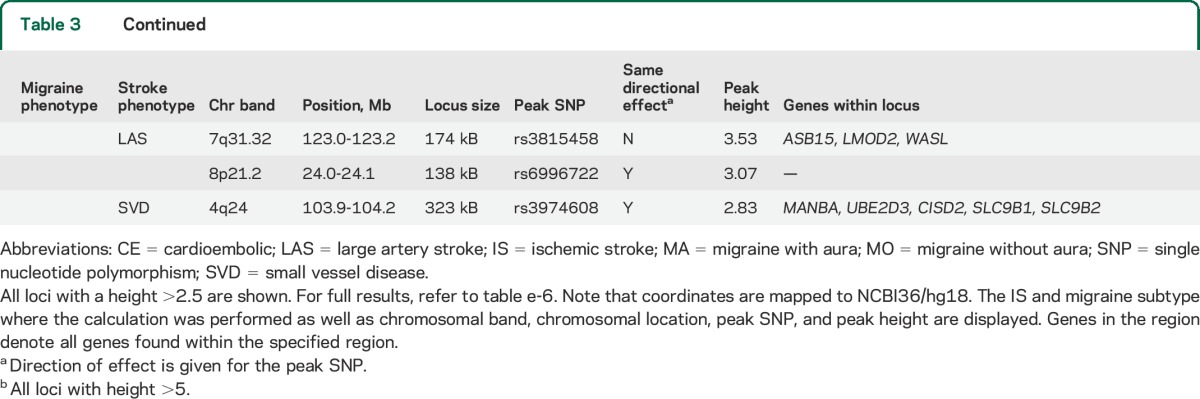
The most significant loci reaching an arbitrary peak height cutoff of 2.5 for CPSM are summarized in table 3 (full results, table e-6). Using this height cutoff, there were 5 shared loci for all IS and all migraine with the strongest signal at chromosome 12q24 (height = 7.2). For LAS and all migraine, we found 3 shared loci, with the LMOD2-WASL region on chromosome 7q31 showing the strongest signal (height = 7.2). CE and SVD showed 8 and 3 shared loci with all migraine, respectively (maximum height, 4.94 for CE and all migraine; 3.99 SVD and all migraine).
Table 3.
Most significant loci detected by the cross-phenotype spatial mapping method
Migraine without aura (MO).
A single variant in the 9p21 region, previously associated with LAS,14 surpassed the Bonferroni-corrected threshold for association with MO (p = 4.0E-5). Focusing on 2 loci previously known to be associated with MO but not MA,11 we identified no variants surpassing the Bonferroni-corrected p value threshold (table e-4).
Polygenic scores.
Scores derived from all IS, LAS, CE, and SVD each showed significant associations with MO (figure 1, tables 1 and e-5); replication p values ranged from 6.43E-28 for LAS polygenic score to 1.47E-5 for SVD polygenic score. The highest percentage of explained variance occurred for scores derived from LAS and CE (0.157% and 0.111%, respectively), and was higher in MO than in all migraine across all p value cutoffs (figure 1, B and C). Conversely, polygenic scores derived from MO significantly associated with all IS, LAS, CE, and SVD (figure 2, tables 1 and e-5) with replication p values between 6E-24 (replication in LAS) and 0.004 (replication in SVD) and an explained variance between 0.015% and 0.198% (figure 2, tables 1 and e-5). Estimates of genetic correlation with IS were markedly higher than observed for all migraine (estimates ranged from 25% for correlation with all IS to 83% for correlation with LAS; table 2).
CPSM analysis.
Using a cutoff of 2.5, we detected 4 shared loci between MO and all IS. MO and LAS shared 3 loci, with the strongest signal at chromosome 9p21 (signal height, 7.7). CE and MO shared 6 loci (maximum height = 3.87), and MO and SVD shared 4 loci (2 loci reaching maximum heights of 6.7 and 6.3). The former was near the CISD2 gene on chromosome 4q24, the latter in a gene-rich region on chromosome 17q21 including the Tau locus (tables 3 and e-6).
Migraine with aura (MA).
None of the variants previously associated with IS surpassed the Bonferroni-corrected p value threshold of 7.69E-5 when tested for association with MA. There were no genome-wide significant loci11 for MA.
Polygenic scores.
Scores derived from IS, LAS, and CE all showed significant associations with MA (figure 1, tables 1 and e-5) with replication p values ranging from 0.002 for the all IS polygenic score to 0.04 for the CE polygenic score. Explained variance ranged from 0.010% (LAS) to 0.012% (IS). Polygenic scores derived from MA significantly associated with all IS and LAS (figure 2, tables 1 and e-5) with replication p values of 0.0017 and 0.0005, and an explained variance of 0.013% and 0.023%, respectively. Estimates of genetic correlation ranged from 8% for correlation with CE to 28% for correlation with LAS (table 2).
CPSM analysis.
We found several shared regions between MA and stroke subtypes. Using a cutoff of 2.5, we found 2 shared loci between all IS and MA with a maximum height of 3.15 and 2 loci shared between LAS and MA with a maximum height of 3.53 in the LMOD2/WASL gene region. CE and MA shared no loci using this cutoff and SVD and MA shared 1 locus with a maximum height of 2.83 (tables 3 and e-6).
DISCUSSION
We demonstrated that the combined contributions of common genetic variants at a number of loci influence risk for both migraine and IS. This is supported by results from 4 investigative approaches: (1) analysis of common variants at loci reaching genome-wide significance for potential signal overlap; (2) investigation of shared genetic load using polygenic score models; (3) estimation of genetic correlation between disease subtypes using data derived from these models; and (4) highlighting regions of shared risk by analysis of covariance patterns between phenotypes using CPSM. We found stronger signal overlap between MO and IS than between MA and IS; overlap is stronger for LA and CE stroke than for SVD. Finally, we identified several individual loci with a strong signal for association with both phenotypes.
Polygenic scores, estimates of genetic correlation, and CPSM results all demonstrated a stronger genetic overlap of IS with MO compared to MA. Polygenic scores from MO replicated in overall IS and IS subtypes across a wide range of p value cutoffs, while scores derived from IS behaved similarly when tested in MO. Scores derived from MA demonstrated weaker association with IS. The variance explained by polygenic scores of each IS subtype was consistently higher for MO (figures 1 and 2). Also, estimates of genetic correlation with IS and its subtypes were consistently higher for MO than for MA (table 2).
Unexpectedly, CPSM revealed that the number of loci reaching a peak height >2.5 was larger for MO and IS than for MA and IS (table 3). Recent epidemiologic studies suggest an association between IS risk and MA but not MO,3,6 but other data suggest that patients with MO are at increased risk of IS.4 One potential explanation is that genetic risk for MA may be more restricted to rare variants not captured by GWAS strategies as suggested by the larger number of genome-wide significant loci for MO compared to MA despite comparable sample sizes.11 However, estimated heritability for MA is as least as high as for MO.25,26 Larger samples together with sequencing efforts or rare variant assays might help to determine whether rare variants indeed influence MA risk and whether the same variants also contribute to IS risk. The same might be true for SVD, for which there are no existing identified genome-wide loci. Hence, we might have underestimated genetic overlap between migraine subtypes and SVD.
We found particularly strong genetic overlap for migraine with LAS and CE. Polygenic scores analyses showed the strongest overlap with LAS for all forms of migraine regardless of whether polygenic scores were derived from LAS and tested in migraine or vice versa. In a recent small population-based study of 360 migraineurs and 617 controls, researchers reported no association between migraine and intima media thickness,27 but more advanced stages of atherosclerosis were not assessed. Most previous studies examining the relationship between migraine and IS did not distinguish among stroke subtypes. Migraineurs display enhanced platelet aggregation,9 which together with other factors might contribute to overlap with LAS.
Analysis of loci previously shown to reach genome-wide significance for association with migraine showed several variants nominally associated with IS or its subtypes and vice versa. In fact, single variants reached a high threshold of statistical significance, e.g., variants on 9p21, a major risk locus for LAS14,15 reaching very low p values in MO (table e-3). However, in all instances index SNPs for the tested phenotype were in poor LD (r2 < 0.4) with published risk SNPs making it unlikely that the same variants confer risk to both stroke and migraine.
CPSM analysis revealed several chromosomal regions with strong evidence for genetic overlap between migraine and IS pointing to shared biological mechanisms, including loci already shown to be associated with either migraine or stroke such as a chromosome 12 locus previously implicated in IS, coronary artery disease, hypertension, diabetes, and blood cell traits including platelet count.19,28–30 Interestingly, MRVI1 on chr11, another locus previously associated with platelet aggregation,31 showed genetic overlap between migraine and IS, adding to previous data suggesting a shared role of platelet dysfunction in migraine and IS.9 Mendelian randomization studies and interventional studies are needed to determine the exact role of platelets in mediating such genetic risk. We also demonstrated genetic overlap between migraine and IS at chromosome 9p21 especially for LAS.
There was genetic overlap at loci not reaching genome-wide significance in migraine or stroke GWAS. A shared locus for all migraine and LAS on 7q31.32 includes the LMOD2-WASL gene region. LMOD2 encodes leiomodin2 that antagonizes tropomodulin, an actin-capping protein.32 WASL is implicated in stabilizing endothelial adherens junctions,33 and is important for synapse development.34 We also found overlapping regions for SVD with MO including a locus on 4q24 that encompasses MANBA, which encodes β-mannosidase. Mutations in MANBA are associated with epileptic encephalopathy35 and leukoencephalopathy.36 This region also contains SLC9B2, previously associated with essential hypertension,37 a major risk factor for SVD. A second shared risk locus between MO and SVD points to MAPT, the gene encoding tau protein on chromosome 17.
We used the largest collections of GWAS data currently available for migraine11 and IS,15 with 4 different but complementary approaches for analysis of genetic overlap including novel methodology (CPSM).38 Polygenic scores reflect multiple variants with very small effect sizes distributed across the whole genome whereas our analysis of known loci and CPSM analysis focus on specific broader regions with highly correlated p values. Overall, results were remarkably consistent. Estimates of genetic correlation between phenotypes were similar in forward and reverse direction as were results of polygenic scores.
Our study also has limitations. First, some patients with MA might have been misdiagnosed with IS and vice versa. However, this should have shifted the results towards a stronger overlap between IS and MA, whereas we found stronger overlap for MO. Thus, diagnostic misclassification is unlikely to contribute substantially to our results. Second, some patients may have had both conditions. We can largely exclude ascertainment bias favoring the selection of patients with comorbidity substantiated by differences in age structure between migraineurs and stroke patients. Third, lacking individual level data, we cannot exclude some overlap in controls. We carefully checked for any potential overlap in controls and excluded samples where appropriate. Bias resulting from overlapping controls would not explain the differences observed between clinical subphenotypes. Finally, we are missing information on clinical subtypes for a substantial proportion of patients, reducing power in subgroup analyses, but this should not result in systematic bias to explain observed differences. Future studies on larger samples should further explore genetic overlap with rare causes of IS such as dissections, which were not considered separately in this study.
Our data provide genetic insights from GWAS meta-analyses into shared mechanisms of migraine and IS and may in part explain the relationship between these 2 common neurovascular disorders.
Supplementary Material
GLOSSARY
- CE
cardioembolic stroke
- CPSM
cross-phenotype spatial mapping
- GWAS
genome-wide association studies
- IHGC
International Headache Genetics Consortium
- IS
ischemic stroke
- LAS
large artery stroke
- LD
linkage disequilibrium
- MA
migraine with aura
- MO
migraine without aura
- SNP
single nucleotide polymorphism
- SVD
small vessel disease
Footnotes
Supplemental data at Neurology.org
Contributor Information
Collaborators: Unnur Thorsteinsdottir, Anita L DeStefano, Christopher Levi, Solveig Gretarsdottir, Peter Donnelly, Ines Barroso, Jenefer M Blackwell, Elvira Bramon, Matthew A Brown, Juan P Casas, Aiden Corvin, Panos Deloukas, Audrey Duncanson, Janusz Jankowski, Hugh S Markus, Christopher G Mathew, Colin NA Palmer, Robert Plomin, Anna Rautanen, Stephen J sawcer, Richard C Trembath F, Ananth C Viswanathan, Nicholas W Wood, Chris CA Spencer, Gavin Band, Celine Bellengues, Colin Freeman, Garrett Hellenthal, Eleni Giannoulatou, Matti Pirinen, Richard Pearson, Amy Strange, Zhan Su, Damjan Vukcevic, Cordelia Langford, Sarah E Hunt, Sarah Edkind, Rhian Gwilliam, Hanah Blackburn, Suzannah J Bumpstead, Serge Dronv, Matthew Gillman, Emma Gray, Naomi Hammond, Alagurevathi Jayakumar, Owen T McCann, Jennifer Liddle, Simon C Potter, Radhi Ravindrarajah, Michelle Rickette, Matthew Waller, Paul Weston, Sara Widaa, Pamela Whittaker, Padhraig Gormley, Francesco Bettella, George McMahon, Unda Todt, Priit Palta, Eija Hamalainen, Stacy Steinberg, Hreinn Stefansson, Markus Färkkilä, Ville Artto, Mari A Kaunisto, Jean Schoenen, Rune R Frants, Guntram Borck, Hartmut Göbel, Axel Heinze, Katja Heinze-Kuhn, and Bertram Müller-Myhsok
AUTHOR AFFILIATIONS
From the Institute for Stroke and Dementia Research (R.M., T.F., M.D.), Ludwig-Maximilians-Universität München, Munich; Abteilung Neurologie mit Schwerpunkt Epileptologie und Hertie Institut für Klinische Hirnforschung (T.F.), Universitätsklinikum Tübingen, Germany; the Department of Neurology and FORMI (B.S.W., J.A.Z.), Oslo University Hospital and University of Oslo, Norway; the Analytical and Translational Genetics Unit, Department of Medicine (B.S.W., V.A., A.P.), the Department of Neurology (J.R., A.P.), and the Psychiatric & Neurodevelopmental Genetics Unit, Department of Psychiatry (A.P.), Massachusetts General Hospital, Boston; the Program in Medical and Population Genetics (V.A., J.R.) and the Stanley Center for Psychiatric Research (V.A., A.P.), Broad Institute of MIT and Harvard, Cambridge; Harvard Medical School (V.A., D.I.C., T.K., J.R.), Boston, MA; the Interdepartmental Program in Computational Biology and Bioinformatics (J.V.H.), Yale University, New Haven, CT; the Department of Clinical Neurosciences (M.T., H.S.M.), University of Cambridge, UK; the Departments of Human Genetics (B.d.V., A.M.J.M.v.d.M.) and Neurology (G.M.T., M.D.F., A.M.J.M.v.d.M.), Leiden University Medical Center, the Netherlands; the University of Newcastle and Hunter Medical Research Institute (E.G.H., J.M.), Newcastle; the Faculty of Health, School of Health Sciences (J.S.), Faculty of Health and Medicine, School of Nursing and Midwifery (J.M.), and the Centre for Translational Neuroscience and Mental Health (J.M.), University of Newcastle; the Department of Neurosciences (J.S.), Central Coast Health District, Gosford, Australia; the Cardiovascular Health Research Unit, Department of Medicine (J.C.B.), and the Departments of Neurology and Epidemiology (W.T.L.), University of Washington, Seattle; the Clinical Trial Service Unit & Epidemiological Studies Unit (J.C.H., R.C.), the Wellcome Trust Centre for Human Genetics (M. Farrall), the Department of Cardiovascular Medicine (M. Farrall), and the Stroke Prevention Research Unit, Nuffield Department of Clinical Neuroscience (P.M.R.), University of Oxford; the Division of Clinical Neurosciences, Centre for Clinical Brain Sciences (K.R., C.S.), University of Edinburgh, UK; the Institute of Genetics (M.W.), Folkhälsan Research Center, Helsinki; the Institute for Molecular Medicine Finland (FIMM) (M.W., J.K., A.P.) and the Department of Public Health (J.K.), University of Helsinki; the Department of Neurology (M.K.), Helsinki University Central Hospital, Finland; the Institute of Human Genetics (C.K.), University Medical Center Hamburg-Eppendorf, Hamburg; the Institute of Human Genetics (C.K.), University of Ulm, Germany; the University of Texas Health Science Center at Houston (M. Fornage), Houston; the Department of Neurology (J.F.M.), Mayo Clinic, Jacksonville, FL; the Department of Clinical Chemistry (T.L.), Fimlab Laboratories and School of Medicine, University of Tampere, Finland; the Division of Preventive Medicine (D.I.C., T.K., M.S.), Brigham and Women's Hospital, Boston, MA; the Department of Medicine (B.D.M.), University of Maryland School of Medicine, Baltimore; the Department of Mental Health and Alcohol Research (J.K.), National Institute for Health and Welfare, Helsinki; the Department of Clinical Physiology and Nuclear Medicine and Research Centre of Applied and Preventive Cardiovascular Medicine (O.T.R.), University of Turku and Turku University Hospital, Finland; the INSERM Research Center for Epidemiology and Biostatistics (U897), Team Neuroepidemiology (T.K.), Bordeaux; the College of Health Sciences (T.K.), University of Bordeaux, France; the Departments of Epidemiology (M.A.I., C.M.v.D.), Neurology (M.A.I.), and Radiology (M.A.I.), Erasmus MC University Medical Center, Rotterdam; the Netherlands Consortium for Healthy Ageing (M.A.I., C.M.v.D.), Leiden, the Netherlands; the Division of Public Health Sciences (A.P.R.), Fred Hutchinson Cancer Research Center, Seattle, WA; the Division of Population Health Sciences and Education (D.P.S.), St George's, University of London; Imperial College Cerebrovascular Research Unit (ICCRU) (P.S.), Imperial College London, UK; the Department of Neurology (S.S.), Boston University School of Medicine, MA; the Department of Twin Research and Genetic Epidemiology (L.Q., L.C.), King's College London, UK; the Department of Neurology (M.S.), University Hospital Essen, Germany; the Center for Human Genetic Research and Division of Neurocritical Care and Emergency Neurology, Department of Biological Psychology (L.L.), VU University, Amsterdam, the Netherlands; the Department of Cerebrovascular Disease (G.B.B.), Fondazione Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Istituto Neurologico Carlo Besta, Milan, Italy; the MRC Integrative Epidemiology Unit at the University of Bristol (G.D.S.), UK; deCODE genetics (K.S.), Reykjavík, Iceland; the Departments of Neurology and Public Health Sciences (B.B.W.), University of Virginia School of Medicine, Charlottesville; the Neurogenetics Laboratory (D.R.N.), QIMR Berghofer, Brisbane, Australia; the Department of Neurology (C.C.), Yale School of Medicine, New Haven, CT; the Wellcome Trust Sanger Institute (A.P.), Wellcome Trust Genome Campus, Cambridge, UK; and the Munich Cluster for Systems Neurology (SyNergy) (M.D.), Germany.
AUTHOR CONTRIBUTIONS
Rainer Malik: drafting/revising the manuscript for content, study concept or design, analysis or interpretation of data, acquisition of data, statistical analysis, accepts responsibility for conduct of research and final approval. Tobias Freilinger: drafting/revising the manuscript for content, study concept or design, acquisition of data, accepts responsibility for conduct of research and final approval. Bendik S. Winsvold: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Verneri Anttila: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Jason Vander Heiden: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Matthew Traylor: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Boukje de Vries: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Elizabeth G. Holliday: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Gisela M. Terwindt: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Jonathan Sturm: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Joshua C. Bis: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Jemma C. Hopewell: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Michel D. Ferrari: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Kristiina Rannikmae: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Maija Wessman: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Mikko Kallela: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Christian Kubisch: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Myriam Fornage: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. James F. Meschia: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Terho Lehtimäki: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Cathie Sudlow: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Robert Clarke: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Dan I. Chasman: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Braxton D. Mitchell: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Jane Maguire: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Jaakko Kaprio: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Martin Farrall: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Olli T. Raitakari: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Tobias Kurth: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. M. Arfan Ikram: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Alex P. Reiner: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Will T. Longstreth: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Peter M. Rothwell: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. David P. Strachan: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Pankaj Sharma: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Sudha Seshadri: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Lydia Quaye: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Lynn Cherkas: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Markus Schürks: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Jonathan Rosand: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Lannie Ligthart: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Giorgio B. Boncoraglio: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. George Davey Smith: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Cornelia M. van Duijn: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Kari Stefansson: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Bradford B. Worrall: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Dale R. Nyholt: drafting/revising the manuscript for content, study concept or design, acquisition of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. Hugh S. Markus: drafting/revising the manuscript for content, study concept or design, acquisition of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. Arn M.J.M. van den Maagdenberg: drafting/revising the manuscript for content, study concept or design, acquisition of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. Chris Cotsapas: drafting/revising the manuscript for content, study concept or design, acquisition of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. John A. Zwart: drafting/revising the manuscript for content, study concept or design, acquisition of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. Aarno Palotie: drafting/revising the manuscript for content, study concept or design, acquisition of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. Martin Dichgans: drafting/revising the manuscript for content, study concept or design, acquisition of data, study supervision or coordination, accepts responsibility for conduct of research and final approval.
STUDY FUNDING
The Australian Stroke Genetics Collaboration (ASGC) Australian population control data were derived from the Hunter Community Study. The authors thank the University of Newcastle for funding and the men and women of the Hunter region who participated in this study. This research was funded by grants from the Australian National and Medical Health Research Council (NHMRC Project Grant ID: 569257), the Australian National Heart Foundation (NHF Project Grant ID: G 04S 1623), the University of Newcastle, the Gladys M Brawn Fellowship scheme, and the Vincent Fairfax Family Foundation in Australia. The Atherosclerosis Risk in Communities Study (ARIC) is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; NIH contract HHSN268200625226C and National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022; and grants R01-HL087641, U01 HL096917 (Mosley), and R01-HL093029 (Fornage). Infrastructure was partly supported by Grant UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. ARIC analyses performed as part of the METASTROKE project were supported by grant HL-093029 to M. Fornage. Bio-Repository of DNA in Stroke (BRAINS) is partly funded by a Senior Fellowship from the Dept. of Health (UK) to Dr. Pankaj Sharma, the Henry Smith Charity and the UK-India Education Research Institutive (UKIERI) from the British Council. Cardiovascular Health Study (CHS) research was supported by National Heart, Lung, and Blood Institute contracts N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85084, N01-HC-85085, N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, and N01-HC-85239, and by HHSN268201200036C and National Heart, Lung, and Blood Institute grants HL080295, HL087652, and HL105756, with additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the National Institute on Aging. See also http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by the National Center for Research Resources, grant UL1RR033176, and is now at the National Center for Advancing Translational Sciences, Clinical and Translational Science Institute grant UL1TR000124, in addition to the National Institute of Diabetes and Digestive and Kidney Disease grant DK063491 to the Southern California Diabetes Endocrinology Research Center. deCODE Genetics Work performed at deCODE was funded in part through a grant from the European Community's Seventh Framework Programme (FP7/2007–2013), the ENGAGE project grant agreement HEALTH-F4-2007-201413. Framingham Heart Study (FHS): this work was supported by the dedication of the Framingham Heart Study participants, the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract Nos. N01-HC-25195 and N02-HL-6-4278), and by grants from the National Institute of Neurological Disorders and Stroke (NS17950), the National Heart, Lung, and Blood Association (HL93029), and the National Institute of Aging (AG033193). The Genetics of Early Onset Stroke (GEOS) Study, Baltimore, was supported by the NIH Genes, Environment and Health Initiative (GEI) Grant U01 HG004436, as part of the GENEVA consortium under the NIH Genes, Environment and Health Initiative, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488); and the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by The Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the NIH to The Johns Hopkins University (contract HHSN268200782096C). Assistance with data cleaning was provided by the GENEVA Coordinating Center (U01 HG 004446; PI Bruce S. Weir). Study recruitment and assembly of datasets were supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control, and by grants from the National Institute of Neurological Disorders and Stroke and the NIH Office of Research on Women's Health (R01 NS45012, U01 NS069208-01). Heart Protection Study (HPS) (ISRCTN48489393) was supported by the UK Medical Research Council, British Heart Foundation, Merck & Co. (manufacturers of simvastatin), and Roche Vitamins Ltd. (manufacturers of vitamins). Genotyping was supported by a grant to Oxford University and CNG from Merck & Co. Jemma C. Hopewell acknowledges support from the British Heart Foundation Centre of Research Excellence, Oxford (RE/08/004). The Heart and Vascular Health Study (HVH) research reported in this article was funded by National Heart, Lung, and Blood Institute grants R01 HL085251 and R01 HL073410. The Ischemic Stroke Genetics Study (ISGS)/Siblings With Ischemic Stroke Study (SWISS): The ISGS/SWISS study was supported in part by the Intramural Research Program of the National Institute on Aging, NIH project Z01 AG-000954-06. ISGS/SWISS used samples and clinical data from the NIH-National Institute of Neurological Disorders and Stroke Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), human subjects protocol numbers 2003-081 and 2004-147. ISGS/SWISS used stroke-free participants from the Baltimore Longitudinal Study of Aging (BLSA) as controls. The inclusion of BLSA samples was supported in part by the Intramural Research Program of the National Institute on Aging, NIH project Z01 AG-000015-50, human subjects protocol number 2003-078. The ISGS study was funded by NIH-National Institute of Neurological Disorders and Stroke Grant R01 NS-42733 (J.F. Meschia, PI). The SWISS study was funded by NIH-National Institute of Neurological Disorders and Stroke Grant R01 NS-39987 (J.F. Meschia, PI). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov). The MGH Genes Affecting Stroke Risk and Outcome Study (MGH-GASROS): GASROS was supported by The National Institute of Neurological Disorders and Stroke (U01 NS069208), the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research 0775010N, the NIH and National Heart, Lung, and Blood Institute's STAMPEED genomics research program (R01 HL087676), and a grant from the National Center for Research Resources. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research resources. Milano–Besta Stroke Register Collection and genotyping of the Milan cases within CEDIR were supported by Annual Research Funding of the Italian Ministry of Health (grant numbers RC 2007/LR6, RC 2008/LR6; RC 2009/LR8; RC 2010/LR8). PROCARDIS control samples were supported by FP6 LSHM-CT-2007-037273. The Rotterdam Study was supported by the Netherlands Organization of Scientific Research (175.010.2005.011), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO), Netherlands Consortium for Healthy Ageing (050-060-810), the Erasmus Medical Center and Erasmus University, Rotterdam, the Netherlands Organization for Health Research and Development, the Research Institute for Diseases in the Elderly, the Ministry of Education, Culture, and Science, the Ministry for Health, Welfare, and Sports, the European Commission, and the Municipality of Rotterdam to the Rotterdam Study. Further funding was obtained from the Netherlands Heart Foundation (Nederlandse Hartstichting) 2009B102. The Wellcome Trust Case-Control Consortium 2 (WTCCC2): The principal funding for the WTCCC2 stroke study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z and WT084724MA). The Stroke Association provided additional support for collection of some of the St George's, London cases. The Oxford cases were collected as part of the Oxford Vascular Study, which is funded by the Medical Research Council, Stroke Association, Dunhill Medical Trust, National Institute of Health Research (NIHR), and the NIHR Biomedical Research Centre, Oxford. The Edinburgh Stroke Study was supported by the Wellcome Trust (clinician scientist award to Dr. Sudlow) and the Binks Trust. Sample processing occurred in the Genetics Core Laboratory of the Wellcome Trust Clinical Research Facility, Western General Hospital, Edinburgh. Much of the neuroimaging occurred in the Scottish Funding Council Brain Imaging Research Centre (www.sbirc.ed.ac.uk), Division of Clinical Neurosciences, University of Edinburgh, a core area of the Wellcome Trust Clinical Research Facility and part of the SINAPSE (Scottish Imaging Network—A Platform for Scientific Excellence) collaboration (www.sinapse.ac.uk), funded by the Scottish Funding Council and the Chief Scientist Office. Collection of the Munich cases and data analysis was supported by the Vascular Dementia Research Foundation. Martin Farrall acknowledges support from the BHF Center of Research Excellence in Oxford and the Wellcome Trust core award (090532/Z/09/Z). Hugh Markus is supported by an NIHR Senior Investigator award. ALSPAC: The authors thank the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The UK Medical Research Council and Wellcome Trust (092731), together with the University of Bristol, provide core support for the ALSPAC study. A grant from the Wellcome Trust funded collection of GWAS data in ALSPAC mothers (WT088806) and a grant from 23andMe, together with support from the Sanger Centre and Centre National de Genotypage funded GWAS data in the offspring. Australian Twin Migraine: The Australian cohort was supported by NIH grants AA07535, AA07728, AA13320, AA13321, AA14041, AA11998, AA17688, DA012854, and DA019951; by grants from the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, and 552498); by grants from the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, and DP0343921); and by the EU‐funded GenomEUtwin (FP5‐QLG2‐CT‐2002‐01254) and ENGAGE (FP7‐HEALTH‐201413) projects. D.R.N. (FT0991022, 613674). The authors thank P. Visscher, D. Duffy, A. Henders, B. Usher, E. Souzeau, A. Kuot, A. McMellon, P.A.F. Madden, M.J. Wright, M.J. Campbell, A. Caracella, L. Bowdler, S. Smith, S. Gordon, B. Haddon, D. Smyth, H. Beeby, O. Zheng, and B. Chapman for their input into project management, databases, phenotype collection, and sample collection, processing, and genotyping. British 58 Birth Cohort: The authors acknowledge use of phenotype and genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02 (http://www.b58cgene.sgul.ac.uk/). Genotyping for the B58C‐WTCCC subset was funded by the Wellcome Trust grant 076113/B/04/Z. The B58C‐T1DGC genotyping utilized resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. B58C‐T1DGC. GWAS data were deposited by the Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research (CIMR), University of Cambridge, which is funded by JDRF, the Wellcome Trust and the National Institute for Health Research Cambridge Biomedical Research Centre; the CIMR is in receipt of a Wellcome Trust Strategic Award (079895). The B58C‐GABRIEL genotyping was supported by a contract from the European Commission Framework Programme 6 (018996) and grants from the French Ministry of Research. Erasmus Rucphen Family: The Erasmus Rucphen Family was supported by grants from The Netherlands Organization for Scientific Research (NWO), Erasmus MC, and the Netherlands Genomics Initiative (NGI)–sponsored Center of Medical Systems Biology (CMSB). The genotyping for the ERF study was supported by European Special Populations Research Network (EUROSPAN) through the European Commission FP6 STRP grant (018947; LSHG‐CT‐2006‐01947). The ERF study was further supported by grants from the Netherlands Organisation for Scientific Research (NWO), Erasmus MC, the Centre for Medical Systems Biology (CMSB1 and CMSB2), and the Netherlands Genomics Initiative (NGI). The authors thank all the patients and their relatives, general practitioners, and neurologists for their contributions and P. Veraart for her help in genealogy, Jeannette Vergeer for the supervision of the laboratory work, and P. Snijders for his help in data collection. FinnTwin: Academy of Finland Center of Excellence in Complex Disease Genetics (grant numbers 213506, 129680), the Academy of Finland (grants 205585 and 141054 to J.K.), ENGAGE—European Network for Genetic and Genomic Epidemiology, FP7‐HEALTH‐F4‐2007, grant agreement number 201413, and U.S.‐ P.H.S. NIH grants AA‐12502, AA‐00145, AA‐09203, AA15416, and K02AA018755. HUNT: The Nord‐Trøndelag Health Study (The HUNT Study) is a collaboration between HUNT Research Centre (Faculty of Medicine, Norwegian University of Science and Technology, NTNU), Nord‐Trøndelag County Council, Central Norway Health Authority, and the Norwegian Institute of Public Institute of Public Health. The current study was supported by the South‐Eastern Norway Regional Health Authority (2010075 and 2011083 to B.S.W. and J.A.Z.), Unger‐Vetlesen Medical Fund (to B.S.W.), and the Ullevaal fund (to B.S.W.). Genotyping was performed by Avazeh Tashakkori‐Ghanbarian, Simon Potter, and Sarah Hunt (calling and quality control) and Douglas Simpkin (production) at Wellcome Trust Sanger Institute. Northern Finland Birth Cohort 1966 (NFBC1966): NFBC1966 received financial support from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, 139900/24300796, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu, Finland (75617), the European Commission (EUROBLCS, Framework 5 award QLG1‐CT‐2000‐01643), NHLBI grant 5R01HL087679‐02 through the STAMPEED program (1RL1MH083268‐01), NIH/NIMH (5R01MH63706:02), ENGAGE project and grant agreement HEALTH‐F4‐2007‐201413, the Medical Research Council, UK (G0500539, G0600705, G0600331, PrevMetSyn/SALVE, PS0476), and the Wellcome Trust (project grant GR069224, WT089549), UK. The DNA extractions, sample quality controls, biobank upkeeping, and aliquotting was performed in the National Public Health Institute, Biomedicum Helsinki, Finland, and supported financially by the Academy of Finland and Biocentrum Helsinki. The authors acknowledge the contribution of the late Academian of Science Leena Peltonen. NTR/NESDA: Funding was obtained from the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW grants 904‐61‐090, 985‐10‐002, 904‐61‐193, 480‐04‐004, 400‐05‐717, Addiction‐31160008 Middelgroot‐911‐09‐032, Spinozapremie 56‐464‐14192), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK (2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI–NL, 184.021.007), the VU University's Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA), the European Science Foundation (ESF, EU/QLRT‐2001‐01254), the European Community's Seventh Framework Program (FP7/2007‐2013), ENGAGE (HEALTHF4‐2007‐201413), the European Science Council (ERC Advanced, 230374), and the NIH (NIH, R01D0042157‐01A). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the US NIH (NIMH, MH081802). The infrastructure for the NESDA study (www.nesda.nl) is funded through the Geestkracht program of the Netherlands Organisation for Health Research and Development (Zon‐Mw, grant number 10‐000‐1002) and is supported by participating universities and mental health care organizations (VU University Medical Center, GGZ inGeest, Arkin, Leiden University Medical Center, GGZ Rivierduinen, University Medical Center Groningen, Lentis, GGZ Friesland, GGZ Drenthe, Scientific Institute for Quality of Healthcare [IQ healthcare], Netherlands Institute for Health Services Research [NIVEL], and Netherlands Institute of Mental Health and Addiction [Trimbos Institute]). Rotterdam studies: The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO Investments (175.010.2005.011, 911‐03‐012). This study is funded by the Research Institute for Diseases in the Elderly (014‐93‐015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project 050‐060‐810. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The Rotterdam Scan Study is supported by the Netherlands Organization of Scientific Research (NWO) project 918‐46‐615, 904‐61‐096, 904‐61‐133, and 948‐00‐010. TwinsUK: The study was funded by the Wellcome Trust, European Community's Seventh Framework Programme (FP7/2007‐2013), ENGAGE project grant agreement (HEALTH‐F4‐2007‐201413). The study also receives support from the Dept. of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St. Thomas' NHS Foundation Trust in partnership with King's College London. Genotyping was performed by The Wellcome Trust Sanger Institute, support of the National Eye Institute via an NIH/CIDR genotyping project. This project was partly funded by the Chronic Disease Research Foundation with money raised by the walking twins, Hazel Green and Christine Dafter. WGHS: Genetic analysis of migraine in the WGHS is supported by a grant from the National Institute of Neurological Disorders and Stroke (NS‐061836). The Women's Health Study and the Women's Genome Health Study are supported by grants from the National Heart, Lung, and Blood Institute (HL‐043851, HL‐080467, and HL‐099355) and the National Cancer Institute (CA‐47988). Genome‐wide genotyping and collaborative scientific support was provided by Amgen. Young Finns: The Young Finns Study has been financially supported by the Academy of Finland: grants 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi), the Social Insurance Institution of Finland, Kuopio, Tampere, and Turku University Hospital Medical Funds (grant 9M048 and 9N035 for TeLeht), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation, and Emil Aaltonen Foundation (T.L). Technical assistance in the statistical analyses by Ville Aalto and Irina Lisinen is acknowledged. Cologne/Kiel/Ulm clinic‐based studies (part of German MA and MO studies): This research was funded by the German Federal Ministry of Education and Research (BMBF) within the framework of the National Genome Research Network (NGFN‐Plus; grants 01GS08120 and 01GS1103 to C.K.; the German Federal Ministry of Education and Research and by the State of Bavaria and supported within the Munich Center of Health Sciences [MC Health] as part of LMUinnovativ) for the KORA research platform, which was initiated by the Helmholtz Center Munich, German Research Center for Environmental Health, the Center for Molecular Medicine Cologne (to C.K.), and the Heinz Nixdorf Foundation for the Heinz Nixdorf Recall study, and the Deutsche Forschungsgemeinschaft (DFG; to C.K.). Finnish clinic‐based study: This work was supported by the Wellcome Trust (grant WT089062) and, among others, by the Academy of Finland (200923 to A.P., 00213 to M.W.); the Academy of Finland Center of Excellence for Complex Disease Genetics; the EuroHead project (LSM‐CT‐2004‐504837); the Finnish Cultural Foundation (to V.A.); the Finnish Neurology Foundation, Biomedicum Helsinki Foundation (to V.A.); the Orion Farmos Research Foundation (to V.A.); the Wellcome Trust (grant number 098051 to A.P.); the Academy of Finland (grant number 251704 to A.P., and 139795 to M.W.); the Academy of Finland, Center of Excellence in Complex Disease Genetics, (grant numbers 213506 and 129680 to A.P. and J.K.); the European Community's Seventh Framework Programme (FP7/2007‐2013), ENGAGE Consortium (grant agreement HEALTH‐F4‐2007‐201413); EU/SYNSYS‐Synaptic Systems (grant number 242167 to A.P.); the Sigrid Juselius Foundation, Finland (to A.P.); the Folkhälsan Research Foundation, Finland (to M.W.); Medicinska Understödsföreningen Liv & Hälsa (to M.W.), and the Helsinki University Central Hospital (to M.K., V.A.). LUMINA clinic‐based studies: Supported by the Netherlands Organization for Health Research and Development (ZonMw) no. 90700217 and VIDI (ZonMw) no. 91711319 (to G.M.T.); the Netherlands Organisation for Scientific Research (NWO) VICI (918.56.602) and Spinoza (2009) grants (to M.D.F.); the EU-funded EuroHead project (LSM‐CT‐2004‐504837) and EUROHEADPAIN (no. 602633); and the Center for Medical Systems Biology (CMSB) established in the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NGI/NWO), project 050‐060‐409 (to M.D.F. and A.M.J.M.v.d.M.). Munich clinic‐based studies (part of German MA and MO studies): Supported by the German Federal Ministry of Education and Research (BMBF) (grant 01GS08121 to M.D. along with support in the context of the German National Genome Research Network, NGFN‐2 and NGFN‐plus, for the Heinz Nixdorf Recall Study) and an unrestricted grant of the Vascular Dementia Research Foundation (to M.D.).
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Launer LJ, Terwindt GM, Ferrari MD. The prevalence and characteristics of migraine in a population-based cohort: the GEM study. Neurology 1999;53:537–542. [DOI] [PubMed] [Google Scholar]
- 2.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 2006;367:1747–1757. [DOI] [PubMed] [Google Scholar]
- 3.Kurth T, Chabriat H, Bousser MG. Migraine and stroke: a complex association with clinical implications. Lancet Neurol 2012;11:92–100. [DOI] [PubMed] [Google Scholar]
- 4.Etminan M, Takkouche B, Isorna FC, Samii A. Risk of ischaemic stroke in people with migraine: systematic review and meta-analysis of observational studies. BMJ 2005;330:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schurks M, Rist PM, Bigal ME, Buring JE, Lipton RB, Kurth T. Migraine and cardiovascular disease: systematic review and meta-analysis. BMJ 2009;339:b3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spector JT, Kahn SR, Jones MR, Jayakumar M, Dalal D, Nazarian S. Migraine headache and ischemic stroke risk: an updated meta-analysis. Am J Med 2010;123:612–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nozari A, Dilekoz E, Sukhotinsky I, et al. Microemboli may link spreading depression, migraine aura, and patent foramen ovale. Ann Neurol 2010;67:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stam AH, Haan J, van den Maagdenberg AM, Ferrari MD, Terwindt GM. Migraine and genetic and acquired vasculopathies. Cephalalgia 2009;29:1006–1017. [DOI] [PubMed] [Google Scholar]
- 9.Zeller JA, Frahm K, Baron R, Stingele R, Deuschl G. Platelet-leukocyte interaction and platelet activation in migraine: a link to ischemic stroke? J Neurol Neurosurg Psychiatry 2004;75:984–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friberg L, Olesen J, Lassen NA, Olsen TS, Karle A. Cerebral oxygen extraction, oxygen consumption, and regional cerebral blood flow during the aura phase of migraine. Stroke 1994;25:974–979. [DOI] [PubMed] [Google Scholar]
- 11.Anttila V, Winsvold BS, Gormley P, et al. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 2013;45:912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freilinger T, Anttila V, de Vries B, et al. Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat Genet 2012;44:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anttila V, Stefansson H, Kallela M, et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet 2010;42:869–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gschwendtner A, Bevan S, Cole JW, et al. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol 2009;65:531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol 2012;11:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353–357. [DOI] [PubMed] [Google Scholar]
- 17.Adams HP, Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke: definitions for use in a multicenter clinical trial: TOAST: Trial of Org 10172 in Acute Stroke Treatment. Stroke 1993;24:35–41. [DOI] [PubMed] [Google Scholar]
- 18.International HapMap Consortium, Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007;449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarborough P, Peto V, Bhatnagar P, et al. Stroke Statistics Oxford. England: British Heart Foundation & Stroke Association; 2009. [Google Scholar]
- 21.Rothwell PM, Coull AJ, Silver LE, et al. Population-based study of event-rate, incidence, case fatality, and mortality for all acute vascular events in all arterial territories (Oxford Vascular Study). Lancet 2005;366:1773–1783. [DOI] [PubMed] [Google Scholar]
- 22.Lovett JK, Coull AJ, Rothwell PM. Early risk of recurrence by subtype of ischemic stroke in population-based incidence studies. Neurology 2004;62:569–573. [DOI] [PubMed] [Google Scholar]
- 23.Bevan S, Traylor M, Adib-Samii P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke 2012;43:3161–3167. [DOI] [PubMed] [Google Scholar]
- 24.Le H, Tfelt-Hansen P, Skytthe A, Kyvik KO, Olesen J. Increase in self-reported migraine prevalence in the Danish adult population: a prospective longitudinal population-based study. BMJ Open 2012;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulrich V, Gervil M, Kyvik KO, Olesen J, Russell MB. Evidence of a genetic factor in migraine with aura: a population-based Danish twin study. Ann Neurol 1999;45:242–246. [DOI] [PubMed] [Google Scholar]
- 26.Gervil M, Ulrich V, Kaprio J, Olesen J, Russell MB. The relative role of genetic and environmental factors in migraine without aura. Neurology 1999;53:995–999. [DOI] [PubMed] [Google Scholar]
- 27.Stam AH, Weller CM, Janssens AC, et al. Migraine is not associated with enhanced atherosclerosis. Cephalalgia 2013;33:228–235. [DOI] [PubMed] [Google Scholar]
- 28.van der Harst P, Zhang W, Mateo Leach I, et al. Seventy-five genetic loci influencing the human red blood cell. Nature 2012;492:369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CARDIoGRAMplusC4D Consortium, Deloukas P, Kanoni S, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson AD, Yanek LR, Chen MH, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet 2010;42:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukada T, Pappas CT, Moroz N, Antin PB, Kostyukova AS, Gregorio CC. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J Cell Sci 2010;123:3136–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajput C, Kini V, Smith M, et al. Neural Wiskott-Aldrich syndrome protein (N-WASP)-mediated p120-catenin interaction with Arp2-Actin complex stabilizes endothelial adherens junctions. J Biol Chem 2013;288:4241–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegner AM, Nebhan CA, Hu L, et al. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem 2008;283:15912–15920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper A, Wraith JE, Savage WJ, Thornley M, Noronha MJ. Beta-mannosidase deficiency in a female infant with epileptic encephalopathy. J Inherit Metabol Dis 1991;14:18–22. [DOI] [PubMed] [Google Scholar]
- 36.Sabourdy F, Labauge P, Stensland HM, et al. A MANBA mutation resulting in residual beta-mannosidase activity associated with severe leukoencephalopathy: a possible pseudodeficiency variant. BMC Med Genet 2009;10:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang M, Feng M, Muend S, Rao R. A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc Natl Acad Sci USA 2007;104:18677–18681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet 2013;9:e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.