Abstract
DNA polymerase eta (Polη) plays unique and pivotal functions in several DNA damage-tolerance pathways. Steady-state level of this short-lived protein is tightly controlled by multiple mechanisms including proteolysis. Here, we have identified the deubiquitinating enzyme, ubiquitin-specific protease 7 (USP7), as a novel regulator of Polη stability. USP7 regulates Polη stability through both indirect and direct mechanisms. Knockout of USP7 increased the steady-state level of Polη and slowed down the turnover of both Polη and p53 proteins through destabilizing their E3 ligase Mdm2. Also, USP7 physically binds Polη in vitro and in vivo. Overexpression of wild-type USP7 but not its catalytically-defective mutants deubiquitinates Polη and increases its cellular steady-state level. Thus, USP7 directly serves as a specific deubiquitinating enzyme for Polη. Furthermore, ectopic expression of USP7 promoted the UV-induced PCNA monoubiquitination in Polη-proficient but not Polη-deficient XPV cells, suggesting that USP7 facilitates UV-induced PCNA monoubiquitination by stabilizing Polη. Taken together, our findings reveal a modulatory role of USP7 in PCNA ubiquitination-mediated stress-tolerance pathways by fine-tuning Polη turnover.
Keywords: USP7, Polη, PCNA, ubiquitination, deubiquitination
Introduction
Translesion DNA synthesis (TLS) carried out by specialized DNA polymerases is a major mechanism for mammalian cells to overcome genotoxic stress caused by blockage of the DNA replication machinery in the S phase of the cell cycle.1,2 Polη, the best characterized TLS polymerase, is unique in its ability to carry out error-free TLS at sites of UV-induced CPD. Polη has recently been found to recruit Rad18 and promote PCNA monoubiquitination at stalled replication forks 3. Besides, Polη also modulates the DNA damage checkpoint and p53 activation.4 Mutations of the human POLH gene, encoding Polη, result in the inherited cancer-propensity syndrome Xeroderma pigmentosum variant (XPV), which is characterized by sun sensitivity and elevated incidence of skin cancer.5
Polη is a low-fidelity enzyme while replicating undamaged DNA 6. Therefore, the activity of Polη is under stringent regulatory control. Indeed, endogenous cellular level of Polη is relatively low due to high turnover rate that is tightly regulated by multiple pathways. In S. cerevisiae, the level of Rad30 (yeast homolog of Polη) protein is shown to turnover via ubiquitination-dependent degradation and is stabilized following UV-irradiation.7 In C. elegans, Polη protein is degraded through CRL4-Cdt2-mediated proteolysis but protected by GEI-mediated sumoylation following UV-irradiation.8 In human cells, a RING finger E3 ligase, Mdm2, mediates Polη polyubiquitination and proteasomal degradation under the native condition and in response to UV irradiation 9. In addition, human POLH gene is a target of p53, and Polη expression can be up-regulated by p53 after genetic stresses.4
Reversal of ubiquitination, or deubiquitination, carried out by specific deubiquitinating enzymes (DUBs), has recently emerged as an important regulatory mechanism for many cellular processes. By reversing the action of ubiquitin ligases, DUBs offer a mechanism to fine-tune the effects of ubiquitination as a post-translational modification. Several DUBs, such as USP1, USP7, and USP28, have been shown to function in DNA damage response.10–15
USP7 deubiquitinates and stabilizes not only p53, but also Mdm2, the primary E3 ubiquitin ligase of p5316,17 Given that both p53 and Mdm2 are known regulators of the steady-level of Polη, we speculated that changes in cellular USP7 levels may also modulate Polη level. In this study, our data show that in conjunction with p53, knocking out USP7 increased the steady-state level and slowed down the turnover of Polη. An in-depth analysis revealed that USP7 deubiquitinates and stabilizes Polη through direct protein-protein interaction. Importantly, USP7-mediated stabilization of Polη was shown to facilitate the critical PCNA monoubiquitination in response to UV irradiation.
Results and Discussions
USP7 Knockout or over-expression increase Polη levels through different mechanisms
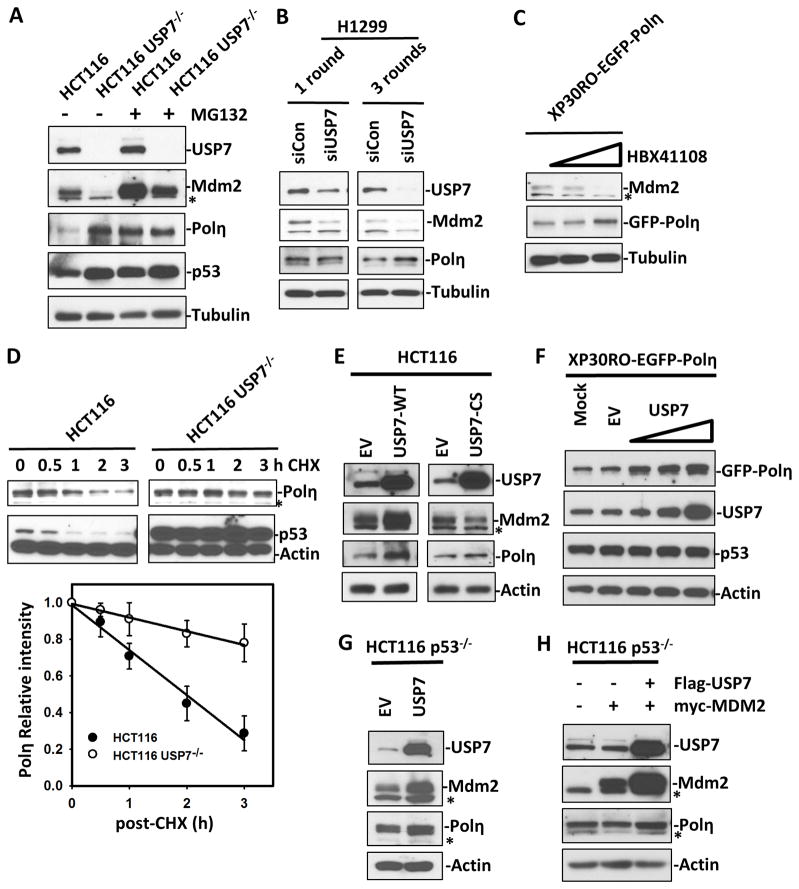
Since both Mdm2 and p53 are known regulators of Polη, we first investigated the effect of cellular USP7 on the Polη levels. We compared the steady-state levels of Polη in HCT116 and HCT116 USP7−/− (USP7-knockout) cells. As expected, USP7 disruption in HCT116 cells resulted in a Mdm2 decrease that led to increased levels of p53 and Polη (Fig. 1A and S1A). Moreover, cells treated in parallel with MG132 for 4 h revealed a distinct accumulation of Mdm2 protein. These observations are consistent with previously reported results of USP7 knockout destabilizing Mdm2 and subsequently stabilizing p53.18 Next, we tested the effect of USP7 ablation or inhibition in other cell types using two different approaches. We examined the consequences of siRNA-mediated reduction of endogenous USP7 in H1299 (a p53-null) cell line. Consistent with previous report,17 three consecutive rounds of transfection with USP7 siRNA resulted in almost complete depletion of USP7 while one round of transfection with USP7 siRNA resulted in only a partial reduction of USP7. Interestingly, severe ablation of USP7 expression diminished Mdm2 but increased Polη (Fig. 1B). Surprisingly, partial reduction of endogenous USP7 resulted in a reduction of Mdm2 but slight change of Polη (Fig. 1B). RT-PCR also revealed that USP7 ablation did not change Polη mRNA level in this p53-null cell line (Fig. S1B). We also used a USP7 specific inhibitor HBX 41108 to inhibit USP7 activity in XP30RO cells that stably express GFP-Polη (XP30RO-EGFP-Polη). As shown in Fig. 1C, high dosage HBX 41108 treatments (6 μM) of cells for 24 h increased Polη levels and completely ablated Mdm2, while low dose of HBX 41108 (3 μM) only partially reduced Mdm2 but did not change Polη levels. We next compared the turnover of Polη and p53 in HCT116 and HCT116 USP7−/− cells upon cycloheximide (CHX) treatment. Turnover of Polη was distinguishably slower in HCT116 USP7−/− than in HCT116 cells (Fig. 1D). The results demonstrated that the reduced Mdm2-mediated protein turnover, resulting from USP7 knockout, increased the steady-state levels of Polη as well as p53.
Figure 1. Either USP7 knockout or overexpression increases the steady-levels of Polη.
The protein levels in cell lysates were detected by Western blotting with their respective antibodies. Asterisk denotes the nonspecific band. S and L stand for short and long film exposures, respectively. (A) HCT116 and HCT116 USP7−/− cells treated with or without MG132 for 4 hours. (B) H1299 cells transiently transfected with control (siControl) or USP7 siRNA (siUSP7) for 1 or 3 rounds. (C). XP30RO-EGFP-Polη treated without or with 3 μM or 6 μM of HBX 41108 for 24 h. (D) HCT116 and HCT116 USP7−/− cells treated with CHX (50 ug/ml) for indicated times. The percentage of intensity was plotted versus time. Error bars represent ±SE of three independent experiments. (E) HCT116 cells were transiently transfected with empty vector (EV) or vector encoding FLAG-tagged USP7-WT or FLAG-tagged USP7-CS for 24 h. (F) XP30RO-EGFP-Polη cells were transiently transfected with empty vector (EV) or vector encoding FLAG-tagged USP7 (1, 2 and 4 ug of DNA) for 24 h. (G) HCT116 p53−/− cells were transiently transfected with empty vector (EV) or vector encoding FLAG-tagged USP7-WT for 24 hours. (H) HCT116 p53−/− cells were transiently transfected with empty vector or vector encoding Myc-MDM2, or/and FLAG-tagged USP7 for 24 hours.
Next, we examined whether ectopic expression of USP7 affects cellular levels of Polη. To this end, FLAG-tagged wild-type USP7 or catalytically inactive USP7 (USP7-CS), which contains a cysteine (C) to serine (S) substitution at amino acid 223,19 was transiently overexpressed in HCT116 cells. Consistent with previous reports, ectopic expression of USP7-WT resulted in accumulation of Mdm2.17 Surprisingly, the levels of Polη were also increased in HCT116 cells upon ectopic expression of USP7-WT (Fig. 1E). Ectopic expression of USP7-CS only resulted in slight accumulation of Polη and slight decrease of Mdm2 (Fig. 1E). To confirm this observation, FLAG-tagged USP7 was transiently expressed in XP30RO-EGFP-Polη cells. The protein levels of GFP-Polη in these cells are relatively high to allow an easy detection of subtle changes of GFP-Polη. Once again, compared to the control (mock or empty vector transfected) cells, we found that the levels of GFP-Polη exhibited a significant dose-dependent increase upon ectopic expression of USP7 (Fig. 1F). By contrast, overexpression of USP7 with the increased transfection dosages did not significantly change the levels of cellular p53 in this SV40-transformed cell line (Fig. 1F). Therefore, it rules out a potential effect of p53 induction of Polη. To validate this, we also showed that Polη protein level but not mRNA level was increased upon ectopic expression of USP7 in p53 knockout HCT116 cells (HCT116 p53−/−) (Fig. 1G and S1C). Moreover, while the levels of Mdm2 were enhanced by USP7 co-transfection, Mdm2-mediated Polη degradation was indeed strongly rescued by USP7 overexpression (Fig. 1H). Collectively, the data suggest that overexpression of USP7 up-regulates steady-levels of Polη independent of p53 and suggested that Polη, like p53, could be a substrate of USP7.
Polη and USP7 interact in vivo and in vitro
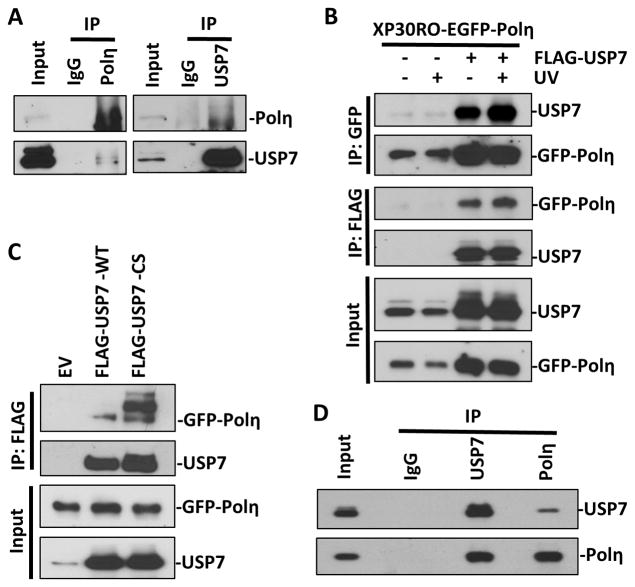
To investigate whether Polη is a substrate of USP7, we tested whether the two proteins interact in vivo. To address this, potential Polη and USP7 complexes in HCT116 cells were immunoprecipitated with anti-Polη and anti-USP7, respectively. As shown in Figure 2A, Polη was detected in anti-USP7 immunoprecipitates. Reciprocally, USP7 was detected in anti-Polη immunoprecipitates. To validate the association between Polη and USP7, FLAG-tagged USP7 was transiently transfected into XP30RO-EGFP-Polη cells and immunoprecipitation was performed using anti-FLAG or anti-GFP beads. Similarly, EGFP-Polη was detected in the anti-FLAG (USP7) immunoprecipitates and ectopically expressed FLAG-USP7 was found in the anti-GFP (Polη) immunoprecipitates (Fig. 2B). In addition, the interaction between the two ectopically expressed proteins was slightly enhanced upon UV irradiation (Fig. 2B).
Figure 2. Polη and USP7 directly interact in vivo and in vitro.
(A) Cell lysates prepared from HCT116 cells were immunoprecipitated with anti-USP7 or anti-Polη or normal IgG antibodies and Protein G/A beads. Bound proteins were recovered and used to detect USP7 and Polη along with whole-cell lysates as input control. (B) Cell lysates, from XP30RO-EGFP-Polη cells transiently transfected with empty vector (EV) or vector encoding FLAG-tagged USP7 for 24 h and treated with or without UV (20 J/m2), were immunoprecipitated with anti-GFP or anti-FLAG beads. Bound proteins were recovered and used to detect USP7 and Polη along with whole-cell lysates as input control. (C) Cell lysates from XP30RO-EGFP-Polη cells, transiently transfected with empty vector or vectors encoding FLAG-tagged wild-type USP7 (WT) or catalytic defective (CS) for 24 h, were immunoprecipitated with anti-FLAG beads. Bound proteins were recovered and used to detect USP7 and Polη. (D) Recombinant His-tagged USP7 and Polη were mixed in in vitro binding buffer and immunoprecipitated with anti-USP7 or anti-Polη antibodies, respectively. Bound proteins were recovered and used to detect USP7 and Polη along with mixed solution as input control.
To further probe the relationship between USP7 and Polη, we assessed the Polη-binding capability of wild-type or catalytically inactive USP7. In such experiments, mutations in catalytic domain enhanced USP7 interactions with EGFP-Polη (Fig. 2C). This is consistent with a substrate-trapping mechanism in which an inability of catalytically inactive USP7 to deubiquitinate substrates would manifest as prolonged binding and thus a tighter interaction. In addition, several slow migrating bands of Polη were co-immunoprecipitated with USP7-CS but not with USP7-WT (Fig. 2C). These low migrating bands are presumably the ubiquitinated forms of Polη.
To determine whether the interaction between USP7 and Polη is direct, we performed in vitro binding assay using purified recombinant proteins. As shown in Fig. 2D, recombinant Polη protein is pulled down with recombinant USP7 by anti-USP7 antibody. Moreover, recombinant USP7 is reciprocally pulled down with recombinant Polη by anti-Polη antibody. These observations suggest that USP7 directly interacts with Polη both in vivo and in vitro.
USP7 deubiquitinates and stabilizes Polη
Polη is polyubiquitinated and its cellular level is subject to regulation through ubiquitin-mediated proteolysis.7–9 Thus, we next tested whether USP7 regulates the levels of Polη via deubiquitination. For this, XP30RO-EGFP-Polη cells were co-transfected with HA-ubiquitin and various USP7 constructs or empty vector and subjected to treatment with MG132 for 4 h. GFP-Polη was immunoprecipitated from cell extracts by anti-GFP beads, and the poly-HA-ubiquitin chains of Polη were detected by anti-HA antibody. In agreement with previous reports,9 polyubiquitinated Polη accumulated in cells treated with proteasome inhibitor MG132. Overexpression of USP7-WT, but not the USP7-CS, significantly suppressed the appearance of polyubiquitinated Polη in cells (Fig. 3A), with decreased ubiquitination coinciding with increased cellular Polη. To further examine the deubiquitination activity of USP7 toward Polη, we utilized a cell-free assay system containing defined interacting components.19 For our in vitro deubiquitination, we used the commercial His-tagged recombinant USP7 against the purified Poly-HA-ub-GFP-Polη from XP30RO-EGFP-Polη cells transiently transfected with HA-tagged ubiquitin, as a substrate. As shown in Fig. 3B, the levels of polyubiquitinated Polη decreased gradually in a dose-dependent manner by increasing the concentration of recombinant USP7, suggesting that deubiquitination of Polη is mediated by USP7.
Figure 3. USP7 deubiquitinates Polη.
(A) Cell lysates from XP30RO-EGFP-Polη cells, transiently co-transfected with vectors encoding HA-tagged ubiquitin (HA-ub) and different FLAG-tagged USP7 (WT, CS) for 24 h, were immunoprecipitated with anti-GFP beads. Recovered proteins were detected by Western blotting with antibodies specific for HA and Polη. (B) HA-Poly-ub-GFP-Polη from XP30RO-EGFP-Polη cells transiently co-transfected with HA-tagged ubiquitin for 24 h were immunoprecipitated with anti-GFP beads, eluted and incubated with different concentration (0, 4, 8 and 16 nM) of recombinant His-tagged USP7. The reaction mixtures were subjected to Western blotting with anti-HA, USP7 and Polη antibodies. (C) Following transient transfection with empty vector or vector encoding FLAG-tagged USP7-WT or -CS for 24 h, HCT116 p53−/− cells were treated with CHX (50 ug/ml) for indicated times.
To further test whether USP7 overexpression actually increases the stability of Polη protein, we compared the turnover of Polη and Mdm2 in HCT116 p53−/− cells transiently transfected with USP7-WT and USP7-CS upon cycloheximide (CHX) treatment. As expected, USP7-WT overexpression delayed the turnover of both Polη and Mdm2, while USP7-CS overexpression slightly accelerated Mdm2 turnover and subsequently slightly delayed turnover of Polη (Fig. 3C). This dominant negative effect also provides an explanation of the phenomenon we previously observed in cells transiently transfected with USP7-CS (Fig. 1E).
Overexpression of USP7 increases UV-induced PCNA ubiquitination through stabilizing Polη
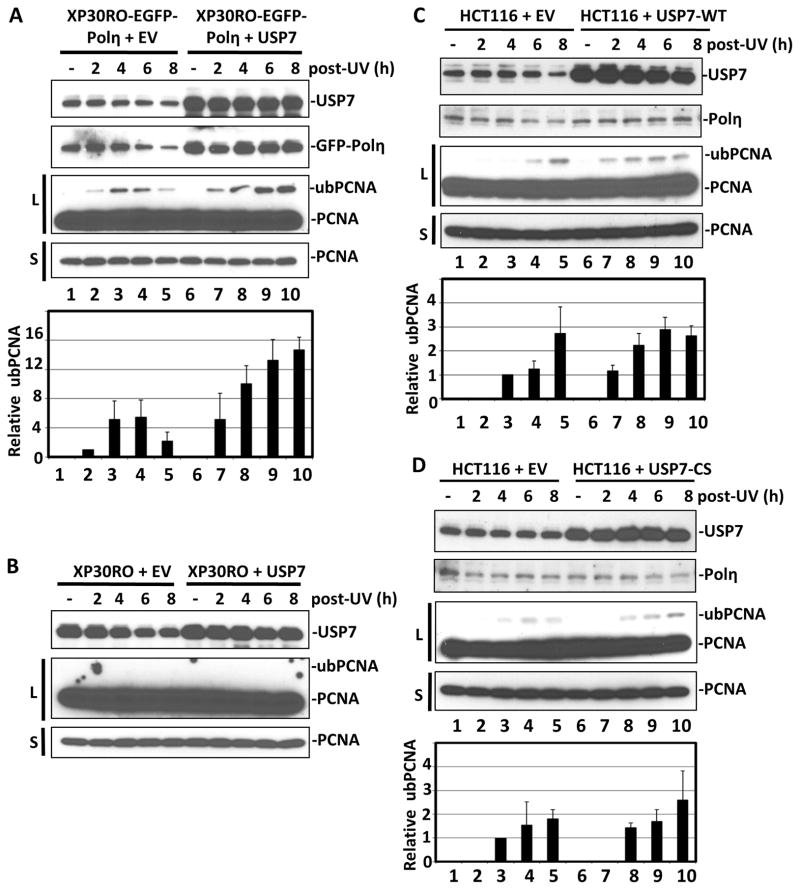
Recent report 3 demonstrated a novel role of Polη in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Since USP7 maintained the steady-levels of Polη, we pondered whether modulation of USP7 activity might also affect PCNA monoubiquitination in response to UV irradiation. To test this, we first compared the time course of UV-mediated PCNA monoubiquitination in XP30RO-EGFP-Polη cells transiently transfected with empty vector or vector encoding FLAG-tagged USP7. As shown in qualitative and quantitative results of Fig. 4A, respectively, overexpression of USP7 in XP30RO-EGFP-Polη cells increased the Polη levels and resulted in stronger and longer-lasting UV-induced PCNA monoubiquitination compared to control cells.
Figure 4. Overexpression of USP7 facilitates UV-induced PCNA ubiquitination by stabilizing Polη.
Cells were transiently transfected with empty vector (EV) or vector encoding FLAG-tagged USP7 for 24 h. The transfected cells, without or with UV (20 J/m2) irradiation, were harvested at indicated post-irradiation time points. The protein levels in cell lysates were detected by Western blotting with their respective antibodies. S and L, respectively, stand for short and long film exposures, which were necessary to depict uniform loading and changes in low abundant ubiquitinated PCNA (ubPCNA). Quantitative analysis of PCNA ubiquitination: The ratio of ubPCNA/PCNA at different time points were normalized to the ratio of XP30RO-EGFP-Polη cells at 2 h post-irradiation, which set as 1. Error bars represent ±SE of three independent Western blot experiments. (A) XP30RO-EGFP-Polη. (B) XP30RO cells. (C) HCT116 cells transfected with empty vector (EV) or vector encoding FLAG-tagged USP7-WT for 24 h. (D) HCT116 cells transfected with empty vector (EV) or vector encoding FLAG-tagged USP7-CS for 24 h.
To determine if the increased UV-induced PCNA monoubiquitination by USP7 overexpression was Polη dependent, we also assessed the time course of UV-induced PCNA monoubiquitination in XP30RO cells transiently transfected with empty vector or vector encoding FLAG-tagged USP7. Despite the most aggressive detection conditions, we were unable to observe any PCNA monoubiquitination in XP30RO cells even after 8 hours of UV irradiation, irrespective of whether USP7 was or was not overexpressed (Fig. 4B). Therefore, the USP7-facilitated UV-induced PCNA ubiquitination is Polη-dependent.
To validate the above observation, the time courses of UV-induced PCNA monoubiquitination in cells with endogenous Polη were assessed. In this experiment, HCT116 rather HCT116 USP7−/− cells were used. HCT116 USP7−/− cells grow extremely slowly because USP7 knockout leads to constitutively high level of p53 and p21.17,18 These cells are also defective for the dissociation of MCM proteins from chromatin in S phase.20 Therefore, HCT116 USP7−/− cells are not a suitable cell line for UV-induced PCNA monoubiquitination experiments. In contrast, USP7 overexpression has no significant effects on cell cycle.10 So, we transiently transfected HCT116 cells with empty vector or vector encoding FLAG-tagged USP7-WT or -CS, to compare the time courses of UV-induced PCNA monoubiquitination. Overexpression of USP7-WT once again resulted in a clear increase of UV-induced PCNA monoubiquitination in HCT116 cells, while overexpression of USP7-CS did not affect a change of UV-induced PCNA monoubiquitination in HCT116 cells (Fig. 4C & D). Taken together, our data suggested that USP7 overexpression facilitates UV-induced PCNA monoubiquitination through stabilizing Polη.
Here, we have identified USP7 as a novel regulator of Polη stability. Interestingly, changes in cellular USP7 levels modulate the endogenous Polη level through direct and indirect mechanisms. USP7 can directly deubiquitinate and stabilize Polη in cells. To our knowledge, this is the first reported specific deubiquitinating enzyme of Polη. Besides its direct effects, USP7 may indirectly affect the steady-level of Polη by stabilizing Mdm2, a known E3 ubiquitin ligase that mediates polyubiquitination and controls protein turnover of Polη and p53. In light of our data alongside previous reports,9 we propose a model in which the coupling of opposing activities of ubiquitination by Mdm2 and deubiquitination by USP7 offers an elegant way to tightly regulate Polη (Fig. S2) as well as its known transcription activator p53. Since USP7 forms a complex with Mdm2 and is required for Mdm2 stability, a knockout of USP7 in cells leads to Mdm2 self-ubiquitination and proteasomal degradation, which in turn results in Polη accumulation. Conversely, overexpression of USP7, directly reducing the polyubiquitination of Polη mediated by Mdm2, also leads to Polη stability. The importance of the model is highlighted by the fact that USP7 as a dual-role regulator, together with Mdm2, comprises a sophisticated regulatory circuitry that dynamically controls Polη turnover.
USP7 have been found to function in both global genomic repair (GGR) and transcription-repair (TCR) with several NER and DNA damage response (DDR) protein as substrates.11,14,21,22 Our finding of Polη as a substrate of USP7 suggests that USP7 is also involved in DNA lesion bypass, an important DDR mechanism of stress tolerance. It is known that PCNA ubiquitination plays very important roles in several genetic stress-tolerance pathways such as TLS. Monoubiquitination of PCNA increases its affinity for the specialized TLS polymerase such as Polη. Recently, Durando et al.3 demonstrated a novel role of Polη in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. They further showed that cellular Polη level is very low in comparison to cellular Rad18 level, which explains why PCNA monoubiquitination is exquisitely sensitive to slight alteration in Polη levels. Therefore, USP7 overexpression would increase stability of Polη and subsequent PCNA monoubiquitination, and in turn facilitates the recruitment of TLS polymerases to bypass DNA lesions. Considering that aberrant USP7 expressions were frequently found in various tumors and elevated TLS is involved in resistance to chemotherapeutic agents, our findings that USP7-mediated stabilization of Polη facilitates PCNA monoubiquitination-mediated stress-tolerance pathways provide unique mechanistic insights for USP7-related tumorigenesis and may be useful for developing future therapeutic strategy.
Supplementary Material
Acknowledgments
We are thankful to Dr. Bert Vogelstein for HCT116, HCT116 p53−/− and HCT116 USP7−/− cells, Dr. Alan Lehmann for XP30RO and XP30RO-GFP-Polη cells, Drs. Yanhui Xu and Yang Shi for FLAG-tagged WT and CS USP7 constructs, Dr. Carol Prives for Myc-Mdm2 construct. This work was supported by grants from the National Institutes of Health (ES012991 and ES023883) to AAW.
Abbreviations
- USP7
ubiquitin-specific protease 7
- Polη
DNA polymerase eta
- TLS
translesion DNA synthesis
- Mdm2
murine double minute 2
- PCNA
proliferating cell nuclear antigen
References
- 1.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Durando M, Tateishi S, Vaziri C. A non-catalytic role of DNA polymerase eta in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 2013;41:3079–3093. doi: 10.1093/nar/gkt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skoneczna A, McIntyre J, Skoneczny M, Policinska Z, Sledziewska-Gojska E. Polymerase eta is a short-lived, proteasomally degraded protein that is temporarily stabilized following UV irradiation in Saccharomyces cerevisiae. J Mol Biol. 2007;366:1074–1086. doi: 10.1016/j.jmb.2006.11.093. [DOI] [PubMed] [Google Scholar]
- 8.Kim SH, Michael WM. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell. 2008;32:757–766. doi: 10.1016/j.molcel.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung YS, Qian Y, Chen X. DNA polymerase eta is targeted by Mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst) 2012;11:177–184. doi: 10.1016/j.dnarep.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J Cell Biol. 2009;184:13–19. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang QE, Wani AA. Ubiquitin-specific Protease 7 Regulates Nucleotide Excision Repair through Deubiquitinating XPC Protein and Preventing XPC Protein from Undergoing Ultraviolet Light-induced and VCP/p97 Protein-regulated Proteolysis. J Biol Chem. 2014;289:27278–27289. doi: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 13.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JH, Demmers JA, Fousteri M, Vermeulen W, Marteijn JA. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44:598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 15.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 16.Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3:689–692. [PubMed] [Google Scholar]
- 17.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 18.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 19.Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, Guo R, Diao J, Lan F, Harper JW, Shi YG, Xu Y, Shi Y. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci U S A. 2012;109:4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagannathan M, Nguyen T, Gallo D, Luthra N, Brown GW, Saridakis V, Frappier L. A role for USP7 in DNA replication. Mol Cell Biol. 2014;34:132–145. doi: 10.1128/MCB.00639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarasin A. UVSSA and USP7: new players regulating transcription-coupled nucleotide excision repair in human cells. Genome Med. 2012;4:44. doi: 10.1186/gm343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, Neilan EG, Honma M, Nohmi T, Yasui A, Tanaka K. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet. 2012;44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.