Abstract
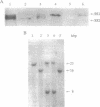
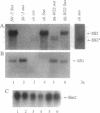
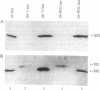
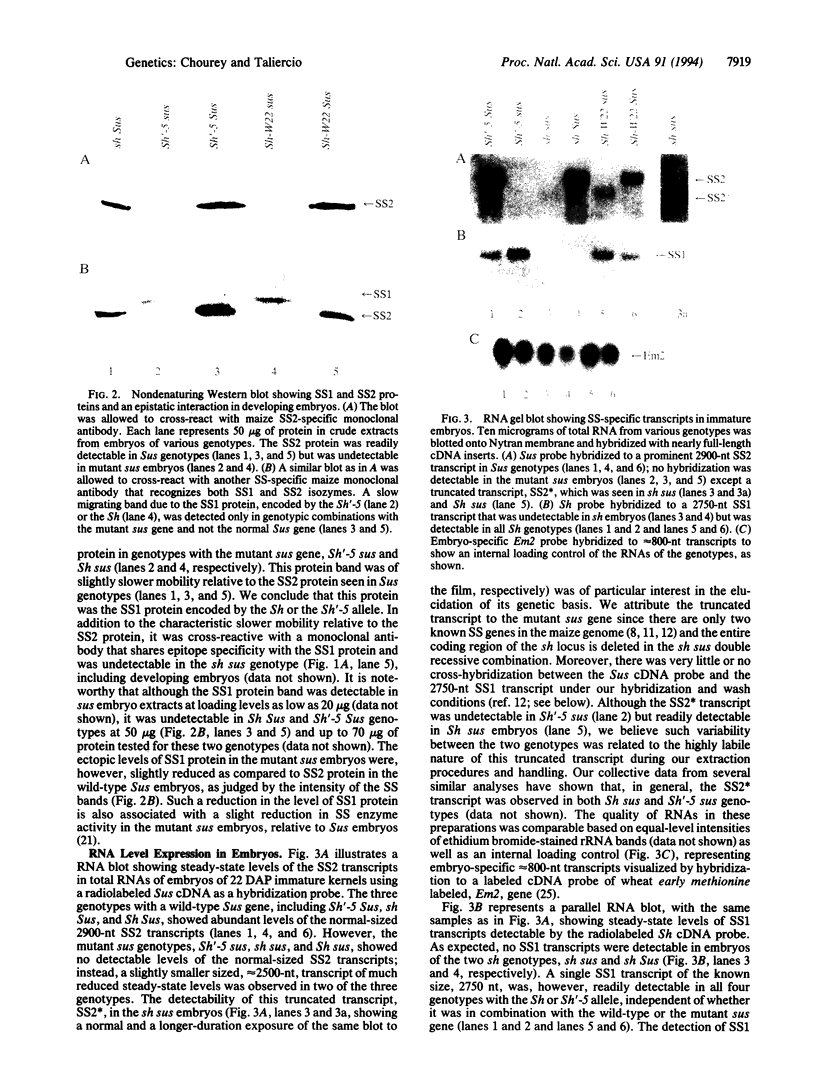
A tissue-specific epistatic mode of gene interaction was observed between molecularly homologous genes Sh1 and Sus1 (hereafter, Sh and Sus), encoding the sucrose synthase (SS) isozymes, SS1 and SS2, respectively. In Sh Sus genotype, both SS genes were expressed simultaneously and approximately equally in young seedlings; however, only the Sus-encoded SS2 protein was seen in the developing embryos. By contrast, the mutant sus genotype, lacking detectable levels of the SS2 protein in various tissues tested, showed expression of the Sh locus as judged by the detection of the SS1 protein in such embryos. Ectopic expression in embryos was seen from two separate Sh alleles, Sh-W22 and Sh'-5 (a revertant allele derived upon Ds excision from sh-m5933). In each case, the Sh expression at the protein level in embryos was unique to genotypes with the mutant sus gene. Based on the observed lack of phenotypic change in the sus mutant, we suggest that the ectopic expression of the Sh in otherwise Sus-specific tissues leads to functional compensation. There was no epistatic interaction of Sh and Sus at the RNA level as SS1 transcripts were detectable in both Sus and sus embryos. Thus, embryo specificity between the two SS genes was determined at posttranscriptional or at translational level of control. We surmise on the basis of these data that metabolic regulatory controls seem to override the normal constraints of tissue and cell specificity of the nonallelic isozyme genes to maintain efficient use of the pathways.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer C. D., Preiss J. Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol. 1981 Jun;67(6):1141–1145. doi: 10.1104/pp.67.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V. L., Radicella J. P., Robbins T. P., Chen J., Turks D. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell. 1989 Dec;1(12):1175–1183. doi: 10.1105/tpc.1.12.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey P. S., Nelson O. E. The enzymatic deficiency conditioned by the shrunken-1 mutations in maize. Biochem Genet. 1976 Dec;14(11-12):1041–1055. doi: 10.1007/BF00485135. [DOI] [PubMed] [Google Scholar]
- Chourey P. S., Taliercio E. W., Kane E. J. Tissue-Specific Expression and Anaerobically Induced Posttranscriptional Modulation of Sucrose Synthase Genes in Sorghum bicolor M. Plant Physiol. 1991 Jun;96(2):485–490. doi: 10.1104/pp.96.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courage-Tebbe U., Döring H. P., Fedoroff N., Starlinger P. The controlling element Ds at the Shrunken locus in Zea mays: structure of the unstable sh-m5933 allele and several revertants. Cell. 1983 Sep;34(2):383–393. doi: 10.1016/0092-8674(83)90372-0. [DOI] [PubMed] [Google Scholar]
- Dickinson D. B., Preiss J. Presence of ADP-Glucose Pyrophosphorylase in Shrunken-2 and Brittle-2 Mutants of Maize Endosperm. Plant Physiol. 1969 Jul;44(7):1058–1062. doi: 10.1104/pp.44.7.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echt C. S., Chourey P. S. A Comparison of Two Sucrose Synthetase Isozymes from Normal and shrunken-1 Maize. Plant Physiol. 1985 Oct;79(2):530–536. doi: 10.1104/pp.79.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J. Control of mRNA Stability in Higher Plants. Plant Physiol. 1993 Aug;102(4):1065–1070. doi: 10.1104/pp.102.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris T., Weber D., Wright S. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics. 1988 Feb;118(2):353–363. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. F., Nguyen-Quoc B., Chourey P. S., Yelle S. Complete nucleotide sequence of the maize (Zea mays L.) sucrose synthase 2 cDNA. Plant Physiol. 1994 Jan;104(1):293–294. doi: 10.1104/pp.104.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer E. E., Rodriguez R. L. Metabolic regulation of rice alpha-amylase and sucrose synthase genes in planta. Plant J. 1992 Jul;2(4):517–523. [PubMed] [Google Scholar]
- Kim H. G., Miller R. F. Rods and cones activate different excitatory amino acid receptors on the mudpuppy retinal horizontal cell. Brain Res. 1991 Jan 4;538(1):141–146. doi: 10.1016/0006-8993(91)90388-c. [DOI] [PubMed] [Google Scholar]
- Koch K. E., Nolte K. D., Duke E. R., McCarty D. R., Avigne W. T. Sugar Levels Modulate Differential Expression of Maize Sucrose Synthase Genes. Plant Cell. 1992 Jan;4(1):59–69. doi: 10.1105/tpc.4.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litts J. C., Colwell G. W., Chakerian R. L., Quatrano R. S. The nucleotide sequence of a cDNA clone encoding the wheat Em protein. Nucleic Acids Res. 1987 Apr 24;15(8):3607–3618. doi: 10.1093/nar/15.8.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas C., Schaal S., Werr W. A feedback control element near the transcription start site of the maize Shrunken gene determines promoter activity. EMBO J. 1990 Nov;9(11):3447–3452. doi: 10.1002/j.1460-2075.1990.tb07552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T., Frommer W. B., Salanoubat M., Willmitzer L. Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. Plant J. 1993 Aug;4(2):367–377. doi: 10.1046/j.1365-313x.1993.04020367.x. [DOI] [PubMed] [Google Scholar]
- McCarty D. R., Shaw J. R., Hannah L. C. The cloning, genetic mapping, and expression of the constitutive sucrose synthase locus of maize. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9099–9103. doi: 10.1073/pnas.83.23.9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson O. E., Chourey P. S., Chang M. T. Nucleoside Diphosphate Sugar-Starch Glucosyl Transferase Activity of wx Starch Granules. Plant Physiol. 1978 Sep;62(3):383–386. doi: 10.1104/pp.62.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Quoc B., Krivitzky M., Huber S. C., Lecharny A. Sucrose Synthase in Developing Maize Leaves: Regulation of Activity by Protein Level during the Import to Export Transition. Plant Physiol. 1990 Oct;94(2):516–523. doi: 10.1104/pp.94.2.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K. K., Fink G. R. Two anthranilate synthase genes in Arabidopsis: defense-related regulation of the tryptophan pathway. Plant Cell. 1992 Jun;4(6):721–733. doi: 10.1105/tpc.4.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Metabolic repression of transcription in higher plants. Plant Cell. 1990 Oct;2(10):1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B., Werr W., Starlinger P., Bennett D. C., Zokolica M., Freeling M. The Shrunken gene on chromosome 9 of Zea mays L is expressed in various plant tissues and encodes an anaerobic protein. Mol Gen Genet. 1986 Dec;205(3):461–468. doi: 10.1007/BF00338083. [DOI] [PubMed] [Google Scholar]
- Taliercio E. W., Chourey P. S. Post-transcriptional control of sucrose synthase expression in anaerobic seedlings of maize. Plant Physiol. 1989 Aug;90(4):1359–1364. doi: 10.1104/pp.90.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thach R. E. Cap recap: the involvement of eIF-4F in regulating gene expression. Cell. 1992 Jan 24;68(2):177–180. doi: 10.1016/0092-8674(92)90461-k. [DOI] [PubMed] [Google Scholar]
- Theodorakis N. G., Cleveland D. W. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol. 1992 Feb;12(2):791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursin V. M., Irvine J. M., Hiatt W. R., Shewmaker C. K. Developmental analysis of elongation factor-1 alpha expression in transgenic tobacco. Plant Cell. 1991 Jun;3(6):583–591. doi: 10.1105/tpc.3.6.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth G. J., Redinbaugh M. G., Scandalios J. G. A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem. 1988 Jul;172(1):279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Wendel J. F., Stuber C. W., Goodman M. M., Beckett J. B. Duplicated plastid and triplicated cytosolic isozymes of triosephosphate isomerase in maize (Zea mays L.). J Hered. 1989 May-Jun;80(3):218–228. doi: 10.1093/oxfordjournals.jhered.a110839. [DOI] [PubMed] [Google Scholar]
- Werr W., Frommer W. B., Maas C., Starlinger P. Structure of the sucrose synthase gene on chromosome 9 of Zea mays L. EMBO J. 1985 Jun;4(6):1373–1380. doi: 10.1002/j.1460-2075.1985.tb03789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]