Abstract
Rationale: The airway transcriptome includes genes that contribute to the pathophysiologic heterogeneity seen in individuals with asthma.
Objectives: We analyzed sputum gene expression for transcriptomic endotypes of asthma (TEA), gene signatures that discriminate phenotypes of disease.
Methods: Gene expression in the sputum and blood of patients with asthma was measured using Affymetrix microarrays. Unsupervised clustering analysis based on pathways from the Kyoto Encyclopedia of Genes and Genomes was used to identify TEA clusters. Logistic regression analysis of matched blood samples defined an expression profile in the circulation to determine the TEA cluster assignment in a cohort of children with asthma to replicate clinical phenotypes.
Measurements and Main Results: Three TEA clusters were identified. TEA cluster 1 had the most subjects with a history of intubation (P = 0.05), a lower prebronchodilator FEV1 (P = 0.006), a higher bronchodilator response (P = 0.03), and higher exhaled nitric oxide levels (P = 0.04) compared with the other TEA clusters. TEA cluster 2, the smallest cluster, had the most subjects that were hospitalized for asthma (P = 0.04). TEA cluster 3, the largest cluster, had normal lung function, low exhaled nitric oxide levels, and lower inhaled steroid requirements. Evaluation of TEA clusters in children confirmed that TEA clusters 1 and 2 are associated with a history of intubation (P = 5.58 × 10−6) and hospitalization (P = 0.01), respectively.
Conclusions: There are common patterns of gene expression in the sputum and blood of children and adults that are associated with near-fatal, severe, and milder asthma.
Keywords: molecular endotyping, genomic, RNA, severe asthma, pathway analysis
At a Glance Commentary
Scientific Knowledge on the Subject
Asthma is a chronic inflammatory disease of the airways that is clinically and physiologically heterogeneous. Patterns that can be captured by analyzing gene expression levels in the airway and circulation could resolve genes and pathways that contribute to this heterogeneity. Comprehensive studies that examine disease heterogeneity by measuring gene expression in the sputum and associate it with important clinical features of asthma are limited.
What This Study Adds to the Field
This study is the first to use noninvasive analysis of sputum gene expression to identify transcriptomic endotypes of asthma clusters that correlate with clinical characteristics of severe disease including a history of hospitalization and near-fatal asthma attack. The transcriptomic endotypes of asthma clusters are associated with a gene signature in the blood and are evident in both children and adults with asthma, which suggests that there are common patterns of gene expression among children and adults with asthma.
Asthma is a chronic inflammatory disease of the airways that will likely afflict more than 10% of the U.S. population by the end of this decade (1). Differences in genetic susceptibility, environmental exposures, and medication compliance are known to contribute to the heterogeneous clinical manifestations of disease (2, 3). However, it is increasingly evident that pathobiologic alterations in asthma are also heterogeneous and that differences in the expression of many biologic pathways underlie differences in the phenotypic expressions of the disease (4). Therefore, asthma could be considered as a collection of airway diseases, each driven by a different set of biologic networks with unique but overlapping genomic, transcriptomic, inflammatory, physiologic, and clinical features of disease. In keeping with this paradigm shift, asthma research efforts have moved to defining subgroups of patients with asthma that have different clinical and physiologic manifestations of disease that may be driven by novel biologic mechanisms or relative differences in the expression of known pathways, such as those driven by IL-13 and IL-5 (5, 6). Ultimately, dissecting these subgroups of disease will enable pathogenesis research, therapeutic development, and clinical management to focus on distinct subsets of asthma and their associated clinical phenotypes, leading to a more personalized approach to disease management (7).
To date, most efforts to define asthma subgroups have relied on clustering individuals by clinical features, such as atopic history, age of onset, lung function, or symptoms of severity. These studies, including the Severe Asthma Research Program and the Childhood Asthma Management Program characterizations of asthma clusters, have generated novel insights, but are driven by analytical approaches that are based on differences in parameters that may be distal to many molecular perturbations associated with the disease (8, 9). In contrast to these clinically biased approaches, unsupervised integrative functional transcriptomics has the potential to discriminate asthma subtypes at a level that is reflective of patterns in gene expression, pathobiology, and common clinical and physiologic features of disease: transcriptional endotypes of asthma (TEA) clusters (10, 11). To this end, we conducted an unsupervised clustering analysis of gene expression in the induced sputum of adults and children with asthma and identified three TEA clusters and their associated clinical features of disease. Some of the results of these studies have been previously reported in the form of an abstract (12).
Methods
Yale Center for Asthma and Airway Diseases Cohort
A cross-sectional analysis was conducted on sputum RNA samples collected from subjects with asthma and control subjects that completed the Yale Center for Asthma and Airway Diseases (YCAAD) phenotyping protocol between September 2009 and June 2012. Subjects were greater than or equal to 12 years of age, nonsmokers, and with less than or equal to 10 pack-years of smoking history. Inclusion criteria for asthma included a history, physical examination, and physiologic testing consistent with a diagnosis of asthma based on National Asthma Education and Prevention Program guidelines. Exclusion criteria included smoking within the past year, a history chronic lung disease other than asthma (i.e., chronic obstructive pulmonary disease, allergic bronchopulmonary aspergillosis, Churg-Strauss syndrome, pulmonary vascular disease, or interstitial lung disease); other severe chronic conditions including congestive heart failure, chronic kidney disease, liver disease, or viral infection; or inability to safely undergo the studies required for participation. The protocol was approved by the Yale University School of Medicine Human Investigation Committee, and all patients or their parents provided informed consent.
YCAAD Phenotyping Protocol
An asthma questionnaire was administered and whole blood was collected in RNA isolation tubes (Life Technologies or Applied Biosystems, Grand Island, NY). Exhaled nitric oxide (FeNO) was measured and spirometry was conducted in adherence with the American Thoracic Society guidelines before and after short-acting bronchodilator administration (13). Sputum induction was performed with hypertonic saline, as previously described (14–17). Mucus plugs were dissected from the sputum sample using a microscope, and the cellular and aqueous compartments separated. Total cell count, viability, and differential were determined by hemocytometer, trypan blue exclusion, and Wright-Giemsa stain, respectively, and cell pellets were stored in All-in-One RNA stabilization buffer (Norgen Biotek, Thorhold, Canada).
Genomic Analysis
Sputum cell pellets were processed using the All-in-One purification kit (Norgen Biotek), checked on an Agilent bioanalyzer (Agilent Technologies, Santa Clara, CA), and, if needed, treated again to remove DNA contamination (Qiagen, Gaithersburg, MD). A total of 10 ng of sputum RNA was amplified using the WT-Ovation Pico RNA amplification System (NuGen, San Carlos, CA) and processed per Affymetrix (Santa Clara, CA) protocols, as previously described (18). Total RNA from the blood was isolated using a column-based system (total-RNA kit; Norgen), and if needed, DNA contamination was removed using a DNA clear kit (Qiagen). Hemoglobin reduction of blood samples was used to remove hemoglobin gene transcripts (GLOBINclear Kit; Ambion, Austin, TX) and samples were checked by Agilent bioanalyzer. Purified RNA from the sputum or blood was processed for gene expression using the Affymetrix HuGene 1.0 ST gene arrays following manufacturer’s protocols as previously described. Samples with RNA integrity numbers less than 4.0 were rejected from the analysis. The data can be obtained from GEO database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE56396.
Computational Analyses
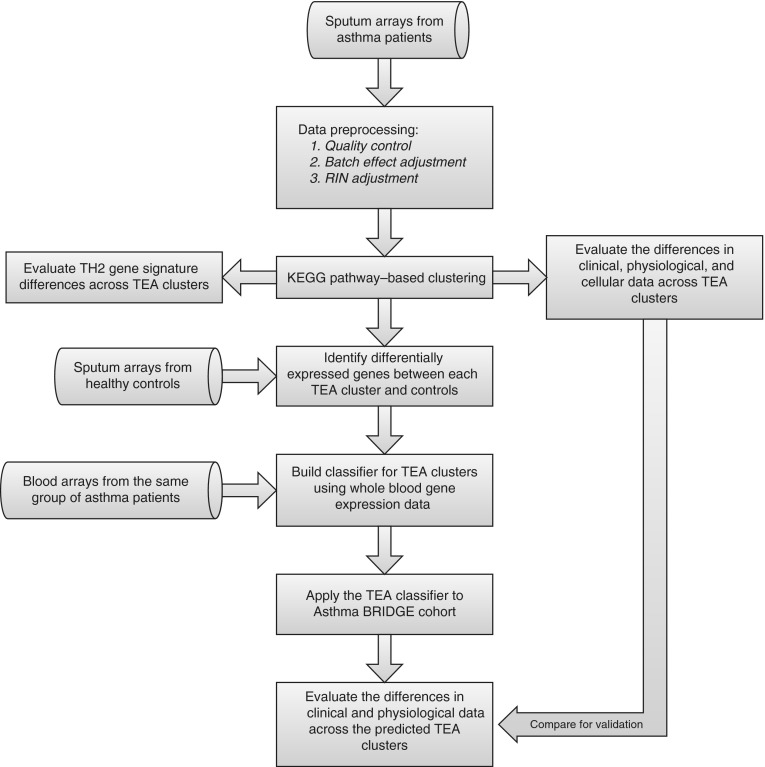
An overview of the computational analysis flow can be found in Figure 1. Raw microarray intensity data were processed using R packages for normalization, quality check, batch effect adjustment, and RIN adjustment. Distances between samples were evaluated using a pathway-based method and three clusters were selected to minimize the connectivity criteria (see online supplement for details) (19). K-means clustering was applied to assign samples into the selected three TEA clusters. Kruskal-Wallis tests were used to assess differences in continuous clinical, physiologic, and inflammatory asthma phenotypes between the clusters, and the chi-square or Cochran-Armitage test was used to assess differences in categorical phenotypes. False discovery rate (FDR) was estimated using a permutation-based method to adjust for the multiple testing error (20). Differentially expressed genes (DEGs) between each TEA cluster and control subjects were identified as genes with an FDR less than 0.05 using the Student t test (21). The online supplement provides more details on computational analyses.
Figure 1.
Identification of transcriptomic endotypes of asthma (TEA) clusters. Diagram showing an overview of the computational analysis flow. BRIDGE = BioRepository for Integrative Genomic Exploration; KEGG = Kyoto Encyclopedia of Genes and Genomes; RIN = RNA integrity number.
Validation Studies
A TEA cluster classifier was built using an L1 regularized logistic regression model in the YCAAD cohort to predict TEA cluster using blood gene expression and visualized by principal component analysis (see online supplement for details) (22, 23). This classifier was applied to 870 whole blood samples selected from the Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE) (24, 25). Cross-platform replication of the TEA cluster clinical phenotypes was conducted (24, 25).
Results
Identification of Sputum TEA Clusters in YCAAD Cohort
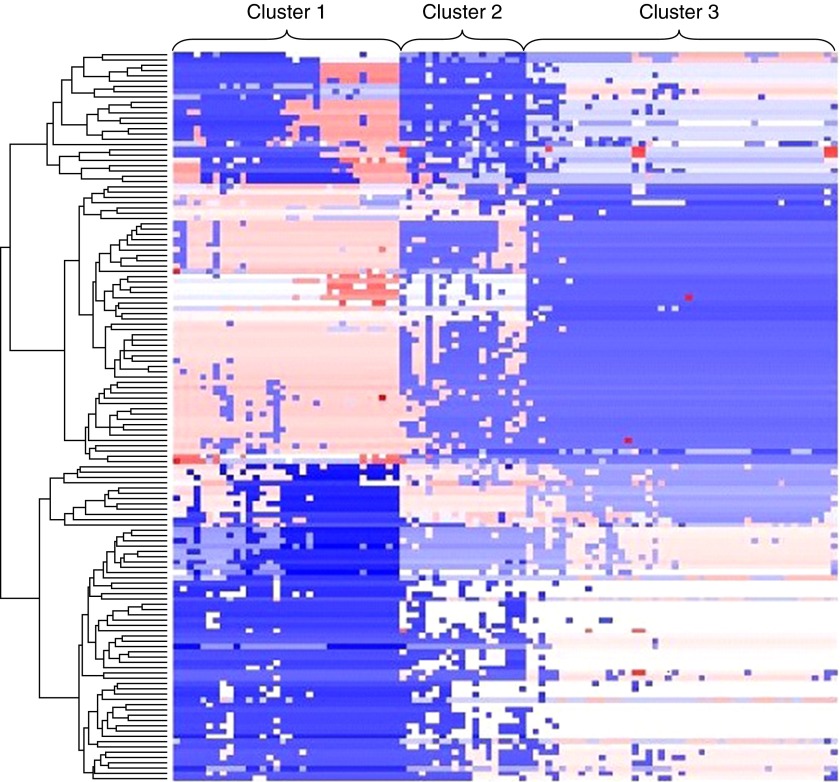
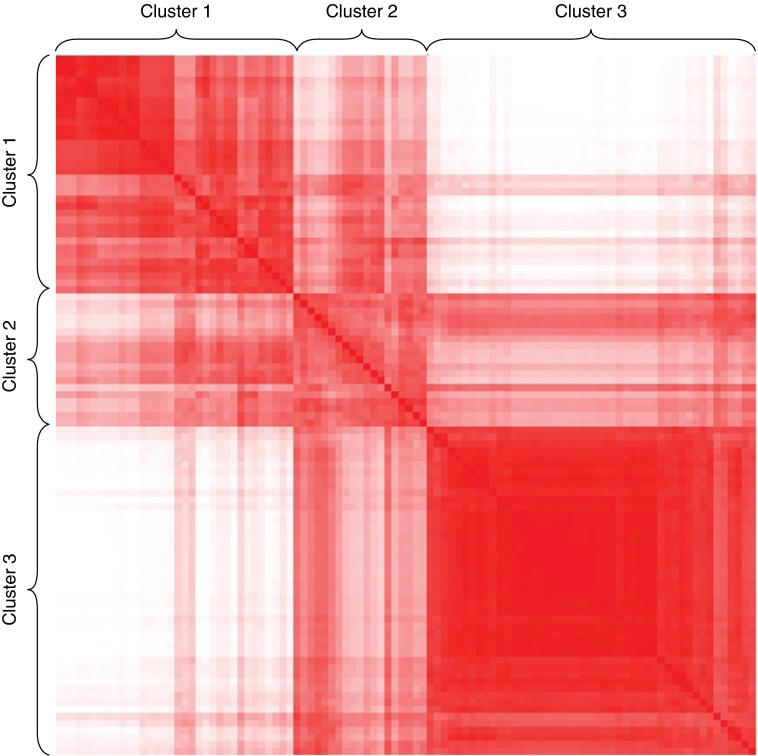
The sputum expression levels of 5,500 genes from 186 Kyoto Encyclopedia of Genes and Genomes pathways were used to assess the pathway-based distance between samples followed by unsupervised K means clustering to define sputum TEA clusters (Figure 2). Three clusters were selected based on the connectivity criteria (see Figure E5 in the online supplement). The “relatedness” of the samples within each cluster was evident on a sample distance matrix (Figure 3). This demonstrated that samples within TEA cluster 3 are the most strongly related (darkest red) and most homogeneous, followed by TEA cluster 1, and then TEA cluster 2, the smallest cluster. The clusters are not associated with differences in the sputum inflammatory cell populations among the clusters. Therefore, using only sputum gene expression as a discriminator, distinct subgroups of disease can be defined within a heterogeneous group of individuals with asthma.
Figure 2.
Heatmap showing the clustering results by Kyoto Encyclopedia of Genes and Genomes pathways using MCLUST. The color represents the clustering assignment of each sample by the Kyoto Encyclopedia of Genes and Genomes pathways.
Figure 3.
Pathway-based distance matrix among the clusters. The color of entry represents the pathway-based distance between the corresponding two samples. Red represents a small distance (samples are strongly related), and white represents a longer distance, showing the strength of the clusters (samples are weakly related). Samples within transcriptomic endotypes of asthma cluster 3 are the most strongly related and most homogeneous, followed by clusters 1 and 2, respectively.
Phenotypic Characteristics of TEA Clusters
To determine whether the defined TEA clusters correspond to distinct phenotypes of asthma, the clinical, physiologic, and biologic parameters were compared among the TEA clusters (Table 1). The subjects in TEA cluster 1 required a higher daily dose of inhaled steroids (mean daily inhaled corticosteroid [ICS] dose, 617 μg/day; P = 0.04) and were more likely to have a history of an intubation for asthma, compared with the other TEA clusters (18%; P = 0.05). In addition, TEA cluster 1 had the lowest prebronchodilator and post- bronchodilator FEV1 (prebronchodilator percent predicted FEV1, 73 ± 24%; P < 0.01), more bronchodilator reversibility (12 ± 12%; P = 0.03), and elevated FeNO levels (mean, 53 ± 43 ppb; P = 0.03), compared with the other TEA clusters (Table 2). The fewest number of individuals (n = 19) were in TEA cluster 2. Compared with the other clusters, TEA cluster 2 had the highest percentage of subjects with no atopy (26%; P = 0.02), subjects of Hispanic origin (P = 0.04), and the highest percentage of individuals that were hospitalized for asthma (68%; P = 0.03). TEA cluster 3, the largest cluster (47%), demonstrated the “mildest” phenotypic characteristics of asthma. This TEA cluster had the lowest percentage of subjects with a history of hospitalization or intubation for asthma and the lowest daily ICS dose. In addition, subjects in TEA cluster 3 had preserved prebronchodilator and post-bronchodilator lung function, minimal bronchodilator reversibility, and the lowest FeNO (mean, 38 ± 27 ppb; P = 0.04) compared with the other TEA clusters (Table 2). There were no between-cluster differences in sputum cell counts, cell differentials, viability, DNase treatment percentage, percentage of patients on ICS, or RNA integrity number, suggesting that the TEA clustering was not related to a particular cell population, sample processing, or treatment (Table 3).
Table 1.
Phenotypic Characteristics of TEA Clusters in the YCAAD Cohort
| Control Subjects (n = 12) | Cluster 1 | Cluster 2 | Cluster 3 | P Value | |
|---|---|---|---|---|---|
| Prevalence, n | 34 | 19 | 47 | 0.003 | |
| Age at visit, yr | 37 ± 14 | 51 ± 13 | 49 ± 16 | 45 ± 17 | 0.32 |
| Female sex, n (%) | 5 (42) | 23 (68) | 15 (79) | 39 (83) | 0.26 |
| Race | 0.58 | ||||
| White, n (%) | 12 (100) | 22 (65) | 14 (74) | 37 (79) | |
| Black, n (%) | 0 (0) | 10 (29) | 4 (21) | 7 (15) | |
| Other, n (%) | 0 (0) | 1 (3) | 1 (5) | 3 (6) | |
| Hispanic origin, n (%) | 0 (0) | 1 (3) | 4 (21) | 7 (15) | 0.04 |
| BMI, kg/m2 | 23.6 ± 2.8 | 30.0 ± 7.2 | 30 ± 7.3 | 29.3 ± 8.0 | 0.84 |
| History of atopy, n (%) | 7 (58) | 33 (97) | 14 (74) | 43 (92) | 0.02 |
| Age of symptom onset | NA | 25.8 ± 19.1 | 29.3 ± 20.4 | 20.7 ± 20.9 | 0.17 |
| Disease duration, yr | NA | 25.2 ± 17.5 | 20.7 ± 16.9 | 24.2 ± 17.3 | 0.70 |
| History of hospitalization, n (%) | NA | 13 (38.1) | 13 (68.4) | 16 (34.0) | 0.03 |
| History of intubations, n (%) | NA | 6.0 (18) | 2.0 (11) | 2.0 (4) | 0.05 |
| OCS tapers in past year, n (%) | NA | 19 (55.9) | 12 (63.2) | 24 (51.1) | 0.67 |
| ACT score | NA | 16 ± 6.4 | 14 ± 6.6 | 18 ± 5.1 | 0.22 |
| ICS dose, μg/d | NA | 617 ± 448 | 530 ± 449 | 396 ± 356 | 0.04 |
| ICS use, yes, n (%) | NA | 27 (79) | 17 (89) | 31 (66) | 0.10 |
| Chronic OCS use, n (%) | NA | 4 (11.8) | 2 (10.5) | 3 (6.4) | 0.68 |
Definition of abbreviations: ACT = Asthma Control Test; BMI = body mass index; ICS = inhaled corticosteroids; NA = not applicable; OCS = oral corticosteroids; TEA = transcriptomic endotypes of asthma; YCAAD = Yale Center for Asthma and Airway Diseases.
Data are means ± SD, except where indicated. P values for comparisons among TEA clusters were determined using Kruskal-Wallis or Cochran-Armitage test. The false discovery rate estimated by the permutation-based method is 11%.
Table 2.
Pulmonary Function of TEA Clusters in YCAAD Cohort
| Control Subjects (n = 12) | Cluster 1 (n = 34) | Cluster 2 (n = 19) | Cluster 3 (n = 47) | P Value | |
|---|---|---|---|---|---|
| FEV1, % of predicted value | |||||
| Pre–β2-agonist use | 96 ± 11 | 73 ± 24 | 76 ± 22 | 86 ± 22 | 0.006 |
| Post–β2-agonist use | 98 ± 14 | 80 ± 24 | 81 ± 20 | 91 ± 22 | 0.493 |
| FVC, % of predicted value | |||||
| Pre–β2-agonist use | 86 ± 22 | 85 ± 22 | 86 ± 18 | 96 ± 19 | 0.09 |
| Post–β2-agonist use | 91 ± 20 | 90 ± 20 | 88 ± 18 | 97 ± 18 | 0.28 |
| FEV1/FVC | |||||
| Pre–β2-agonist use | 0.77 ± 0.62 | 0.67 ± 0.13 | 0.70 ± 0.11 | 0.72 ± 0.10 | 0.13 |
| Post–β2-agonist use | 0.79 ± 0.50 | 0.70 ± 0.13 | 0.72 ± 0.12 | 0.80 ± 0.10 | 0.06 |
| BDR, % | 2 ± 6.2 | 12 ± 12 | 9 ± 13 | 6 ± 7 | 0.03 |
| FeNO, ppb | 20 ± 9.7 | 53 ± 43 | 52 ± 42 | 38 ± 27 | 0.04 |
Definition of abbreviations: BDR = bronchodilator response; FeNO = exhaled nitric oxide; TEA = transcriptomic endotypes of asthma; YCAAD = Yale Center for Asthma and Airway Diseases.
Data are means ± SD. P values for comparisons among TEA clusters were determined using Kruskal-Wallis or Cochran-Armitage test. The false discovery rate estimated by the permutation-based method is 5.6%.
Table 3.
Sputum Characteristics of TEA Clusters in the YCAAD Cohort
| Control Subjects (n = 12) | Cluster 1 (n = 34) | Cluster 2 (n = 19) | Cluster 3 (n = 47) | P Value | |
|---|---|---|---|---|---|
| Mucus cell concentration* | 40.86 ± 20.98 | 83.02 ± 105.75 | 89.23 ± 143.61 | 73.72 ± 62.48 | 0.63 |
| Squamous, % | 8.2 ± 6.7 | 7.9 ± 7.0 | 8.0 ± 5.9 | 9.2 ± 6.9 | 0.60 |
| Viability, % | 58.1 ± 9.6 | 56.5 ± 16.1 | 64.4 ± 11.9 | 61.7 ± 17.8 | 0.14 |
| Neutrophils, % | 34.6 ± 10.0 | 41.5 ± 13.0 | 41.9 ± 15.2 | 37.8 ± 14.6 | 0.34 |
| Eosinophil, % | 1.5 ± 1.8 | 5.8 ± 6.7 | 4.7 ± 5.9 | 5.2 ± 7.7 | 0.91 |
| Macrophage, % | 61.3 ± 11.8 | 50.9 ± 13.0 | 50.9 ± 16.0 | 55.4 ± 15.4 | 0.31 |
| Lymphocyte, % | 1.0 ± 0.9 | 1.3 ± 1.5 | 1.2 ± 1.0 | 1.3 ± 1.4 | 0.90 |
| Bronchial epithelial cell, % | 1.6 ± 4.3 | 0.8 ± 1.5 | 1.3 ± 3.3 | 0.4 ± 1.0 | 0.26 |
| RIN, mean | 7.6 ± 1.1 | 7.4 ± 1.2 | 7.5 ± 1.0 | 7.7 ± 1.4 | 0.1 |
Definition of abbreviations: RIN = RNA integrity number; TEA = transcriptomic endotypes of asthma; YCAAD = Yale Center for Asthma and Airway Diseases.
Data are means ± SD. P values for comparisons among TEA clusters were determined using Kruskal-Wallis or Cochran-Armitage test.
Cells per microliter × 104.
DEGs in the Airway among TEA Clusters
We considered each TEA cluster as a unique pathobiologic process and compared the gene expression in the sputum of each TEA cluster with control subjects without asthma to determine the genes associated with each of the TEA clusters. Using an FDR threshold of 0.05, there were 31 significant DEGs in TEA cluster 1, a total of 0 DEGs in TEA cluster 2 (15 DEGs had an FDR <0.25), and 27 DEGs in TEA cluster 3 compared with control subjects without asthma (the top 10 most significant DEGs are shown in Tables 4 and 5). In TEA cluster 1, expression of l-histidine decarboxylase, an enzyme in the histamine metabolism pathway that converts histidine to histamine, was increased in individuals with asthma compared with control subjects with no asthma (26). Two DEGs (EXOSC9 and SNAPC5) that code for proteins involved in RNA processing were down-regulated compared with control subjects with no asthma (27). Three DEGs (NRCAM, PCLO, and SLC4A4) that are associated with neuron function were significantly increased in TEA cluster 1 compared with control subjects with no asthma (28–30). In TEA cluster 3, all of the 27 DEGs are up-regulated compared with control subjects. These included DNAH17, a force-generating dynein heavy chain motor protein in respiratory epithelium, and defensin β1 (DEFB1), an antimicrobial peptide. Both genes have been previously associated with asthma and primary ciliary dyskinesia, respectively (31–33). Gene set enrichment analysis of DEGs using GeneGO MetaCore for TEA cluster 1 and 3 shows that the strongest pathway enrichment was in TEA cluster 3 (see Table E2).
Table 4.
Top 10 Differentially Expressed Genes between TEA Cluster 1 and Control Subjects
| Gene Name | Gene Symbol | Biologic Processes | Function | Fold Change | P Value | FDR | PubMed References |
|---|---|---|---|---|---|---|---|
| Hemogen | HEMGN | Apoptosis | Transcription factor | 1.25 | 2.79 × 10−7 | 4.01 × 10−3 | 16 |
| Piccolo (presynaptic cytomatrix protein) | PCLO | Cytoskeleton organization, regulation of exocytosis | Protein | 1.21 | 6.26 × 10−7 | 4.01 × 10−3 | 26 |
| Cartilage-associated protein | CRTAP | Extracellular matrix organization | Scaffolding protein | −1.48 | 7.87 × 10−7 | 4.01 × 10−3 | 25 |
| Exosome component 9 | EXOSC9 | RNA processing | RNase complex component | −2.10 | 1.11 × 10−6 | 4.01 × 10−3 | 32 |
| Chromosome 9 open reading frame 173 | C9orf173 | NA | NA | 1.36 | 1.23 × 10−6 | 4.01 × 10−3 | 2 |
| Small nuclear RNA activating complex, polypeptide 5, 19 kD | SNAPC5 | DNA-dependent regulation of transcription | Transcription factor | −1.08 | 1.23 × 10−6 | 4.01 × 10−3 | 9 |
| Impact RWD domain protein | IMPACT | Negative regulation of protein phosphorylation | Enzyme | −2.12 | 1.44 × 10−6 | 4.01 × 10−3 | 11 |
| Solute carrier family 4, sodium bicarbonate cotransporter, member 4 | SLC4A4 | Sodium ion transport | Membrane transporter protein | 1.22 | 1.45 × 10−6 | 4.01 × 10−3 | 61 |
| Histidine decarboxylase | HDC | Histamine biosynthesis | Enzyme | 1.39 | 1.94 × 10−6 | 4.78 × 10−3 | 48 |
| Neuronal cell adhesion molecule | NRCAM | Neuron migration | Cell adhesion molecules | 1.19 | 2.29 × 10−6 | 5.08 × 10−3 | 40 |
Definition of abbreviations: FDR = false discovery rate; NA = not applicable; TEA = transcriptomic endotypes of asthma.
Table 5.
Top 10 Differentially Expressed Genes between TEA Cluster 3 and Control Subjects
| Gene Name | Gene Symbol | Biologic Processes | Functions | Fold Change | P Value | FDR | PubMed References |
|---|---|---|---|---|---|---|---|
| Heart development protein with EGF-like domains 1 | HEG1 | Endothelial cell angiogenesis | Cell adhesion molecules | 1.78 | 1.09 × 10−7 | 2.42 × 10−3 | 7 |
| Small nucleolar RNA, C/D box 104 | SNORD104 | RNA modification | Noncoding RNA | 2.58 | 2.46 × 10−7 | 2.73 × 10−3 | 2 |
| Dynein, axonemal, heavy chain 17 | DNAH17 | Ciliary motility | Microtubule-associated motor protein | 1.88 | 2.61 × 10−6 | 1.18 × 10−2 | 7 |
| Cbl proto-oncogene, E3 ubiquitin protein ligase B | CBLB | Regulation of T-cell anergy | Ubiquitin protein ligase | 2.04 | 2.95 × 10−6 | 1.18 × 10−2 | 93 |
| Defensin, β1 | DEFB1 | Innate immune response | Antimicrobial peptide | 2.85 | 2.97 × 10−6 | 1.18 × 10−2 | 133 |
| Nonprotein coding RNA 204 | NCRNA204 | NA | Noncoding RNA | 2.20 | 3.55 × 10−6 | 1.18 × 10−2 | 1 |
| Transcription elongation factor A (SII) N-terminal and central domain containing | TCEANC | RNA elongation | Transcription elongation factor | 2.22 | 6.58 × 10−6 | 1.82 × 10−2 | 10 |
| Radical S-adenosyl methionine domain containing 2 | RSAD2 | Regulation of Toll-like receptor 9 signaling pathway | Antiviral protein | 1.57 | 1.04 × 10−5 | 2.07 × 10−2 | 28 |
| Purinergic receptor P2Y, G-protein coupled, 14 | P2RY14 | Regulation of inflammation | UDP-glucose receptor | 5.14 | 1.04 × 10−5 | 2.07 × 10−2 | 31 |
| Malignant fibrous histiocytoma amplified sequence | MFHAS1 | Cell cycle | Protein | 4.14 | 1.11 × 10−5 | 2.07 × 10−2 | 11 |
Definition of abbreviations: EGF = epidermal growth factor; FDR = false discovery rate; NA = not applicable; TEA = transcriptomic endotypes of asthma.
Validation of Sputum TEA Clusters Using Blood Gene Expression
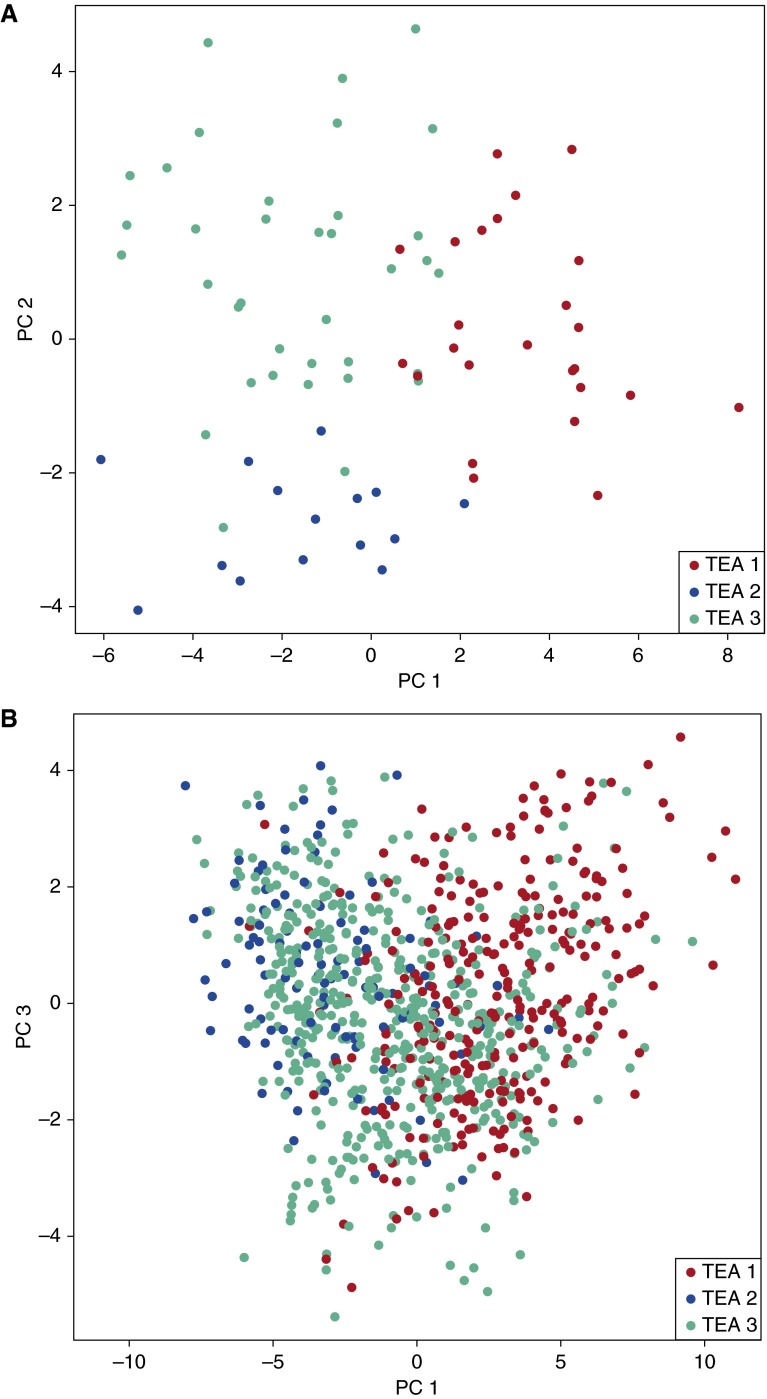
Because additional, sufficiently powered datasets of genome-wide sputum gene expression were not available for validation of the TEA cluster model, we turned to a second compartment, the peripheral blood, to validate the TEA clusters. First, using 76 YCAAD subjects for whom both sputum and peripheral blood expression data were available, we identified 53 gene expression signatures in the peripheral blood using L1 regularized logistic regression that predicts an individual’s sputum TEA cluster assignment (see Table E3). Principal components analysis (PCA) of the peripheral blood expression levels of these 53 genes shows that the first two components separate the population into clusters that closely recapitulate those defined with the sputum data (Figure 4A).
Figure 4.
Data visualization of the transcriptomic endotypes of asthma (TEA) clusters using the 53 blood expression. (A) Principal components analysis plot of the 76 matched blood arrays in the Yale Center for Asthma and Airway Diseases cohort. (B) Principal components analysis plot of the blood arrays from the Asthma BioRepository for Integrative Genomic Exploration cohort. TEA cluster assignment was predicted using the TEA cluster classifier built in the Yale Center for Asthma and Airway Diseases cohort. PC = principal component.
Next, to validate the clinical phenotypes of the TEA clusters in a separate cohort, we applied the blood TEA classifier to 870 whole blood samples from the Asthma BRIDGE cohort using the 53 transcripts common to both microarray platforms (see online supplement for details) (24, 25). Similar to the patterns observed in the YCAAD cohort, the PCA plot of the blood arrays from the Asthma BRIDGE cohort in Figure 4B shows strong separation between TEA 1 and TEA 3. TEA 2 in the Asthma BRIDGE cohort, however, mingled with TEA 3. The prevalence of each TEA cluster in the Asthma BRIDGE cohort was similar to the YCAAD cohort (31%, 12%, 57% for TEA cluster 1, 2, and 3, respectively; P = 2.2 × 10−16). This demonstrated that a blood signature was able to discriminate subgroups of children with asthma (Asthma BRIDGE) with a prevalence similar to the adults with asthma (YCAAD).
To determine if the childhood and adult TEA clusters have similar clinical features, differences among the clinical phenotypes were evaluated in the TEA clusters in the Asthma BRIDGE cohort (Table 6). Consistent with the clinical phenotypes of the YCAAD cohort, TEA cluster 1 subjects in the Asthma BRIDGE cohort were significantly more likely to have a history of intubation for asthma (8%; P = 5.58 × 10−6) and TEA cluster 2 subjects were the most likely to have a history of hospitalization for asthma (35%; P = 0.01), compared with the other TEA clusters. Therefore, in the Asthma BRIDGE cohort, the prevalence of the clusters, the association of TEA cluster 1 with near-fatal disease, and TEA cluster 2 with severe asthma was the same as the YCAAD cohort. Although TEA cluster 1 in the Asthma BRIDGE cohort did not have lower lung function compared with the other TEA clusters (data not shown), 8% of these children had a history of near-fatal asthma attacks.
Table 6.
Phenotypic Characteristics of TEA Clusters in the Asthma BRIDGE Cohort
| Cluster 1 (n = 266) | Cluster 2 (n = 105) | Cluster 3 (n = 499) | P Value | |
|---|---|---|---|---|
| Prevalence in cohort | 31% | 12% | 57% | <2.2 × 10−16 |
| Age at visit, yr | 14.8 ± 8 | 10.1 ± 5 | 12.6 ± 7 | 3.27 × 10−8 |
| Sex, n (%) female | 128 (48) | 45 (43) | 212 (43) | 0.31 |
| Race | 1.68 × 10−7 | |||
| White, n (%) | 102 (38) | 79 (75) | 284 (57) | |
| Black, n (%) | 105 (40) | 9 (9) | 124 (25) | |
| Other, n (%) | 29 (11) | 7 (7) | 37 (7) | |
| Hispanic origin, n (%) | 30 (11) | 10 (10) | 54 (11) | 0.09 |
| History of atopy, n (%) | 69 (26) | 40 (38) | 113 (23) | 0.0013 |
| Age of symptom onset | 3.50 ± 3.21 | 3.32 ± 2.89 | 3.48 ± 2.80 | 0.86 |
| History of hospitalization, n (%) | 91 (34) | 37 (35) | 128 (26) | 0.011 |
| History of intubations, n (%) | 21 (8) | 0 (0) | 9 (2) | 5.58 × 10−6 |
| ACT score | 14 ± 4 | 12 ± 3 | 13 ± 3 | 8.79 × 10−7 |
Definition of abbreviations: ACT = Asthma Control Test; BRIDGE = BioRepository for Integrative Genomic Exploration; TEA = transcriptomic endotypes of asthma.
Data are means ± SD, except where indicated.
Discussion
Noninvasive analysis of the sputum transcriptome conducted in these studies identified three TEA clusters with different clinical and physiologic characteristics of disease. Two TEA clusters are associated with phenotypes of severe disease: a history of a near-fatal asthma attack and a history of hospitalization for asthma. These phenotypes were replicated in the TEA clusters of a second cohort of children with asthma that were determined using a unique 53-gene transcriptomic profile in whole blood that is associated with the sputum transcriptome. Taken together, these data suggest that there are common, stable patterns of gene expression in individuals with asthma that are independent of age, age of disease onset, or duration. These TEA clusters are associated with severe phenotypes of asthma and gene signatures in the blood. This indicates that there are systemic alterations in the gene expression that link tissue compartments in patients with severe asthma that can be used to identify subgroups of disease that could be clinically relevant.
The generalizable clinical features associated with each TEA cluster suggest that unsupervised transcriptomic clustering generates disease subgroups that overlap with guideline-defined or Th2 gene level–defined disease severity (see Figure E6). Although this shows that there is some biologic overlap between the TEA clusters and Th2 biology, the link is relatively weak because allergic inflammation Kyoto Encyclopedia of Genes and Genomes pathway was not a pathway that significantly contributed to the clustering result. This suggests that the TEA clusters are driven by biologic phenomena that are upstream or possibly parallel to Th2 inflammation (34).
TEA cluster 1 has the highest percentage of subjects with a history of near-fatal asthma in both the YCAAD and Asthma BRIDGE cohorts, despite a large difference in age among the individuals. This cluster also has the lowest baseline lung function, the highest bronchodilator reversibility, the highest FeNO levels, and the highest doses of ICS: all characteristics that are associated with near-fatal asthma (35). Given that the adults and children in TEA cluster 1 are linked by a common transcriptomic signature in the airway that is associated with epithelial cell differentiation (EXOSC9 and SNAPC5), neurohumoral hemostasis (NRCAM and PCLO), and histamine synthesis (DNAH17 and DEFB1), it is plausible that these genes contribute to a greater risk of severe bronchospasm and near-fatal asthma associated with this cluster (26–30).
TEA cluster 2 is the least common TEA cluster in both cohorts (19% of YCAAD and 12% of Asthma BRIDGE) and has the most within-cluster heterogeneity, compared with the other TEA clusters (color heterogeneity seen in Figure 3). These individuals also have severe disease and are more likely to have been hospitalized for asthma. Although the evaluation of larger cohorts is required to further define this cluster and its associated DEGs (no significant genes associated with this TEA cluster were identified by an FDR cutoff of 0.05 in part because of the small number of individuals in this cluster), the transcriptomic discrimination of this cluster suggests that there are distinct pathobiologic differences between patients that have near-fatal asthma attacks and those that have severe exacerbations requiring hospitalization. Consistent with this concept is the fact that patients in TEA cluster 2 have the highest levels of YKL-40 in the sputum among the clusters (P = 0.03, data not shown), findings consistent with a possible YKL-40 endotype/phenotype with increased risk of exacerbations and abnormal post-bronchodilator FEV1 (36, 37).
Individuals in TEA cluster 3 have clinical features most consistent with mild disease including increased expression of DEFB1, a gene that has been associated with mild asthma in multiple studies (31). Compared with the other TEA clusters, these individuals have preserved lung function, the lowest ICS dose, and the lowest FeNO level and are less likely to have been hospitalized or intubated for asthma. TEA cluster 3 is also the most strongly related with the least within-cluster heterogeneity (consistent red color of this cluster in Figure 3). It is also the most common cluster with a prevalence of approximately 50% in children and adults. Interestingly, PCA analysis of the blood transcriptome shows that in children, TEA cluster 3 overlaps with cluster 1 and 2 (Asthma BRIDGE cohort, Figure 4B), but is distinct from the other TEA clusters in adults (Figure 4A). This suggests that the transcriptome (and clinical phenotype) of the individuals in this TEA cluster could change over time.
These studies also demonstrate that the unsupervised clustering analysis of gene expression in the sputum and/or blood has the capacity to discriminate subgroups of asthma that are independent of clinical characteristics typically used to study severe asthma (i.e., body mass index, atopy, FEV1). This is caused, in part, by the clustering approach we developed that is distinctly different compared with conventional approaches that use clinical features, and large differences in gene expression compared with subjects with no asthma to select genes for the clustering analysis (5, 9–11). In contrast, the analytical approach used herein was not biased by clinical phenotypes and is solely based on ontologically derived, pathway-based gene expression. This approach results in reduced background from random statistical events interfering with the clustering algorithm and the overwhelming effects of analyzing gene expression that is different between patients with asthma and control subjects: eliminating gene expression signals that are associated with disease heterogeneity. Ultimately we found two different sets of genes in the blood and sputum that are associated with the same or similar clinical phenotypes. Because clustering analysis using only blood gene expression identified clusters without unique clinical features (data not shown), we believe that the evaluation of gene expression in the primary organ of involvement is essential to dissect disease heterogeneity of chronic inflammatory airway disease.
Although TEA clusters are clearly distinguished, there remains heterogeneity within each cluster, especially within TEA clusters 1 and 2. This suggests that analysis of larger populations of patients will define additional TEA clusters that are biologically similar within the clusters we have defined. These may reveal additional novel molecular phenomena that further define the heterogeneity of disease. In addition, longitudinal studies of patients with asthma currently underway will determine how stable and robust the TEA clusters are over time, and will define the potential of unsupervised transcriptomic analysis of the blood and sputum to identify patients at risk of adverse outcomes, such as near-fatal and severe asthma exacerbations early in the course of their disease. Ultimately, these studies will determine if transcriptomic signatures in the blood and/or airway have the capacity to personalize approaches to the management of asthma, enhance outcomes and selection for existing and emerging treatments, or will be most useful to advance the pathogenesis research in asthma and other complex diseases.
Acknowledgments
Acknowledgment
The authors thank all the subjects who participated in this study, Donna Cook for data management and support enrolling subjects, and Arron Mitchell for development of the Yale Center for Asthma and Airway Diseases database.
Footnotes
Supported by National Institutes of Health (NIH) grants R01HL095390-04 and R01HL118346-01 (G.L.C.); NIH training grants T32HL007778-18 and T15LM007056-26 and FAMRI Young Clinical Scientist Award (J.G.); NIH/NHLBI grant K99HL114651-01A1 (J.-H.C.); Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (S.J.L.); the Mary Beryl Patch Turnbull Scholar Program (K.C.B.); and NCATS grant UL1 3 TR000142 (X.Y.).
Author Contributions: G.L.C. conceived of and designed experiments. X.H., C.H., J.G., M.K., M.F.P., L.C., and S.M. collected the samples and performed the experiments. X.Y. and H.Z. analyzed the data. J.-H.C., F.D.M., C.O., D.L.N., K.C.B., S.J.L., F.G., S.T.W., and B.A.R. provided the Asthma BioRepository for Integrative Genomic Exploration cohort data. X.Y. and G.L.C. wrote the manuscript.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201408-1440OC on March 12, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Moorman JE, Zahran H, Truman BI, Molla MT Centers for Disease Control and Prevention (CDC) Current asthma prevalence - United States, 2006-2008. MMWR Surveill Summ. 2011;60:84–86. [PubMed] [Google Scholar]
- 2.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 3.American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000;162:2341–2351. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 4.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO GABRIEL Consortium. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Koth LL, Arron JR, Fahy JV. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters MC, Mekonnen ZK, Yuan S, Bhakta NR, Woodruff PG, Fahy JV. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol. 2014;133:388–394. doi: 10.1016/j.jaci.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, Calhoun WJ, Castro M, Chung KF, Clark MP, et al. National Heart, Lung, Blood Institute’s Severe Asthma Research Program. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howrylak JA, Fuhlbrigge AL, Strunk RC, Zeiger RS, Weiss ST, Raby BA Childhood Asthma Management Program Research Group. Classification of childhood asthma phenotypes and long-term clinical responses to inhaled anti-inflammatory medications. J Allergy Clin Immunol. 2014;133:1289–1300, e1–e12. doi: 10.1016/j.jaci.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, D’Agostino R, Jr, Castro M, Curran-Everett D, Fitzpatrick AM, et al. National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haldar P, Pavord ID, Shaw DE, Berry MA, Thomas M, Brightling CE, Wardlaw AJ, Green RH. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127:153–60, e1–e9. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Chupp GL, Perez MF, Gomez-Arroyo JG, He S, Holm C, Cook D, Zhao H, Cohn LE, Yan X. Determination of asthma phenotypes by unsupervised transcriptomic cluster analysis of induced sputum [abstract] Am J Respir Crit Care Med. 2013;187:A2328. [Google Scholar]
- 13.American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 14.Covar RA, Spahn JD, Martin RJ, Silkoff PE, Sundstrom DA, Murphy J, Szefler SJ. Safety and application of induced sputum analysis in childhood asthma. J Allergy Clin Immunol. 2004;114:575–582. doi: 10.1016/j.jaci.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 15.Gelder CM, Thomas PS, Yates DH, Adcock IM, Morrison JF, Barnes PJ. Cytokine expression in normal, atopic, and asthmatic subjects using the combination of sputum induction and the polymerase chain reaction. Thorax. 1995;50:1033–1037. doi: 10.1136/thx.50.10.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grootendorst DC, van den Bos JW, Romeijn JJ, Veselic-Charvat M, Duiverman EJ, Vrijlandt EJ, Sterk PJ, Roldaan AC. Induced sputum in adolescents with severe stable asthma: safety and the relationship of cell counts and eosinophil cationic protein to clinical severity. Eur Respir J. 1999;13:647–653. doi: 10.1183/09031936.99.13364799. [DOI] [PubMed] [Google Scholar]
- 17.Green RH, Brightling CE, Woltmann G, Parker D, Wardlaw AJ, Pavord ID. Analysis of induced sputum in adults with asthma: identification of subgroup with isolated sputum neutrophilia and poor response to inhaled corticosteroids. Thorax. 2002;57:875–879. doi: 10.1136/thorax.57.10.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handl J, Knowles J, Kell DB. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21:3201–3212. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- 20.Millstein J, Volfson D. Computationally efficient permutation-based confidence interval estimation for tail-area FDR. Front Genet. 2013;4:179. doi: 10.3389/fgene.2013.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 22.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Stat. 2004;32:407–451. [Google Scholar]
- 23.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Series B Stat Methodol. 1996;58:267–288. [Google Scholar]
- 24.Raby BA BK, Beaty TH, Bosco A, Carey VJ, Castro M, Cheadle C, Gilliland FD, Islam KTS, Salam MT, Kelly R, et al. Asthma BRIDGE: the Asthma BioRepository for Integrative Genomic Exploration. Am J Respir Crit Care Med. 2010;183:A6189. [Google Scholar]
- 25.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 26.Koarai A, Ichinose M, Ishigaki-Suzuki S, Yamagata S, Sugiura H, Sakurai E, Makabe-Kobayashi Y, Kuramasu A, Watanabe T, Shirato K, et al. Disruption of L-histidine decarboxylase reduces airway eosinophilia but not hyperresponsiveness. Am J Respir Crit Care Med. 2003;167:758–763. doi: 10.1164/rccm.200206-619OC. [DOI] [PubMed] [Google Scholar]
- 27.Mistry DS, Chen Y, Sen GL. Progenitor function in self-renewing human epidermis is maintained by the exosome. Cell Stem Cell. 2012;11:127–135. doi: 10.1016/j.stem.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoki T, Matsumoto Y, Hirata K, Ochiai K, Okada M, Ichikawa K, Shibasaki M, Arinami T, Sumazaki R, Noguchi E. Expression profiling of genes related to asthma exacerbations. Clin Exp Allergy. 2009;39:213–221. doi: 10.1111/j.1365-2222.2008.03186.x. [DOI] [PubMed] [Google Scholar]
- 29.Laprise C. The Saguenay-Lac-Saint-Jean asthma familial collection: the genetics of asthma in a young founder population. Genes Immun. 2014;15:247–255. doi: 10.1038/gene.2014.12. [DOI] [PubMed] [Google Scholar]
- 30.Dorfman R, Li W, Sun L, Lin F, Wang Y, Sandford A, Paré PD, McKay K, Kayserova H, Piskackova T, et al. Modifier gene study of meconium ileus in cystic fibrosis: statistical considerations and gene mapping results. Hum Genet. 2009;126:763–778. doi: 10.1007/s00439-009-0724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy H, Raby BA, Lake S, Tantisira KG, Kwiatkowski D, Lazarus R, Silverman EK, Richter B, Klimecki WT, Vercelli D, et al. Association of defensin beta-1 gene polymorphisms with asthma. J Allergy Clin Immunol. 2005;115:252–258. doi: 10.1016/j.jaci.2004.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park YD, Lyou YJ, Lee KJ, Lee DY, Yang JM. Towards profiling the gene expression of fibroblasts from atopic dermatitis patients: human 8K complementary DNA microarray. Clin Exp Allergy. 2006;36:649–657. doi: 10.1111/j.1365-2222.2006.02480.x. [DOI] [PubMed] [Google Scholar]
- 33.Chodhari R, Mitchison HM, Meeks M. Cilia, primary ciliary dyskinesia and molecular genetics. Paediatr Respir Rev. 2004;5:69–76. doi: 10.1016/j.prrv.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 34.National Asthma Education and Prevention Program. Expert Panel Report 3 (EPR-3): guidelines for the diagnosis and management of asthma—summary report 2007. J Allergy Clin Immunol. 2007;120(5 Suppl):S94–S138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 35.Wenzel SE, Szefler SJ, Leung DY, Sloan SI, Rex MD, Martin RJ. Bronchoscopic evaluation of severe asthma. Persistent inflammation associated with high dose glucocorticoids. Am J Respir Crit Care Med. 1997;156:737–743. doi: 10.1164/ajrccm.156.3.9610046. [DOI] [PubMed] [Google Scholar]
- 36.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, Dziura JD, Reed J, Coyle AJ, Kiener P, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 37.Cunningham J, Basu K, Tavendale R, Palmer CN, Smith H, Mukhopadhyay S. The CHI3L1 rs4950928 polymorphism is associated with asthma-related hospital admissions in children and young adults. Ann Allergy Asthma Immunol. 2011;106:381–386. doi: 10.1016/j.anai.2011.01.030. [DOI] [PubMed] [Google Scholar]