Abstract
A rapid array-based protocol is presented by which a modest affinity protein-binding small molecule can be appended to a library of peptoids via Click chemistry. The array can then be screened for improved ligands that exhibit a higher affinity for the protein target.
Graphical Abstract
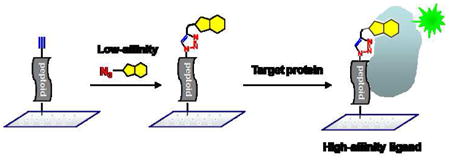
There is a great interest in identifying synthetic molecules that bind to proteins with high affinity and specificity for applications in drug discovery, biology and proteomics. To date, most protein binding molecules have been obtained from high-throughput screens of combinatorial libraries or compound collections. However, these initial hits generally have only modest affinity or potency. The standard approach to improving the properties of these hits is to carry out relatively tedious analysis of a variety of structural analogues. An attractive alternative approach is to instead employ bivalent ligands, in which two non-competitive ligands coupled via an appropriate linker cooperate to deliver high affinity1–6. However, this approach is not without its practical hurdles. The typical approach to bivalent ligands requires not only the isolation of more than one modest affinity ligand, but also that competition binding assays be carried out to ensure that one chooses non-competitive compounds with which to furnish a bivalent compound. Optimization of the linker can also be a time-consuming process.
An alternative route to improved ligands that we7 and others8–13 have explored is to append the lead compound to a new collection of compounds and then screen for higher affinity or potency, with the hope of better filling the binding site on the protein or, possibly, reaching over to engage a new surface of the protein as well. Previously, we demonstrated the feasibility of this approach by appending lead compounds to the terminus of “one bead one compound” (OBOC) libraries of peptoids7. These capped peptoid libraries were screened under conditions where the modest affinity lead compound did not have sufficient affinity for the target protein to retain it on the bead, thus demanding that the hits show a significant improvement over the lead compound. However, the throughput of this approach is limited by the need to synthesize a new bead-based library for each lead compound. In this report, we describe an extension of this concept that makes this approach to improved ligand discovery far more practical. We show that an azide-containing lead compound can be appended efficiently to thousands of alkyne-terminated peptoids arrayed on a chemically-modified glass microscope slide (Figure 1)14, 15. Since a single bead library is sufficient for the construction of thousands of microarrays, this chemistry significantly improves the throughput of this approach.
Figure 1.
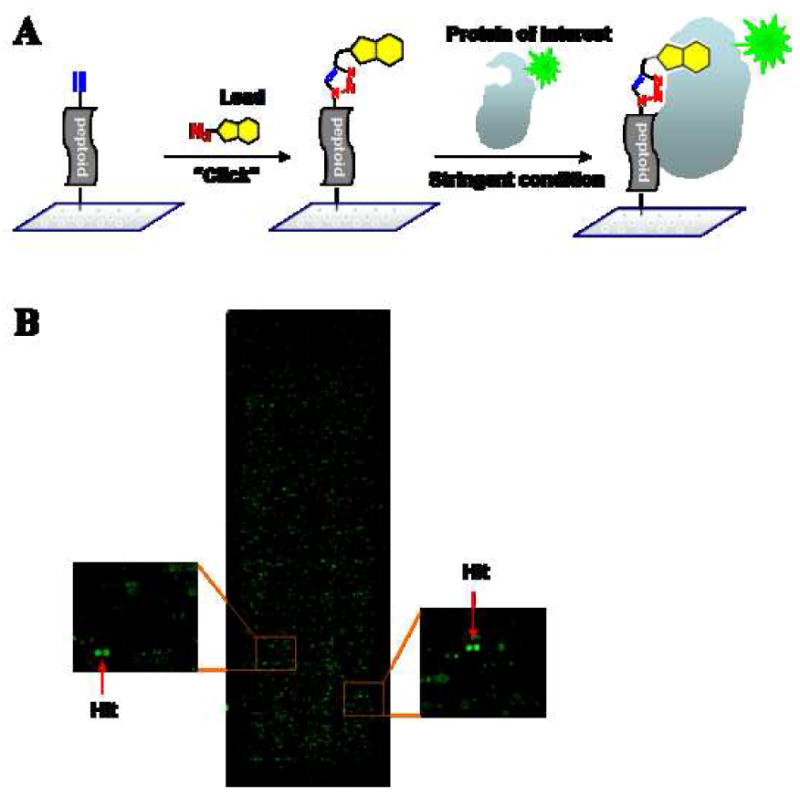
(A) Schematic representation of a strategy for isolating improved ligands. A modest-affinity lead compound is conjugated to each peptoid on a microarray by click chemistry. The slide is subsequently incubated with a target protein under stringent condition to provide a high-affinity ligand. (B) Microarray images of the hit compounds (spotted in duplicate) were obtained by incubating fluorescently labeled KIX-His6 on a microarray in which each peptoid molecule is capped with BHK2.
To test this idea, we chose a six residue peptoid called BHK216, 17 as a lead compound. BHK2 was previously isolated from a high-throughput screen of peptoids that bind the KIX domain of the transcription coactivator CREB-binding protein (CBP). The BHK2 peptoid binds a His6-tagged derivative of the KIX domain with a KD of approximately 195 μM. To couple BHK2 to an existing array of peptoids, we chose to explore Click chemistry18, since this chemistry has been used previously for affixing molecules to arrays19. An azide moiety was incorporated on the C-terminus of BHK2 (Figure 2A). An OBOC 6-mer peptoid library was synthesized by a conventional split and pool method on 500 μm polystyrene macrobeads using eight different amines (see Figure 2B)20, 21 Each peptoid molecule in the library had cysteine on its C-terminal end and a propargyl amine group on its N-terminal end. The cysteine residue enabled covalent immobilization of the peptoid onto the maleimidefunctionalized glass slide14 and the propargyl group allows for conjugation to any lead compound having an azide group by click chemistry. Several thousand individual beads were separated into the wells of a microtiter plates, the peptoids were cleaved from the beads and microarrays were constructed as described previously14. For the experiments reported here, 4,000 different acetylene-capped peptoids were spotted in duplicate onto the slide.
Figure 2.
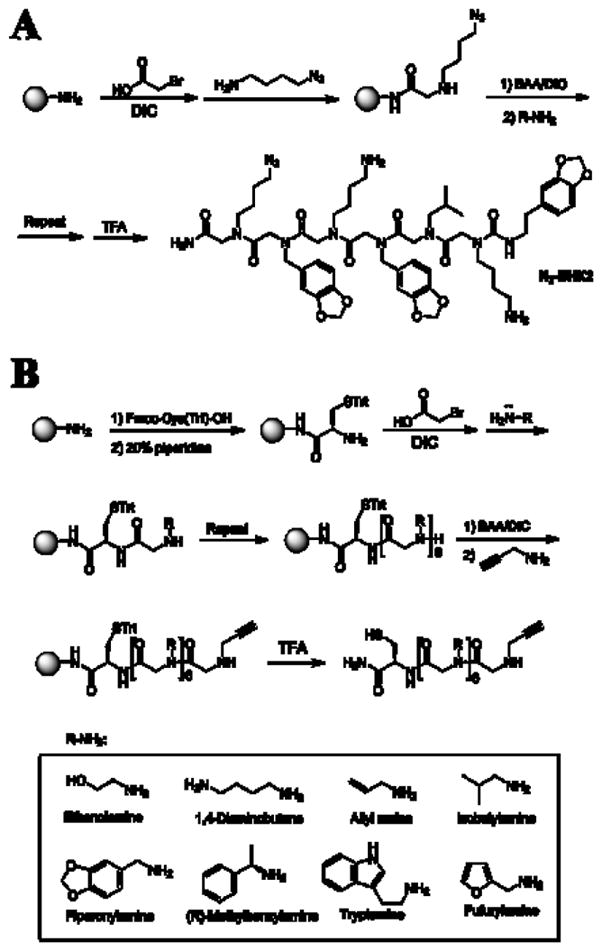
Structure and synthesis of the compounds used in this study. (A) Azide-modified BHK2. (B) The peptoid library that was immobilized on the microarray.
We then coupled N3-BHK2 to each peptoid on the microarray by copper-catalyzed Click chemistry. The alkyne-peptoid slide was placed in a hybridization chamber and treated with a solution of N3-BHK2 (1.53 μmol), CuSO4, and sodium ascorbate in tert-butyl alcohol and water (2 mL final volume) with gentle shaking at room temperature overnight. After washing and drying, the slide was used for screening (see Supporting Information).
To isolate higher affinity KIX domain ligands, we employed more stringent conditions (25 nM His6-KIX and 1,000-fold excess of proteins from a crude E. coli lysate as competitor to compete non-specific binding events) than those used originally to isolate BHK2 (500 nM KIX-His6 and 200-fold excess E. coli lysate)16. Fluorescently labeled KIX-His6 was incubated with an array of BHK2-capped peptoids as well as with an array displaying the same library without BHK2 capping and the results were compared. Two compounds on the BHK2-capped slides were selected as hits based on the fact that they exhibited a higher fluorescence intensity (3- to 4-fold) than the same spots on the slides without the capping molecule, BHK2 (Figure 1B and Figure S3). MS/MS-based sequencing revealed the structures of the compounds, 3 and 4 (Scheme 1 and Supporting Information).
Scheme 1.
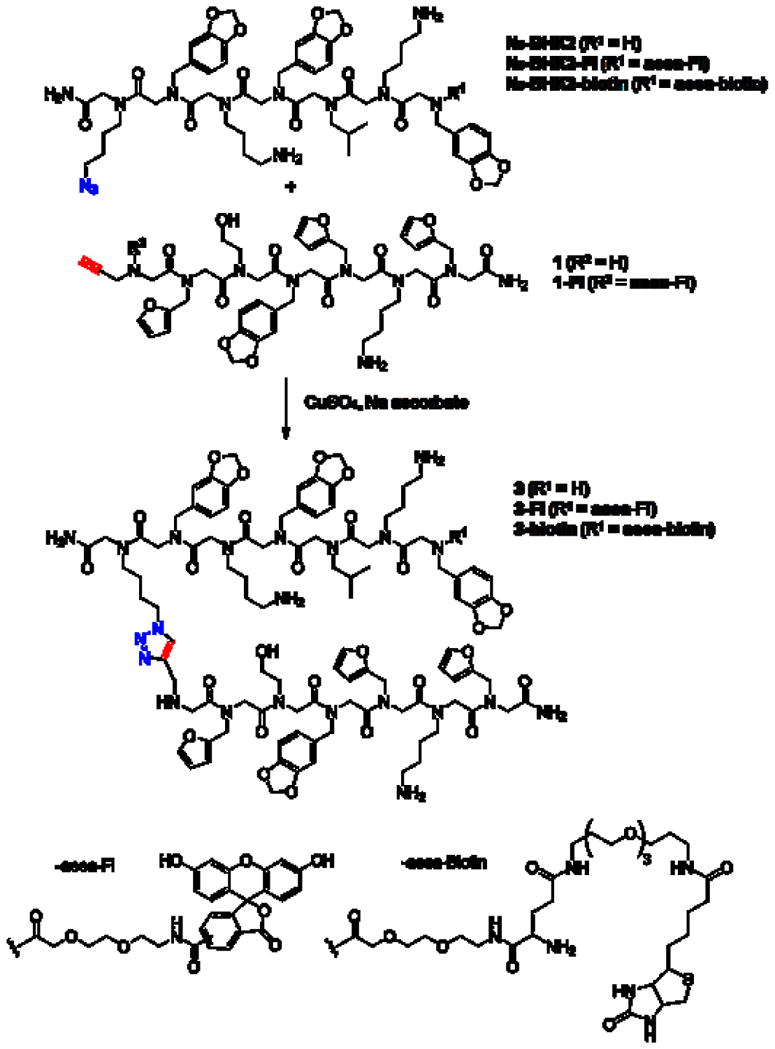
Structures and synthesis of compounds used in this study.
To examine whether the compounds isolated from the microarray screen indeed bind more tightly than the BHK2 lead compound to the KIX domain, we synthesized fluoresceinated derivatives of BHK2, 3 and 4 (Scheme 1 and Supporting Information) and carried out titration experiments monitored by fluorescence polarization spectroscopy. As shown in Table 1 and Supplementary Figure S3, compounds 3 and 4 indeed showed considerably improved binding affinities compared to the lead compound, BHK2. 3 and 4 bound to KIX-His6 with KDs of 4.6 μM and 8.3 μM respectively, representing a 42- and 23-fold improvement, respectively over the binding affinity of the BHK2 lead compound (KD = 195 μM). Not surprisingly, the peptoids isolated from the microarray library, when tested without tethered BHK2 (1 and 2) bound more weakly to the His6-KIX protein with KD values of 47.0 μM and 72.8 μM, respectively.
Table 1.
Dissociation constants for monomeric and dimeric compounds as determined by fluorescence polarization.
| BHK2 | 1 | 2 | |
|---|---|---|---|
| KD (μM) | 195.0 ± 25.5 | 47.0 ± 7.1 | 4.6 ± 1.0 |
Given that peptoids 3 and 4 were isolated from a screen carried out in the presence of a 1000-fold excess of E. coli proteins derived from a crude cell lysate, we anticipated that these molecules would be highly specific ligands for the KIX domain. If so, peptoids such as 3 or 4 might be useful reagents for the affinity purification of their target proteins from complex mixtures.
To examine this, biotinylated derivatives of 3 and BHK2 were synthesized and immobilized on a Streptavidin agarose bead (Supporting Information). After incubation with a crude lysate obtained from bacteria expressing KIX-His6 and thorough washing, the remaining proteins were analyzed by Western blotting with an anti-His6 antibody. As shown in Figure 3A, His6-KIX protein was retained by immobilized peptiod 3 (lane 2) while the beads lacking a peptoid (lane 1) and immobilized BHK2 (lane 3) did not pull down detectable amounts of His6-KIX protein under the same conditions. More importantly, Coomassie Blue staining of the gel showed that His6-KIX is purified to essential homogeneity by immobilized 3 (Figure 3B, lane 2) consistent with the idea that bivalent peptoid ligands may be useful reagents for the purification of unmodified proteins.
Figure 3.
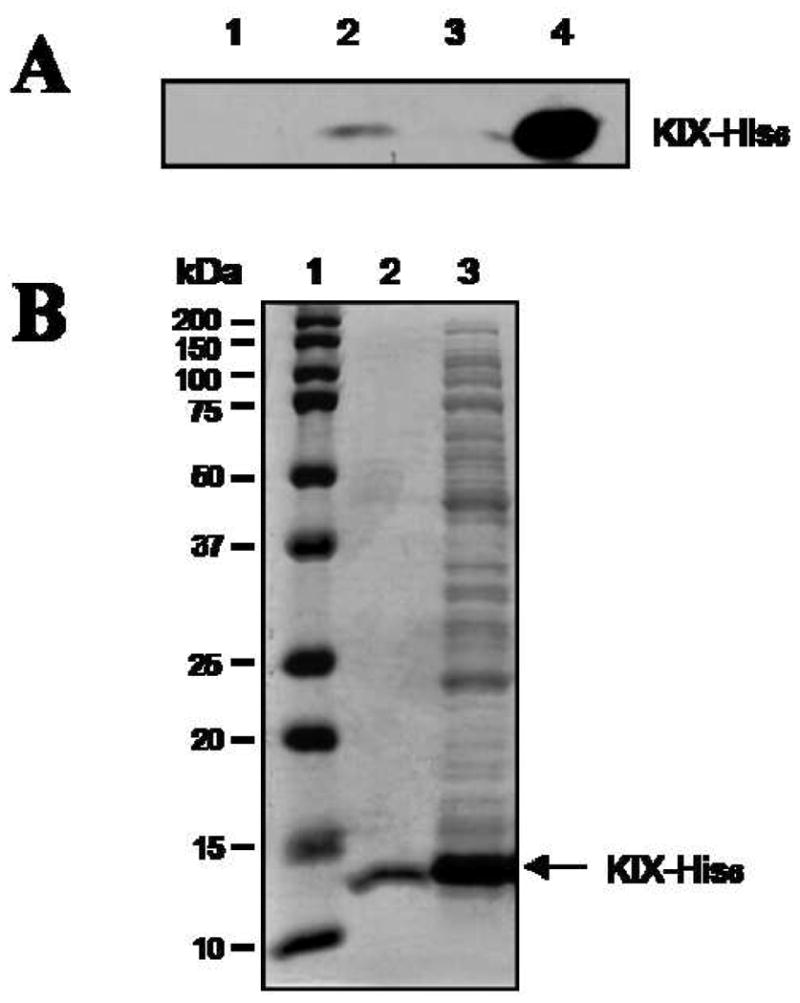
Dimeric compound 3 retains His6-KIX protein specifically from a whole cell lysate. (A) Streptavidin beads were treated with DMSO (lane 1), 3-biotin (lane 2), and BHK2-biotin (lane 3) and then incubated with a cell lysate obtained from E. coli cells that overexpress His6-KIX. After washing, bound proteins were analyzed by Western blot with anti-His6 antibody. Lane 4: input. Note that this experiment was done under conditions of a large excess of KIX-His6 protein over immobilized peptoid and thus only a fraction if the input KIX domain could be retained by biotinylated 3. (B) Bound proteins were analyzed by Coomassie Blue staining. Lane 1: protein markers, lane 2: 3-biotin treated, lane 3: input (whole cell extract).
In summary, we have developed a straightforward microarray-based method to rapidly identify improved protein ligands starting from a modest affinity lead compound. Efficient conditions have been developed that allows an azide-containing lead molecule to be appended to thousands of alkyne-terminated peptoids displayed on the surface of a chemically modified glass slide. Thousands of such microarrays can be made from a single OBOC library.10 Thus, a single peptoid library could be used to couple with a large number of different lead molecules, providing a route the rapid isolation of higher affinity ligands for a large number of proteins. Such efforts are underway.
Supplementary Material
Acknowledgments
This work was supported by a contract from the National Heart, Lung, and Blood Institute (NO1-HV-28185) for the UT Southwestern Center for Proteomics Research and a grant from the Welch Foundation (I-1299).
Footnotes
Supporting Information Available: Detailed experimental procedures and supplementary figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Science. 1996;274:1531. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 2.Maly DJ, Choong IC, Ellman JA. Proc Natl Acad Sci USA. 2000;97:2419. doi: 10.1073/pnas.97.6.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erlanson DA, Wells JA, Braisted AC. Ann Rev Biophys Biomol Structure. 2004;33:199. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 4.Profit AA, Lee TR, Lawrence DS. J Amer Chem Soc. 1999;121:280. [Google Scholar]
- 5.Cussac D, Vidal M, Leprince C, Liu W-q, Cornille F, Tiraboschi G, Roques BP, Garbay C. FASEB J. 1999;13:31. doi: 10.1096/fasebj.13.1.31. [DOI] [PubMed] [Google Scholar]
- 6.Kitov PI, Shimizu H, Homans SW, Bundle DR. J Amer Chem Soc. 2003;125:3284. doi: 10.1021/ja0258529. [DOI] [PubMed] [Google Scholar]
- 7.Reddy MM, Bachhawatt-Sikder K, Kodadek T. Chem & Biol. 2004;11:1127. doi: 10.1016/j.chembiol.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson KA, Kirk KL, Padgett WL, Daly JW. Mol Pharmacol. 1986;29:126. [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson KA, Marr-Leisy D, Rosenkranz RP, Verlander MS, Melmon KL, Goodman M. J Med Chem. 1983;26:492. doi: 10.1021/jm00358a007. [DOI] [PubMed] [Google Scholar]
- 10.Verlander MS, Jacobson KA, Rosenkranz RP, Melmon KL, Goodman M. Biopolymers. 1983;22:531. doi: 10.1002/bip.360220166. [DOI] [PubMed] [Google Scholar]
- 11.Combs AP, Kapoor TM, Feng S, Chen JK, Daude-Snow LF, Schreiber SL. J Amer Chem Soc. 1996;118:287. [Google Scholar]
- 12.Lawrence DS. Biochim Biophys Acta. 2005;1754:50. doi: 10.1016/j.bbapap.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 13.Agnew HD, Rohde RD, Millward SW, Nag A, Yeo WS, Hein JE, Pitram SM, Tariq AA, Burns VM, Krom RJ, Fokin VV, Sharpless KB, Heath JR. Angew Chem Int Ed Engl. 2009 doi: 10.1002/anie.200900488. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy MM, Kodadek T. Proc Natl Acad Sci U S A. 2005;102:12672. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacBeath G, Koehler AN, Schreiber SL. J Amer Chem Soc. 1999;121:7967. [Google Scholar]
- 16.Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. J Comb Chem. 2007;9:592. doi: 10.1021/cc070023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. Angew Chem Int Ed Engl. 2007;46:2865. doi: 10.1002/anie.200604485. [DOI] [PubMed] [Google Scholar]
- 18.Kolb HC, Sharpless KB. Drug Discovery Today. 2003;8:1128. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 19.Liang PH, Wang SK, Wong CH. J Am Chem Soc. 2007;129:11177. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- 20.Figliozzi GM, Goldsmith R, Ng SC, Banville SC, Zuckermann RN. Methods Enzymol. 1996;267:437. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- 21.Alluri PG, Reddy MM, Bacchawat-Sikder K, Olivos HJ, Kodadek T. J Amer Chem Soc. 2003;125:13995. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


