Abstract
Background
The molecular signature of atopic dermatitis/AD lesions is associated with Th2 and Th22 activation, and epidermal alterations. However, the epidermal and dermal AD transcriptomes and their respective contributions to abnormalities in respective immune and barrier phenotypes are unknown.
Objective
To establish the genomic profile of the epidermal and dermal compartments of lesional/LS and non-lesional/NL AD, as compared with normal skin.
Methods
Laser capture micro-dissection/LCM was performed to separate epidermis and dermis of LS and NL skin from AD patients and normal skin from healthy volunteers followed by gene expression (microarrays and RT-PCR) and immunostaining studies.
Results
Our study identified novel immune and barrier genes, including the IL-34 cytokine and claudins 4 and 8, and showed increased detection of key AD genes usually undetectable on arrays (i.e. IL-22, TSLP, CCL22, and CCL26). Overall, the combined epidermal and dermal transcriptomes enlarged the AD transcriptome adding 674 up-regulated and 405 down-regulated differentially expressed genes between LS and NL skin to the AD transcriptome. We were also able to localize individual transcripts as primarily epidermal (DEFB4A) or dermal (IL-22, CTLA4, and CCR7), and link their expressions to possible cellular sources.
Conclusions
This is the first report that establishes robust epidermal and dermal genomic signatures of LS, NL AD and normal/N skin, as compared with whole tissues. These data establish the utility of LCM to separate different compartments and cellular subsets in AD, allowing localization of key barrier or immune molecules, and enable detection of gene products usually not detected on arrays.
Keywords: atopic dermatitis, laser capture micro-dissection, IL-34, claudins 8 and 4, immune, barrier
Introduction
Atopic dermatitis/AD is the most common inflammatory skin disease.1,2 Although its pathogenesis is not fully understood, both barrier and immune components have been suggested to play key roles in AD, as indicated by the “outside-in” and “inside-out” hypotheses.3,4,5,6,7,8,9,10 Whereas barrier-related molecules are largely epidermal, inflammatory responses are derived from both the epidermal (i.e. keratinocytes/KCs, Langerhans cells/LCs) and dermal (i.e. T-cells and dendritic cells/DCs) compartments.
Using genomic analyses on whole tissue/bulk samples we have previously shown that the AD phenotype/transcriptome is associated with polar immune activation of Th2/Th22, as well as Th1 and Th17 pathways, and corresponding epidermal alterations (epidermal hyperplasia and abnormal differentiation).11,12,13,14 However, bulk sample genomic analysis (by microarrays and RT-PCR) presents some limitations: 1) It is difficult to determine whether altered gene expression is due to expansion (hypertrophy) of one tissue compartment vs. altered gene expression at the cellular level; 2) It cannot localize a particular gene/transcript to an epidermal/dermal compartment; 3) Low abundance genes are often below detection level on conventional microarrays due to dilution of mRNAs within full-thickness samples with more dominant products.
Laser capture micro-dissection/LCM is an established technique for procuring sub-populations of tissues/cells of interest under direct microscopic visualization to study alterations in disease states.15 Our group has previously demonstrated that epidermal and dermal separation of lesional/LS and non-lesional/NL psoriasis and normal samples using LCM complemented by microarrays largely increases the detection of low abundance genes compared to whole-tissue analyses.16,17 Despite the pathogenic relevance of separating the epidermal and dermal compartments, such studies are unavailable in AD.
In this study, we sought to determine the molecular phenotypes of the epidermal and dermal compartments of LS and NL AD skin (compared with skin from healthy individuals). Overall, our results: 1. Enlarged the AD transcriptome; 2. Detected low abundance genes (which are usually below detection levels on whole-tissue microarrays, i.e. IL-22 and TSLP); and 3. Identified novel immune and barrier genes (i.e. IL-34, claudin 4 and claudin 8) and suggested possible cellular sources of immune markers (i.e. CCR7).
Methods
Skin samples
Paired NL and LS AD skin biopsies were collected from 5 patients with moderate-to-severe chronic AD (3 males and 2 females, age: 27–59 years; mean age: 39.4 years) (see ETable1 in this article’s Online Repository at www.jacionline.org) under Institutional Review Board approved protocols. Normal skin samples from healthy volunteers (n=10), previously collected for a prior LCM publication,18 were also included. Paired epidermal, dermal, and full thickness LS and NL samples were used for RT-PCR and microarray analysis (n=5 in each group). LS and NL expression values were compared to 10 epidermal, 6 dermal, and 6 bulk corresponding normal samples. For RT-PCR confirmation, 3 normal paired epidermis and dermis samples were used, due to the limited available mRNA.
Slide preparation and LCM
LCM was performed following the manufacturer's protocol for CellCut system (Molecular Machines & Industries, Haslett, MI) (see the Methods section in the Online Repository).
RNA extraction
Total RNA was extracted using RNeasy Micro Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol with on-column DNase digestion.
Sample preparation for gene chip analysis
Target amplification and labeling was performed according to the Affymetrix protocols for two-cycle cDNA synthesis, using Affymetrix Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA) as previously reported.16
Sample preparation for quantitative real-time PCR
Reverse transcription to cDNA from RNA of LCM samples was carried out using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems), and cDNA was amplified using TaqMan® PreAmp Master Mix (Applied Biosystems), and preamplified cDNA product was analyzed using TaqMan® Gene Expression Master Mix according to the manufacturer's instructions. The RT-PCRs for each assay were run in triplicate, and all data were normalized to hARP. Primers and probes used in this study are listed in ETable2.
Immunohistochemistry (IHC) and immunofluorescence (IF)
IHC and IF were performed on frozen skin sections as previously described.19 The antibodies used in this study are listed in ETable3.
Statistical Analysis
Preprocessing and statistical analysis of microarray data was conducted using R (R-project.org) and Bioconductor packages20. Full details of the pipeline and downstream analysis are described in the Methods section of the Online Repository. Succinctly, the Harshlight package21 was used to QC the images, and expression values were obtained using the GCRMA algorithm.22 Batch adjustments were carried out using the ComBat algorithm and mixed-effect models in the limma package were used to model differential expression.23,24,25 Genes with FDR<0.05 and FCH>2 were considered significantly differentially expressed. Similar models were used to analyze log2-transformed values of the normalized RT-PCR data.
Results
LCM localizes genes selectively expressed in the dermis and epidermis
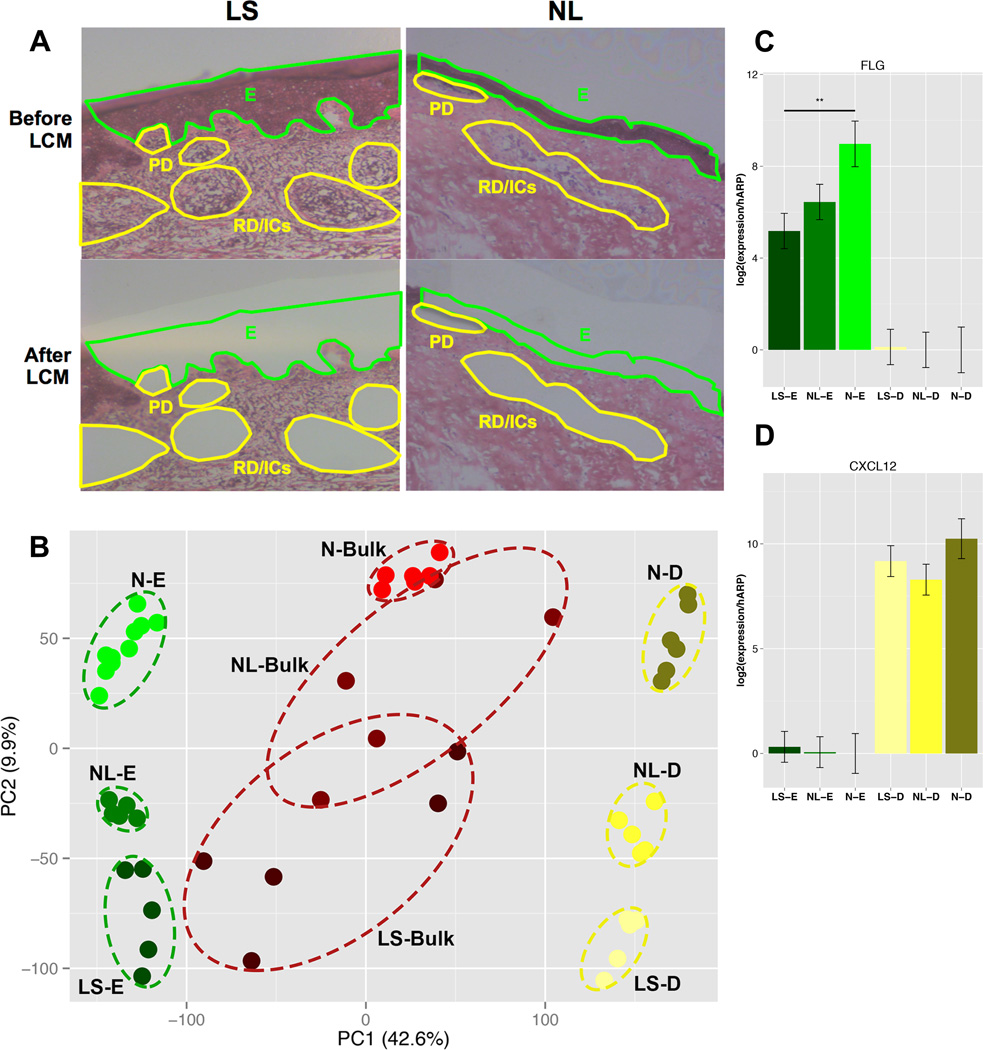
We performed laser capture micro-dissection/LCM to collect cells from the epidermis and dermis (papillary, reticular, and inflammatory aggregates) cells in frozen sections of lesional/LS, non-lesional/NL, and normal skin samples as shown in Fig.1A. To define the epidermal and dermal transcriptomes, microarray profiling of LS, NL, and normal epidermal and dermal tissues was performed using Affymetrix HGU133Plus2.0 microarrays. A heatmap of epidermal- and dermal-specific genes shows clear separation of differentially expressed genes/DEGs localized to the epidermis and dermis across LS, NL, and normal samples (EFig.1). A principal component analysis/PCA of expression values illustrates the lack of outliers and that samples cluster in accordance with tissue type (Fig.1B). Markers primarily considered dermal (CXCL12, CD93, collagens [COL1A2, CLO6A3]) were highly represented in the dermis (ETables4A,5A) and epidermal markers (FLG, LOR, late cornified envelope/LCEs, small proline-rich protein/SPRRs) were localized to the epidermis, consistent with the clear epidermal-dermal separation (ETables4B,5B). The accuracy of the LCM separation was validated using RT-PCR, with >69.7-fold and >846.2-fold enrichment of the mRNA expressions of filaggrin/FLG and CXCL12 in the epidermis vs. dermis, respectively, in normal tissues (Fig.1C–D).
Figure 1.
A. Representative H&E staining of lesional/LS and non-lesional/NL AD skin. LCM was performed in the zones indicated (green: epidermis/E, yellow: papillary dermis/PD, reticular dermis/RD, and inflammatory aggregates/ICs). B. A principal component analysis/PCA plot showing clear separation of groups. C,D. mRNA expression of filaggrin/FLG (C) and CXCL12 (D) in epidermis/E and dermis/D of lesional/LS and non-lesional/NL AD and normal/N skin. Mean±SD.
LCM enlarges the AD transcriptome and increases detection of low abundance immune genes on microarrays
To characterize the AD phenotype within each compartment, we defined the LS epidermal and dermal transcriptomes as the set of DEGs between LS and NL tissue within each compartment, respectively, using the classical FCH>2.0 and FDR<0.05 criteria. Combining the epidermal and dermal transcriptomes, many more compartment-specific DEGs were identified (860 up and 495 down; EFIg.2), adding 674 up-regulated and 405 down-regulated DEGs to the recently defined AD transcriptome.11,12,27 The bulk transcriptome consists of 710 up- and 487 down-regulated DEGs, whereas the LCM epidermal transcriptome contains 566 up- and 268 down-regulated DEGs, and the LCM dermal transcriptome has 330 up- and 244 down-regulated DEGs. Little overlap was observed between the epidermal and dermal transcriptomes (only 36 up- and 17 down-regulated DEGs detected in both).
Overall, the top 25 up- and down-regulated genes in each of the epidermal and dermal LS transcriptomes included many genes that have been shown to contribute to the AD phenotype (ETables6A–B,7A–B).11,12 The top 25 up-regulated genes in the LS epidermal transcriptome consisted of proliferation-related (KRT6A, KRT6B, KRT16), epidermal differentiation complex/EDC (S100s), inflammatory (MMP12), and anti-microbial peptide/AMP derived (DEFB4A, PI3/Elafin) genes (ETable6A). The top 25 up-regulated genes in the LS dermal transcriptome included T-cell activation (GZMB, IL2RA), Th2-related (CCL17, CCL22, CCL26), Th22/Th17-related (IL22, S100A8, CXCL1, CXCL2), and collagen (COL6A6, COL6A5, COL4A3) genes (ETable6B). IL-22 was significantly increased in the LS dermal transcriptome (12.46 FCHs, FDR<0.001), whereas it was below detection levels in previous reports,11,12,27 indicating that LCM is a useful method to increase the sensitivity of detecting low abundance genes.
Key inflammatory and barrier genes were uniquely detected in the LCM epidermal (ETable8A) and dermal (ETable8B) transcriptomes. The unique LS epidermal transcriptome included IL-17 related (DEFB4A, CCL20), and inflammatory (CXCR4, STAT3, IL8) genes (ETable8A). Key Th2 (CCL17, CCL22, CCL26), Th22 (IL22) markers, and T-cell migration/activation markers (CCR7, ICOS) were found within the unique LS dermal transcriptome (ETable8B). Although the LCM approach detected an appreciably larger number of genes within bulk tissues, there was a subset of DEGs that was not identified in the corresponding epidermal and dermal transcriptomes (429 up-regulated and 381 down-regulated genes; Fig.2, ETable8C). A deeper analysis of these data suggests that most of this discrepancy is created by increased or decreased gene abundance due to differing contributions of epidermis and dermis in LS vs. NL skin (see Supplementary Results, EFig.3A–C).
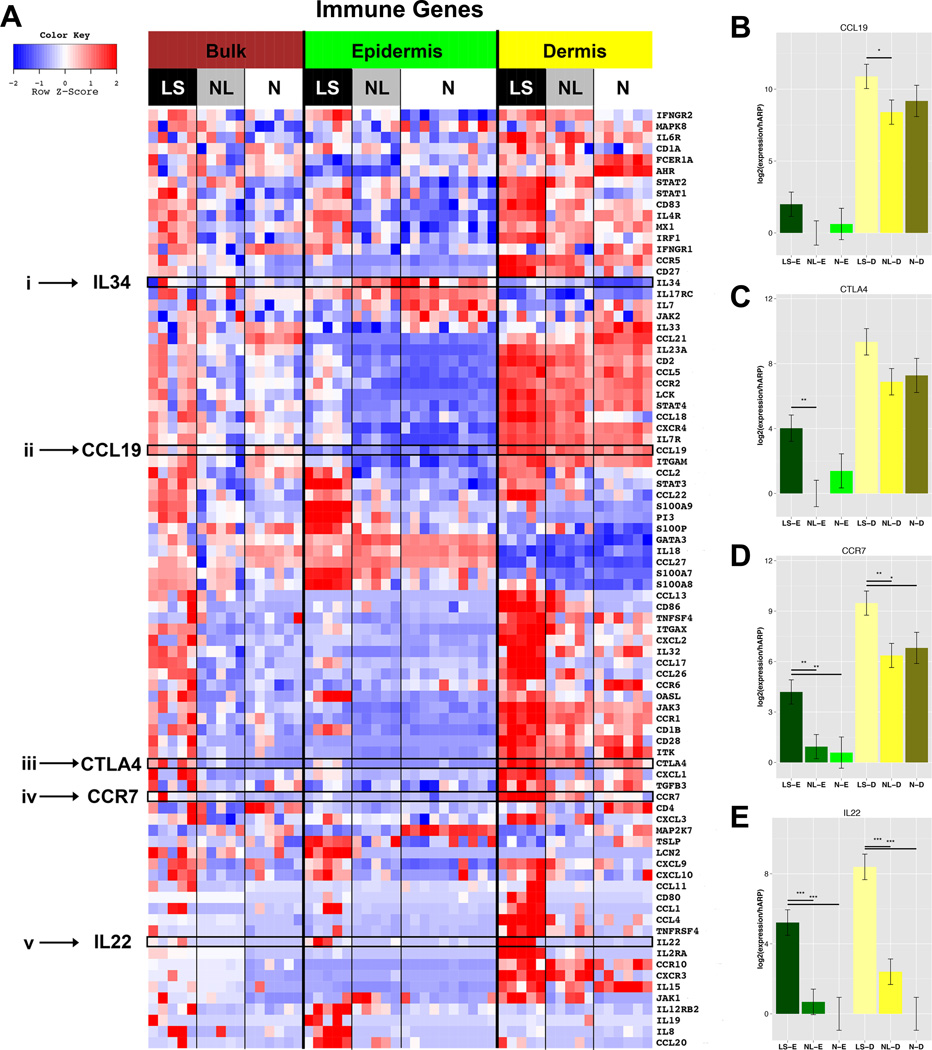
Figure 2.
A. Heatmap of immune-related genes organized by compartment. FCHs comparing lesional/LS; non-lesional/NL and normal/N skin in bulk/B, epidermis/E, dermis/D, (n=5 AD; n=6, 10, 6 normal bulk, epidermis, dermis, respectively) B–E. mRNA expression by RT-PCR of CCL19 (B), CTLA4 (C), CCR7 (D), IL22 (E) in epidermis/E and dermis/D of LS, NL AD and N skin. Mean±SD (B–E). *p<0.05; **p<0.01;***p<0.001.
We further explored the compartmental distribution of immune DEGs in LS, NL, and normal skin, using our previously curated and reported immune-gene subset,19,27,28 as shown in a heatmap (Fig.2A, ETable9). Among significantly up-regulated genes in the LS dermal transcriptome were T-cell (CTLA4, CD28), DC (ITGAX/CD11c, CD1A), lymphoid organizing chemokines (CCR7 and its ligands [CCL19, CCL21]), and Th2 (CCL11, CCL13, CCL17, CCL22, CCL26, TNFRSF4/OX40, IL4R) related genes. Significant increases in Th2-related (IL7R), Th17-related (PI3, LCN2, CCL20, STAT3), Th22/Th17-related (IL22, S100A8, S100A9), and Th1-related (OASL, MX1, IL12RB2, IFNGR2, IRF1) products were found. We validated, by RT-PCR, the primarily dermal mRNA expression of selected markers, including IL-22, CTLA4, CCR7, and CCL19. High IL-22 mRNA levels were characteristic of only LS skin (Fig.2B–E). A list of DEGs in all comparisons is presented in ETable8A–D.
IL-34: a novel cytokine identified by LCM
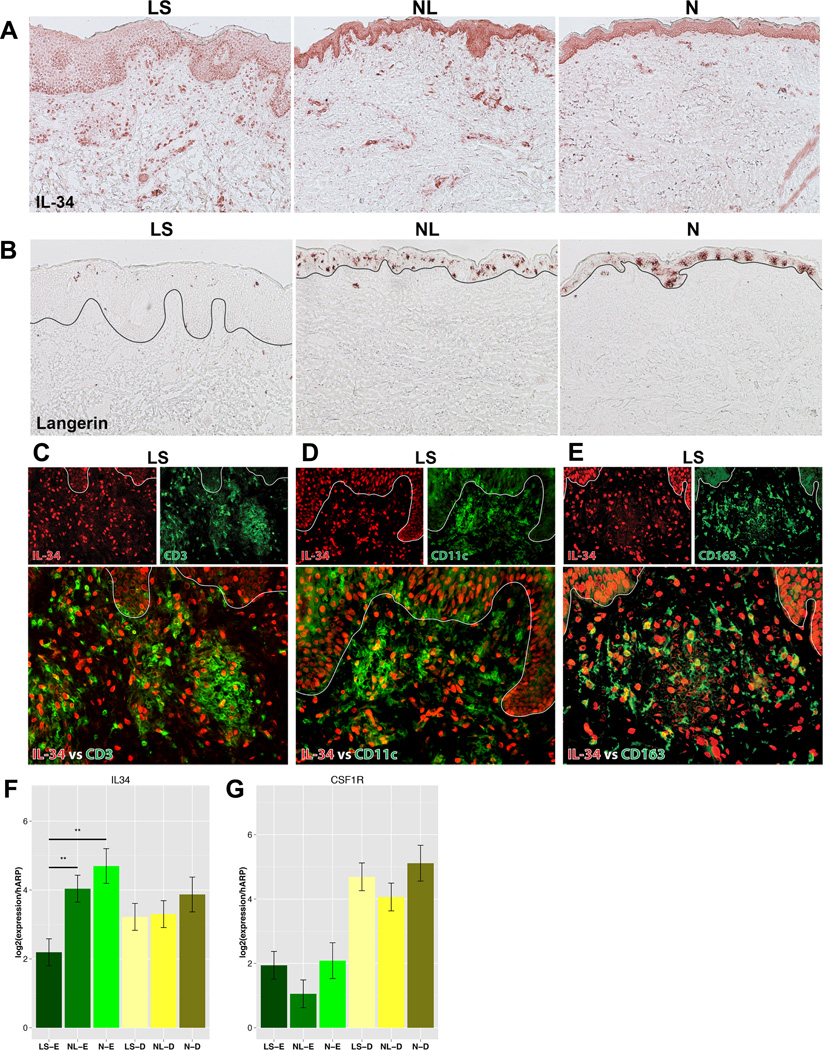
IL-34 was significantly down-regulated in LS compared to both NL and normal epidermis (Fig.2A, ETable9). IL-34 is a recently identified cytokine in mice and normal human skin,29 suggested to regulate LC differentiation in steady states. It has been identified as an alternative ligand to the CSF-1 receptor/CSF-1R, which has been shown to be expressed in epidermal LCs, dermal monocytes, and DCs.30 IL-34 has not been previously reported in human skin diseases. We performed immunohistochemical staining of IL-34, which showed stronger epidermal staining in normal and NL skin compared with LS skin and stronger dermal staining in LS skin (Fig.3A).
Figure 3.
Representative staining of IL-34 (A) and langerin (B) in lesional/LS, non-lesional/NL AD and normal/N skin. C–E. Representative double immunofluorescence for co-expression of IL-34 (red) vs. CD3 (green) (C), CD11c (green) (D), and CD163 (green) (E) in LS skin. F–G. mRNA expression of IL-34 (F) and its receptor/CSF-1R (G) in epidermis/E and dermis/D of lesional/LS, non-lesional/NL, and normal/N skin. Mean±SD. **p<0.01;***p<0.001.
Since IL-34 was reported to induce LC differentiation only in steady states,30 langerin (CD207) staining for LCs was performed. Many fewer LC cells were detected in LS compared to both NL and normal skin (Fig.3B). To identify the cellular distribution of IL-34 expression in the dermis, we performed double immunofluorescence for IL-34 (red) with CD3+320 /T-cells (green; EFig.4A), CD11c+/DCs (green; EFig.4B), and CD163+321 /macrophages (green; EFig.4C) in normal, NL, and LS skin. Few double positive IL-34+/CD3+ cells were found. Many IL-34+ cells co-localized with CD11c+ and CD163 + cells (Fig.3C–E). Thus, IL-34 in the dermis is preferentially expressed by myeloid DCs and macrophages.
We validated the lower expression of IL-34 in LS epidermis by RT-PCR, with significantly lower IL-34 mRNA expression observed in LS vs. both NL and normal epidermis and with slightly lower dermal mRNA expression (Fig.3F). We also measured CSF-1R mRNA expression, which was significantly higher in the dermis, regardless of tissue (Fig.3G).
LCM highlights tight junction defects in AD
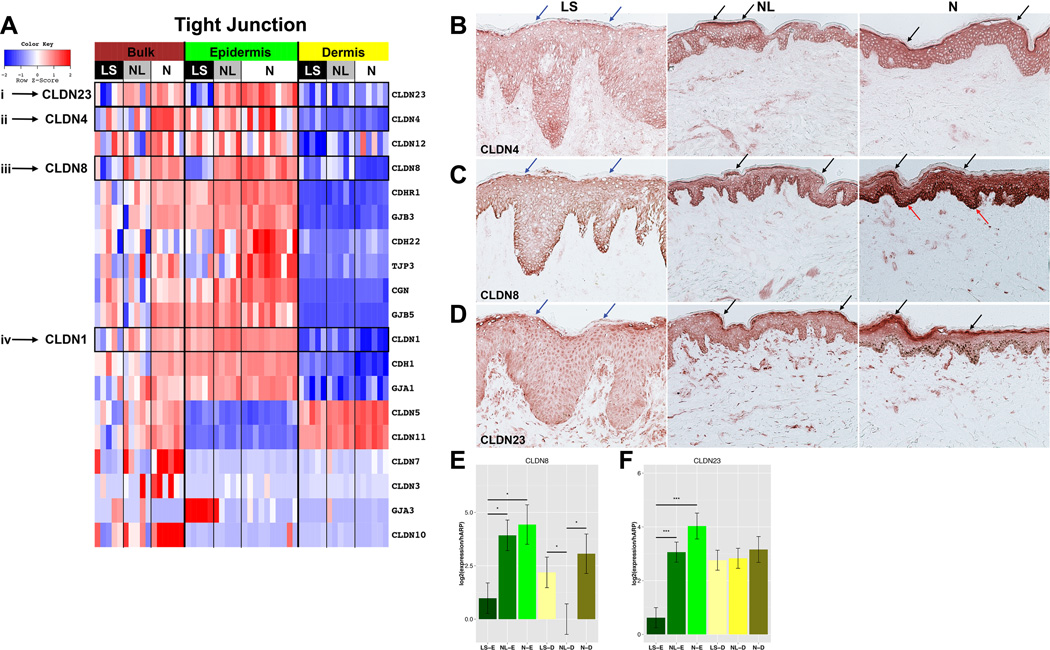
Since defective barrier function is a hallmark of AD,1 we evaluated for EDC and cornified envelope/CE markers. A heatmap of EDC and CE genes (including S100As) is shown in EFig.5. The majority of S100s (S100A8/A9/A12/A13), IL-17-induced PI3/Elafin, and SPRRs (SPRR1A) showed a primarily epidermal expression and higher expression in LS vs. both NL and normal skin (FDR<0.001 for most markers), whereas a few S100s (S100A4/A6/A10) showed predominantly dermal lesional expression. As previously reported in bulk tissues,27 the differentiation genes (late cornified envelope/LCEs, LOR, FLG, and periplakin/PPL) showed decreased expression in LS vs. NL and normal epidermis (EFig.5, ETable10). Another crucial barrier component, claudins, is essential for tight junction/TJ formation. 23 claudins have been identified in humans, however their contribution to AD is not fully defined.30,31,32 To gain insight into which claudins are expressed in AD, we evaluated a subset of TJ genes, the expression profiles of which are visualized in a heatmap (Fig.4A, ETable11). Down-regulation of claudins 1 and 23 was observed in LS vs. NL and normal epidermis as well as down-regulation of claudins 4 and 8, that were previously unreported in AD33 (FDR<0.001). We also detected dermal claudins (5 and 11), which were significantly down-regulated in the LS vs. normal skin comparison, previously not reported in AD.
Figure 4.
A. Heatmap of barrier-related genes FCHs comparing lesional/LS; non-lesional/NL, normal/N skin in bulk, epidermis, dermis (n=5 AD; n=6, 10, 6 normal bulk, epidermis, dermis, respectively). B–D. Representative claudin/CLDN 4, (B) 8 (C), and 23 (D) staining in lesional/LS, non-lesional/NL, and normal/N skin. Arrows: increased stratum corneum (black), decreased LS stratum corneum (blue), increased normal skin basal layer (red) staining. E–F. Expression of CLDN8 (E), CLDN23 (F) in epidermis/E, dermis/D of LS, NL, normal skin. Mean±SD. **p<0.01;***p<0.001.
The differential expression of claudin genes in the epidermal and dermal compartments was validated by IHC and RT-PCR. Claudins 4, 8, and 23 stained all epidermal layers, except stratum corneum (Fig.4B–D), with highly attenuated staining in LS vs. both NL and normal epidermis. The reduced staining was particularly evident in the granular layer. Claudin 8 showed increased intensity of basal layer staining in normal epidermis (Fig.4C). The mRNA expression of claudins 8 and 23 was also significantly reduced in LS epidermis (Fig.4E–F; p<0.05).
Markers associated with activated T cells and other inflammatory cells are enriched by LCM
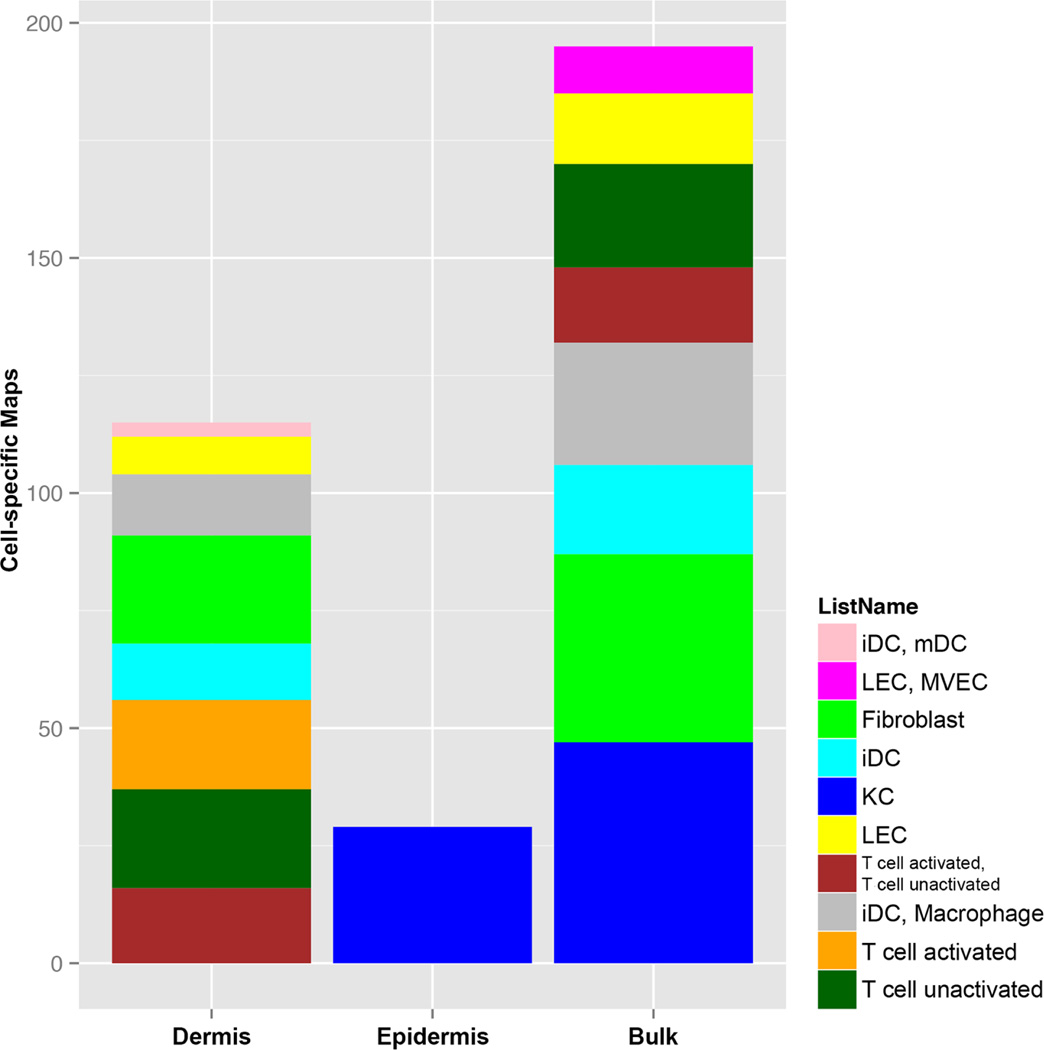
Using individual cell-culture expression data (keratinocytes/KCs, fibroblasts, activated and unactivated T-cells, DCs, LCs),34,35,36 we explored the distribution of DEGs identified in epidermal, dermal, and bulk AD transcriptomes28 using a Gene Set Enrichment Analysis/GSEA. Whereas only the KC-gene subset was enriched in the epidermal transcriptome (Fig.5, ETable12A), the dermal transcriptome showed enrichment of key inflammatory subsets, including activated and unactivated T-cells, various DC-subsets (immature DCs/iDCs and mature DCs/mDCs), macrophages, fibroblasts, and lymphatic endothelial cells/LECs (Fig.5, ETable12B). The bulk transcriptome showed a significant enrichment for KCs, fibroblasts, and inflammatory subsets, with no enrichment for activated T-cells and mDCs (Fig.5, ETable12C).
Figure 5.
The cell-specific map of the LCM epidermal and dermal transcriptomes and bulk transcriptome. The cellular source of the epidermal transcripome was mainly keratinocytes/KCs, whilte activated T-cells were only significantly enriched in the LCM dermal transcriptome. (KC/keratinocyte, iDC/immature dendritic cell, LEC/lymphatic endothelial cell, MVEC/microvascular endothelial cell, mDC/mature dendritic cell).
The GSEA analysis also linked key mediators (IL-22, TNFSF4/OX40L, CTLA4) detected in the dermal transcriptome to the gene signature of activated T-cells (ETableE12B). Among the genes found to be associated with both activated and un-activated T-cells in the dermal transcriptome were GZMB, ICOS, and CD27.
CCR7, which is associated with T-cell trafficking and cutaneous lymphoid aggregates,37,38 was shown to be expressed by various cells types, including T-cells and DCs. To investigate the predominant cellular expression of CCR7, we performed double immunofluorescence staining for CD3+ T-cells (green) or CD11c+ myeloid DCs (green) with CCR7 (red) in LS, NL, and normal skin. CCR7 co-localized mainly with CD11c, showing increased expression in LS vs. NL and normal dermis, and some co-localization with CD3, primarily in LS dermis (Figs. E6A–B). Thus, CCR7 is preferentially expressed by myeloid DCs, rather than T-cells.
Discussion
Evolving disease concepts associate the atopic dermatitis/AD phenotype with barrier and immune abnormalities.1,2 In this model, altered proliferation and differentiation of keratinocytes result from cytokines derived from distinct T-cell subsets.2,12 In order to fully understand the pathogenic mechanisms driving AD, it is important to understand the relative contributions of the epidermal and dermal compartments in creating the abnormal lesional/LS phenotype.27
This is the first report that establishes robust epidermal and dermal genomic signatures of LS, NL AD and normal skin, as compared with corresponding whole tissue fingerprinting. Using bulk skin, we have associated the AD phenotype with Th2/Th22 immune activation and abnormal epidermal differentiation.11,12,13,14 However, in bulk genomic profiling we could not determine which compartments are responsible for individual gene expression, or locate their cellular sources.4,12,28 Furthermore, due to a dilution effect in bulk tissues, which is composed of functionally heterogeneous cells, many genes linked to AD pathogenesis are below detection level on microarrays.11,12,27,28 Additionally, because a large subset of genes are expressed in either the epidermis or dermis, but not the other, some of the measured differential gene expression between LS and NL skin appears to be an artifact of the unequal contribution of epidermal expansion in LS skin, rather than due to true changes in gene expression at the cellular level. The previous disease model for AD, which has relied on bulk tissue genomic profiling, is limited by the inability to distinguish between these differences. A large proportion of DEGs found only in bulk tissue seem to derive from unequal contributions of LS epidermis and dermis compared to NL epidermis and dermis. Overall, we believe that the differences in cellular physiology created by AD are best detected in the LCM-generated DEGs.
Using the laser capture micro-dissection/LCM method followed by genomic and cellular studies, we have identified a largely increased AD transcriptome, with an additional 674 up- and 405 down-regulated genes compared with prior reports.11,12,27,28 By separating the two compartments we have also identified key immune and barrier markers that are usually undetectable on arrays (i.e. IL-22 and TSLP), and obtained more robust genomic differences for most immune genes when comparing LS, NL and normal tissues. Through this approach and previously acquired pathways and cell-specific genomic maps,11,12,13,14,27,28 we have localized many immune and barrier genes to the epidermis or dermis (or both), leading to a deeper understanding of inflammatory circuits and cellular subset involved in creating the AD phenotype. For example, dermal expression of CCL19 and CL21 likely attracts CCR7+/CD11c+ DCs and may organize lymphoid tissue in the dermis. Our LCM and genomic approach also led to enrichment of gene products associated with activated T-cells and inflammatory DCs that play a role in effector responses in AD.38
Our data is the first to identify IL-34, the newly identified cytokine in mice models, and its receptor colony-stimulating factor-1 receptor/CSF-1R, in a human skin disease. The mice studies demonstrated a critical role for IL-34 in differentiation and proliferation of Langerhans cells/LCs in the epidermis during steady states, while repopulation of LCs in inflammatory states was independent of IL-34.29 Although IL-34 is mainly produced by KCs, its receptor CSF-1R originates from dermal macrophages and mononuclear cells.32 Our data shows decreased epidermal expression of IL-34 in LS compared to both NL AD and normal epidermis (Figs.3B–C). This confirms the role of IL-34 in the maintenance phase, corresponding to prior mouse data.29 Thus, IL-34 cytokine might function as a negative regulator, and its induction in NL skin might inhibit the propagation of the inflammatory cascade towards development of active skin lesions. Additionally, we found that IL-34 expression in LS dermis co-localizes most commonly with myeloid DCs and macrophages. Future studies are needed to evaluate possible functions of the IL-34/ CSF-1R cytokine-receptor complex in background and diseased AD skin, and whether strategies of increasing this cytokine might be able to prevent development of skin lesions.
Separating the two compartments, we also identified two novel barrier genes, claudin 4 and 8, for the first time in the AD transcriptome. Claudins are pivotal for tight junction/TJ formation.39 Prior mouse and human skin equivalent models demonstrated that claudin 4 co-localized with claudin 1 to the epidermal granular layer,40,41 while claudin 8 was previously shown in human kidneys and intestines.42,43 Nevertheless, our knowledge of claudins and their involvement in the barrier alterations in AD remains incomplete.32 De Benedetto et al. reported that claudins 1 and 23 show significantly reduced expression in NL AD compared with healthy skin.33 In our study, claudins 8 and 23 showed significantly reduced expression in LS compared to NL and normal epidermis by microarrays (Fig.4A, ETable11), with respective fainter epidermal staining of Claudins 4, 8, and 23 in LS versus NL AD and normal epidermis (Figs.4B–D). Claudin 11 was identified as a novel dermal claudin, previously shown in other tissues (brain, testis, and cochlea).44,45,46 Future experiments are needed to investigate the function of the newly identified claudins 4 and 8, but together with claudins 1 and 23 these might contribute to the barrier defect associated with the AD phenotype.
Our study also presents some limitations: 1. LCM is a labor-intensive method, usually allowing analyses of small number of samples as in our study (n=5 of LS and NL each: n=3 of normal samples separated into epidermis and dermis), possibly resulting in weaker statistical power, particularly for bulk tissue comparisons; 2. Although our LCM approach detected novel barrier genes, the down-regulation of key differentiation markers (i.e. FLG, LOR, and IVL) in the LS and NL (versus N) epidermal transcriptomes was not as impressive as in prior bulk data,13,27 contrary to the enrichment of FLG and LOR in a prior normal skin epidermis LCM study,17,48 perhaps due to lower recovery of granular layer products using the epidermal-dermal separation approach.47 3. Even though the combined LCM epidermal and dermal transcriptomes were larger than the bulk tissue transcriptome, many genes were only detected in the bulk transcriptome (EFig.2 and ETable8C). This might be explained by a) the unequal contribution of LS epidermis and dermis compared to their counterparts; b) possible inclusion of subcutaneous tissue in bulk biopsies; c) by platform/technical issues as described in a previous LCM paper separating epidermis and dermis from only 3 psoriasis (LS and NL) and 3 normal tissues;16 and d) this analysis was restricted to patients with severe AD, in which NL skin has a more abnormal phenotype.49 Thus, many genes might not pass defined thresholds of FCH and significance when comparing LS and NL skin.
Our study establishes the utility of LCM in AD to separate different skin compartments and cellular infiltrates. It provides complementary information to bulk analysis, allowing regional and/or cellular localization of key barrier or immune molecules, and enables detection of genes that are not usually detected on arrays due to the mixture of transcripts within the heterogeneous bulk tissue.50,51 LCM is particularly beneficial for AD, in which a complex network of immune and barrier abnormalities results in the global phenotypes of active and “normal-appearing” skin, and dissecting the individual skin components and cells is crucial to unraveling their respective contributions to pathogenesis. Our combined LCM and genomic approach can be useful in future studies aimed at dissecting the relative roles of barrier versus immune activation of different AD phenotypes (i.e. intrinsic and extrinsic AD)11 and for dissecting therapeutic responses to various agents that are now in clinical trials for AD patients.52
Supplementary Material
Highlights.
-
-
Previous microrarray expression studies are limited to bulk tissue Atopic Dermatitis (AD) phenotyping
-
-
This study defines the epidermal and dermal AD phenotypes
-
-
We established compartmental and cellular localization of key AD markers
-
-
We identified novel cytokines and barrier products that might play a role in AD
Acknowledgements
JGK and MSF were supported by grant number 5UL1RR024143-02 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. E.G.Y was supported by the Dermatology Foundation Physician Scientist Career Development Award and by Leo Pharma and D.A.E is a joint PhD student of Leo Pharma and DTU, and partly funded by the Danish Ministry of Higher Education and Science.
Abbreviations
- AD
Atopic dermatitis
- CADM3
Cell adhesion molecule 3
- CCNB2
Cyclin B2
- CCL
Chemokine (C-C motif) ligand
- CCR
Chemokine (C-C motif) receptor
- CDC
Cell division cycle
- CE
Cornified envelope
- CLDN
Claudin
- COL
Collagen
- CSF-1R
Colony-stimulating factor 1 receptor
- CTLA4
Cytotoxic T-lymphocyte antigen 4
- CXCL
Chemokine (C-X-C motif) ligand
- DC
Dendritic cell
- DEFB4A
Defensin, beta 4A
- DEG
Differentially expressed gene
- EDC
Epidermal differentiation complex
- FCH
Fold change
- FDR
False discovery rate
- FLG
Filaggrin
- GHR
Growth hormone receptor
- GJA3
Gap junction protein, alpha 3
- GZMB
Granzyme B
- hARP
human acidic ribosomal protein P0
- ICOS
Inducible T-Cell Co-Stimulator
- iDC
Immature dendritic cell
- IF
Immunofluorescence
- IFNGR2
Interferon gamma receptor 2 (interferon gamma transducer 1)
- IHC
Immunohistochemistry
- IL
Interleukin
- IL-12RB2
Interleukin 12 receptor, beta 2
- IL-2RA
Interleukin 2 receptor, alpha
- IL-4R
Interleukin 4 receptor
- IL-7R
Interleukin 7 receptor
- IRF1
Interferon regulatory factor 1
- ITGAX
Integrin, alpha X (complement component 3 receptor 4 subunit)
- ITGBL1
Integrin, beta-like 1 (with EGF-like repeat domains)
- IVL
Involucrin
- KC
Keratinocyte
- KRT
Keratin
- LC
Langerhans cell
- LCE
Late cornified envelope
- LCM
Laser capture micro-dissection
- LCN2
Lipocalin 2
- LEP
Leptin
- LOR
Loricrin
- LPL
Lipoprotein lipase
- LS
Lesional
- mDC
Mature dendritic cell
- MMP12
Matrix metalloproteinase 12
- MX1
Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse)
- NL
Non-lesional
- OASL
2'-5'-oligoadenylate synthetase-like
- PCA
Principal Component Analysis
- PI3
Peptidase inhibitor 3, skin-derived
- PPL
Periplakin
- RT-PCR
Real time PCR
- SELE
Selectin E
- SPRR
Small proline-rich protein
- STAT3
Signal transducer and activator of transcription 3 (acute-phase response factor)
- TJ
Tight junction
- TNFRSF4/OX40
Tumor necrosis factor receptor superfamily, member 4
- TNFSF4/OX40L
Tumor necrosis factor superfamily, member 4
- TSLP
Thymic stromal lymphopoietin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have declared that they have no conflict of interest.
Clinical Implications: Our approach can be useful to differentiate ability of targeted treatments to reverse epidermal and immune alterations in atopic dermatitis skin.
References
- 1.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358(14):1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 2.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part I: Clinical and pathologic concepts. J Allergy Clin Immunol. 2011;127(5):1110–1118. doi: 10.1016/j.jaci.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 3.Boguniewicz M, Leung DY. Recent insights into atopic dermatitis and implications for management of infectious complications. J Allergy Clin Immunol. 2010;125:4–15. doi: 10.1016/j.jaci.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nograles KE, Zaba LC, Shemer A, Fuentes-Duculan J, Cardinale I, Kikuchi T, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol. 2009;123:1244–1252. doi: 10.1016/j.jaci.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong PY, Leung DY. Immune dysregulation in atopic dermatitis. Curr Allergy Asthma Rep. 2006;6:384–389. doi: 10.1007/s11882-996-0008-5. [DOI] [PubMed] [Google Scholar]
- 6.Hatano Y, Terashi H, Arakawa S, Katagiri K. Interleukin-4 suppresses the enhancement of ceramide synthesis and cutaneous permeability barrier functions induced by tumor necrosis factor-alpha and interferon-gamma in human epidermis. J Invest Dermatol. 2005;124:786–792. doi: 10.1111/j.0022-202X.2005.23651.x. [DOI] [PubMed] [Google Scholar]
- 7.Elias PM, Hatano Y, Williams ML. Basis for the barrier abnormality in atopic dermatitis: outside-inside-outside pathogenic mechanisms. J Allergy Clin Immunol. 2008;121:1337–1343. doi: 10.1016/j.jaci.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onoue A, Kabashima K, Kobayashi M, Mori T, Tokura Y. Induction of eosinophil- and Th2-attracting epidermal chemokines and cutaneous late-phase reaction in tape-stripped skin. Exp Dermatol. 2009;18:1036–1043. doi: 10.1111/j.1600-0625.2009.00899.x. [DOI] [PubMed] [Google Scholar]
- 9.Oyoshi MK, Larson RP, Ziegler SF, Geha RS. Mechanical injury polarizes skin dendritic cells to elicit a T(H)2 response by inducing cutaneous thymic stromal lymphopoietin expression. J Allergy Clin Immunol. 2010;126:976–984. 984.e1–984.e5. doi: 10.1016/j.jaci.2010.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demehri S, Morimoto M, Holtzman MJ, Kopan R. Skin-derived TSLP triggers progression from epidermal-barrier defects to asthma. PLoS Biol. 2009;7:e1000067. doi: 10.1371/journal.pbio.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suarez-Farinas M, Dhingra N, Gittler J, Shemer A, Cardinale I, de Guzman Strong C, et al. Intrinsic atopic dermatitis shows similar TH2 and higher TH17 immune activation compared with extrinsic atopic dermatitis. J Allergy Clin Immunol. 2013;132(2):361–370. doi: 10.1016/j.jaci.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gittler JK, Shemer A, Suarez-Farinas M, Fuentes-Duculan J, Gulewicz KJ, Wang CQ, et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol. 2012;130(6):1344–1354. doi: 10.1016/j.jaci.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman-Yassky E, Suarez-Farinas M, Chiricozzi A, Nograles KE, Shemer A, Fuentes-Duculan J, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol. 2009;124(6):1235. doi: 10.1016/j.jaci.2009.09.031. 1244.e58. [DOI] [PubMed] [Google Scholar]
- 14.Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Whynot J, Novitskaya I, Cardinale I, et al. Major differences in inflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. J Allergy Clin Immunol. 2007;119(5):1210–1217. doi: 10.1016/j.jaci.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, et al. Laser-capture microdissection. Nat Protoc. 2006;1(2):586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 16.Mitsui H, Suarez-Farinas M, Belkin DA, Levenkova N, Fuentes-Duculan J, Coats I, et al. Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J Invest Dermatol. 2012;132(6):1615–1626. doi: 10.1038/jid.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulati N, Krueger JG, Suarez-Farinas M, Mitsui H. Creation of differentiation-specific genomic maps of human epidermis through laser capture microdissection. J Invest Dermatol. 2013;133(11):2640–2642. doi: 10.1038/jid.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ning H, Mitsui H, Wang C, Suarez-Farinas M, Gonazleaz J, Shah K, et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in Basal Cell Carcinoma. Oncotarget. 2013;4(12):2237–2248. doi: 10.18632/oncotarget.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhingra N, Shemer A, Correa da Rosa J, Rozenblit M, Suarez-Farinas M, Fuentes-Duculan J, et al. Molecular profiling of contact dermatitis skin identifies allergen-dependent differences in immune response. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.03.009. (in press). [DOI] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suarez-Farinas M, Pellegrino M, Wittkowski K, Magnasco M. Harshlight: a “corrective make-up” program for microarray chips. BMC Bioinformatics. 2005;6(294) doi: 10.1186/1471-2105-6-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99(468):909–917. [Google Scholar]
- 23.Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer-Verlag; pp. 397–420. [Google Scholar]
- 24.Boedigheimer MJ, Wolfinger RD, Bass MB, Bushel PR, Chou JW, Cooper M, et al. Sources of variation in baseline gene expression levels from toxicogenomics study control animals across multiple laboratories. BMC Genomics. 2008;12(9):285. doi: 10.1186/1471-2164-9-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, et al. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS One. 2011;6(2):e17238. doi: 10.1371/journal.pone.0017238. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 27.Suarez-Farinas M, Tintle SJ, Shemer A, Chiricozzi A, Nograles K, Cardinale I, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127(4):954, 64.e1–64.e4. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khattri S, Shemer A, Rozenblit M, Dhingra N, Czarnowicki T, Finney R, et al. Cyclosporine in patients with atopic dermatitis modulates activated inflammatory pathways and reverses epidermal pathology. J Allergy Clin Immunol. 2014;133(6):1626–1634. doi: 10.1016/j.jaci.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012;37(6):1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2(4):285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 31.Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10(8):235. doi: 10.1186/gb-2009-10-8-235. 2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, et al. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101(3):1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- 33.De Benedetto A, Rafaels NM, McGirt LY, Ivanov AI, Georas SN, Cheadle C, et al. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol. 2011;127(3):773, 86.e1–86.e7. doi: 10.1016/j.jaci.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiricozzi A, Nograles KE, Johnson-Huang LM, Fuentes-Duculan J, Cardinale I, Bonifacio KM, et al. IL-17 induces an expanded range of downstream genes in reconstituted human epidermis model. PLoS One. 2014;9(2):e90284. doi: 10.1371/journal.pone.0090284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nograles KE, Suarez-Farinas M, Shemer A, Fuentes-Duculan J, Chiricozzi A, Cardinale I, et al. Atopic dermatitis keratinocytes exhibit normal T(H)17 cytokine responses. J Allergy Clin Immunol. 2010;125(3):744, 6, 746.e1–746.e2. doi: 10.1016/j.jaci.2009.12.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujita H, Nograles KE, Kikuchi T, Gonzalez J, Carucci JA, Krueger JG. Human langerhans cells induce distinct IL-22-producing CD4+ T cells lacking IL-17 production. Proc Natl Acad Sci U S A. 2009;106(51):21795–21800. doi: 10.1073/pnas.0911472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: Balancing immunity and tolerance. Nat Rev Immunol. 2008;8(5):362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 38.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part II: Immune cell subsets and therapeutic concepts. J Allergy Clin Immunol. 2011;127(6):1420–1432. doi: 10.1016/j.jaci.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 39.Agrawal R, Woodfolk JA. Skin barrier defects in atopic dermatitis. Curr Allergy Asthma Rep. 2014;14(5):433. doi: 10.1007/s11882-014-0433-9. 014-0433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J Cell Biol. 2002;156(6):1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuki T, Komiya A, Kusaka A, Kuze T, Sugiyama Y, Inoue S. Impaired tight junctions obstruct stratum corneum formation by altering polar lipid and profilaggrin processing. J Dermatol Sci. 2013;69(2):148–158. doi: 10.1016/j.jdermsci.2012.11.595. [DOI] [PubMed] [Google Scholar]
- 42.Juric M, Xiao F, Amasheh S, May O, Wahl K, Bantel H, et al. Increased epithelial permeability is the primary cause for bicarbonate loss in inflamed murine colon. Inflamm Bowel Dis. 2013;19(5):904–911. doi: 10.1097/MIB.0b013e3182813322. [DOI] [PubMed] [Google Scholar]
- 43.Hou J, Renigunta A, Yang J, Waldegger S. Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc Natl Acad Sci U S A. 2010;107(42):18010–18015. doi: 10.1073/pnas.1009399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bronstein JM, Micevych PE, Chen K. Oligodendrocyte-specific protein (OSP) is a major component of CNS myelin. J Neurosci Res. 1997;50(5):713–720. doi: 10.1002/(SICI)1097-4547(19971201)50:5<713::AID-JNR8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 45.Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, et al. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell. 1999;99(6):649–659. doi: 10.1016/s0092-8674(00)81553-6. [DOI] [PubMed] [Google Scholar]
- 46.Kitajiri SI, Furuse M, Morita K, Saishin-Kiuchi Y, Kido H, Ito J, et al. Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear Res. 2004;187(1–2):25–34. doi: 10.1016/s0378-5955(03)00338-1. [DOI] [PubMed] [Google Scholar]
- 47.Radoja N, Gazel A, Banno T, Yano S, Blumenberg M. Transcriptional profiling of epidermal differentiation. Physiol Genomics. 2006;27(1):65–78. doi: 10.1152/physiolgenomics.00031.2006. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy-Crispin M, Billick E, Mitsui H, Gulati N, Fujita H, Gilleaudeau P, et al. Human keratinocytes' response to injury upregulates CCL20 and other genes linking innate and adaptive immunity. J Invest Dermatol. 2012;132(1):105–113. doi: 10.1038/jid.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suarez-Farinas M, Tintle SJ, Shemer A, Chiricozzi A, Nograles K, Cardinale I, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127(4):954–964. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu A. Laser capture microdissection in the tissue biorepository. J Biomol Tech. 2010;21(3):120–125. [PMC free article] [PubMed] [Google Scholar]
- 51.Decarlo K, Emley A, Dadzie OE, Mahalingam M. Laser capture microdissection: Methods and applications. Methods Mol Biol. 2011;755:1–15. doi: 10.1007/978-1-61779-163-5_1. [DOI] [PubMed] [Google Scholar]
- 52.Guttman-Yassky E, Dhingra N, Leung DY. New era of biologic therapeutics in atopic dermatitis. Expert Opin Biol Ther. 2013;13(4):549–561. doi: 10.1517/14712598.2013.758708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.