Abstract
Background
The Wnt/β-catenin pathway regulates liver growth, repair, and regeneration. Chronic ethanol exposure blunts normal liver regenerative responses, in part by inhibiting insulin/IGF signaling, and correspondingly, previous studies showed that ethanol-impaired liver regeneration could be restored by insulin sensitizer (PPAR-δ agonist) treatment. Since Wnt/β-catenin functions overlap and crosstalk with insulin/IGF pathways, we investigated the effects of ethanol exposure and PPAR-δ agonist treatment on Wnt pathway gene expression in relation to liver regeneration.
Methods
Adult male Long Evans rats were fed with isocaloric liquid diets containing 0% or 37% ethanol for 8 weeks, and also treated with vehicle or a PPAR-δ agonist during the last 3 weeks of the feeding regimen. The rats were then subjected to 70% partial hepatectomy (PH) and livers harvested at various post-PH time points were used to quantify expression of 19 Wnt pathway genes using Quantigene 2.0 Multiplex Assay.
Results
Ethanol broadly inhibited expression of Wnt/β-catenin signaling-related genes, including down-regulation of Wnt1, Fzd3, Lef1, and Bcl9 throughout the post-PH time course (0-72 h), and suppression of Wnt7a, Ccnd1, Fgf4, Wif1, Sfrp2, and Sfrp5 at 18, 24 hours post-PH time points. PPAR-δ agonist treatments rescued the ethanol-induced suppression of Wnt1, Wnt7a, Fzd3, Lef1, Bcl9, Ccnd1, and Sfrp2 gene expression in liver, corresponding with the improvements in DNA synthesis and restoration of hepatic architecture.
Conclusions
Chronic high-dose ethanol exposures inhibit Wnt signaling, which likely contributes to the impairments in liver regeneration. Therapeutic effects of PPAR-δ agonists extend beyond restoration of insulin/IGF signaling mechanisms and are mediated in part by enhancement of Wnt pathway signaling. Future studies will determine the degree to which targeted restoration of Wnt signaling is sufficient to improve liver regeneration and remodeling in the context of chronic ethanol exposure.
Keywords: Alcoholic liver disease, Wnt, peroxisome proliferator-activated receptor agonist
INTRODUCTION
Chronic alcoholic liver disease (ALD) is one of the major causes of illness and death in the USA (Addolorato et al., 2013; Schwartz et al., 2012). ALD progresses through stages of steatohepatitis, fibrosis, cirrhosis, and finally end-stage liver disease, and culminates in liver failure. Many of the long-term adverse effects of ethanol on hepatocellular growth, survival, energy metabolism, and gene expression, are due to impairments in insulin/IGF signaling through growth and survival networks (de la Monte et al., 2012). These effects of ethanol are mediated by reduced phosphorylation of the insulin receptor and insulin receptor substrate 1 (IRS1), activation of insulin receptor tyrosine kinase, and reduced downstream signaling through PI3 Kinase/Akt, and Erk MAPK (Banerjee et al., 1998; He et al., 2007; Sasaki and Wands, 1994). One of the fundamental problems contributing to insulin pathway inhibition is hepatic insulin resistance (Longato et al., 2012; de la Monte et al., 2008; Pang et al., 2009). A second consideration is the adverse effect of insulin resistance on larger networks of inter-related signaling that also regulate hepatocellular growth, repair, regeneration, and metabolic functions. In this regard, several studies have shown that Wnt and insulin/IGF pathways crosstalk to regulate various cellular functions (Chiang et al., 2013; Kim et al., 2013; Tomimaru et al., 2013a).
The Wnt/β-catenin signaling pathway is an evolutionarily conserved, highly complex, and critical for liver development. Wnt/β-catenin signaling is activated by Wnt protein interactions with frizzled (FZD) cell surface receptors, which results in stabilization and accumulation of β-catenin in the cytoplasm, followed by its translocation to the nucleus, where it binds to T-cell factor (Tcf)/lymphoid enhancer-binding factor (Lef) family protein to initiate transcription of target genes. However, in the absence of Wnt signaling, cytoplasmic β-catenin levels are kept low due to its phosphorylation by the APC-Axin-GSK-3β destruction complex, which targets β-catenin for proteasomal degradation (Saito-Diaz et al., 2013).
Beyond its effects on development, recent studies have highlighted the importance of Wnt/β-catenin pathway in relation to regeneration, metabolism and homeostatic functions in the adult liver (Kuncewitch et al., 2013). In particular, Wnt1 and β-catenin proteins are crucial in liver regeneration after 70% partial hepatectomy (PH), and β-catenin activation appears to be essential for timely stimulation of liver regeneration following acetaminophen-induced acute liver failure (Apte et al., 2009). Furthermore, Wnt/β-catenin signaling promotes growth of adult hepatic stem cells following chemical injury and PH (Apte et al., 2008). However, given the clinical and epidemiological importance of alcoholic liver disease in relation to liver degeneration and failure, it would be important to determine if ethanol-mediated impairments in liver regeneration were also mediated by inhibition of Wnt/β-catenin signaling, because this information could expand therapeutic approaches to this disease.
Accordingly, we hypothesized that chronic ethanol exposure alters hepatic expression of Wnt pathway-related genes needed for growth, remodeling, and homeostasis, including during liver regeneration. Moreover, we explored the concept that ethanol-mediated impairments in liver regeneration were mediated in part via crosstalk suppression of Wnt/β-catenin, and therefore, treatment with insulin sensitizer agents such as peroxisome proliferator-activated receptor (PPAR) would rectify Wnt/β-catenin signaling mechanisms vis-à-vis chronic ethanol exposure. PPARs are nuclear hormone receptors that bind to DNA and regulate gene transcription (Gyamfi et al., 2010; Yessoufou et al., 2010). There are 3 distinct PPAR isoforms, namely PPAR-α, PPAR-β/δ, and PPAR-γ (Berger and Moller, 2002; Chang et al., 2007; Tontonoz and Spiegelman, 2008). Previously, a PPAR-δ agonist was shown to be more effective than PPAR-β/δ and PPAR-γ agonists for reducing hepatic injury, oxidative stress, and lipid peroxidation (de la Monte et al., 2008), and effective in restoring insulin signaling, regeneration, and repair following PH in chronic ethanol-fed rats (Pang et al., 2009). In the present study, we examined the effects of chronic ethanol exposure and PPAR-δ agonist treatment on the expression of Wnt pathway genes at baseline and over the time course of liver regeneration following PH.
METHODS
Chronic ethanol exposure model
Adult male Long Evans rats (Harlan Sprague Dawley, Indianapolis, IN, USA) were pair-fed isocaloric liquid diets (Bioserv, Frenchtown, NJ, USA) containing 0% (control) or 37% ethanol by caloric content (9.2% v/v) for 8 weeks. Rats were monitored daily to ensure equivalent food consumption and maintenance of body weight. During the last 3 weeks, rats were administered twice weekly with intraperitoneal injections of saline or a PPAR-δ agonist (L-165,041; 2 μg/kg) (Calbiochem, Carlsbad, CA). The rats were then subjected to 2/3 PH. Livers were harvested under deep vaporized isofluorane (SurgiVet, Waukesha, WI, USA) anesthesia at 0, 18, 24, 30, 48, 72, and 168 hours post-PH. Liver tissue was snap-frozen in liquid nitrogen and stored at −80°C. All experiments and protocols conformed to guidelines established by the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee at the Lifespan Rhode Island Hospital.
Microsphere-based multiplex-branched DNA assay of gene expression
Total RNA was extracted using the Qiagen RNeasy Mini kit according to the manufacturer's protocol (Qiagen, ON, Canada). RNA concentrations and purity were determined from the absorbances measured at 260 and 280 nm. Quality of RNA was assessed by gel electrophoresis. Messenger RNA levels were quantified using a customized Quantigene 2.0 Multiplex (QGP) Assay (Affymetrix Inc., Fremont, CA, USA). Cooperative hybridization and subsequent quantification were performed following the manufacturer's recommended procedure for frozen animal tissue. Briefly, the assay allows for direct quantification of mRNA using xMAP Luminex beads and reporter signal amplification. A working bead mix containing lysis mixture, blocking reagent, capture beads, and 2.0 probe set was prepared and distributed to the plate. Total RNA (1 μg) was added to the sample wells and incubated with the xMAP fluorescent beads overnight. For background control, sterile nuclease-free water was used. A set of oligonucleotide probes (pre-amplifier, amplifier, and biotin-labeled probe) followed by SAPE (streptavidin-conjugated R-Phycoerythrin) was incubated with samples subsequently. The resultant fluorescent signal was detected by using the BioRad BioPlex 200 instrument (Bio-Rad, Hercules, CA, USA). It was verified that the level of SAPE fluorescence was proportional to the amount of RNA transcripts captured by the beads. We analyzed 19 Wnt signaling-related genes, including Wnt ligands, Fzd receptors, transcription factors, downstream target genes, and ligand binding proteins (Table 1). We also measured Hprt1, Gapdh, and Polr2a expression as internal controls for sample loading.
Table 1.
List of Quantigene probe for targets of Wnt signaling included in the Quantigene multiplex panel
| Group | Gene | Probe set region | Accession No |
|---|---|---|---|
| Ligands & related genes | Wnt1 | 561-2114 | NM_001105714 |
| Wnt3 | 250-658 | NM_001105715 | |
| Wnt7a | 361-1609 | XM_342723 | |
| Porcn | 162-579 | XM_228781 | |
| Receptors | Fzd3 | 485-1075 | NM_153474 |
| Fzd6 | 1872-2250 | NM_001130536 | |
| Transcription regulators | Lef1 | 134-587 | NM_130429 |
| Tcf7l2 | 535-932 | NM_001191052 | |
| Bcl9 | 920-1411 | XM_003753641 | |
| Target genes | Axin2 | 1297-1764 | NM_024355 |
| Ccnd1 | 283-668 | NM_171992 | |
| Jun | 889-1408 | NM_021835 | |
| Wisp1 | 230-777 | NM_031716 | |
| Ligand binding proteins | Sfrp2 | 549-1007 | NM_001100700 |
| Sfrp5 | 714-1087 | NM_001107591 | |
| Wif1 | 767-1171 | NM_053738 | |
| Other components | Apc2 | 18-501 | NM_001106769 |
| Senp2 | 375-805 | NM_023989 | |
| Fgf4 | 112-530 | NM_053809 | |
| Housekeeping genes | Hprt1 | 81-786 | NM_012583 |
| Gapdh | 214-456 | NM_017008 | |
| Polr2a | 2664-3142 | XM_343922 | |
Immunohistochemical staining
Paraffin-embedded slides were immunostained with β-catenin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) as previously described (Tomimaru et al., 2013b).
Cells
Immortalized human hepatocytes (OUMS29, kindly provided by Dr. Hironori Koga, Kurume University, Japan) were grown in DMEM Dulbecco's Modified Eagle Medium with 10% fetal bovine serum, L-glutamine (Life Technologies, Gaithersburg, MD), and MEM nonessential amino acids (Sigma Chemical Co, St Louis, MO).
Western blot analysis
Western blot analysis was carried out as previously described (Tomimaru et al., 2013a). In brief, immunoblots were probed with primary antibodies generated against β-catenin, β-actin (Cell Signaling Technology, Beverly, MA), cyclin D1, and PCNA (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and detected with HRP-labeled anti-mouse or anti-rabbit IgG (Amersham Pharmacia Biotech, Buckinghamshire, UK) using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, Rockford, IL). Positive signals were captured by the Image analyzer LAS-1000 plus (Fujifilm, Tokyo, Japan) and the band intensity of protein was determined using Image Gauge version 3.45 (Fujifilm).
Statistics
Data are expressed as mean ± SEM or SD. Inter-group differences were assessed by Mann Whitney test, unpaired t test with Welch's correction, or two-way repeated measures ANOVA (GraphPad Prism 5 software, San Diego, CA). P<0.05 was considered to be statistically significant.
RESULTS
Chronic ethanol exposure reduces expression of Wnt pathway-related genes
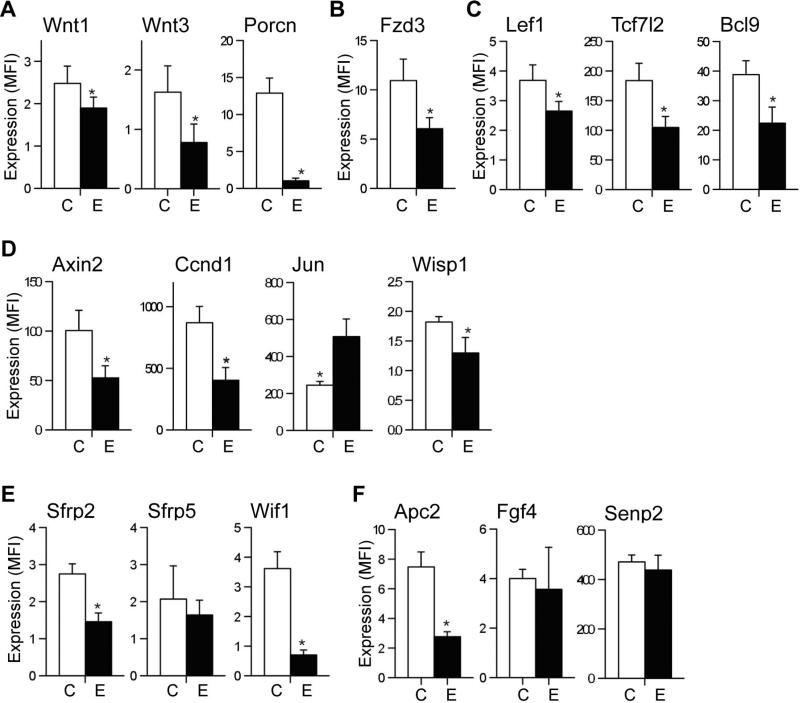
We used a custom QGP assay to simultaneously measure 19 different mRNAs involved in Wnt/β-catenin signaling. Chronic ethanol feeding significantly decreased mRNA expression of Wnt1 and Wnt3 ligands and Fzd3 receptor (Fig. 1A and 1B). These results are consistent with the findings in an exploratory study in which we used a Wnt pathway-focused gene expression array (RT2 Profiler PCR Array system, Qiagen) to provisionally identify Wnt genes targeted by ethanol in the liver (Supplementary Table 1 and 2). In contrast, Wnt7a and Fzd6 expressions were not significantly modulated by ethanol (data not shown). Porcupine (Porcn) is a membrane bound O-acyltransferase in the ER that is required for and dedicated to palmitoylation of Wnt ligands, a necessary step in processing of Wnt ligands secretion (Biechele et al., 2011). Consistent with reduced expression of Wnt ligands, Porcn was significantly downregulated in ethanol-exposed livers (Fig. 1A). As shown in Fig. 1C, levels of Lef1, Tcf7l2 and Bcl9 mRNA were significantly reduced in chronic ethanol-fed rat liver. Lef1 and Tcf7l2 are transcriptional factor proteins that bind to the WRE (Wnt responsive element) region of Wnt responsive target genes. Bcl9 is a co-activator of Wnt signaling and associates with other co-activators including Pygopus and Legless in the signaling cascade (Brembeck et al., 2011).
Fig. 1.
Effects of chronic ethanol feeding on hepatic expression of Wnt pathway-related genes. Adult male Long Evans rats were fed with isocaloric liquid diets containing 0% (C) or 37% ethanol (E) by caloric content for 8 weeks. A Quantigene 2.0 Multiplex Assay was used to quantify mRNA levels of (A) Wnt1 and Wnt3 ligands and Porcn, (B) Fzd3 receptor, (C) Lef1, Tcf7l2 Wnt transcription factors and Bcl9 co-factor, (D) Axin2, Ccnd1, Jun, and Wisp1 Wnt target genes, (E) Wif1, Sfrp2 and Sfrp5 Wnt ligand binding proteins, and (F) Apc2, Fgf4 andSenp2. Gene expression level is reflected by mean fluorescent intensity (MFI; arbitrary units). Graphs depict the net MFI ± S.D. with results normalized to housekeeping genes. *p < 0.05 vs. control-fed rats (n=3-5).
We furthered our analysis by measuring Wnt-inducible target gene expression including Axin2, Ccnd1, Jun and Wnt1 inducible signaling pathway protein 1 (Wisp1) (Fig. 1D). Axin2 functions as a Wnt antagonist and negative regulator of Wnt/β-catenin signaling, despite its elevated levels of expression in various cancers (Koch, et al., 2005; Yan et al., 2001). Ccnd1 regulates cell cycle progression to the proliferative stage (Hsu et al., 2006), and Wisp1, a matricellular protein, regulates cell proliferation, adhesion, and migration (Berschneider and Konigshoff, 2011). Correspondingly, chronic ethanol exposure decreased expression of Axin2, Ccnd1, and Wisp1. In contrast, Jun expression was increased by chronic ethanol exposure.
Although Wnt inhibitory factor 1 (Wif1), secreted frizzled-related protein 2 (Sfrp2), and Sfrp5 function as Wnt antagonists (Kaur et al., 2012), Sfrps can also stimulate Wnt signaling by either signaling through Fzd receptors and activating β-catenin, or enhancing diffusion and transport of Wnt ligands by mimicking (Kress et al., 2009; Mii and Taira, 2011). Moreover, down-regulation of Sfrp5 causes insulin resistance by inhibiting IRS-1 activation (Ouchi et al., 2010). The QGP assay results showed that ethanol downregulates Wif1 and Sfrp2 expression in liver but has no significant effect on Sfrp5 (Fig. 1E).
Adenomatous polyposis coli (APC) is a component of β-catenin destruction complex and known as a tumor suppressor, but its function is still remained elusive (Stamos and Weis, 2013). As shown in Fig. 1F, Apc2 mRNA was significantly downregulated by ethanol exposure, while there are no significant effect on the levels of fibroblast growth factor 4 (Fgf4) and SUMO-specific protease 2 (Senp2). Fgf4 is known to crosstalk with Wnt/β-catenin pathway to regulate cell proliferation and survival (Katoh and Katoh, 2006). Senp2 is a de-SUMOylation protease family member and promotes degradation of β-catenin (Lee et al., 2012; Tan et al, 2013).
Taken together, these results suggest that chronic ethanol exposure broadly inhibits Wnt/β-catenin signaling by down-regulating of Wnt signaling mediators at multiple levels of the cascade, from ligand/receptor expression and β-catenin stabilization to Wnt/β-catenin responsive target genes expression.
Effects of chronic ethanol exposure on Wnt signaling during liver regeneration
A potential role for Wnt/β-catenin signaling as a mediator of liver regeneration after PH was suggested by the findings that Wnt1 and β-catenin protein expression increased after 2/3 PH (Apte et al., 2008), and knockdown of β-catenin expression impaired liver regeneration (Sodhi et al., 2005) in rats. Sekine et al. reported that, although β-catenin is critical for proper regulation of hepatocyte proliferation and liver regeneration, the magnitude of the proliferative response is not tightly correlated with the levels of β-catenin/TCF signaling, indicating that this regulation may be indirect (Sekine et al., 2007). Recently, a comprehensive expression profiling analysis of long noncoding RNAs (lncRNAs) in post-PH livers showed that lncRNA-LALR1 functions as regulator of liver regeneration by facilitating cyclin D1 expression through the activation of Wnt/β-catenin signaling (Xu et al., 2013). Herein, we sought to further characterize changes in Wnt/β-catenin pathway gene expression in relation to the time course of liver regeneration following PH and chronic ethanol exposure. In interpreting the results, it is important to bear in mind that the curve corresponding to after PH DNA synthesis during liver regeneration is biphasic. The first peak (24 hours after PH) reflects DNA synthesis in hepatocytes and bile duct epithelial cells (Koniaris et al., 2003). The second peak (72 hours after PH) reflects less distinct wave of hepatocyte mitoses as well as Kupper and stellate cells caused by 2/3 PH (Koniaris et al., 2003). Notably, the degree of hepatocyte proliferation is directly proportional to the degree of injury; i.e., a minor liver insult will result in local mitotic reaction but any injury greater than about 10% will result in proliferation of cells throughout the liver and with greater than 50% resection a second wave can be observed at 3 days after PH in the rat (Koniaris et al., 2003).
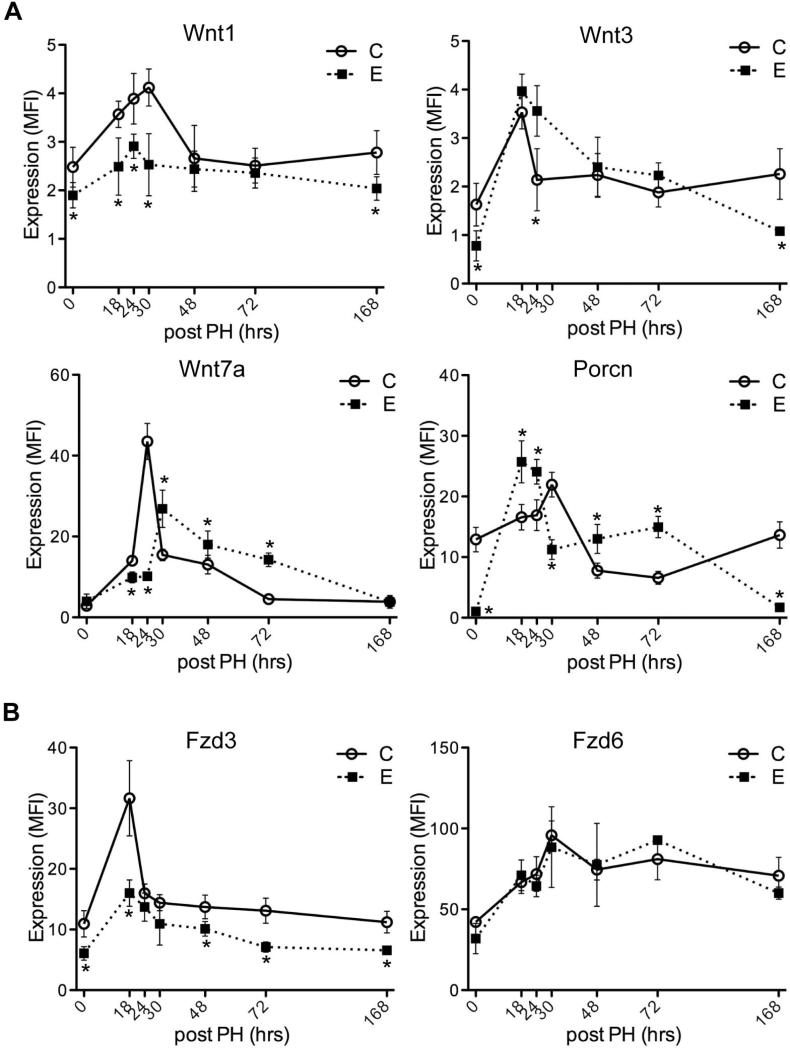
Previously, we showed that ethanol delayed the time course and blunted the peak levels of DNA synthesis in regenerating livers, and that these effects were largely abrogated by treatment with the PPAR-δ agonist (Pang et al., 2009). The present work exhibits that ethanol suppresses expression of Wnt-related gene expression during liver regeneration. Although both Wnt1 and Wnt3 ligands were downregulated in chronic ethanol-fed liver, ethanol suppresses Wnt1 expression, not Wnt3 during liver regeneration. On the other hand, the expression level of Wnt7a, which had no change by chronic ethanol exposure, exhibited the delayed peak at 30 hours post-PH. Interestingly, Porcn that involved in processing and maturation of Wnt ligands showed a similar pattern of Wnt3 expression in ethanol-fed rat liver (a peak at 18 hours post-PH), while it is correlated with Wnt1 expression in control-fed liver (a peak at 30 hours post-PH) (Fig. 2A). Fzd3 receptor was downregulated by ethanol treatment through liver regeneration, while ethanol has no effect on Fzd6 (Fig. 2B).
Fig. 2.
Effects of chronic ethanol feeding on Wnt ligands and receptors expression following PH. Adult male Long Evans rats were fed with isocaloric liquid diets containing 0% (C) or 37% ethanol (E) for 8 weeks. Livers harvested 0, 18, 24, 30, 48, 72, or 168 hours post-PH were obtained to measure time-course dependent shifts in (A) Wnt1, Wnt3, Wnt7a and Porcn, (B) Fzd3 and Fzd6 mRNA expression using the Quantigene 2.0 Multiplex Assay. Gene expression level is reflected by mean fluorescent intensity (MFI; arbitrary units). Graphs depict time-dependent shifts in net MFI ± S.D. with results normalized to housekeeping genes. Inter-group comparisons were made using two-way repeated measures ANOVA with Bonferroni's pos-hoc test, *p < 0.05 vs. control-fed rats (n = 3-5).
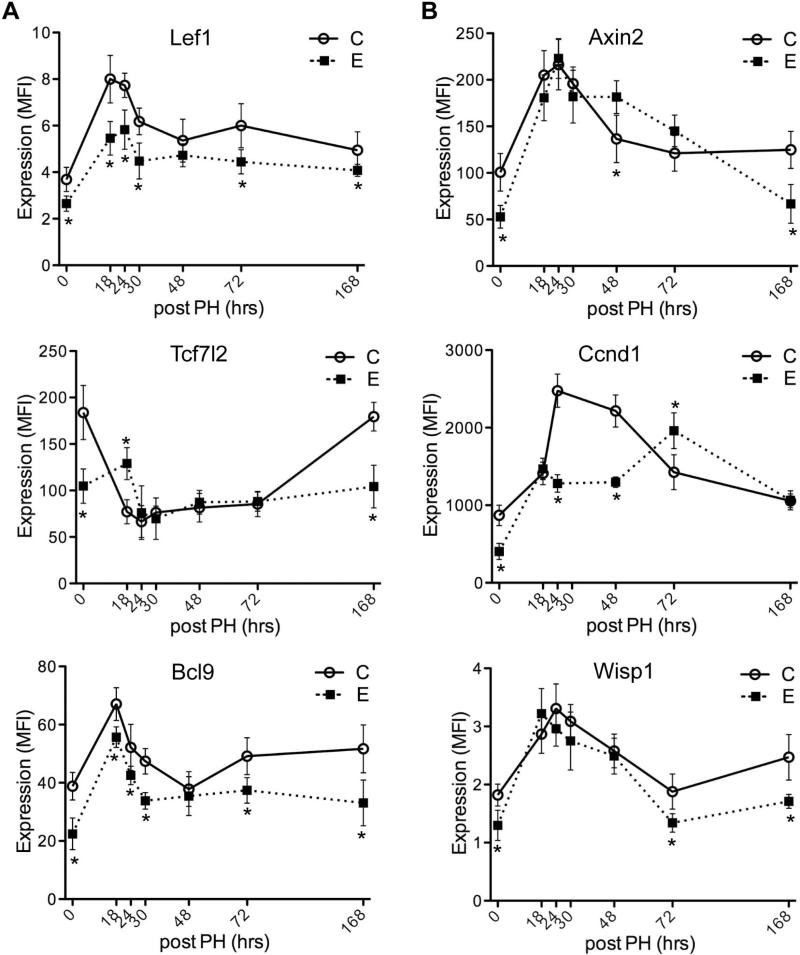
Levels of Lef1 transcription factor and Bcl9 co-factor expression were suppressed during liver regeneration by ethanol, but not Tcf7l2 (TCF-4) (Fig. 3A). Among Wnt/β-catenin-responsive target genes, Ccnd1 level was decreased at 24, 48 hours post-PH and delayed peak stimulation at 72 hour time point post-PH, whereas levels of Axin2 and Wisp1 were not affected by ethanol during liver regeneration (Fig. 3B).
Fig. 3.
Effects of chronic ethanol feeding on Wnt pathway transcription regulators and responsive target genes expression during liver regeneration following PH. Livers harvested at different time point post-PH from control (C) or ethanol (E) fed rats were obtained to measure time-course dependent shifts in (A) Lef1, Tcf7l2 and Bcl9, (B) Axin2, Ccnd1 and Wisp1 mRNA expression using the Quantigene 2.0 Multiplex Assay. Gene expression level is reflected by mean fluorescent intensity (MFI; arbitrary units). Graphs depict time-dependent shifts in net MFI ± S.D. with results normalized to housekeeping genes. Inter-group comparisons were made using two-way repeated measures ANOVA with Bonferroni's pos-hoc test, *p < 0.05 vs. control-fed rats (n = 3-5).
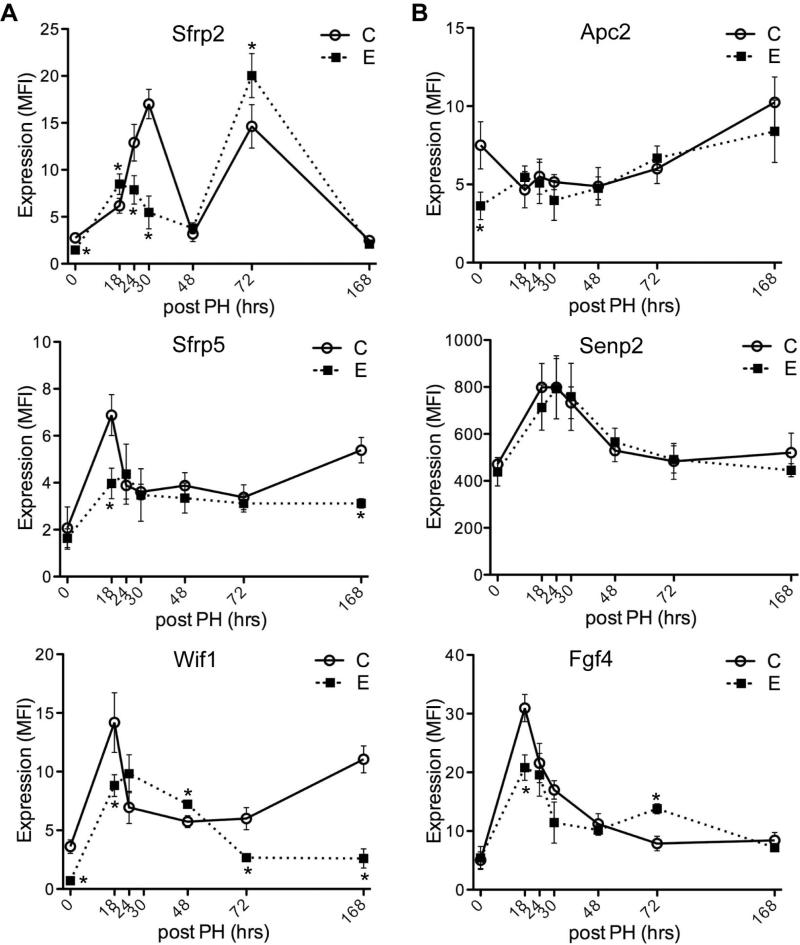
Expression of Sfrp2 exhibited biphasic peaks at 24-30 and 72 hours post-PH. Interestingly, ethanol exposure decreased Sfrp2 level at the first peak, whereas increased at the second peak (Fig. 4A). Level of Sfrp5 was downregulated during liver regeneration in ethanolfed liver. Reduced expression of Wif1 by ethanol exposure showed a delayed peak expression. In contrast, chronic ethanol feeding had no significant effects on Apc2 or Senp2 expression during liver regeneration. Ethanol reduced expression of Fgf4 at the first peak, but transiently increased at 72-hour time point post-PH (Fig. 4B).
Fig. 4.
Effects of chronic ethanol feeding on Wnt ligand binding proteins and other components expression following PH. Adult male Long Evans rats were fed with isocaloric liquid diets containing 0% (C) or 37% ethanol (E) for 8 weeks, after which they underwent 2/3 PH. Livers harvested 0, 18, 24, 30, 48, 72, or 168 hours post-PH were obtained to measure time-course dependent shifts in (A) Sfrp2, Sfrp5 and Wif1, (B) Apc2, Senp2 and Fgf4 mRNA expression using the Quantigene 2.0 Multiplex Assay. Gene expression level is reflected by mean fluorescent intensity (MFI; arbitrary units). Graphs depict time-dependent shifts in net MFI ± S.D. with results normalized to housekeeping genes. Inter-group comparisons were made using two-way repeated measures ANOVA with Bonferroni's pos-hoc test, *p < 0.05 vs. control-fed rats (n = 3-5).
PPAR-δ agonist abrogates chronic ethanol-induced suppression of Wnt-related genes during liver regeneration
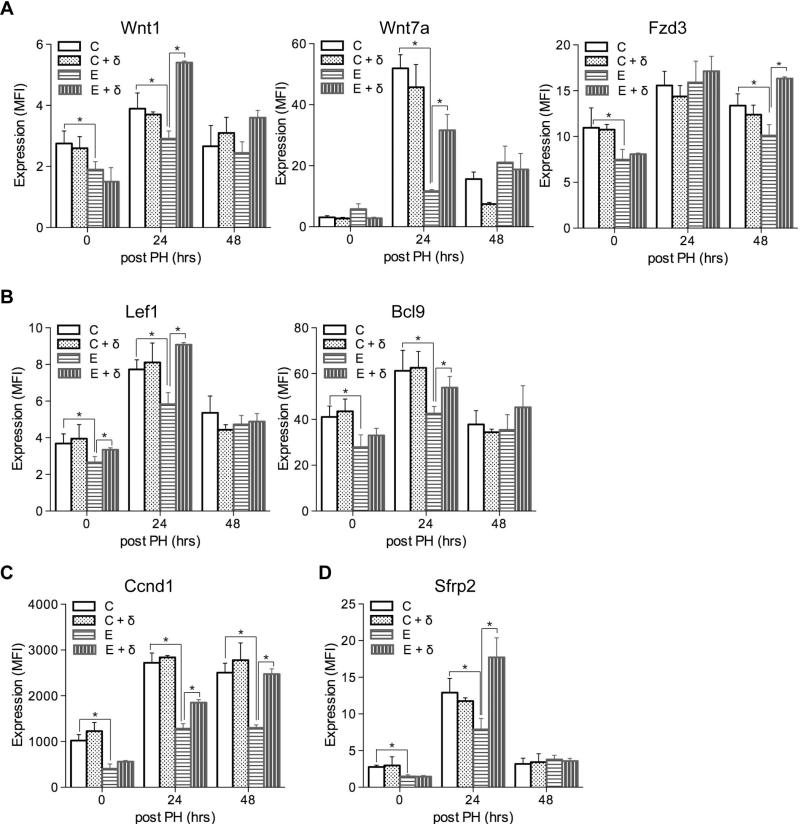
Previous report showed that PPAR-δ agonist attenuated the severity of chronic ethanol-induced liver injury and ethanol's adverse effects on liver regeneration and insulin responsiveness (Pang et al., 2009). As shown in Suppl Fig. 1E, PPAR-δ agonist treatment of ethanol-exposed livers normalized hepatic architecture during regeneration, reduced cytoplasmic vacuolation, foci of necrosis, and apoptotic bodies compared to regenerating livers in chronic ethanol fed rats (Suppl Fig. 1E). Furthermore, we observed that PPAR-δ agonist treatments significantly increased Wnt1, Wnt7a, Lef1, Bcl9, and Sfrp2 expression in ethanol-exposed livers at the 24 hours post-PH time point (Fig. 5), during peak DNA synthesis. The stimulatory effects of the PPAR-δ agonist on Wnt pathway genes were either modestly or significantly sustained through the 48 hours post-PH time point for Wnt1, Fzd3, Bcl9, and Ccnd1 (Fig. 5). In contrast, PPAR-δ agonist treatments of control rats did not significantly alter Wnt pathway gene expression relative to corresponding vehicle-treated controls.
Fig. 5.
Effects of PPAR-δ agonist treatment on hepatic Wnt pathway gene expression following PH in control and chronic ethanol-fed rats. Adult male Long Evans rats were fed with isocaloric liquid diets containing 0% or 37% ethanol for 8 weeks. During the last 3 weeks of the feeding regimen, rats were administered twice weekly i.p. injections of saline or a PPAR-δ agonist. The rats were then subjected to PH and livers were harvested 0, 24, or 48 hours later to measure Wnt pathway gene expression using the Quantigene 2.0 Multiplex Assay. Gene expression levels were indexed in arbitrary mean fluorescence intensity (MFI) units, and results were normalized to a housekeeping gene that was not modulated by PH or liver regeneration. Graphs depict results obtained for (A) Wnt1, Wnt7a, and Fzd3; (B) the Lef1 and Bcl9 Wnt transcription factors; (C) the Ccnd1 Wnt target gene; and (D) the Sfrp2 Wnt ligand binding protein. C, control + saline; C + δ, control + PPAR-δ; E, ethanol + saline; E + δ, ethanol + PPAR-δ. Inter-group comparisons were made by two-way repeated measures ANOVA tests with Bonferroni's pos-hoc test using GraphPad Prism 5. *p < 0.05.
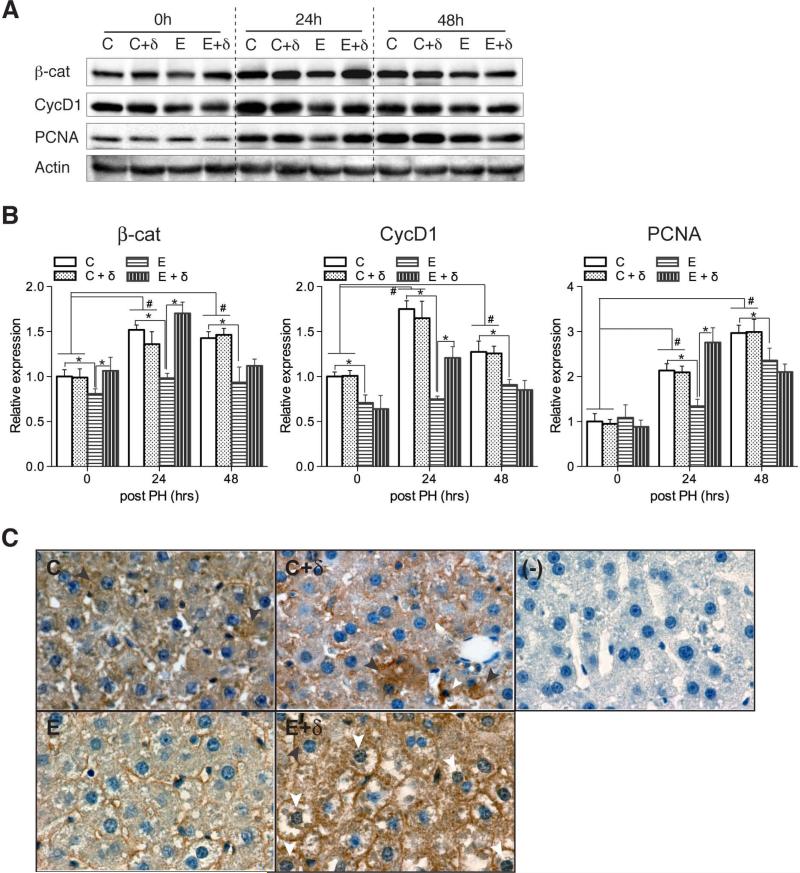
To further demonstrate effects of ethanol and PPAR-δ agonist treatments on Wnt signaling activity, we measured β-catenin, cyclin D1, and PCNA expression by Western blot analysis. Cyclin D1 is a Wnt target gene, and PCNA is expressed in the late G1 and S phases of replication (Hsu et al., 2006). Western blot analysis of control livers demonstrated increased hepatic levels of β-catenin, Cyclin D1, and PCNA at 24 and 48 hours relative to 0 hour post-PH (Figs. 6A and 6B). In addition, the responses in the vehicle and PPAR-δ agonist treated livers were similar over the time period studied in the control-fed rat. Chronic ethanol feeding reduced hepatic expression of β-catenin and cyclin D1 relative to control at the 0, 24, and 48 hours time points, and PCNA expression at the 24 hours post-PH. PPAR-δ agonist treatment of ethanol-fed rats significantly increased β-catenin expression at the 0 and 24 hours time points, and cyclin D1 and PCNA expression at the 24 hours post-PH time points relative to ethanol+vehicle treatment. The mean levels of β-catenin and PCNA measured in the ethanol+PPAR-δ agonist treated livers were comparable to or higher than in control livers, reflecting abrogation of ethanol's inhibitory effects on Wnt signaling. Correspondingly, we found that PPAR-δ agonist treatment induced nuclear localization of β-catenin in hepatocytes, especially ethanol-fed livers (Fig. 6C, E+δ). Taken together, effects of PPAR-δ agonist on chronic ethanol-induced liver injury and liver regeneration may be mediated in part by restoration of Wnt signaling.
Fig. 6.
Effects of PPAR-δ agonist treatment on hepatic Wnt pathway protein expression following PH in control and ethanol-fed rats. Adult male Long Evans rats were fed with isocaloric liquid diets containing 0% or 37% ethanol for 8 weeks. During the last 3 weeks of the feeding regimen, rats were administered twice weekly i.p. injections of saline or a PPAR-δ agonist. The rats were then subjected to PH and livers were harvested 0, 24, or 48 hours later to measure (A) β-catenin, Cyclin D1, PCNA, and Actin by Western blot analysis. (B) Graphs show relative levels of immunoreactivity (mean ± S.E.M.) normalized to control+vehicle values measured at the corresponding time point. Data were obtained by subjecting the Western blot signals to densitometric analysis (arbitrary units). Signal intensities were normalized to Actin. Inter-group comparisons were made by two-way repeated measures ANOVA tests with Bonferroni's pos-hoc test using GraphPad Prism 5. *, # p < 0.05. (C) Localization of β-catenin by immunohistochemistry. Panels depict representative β-catenin staining of livers from 24 hour post partial hepatectomy. (C) and (C + δ) had cytoplasmic expression (black arrowhead), whereas (E + δ) liver exhibited nuclear localization of β-catenin (white arrowhead).
DISCUSSION
Despite Wnt signaling is known to be important to regulate many critical organ functions, few studies to date have shown the links between alcohol and Wnt/β-catenin signaling. In particular, ethanol exposure resulted in suppression of Wnt signaling proteins in differentiating human neural stem cells (Vangipuram et al, 2012). Lauing et al suggested that the canonical Wnt pathway is a target for alcohol in bone and acute alcohol exposure decreased Wnt transcriptional activation, which may cause a significant impairment of fracture callus tissue formation (Lauing et al., 2012). However, the effect of ethanol on Wnt/β-catenin signaling in association with hepatocellular growth and function are poorly characterized yet. Here we utilized a customized Quantigene multiplex (QGP) assay to investigate an impairment of Wnt signaling caused by chronic ethanol exposure and the effect of insulin sensitizer (PPAR-δ agonist) during liver regeneration. In order to select the most appropriate Wnt-pathway related genes for QGP assay, we performed a preliminary experiment using WNT Signaling Pathway RT2 profiler PCR array containing 84 different genes related to Wnt pathway and 5 housekeeping genes (Supplemental Table 1). According to the result of the assay (Supplemental Table 2) and functional relevance of liver, 19 different Wnt-pathway related genes (Table 1) have been assessed for the present study. Fourteen out of 19 genes showed significantly reduced mRNA expression in chronic ethanol-fed rat liver, which is consistent with the preliminary experiment. On the other hand, expression levels of 4 genes (Wnt7a, Fzd6, Fgf4 and Senp2) revealed no change between control-fed and ethanol-fed group. The discrepancy of these results might be due to different techniques to quantify mRNA level. RT2 Profiler PCR Array system is based on PCR, whereas QGP assay utilizes a direct quantification of mRNA using xMAP luminex beads to avoid artifacts during RT-PCR and PCR. Therefore, the data from QGP assay may be more reliable.
We found that chronic ethanol exposure increased expression of Jun, which is one of Wnt-responsive target genes (Fig. 1D) regardless the inhibition of Wnt signaling mediated by chronic ethanol exposure. It is not unexpected since it has been reported that chronic ethanol treatment increased c-Jun mRNA expression (Park et al., 2012) and acute ethanol-induced apoptosis was mediated via c-Jun N-terminal kinase (JNK) pathway with enhanced JNK activity (Luo et al., 2013). Thus, Jun expression may be affected by other signaling rather than Wnt pathway in this experimental condition.
During liver regeneration after PH, the first peak for DNA synthesis in hepatocytes occurs at 24 hours in rat liver (Koniaris et al., 2003). Intriguingly, the expression levels of Wnt3, Fzd3, Lef1, Bcl9, Sfrp5, Wif1 and Fgf4 exhibited the peak at 18 hours rather than 24 hours post-PH in control-fed rat liver, which may represent that the activation of Wnt signaling is necessary prior to hepatocyte proliferation and DNA synthesis.
Both Wnt1 and Wnt3 ligands activate the Wnt/β-catenin pathways (Kim et al., 2008), however, their expression patterns during liver regeneration exhibited differentially (Fig. 2A). Consistent with a previous study demonstrated that Wnt1 protein is critical for liver regeneration after 70% PH, Wnt1 mRNA expression was increased during liver regeneration at the first peak of hepatocyte proliferation (18 through 30 hours post-PH) and suppressed by ethanol treatment. However, there was no information of other Wnt ligands expression during liver regeneration so far. Here, we found that Wnt3 expression was increased transiently at 18 hours post-PH in control-fed rat liver. Moreover, chronic ethanol exposure resulted in moderate or significantly enhanced Wnt3 expression during liver regeneration (Fig. 2A). On the other hand, increased level of Wnt7a, which activates non-canonical Wnt signaling, was sustained longer until 48 hours after PH in control-fed, but it was delayed peak stimulation in chronic ethanol-fed rat liver. Our finding implies that Wnt1, Wnt3 and Wnt7a ligands may have distinct role on liver regeneration in the context of different cell types. Considering that the first peak of DNA synthesis in the parenchymal cells (hepatocytes and bile duct epithelial cells) occurs at 24 hours post-PH, Wnt3 and Wnt7a may have major role for parenchymal cells proliferation. On the other hand, non-parenchymal cells display a first peak of proliferation at 36 hours post-PH (Fabrikant, 1968), Wnt1 may have important role on non-parenchymal cells as well as parenchymal cells proliferation. Expression level of Wnt3 was upregulated at the first peak of DNA synthesis in ethanol-fed compared to control-fed groups post-PH, despite chronic ethanol feeding induced a decreased Wnt3 expression at 0 as well as 1 week after PH. Thus, Wnt3 signal may have different role on liver cells proliferation, i.e., homeostatic liver function vs. injured liver repair post-PH. Porcn showed two distinct expression profiles during liver regeneration. As shown in Fig. 2A, the peak expression appeared at 30 hours post-PH in control feeding which is similar to that of Wnt1, while it showed at 18 hours post-PH in ethanol-fed which is similar to that of Wnt3. Considering Porcn is specific and dedicated to Wnt post-translational acylation (palmitoylation), which is required for subsequent Wnt secretion, the activity of Wnt1 and Wnt3 proteins would be affected mainly by expression level of Porcn.
TCF-4 (Tcf7l2) expression in both control and ethanol-fed rats were downregulated during the time of peak cell proliferation post-PH (Fig. 3A, middle panel). The probe set region (535-932, Table 1) of rat TCF-4 includes a predicted exon 4 (854-923) of TCF-4. Recent our studies demonstrated that exon 4 confers a repressive property to human TCF-4, characterized by reduced TCF transcriptional activity, cell-proliferation, cell cycle progression, and expression of Wnt-target genes (Tomimaru et al., 2013b). In this context, a down-regulation of this repressive form of TCF-4 may represent an increased Wnt signaling to orchestrate the regenerative response post-PH. More important, level of TCF-4 in control was lower than that in ethanol-fed rat at 18 hours post-PH and exhibited a reverse relation with expression of Wnt-related genes including Fzd3, Lef1, Bcl9, Fgf4, Wif1, and Sfrp5. This concern, however, is not applied at time 0 and 1 week post-PH since the level of TCF-4 is upregulated in control compared to ethanol-fed rats. The liver is a largely quiescent organ in terms of cell proliferation, with only 0.0012% to 0.01% of hepatocytes undergoing mitosis at any given time, despite its large metabolic load. This low cell turnover in healthy liver tissue can be altered by toxic liver injury or surgical resection such as 2/3 PH in an adult rat. The remnant rat liver post-PH undergoes almost complete restoration of the lost mass and function by about 1 week's time (Diehl and Rai, 1996; Michalopoulos et al, 1997). Accordingly, our observations suggest that TCF-4 transcription factor protein may activate different target genes mediated by different TCF-4 isoforms depending on normal hepatocyte turnover vs. massive hepatocyte proliferation during regeneration post-PH. Of note, TCF-4B isoform was dominantly expressed in human normal liver while other isoforms were found in HCC tissue (Tsedensodnom et al., 2011). It would be great of interest to define specific TCF-4 isoform-responsive target genes at different environment in the future.
Previous study showed that PPAR-δ agonist treatments significantly increased DNA synthesis and nearly normalized the liver histology in ethanol-fed rats (Pang et al., 2009). Given these reports, we hypothesized that PPAR-δ treatment might, in addition to improving insulin sensitivity, restore an impaired Wnt signaling caused by chronic ethanol exposure. Indeed, treatment with PPAR-δ agonist improves the expression of Wnt-related genes (Wnt1, Wnt7a, Fzd3, Lef1, Bcl9, Ccnd1, and Sfrp2) over a PH time course and also restored the expression of cyclinD1 and PCNA at 24 hours post-PH in the ethanol group. On the other hand, the expression levels of Wnt3, Porcn, Sfrp5, and Fgf4 increased with PPAR-δ agonist treatment in both control and ethanol-fed groups (data not known), suggesting that other pathways other than Wnt/β-catenin signaling may have mediated this response. PPAR-δ agonist has been suggested to be involved in neural stem cell and progenitor cell proliferation, and protection against apoptosis (Glazer et al., 2008). Thus, it would be interested to identify the effect of ethanol and PPAR-δ agonist in the various liver cell types, considering differential expression profile of Wnt pathway components.
Liver/body weight ratio and BrdU positivity can be often used as parameters for liver proliferation. Previously we showed that PPAR-δ agonist treatments partially reversed ethanol's inhibitory effects on DNA synthesis as demonstrated by the 4- to 5-fold increases in BrdU immunohistochemical staining relative to vehicle-treatment at 24 hours after partial hepatectomy (Pang et al., 2009). While PPAR-δ agonist treatment increased the liver/body weight ratios in ethanol fed group compared to control (Suppl Fig. 2 and Suppl Table 3). The elevated liver/body ratio may be reflected liver lipid levels (triglyceride, glycerol, and cholesterol) which can be observed in alcohol-fed mice.
As we showed in Fig 5, PPAR-δ agonist induced levels of Wnt1 and 7a in the ethanol fed livers, we explored whether PAPR-δ agonist treatment modulated expression levels of Wnt1 and Wnt7a through transcriptional regulation in in vitro system. Human hepatocytes were treated with PPAR-δ agonist for 8 hours to preclude its 2nd and indirect effects, since some transcriptional factor proteins may be activated after 8 hours of treatment. There was no significant change in the expression levels of Wnt1 and Wnt7a (Suppl Fig 3A), but we found that PPAR-δ agonist reduced phosphorylation of β-catenin (Suppl Fig. 3B). Thus, the effect of PAPR-δ agonist on Wnt1 and 7a expression may not be a direct transcriptional regulation. Since regulation mechanism of Wnt ligands expression is not defined yet, it would be great interest to study in the future. Nevertheless, the reduced level of phospho-β-catenin by PAPR-δ agonist is consistent with in vivo data shown in Fig. 6. Furthermore, PPAR-δ agonist treatment could significantly increase human hepatocyte growth (Suppl Fig. 4), implying that PPAR-δ agonist promoted hepatocyte proliferation through β-catenin signaling in vitro as well as in vivo.
Our observations have enhanced knowledge about the long-term adverse effects of ethanol on hepatocellular growth and regeneration. In particular, chronic high-dose ethanol exposures inhibit Wnt signaling, which likely contributes to the impairments in liver regeneration. Although hepatic insulin/IGF resistance is mainly responsible for ethanol-mediated impairments in liver regeneration and repair following injury, it is widely accepted that a larger network of inter-related signaling pathways involves in hepatocellular growth, repair and regeneration, and restoration of liver function. Therefore, novel functions of Wnt signaling on chronic ethanol-induced liver injury via crosstalk with insulin/IGF signaling pathway could develop new therapeutic option to the treatment of chronic ALD. Future studies will determine the degree to which targeted restoration of Wnt signaling is sufficient to improve liver regeneration and remodeling in the context of chronic ethanol exposure.
Supplementary Material
Acknowledgments
All sources of support: This work was supported by AA020587 from the National Institute of Health
Footnotes
Conflict of Interest: No
Alcoholism: Clinical & Experimental Research
REFERENCES
- Addolorato G, Mirijello A, Leggio L, Ferrulli A, Landolfi R. Management of alcohol dependence in patients with liver disease. CNS Drugs. 2013;27:287–299. doi: 10.1007/s40263-013-0043-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte U, Thompson MD, Cui S, Liu B, Cieply B, Monga SP. Wnt/beta-catenin signaling mediates oval cell response in rodents. Hepatology. 2008;47:288–295. doi: 10.1002/hep.21973. [DOI] [PubMed] [Google Scholar]
- Apte U, Singh S, Zeng G, Cieply B, Virji MA, Wu T, Mong SP. Beta-catenin activation promotes liver regeneration after acetaminophen-induced injury. Am J Pathol. 2009;175:1056–1065. doi: 10.2353/ajpath.2009.080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Mohr L, Wands JR, de la Monte SM. Ethanol inhibition of insulin signaling in hepatocellular carcinoma cells. Alcohol Clin Exp Res. 1998;22:2093–2101. [PubMed] [Google Scholar]
- Berger J, Moller DE. The mechanisms of action of PPARs. Ann Rev Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- Berschneider B, Konigshoff M. Wnt1 inducible signaling pathway protein 1 (WISP1): a novel mediator linking development and disease. Int J Biochem Cell Biol. 2011;43:306–309. doi: 10.1016/j.biocel.2010.11.013. [DOI] [PubMed] [Google Scholar]
- Biechele S, Cox BJ, Rossant J. Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos. Dev Biol. 2011;355:275–285. doi: 10.1016/j.ydbio.2011.04.029. [DOI] [PubMed] [Google Scholar]
- Brembeck FH, Wiese M, Zatula N, Grigoryan T, Dai Y, Fritzmann J, Birchmeier W. BCL9-2 promotes early stages of intestinal tumor progression. Gastroenterology. 2011;141:1359–70. 1370, e1–3. doi: 10.1053/j.gastro.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Chang F, Jaber LA, Berlie HD, O'Connell MB. Evolution of peroxisome proliferator-activated receptor agonists. Ann Pharmacother. 2007;41:973–983. doi: 10.1345/aph.1K013. [DOI] [PubMed] [Google Scholar]
- Chiang YA, Jin T. P21-activated protein kinases and their emerging roles in glucose homeostasis. Am J Physiol Endocrinol Metab In press. 2013 doi: 10.1152/ajpendo.00506.2013. doi:10.1152. [DOI] [PubMed] [Google Scholar]
- Diehl AM, Rai RM. Liver regeneration 3: regulation of signal transduction during liver regeneration. FASEB J. 1996;10:215–227. doi: 10.1096/fasebj.10.2.8641555. [DOI] [PubMed] [Google Scholar]
- Fabrikant JI. The kinetics of cellular proliferation in regenerating liver. J Cell Biol. 1968;36:551–565. doi: 10.1083/jcb.36.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer RI, Yuan H, Xie Z, Yin Y. PPARgamma and PPARdelta as Modulators of Neoplasia and Cell Fate. PPAR Res. 2008;2008:247379. doi: 10.1155/2008/247379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyamfi MA, Wan YJ. Pathogenesis of alcoholic liver disease: the role of nuclear receptors. Exp Biol Med. 2010;235:547–560. doi: 10.1258/ebm.2009.009249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, de la Monte S, Wands JR. Acute ethanol exposure inhibits insulin signaling in the liver. Hepatology. 2007;46:1791–1800. doi: 10.1002/hep.21904. [DOI] [PubMed] [Google Scholar]
- Hsu MK, Qiao L, Ho V, Zhang BH, Zhang H, Teoh N, Dent P, Farrel GC. Ethanol reduces p38 kinase activation and cyclin D1 protein expression after partial hepatectomy in rats. J Hepatol. 2006;44:375–382. doi: 10.1016/j.jhep.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5:1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- Kaur P, Mani S, Cros MP, Scoazec JY, Chemin I, Hainaut P, Herceg Z. Epigenetic silencing of sFRP1 activates the canonical Wnt pathway and contributes to increased cell growth and proliferation in hepatocellular carcinoma. Tumour Biol. 2012;33:325–336. doi: 10.1007/s13277-012-0331-5. [DOI] [PubMed] [Google Scholar]
- Kim M, Lee HC, Tsedensodnom O, Hartley R, Lim Y-S, Yu E, Merle P, Wands JR. Functional interaction between Wnt3 an Frizzled-7 leads to activation of the Wnt/β-catenin signaling pathway in hepatocellular carcinoma cells. J Hepatol. 2008;48:780–791. doi: 10.1016/j.jhep.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-H, Hong S-H, Lee M-K. Insulin receptor-overexpressing b-cells ameliorate hyperglycemia in diabetic rats through Wnt signaling activation. PLoS ONE. 2013;8:e67802. doi: 10.1371/journal.pone.0067802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Waha A, Hartmann W, Hrychyk A, Schuller U, Wharton KA, Jr, Fuchs SY, von Schweinitz D, Pietsch T. Elevated expression of Wnt antagonists is a common event in hepatoblastomas. Clin Can Res. 2005;11:4295–4304. doi: 10.1158/1078-0432.CCR-04-1162. [DOI] [PubMed] [Google Scholar]
- Koniaris LG, Mckillop IH, Schwartz SI, Zimmers TA. Liver regeneration. J Am Coll Surg. 2003;197:634–659. doi: 10.1016/S1072-7515(03)00374-0. [DOI] [PubMed] [Google Scholar]
- Kress E, Rezza A, Nadjar J, Samarut J, Plateroti M. The frizzled-related sFRP2 gene is a target of thyroid hormone receptor alpha1 and activates beta-catenin signaling in mouse intestine. J Biol Chem. 2009;284:1234–1241. doi: 10.1074/jbc.M806548200. [DOI] [PubMed] [Google Scholar]
- Kuncewitch M, Yang WL, Molmenti E, Nicastro J, Coppa GF, Wang P. Wnt agonist attenuates liver injury and improves survival after hepatic ischemia/reperfusion. Shock. 2013;39:3–10. doi: 10.1097/SHK.0b013e3182764fe8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauing KL, Roper PM, Nauer RK, Callaci JJ. Acute alcohol exposure impairs fracture healing and deregulates β-catenin signaling in the facture callus. Alcol Clin Exp Res. 2012;36(12):2095–2103. doi: 10.1111/j.1530-0277.2012.01830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Lee MH, Park JH, Kang SH, Yoo HM, Ka SH, Oh YM, Jeon YJ, Chung CH. SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 2012;31:4441–4452. doi: 10.1038/emboj.2012.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longato L, Ripp K, Setshedi M, Dostalek M, Akhlaghi F, Branda M, Wands JR, de la Monte SM. Insulin resistance, ceramide accumulaton, and endoplasmic reticulum stress in human chronic alcohol-related liver disease. Oxid Med Cell Longev. 2012;2012:479348. doi: 10.1155/2012/479348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X-J, Liu B, Dai Z, Li T-B, Li N-S, Zhang X-J, Yang Z-C, Li Y-J, Peng J. Expression of apoptosis-associated microRNAs in ethanol-induced acute gastric mucosal injury via JNK pathway. Alcohol. 2013;47:481–493. doi: 10.1016/j.alcohol.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Mii Y, Taira M. Secreted Wnt “inhibitors” are not just inhibitors: regulation of extracellular Wnt by secreted Frizzled-related proteins. Dev Growth Differ. 2011;53:911–923. doi: 10.1111/j.1440-169X.2011.01299.x. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Yeon JE, Tong M, Longato L, Chaudhry R, Pang MY, Duan K, Wands JR. Insulin resistance in experimental alcohol-induced liver disease. J Gastroenterol Hepatol. 2008;23:e477–486. doi: 10.1111/j.1440-1746.2008.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte S, Derdak Z, Wands JR. Alcohol, insulin resistance and the liver-brain axis. J Gastroenterol Hepatol. 2012;27(Suppl. 2):33–41. doi: 10.1111/j.1440-1746.2011.07023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi N, Higuchi A, Ohashi K, Oshima Y, Gokce N, Shibata R, Akasaki Y, Shimono A, Walsh K. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science. 2010;329:454–457. doi: 10.1126/science.1188280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M, de la Monte SM, Longato L, Tong M, He J, Chaudhry R, Duan K, Ouh J, Wands JR. PPARdelta agonist attenuates alcohol induced hepatic insulin resistance and improves liver injury and repair. J Hepatol. 2009;50:1192–1201. doi: 10.1016/j.jhep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P-H, Lim RW, Shukla SD. Gene-selective Histone H3 acetylation in the absence of increase in global histon acetylation in liver of rats chronically fed alcohol. Alcol Alcol. 2012;47:233–239. doi: 10.1093/alcalc/ags004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Diaz K, Chen TW, Wang X, Thorne CA, Wallace HA, Page-McCaw A, Lee E. The way Wnt works: components and mechanism. Growth Factors. 2013;31:1–31. doi: 10.3109/08977194.2012.752737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Wands JR. Ethanol impairs insulin receptor substrate-1 mediated signal transduction during rat liver regeneration. Biochem Biophys Res Commun. 1994;199:403–409. doi: 10.1006/bbrc.1994.1243. [DOI] [PubMed] [Google Scholar]
- Schwartz JM, Reinus JF. Prevalence and natural history of alcoholic liver disease. Clin Liver Dis. 2012;16:659–666. doi: 10.1016/j.cld.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Sekine S, Gutierrez PJ, Lan BY, Feng S, Hebrok M. Liver-specific loss of beta-catenin results in delayed hepatocyte proliferation after partial hepatectomy. Hepatology. 2007;45:361–368. doi: 10.1002/hep.21523. [DOI] [PubMed] [Google Scholar]
- Sodhi D, Micsenyi A, Bowen WC, Monga DK, Talavera JC, Monga SP. Morpholino oligonucleotide-triggered beta-catenin knockdown compromises normal liver regeneration. J Hepatol. 2005;43:132–141. doi: 10.1016/j.jhep.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Stamos JL, Weis WI. The b-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M-Y, Mu X-Y, Liu B, Wang Y, Bao E-D, Qiu J-X, Fan Y. SUMO-specific protease 2 suppresses cell migration and invasion through inhibiting the expression of MMP12 in bladder cancer cells. Cell Physiol Biohem. 2013;32:542–548. doi: 10.1159/000354458. [DOI] [PubMed] [Google Scholar]
- Tomimaru Y, Koga H, Yano H, de la Monte S, Wands JR, Kim M. Upregulation of T-cell factor-4 isoform-responsive target genes in hepatocellular carcinoma. Liver Int. 2013a;33:1100–1112. doi: 10.1111/liv.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimaru Y, Xu CQ, Nambotin SB, Yan T, Wands JR, Kim M. Loss of exon 4 in a human T-cell factor-4 isoform promotes hepatic tumourigenicity. Liver Int. 2013b;33:1536–1548. doi: 10.1111/liv.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Ann Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Tsedensodnom O, Koga H, Rosenberg SA, Nambotin SB, Carroll JJ, Wands JR, Kim M. Identification of T-cell factor-4 isoforms that contribute to the malignant phenotype of hepatcellular carcinoma cells. Exp Cell Res. 2011;317:920–931. doi: 10.1016/j.yexcr.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangipuram SD, Lyman WD. Ethanol affects differentiation-related pathways and suppresses Wnt signaling protein expression in human neural stem cells. Alcol Clin Exp Res. 2012;36(5):788–797. doi: 10.1111/j.1530-0277.2011.01682.x. [DOI] [PubMed] [Google Scholar]
- Xu D, Yang F, Yuan J, Zhang L, Bi H, Zhou C, Liu F, Wang F, Sun S. Long noncoding RNAs Associated With Liver Regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/β-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L, Sakamoto D, Caothien RH, Fuller JH, Reinhard C, Garcia PD, Randazzo FM, Escobedo J, Fantl WJ, Williams LT. Elevated expression of axin2 and hnkd mRNA promotes evidence that Wnt/beta-catenin signaling is activated in human colon tumors. Proc Natl Acad Sci USA. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yessoufou A, Wahli W. Multifaceted roles of peroxisome proliferator-activated receptors (PPARs) at the cellular and whole organism levels. Swiss Med Wkly. 2010;140:W13071. doi: 10.4414/smw.2010.13071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.