Abstract
Secreted proteins are the frontline between the host and pathogen. In mammalian hosts, secreted proteins enable invasive infection and can modulate the host immune response. Cryptococcosis, caused by pathogenic Cryptococcus species, begins when inhaled infectious propagules establish to produce pulmonary infection, which, if not resolved, can disseminate to the central nervous system to cause meningoencephalitis. Strains of Cryptococcus species differ in their capacity to cause disease, and the mechanisms underlying this are not well understood. To investigate the role of secreted proteins in disease, we determined the secretome for three genome strains of Cryptococcus species, including a hypovirulent and a hypervirulent strain of C. gattii and a virulent strain of C. neoformans. Sixty-seven unique proteins were identified, with different numbers and types of proteins secreted by each strain. The secretomes of the virulent strains were largely limited to proteolytic and hydrolytic enzymes, while the hypovirulent strain had a diverse secretome, including non-conventionally secreted canonical cytosolic and immunogenic proteins that have been implicated in virulence. The hypovirulent strain cannot establish pulmonary infection in a mouse model, but strains of this genotype have caused human meningitis. To directly test brain infection, we used intracranial inoculation and found that the hypovirulent strain was substantially more invasive than its hypervirulent counterpart. We suggest that immunogenic proteins secreted by this strain invoke a host response that limits pulmonary infection but that there can be invasive growth and damage if infection reaches the brain. Given their known role in virulence, it is possible that non-conventionally secreted proteins mediate this process.
INTRODUCTION
Protein secretion is an essential process for all cells. Secretion has roles in various aspects of cell physiology and lifestyle, including nutrient acquisition, cell wall remodeling, signaling, quorum sensing, and defense against other organisms (1). For pathogenic organisms, the secretion of specific proteins can be key to disease progression, allowing the pathogen both to invade and obtain nutrients and to directly modulate the host organism's immune response.
The ability to secrete proteins is of particular importance to fungal organisms. Fungi use exodigestion and absorptive nutrition to acquire nutrients and have evolved a complex suite of secreted proteins to degrade the diverse biopolymers encountered in the host or abiotic environment. Comparisons of characterized fungal secretomes suggest that their basal secretome consists of a core set of degradation enzymes. These include polysaccharide-active enzymes, including glycoside hydrolases, carbohydrate esterases, and polysaccharide lyases, which can degrade the major components of plant cell walls such as cellulose and pectin (2). Other degradative enzymes include proteases, for example, secreted aspartyl proteases (SAPs), which have been associated with pathogenicity in the human commensal yeast Candida albicans (3, 4).
Many proteins destined for secretion are tagged with a signal peptide that can be used to predict secretion via “classical” pathways (5). However, extracellular proteins that lack any known signal peptide have frequently been identified and are generally referred to as non-classically or unconventionally secreted proteins (6). A number of unconventional protein secretion mechanisms are known, and these include both vesicle-dependent and vesicle-independent pathways (7). Many proteins secreted via nonclassical pathways have been described as “moonlighting” proteins, which, in contrast to those proteins characterized as having a single primary core function, are multifunctional, often with secondary roles that are unrelated to their core function (8–11).
Species of Cryptococcus are environmental fungi capable of causing disease in immunocompromised and immunocompetent individuals. The two predominant pathogenic species, Cryptococcus neoformans and Cryptococcus gattii, can cause cryptococcosis in animals and humans, with outcomes ranging from an asymptomatic state to severe, fatal meningitis (12). Notable differences between C. gattii and C. neoformans include the preferred environmental niche, basidiospore morphology, drug susceptibility, epidemiology, the clinical manifestations of associated disease, and host susceptibility (13). Additional differences have been observed within Cryptococcus species. In C. gattii, a hypervirulent subgenotype designated VGIIa caused a recent significant outbreak of cryptococcosis on Vancouver Island in British Columbia, Canada, and in the Pacific Northwest of the United States. C. gattii VGIIa occurs sympatrically with subgenotype VGIIb, which is globally distributed and is considered to be hypovirulent (14). These differences in virulence and epidemiology between Cryptococcus species and subgenotypes provide an opportunity for understanding pathogenicity and disease progression by what are otherwise very genetically similar fungal organisms.
The current study was designed to analyze the secretomes of three Cryptococcus strains that differ in virulence. Previous analyses of the protein cohort secreted by Cryptococcus spp. have used acapsular mutant strains or nutrient-replete culture media to suppress capsule formation (15–17). However, this may induce a secretion profile different from that seen under the nutrient-limited conditions encountered in the host (6, 15). In the current study, we set out to analyze secretion by encapsulated, wild-type Cryptococcus strains under conditions designed to be as similar as possible to those encountered within the host. As low-nutrient conditions provoke excess production and shedding of extracellular polysaccharide capsule (18), we developed a novel method of capturing secreted proteins using ProteoMiner (Bio-Rad) beads. We report here that different strains of Cryptococcus secrete distinctly different sets of proteins, with the C. gattii VGIIb isolate secreting many more proteins predicted to be non-classically secreted and immunogenic than the hypervirulent C. gattii VGIIa and C. neoformans strains. Infection of mice by intracerebral inoculation revealed that the VGIIb isolate produced rapidly fatal meningitis with substantially more growth than the C. gattii VGIIa strain. Given their role in infection and immunity, it is possible that non-classically secreted and nonimmunogenic proteins play a role in modulating cryptococcosis, on the one hand enhancing immune clearance from the lungs but on the other promoting disease following passage across the blood-brain barrier (BBB).
MATERIALS AND METHODS
Strains and culture conditions.
Cryptococcus strains included C. gattii R265 (genotype VGIIa) and C. gattii R272 (genotype VGIIb) and C. neoformans KN99α (secretome analysis) and H99 (congenic with KN99α; infection model, genotype VNI). Strains were retrieved from storage at −80οC and streaked onto yeast extract-peptone-dextrose (YPD) plates, which were incubated at 30°C for 2 days. Single yeast colonies were inoculated into a starter culture containing RPMI minimal media (Invitrogen), supplemented with 1× minimal essential medium (MEM) complete amino acid mix (Sigma-Aldrich), 2.5 M MOPS (morpholinepropanesulfonic acid) buffer (Sigma-Aldrich) (pH 7.2), 0.2% glucose, and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Life Technologies). Cultures were grown for 2 days at 37°C in an incubator with orbital shaking at 120 rpm. Cells were counted using an improved Bright-Line hemocytometer (Neubauer). Approximately 108 cells from the starter culture were inoculated into each of two 1-liter flasks containing the same medium as that described above. Cells were grown for a further 7 days at 37°C with shaking at 120 rpm. On day 7, all cultures were assayed for cell viability. A 10-ml aliquot was collected from each culture, and the cells were pelleted, resuspended in 500 μl phosphate-buffered saline, and stained with trypan blue, which is excluded from live but not dead cells. The level of viability was assessed by microscopic examination.
Isolation of secreted protein.
For each Cryptococcus strain, the two 1-liter cultures produced as described above were pooled and centrifuged at 4,500 × g for 10 min to pellet yeast cells, and the supernatant was decanted and saved. For KN99α, the culture was centrifuged twice due to the presence of residual cells in the medium after the first spin. Secreted proteins were then isolated from the cell-free supernatant using ProteoMiner beads (Bio-Rad Laboratories). The beads were prepared according to the instructions of the manufacturers before being transferred to a 9-cm-long Poly-Prep gravity flow chromatography column (Bio-Rad Laboratories) connected to a system of sterile silicon tubing. This tubing started and terminated in a flask containing the culture supernatant, which was circulated through the tubing via the action of a peristaltic pump (LongerPump; BT100-2J). This apparatus allowed the supernatant to be continually recirculated across the ProteoMiner beads at a flow rate of 12 ml/min for 2 days. The entire process was carried out at 4°C to minimize proteolysis. Captured proteins were eluted by transferring the beads back into the original column and adding 100 μl of 2× sample buffer (20% glycerol, 200 mM Tris-HCl [pH 6.8], 4% SDS, 5% beta-mercaptoethanol, bromophenol blue) before the column was placed at 100°C for 10 min. This was repeated 4 times to ensure removal of all protein from the beads. A 10-μl aliquot of the eluted protein sample was electrophoresed in a 4%-to-12% XT Bis-Tris Criterion gel (Bio-Rad Laboratories) and quantified by comparison with a dilution series of bovine serum albumin standards. The remaining protein samples were stored at −20°C.
Preparation of protein for liquid chromatography-tandem mass spectrometry (LC-MS/MS).
The entire remaining protein sample was reduced to approximately 45 μl by vacuum centrifugation, boiled for 10 min, and electrophoresed in a 4%-to-12% Bis-Tris Criterion gel (Bio-Rad Laboratories) at 160 V for 10 min. Gels were then fixed for 30 min in 40% methanol plus 10% acetic acid and stained with Bio-Safe Coomassie stain (Bio-Rad Laboratories). Lanes containing proteins were excised from the stained gels, finely diced into approximately 1-mm-by-1-mm pieces, and then subjected to trypsin in-gel digestion for mass spectrometry, as previously described (19).
LC-MS/MS analysis.
Peptides were separated using liquid chromatography (LC) and analyzed via tandem mass spectrometry (MS/MS). Briefly, an Eksigent AS-1 autosampler was used to load the peptide samples onto a Tempo nanoLC system (Eksigent, USA) at a flow rate of 20 μl/min with MS loading solvent (2% acetonitrile–0.2% trifluoroacetic acid) onto a C8 trap column (CapTrap; Michrom Biosciences, USA). After washing for 3 min was performed, the peptides were washed off the C8 trap at a flow rate of 300 nl/min with MS solvent A (2% acetonitrile–0.2% formic acid) and collected onto a PicoFrit column (75 μm by 150 mm) packed with Magic C18AQ resin (Michrom Biosciences). The LC-separated peptides were eluted from the column and into the source of a QStar Elite hybrid quadrupole time of flight mass spectrometer (Applied Biosystems/MDS Sciex) using the following program: 5% to 50% MS solvent B (98% acetonitrile–0.2% formic acid) over 60 min, 50% to 80% MS solvent B over 5 min, 80% MS solvent B for 2 min, and, finally, 80% to 5% MS solvent B for 3 min. The eluted peptides were ionized from the PicoFrit column at 2,300 V. An intelligent data acquisition (IDA) experiment was performed, with a mass range of 375 to 1,500 Da continuously scanned for peptides of charge state 2+ to 5+ and with an intensity of more than 30 counts/s. Selected peptides were fragmented, and the product ion fragment masses were measured over a mass range of 100 to 1,500 Da. The mass of the precursor peptide was then excluded for 1 min.
Generation of LC-MS/MS data.
The MS/MS data files produced by a QStar Elite hybrid quadrupole time of flight mass spectrometer were searched using Mascot Daemon (version 2.2.2, provided by the Walter and Eliza Hall Institute) (20) and the LudwigNR database (comprised of the UniProt, plasmoDB, and Ensembl databases). The searched parameters were as follows: variable modifications of propionamide, oxidized methionine, and deamidated asparagine and a semi-trypsin digest, with a maximum of three missed cleavages, namely, a peptide mass tolerance of 100 ppm, a MS/MS mass tolerance of 0.2 Da, and charge states of 2+, 3+, and 4+. The results of the search were then filtered by including only protein hits with at least one unique peptide and a Mascot score of >60. Single peptide identifications were further validated by manual inspection of the MS/MS spectra for the peptide to ensure that the b-ion and y-ion series were sufficiently extensive for an accurate identification, with the y1 ion identified as either arginine or lysine, as the samples were trypsin digested. The list was filtered again by keeping only those proteins that were identified in three or more biological replicates and were identified as predicted proteins of Cryptococcus spp. via the LudwigNR database.
Bioinformatic analysis of identified proteins.
Biological functions for protein identifications were assigned using the UniProtKB and InterPro databases (21, 22). UniProtKB uses the International Nucleotide Sequence Database Collaboration, which encompasses all sequences submitted to the EMBL-Bank/GenBank/DDBJ databases. These resources were used to assign putative functions to uncharacterized proteins. All identified proteins were analyzed for secretory signals using the FunSecKB2 database (23). This resource combines multiple search algorithms, including SignalP, WolfPsort, Phobius, and FragAnchor, to interrogate all fungal protein data in the NCBIRefSeq database to predict secreted fungal proteins. The online tool SecretomeP was further used to predict protein secretion by both classical and nonclassical mechanisms. This program produces ab initio predictions, i.e., those generated without reference to previous secretion predictions, of nonclassical protein secretion. It does this by compiling information from feature prediction servers on posttranslational and localization aspects of a protein and using this information to produce a secretion prediction. A protein with a SecretomeP score of >0.5 is predicted to be secreted via a nonclassical pathway (24).
Intracranial cryptococcal meningitis infection model.
This study was conducted in conformity with the Australian Code for the Care and Use of Animals for Scientific Purposes and with the review and approval of the University of Sydney Animal Ethics Committee (project number L02/4-2013/1/5887). Cryptococcus infection in the brain was studied in mice using an intracranial infection model (25). Six-week-old male C57BL/6 mice (n = 3) and SCID mice (n = 3) (Animal Resources Centre, Western Australia) were infected with each Cryptococcus strain. Animals were anesthetized, and 1,000 yeast cells were injected directly into the brain in a volume of 0.02 ml (5 × 104 cells/ml inoculum) using a 26-gauge needle inserted through the midline of the dorsal surface of the skull. Mice were monitored daily for weight loss and symptoms of illness, including excessive lethargy and ruffled fur. Once weight loss exceeded 15% of the weight of the animal at the time of infection, the mouse was euthanized. All surviving animals were euthanized at 14 days postinfection. At that time, the brain was harvested and immediately fixed in 4% paraformaldehyde. Statistical differences between survival curves were assessed by the Mann-Whitney U test.
Histochemistry and immunohistochemistry of cryptococcal brain infection.
Longitudinal sections were prepared from fixed brain tissue for each of the infecting cryptococcal strains and mouse strains. Hematoxylin and eosin stains were performed and counterstained with alcian blue to visualize cryptococcal cells. Alcian blue stains mucin and reacts strongly with cryptococcal capsular material. Immunohistochemistry was performed on unstained slides using an anti-glial fibrillary acidic protein (anti-GFAP) antibody (DAKOCytomation) for visualization of astrocytes and anti-Iba1 (Wako) to visualize microglial cells. Briefly, paraformaldehyde-fixed tissue sections were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol. Prior to antibody hybridization, for antigen retrieval, slides were treated either with Tris-HCl buffer (25 mM; pH 8.5) containing 0.05% SDS (wt/vol) and EDTA (1 mM) or with sodium citrate buffer (10 mM; pH 8.5) for 40 min at 97°C. Endogenous peroxidases were then blocked by incubation of slides for 5 min in 1% hydrogen peroxide and then blocked with 5% goat serum–Tris-buffered saline–0.1% Tween 20 (TBS-T). Slides treated with Tris-SDS-EDTA were incubated overnight at 4°C with anti-Iba1 antibody (diluted 1:1,000), while slides treated with sodium citrate buffer were incubated under similar conditions with anti-GFAP antibody (diluted 1:1,000). Following the primary antibody incubation, all slides were incubated with biotinylated secondary antibodies (Vector Laboratories) (diluted 1:200; 45 min at room temperature [RT]) and then with horseradish peroxidase (HRP)-coupled streptavidin (Vector Laboratories) (diluted 1:200; 45 min at RT). Visualization of the immunohistochemistry was performed using diaminobenzidine peroxidase substrate (Vector Laboratories). Finally, sections were counterstained with hematoxylin (Sigma-Aldrich) for 3 min before being dehydrated in graded ethanol and xylene. Stained sections were examined under a DM4000B bright-field microscope (Leica, Wetzlar, Germany), and images were captured using a Spot Flex camera and Spot V4.5 software (Diagnostic Instruments, USA).
RESULTS
Proteins secreted by Cryptococcus strains of high (C. neoformans and C. gattii VGIIa) and low (C. gattii VGIIb) virulence were identified using a novel method of protein capture, followed by liquid chromatography-mass spectroscopy. FunSecKB:v2 and SecretomeP were used to determine if proteins that were present in the fungal secretomes were likely to be classically or nonclassically secreted. It was hypothesized that differences in the secretomes might contribute to differences in host response and pathology, and this was tested in a murine intracranial infection model.
ProteoMiner beads allow isolation of proteins secreted from Cryptococcus species under low-nutrient conditions designed to approximate those in the mammalian host lung.
Species of Cryptococcus produce and shed large quantities of extracellular polysaccharide into culture medium, particularly when the availability of nutrients is limited. This complicates the isolation and analysis of secreted Cryptococcus proteins when culture conditions are designed to be similar to those encountered within the mammalian host. To overcome this, we supplemented protein-free RPMI tissue-culture media with amino acids, which we have found to limit the production of excessive shed capsule (D. A. Carter, unpublished data). To maximize the capture of secreted proteins from the culture supernatant, we developed a novel method using ProteoMiner beads (Bio-Rad Laboratories, USA). These beads contain randomly generated hexapeptide ligands designed to capture proteins following direct contact. This process yielded 50 to 60 μg of protein from each 2-liter biological replicate culture of C. neoformans and 40 to 50 μg from the 2-liter biological replicate cultures of the C. gattii strains.
To ensure that proteins collected from the supernatants had not been released as a result of cell death and lysis, cultures were assayed throughout the experimental process using trypan blue viability staining (see Fig. S1 in the supplemental material). All strains appeared similar, and there was no appreciable cell death or debris observed. This was further confirmed by subsequent mass spectroscopy, as ribosomal proteins, which would be present as a result of significant cell lysis, were not identified in any of the secretomes (see Table S1).
Different Cryptococcus strains secrete different numbers and types of proteins with limited overlap among strains.
After manual validation of the LC-MS/MS protein identifications, a total of 67 cryptococcal proteins were identified. Of these, 17 had characterized functions and 24 had putative characterizations according to the UniProt KnowledgeBase. The remaining 26 proteins were annotated as “putative uncharacterized protein” (n = 25) or as “expressed protein” (n = 1) and were assigned functional roles according to predicted domains using InterPro and by gene ontology using UniProtKB. Using these approaches, putative functions were assigned to all but seven of the uncharacterized proteins (Table 1; see also Table S1 in the supplemental material).
TABLE 1.
Proteins identified from the Cryptococcus secretomes
| Protein name | Strain(s) in which the protein was identified | Molecular function or predicted functiona | Functional grouping | Protein accession no. |
|---|---|---|---|---|
| Classically secreted hydrolytic and proteolytic proteins | ||||
| Glycosyl hydrolase, putative | All strains | Hydrolase | Hydrolysis | E6QY56 |
| Carboxypeptidase D, putative | C. gattii VGIIa & VGIIb | Serine-type carboxypeptidase activity | Proteolysis | E6RCV7 |
| Peptidase, putative | C. gattii VGIIa & VGIIb | Serine-type endopeptidase activity | Proteolysis | E6R030 |
| Chitin deacetylase | C. neoformans & C. gattii VGIIa | Chitin deacetylase activity | Hydrolysis | P82476 |
| Putative uncharacterized protein | C. neoformans & C. gattii VGIIa | Glycoside hydrolase | Hydrolysis | Q55ZC4 |
| Alpha-amylase AmyA, putative | C. neoformans & C. gattii VGIIb | Alpha-amylase activity | Hydrolysis | E6RCN1 |
| Chitinase, putative | C. gattii VGIIa | Chitinase activity | Hydrolysis | E6RB39 |
| Cellulase, putative | C. gattii VGIIa | Hydrolyzing O-glycosyl compounds | Hydrolysis | E6R762 |
| Endopeptidase, putative | C. gattii VGIIb | Aspartyl protease | Proteolysis | E6QY25 |
| Serine-type endopeptidase, putative | C. gattii VGIIb | Serine-type endopeptidase activity | Proteolysis | E6R505 |
| Putative uncharacterized protein | C. gattii VGIIb | Glycoside hydrolase | Hydrolysis | E6QYQ1 |
| Putative uncharacterized protein | C. gattii VGIIb | Glycoside hydrolase | Hydrolysis | E6R0L9 |
| Putative uncharacterized protein | C. neoformans | Glycoside hydrolase | Hydrolysis | E6QZI2 |
| Putative uncharacterized protein | C. neoformans | Glycoside hydrolase | Hydrolysis | E6R597 |
| Putative uncharacterized protein | C. neoformans | Alpha amylase | Hydrolysis | F5HAT9 |
| Putative uncharacterized protein | C. neoformans | Peptidase | Proteolysis | Q560V8 |
| Putative uncharacterized protein | C. neoformans | Glucanase | Hydrolysis | Q5KA52 |
| Putative uncharacterized protein | C. neoformans | Carboxylesterase | Hydrolysis | Q5KA58 |
| Putative uncharacterized protein | C. neoformans | Glycoside hydrolase | Hydrolysis | Q5KN45 |
| Putative uncharacterized protein | C. neoformans | Glycoside hydrolase | Hydrolysis | Q55S66 |
| Putative uncharacterized protein | C. neoformans | Glycoside hydrolase | Hydrolysis | Q5KPL2 |
| Classically secreted nondegradative proteins | ||||
| Transmembrane receptor, putative | C. gattii VGIIa & VGIIb | Receptor | Signaling | E6QYF2 |
| Cytokine inducing-glycoprotein, putative | C. gattii VGIIa & VGIIb | Cytokine inducing-glycoprotein | Unknown | E6R316 |
| Putative uncharacterized protein | C. gattii VGIIa & VGIIb | Unknown/lyase activity | Unknown | E6R9N5 |
| Putative uncharacterized protein | C. gattii VGIIa & VGIIb | Oxidoreductase | Redox | E6RF45 |
| Meiotic recombination-related protein | C. neoformans & C. gattii VGIIb | Stress response nuclear envelope protein | Stress response | E6R3P7 |
| Putative uncharacterized protein | C. gattii VGIIa | Beta-glucan synthesis | Metabolism | Q5K715 |
| Glycoprotein, putative | C. gattii VGIIa | Unknown | Unknown | Q5K852 |
| Sterol-binding protein | C. neoformans | Sterol binding | Transport | E6RAN9 |
| Expressed protein | C. neoformans | Beta-glucan synthesis | Metabolism | Q5KIN3 |
| Transmembrane receptor | C. neoformans | Receptor | Signaling | Q5KNE4 |
| Non-classically secreted proteins | ||||
| G protein beta subunit Gib2 | C. gattii VGIIb | cAMP signaling | Signaling | A0AUJ0 |
| Fructose-bisphosphate aldolase, putative | C. gattii VGIIb | Fructose-bisphosphate aldolase activity | Metabolism | E6R1G7 |
| Aminotransferase, putative | C. gattii VGIIb | Transaminase activity | Metabolism | E6R1V5 |
| Actin-binding protein Cofilin, putative | C. gattii VGIIb | Actin filament depolarization | Organelle organization | E6R286 |
| Orotidine 5′-phosphate decarboxylase | C. gattii VGIIb | Decarboxylase activity | Metabolism | Q5K890 |
| ATP synthase subunit beta | C. gattii VGIIb | ATP binding | Transport | E6R8N5 |
| Phosphoglycerate kinase | C. gattii VGIIb | Phosphoglycerate kinase activity | Metabolism | E6R9I3 |
| Phosphomannomutase, putative | C. gattii VGIIb | Phosphomannomutase activity | Metabolism | E6RA79 |
| Guanosine-diphosphatase, putative | C. gattii VGIIb | Guanosine-diphosphatase activity | Metabolism | Q5KLL8 |
| Oxidoreductase, putative | C. gattii VGIIb | Oxidoreductase activity | Redox | Q5KLM8 |
| Dihydrolipoyl dehydrogenase | C. gattii VGIIb | Oxidoreductase | Redox | Q5Y229 |
| Dihydrolipoyl dehydrogenase | C. gattii VGIIb | Oxidoreductase | Redox | Q8J0Z3 |
| Putative uncharacterized protein | C. gattii VGIIb | Unknown | Unknown | E6RDV2 |
| Putative uncharacterized protein | C. gattii VGIIb | TolB-like propeller | Unknown | F5HD57 |
| Putative uncharacterized protein | C. gattii VGIIb | Unknown | Unknown | E6RET6 |
| UDP-glucose 6-dehydrogenaseb | C. gattii VGIIb | Oxidoreductase | Redox | E6R5A2 |
| Thioredoxin peroxidase, putativeb | C. gattii VGIIb | Peroxiredoxin activity | Redox | E6RE09 |
| Citrate synthaseb | C. gattii VGIIb | Citrate (Si)-synthase activity | Metabolism | Q5KQ45 |
| Putative uncharacterized protein | C. neoformans | Glutaredoxin/oxidoreductase activity | Redox | E6QYZ1 |
| Putative uncharacterized protein | C. neoformans | Unknown | Unknown | Q5KCM3 |
| Putative uncharacterized protein | C. neoformans | Unknown | Unknown | Q5KD42 |
| Nondegradative proteins secreted via unknown mechanisms | ||||
| 3-Isopropylmalate dehydrogenaseb | C. gattii VGIIb | Oxidoreductase | Redox | E6QXQ4 |
| Enolaseb,c | C. gattii VGIIb | Phosphopyruvate hydratase activity | Metabolism | Q55UX4 |
| Glutamate dehydrogenaseb | C. gattii VGIIb | Oxidoreductase | Redox | E6R2U4 |
| Glyceraldehyde-3-phosphate dehydrogenaseb,c | C. gattii VGIIb | NAD/P binding | Metabolism | E6R7Z5 |
| 6-Phosphogluconate dehydrogenaseb,c | C. gattii VGIIb | NADP binding | Metabolism | E6RDR8 |
| Heat shock protein, putativeb | C. gattii VGIIb | ATP binding | Stress response | E6RFH1 |
| Putative uncharacterized proteinb | C. gattii VGIIb | HSP90-like | Stress response | F5HDC9 |
| Putative uncharacterized proteinb | C. gattii VGIIb | 14-3-3-Like | Signaling | F5HI88 |
| Superoxide dismutase (Cu-Zn)b,c | C. neoformans | Oxidoreductase activity | Redox | Q9C0S4 |
| Eukaryotic ADP/ATP carrier | C. gattii VGIIb | Transporter activity | Transport | Q2XPZ3 |
| Glucose-6-phosphate isomerase | C. gattii VGIIb | Isomerase | Metabolism | E6QZC4 |
| Transaldolase | C. gattii VGIIb | Transaldolase activity | Metabolism | E6RD54 |
| Putative uncharacterized protein | C. gattii VGIIb | Unknown | Unknown | E6R123 |
| Putative uncharacterized protein | C. gattii VGIIb | AAA+ ATPase | Transport | F5HD04 |
| Cytokine-inducing glycoprotein | C. neoformans | Unknown | Unknown | Q96UH1 |
Italic entries represent predicted or unknown functions.
Reported in association with microvesicles (26).
Potentially immunogenic.
The 67 secreted proteins were unevenly distributed across the three Cryptococcus strains, with significant differences with respect to both the number and type of proteins secreted (Table 1 and Fig. 1). A smaller number of less diverse proteins were identified in the secretomes of the high-virulence C. neoformans and C. gattii VGIIa strains, with 22 and 13 identified secreted proteins, respectively, compared to those identified in the secretome of the lower-virulence C. gattii VGIIb strain with 44 identified proteins.
FIG 1.
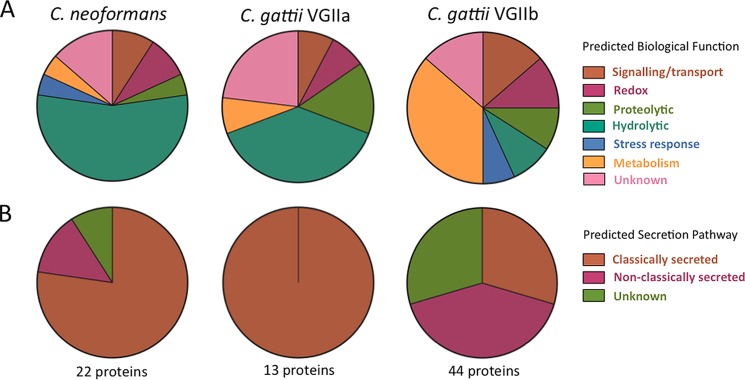
Predicted functional groups and secretory pathways for the secreted Cryptococcus proteins. (A) The secretomes of high-virulence C. neoformans and C. gattii VGIIa are dominated by hydrolytic and proteolytic proteins, while the secretome of C. gattii VGIIb contains a large number of proteins involved in metabolism. (B) C. gattii VGIIb has a much greater proportion of proteins secreted by nonclassical pathways than C. neoformans and C. gattii VGIIa.
Eleven proteins were identified in the secretomes of two or more of the three strains. One protein, a putative glycosyl hydrolase (E6QY56) predicted to belong to the glycosyl hydrolase 61 family, was found in all three strains. The C. gattii strains had six proteins in common, including a putative transmembrane receptor (E6QYF2), a putative cytokine-inducing glycoprotein (E6R316), a putative peptidase (E6R030), and a putative carboxypeptidase D (E6RCV7). Two proteins were uncharacterized but were predicted to have lyase (E6R9N5) and oxidoreductase (E6RF45) activity. Four proteins were common to one of the C. gattii strains and the C. neoformans strain. C. neoformans and the high-virulence C. gattii VGIIa strain shared a chitin deacetylase (P82476) and a putative uncharacterized protein (Q55ZC4). C. neoformans and the low-virulence C. gattii VGIIb strain both secreted a putative meiotic recombination-related protein (E6R3P7) and an α-amylase (E6RCN1). The latter has been linked to virulence in some fungi, including Aspergillus flavus and Histoplasma capsulatum (27, 28).
All secretomes contained a core set of hydrolytic and proteolytic proteins with potential nutrient-scavenging roles.
Approximately half of the proteins identified in the secretomes of the high-virulence C. gattii VGIIa and C. neoformans strains (7/13 and 13/22 proteins, respectively) are known or predicted to have hydrolytic or proteolytic functions (Fig. 1). These enzymes are secreted by a wide range of fungi and have a role in nutrient scavenging via exodigestion (2). In the C. gattii VGIIa secretome, these proteins included a putative cellulase (E6R762) and a chitinase (E6RB39), while those identified in the C. neoformans secretome were generally uncharacterized proteins with putative glycoside hydrolase, peptidase, or carboxyesterase activity (Table 1). The C. gattii VGIIb secretome contained a number of proteins with hydrolytic and proteolytic functions similar to those seen with the other two strains, but these comprised a substantially smaller fraction of the total proteins in its secretome (8/44 [18%]). These hydrolytic and proteolytic proteins may represent a “core set” of degradative enzymes, similar to those known to be secreted by many fungal species (2).
Extracellular Cryptococcus proteins are secreted by both classical and nonclassical mechanisms.
FunSecKB:v2 and SecretomeP were used to predict if proteins identified in the cryptococcal secretomes were secreted via classical or nonclassical mechanisms. Of the 67 unique proteins identified across the three secretomes, 32 were predicted as being classically secreted. These included the majority of the proteins identified in the secretomes of the virulent strains, with 13/13 of the C. gattii VGIIa and 18/22 of the C. neoformans proteins predicted to be secreted via classical mechanisms. Four of these classically secreted proteins were predicted to have glycosylphosphatidylinositol (GPI)-anchor signal sequences, suggesting an interaction with the fungal cell wall (Fig. 1; see also Table S1 in the supplemental material). In contrast, less than a third (13/44) of the proteins identified in the C. gattii VGIIb secretome were predicted to be classically secreted. All of the hydrolytic and proteolytic proteins across the three secretomes, which potentially belong to a core suite of nutrient-scavenging enzymes, were predicted to be classically secreted, consistent with a role in fungal exodigestion.
In the C. gattii VGIIb and the C. neoformans secretomes, 18 and 3 proteins, respectively, were predicted as being secreted via alternative or nonclassical mechanisms (Table 1). These proteins included dihydrolipoyl dehydrogenase (Q5Y229 and Q8J0Z3) and a putative thioredoxin peroxidase (E6RE09), both involved in redox homeostasis within the cell, the G-protein Gib2 beta subunit (A0AUJ0) involved in cyclic AMP (cAMP) signaling, and metabolic proteins, including a putative fructose-bisphosphate aldolase (E6R1G7), a putative aminotransferase (E6R1V5), and a UDP-glucose 6-dehydrogenase (E6R5A2).
Fifteen proteins were not predicted by FunSecKB or SecretomeP to be secreted by either classical or nonclassical mechanisms. Cryptococcus species are known to secrete microvesicles, which are analogous to mammalian exosomes and are described as an alternative, nonclassical secretion mechanism (29). Of the 67 proteins identified as secreted in this study, 12 have been reported as contained within Cryptococcus microvesicles, and nine of these were among the 15 proteins with no predicted secretion mechanism, with the remaining three predicted to be non-classically secreted. These microvesicle-associated proteins included the common intracellular proteins enolase (Q55UX4), glyceraldehyde-3-phosphate dehydrogenase (E6R7D5), 6-phosphogluconate dehydrogenase (E6RDR8), and Cu-Zn superoxide dismutase (Q9C0S4) as well as a putative uncharacterized protein (F5HI88) with a predicted 14-3-3 regulatory-protein-like function.
The C. gattii VGIIb secretome contains a unique set of secreted proteins, some of which may have “moonlighting” or immunogenic functions.
Thirty-five of the 67 proteins identified across the three cryptococcal secretomes analyzed in this study were unique to the secretome of the low-virulence C. gattii VGIIb strain (Table 1). These proteins are primarily involved in metabolism, signaling/transport, glycolysis, and redox processes and are canonical intracellular proteins. A number of these have orthologues that have previously been identified in the extracellular milieu of various cell types from other organisms (30–33). Their presence in this alternative environment, where interacting protein partners or substrates are typically absent, suggests they may have alternative or “moonlighting” functions when secreted. In addition, some of the proteins secreted by the C. gattii VGIIb strain have orthologues that can initiate an immune response in the host. These included the glycolytic proteins glyceraldehyde-3-phosphate dehydrogenase (E6R7D5), enolase (Q55UX4), and 6-phosphogluconate dehydrogenase (E6RDR8) and the stress response protein Cu-Zn superoxide dismutase (Q9C0S4) (Table 1).
C. gattii high-virulence and low-virulence strains have reversed pathogenicity characteristics in an intracranial infection model of cryptococcal meningitis.
Past epidemiological data and studies using a murine inhalation model of virulence have suggested that the C. gattii VGIIa genotype is more virulent than the VGIIb genotype (14, 34). However, the results of a recent epidemiological study of the Vancouver Island cryptococcal outbreak indicated that, while a much smaller number of patients had been infected with the VGIIb genotype, a greater proportion of these progressed to cerebral sequelae resulting in death. In contrast, C. gattii VGIIa infections were more likely to be pulmonary (35). The difference in the levels of immunogenicity of the proteins secreted by the VGIIa and VGIIb strains noted here suggested that differences between these genotypes with respect to virulence and pathology might be mediated in part by the fungal secretome, which on the one hand might trigger a more rapid immune clearance of C. gattii VGIIb strains but on the other might result in more-severe symptoms if the immune system were breached. To test this, we bypassed the lung, using an intracranial infection model with direct injection of 1,000 CFU of Cryptococcus yeast cells into the mouse brain. C. neoformans strain H99, which is widely used as a model for virulent cryptococcosis, was included for comparison. Two different strains of mice were used for intracerebral infection, inbred wild-type C57BL/6 mice and immunocompromised SCID mice, which are T cell deficient and are often used as an animal model for AIDS. The survival curve following infection is shown in Fig. 2. In contrast to the survival profile seen following inhalation, where mice infected with C. neoformans H99 and C. gattii VGIIa strains are rapidly killed while those infected with C. gattii VGIIb survive (14), infection by C. gattii VGIIb caused significantly more rapid death than infection by C. gattii VGIIa (P < 0.05).
FIG 2.
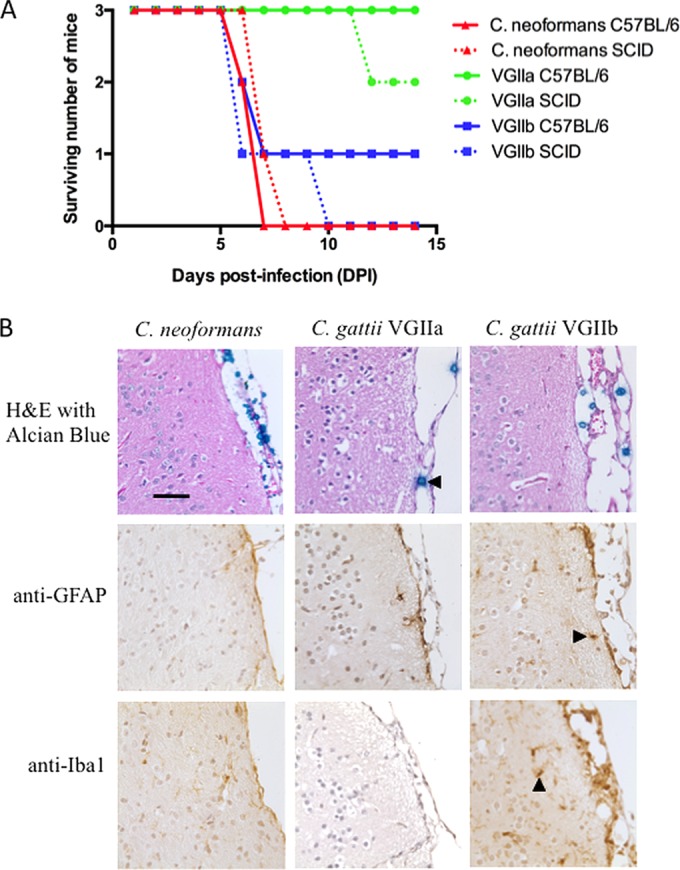
Survival curve, histochemistry, and immunohistochemistry for C57BL/6 and SCID mice infected intracranially with Cryptococcus strains. (A) Intracranial infection results in rapid death of mice infected with C. neoformans and C. gattii strain R272, which is considered hypovirulent in a pulmonary infection model. In contrast, most mice survive infection with the “hypervirulent” C. gattii VGIIa strain. (B) Histochemistry and immunohistochemistry for representative infected C57BL/6 mice. (Top panel) alcian blue counterstaining showing cryptococcal cells in the mice meninges (left-pointing arrowhead). The fungal burden is considerably lower in mice infected with C. gattii VGIIa (see also Fig. S2 in the supplemental material for a zoomed-out field of view). (Middle panel) Anti-GFAP antibody staining showing astrocytes (right-pointing arrowhead). (Bottom panel) Anti-Iba1 visualizing microglial cells (upward-pointing arrowhead). The immune response is extremely limited, particularly for C. neoformans infection, despite the significant fungal burden. There was a greater microglial response in mice infected with C. gattii VGIIb than in those infected with C. neoformans and C. gattii VGIIa. Bar, 50 μm. H&E, hematoxylin and eosin.
Although the severely immunocompromised SCID mice succumbed slightly earlier than the immunocompetent mice, the results were not significantly different for any of the infecting Cryptococcus strains (P > 0.05), suggesting that the host immune status was having little influence on cryptococcal infection once this had established in the brain. To assess pathology, we examined embedded sections of brain tissue from infected C57BL/6 mice by histochemistry and immunohistochemistry. Brains infected with the C. gattii VGIIb and C. neoformans strains contained a large number of Cryptococcus cells located predominantly in the meninges, with evidence of extensive polysaccharide capsule production. In contrast, the brain sections from mice infected with the C. gattii VGIIa strain had very few visible Cryptococcus cells (Fig. 2; see also Fig. S2 in the supplemental material). Interestingly, while the C. gattii cells visible in the meninges were relatively large (∼10 to 20 μm) and uniform in size, the C. neoformans cells were very heterogeneous, with a large number of very small (<5-μm-diameter) cells. An observable effect on the meninges was apparent only with the VGIIb strain, where a slight to moderate thickening was apparent. Immunohistochemistry was performed on tissue sections from the C57BL/6 mice to determine the presence and activation state of astrocytes, which respond rapidly to tissue damage in the brain, and of microglia, the immune cells resident in the central nervous system (CNS). Activation of some astrocyte and microglial cells was observed adjacent to the meninges in the VGIIb-infected tissue; however, compared to the results seen with other intracerebral infection models, this response was very weak (36, 37). There was no apparent response to infection seen in brain tissue from mice infected with the C. gattii VGIIa or the C. neoformans strain.
DISCUSSION
In our study, we compared individual proteins secreted from Cryptococcus strains with different virulence attributes. Our aims were to identify proteins likely to be secreted by the Cryptococcus strains during mammalian infection and to determine if these differed between strains known to have different degrees of pathogenicity. Our results suggest that the secretomes of the strains differ and may mediate a complex interplay between host recognition and clearance on the one hand and invasion and pathogenic disease on the other.
The most striking finding in this study was that the proteins secreted by C. gattii VGIIa and VGIIb and C. neoformans were very different, with little overlap among strains. However, in terms of number and function of proteins, there was substantially greater similarity between the secretomes of the two physiologically related high-virulence strains than between the two genetically related C. gattii strains. We took a very conservative approach to calling protein hits, with a requirement of identification in at least three independent biological replicates; in most cases, however, proteins were either present in the majority of samples or entirely absent, and few appeared in only one or two samples (see Table S1 in the supplemental material). At the outset of this study, we hypothesized that the higher-virulence strains would have distinctive, highly active cell-damaging enzymes that might promote host invasion. While we did identify secreted enzymes with predicted hydrolytic or proteolytic functions, these were present in all three cryptococcal secretomes and were not restricted to the virulent strains. Similar enzymes have been described in a range of fungi, and they may be a fundamental component of fungal secretomes, regardless of ecological niche or lifestyle (2). Although their primary function is to degrade polymers in the environment for nutrition, these enzymes can have a crossover function in fungal pathogenesis, where they can promote invasion and cause cell damage (3, 4, 27, 28).
A second striking difference was that the secretome of the “hypovirulent” VGIIb strain included a suite of canonical intracellular proteins, many with known immunogenic properties, which were absent from the secretomes of the virulent strains. These types of proteins have been observed in the secretomes of many different organisms; however, they have been noted as a particular feature of the secretomes of pathogenic fungal species compared with nonpathogenic fungi (2). Further, in fungal pathogens, these proteins have been associated with secretion during growth within the host. For example, the plant pathogen Fusarium graminearum secretes a large number of canonical intracellular proteins when grown in planta that are not secreted when the fungus is cultured in vitro (38).
Orthologues of many of the canonical intracellular proteins secreted by the VGIIb strain are known to have alternative extracellular “moonlighting” functions that appear unrelated to their intracellular roles (8). These proteins do not contain secretory signal motifs and hence are generally not predicted to be extracellular. The moonlighting roles of similar proteins in different organisms differ greatly and appear to be organism specific, making it difficult to extrapolate their functions among even closely related species (8, 30). It has been noted, however, that certain functional groups are prominent among known moonlighting proteins. Glycolytic proteins are one such group, with a large number of the enzymes involved in glycolysis and gluconeogenesis found to have moonlighting roles, including enolase and GAPDH (glyceraldehyde 3-phosphate dehydrogenase), which were both present in the VGIIb secretome (31).
Enolase is one of the most abundant cytosolic proteins, with a key function in the pentose phosphate pathway. This protein is known to be multifunctional and has been identified in the secretomes of many different organisms and cell types (32). In the fungal pathogens Candida albicans, Pneumocystis jirovecii, and Aspergillus fumigatus, cell wall-associated enolase has been shown to be immunogenic, to bind to and activate host plasminogen, and to promote invasive disease (39–42). C. neoformans enolase has been implicated in plasminogen activation, allowing Cryptococcus strains to traverse endothelial cells and potentially cross the blood-brain barrier (BBB) (43, 44). Extracellular GAPDH appears to have different roles in the progression of infection by a wide range of pathogens, including plasminogen binding in parasites (45), adherence to host cells by pathogenic Escherichia coli, Neisseria meningitides, Bacillus anthracis, Paracoccidioides brasiliensis, and Candida albicans (33), and complement and fibronectin binding in helminths (46, 47) and as an immunogen in C. albicans (26). Given these diverse roles, cryptococcal GAPDH secreted by the C. gattii VGIIb strain has the potential to trigger a host response leading to clearance from the lung or to aid disease progression, leading to cryptococcal meningitis.
Intracellular vesicles are increasingly implicated in nonclassical secretion by pathogens, with microvesicles, secretory lysosomes, and autophagosomes involved in this process (7, 48–51). A key role for microvesicles in the host-pathogen interface was recently described for the malaria parasite Plasmodium falciparum in a study in which infected host red blood cells produced microvesicles containing both host and parasite proteins, facilitating communication within the parasite population and between the parasite and the host immune system (52). Cryptococcus species are known to produce microvesicles, and these have been shown to be immunologically active, stimulating macrophages to produce cytokines and other antimicrobial compounds (29, 53). Microvesicles have also been observed during in vivo infection, and proteins derived from them are immunoreactive to sera from cryptococcosis patients (29, 54). Microvesicles appear to facilitate the traversal of Cryptococcus cells across the BBB and to aggregate at lesion sites during brain infection (55). There is considerable overlap between some of the non-classically secreted proteins released by the VGIIb strain shown here and a number of proteins previously identified from cryptococcal microvesicles; in particular, a 14-3-3 protein is considered a marker of microvesicle presence (Table 1) (29). Thus, although indirectly, our secretome data suggest that microvesicles may be associated with VGIIb infection, and this could be a further factor provoking more-severe CNS infection by this genotype (35).
Many non-classically secreted proteins, in addition to their moonlighting roles, are known to be immunogenic. In addition to the results seen with enolase and GAPDH described above, the C. gattii VGIIb strain secreted glucose-6-phosphate isomerase, which has been linked to the development of rheumatoid arthritis following Aspergillus infection (56). Further, some non-classically secreted proteins identified in this study, including fungal serine-type endopeptidases (57) and transaldolase, an immunodominant protein in C. neoformans (58–60), have been found to bind IgE. The presence of immunogenic proteins in the VGIIb secretome might be expected to provoke rapid induction of the innate immune response, preventing the initiation of disease. In contrast, high-virulence strains that lack these secreted immune-stimulating proteins may bypass the host response and establish lung infection.
As our data suggested that a significant factor contributing to the differences in virulence seen between Cryptococcus strains may be the presence of immunogenic and invasion-promoting secreted proteins that on the one hand prevent low-virulence strains from establishing infection in the lung but on the other might provoke more-serious disease if the pulmonary immune response were to be bypassed, we performed intracranial infections in a mouse model (25). In agreement with our hypothesis, the hypovirulent C. gattii VGIIb strain was able to establish infection and was in fact considerably more pathogenic than the VGIIa strain, where all infected immunocompetent mice survived (Fig. 2). This is consistent with epidemiological evidence that VGIIb infections, while much more infrequent, are significantly more likely to progress to cryptococcal meningitis than VGIIa infections, where severe pulmonary disease is the most common cause of death (35). Analysis of the brains of infected mice revealed substantial colonization of the meninges in mice infected with C. gattii VGIIb and C. neoformans, while VGIIa cells were almost completely cleared (Fig. 2; see also Fig. S2 in the supplemental material). There was a surprising lack of immune response or associated cellular activation in the meninges and adjacent tissue in all mice regardless of which Cryptococcus strain was used for infection, although the anti-Iba1 stain indicated greater activation of microglia following VGIIb infection (Fig. 2). C. neoformans H99 was an effective pathogen in both the pulmonary and meningitis models of infection, consistent with its ability to cause fatal lung and CNS infection.
There are a number of limitations to this study that may be addressed with further work. First, only three Cryptococcus strains were analyzed, and follow-up work that includes more strains of the different species and genotypes is required to confirm our observations. Second, while we attempted to simulate the host lung using RPMI tissue culture medium, such attempts cannot recapitulate the complexities of the host environment. In vivo proteomics in a mammalian host is extremely challenging; however, the use of antibodies or new mass spectroscopy-based methods such as selected reaction monitoring (SRM), which is designed to detect individual proteins from a heterogenous mixture, may be able to validate whether selected proteins are secreted during infection. Third, the use of ProteoMiner beads limited us to a qualitative analysis, and we had no way of assessing the relative abundances of the various secreted proteins, which may also be very important in determining the virulence of a strain. Finally, although our analysis found interesting correlations between the nature of the secretome and the ability to produce pulmonary and CNS infection, we cannot assume causation, and for now, this link remains an interesting but untested hypothesis.
In conclusion, the three Cryptococcus secretomes contained a core set of hydrolytic and proteolytic functions with various proteins that are predicted to have roles in nutrient scavenging. The secretome of the VGIIb strain, which is hypovirulent in a lung infection model, included additional, non-classically secreted proteins, with many reported previously to be associated with microvesicles. As a number of these are immunogenic, their absence from the C. neoformans and C. gattii VGIIa secretomes suggests that these strains may evade host immune detection, allowing cryptococcosis to be initiated in the lung. However, microvesicles and their associated proteins may also facilitate dissemination to the CNS, consistent with the ability of the VGIIb genotype to cause cerebral infection with a poor outcome, despite a lower incidence of disease. We propose that the activity of secreted cryptococcal proteins and their interactions with the host immune system are likely to play an important role in cryptococcal disease progression. If particular secreted proteins can be associated with disease outcome, these could be useful as prognostic markers for disease progression. Secreted proteins may also be useful targets for diagnostics, vaccines, or antifungal therapy, which remain underdeveloped for fungal diseases.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by Australian National Health and Medical Research Council grant no. 971354.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00052-15.
REFERENCES
- 1.Witzany G. 2010. Uniform categorization of biocommunication in bacteria, fungi and plants. World J Biol Chem 1:160–180. doi: 10.4331/wjbc.v1.i5.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girard V, Dieryckx C, Job C, Job D. 2013. Secretomes: the fungal strike force. Proteomics 13:597–608. doi: 10.1002/pmic.201200282. [DOI] [PubMed] [Google Scholar]
- 3.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E, Weindl G, Tappuni AR, Rodgers CA, Woodman AJ, Challacombe SJ, Schaller M, Hube B. 2008. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology 154:3266–3280. doi: 10.1099/mic.0.2008/022293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouws H, Wattenberg A, Zorn H. 2008. Fungal secretomes—nature's toolbox for white biotechnology. Appl Microbiol Biotechnol 80:381–388. doi: 10.1007/s00253-008-1572-5. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal GK, Jwa N-S, Lebrun M-H, Job D, Rakwal R. 2010. Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10:799–827. doi: 10.1002/pmic.200900514. [DOI] [PubMed] [Google Scholar]
- 7.Rabouille C, Malhotra V, Nickel W. 2012. Diversity in unconventional protein secretion. J Cell Sci 125:5251–5255. doi: 10.1242/jcs.103630. [DOI] [PubMed] [Google Scholar]
- 8.Jeffery CJ. 1999. Moonlighting proteins. Trends Biochem Sci 24:8–11. doi: 10.1016/S0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 9.Jeffery CJ. 2003. Moonlighting proteins: old proteins learning new tricks. Trends Genet 19:415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- 10.Jeffery CJ. 2009. Moonlighting proteins—an update. Mol Biosyst 5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- 11.Jeffery CJ. 2011. Proteins with neomorphic moonlighting functions in disease. IUBMB Life 63:489–494. doi: 10.1002/iub.504. [DOI] [PubMed] [Google Scholar]
- 12.Lin X, Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol 60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 13.Springer DJ, Chaturvedi V. 2010. Projecting global occurrence of Cryptococcus gattii. Emerg Infect Dis 16:14–20. doi: 10.3201/eid1601.090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrnes EJ III, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen LC, Pirofski LA, Casadevall A. 1997. Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect Immun 65:2599–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biondo C, Mancuso G, Midiri A, Bombaci M, Messina L, Beninati C, Teti G. 2006. Identification of major proteins secreted by Cryptococcus neoformans. FEMS Yeast Res 6:645–651. doi: 10.1111/j.1567-1364.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 17.Eigenheer RA, JL Y, Blumwald E, Phinney BS, Gelli A. 2007. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res 7:499–510. doi: 10.1111/j.1567-1364.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- 18.Sivell M. 2014. Cryptococcosis: a proteomic investigation of an emerging fungal disease. Ph.D. thesis University of Sydney. [Google Scholar]
- 19.Jobbins SE, Hill CJ, D'Souza-Basseal JM, Padula MP, Herbert BR, Krockenberger MB. 2010. Immunoproteomic approach to elucidating the pathogenesis of cryptococcosis caused by Cryptococcus gattii. J Proteome Res 9:3832–3841. doi: 10.1021/pr100028t. [DOI] [PubMed] [Google Scholar]
- 20.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 21.The UniProt Consortium. 2013. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res 41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong S-Y, Bateman A, Punta M, Attwood TK, Sigrist CJA, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lum G, Min XJ. 2011. FunSecKB: the Fungal Secretome KnowledgeBase. Database (Oxford) 2011:bar001. doi: 10.1093/database/bar001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S. 2004. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 25.Thompson GR, Wiederhold NP, Najvar LK, Bocanegra R, Kirkpatrick WR, Graybill JR, Patterson TF. 29 February 2012, posting date A murine model of Cryptococcus gattii meningoencephalitis. J Antimicrob Chemother doi: 10.1093/jac/dks060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gil-Navarro I, Gil ML, Casanova M, O'Connor JE, Martínez JP, Gozalbo D. 1997. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J Bacteriol 179:4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown RL, Chen Z-Y, Cleveland TE, Cotty PJ, Cary JW. 2001. Variation in in vitro α-amylase and protease activity is related to the virulence of Aspergillus flavus isolates. J Food Prot 64:401–404. [DOI] [PubMed] [Google Scholar]
- 28.Marion CL, Rappleye CA, Engle JT, Goldman WE. 2006. An α-(1,4)-amylase is essential for α-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol Microbiol 62:970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell 7:58. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores C-L, Gancedo C. 2011. Unraveling moonlighting functions with yeasts. IUBMB Life 63:457–462. doi: 10.1002/iub.454. [DOI] [PubMed] [Google Scholar]
- 31.Gómez-Arreaza A, Acosta H, Quiñones W, Concepción JL, Michels PAM, Avilán L. 2014. Extracellular functions of glycolytic enzymes of parasites: unpredicted use of ancient proteins. Mol Biochem Parasitol 193:75–81. doi: 10.1016/j.molbiopara.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, Lopez-Alemany R. 2012. alpha-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol 2012:156795. doi: 10.1155/2012/156795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egea L, Aguilera L, Giménez R, Sorolla MA, Aguilar J, Badía J, Baldoma L. 2007. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int J Biochem Cell Biol 39:1190–1203. doi: 10.1016/j.biocel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 34.MacDougall L, Fyfe M, Romney M, Starr M, Galanis E. 2011. Risk factor for Cryptococcus gattii infection, British Columbia, Canada. Emerg Infect Dis 17:193–199. doi: 10.3201/eid1702.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galanis E, MacDougall L. 2010. Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999–2007. Emerg Infect Dis 16:251–257. doi: 10.3201/eid1602.090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofer MJ, Li W, Manders P, Terry R, Lim SL, King NJC, Campbell IL. 2012. Mice deficient in STAT1 but Not STAT2 or IRF9 develop a lethal CD4+ T-cell-mediated disease following infection with lymphocytic choriomeningitis virus. J Virol 86:6932–6946. doi: 10.1128/JVI.07147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Getts DR, Terry RL, Getts MT, Müller M, Rana S, Shrestha B, Radford J, Van Rooijen N, Campbell IL, King NJC. 2008. Ly6c+ “inflammatory monocytes” are microglial precursors recruited in a pathogenic manner in West Nile virus encephalitis. J Exp Med 205:2319–2337. doi: 10.1084/jem.20080421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paper JM, Scott-Craig JS, Adhikari ND, Cuomo CA, Walton JD. 2007. Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics 7:3171–3183. doi: 10.1002/pmic.200700184. [DOI] [PubMed] [Google Scholar]
- 39.Jong AY, Chen SHM, Stins MF, Kim KS, Tuan T-L, Huang S-H. 2003. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J Med Microbiol 52:615–622. doi: 10.1099/jmm.0.05060-0. [DOI] [PubMed] [Google Scholar]
- 40.Fox D, Smulian AG. 2001. Plasminogen-binding activity of enolase in the opportunistic pathogen Pneumocystis carinii. Med Mycol 39:495–507. doi: 10.1080/mmy.39.6.495.507. [DOI] [PubMed] [Google Scholar]
- 41.Eroles P, Sentandreu M, Elorza MV, Sentandreu R. 1997. The highly immunogenic enolase and Hsp70p are adventitious Candida albicans cell wall proteins. Microbiology 143:313–320. doi: 10.1099/00221287-143-2-313. [DOI] [PubMed] [Google Scholar]
- 42.Lai HY, Tam MF, Tang RB, Chou H, Chang CY, Tsai JJ, Shen HD. 2002. cDNA cloning and immunological characterization of a newly identified enolase allergen from Penicillium citrinum and Aspergillus fumigatus. Int Arch Allergy Immunol 127:181–190. doi: 10.1159/000053862. [DOI] [PubMed] [Google Scholar]
- 43.Stie J, Bruni G, Fox D. 2009. Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion. PLoS One 4:e5780. doi: 10.1371/journal.pone.0005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stie J, Fox D. 2012. Blood-brain barrier invasion by Cryptococcus neoformans is enhanced by functional interactions with plasmin. Microbiology 158:240–258. doi: 10.1099/mic.0.051524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figuera L, Gómez-Arreaza A, Avilán L. 2013. Parasitism in optima forma: exploiting the host fibrinolytic system for invasion. Acta Trop 128:116–123. doi: 10.1016/j.actatropica.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 46.Lama A, Kucknoor A, Mundodi V, Alderete JF. 2009. Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis. Infect Immun 77:2703–2711. doi: 10.1128/IAI.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahoo S, Murugavel S, Devi IK, Vedamurthy GV, Gupta SC, Singh BP, Joshi P. 2013. Glyceraldehyde-3-phosphate dehydrogenase of the parasitic nematode Haemonchus contortus binds to complement C3 and inhibits its activity. Parasite Immunol 35:457–467. doi: 10.1111/pim.12058. [DOI] [PubMed] [Google Scholar]
- 48.Andrei C, Dazzi C, Lotti L, Torrisi MR, Chimini G, Rubartelli A. 1999. The secretory route of the leaderless protein Interleukin 1β involves exocytosis of endolysosome-related vesicles. Mol Biol Cell 10:1463–1475. doi: 10.1091/mbc.10.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, Surprenant A. 2001. Rapid secretion of interleukin-1β by microvesicle shedding. Immunity 15:825–835. doi: 10.1016/S1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 50.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manjithaya R, Subramani S. 2010. Role of autophagy in unconventional protein secretion. Autophagy 6:650–651. doi: 10.4161/auto.6.5.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mantel P-Y, Hoang Anh N, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, Ghiran I, Toner M, Irimia D, Ivanov Alexander R, Barteneva N, Marti M. 2013. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13:521–534. doi: 10.1016/j.chom.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliveira DL, Freire-de-Lima CG, Nosanchuk JD, Casadevall A, Rodrigues ML, Nimrichter L. 2010. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect Immun 78:1601–1609. doi: 10.1128/IAI.01171-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang S-H, Wu C-H, Chang YC, Kwon-Chung KJ, Brown RJ, Jong A. 2012. Cryptococcus neoformans derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS One 7:e48570. doi: 10.1371/journal.pone.0048570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pizzolla A, Wing K, Holmdahl R. 2013. A glucose-6-phosphate isomerase peptide induces T and B cell–dependent chronic arthritis in C57BL/10 mice: arthritis without reactive oxygen species and complement. Am J Pathol 183:1144–1155. doi: 10.1016/j.ajpath.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 57.Shen HD LW, Tam MF, Wang SR, Tsai JJ, Chou H, Han SH. 1998. Alkaline serine proteinase: a major allergen of Aspergillus oryzae and its cross-reactivity with Penicillium citrinum. Int Arch Allergy Immunol 116:29–35. doi: 10.1159/000023921. [DOI] [PubMed] [Google Scholar]
- 58.Chou H, Tam MF, Chiang CH, Chou CT, Tai HY, Shen HD. 2011. Transaldolases are novel and immunoglobulin E cross-reacting fungal allergens. Clin Exp Allergy 41:739–749. doi: 10.1111/j.1365-2222.2011.03698.x. [DOI] [PubMed] [Google Scholar]
- 59.Chou H, Wu K-G, Yeh C-C, Tai H-Y, Tam MF, Chen Y-S, Shen H-D. 2014. The transaldolase, a novel allergen of Fusarium proliferatum, demonstrates IgE cross-reactivity with its human analogue. PLoS One 9:e103488. doi: 10.1371/journal.pone.0103488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young M, Macias S, Thomas D, Wormley FL. 2009. A proteomic-based approach for the identification of immunodominant Cryptococcus neoformans proteins. Proteomics 9:2578–2588. doi: 10.1002/pmic.200800713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


