ABSTRACT
Transmission of malaria occurs during Anopheles mosquito vector blood meals, when Plasmodium sporozoites that have invaded the mosquito salivary glands are delivered to the mammalian host. Sporozoites display a unique form of motility that is essential for their movement across cellular host barriers and invasion of hepatocytes. While the molecular machinery powering motility and invasion is increasingly well defined, the signaling events that control these essential parasite activities have not been clearly delineated. Here, we identify a phosphodiesterase (PDEγ) in Plasmodium, a regulator of signaling through cyclic nucleotide second messengers. Reverse transcriptase PCR (RT-PCR) analysis and epitope tagging of endogenous PDEγ detected its expression in blood stages and sporozoites of Plasmodium yoelii. Deletion of PDEγ (pdeγ−) rendered sporozoites nonmotile, and they failed to invade the mosquito salivary glands. Consequently, PDEγ deletion completely blocked parasite transmission by mosquito bite. Strikingly, pdeγ− sporozoites showed dramatically elevated levels of cyclic GMP (cGMP), indicating that a perturbation in cyclic nucleotide balance is involved in the observed phenotypic defects. Transcriptome sequencing (RNA-Seq) analysis of pdeγ− sporozoites revealed reduced transcript abundance of genes that encode key components of the motility and invasion apparatus. Our data reveal a crucial role for PDEγ in maintaining the cyclic nucleotide balance in the malaria parasite sporozoite stage, which in turn is essential for parasite transmission from mosquito to mammal.
IMPORTANCE
Malaria is a formidable threat to human health worldwide, and there is an urgent need to identify novel drug targets for this parasitic disease. The parasite is transmitted by mosquito bite, inoculating the host with infectious sporozoite stages. We show that cellular signaling by cyclic nucleotides is critical for transmission of the parasite from the mosquito vector to the mammalian host. Parasite phosphodiesterase γ is essential for maintaining cyclic nucleotide balance, and its deletion blocks transmission of sporozoites. A deeper understanding of the signaling mechanisms involved in transmission might inform the discovery of novel drugs that interrupt this essential step in the parasite life cycle.
INTRODUCTION
Malaria, a disease caused by Plasmodium parasites, is a formidable threat to human health, especially in resource-poor regions of the world (1). The complex life cycle of the malaria parasites provides numerous opportunities for points of intervention that pursue distinct goals such as treatment of disease or prevention of parasite transmission (2). Transmission of Plasmodium parasites is initiated in the mosquito when Anopheles vectors take a blood meal from an infected mammalian host that contains male and female gametocytes. The gametocytes differentiate into gametes in the mosquito midgut and undergo fertilization to form a zygote. Through a series of developmental steps, the zygote differentiates into sporozoites, which migrate from the midgut via the hemolymph and invade the mosquito salivary glands. Sporozoite motility and invasiveness are essential for successful completion of the Plasmodium life cycle in the mosquito as well as transmission to and infection of the mammalian host. The signaling events that regulate sporozoite motility and host cell infection have not been broadly studied on the molecular level, but if better understood, they might provide targets for prevention of infection.
Sporozoite invasion of Anopheles salivary glands is mediated by specific interactions between receptors on the salivary gland epithelium and their respective ligands on the sporozoite surface (3, 4). To invade the salivary gland, sporozoites first penetrate the basal lamina and then enter epithelial cells within a parasitophorous vacuole (PV) (3), which disintegrates soon after invasion (5). Sporozoites exit the apical end of invaded epithelial cells and are released into the central secretory cavity of the gland from where they are delivered to the mammalian host during a blood meal (6). Upon delivery into the mammalian skin, sporozoites display robust motility, which is also observed in vitro (7). This interaction causes a spike in sporozoite intracellular levels of the cyclic nucleotide cyclic AMP (cAMP) (7). Motile sporozoites invade dermal capillaries and are transported to the liver, where they exit the blood by traversing the endothelial barrier, before productively invading and establishing infection in a hepatocyte. Sporozoite motility and infection of hepatocytes require a regulated release of micronemal proteins from the apical end of the sporozoite. This apical exocytosis is cAMP dependent (8). Thus, cyclic nucleotides play a critical role in sporozoite transmission and infection.
The cyclic nucleotides cAMP and cyclic GMP (cGMP) function as signaling second messengers downstream of surface receptor-ligand interactions by activating cAMP-dependent protein kinase (PKA) and cGMP-dependent protein kinase (PKG), respectively (9). Signaling through cAMP and cGMP is regulated by phosphodiesterases (PDEs), metal ion-dependent enzymes that hydrolyze the 3′-phosphoester bond of cAMP and cGMP (9). The Plasmodium genome encodes four PDEs (α, β, γ, and δ), and the essentiality of PDEs (and therefore cyclic nucleotide-based signaling) in cellular homeostasis has fueled interest in PDEs as potential antimalarial drug targets (10, 11). Indeed, studies have shown that Plasmodium PDEs are important in a variety of cellular processes, including P. gallinaceum male gametocyte exflagellation (12), P. falciparum gametocytogenesis (13), cell cycle regulation (14), and P. berghei ookinete maturation (15).
Here, we show, through the creation of a P. yoelii PDEγ deletion mutant, an essential role for PDEγ in sporozoite transmission. P. yoelii pdeγ− sporozoites were nonmotile, failed to invade the salivary glands, and exhibited dramatically elevated levels of cGMP. These findings demonstrate a vital role for PDEγ in maintaining the cyclic nucleotide balance in the malaria sporozoites, which is critical for parasite transmission.
RESULTS
PDEγ is transcribed in blood stages and sporozoites.
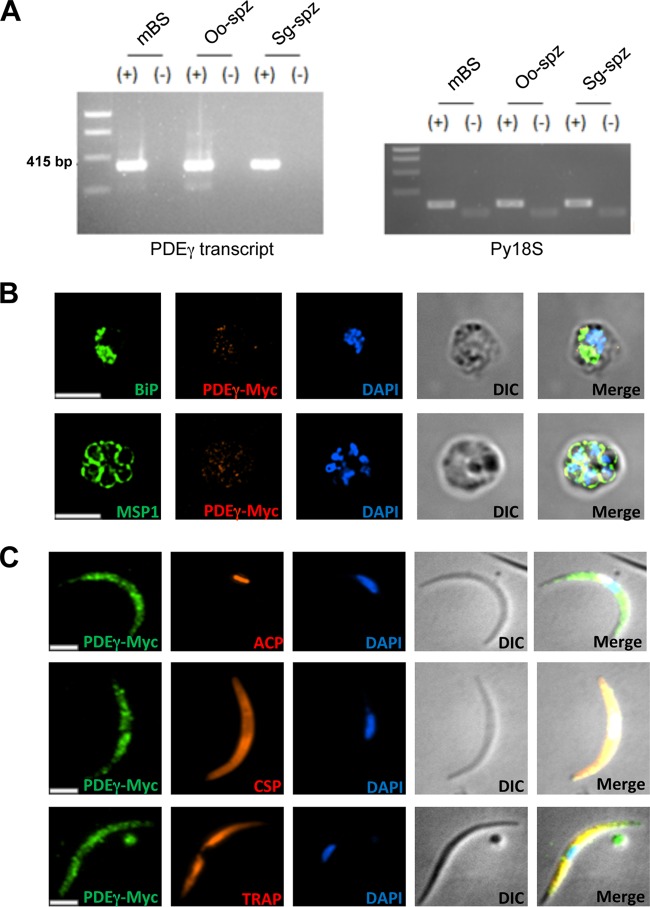
Plasmodium yoelii PDEγ (identifier [ID] PY17X_1421600; gene information available on http://plasmodb.org/plasmo/) is predicted to be a 782-amino-acid type II membrane protein with six transmembrane domains. A search for the presence of domains using the PDEγ sequence on Prosite (http://prosite.expasy.org/) predicted the protein to possess the conserved catalytic domain amino acid signature H-D-I-g-H-f-G-r-t-N-m-F for PDEs (16). To determine the stage of the P. yoelii life cycle during which PDEγ is expressed, RNA was extracted from P. yoelii 17XNL strain mixed blood stages (BS), oocyst and salivary gland sporozoites isolated from mosquitoes, and liver samples collected from BALB/cJ mice 24 h and 44 h after injection with salivary gland sporozoites. Complementary DNA was synthesized, and reverse transcriptase PCR (RT-PCR) was performed using PDEγ cDNA-specific primers. PDEγ transcript was detected in mixed blood stages (BS), oocyst sporozoites, and salivary gland sporozoites (Fig. 1A). To analyze protein expression, the endogenous copy of P. yoelii PDEγ was replaced with a tagged version encoding PDEγ with four C-terminal copies of the c-Myc (EQKLISEEDL) epitope. Immunofluorescence assays showed PDEγ expression to be low in BS (Fig. 1B) and high in salivary gland sporozoites (Fig. 1C). The expression in BS appeared internal and partially colocalized with the endoplasmic reticulum (ER) marker BiP (Fig. 1B). The protein also displayed an intracellular localization in sporozoites (Fig. 1C).
FIG 1 .
Expression analysis of P. yoelii PDEγ by RT-PCR and immunofluorescence assay. (A) RT-PCR for P. yoelii PDEγ transcripts in mixed blood stages (mBS), oocyst sporozoites (Oo-spz), and salivary gland sporozoites (Sg-spz) of P. yoelii WT parasites. 18S rRNA of P. yoelii (Py18S) was used as a positive control. + or − indicates cDNA synthesis with or without reverse transcriptase, respectively. (B) Immunofluorescence assay of mixed blood stages stained with anti-Myc antibody and costained with antibody against either the ER marker BiP (top panel) or the parasite plasma membrane marker MSP1 (bottom panel). (C) Immunofluorescence assay of salivary gland sporozoites stained with anti-Myc antibody and costained with antibody against either the apicoplast marker ACP (top panel), sporozoite surface marker CSP (middle panel), or IMC marker TRAP (bottom panel). Nucleus was visualized using DAPI. Bars, 2.5 µm. DIC, differential interference contrast.
PDEγ deletion partially affects blood-stage growth.
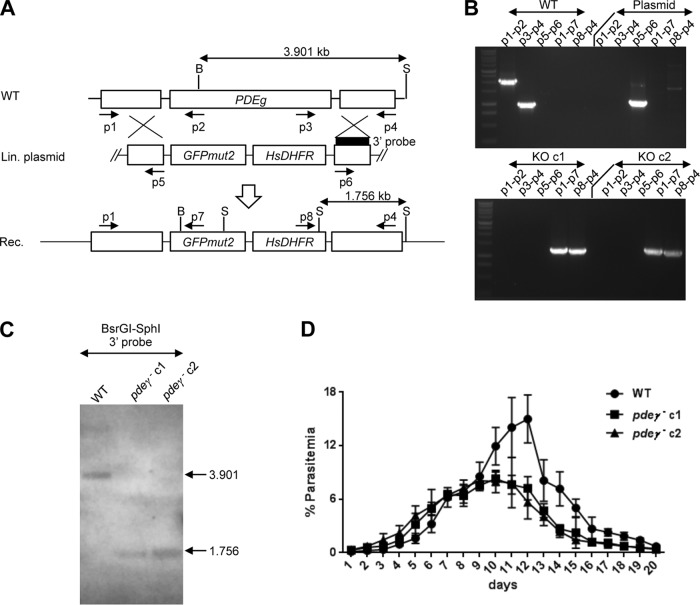
To analyze whether PDEγ is important for parasite life cycle progression, a double-crossover homologous recombination strategy was used to delete PDEγ in P. yoelii (Fig. 2A). The gene was not refractory to deletion, and several clones of knockout (pdeγ−) parasites were successfully generated, showing that PDEγ was not essential for BS replication. Two clones of pdeγ− parasites were selected and genotyped by PCR (Fig. 2B) and Southern blotting (Fig. 2C) to confirm the purity of the clones. A BS growth assay was performed to determine whether there was a difference in the kinetics of growth between wild-type (WT) and pdeγ− parasites. Swiss Webster (SW) mice were injected intravenously (i.v.) with WT and pdeγ− clones, and parasitemia was analyzed daily by microscopic examination of Giemsa-stained thin blood smears for 20 days. Parasitemias were comparable between WT and both pdeγ− clones during the initial phase of BS growth (from days 1 through 9) (Fig. 2D). However, WT and pdeγ− parasites differed in growth kinetics from days 10 through 15. The peak average parasitemia in mice infected with WT parasites was ~16%, which was ~2-fold higher than the peak average parasitemia of ~8% in mice infected with the pdeγ− clones. During later stages of growth (day 12 onward), the pdeγ− clones were cleared from mice earlier than WT parasites (Fig. 2D). These data show that PDEγ is not essential for BS parasite growth and replication.
FIG 2 .
Deletion of P. yoelii PDEγ and characterization of pdeγ− parasites. (A) Schematic of the strategy for deleting PDEγ in P. yoelii 17XNL by homologous recombination using a linearized plasmid. Primers used for genotyping PCR and enzymes (B, BsrGI; S, SphI) and probe (black bar) used for Southern blotting are shown. Sizes of genomic DNA fragments distinguishing WT from knockout clones are indicated in kilobases. (B) Genotyping PCR of two pdeγ− clones (c1 and c2) with WT and plasmid controls. (C) Southern blotting of AflIII-digested genomic DNA from WT and two pdeγ− clones with a 3′ probe yielded a 3.901-kb band for WT and a 1.756-kb band for the two pdeγ− clones, respectively. (D) Comparison of asexual blood-stage growth rates between WT and pdeγ− sporozoites as measured by increase in parasitemia in infected mice over time. Parasitemias are plotted as means ± standard deviations.
PDEγ deletion abrogates sporozoite infection of the mosquito salivary glands.
Next, we analyzed whether there were differences in gametocytogenesis and mosquito infections between WT and pdeγ− parasites. No significant difference (data not shown) was observed. Mosquitoes were then allowed to feed on mice infected with WT or pdeγ− parasites, and oocyst sporozoites and salivary gland sporozoites were enumerated on days 10 and 14 postfeeding, respectively. No statistically significant difference was observed for the number of oocyst sporozoites per mosquito between WT (48,942 ± 17,497) and pdeγ− clones c1 (36,112 ± 16,521; P = 0.6058 compared to WT) and c2 (32,202 ± 18,200; P = 0.5232 compared to WT) (Fig. 3A). In contrast, salivary gland sporozoite numbers in mosquitoes that were infected with pdeγ− clones were on average ~55-fold lower (304 ± 110 for c1, P = 0.0144 versus WT, and 358 ± 143 for c2, P = 0.0146 versus WT) than salivary gland sporozoite numbers from mosquitoes that were infected with WT parasites (18,114 ± 5,233) (Fig. 3B). These data show that pdeγ− sporozoites failed to invade the salivary glands of mosquitoes.
FIG 3 .
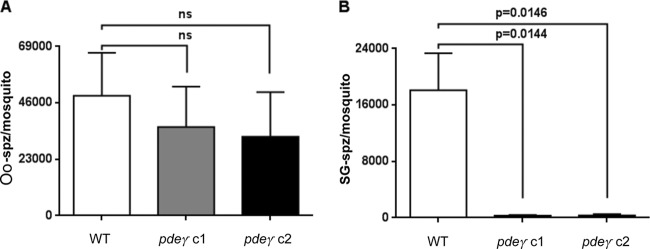
Quantification of oocyst sporozoites and salivary gland sporozoites. (A) Oocyst sporozoite numbers from mosquitoes infected with WT or pdeγ− clones on day 10 after infectious blood meal. (B) Salivary gland sporozoite numbers from mosquitoes infected with WT or pdeγ− clones on day 14 after infectious blood meal. Both oocyst and salivary gland sporozoite numbers were determined multiple times with independent infections. Bar graphs represent means ± standard errors of the means. Unpaired t test with Welch’s correction was used for statistical analysis.
PDEγ deletion blocks sporozoite transmission by mosquito bite.
Given the apparent failure of pdeγ− sporozoites to invade the mosquito salivary glands, we tested whether they were transmissible to mice through mosquito bite. An initial experiment with 20 pdeγ− sporozoite-infected mosquitoes per mouse did not result in patent BS infection in any mice (Table 1). Strikingly, no BS patency was observed even with 45 or 100 pdeγ− sporozoite-infected mosquito bites per mouse. In contrast, WT parasite-infected mosquito bites consistently caused patent BS infection in all mice (Table 1). We next set out to determine whether pdeγ− sporozoites that were associated with mosquito salivary glands could cause infection in mice when delivered intravenously. Injection of 1,000 pdeγ− salivary gland-associated sporozoites did not result in BS parasitemia in mice, whereas all mice injected with WT salivary gland sporozoites became patent, as expected (Table 2). However, injection of a higher dose of 10,000 pdeγ− salivary gland-associated sporozoites caused blood-stage patency in a fraction of the mice (~63%) around day 4. In comparison, 100% of 10,000 WT sporozoite-injected mice became patent around day 3 (Table 2). We also compared the infectivity with 10,000 WT and pdeγ− sporozoites extracted from the mosquito hemolymph by intravenous injection. Once again, only a fraction of mice (40%) injected with pdeγ− sporozoites developed blood-stage patency, whereas 100% of mice injected with WT sporozoites became BS patent. As with salivary gland sporozoites, pdeγ− hemolymph sporozoites caused patency with a 1-day delay compared to WT (day 4.5 for pdeγ− sporozoites versus day 3.5 for WT sporozoites) (Table 2).
TABLE 1 .
Infections of BALB/cJ mice with WT or pdeγ− parasites via mosquito bite
| No. of mosquitoes/mouse | Genotype | No. of mice/no. patent |
|---|---|---|
| ~20 | WT | 3/3 |
| pdeγ− c1 | 3/0 | |
| pdeγ− c2 | 3/0 | |
| ~45 | WT | 3/3 |
| pdeγ− c1 | 3/0 | |
| pdeγ− c2 | 3/0 | |
| ~100 | WT | 5/3 |
| pdeγ− c1 | 5/0 | |
| pdeγ− c2 | 5/0 |
TABLE 2 .
Infections of BALB/cJ mice with WT or pdeγ− parasites via intravenous sporozoite injectionsa
| Sporozoite dose | Sporozoite source | Genotype | No. of mice/no. patent | No. of days to patency |
|---|---|---|---|---|
| 1000 | SG | WT | 6/6 | 4.3 |
| SG | pdeγ− c1 | 6/0 | NP | |
| SG | pdeγ− c2 | 6/0 | NP | |
| 10,000 | HL | WT | 5/5 | 3.5 |
| HL | pdeγ− | 5/2 | 4.5 | |
| SG | WT | 6/6 | 3 | |
| SG | pdeγ− | 8/5 | 4.2 |
Abbreviations: HL, hemolymph; SG, salivary gland; NP, not patent.
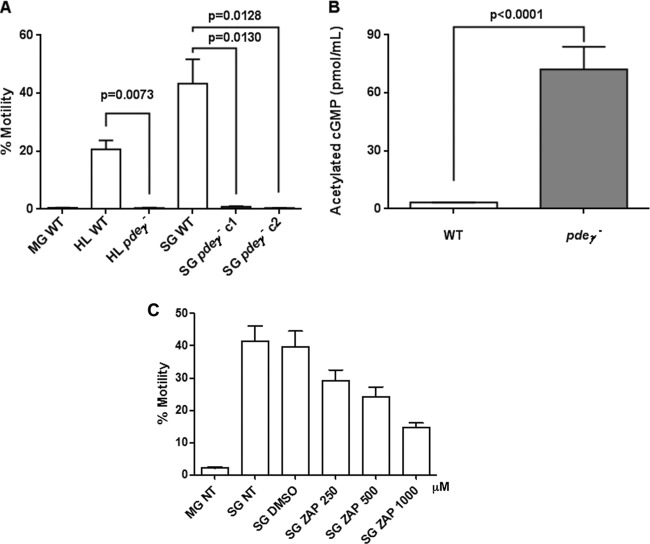
pdeγ− salivary gland sporozoites are defective in substrate-dependent gliding motility.
We hypothesized that the failure of pdeγ− sporozoites to invade mosquito salivary glands might be due to a defect in their motility. The average frequency of motility exhibited by WT salivary gland sporozoites was ~43% ± 8%. Strikingly, pdeγ− salivary gland-associated sporozoites were nearly immotile. The percentages of sporozoites with circumsporozoite protein (CSP) trails for pdeγ− clones were 0.8% ± 0.2% and 0.4% ± 0.07%, on average ~72-fold lower than those for WT salivary gland sporozoites (P = 0.0130 for pdeγ− c1 versus WT and P = 0.0128 for pdeγ− c2 versus WT) (Fig. 4A). This near lack of motility was comparable to WT oocyst sporozoites, which are known to display little motility (~0.5% ± 0.1%). To more accurately compare motility of sporozoites, we collected hemolymph sporozoites from mosquitoes infected with either WT or pdeγ− parasites. While ~21% (20.67% ± 1.76%) of WT hemolymph sporozoites generated CSP trails and thus were motile, no motility was observed with pdeγ− hemolymph sporozoites. The percentage of pdeγ− hemolymph sporozoites with CSP trails was 0.35% ± 0.08%, on average ~60-fold lower than (P = 0.0073 versus WT) that of WT hemolymph sporozoites (Fig. 4A).
FIG 4 .
Sporozoite motility assay and quantification of cGMP level in sporozoites. (A) Percentages of day 10 WT oocyst (MG), day 12 WT and pdeγ− hemolymph (HL), and day 14 WT and pdeγ− salivary gland (SG) sporozoites to assess substrate-dependent motility by deposition of CSP trails. Motility was assessed multiple times with independent sporozoite preparations. (B) Acetylated cGMP levels in extracts prepared from day 10 WT and pdeγ− oocyst sporozoites. Bar graphs represent means ± standard errors of the means. Unpaired t test with Welch’s correction was used for statistical analysis. (C) Percentages of day 10 WT oocyst (MG NT) and day 14 salivary gland WT (SG NT) salivary gland sporozoites to assess substrate-dependent motility compared to day 14 salivary gland sporozoites treated with zaprinast (ZAP), a PDE inhibitor at different concentrations. SG DMSO, salivary gland sporozoites treated with dimethyl sulfoxide (DMSO) as a control for zaprinast solvent.
pdeγ− sporozoites exhibit elevated cGMP levels.
To address whether cyclic nucleotide levels were perturbed in pdeγ− parasites, cGMP concentration was measured in WT and pdeγ− oocyst sporozoites. The concentration of cGMP was found to be ~18-fold higher (P < 0.0001) in pdeγ− sporozoites (72 ± 12 pmol/ml) than in WT parasites (4 ± 0.1 pmol/ml) (Fig. 4B). In order to assess whether inhibiting PDE would have an effect on the motility of salivary gland sporozoites, we used zaprinast (an inhibitor of PDE that hydrolyzes cGMP) in the motility assay. At 250, 500, and 1,000 µM, zaprinast caused 28, 37, and 43% decreases in sporozoite motility, respectively, compared to control (Fig. 4C). Together, the data suggest that PDEγ regulates cyclic GMP levels, which in turn regulate signaling events that control sporozoite motility.
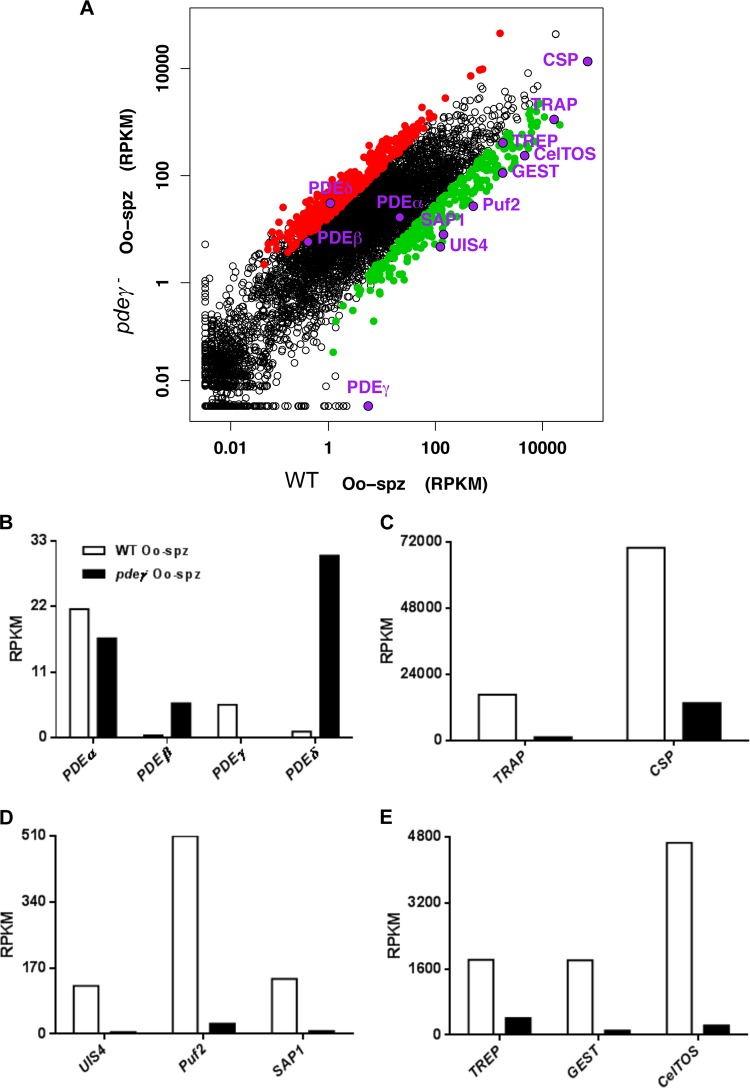
Transcript abundance for proteins involved in motility and invasion is downregulated in pdeγ− sporozoites.
Given the involvement of PDEs in regulating signaling and gene expression, we performed RNA-Seq analysis to analyze the effect of PDEγ deletion on transcript abundance in sporozoites. RNA was extracted from day 10 WT and pdeγ− oocyst sporozoites (three independent mosquito infections were used to produce sporozoites that were pooled for RNA-Seq) that had been purified using DE52 columns to remove mosquito debris (17). Differentially expressed genes were determined (at least 2-fold, with Bonferroni-corrected P value of <0.05) (Fig. 5A). As expected, no PDEγ transcript was detected in pdeγ− sporozoites (Fig. 5B). Interestingly, expression analysis of other cyclic nucleotide PDEs in pdeγ− sporozoites showed upregulation of PDEδ and PDEβ, respectively (as determined by comparing normalized RPKM [read per kilobase per million] values), whereas PDEα levels were relatively unaffected (Fig. 5B). Given the pdeγ− sporozoite defects in gliding motility and mosquito salivary gland invasion as well as mammalian host liver infection, we also analyzed transcript levels of genes known to be involved in sporozoite motility and invasion. We found transcripts for TRAP and CSP to be downregulated ~15-fold and ~5-fold, respectively (Fig. 5C). Other transcripts of relevance that were downregulated in pdeγ− sporozoites included UIS4 (~26-fold), Puf2 (~19-fold), and SAP1 (~18-fold) (Fig. 5D), TREP (~4.5-fold), GEST (~16-fold), and CelTOS (~20-fold) (Fig. 5E). RPKM values for all transcripts are shown in Table S1 in the supplemental material.
FIG 5 .
Comparison of transcript abundance (shown as reads per kilobase per million [RPKM] values) for representative genes in P. yoelii WT and pdeγ− oocyst sporozoites. (A) MA plot (or scatter plot) of comparative expression of 6,053 P. yoelii genes from PDEγ− and wild-type (WT) samples. Significantly differentially expressed genes (at least 2-fold, with Bonferroni-corrected P value of <0.05) are highlighted in color: 428 genes upregulated in PDEγ− sporozoites shown in red, 271 genes upregulated in WT shown in green. (B to E) Comparisons of transcript abundances: PDEα, PDEβ, PDEγ, and PDEδ (B); TRAP and CSP (C); UIS4, Puf2, and SAP1 (D); and TREP, GEST, and CelTOS (E).
DISCUSSION
Previous studies have indicated the importance of cyclic nucleotide-based signaling in Plasmodium parasites, including a role of cAMP in gametocyte biology (12, 13, 18), asexual blood-stage cell cycle synchronization (14), and merozoite egress (19). In this report, we have demonstrated that P. yoelii PDEγ is a cGMP-capable phosphodiesterase which is critical for maintaining cGMP balance in sporozoites. Deleting PDEγ dramatically increased cGMP levels, rendering sporozoites immotile and unable to invade the mosquito salivary glands. Consequently, the pdeγ− parasites failed to transmit to the mammalian host by mosquito bite.
PDEγ transcripts were detectable by RT-PCR in blood stages, oocyst sporozoites, and salivary gland sporozoites. A previous proteomic analysis detected P. yoelii PDEγ in whole-cell lysates of WT salivary gland sporozoites (20). Expression was demonstrated here by immunofluorescence analysis of Myc-tagged PDEγ salivary gland sporozoites, revealing intracellular PDEγ expression with a granular distribution. PDEγ also showed weak expression in asexual blood stages. Deletion of PDEγ reduced peak blood-stage parasitemia, which suggests a nonessential role of PDEγ during blood-stage growth/replication. It is possible that other PDEs partially compensated for the loss of PDEγ, enabling pdeγ− parasites to maintain asexual blood-stage growth. In this context, it is of note that P. falciparum PDEγ can also be deleted, but an effect on blood-stage growth was not reported (21). In P. falciparum blood stages, PDEα and PDEβ are the predominant PDE transcripts (22), but the essentiality for blood-stage replication has been determined only for PDEα. PDEδ is not essential for P. falciparum (21) and P. berghei (15) blood-stage replication. Other PDE family members may complement single PDE gene deletions, although no changes in expression levels of PDEs were observed upon deleting PDEα in P. falciparum (22).
We found that deleting P. yoelii PDEγ did not affect gametocytogenesis, exflagellation of male gametes, oocyst development, and oocyst sporozoite differentiation. This observation is in agreement with an earlier report in which P. berghei PDEγ deletion mutants were viable with no discernible phenotype up to and including the oocyst stage (15). In stark contrast to normal oocyst sporozoite development, pdeγ− sporozoite salivary gland infection was severely affected, suggesting that PDEγ is essential for parasite entry into the glands. In addition, pdeγ− sporozoites failed to transmit to the mammalian host via mosquito bite. Interestingly, artificial transmission by intravenous injection of large numbers of pdeγ− hemolymph or salivary gland sporozoites resulted in blood-stage patency in a fraction of challenged mice, but the mice that became infected did so with delayed patency. A similar severe-infection phenotype was previously observed with P. berghei trap− sporozoites, and the reason for a fraction of animals becoming patent was attributed to stochastic events such as uptake by host cells of noninvasive sporozoites (23). Similar events may explain the breakthrough infections observed with pdeγ− sporozoites.
It has been hypothesized that plasmodial PDEs are structurally and catalytically more adept at metabolizing cGMP rather than cAMP (24), and biochemical assays have indeed demonstrated that Plasmodium PDEα and PDEδ have a specificity for cGMP (22, 25), whereas PDEβ likely has dual specificity (22, 25). The cyclic nucleotide specificity of PDEγ, however, has not previously been defined, and we surmised that cyclic nucleotide levels in pdeγ− sporozoites were perturbed. Indeed, cGMP levels in P. yoelii pdeγ− sporozoites were dramatically elevated compared to WT parasites. These data indicate that PDEγ has strong capability for hydrolyzing cGMP. Inhibition of sporozoite motility by zaprinast, a cGMP-specific PDE inhibitor, is in agreement with this notion.
Cyclic GMP is a key regulator of several physiological processes, including regulation of gene expression at both transcriptional and posttranscriptional levels (26). The latter includes splicing, mRNA stability, and translation (26). By dampening cyclic nucleotide signal transduction, PDEs control cyclic nucleotide-based regulation of gene expression (26–28). Thus, we assessed whether the perturbation of cGMP levels in pdeγ− oocyst sporozoites affected gene expression on a global level by RNA-Seq comparisons of WT and pdeγ− sporozoites. We initially examined PDE transcription and found that PDEδ and PDEβ transcript levels were upregulated in pdeγ− sporozoites although PDEα transcript levels were unaffected. In light of the strong defect in salivary gland invasion, we next determined whether deleting PDEγ altered the levels of transcripts encoding proteins involved in this process and interestingly found a downregulation of TRAP, CSP, and TREP transcripts in pdeγ− sporozoites. This downregulation could be the functional cause for the lack of pdeγ− sporozoite invasion into salivary glands. TRAP, CSP, and TREP are also important for sporozoite motility (29). Thus, reduction of transcript abundance for these genes in pdeγ− sporozoites would affect motility. Indeed, pdeγ− sporozoites were defective in substrate-dependent gliding motility and CSP shedding. These data are in agreement with the roles of TRAP, CSP, and TREP and point to a critical role for PDEγ in sporozoite invasion and motility via regulating gene expression for key sporozoite invasion and motility-related proteins.
Interestingly, transcript abundance for UIS4, Puf2, and SAP1, which encode proteins that are critical for sporozoite infectivity of the mammalian host, was also strongly diminished in pdeγ− sporozoites. UIS4 is expressed in infective sporozoites and liver stages, and deleting UIS4 causes impaired liver-stage development (30). Deleting Puf2 results in premature initiation of sporozoite transformation into liver stages in mosquito salivary glands and reduced sporozoite gliding motility, cell traversal, and hepatocyte infectivity (31). P. yoelii SAP1 is essential for a productive liver-stage infection, and deleting SAP1 causes the downregulation of several sporozoite transcripts, including UIS4 and Puf2 but not CSP and TRAP (32). Both SAP1 and Puf2 are involved in the storage and protection of mRNAs that code for proteins critical in mammalian host infection. We showed that deleting P. yoelii PDEγ leads to perturbed cGMP levels in sporozoites. Given the role of cGMP in regulating gene expression at the transcriptional and posttranscriptional levels (26), uninhibited signaling downstream of cGMP in pdeγ− sporozoites could explain the perturbations and phenotypes that we observed.
In light of the elevated cGMP levels and the resulting lack of motility of pdeγ− sporozoites, it is noteworthy that the interaction of WT sporozoites with albumin causes a spike in intracellular levels of cAMP (and Ca2+) and the release of the micronemal proteins TRAP and CSP, necessary for motility (7). If cAMP formation is inhibited, however, motility is lost (7). Thus, it is possible that cAMP- and cGMP-based signaling pathways perform opposing functions in the sporozoite and that maintaining a balance between cAMP and cGMP levels is critical for sporozoite motility and infectivity. Further experiments are required to assess whether cAMP levels are perturbed in pdeγ− sporozoites and whether there is cross talk between cAMP and cGMP signaling.
Our data indicate that PDEγ plays a critical role in maintaining cGMP balance in sporozoites. Skewing this balance causes downregulation of transcripts that code for proteins involved in sporozoite motility and invasion. Currently, there is a knowledge gap concerning the specific parasite-host interactions that activate downstream cyclic nucleotide/PDE-dependent signaling pathways. Further experiments are required to understand how cGMP-mediated signaling and PDE activity translate external signals into intracellular gene regulatory responses in malaria parasites.
MATERIALS AND METHODS
Experimental animals and parasite production.
Six- to 8-week-old female Swiss Webster (SW) mice from Harlan (Indianapolis, IN) were used for production of transgenic parasites and for parasite life cycle maintenance. Six- to 8-week-old female BALB/cJ mice from the Jackson Laboratory (Bar Harbor, ME) were used for assessment of parasite infectivity. P. yoelii 17XNL nonlethal WT and transgenic parasites were cycled between SW mice and Anopheles stephensi mosquitoes. Infected mosquitoes were maintained on sugar water at 24°C and 70% humidity. This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (42). The Seattle Biomedical Research Institute has an OLAW Animal Welfare Assurance (A3640-01). The protocol was approved by Seattle BioMed’s Institutional Animal Care and Use Committee.
Generation of pdeγ− parasites.
Gene targeting constructs for transgenic parasite production were designed as previously described (33). P. yoelii 17XNL genomic DNA was used to amplify a 658-bp fragment of the 3′ untranslated region (UTR) using oligonucleotide primers 5′ GCGAGCTCGGTACCTATGCGTATAATATTATATGAATAAGAC (sense) and 5′ CATGATATATAACCCTGCAGGTTAACTTGTTTTTATGGAAATTTAAAACC (antisense) and a 566-bp fragment of the 5′ UTR with primers 5′ CATAAAAACAAGTTAACCTGCAGGGTTATATATCATGGAATTTCGTTGCAC (sense) and 5′ ATGCGGCCGCTATCGTTTGACACGAATAAATTTAATCG (antisense) of P. yoelii PDEγ (PyPDEγ). The two PCR products were fused by sequence overlap extension PCR (SOE PCR) (33). The SOE PCR product was cloned into pCR-Blunt (Life Technologies), sequenced, digested with KpnI and NotI, gel purified using a gel extraction kit (Qiagen), and cloned into a modified version of plasmid pL0005 (MR4: MRA-774) containing GFPmut2 under the control of the constitutive P. berghei elongation factor 1 alpha promoter. The final plasmid was linearized with SbfI. Transfection of P. yoelii 17XNL parasites using the Amaxa Nucleofector device (Lonza) and selection of resistant parasites were conducted as previously described (34).
Genotyping of transgenic parasites.
Transgenic parasites were cloned by limiting dilution infection of female SW mice, and two independent clones were selected for phenotypic analysis. The presence of transgenic parasites was assessed by genotyping PCR using primers 5′ TTCAATATTTGTAGTTGATAGTTTTTGC (sense) and 5′ AAACATGTTTGTAAACATTTGTTAATATC (antisense) for the 5′ end and primers 5′ TAACCCATTATTTGATCGAAAAGCTC (sense) and 5′ GCAAAAATGCTCAAACCAAACATTGG (antisense) for the 3′ end of the WT locus, primers 5′ CAACTCCAGTGAAAAGTTCTTCTCC (sense) and 5′ AAACATGTTTGTAAACATTTGTTAATATC (antisense) for the 5′ end and primers 5′ TAAGTACAAATTTGAAGTATATGAGAAG (sense) and 5′ AAACGAAAAACTATTATAAAGTATATACG (antisense) for the 3′ end of the pdeγ− locus, and primers 5′ GCGAGCTCGGTACCTATGCGTATAATATTATATGAATAAGAC (sense) and 5′ ATGCGGCCGCTATCGTTTGACACGAATAAATTTAATCG (antisense) for the episomal plasmid. Southern blotting for the PDE locus was performed by hybridizing BsrGI-SphI-digested genomic DNA from WT and pdeγ− clones with a 3′ probe generated using primers 5′ GCGAGCTCGGTACCTATGCGTATAATATTATATGAATAAGAC (sense) and 5′ CATGATATATAACCCTGCAGGTTAACTTGTTTTTATGGAAATTTAAAACC (antisense). Digested DNA was run on a 0.7% Tris-acetate-EDTA agarose gel at 55 V and transferred to a Hybond-N membrane (Amersham, GE Healthcare Life Sciences, Pittsburgh, PA) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight at room temperature. DNA was UV cross-linked to the membrane and hybridized with digoxigenin (DIG)-labeled probes prepared using the DIG kit (Roche Diagnostics, Indianapolis, IN).
Blood-stage growth assay.
Groups of three SW mice each were infected intravenously with 1 × 10e6 infected red blood cells (iRBCs) of WT and two pdeγ− clones. Blood smears were prepared and stained with Giemsa stain, and parasitemia was checked each day for 20 days.
Reverse transcriptase PCR (RT-PCR).
Samples for RNA extraction were stored in TRIzol (Life Technologies) at −80°C until used. Total RNA was extracted using the Direct-zol MiniPrep kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. Complementary DNA synthesis was performed using the QuantiTect reverse transcription kit (Qiagen) according to the manufacturer’s instructions. PCR cycling conditions used for amplification of cDNA were 92°C for 30 s for DNA denaturation, 54°C for 30 s for primer annealing, and 62°C for 1 min for extension (35 cycles). P. yoelii PDEγ was amplified using primers 5′ TTAAGGAAAAAGACGAAGAAACTCTG (sense) and 5′ GGATCTATTACCAATTGTTGTAAACG (antisense). P. yoelii 18S rRNA was amplified using primers 5′ GGGGATTGGTTTTGACGTTTTTGCG (sense) and 5′ AAGCATTAAATAAAGCGAATACATCCTTAT (antisense).
Epitope tagging.
The tagging construct was designed to replace the endogenous locus with the tagged version of P. yoelii PDEγ by double-crossover homologous recombination. P. yoelii 17XNL genomic DNA was used to amplify a 669-bp fragment of the 3′ end of the coding sequence without the stop codon of P. yoelii PDEγ using oligonucleotide primers 5′ GATAAATGACAAATTTACGGCCGAATCAATATTAGAGAATTATCATTGCTC (sense) and 5′ ATACTAGTTAATTTATATATATTAAGATTTGGTGCATAAAC (antisense) and a 630-bp fragment of the 3′ UTR with primers 5′ ATCCGCGGCATGGAAAATTGTTTATGCACCAAATC (sense) and 5′ CTAATATTGATTCGGCCGTAAATTTGTCATTTATCATATATATACATG (antisense). The two PCR products were fused by sequence overlap extension PCR (SOE PCR) (33). The SOE PCR product was cloned into pCR-Blunt (Life Technologies), sequenced, digested with SacII and SpeI, gel purified using a gel extraction kit (Qiagen), and cloned into a modified version of plasmid pL0005 (MR4: MRA-774), pL0005-cMyc, which allowed tagging of proteins with a C-terminal quadruple Myc (4× Myc) tag. The final plasmid was linearized with EagI. Transfection of P. yoelii 17XNL parasites and selection of resistant parasites were conducted as previously described (33).
Immunofluorescence assays.
Oocyst or salivary gland sporozoites were isolated by microdissection and fixed with 4% paraformaldehyde (PFA) for 15 min. Sporozoites were washed twice with phosphate-buffered saline (PBS), permeabilized with 0.1% Triton X-100 in PBS for 15 min, and blocked with 3% bovine serum albumin (BSA) in PBS for 1 h. Sporozoites were stained with antibodies against BiP, acyl carrier protein (ACP), CSP (2F6), TRAP, or c-Myc (SC-789; Santa Cruz) for 1 h. Sporozoites were washed and incubated with fluorophore-conjugated secondary antibodies specific to rabbit or mouse IgG for 1 h. Sporozoites were then washed and stained for the DNA with DAPI (4′,6-diamidino-2-phenylindole) for 5 min. Sporozoites were applied to glass slides and mounted with antifade reagent (Vectashield; Vector Laboratories). All steps were performed at room temperature. Images were acquired using an OlympusIX70 DeltaVision microscope equipped with deconvolution software.
Analysis of sporozoite motility.
Sporozoites were tested for in vitro substrate-dependent motility using coverslips precoated with anti-circumsporozoite protein (anti-CSP) antibodies. Motility was assessed by determining the percentage of sporozoites that were able to generate CSP trails. For zaprinast inhibitor (Sigma) experiments, the inhibitor was resuspended in dimethyl sulfoxide (DMSO) (Sigma) and added during the gliding assay.
Analysis of sporozoite infectivity by mosquito bite infection.
Groups of BALB/cJ mice (3 to 5 per group) were anesthetized and individually exposed to the bites of 20 to 100 WT or pdeγ− sporozoite-infected mosquitoes. Mosquitoes were allowed to feed for a total of 7.5 min, with rotation of the mice between five mosquito cages every 1.5 min. Before performing mosquito bite infections, salivary glands of mosquitoes were checked for the presence of sporozoites. The time to blood-stage patency was determined microscopically by Giemsa-stained thin blood smears. All mice were checked either until patency was observed or for 14 days, whichever was earlier.
Analysis of sporozoite infectivity by i.v. injection.
Sporozoites were isolated by microdissecting mosquito salivary glands 14 or 15 days after the infectious blood meal. Sporozoites were injected intravenously (i.v.) into the tail vein of BALB/cJ mice. The time to blood-stage patency (defined as >1 infected erythrocyte/10,000 erythrocytes) was determined microscopically using Giemsa-stained thin blood smears. All mice were checked either until patency was observed or for 14 days, whichever was earlier.
cGMP assay.
The assay for determining cGMP levels in sporozoites was performed using the cyclic GMP enzyme immunoassay (EIA) kit (catalog no. 581021; Cayman Chemical) per the manufacturer’s instructions. Sporozoites for the assay were purified on an Accudenz gradient to eliminate mosquito debris (17), and sporozoite extracts were prepared by two rounds of freezing on dry ice-ethanol, thawing on ice, and passaging through a 28-gauge needle from the same number of sporozoites for each line. Equal volumes of extract from WT and pdeγ− sporozoites were used to assay for cGMP.
Transcription abundance from RNA-Seq sequencing data.
Raw FASTQ read data were processed using in-house R package DuffyNGS as originally described (35). Briefly, raw reads pass through a 3-stage alignment pipeline: (i) a prealignment stage to filter out unwanted transcripts, such as rRNA, mitochondrial RNA, albumin, and globin; (ii) a main genomic alignment stage against the genome(s) of interest; and (iii) a splice junction alignment stage against an index of standard and alternative exon splice junctions. All alignments were performed with Bowtie2 (36), using the command line option “very-sensitive.” BAM files from stages 2 and 3 are combined into read depth wiggle tracks that record both uniquely mapped and multiply mapped reads to each of the forward and reverse strands of the genome(s) at single-nucleotide resolution. Multiply mapped reads are prorated over all highest-quality aligned locations. Gene transcript abundance is then measured by summing total reads landing inside annotated gene boundaries, expressed as both RPKM (37) and raw read counts. Two stringencies of gene abundance are provided using all aligned reads and by just counting uniquely aligned reads.
Differential transcription.
To minimize biases from the choice of algorithm for calling differential expressed (DE) genes, a panel of 5 DE tools was utilized. They included (i) RoundRobin (in-house); (ii) RankProduct (38); (iii) significance analysis of microarrays (SAM) (39); (iv) EdgeR (40); and (v) DESeq (41) (see Table S1 in the supplemental material). Each DE tool was called with appropriate default parameters and operated on the same set of transcription results, using RPKM abundance units for RoundRobin, RankProduct, and SAM and raw read count abundance units for DESeq and EdgeR. All 5 DE results were then synthesized, by combining gene DE rank positions across all 5 DE tools. Specifically, a gene’s rank position in all 5 results was averaged, using a generalized mean to the 1/2 power, to yield the gene’s final net rank position. Each DE tool’s explicit measurements of differential expression (fold change) and significance (P value) were similarly combined via appropriate averaging (arithmetic and geometric mean, respectively). The final DE result was sorted by gene net rank position such that the top genes were those found in common by all DE tools.
SUPPLEMENTAL MATERIAL
RPKM values for all transcripts
ACKNOWLEDGMENTS
This research was funded by a grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative (grant 1481).
The authors declare no conflict of interest.
Footnotes
Citation Lakshmanan V, Fishbaugher ME, Morrison B, Baldwin M, Macarulay M, Vaughan AM, Mikolajczak SA, Kappe SHI. 2015. Cyclic GMP balance is critical for malaria parasite transmission from the mosquito to the mammalian host. mBio 6(2):e02330-14. doi:10.1128/mBio.02330-14.
REFERENCES
- 1.World Health Organization, Malaria Control Department . 2013. Malaria report 2013. World Health Organization, Geneva, Switzerland: http://www.who.int/malaria/publications/world_malaria_report_2013/en. [Google Scholar]
- 2.Greenwood BM, Fidock DA, Kyle DE, Kappe SH, Alonso PL, Collins FH, Duffy PE. 2008. Malaria: progress, perils, and prospects for eradication. J Clin Invest 118:1266–1276. doi: 10.1172/JCI33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh AK, Jacobs-Lorena M. 2009. Plasmodium sporozoite invasion of the mosquito salivary gland. Curr Opin Microbiol 12:394–400. doi: 10.1016/j.mib.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Zhang Y, Zhao YO, Li MW, Zhang L, Dragovic S, Abraham NM, Fikrig E. 2013. Anopheles gambiae circumsporozoite protein-binding protein facilitates plasmodium infection of mosquito salivary glands. J Infect Dis 208:1161–1169. doi: 10.1093/infdis/jit284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez MH, Hernández-Hernández FDLC. 2004. Insect-malaria parasites interactions: the salivary gland. Insect Biochem Mol Biol 34:615–624. doi: 10.1016/j.ibmb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Pimenta PF, Touray M, Miller L. 1994. The journey of malaria sporozoites in the mosquito salivary gland. J Eukaryot Microbiol 41:608–624. doi: 10.1111/j.1550-7408.1994.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 7.Kebaier C, Vanderberg JP. 2010. Initiation of plasmodium sporozoite motility by albumin is associated with induction of intracellular signalling. Int J Parasitol 40:25–33. doi: 10.1016/j.ijpara.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono T, Cabrita-Santos L, Leitao R, Bettiol E, Purcell LA, Diaz-Pulido O, Andrews LB, Tadakuma T, Bhanot P, Mota MM, Rodriguez A. 2008. Adenylyl cyclase alpha and cAMP signaling mediate plasmodium sporozoite apical regulated exocytosis and hepatocyte infection. PLoS Pathog 4:e1000008. doi: 10.1371/journal.ppat.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conti M, Beavo J. 2007. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem 76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 10.Beghyn TB, Charton J, Leroux F, Henninot A, Reboule I, Cos P, Maes L, Deprez B. 2012. Drug-to-genome-to-drug, step 2: reversing selectivity in a series of antiplasmodial compounds. J Med Chem 55:1274–1286. doi: 10.1021/jm201422e. [DOI] [PubMed] [Google Scholar]
- 11.Beghyn TB, Charton J, Leroux F, Laconde G, Bourin A, Cos P, Maes L, Deprez B. 2011. Drug to genome to drug: discovery of new antiplasmodial compounds. J Med Chem 54:3222–3240. doi: 10.1021/jm1014617. [DOI] [PubMed] [Google Scholar]
- 12.Martin SK, Miller LH, Nijhout MM, Carter R. 1978. Plasmodium gallinaceum: induction of male gametocyte exflagellation by phosphodiesterase inhibitors. Exp Parasitol 44:239–242. doi: 10.1016/0014-4894(78)90104-2. [DOI] [PubMed] [Google Scholar]
- 13.Trager W, Gill GS. 1989. Plasmodium falciparum gametocyte formation in vitro: its stimulation by phorbol diesters and by 8-bromo cyclic adenosine monophosphate. J Protozool 36:451–454. doi: 10.1111/j.1550-7408.1989.tb01079.x. [DOI] [PubMed] [Google Scholar]
- 14.Beraldo FH, Almeida FM, da Silva AM, Garcia CR. 2005. Cyclic AMP and calcium interplay as second messengers in melatonin-dependent regulation of Plasmodium falciparum cell cycle. J Cell Biol 170:551–557. doi: 10.1083/jcb.200505117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon RW, Taylor CJ, Bex C, Schepers R, Goulding D, Janse CJ, Waters AP, Baker DA, Billker O. 2009. A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS Pathog 5:e1000599. doi: 10.1371/journal.ppat.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charbonneau H, Beier N, Walsh KA, Beavo JA. 1986. Identification of a conserved domain among cyclic nucleotide phosphodiesterases from diverse species. Proc Natl Acad Sci U S A 83:9308–9312. doi: 10.1073/pnas.83.24.9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy M, Fishbaugher ME, Vaughan AM, Patrapuvich R, Boonhok R, Yimamnuaychok N, Rezakhani N, Metzger P, Ponpuak M, Sattabongkot J, Kappe SH, Hume JC, Lindner SE. 2012. A rapid and scalable density gradient purification method for plasmodium sporozoites. Malar J 11:421. doi: 10.1186/1475-2875-11-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamoto F, Alejo-Blanco R, Fleck SL, Kawamoto Y, Sinden RE. 1990. Possible roles of Ca2+ and cGMP as mediators of the exflagellation of Plasmodium berghei and Plasmodium falciparum. Mol Biochem Parasitol 42:101–108. doi: 10.1016/0166-6851(90)90117-5. [DOI] [PubMed] [Google Scholar]
- 19.Collins CR, Hackett F, Strath M, Penzo M, Withers-Martinez C, Baker DA, Blackman MJ. 2013. Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog 9:e1003344. doi: 10.1371/journal.ppat.1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ Jr., Kappe SH. 2013. Perturbations of plasmodium Puf2 expression and RNA-seq of Puf2-deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol 15:1266–1283. doi: 10.1111/cmi.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor CJ, McRobert L, Baker DA. 2008. Disruption of a Plasmodium falciparum cyclic nucleotide phosphodiesterase gene causes aberrant gametogenesis. Mol Microbiol 69:110–118. doi: 10.1111/j.1365-2958.2008.06267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wentzinger L, Bopp S, Tenor H, Klar J, Brun R, Beck HP, Seebeck T. 2008. Cyclic nucleotide-specific phosphodiesterases of Plasmodium falciparum: PfPDEalpha, a non-essential cGMP-specific PDE that is an integral membrane protein. Int J Parasitol 38:1625–1637. doi: 10.1016/j.ijpara.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Sultan AA, Thathy V, Frevert U, Robson KJ, Crisanti A, Nussenzweig V, Nussenzweig RS, Ménard R. 1997. TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell 90:511–522. doi: 10.1016/S0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- 24.Howard BL, Thompson PE, Manallack DT. 2011. Active site similarity between human and Plasmodium falciparum phosphodiesterases: considerations for antimalarial drug design. J Comput Aided Mol Des 25:753–762. doi: 10.1007/s10822-011-9458-5. [DOI] [PubMed] [Google Scholar]
- 25.Hopp CS, Bowyer PW, Baker DA. 2012. The role of cGMP signalling in regulating life cycle progression of plasmodium. Microbes Infect 14:831–837. doi: 10.1016/j.micinf.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilz RB, Broderick KE. 2005. Role of cyclic GMP in gene regulation. Front Biosci 10:1239–1268. doi: 10.2741/1616. [DOI] [PubMed] [Google Scholar]
- 27.Bastian R, Dawe A, Meier S, Ludidi N, Bajic VB, Gehring C. 2010. Gibberellic acid and cGMP-dependent transcriptional regulation in Arabidopsis thaliana. Plant Signal Behav 5:224–232. doi: 10.4161/psb.5.3.10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gancedo JM. 2013. Biological roles of cAMP: variations on a theme in the different kingdoms of life. Biol Rev Camb Philos Soc 88:645–668. doi: 10.1111/brv.12020. [DOI] [PubMed] [Google Scholar]
- 29.Montagna GN, Matuschewski K, Buscaglia CA. 2012. Plasmodium sporozoite motility: an update. Front Biosci 17:726–744. doi: 10.2741/3954. [DOI] [PubMed] [Google Scholar]
- 30.Mueller AK, Camargo N, Kaiser K, Andorfer C, Frevert U, Matuschewski K, Kappe SH. 2005. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc Natl Acad Sci U S A 102:3022–3027. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes-Santos CS, Braks J, Prudêncio M, Carret C, Gomes AR, Pain A, Feltwell T, Khan S, Waters A, Janse C, Mair GR, Mota MM. 2011. Transition of plasmodium sporozoites into liver stage-like forms is regulated by the RNA binding protein Pumilio. PLoS Pathog 7:e1002046. doi: 10.1371/journal.ppat.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aly AS, Lindner SE, MacKellar DC, Peng X, Kappe SH. 2011. SAP1 is a critical post-transcriptional regulator of infectivity in malaria parasite sporozoite stages. Mol Microbiol 79:929–939. doi: 10.1111/j.1365-2958.2010.07497.x. [DOI] [PubMed] [Google Scholar]
- 33.Mikolajczak SA, Aly AS, Dumpit RF, Vaughan AM, Kappe SH. 2008. An efficient strategy for gene targeting and phenotypic assessment in the Plasmodium yoelii rodent malaria model. Mol Biochem Parasitol 158:213–216. doi: 10.1016/j.molbiopara.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Jongco AM, Ting LM, Thathy V, Mota MM, Kim K. 2006. Improved transfection and new selectable markers for the rodent malaria parasite Plasmodium yoelii. Mol Biochem Parasitol 146:242–250. doi: 10.1016/j.molbiopara.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Vignali M, Armour CD, Chen J, Morrison R, Castle JC, Biery MC, Bouzek H, Moon W, Babak T, Fried M, Raymond CK, Duffy PE. 2011. NSR-seq transcriptional profiling enables identification of a gene signature of Plasmodium falciparum parasites infecting children. J Clin Invest 121:1119–1129. doi: 10.1172/JCI43457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wold B, Myers RM. 2008. Sequence census methods for functional genomics. Nat Methods 5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 38.Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 39.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, Smyth GK. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 41.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC: http://www.ncbi.nlm.nih.gov/books/NBK54050/. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RPKM values for all transcripts