ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) USA300 is a successful S. aureus clone in the United States and a common cause of skin and soft tissue infections (SSTIs). We performed whole-genome sequencing (WGS) of 146 USA300 MRSA isolates from SSTIs and colonization cultures obtained from an investigation conducted from 2008 to 2010 in Chicago and Los Angeles households that included an index case with an S. aureus SSTI. Identifying unique single nucleotide polymorphisms (SNPs) and analyzing whole-genome phylogeny, we characterized isolates to understand transmission dynamics, genetic relatedness, and microevolution of USA300 MRSA within the households. We also compared the 146 USA300 MRSA isolates from our study with the previously published genome sequences of the USA300 MRSA isolates from San Diego (n = 35) and New York City (n = 277). We found little genetic variation within the USA300 MRSA household isolates from Los Angeles (mean number of SNPs ± standard deviation, 17.6 ± 35; π nucleotide diversity, 3.1 × 10−5) or from Chicago (mean number of SNPs ± standard deviation, 12 ± 19; π nucleotide diversity, 3.1 × 10−5). The isolates within a household clustered into closely related monophyletic groups, suggesting the introduction into and transmission within each household of a single common USA300 ancestral strain. From a Bayesian evolutionary reconstruction, we inferred that USA300 persisted within households for 2.33 to 8.35 years prior to sampling. We also noted that fluoroquinolone-resistant USA300 clones emerged around 1995 and were more widespread in Los Angeles and New York City than in Chicago. Our findings strongly suggest that unique USA300 MRSA isolates are transmitted within households that contain an individual with an SSTI. Decolonization of household members may be a critical component of prevention programs to control USA300 MRSA spread in the United States.
IMPORTANCE
USA300, a virulent and easily transmissible strain of methicillin-resistant Staphylococcus aureus (MRSA), is the predominant community-associated MRSA clone in the United States. It most commonly causes skin infections but also causes necrotizing pneumonia and endocarditis. Strategies to limit the spread of MRSA in the community can only be effective if we understand the most common sources of transmission and the microevolutionary processes that provide a fitness advantage to MRSA. We performed a whole-genome sequence comparison of 146 USA300 MRSA isolates from Chicago and Los Angeles. We show that households represent a frequent site of transmission and a long-term reservoir of USA300 strains; individuals within households transmit the same USA300 strain among themselves. Our study also reveals that a large proportion of the USA300 isolates sequenced are resistant to fluoroquinolone antibiotics. The significance of this study is that if households serve as long-term reservoirs of USA300, household MRSA eradication programs may result in a uniquely effective control method.
INTRODUCTION
Staphylococcus aureus is the most common cause of human skin and soft tissue infections (SSTIs) and is also a common cause of osteomyelitis, endocarditis, and pneumonia (1). Methicillin-resistant S. aureus (MRSA) strains are resistant to all β-lactam antibiotics, with the exception of new cephalosporins, and have posed therapeutic challenges since their first description more than 50 years ago (2). In the 1990s, an epidemic of MRSA infections in the United States began outside health care facilities (3). With this shift in epidemiology, the majority of patients who seek care for S. aureus SSTIs in United States emergency departments (4), jails (5), large medical centers (6), and community primary-care offices (7) are infected with MRSA.
By 2004, nearly all of the MRSA isolates from community-associated SSTIs in the United States (>97%) had a common pulsed-field gel electrophoresis (PFGE) type, known as USA300 (6, 8). In these strains, the Panton-Valentine leukocidin (PVL) toxin genes (lukS-PV and lukF-PV), the arginine catabolic mobile element (ACME), and staphylococcal cassette chromosome mec (SCCmec) type IV were almost uniformly present. In contrast, these characteristics were rarely found in health care-associated MRSA (HA-MRSA) strains (9). USA300 MRSA (referred to here as USA300), in addition to causing SSTIs, has emerged as a common cause of invasive infections (10). The success of USA300 in both community and health care settings has been attributed to overexpression of the global transcriptional regulator agr and sae, leading to increased expression of toxin genes, including PVL (11–13). Also in USA300, the presence of ACME likely enhances the survival of MRSA on the skin (14).
A critical reservoir of USA300, as for all S. aureus strains, is asymptomatic colonization of the human body. Studies have been performed among household contacts of patients with S. aureus infections to assess the frequency of asymptomatic colonization (15, 16). S. aureus colonization of more than one individual in the household of a patient already infected has been identified, but until recently, studies have either not assessed the genetic relatedness of strains or have used sequence-based techniques with limited discriminatory power (17).
Whole-genome sequencing (WGS) has come into general use in bacterial epidemiological studies as it offers the ultimate level of sensitivity in the genetic discrimination of closely related strains and the identification of genetic markers associated with virulence and antibiotic resistance (18–20). We set out to determine if WGS could identify single nucleotide polymorphisms (SNPs) among USA300 isolates that would cluster by household or city of origin. Using a large number of isolates collected in two different geographic regions, we provide strong evidence that USA300 spreads within households and persists for a period of several years. Furthermore, we show that a large number of the USA300 isolates, predominantly from California, had acquired mutations associated with fluoroquinolone resistance, whereas the prevalence of resistance remained low in Chicago. This study reveals the microevolutionary processes that are shaping the USA300 epidemic.
RESULTS
Long-term persistence of USA300 MRSA in households.
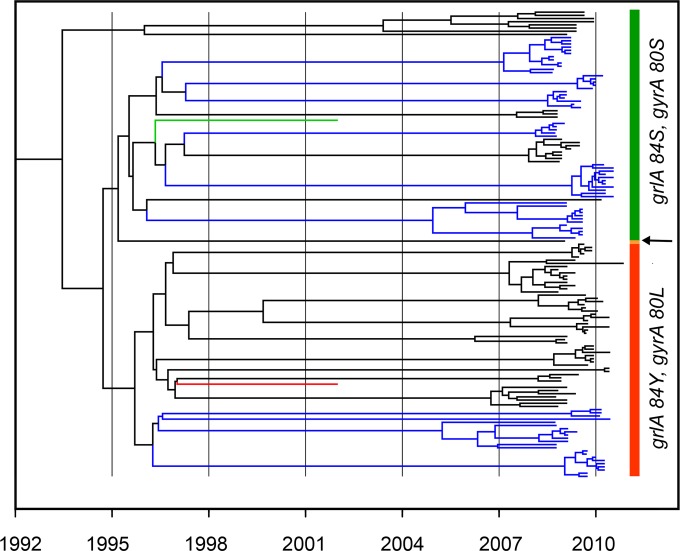
We sequenced 146 USA300 isolates to an ~200× median depth of coverage per strain. The strains were reconfirmed as belonging to sequence type 8 (ST8) from the WGS data by using the Short Read Sequence Typing (SRST) tool (21). The 2,203,292-bp core nucleotide sequence (of the 2.9-Mb genome) common to all USA300 isolates extracted from the progressiveMauve alignment was used for a Bayesian coalescent analysis implemented in BEAST (Bayesian evolutionary analysis by sampling trees) to derive estimated dates for the common ancestries of the strains. Using a strict molecular clock and a Bayesian skyline coalescent model, we estimated an average mutation rate of 1.25 × 10−6 (95% confidence interval, 1.02 × 10−6 to 1.52 × 10−6) mutations per site per year, similar to that reported previously in USA300 and other S. aureus genetic types (17, 22–25). Uhlemann et al. reported a mutation rate of 1.22 × 10−6 (95% confidence interval, 6.04 × 10−7 to 1.86 × 10−6) mutations per site per year in the USA300 isolates analyzed from the New York City (17). Similarly, a mean nucleotide substitution rate of 2.0 × 10−6 (95% confidence interval, 1.2 × 10−6 to 2.9 × 10−6) per site per year in ST225 S. aureus strains of Central Europe was estimated (22). The average estimate of the time to the most recent common ancestor (TMRCA) of the USA300 isolates indicates a divergence at around 1993, consistent with observations of the start of the epidemic of community-associated MRSA (CA-MRSA) infections caused by this strain type in the United States (26). In addition, our analysis revealed that the fluoroquinolone-resistant strains emerged in the mid-1990s (Fig. 1). The BEAST analysis demonstrated a range of mean TMRCAs based on the final date of isolation of bacteria in 2010 from households of 2.33 years (household 9131) to 8.35 years (household 8113) (see Table S1 in the supplemental material). However, the 95% confidence intervals for these dates fail to overlap only for the two isolates at the maximum and minimum ends of the scale. We also constructed a well-supported root-to-tip regression plot of the date of isolation of these strains and increased genetic diversity by using Path-O-Gen (correlation coefficient, 0.351; r2 = 0.123) (see Fig. S1 in the supplemental material). The root-to-tip regression analysis suggested a rate estimate (1.21 × 10−6 per site per year) and TMRCA of the USA300 isolates very similar to those obtained in the Bayesian coalescent analysis.
FIG 1 .
Maximum clade credibility tree resulting from BEAST analysis of the core genome alignment of 146 USA300 isolates from Los Angeles and Chicago. Bayesian analysis was run under a strict molecular clock and with an HKY model of nucleotide substitution assuming a Bayesian skyline demographic model. Blue branches are isolates from Chicago households, and black ones are from Los Angeles households. The green and red branches, respectively, are the USA300 TCH1516 and FPR3757 reference strains. The fluoroquinolone-susceptible (with grlA 84S and gyrA 80S) and -resistant (with grlA 84Yand gyrA 80L) clades are indicated by vertical green and red bars, respectively. The arrow indicates a single strain with the grlA 84F gyrA 80L genotype.
Continuous reshaping of the USA300 pangenome.
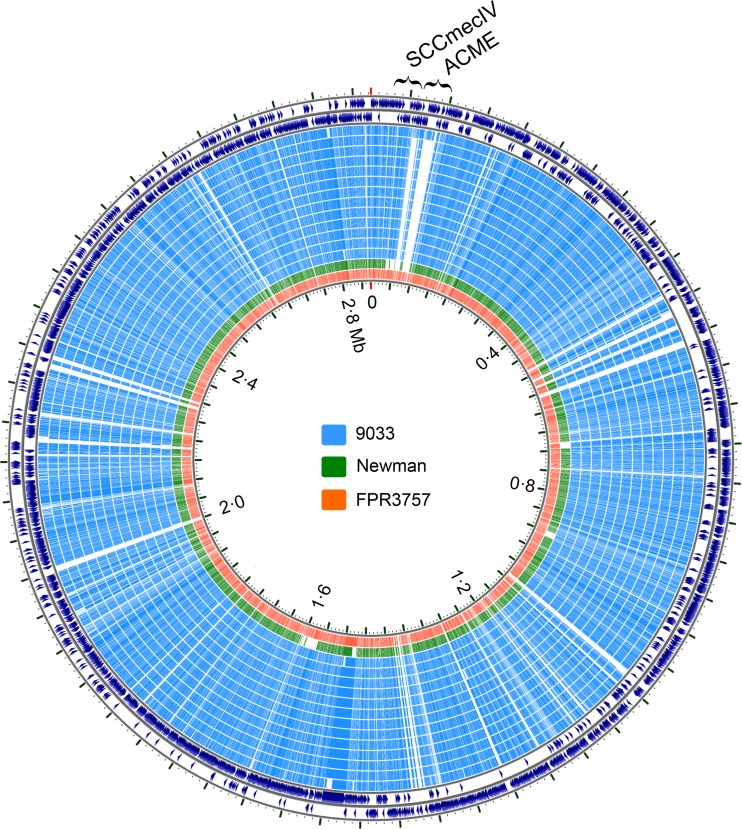
We investigated the pangenome composition of the 146 USA300 draft sequenced strains and 49 completely sequenced genomes of S. aureus available in the National Center for Biotechnology Information (NCBI) refSeq database (see Table S2 in the supplemental material). Unsupervised hierarchal clustering based on patterns of the presence or absence of gene families (27) grouped all of the USA300 strains in the same clade (see Fig. S2 in the supplemental material). Twelve strains from Chicago household 9033 were missing the ACME cassette (Fig. 2). The position of the ACME-negative strains in the phylogeny suggested the deletion of a preexisting island. Interestingly, the index infection isolate of this household (strain 120381) is from a phylogenetically distinct ACME-positive lineage. This genotype was not subsequently detected in household 9033 at 3 or 6 months. Our study design precluded an analysis of the loss or gain of PVL genes because the presence of the PVL genes was a criterion for isolate inclusion in this study.
FIG 2 .
BLAST map showing ACME deletion in the USA300 isolates from Chicago household 9033. The map was generated by the CCT, which uses all-versus-all BLAST and arranges the genomes compared according to their homology with reference genomes (the reference genome here is TCH1516, which is shown in the outermost two rings). Each ring is a genome (isolate), as indicated. As shown, one isolate in household 9033 (the outermost blue ring) was ACME positive, while the remaining isolates had this element deleted. Strain Newman also lacks ACME (shown as the green ring). FPR3757 was included here as another ACME-positive USA300 reference strain.
There was evidence that USA300 clones, when persisting in households, continued to acquire extraneous DNA by horizontal gene transfer (HGT) and recombination. A few isolates acquired new genes on phages (e.g., isolate 119700 in household 8012) compared with others within the same household. This event could only have occurred after the strain entered the household, and thus, the DNA must have originated from coresident Staphylococcus species (28) on the colonized human host. Likewise, there are at least two SNP patterns that are best explained by an exchange of short segments of DNA through homologous recombination with a non-USA300 S. aureus isolate. In strain 119720, from household 8182 in Los Angeles, there are five SNPs in a short intergenic region (coordinates 1996070 to 1996129 bp of the TCH1516 genome) that match the ancestral S. aureus genome rather than the typical USA300 genome. This suggests that a homologous recombination event may have taken place prior to the introduction of USA300 into household 8182.
Clustering of USA300 within households.
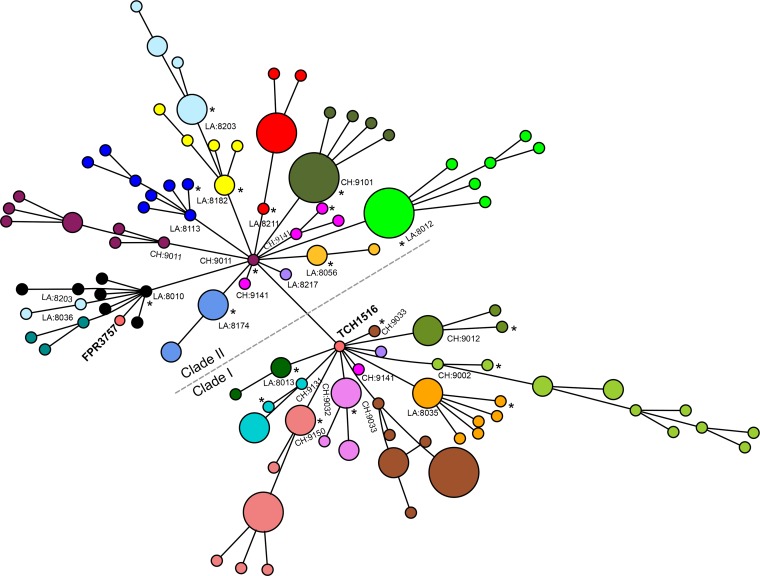
To investigate the clustering of strains within households, we constructed a minimum spanning tree (MST) based on an alignment matrix of 1,629 core protein clusters (1,335,849 bp) of USA300 as determined by OrthoMCL. Similar to BEAST phylogeny, strains in the MST were split into two clades (Fig. 3). Also, 18 of 21 households contained USA300 isolates clustered into closely related monophyletic groups, which suggested the introduction into each household of a single USA300 ancestor strain. The index infection isolate haplotype was the connection of the household-specific branch to the majority of the strains in the MST (12 out of 21 households, P = 1.0, Fisher’s exact test). This suggests that in many households the index cases’ infection isolates were not responsible for the introduction of USA300 into these households; instead, the infecting isolate was derived from isolates that were already present in the household.
FIG 3 .
Minimum spanning tree showing genetic relationships among all household USA300 isolates. Each circle represents an individual genotype based on a 1,335,849-bp core gene alignment, and each color represents a different household. The size of the circle is proportional to the number of isolates with the genotype indicated. The household numbers are prefixed with CH (for Chicago) or LA (for Los Angeles). The index infection isolate in the household is indicated by an asterisk. The clades of USA300 strains with and without the grlA 80Y and gyrA 84L mutations are labeled clades I and II, respectively. As shown, the isolates from Los Angeles household 8203 split into two clusters but still remained in larger clade II. The isolates from Chicago household 9141 separated into two different clades with the presence or absence of grlA and gyrA mutations.
In addition, households could generally be connected to the root of the phylogenetic tree without passing through another household. This finding supports the hypothesis that, for the samples in this study, transmission occurred primarily within the households after a single introduction and does not support the alternative explanation that there were frequent or repeated introductions of USA300 from another reservoir outside the household. Overall, the levels of genetic diversity within the Los Angeles and Chicago isolates were similar, with medians of six (mean and standard deviation of 17.6 ± 35 [range, 0 to 199]) and five (mean and standard deviation of 12 ± 19 [range, 0 to 102]) SNPs per household, respectively (see Fig. S3 and Table S1 in the supplemental material). The nucleotide diversity (π, the average number of nucleotide differences per site between two sequences) was also the same (π = 3.1 × 10−5) in both Los Angeles and Chicago isolates and was similar to the diversity reported for S. aureus isolates of the ST225 genetic background from Central Europe (22). The theta (θ) diversities of Chicago and Los Angeles isolates, respectively, were estimated to be 5.4 × 10−5 and 7.1 × 10−5. A core protein alignment-based maximum-likelihood (ML) phylogeny resulted in similar clustering of the isolates within the households studied (see Fig. S4 in the supplemental material).
Evolution of fluoroquinolone-resistant USA300.
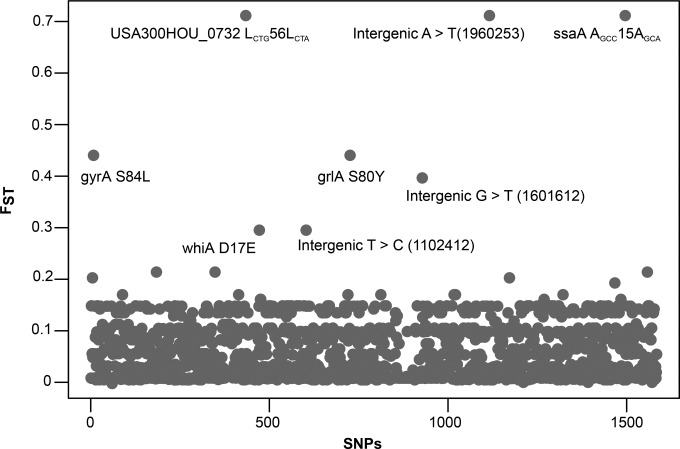
To identify genetic changes driving the clustering of strains within the two larger clades, we estimated genome-wide Weir and Cockerham’s FST (genetic differentiation) between the bacterial populations identified in Los Angeles and Chicago. We found eight SNPs (three nonsynonymous, two synonymous, and three intergenic) that were highly differentiated between Chicago and Los Angeles isolates (Fig. 4). Two of the three nonsynonymous SNPs within the gyrase A (gyrA S84L) and topoisomerase IV (grlA S80Y) genes were associated with fluoroquinolone resistance in S. aureus (17, 29) and several other bacterial species (30–32). The third nonsynonymous SNP was in a poorly characterized gene called whiA (whiA D17E) located close to the essential walKR cell wall two-component signal transduction system.
FIG 4 .
Weir and Cockerham FST at 1,595 SNP positions identified in 146 USA300 MRSA isolates analyzed in this study. Each dot represents an SNP, and those with elevated FST values are indicated (FST across all SNPs, 0.15). Numbers in parentheses are genomic coordinates corresponding to the TCH1516 reference genome.
Phenotypic ciprofloxacin resistance segregated with the presence of the gyrA and grlA SNPs. All of the isolates from the households in clade II harbored the mutations gyrA S84L and grlA S80Y, conferring fluoroquinolone resistance, whereas those in clade I were fluoroquinolone susceptible (Fig. 1 and 3; Table 1). With the exception of one household (no. 9141), fluoroquinolone resistance also clustered by household; all of the isolates within a household were either susceptible or resistant. The four fluoroquinolone-resistant strains in household 9141 segregated with clade II, while only one susceptible strain segregated with clade I (Fig. 3). In an example of parallel evolution, the same two mutations were observed in EMRSA-15, an epidemic health care-associated ST22 strain that emerged in the United Kingdom in the 1980s (25). Fluoroquinolone resistance has also been associated with the rapid intercontinental spread of another Gram-positive pathogen, Clostridium difficile (32).
TABLE 1 .
Ciprofloxacin susceptibility profiles of the Los Angeles and Chicago USA300 household MRSA isolates and SNPs in their grlA and gyrA genes
| Household | Region | na | Ciprofloxacin susceptibilityb (MIC [μg/ml]) | grlA position 80 nucleotide | gyrA position 84 nucleotide |
|---|---|---|---|---|---|
| 8010 | Los Angeles | 7 | R (>4.0) | Y | L |
| 8012 | Los Angeles | 11 | R (>4.0) | Y | L |
| 8013 | Los Angeles | 3 | S (1.0) | S | S |
| 8035 | Los Angeles | 8 | S (1.0) | S | S |
| 8036 | Los Angeles | 3 | R (>4.0) | Y | L |
| 8056 | Los Angeles | 3 | R (>4.0) | Y | L |
| 8113 | Los Angeles | 8 | R (>4.0) | Y | L |
| 8174 | Los Angeles | 5 | R (>4.0) | Y | L |
| 8182 | Los Angeles | 6 | R (>4.0) | Y | L |
| 8203 | Los Angeles | 9 | R (>4.0) | Y | L |
| 8211 | Los Angeles | 7 | R (>4.0) | Y | L |
| 8217 | Los Angeles | 1 | NDc | Y | L |
| 9002 | Chicago | 12 | S (0.5) | S | S |
| 9011 | Chicago | 9 | R (>4.0) | Y | L |
| 9012 | Chicago | 5 | S (0.5) | S | S |
| 9032 | Chicago | 6 | S (0.5) | S | S |
| 9033d | Chicago | 13 | S (0.5) | S | S |
| 9101 | Chicago | 9 | R (>4.0) | Y | L |
| 9131 | Chicago | 5 | S (1.0) | S | S |
| 9141e | Chicago | 5 | R (>4.0) | Y | L |
| 9150 | Chicago | 11 | S (0.5) | S | S |
Number of isolates sequenced in the household.
R, resistant; S, susceptible.
ND, MIC not determined.
One isolate (120381) in household 9033 had grlA 80F and gyrA 84L SNPs and was ciprofloxacin sensitive.
One isolate (119813) in household 9141 did not have these SNPs.
Fluoroquinolone resistance in USA300 is not geographically constrained.
A high-resolution ML phylogeny based on core genome alignment of 460 USA300 strains from San Diego (23), Los Angeles (this study), Chicago (this study), and New York City (17) was constructed in REALPHY (33). As shown in Fig. 5, the isolates with and without fluoroquinolone resistance-conferring mutations gyrA S84L and grlA S80Y were grouped into two clades (Fig. 5). Fluoroquinolone-resistant USA300 strains, however, were more widespread in San Diego (28/35, 80.0%), Los Angeles (59/70, 84.2%), and New York (186/277, 67.1%) than in Chicago (23/75, 30.6%) (P < 0.0001 for each comparison).
FIG 5 .
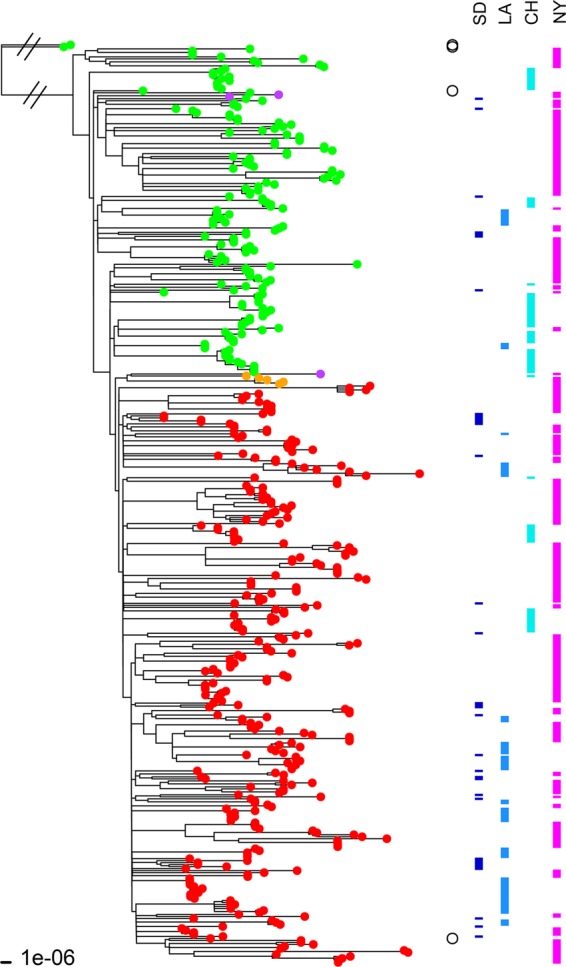
ML tree of 460 USA300 MRSA isolates along with two non-USA300 strains (Newman and COL) included as outgroups. The ML tree was constructed with the 2,104,213-bp core sequence alignment of these isolate genomes in REALPHY. The fluoroquinolone-susceptible isolates (i.e., with no mutations in the grlA and gyrA genes) are indicated by green branch tips, whereas fluoroquinolone-resistant strains (i.e., with the grlA 80Y and gyrA 84L mutations) are indicated by red branch tips. Five fluoroquinolone-resistant isolates had a different set of mutations (grlA 80F and gyrA 84L) and are indicated by orange branch tips. Three isolates indicated by purple tips had only one mutation in the grlA gene (80F). The two non-USA300 strains, Newman (grlA 80S and gyrA 84S) and COL (grlA 80S and gyrA 84S), and two USA300 reference strains, TCH1516 (grlA 80S and gyrA 84S) and FPR3757 (grlA 80Y and gyrA 84L), are indicated by open circles to the right of the tree. USA300 strains TCH1516 and FPR3757 segregated with their fluoroquinolone-susceptible and -resistant clades, respectively. Isolates from San Diego (SD), Los Angeles (LA), Chicago (CH), and New York (NY) are indicated by the bars on the right.
DISCUSSION
Utilizing WGS of USA300 isolates from households in two cities, Chicago and Los Angeles, we have demonstrated that households serve as major sites of MRSA transmission. A recent study by Uhlemann et al. observed similar results in household isolates from New York City (17). With time, strains in the households that we studied acquired a small number of point mutations and DNA from HGT from other bacteria (likely Staphylococcus species or other close relatives of S. aureus). It would be useful in the context of USA300 epidemiology to consider whether the virulence potential of strains decreases over time in a small human population. We found no genome-wide patterns of nucleotide substitution or HGT to distinguish strains colonizing individuals from those causing infections, unlike in other studies (23, 34). There were few SNPs in Los Angeles and Chicago household isolates, similar to findings in earlier studies, suggesting a recent clonal expansion and diversification of USA300 clones (17, 23, 35). We also observed that the USA300 isolates from different regions of the United States belonged to the same clade of the whole-genome ML phylogenetic tree, suggesting frequent migration of strains between geographical regions.
Importantly, we showed in this study that USA300 clonal lineages persisted within households for about 2 to 8 years before a symptomatic patient was admitted to a hospital and continued for at least another year afterward. This could be caused by a combination of long-term asymptomatic human carriage (36) and/or frequent reinfections from other household members, pets, or fomites. We need to understand how MRSA persists (the reasons may be different in different households) to design decolonization strategies and public health programs aimed at controlling the spread of USA300. Interventions may need to address all of the symptomatically infected and asymptomatically colonized individuals in a household.
We found that households were common sites of USA300 transmission and that once USA300 was introduced, within-household transmission was more common than repeated reintroduction of this S. aureus genetic background. While other reservoirs of USA300 such as health care settings, jails, gyms, schools, and other public institutions may be sites of transmission and sources of isolates of this genetic background in U.S. households, the WGS data in our study provided the resolution to support the hypothesis that within-household transmission creates the conditions for a long-term reservoir and for CA-MRSA persistence. Our results are supported by the findings of other studies (34, 37).
The presence of ACME has been postulated to contribute to the success of the USA300 clone (14). ACME contains the arc operon thought to be important for arginine catabolism, enhancing survival in acidic environments on the skin. In USA300, ACME includes speG, which encodes spermidine acetyltransferase, which decreases the toxicity of spermidine, which is secreted as a defense molecule on mammalian skin. The recent addition of speG to ACME is thought to have been an important factor in the emergence of the USA300 epidemic in the late 1990s (14). In our study, as in other studies of USA300 (17, 23, 35), ACME was not universally present, and we found that isolates lacking ACME were still able to spread in a household and colonize people stably. The importance of ACME in the fitness of USA300 thus requires further study.
A phylogeny of 460 strains from Los Angeles, Chicago, San Diego, and New York City revealed that USA300 strains form two clades according to their fluoroquinolone resistance phenotypes (with or without gyrA 84L plus grlA 80Y/F mutations). These findings echo other recent North American studies (17, 38). We cannot ascertain at this stage whether clade II is replacing the older fluoroquinolone-sensitive lineage or what the significance of the higher frequency of fluoroquinolone resistance in Los Angeles than in Chicago is. Unlike ST22 (EMRSA-15) (25), another clade that has acquired and frequently exhibits fluoroquinolone resistance and is primarily HA-MRSA, USA300 is primarily CA-MRSA (all of the strains in this study were epidemiologically characterized as CA-MRSA). Cheng et al. showed that fluoroquinolone use might increase the rate of nasal colonization by MRSA (39). The fluoroquinolone-resistant strains may have been selected for by antibiotic treatment for other conditions. This raises the question of what the selection pressures are that allow long-term maintenance of the gyrA/grlA mutations within households. Presumably, there is little fitness cost to the mutations and/or regular antibiotic exposure is occurring.
We believe the next step to answering the questions raised in this work is a longitudinal evaluation of the USA300 isolates within households to examine the patterns of person-to person transmission, the persistence of fluoroquinolone resistance, and whether mutations, over time, lead to enhanced human colonization or to enhancement of the potential of a USA300 strain to cause an infection.
MATERIALS AND METHODS
Household survey and strain collection.
In our household contact study, 350 evaluable index patients with SSTIs were enrolled between August 2008 and June 2010 at the University of Chicago Medical Center (n = 177) or at the Harbor-UCLA Medical Center (n = 173). Details of this study have been described previously (16). Each member of the household of each consenting index patient was visited on three occasions, <21 days after the treatment of the SSTI (baseline) and 3 and 6 months after the baseline visit. At each visit, the index patient and each household contact were tested for S. aureus colonization by culturing samples from the nares, the oropharynx, and the inguinal region. All of the S. aureus isolates obtained from the index SSTI (the index infection isolate) and from colonization cultures from index patients and household contacts from all three household visits underwent genotyping by multilocus sequence typing (MLST), SCCmec typing, and assessment for the presence of PVL genes. Isolates were considered to be USA300 if they were of ST8 and carried SCCmec type IV and the PVL genes (16), as shown in a previous validation study (40). Susceptibility to ciprofloxacin was tested by the broth microdilution method as recommended by the Clinical and Laboratory Standards Institute (41).
Among 1,162 persons enrolled (350 index patients and 812 household members), S. aureus colonized 40% (137/350) of the index patients and 50% (405/812) of their household contacts at one or more sites. Factors independently associated (P < 0.05) with the index infection isolate strain type being present colonizing a household contact were recent skin infection in the contact, recent cephalexin use, and USA300 genetic background of the index infection isolate (see Table S3 in the supplemental material) (16). USA300 was the predominant S. aureus type identified among the infecting (53%) and colonizing (29%) isolates. We randomly selected 21 households, 12 from Los Angeles and 9 from Chicago (a total of 146 isolates) for WGS that met the following criteria. (i) The index infection isolate was USA300, and (ii) at least two household members (one of whom could be the index patient) were colonized with USA300 at one or more body sites at the baseline visit (see Table S3 in the supplemental material). Additional information on recruitment has been previously published (16).
WGS, SNP calling, genome assembly, and annotation.
DNA was extracted from each isolate with the Qiagen genome preparation kit according to the manufacturer’s protocol. Sequencing libraries were prepared with 1 to 2 µg of DNA by following the standard Illumina protocols and chemistry and paired-end sequencing was performed on the Illumina HiSeq 2000 platform (Illumina Inc., San Diego, CA), generating 100-bp sequence reads. The MLST genetic background of the isolates was reconfirmed from the WGS data by using the SRST tool (21). To minimize errors and artifacts in the downstream analyses, sequence reads were preprocessed with PRINSEQ (version 0.20.3) (42). The preprocessing of the reads involved two sequential steps. In the first step, we simply discarded any reads with two or more N’s or a mean Phred quality score of <20. In the second step, bases with low Phred quality scores (≤19) were trimmed from the 3′ end, and if the length of the trimmed reads fell below 70 bp, they were removed from further analysis. The resulting high-quality reads were aligned against the S. aureus USA300 TCH1516 reference genome (GenBank accession number NC_010079; 2,872,915-bp length) with the Burrows-Wheeler Aligner (version 0.7.2) with a mismatch penalty of 3 and a gap open penalty of 5 (43). The programs SAMtools and Picard Tools were used to format and reformat the intermediate-alignment files, and variant SNPs and insertions-deletions (indels) were identified with the Genome Analysis Toolkit UnifiedGenotyper (44, 45). We considered only those variant positions that were covered by at least 10 reads, and 90% or more sequence reads supported mutation (an allele different from the TCH1516 reference genome). We also excluded the variants if they were in the homopolymer tract regions or were ambiguous in or missing from any of the isolates.
Velvet (version 1.2.06) was used for de novo genome assembly (46). The assembled contigs larger than 200 bp were ordered and converted into a single pseudocontig with Abacas (47) and finally annotated with the bacterial genome annotation tool Prokka (48).
Orthologous gene clustering, pangenome analysis, and core genome phylogeny.
The predicted proteins from each isolate were categorized into orthologous clusters by OrthoMCL (with the option −E value for BLAST alignments set at 1e-05 and −C set at 75%) as implemented in GET_HOMOLOGUES (27, 49, 50). The pangenome matrix (presence and absence of genes/proteins) produced from the OrthoMCL step was used for hierarchical gene clustering analysis with the R heatmap2 function (51). Deletions of ACME or any other large region of the genome were examined and visualized with the CGView comparison tool (CCT) (52), which uses all-versus-all BLAST to compare the genomes and presents the homology as a circular map. The 1,629 core protein clusters identified in 148 USA300 isolates (proteins present in all 148 isolates), which included the two reference strains TCH1516 and FPR3757, by OrthoMCL were individually aligned with Muscle (53), edited in trimAl (54) to eliminate positions containing gaps and poorly aligned regions, and finally concatenated to generate a single alignment (445,283 amino acids) of the core protein clusters. The nucleotide sequence alignments corresponding to each core protein cluster were retrieved with PAL2NAL (55) and concatenated to create a nucleotide alignment matrix (1,335,849 bp) of core gene clusters.
Phylogenetic analysis with the above core protein and core gene alignment matrixes was performed to investigate genetic relationships among household strains by two different methods. An MST based on the alignment matrix of core gene clusters was generated in a Bioconductor library Rgraphviz (version 2.6.0) (56). ML trees based on the core gene and core protein alignment matrixes were constructed in RAxML (version 7.0.4) (57) under GTRGAMMA and HIVWF substitution models, respectively. The best-fit nucleotide and amino acid substitution models, respectively, were determined with jModelTest (v.0.1.1) (58, 59) and ProtTest (60) on the basis of the Akaike information criterion score and the number of estimated parameters. The node support of the ML tree was assessed by the nonparametric bootstrapping method with 200 replicates.
Estimation of FST.
To identify the SNP loci highly differentiated between Los Angeles and Chicago USA300 MRSA isolates, the Weir and Cockerham genetic statistics (FST) (61) across all 1,585 SNP loci identified in USA300 isolates with reference to the TCH1516 USA300 genome (obtained at the SNP calling step as described above) was estimated with the diveRsity R package (version 1.7.6) (62).
Estimation of substitution rate and duration of presence within households.
The rate of substitution within the USA300 genome and age of infection within each household were estimated with BEAST, version 2.0.2 (63). For this analysis, we aligned 146 USA300 draft genomes produced in this study along with two completed genomes of USA300 strains TCH1516 and FPR3757 with the progressiveMauve algorithm of Mauve (64). The core alignment blocks longer than 500 bp, shared by all of the genomes (locally collinear blocks) were concatenated to produce a 2,203,292-bp alignment matrix. An ML tree was constructed with this alignment under the GTRGAMMA substitution model in RAxML as described above. Linear regression analysis was performed to assess the relationship between root-to-tip branch length and the date of isolation of the isolates in Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/). Three independent runs of BEAST, each with 300 million Markov chain Monte Carlo (MCMC) generations, sampling at every 10,000 generations were performed with the following settings: a strict molecular clock, the HKY model of nucleotide substitution, and a Bayesian skyline coalescent, as used by Uhlemann et al. (17) and Nübel et al. (22). The date of isolation of the USA300 strains for this analysis were specified in day, month, and year. The log files resulting from all three runs were analyzed to assess convergence by checking the effective sample sizes (ESSs) of different parameters in Tracer (version 1.5). After confirming that all three runs had converged on the same posterior distribution in the MCMC run with almost identical marginal density distributions and all of the parameters had an ESS of >200, they were combined in LogCombiner (version 2.0.2), discarding the first 25% of the MCMC generations as a burn-in phase. A single maximum clade credibility tree was summarized in TreeAnnotator (version 2.0.9) and visualized in FigTree (version 1.4.0). We also conducted another BEAST run by using a strict molecular clock, the HKY nucleotide substitution model, and a constant size coalescent to rule out the impact of demographic models on our estimates (see Table S1 in the supplemental material).
Phylogenetic structures of all available USA300 genomes.
In order to investigate the high-resolution phylogenetic structures of all of the available USA300 isolates sequenced to date, we reanalyzed 146 strains sequenced in this study along with 312 previously published sequences of USA300 isolates from San Diego, CA (n = 35) (23), and New York, NY (n = 277) (17). In addition, we included the completed genomes of USA300 strains TCH1516 and FPR3757. An ML phylogeny of these 460 USA300 isolates was constructed on the basis of their core genome alignment (2,104,213 bp) with PhyML as implemented in REALPHY (33).
Nucleotide sequence accession numbers.
The raw sequence reads analyzed in this study have been deposited in the NCBI Sequence Read Archive database under accession number SRP039020. The GenBank accession numbers of all of the completed S. aureus genomes used in this study are listed in Table S2 in the supplemental material.
SUPPLEMENTAL MATERIAL
Details of the genetic diversity and Bayesian analysis of 146 USA300 MRSA strains.
GenBank accession numbers of the S. aureus completed genomes included in this study.
Details of the USA300 MRSA isolates analyzed in this study.
Linear regression plot showing correlation between the root-to-tip genetic distance and the isolation date of S. aureus isolates inferred from the ML tree. The ML tree was constructed in RAxML under the GTRGAMMA substitution model with the core genome alignment produced by progressiveMauve. The blue and red dots represent USA300 reference strains TCH1516 and FPR3757, respectively. The remaining dots represent isolates from this study. Download
Hierarchical clustering analysis of the pangenome of 196 S. aureus strains. The rows represent 196 S. aureus genomes, and the columns represent 3,455 core (genes present in all of the strains compared) and noncore (genes present in only a subset of the strains compared) genes identified among the isolates compared. Red and blue blocks indicate the presence and absence of gene clusters, respectively. Columns with all red blocks represent core genes, whereas those with mixed colors represent noncore genes. Of the 196 isolates, 148 are USA300 (146 from this study, TCH1516 [blue dot], and FPR3757 [red dot]) and the remaining 48 are non-USA300 isolates, as listed in Table S2 in the supplemental material. As shown (designated I), the USA300 and non-USA300 isolates formed two separate clades based on the presence or absence of the gene clusters. The USA300 isolates from the same household tend to cluster together, as indicated by the color code (designated II). The color codes used to indicate households are the same as in Fig. 3. Download
Box plot showing the number of SNPs per household as determined from the core genome alignment produced by progressiveMauve. The color scheme used to indicate households is the same as in Fig. 3. Download
ML tree constructed by alignment of 1,629 core protein clusters identified in Los Angeles and Chicago USA300 S. aureus isolates. The branch tips are colored on the basis of the mutations within the grlA and gyrA genes, as indicated. (A) Rooted tree. The color scheme in column I is as follows: blue, Chicago strains; orange, Los Angeles strains; red, Newman/COL; grey, TCH1516/FPR3757. The color scheme in column II indicates households and is the same as in Fig. 3. (B) Unrooted tree. Download
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI091827 to T.D.R., AI067584-01 to R.S.D., and AI095361-01 to M.Z.D. The support of the Emory Genome Center (EGC) is gratefully acknowledged. Equipment used by EGC was funded by the Atlanta Clinical and Translational Sciences Institute, the Department of Human Genetics, and the School of Medicine.
We thank Jessica Peterson for assistance and support during this study. We also thank Julia Sieth for assistance with genotyping and organization of isolates.
Footnotes
Citation Alam MT, Read TD, Petit RA, III, Boyle-Vavra S, Miller LG, Eells SJ, Daum RS, David MZ. 2015. Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. mBio 6(2):e00054-15. doi:10.1128/mBio.00054-15
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Jevons MP. 1961. Celbenin-resistant staphylococci. BMJ 1:124–125. [Google Scholar]
- 3.Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 4.Talan DA, Krishnadasan A, Gorwitz RJ, Fosheim GE, Limbago B, Albrecht V, Moran GJ, EMERGEncy ID Net Study Group . 2011. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis 53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- 5.Pan ES, Diep BA, Carleton HA, Charlebois ED, Sensabaugh GF, Haller BL, Perdreau-Remington F. 2003. Increasing prevalence of methicillin-resistant Staphylococcus aureus infection in California jails. Clin Infect Dis 37:1384–1388. doi: 10.1086/379019. [DOI] [PubMed] [Google Scholar]
- 6.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med 144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 7.Labreche MJ, Lee GC, Attridge RT, Mortensen EM, Koeller J, Du LC, Nyren NR, Treviño LB, Treviño SB, Peña J, Mann MW, Muñoz A, Marcos Y, Rocha G, Koretsky S, Esparza S, Finnie M, Dallas SD, Parchman ML, Frei CR. 2013. Treatment failure and costs in patients with methicillin-resistant Staphylococcus aureus (MRSA) skin and soft tissue infections: a South Texas ambulatory research network (STARNet) study. J Am Board Fam Med 26:508–517. doi: 10.3122/jabfm.2013.05.120247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA, EMERGEncy ID Net Study Group . 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 9.Daum RS, Ito T, Hiramatsu K, Hussain F, Mongkolrattanothai K, Jamklang M, Boyle-Vavra S. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J Infect Dis 186:1344–1347. doi: 10.1086/344326. [DOI] [PubMed] [Google Scholar]
- 10.Limbago B, Fosheim GE, Schoonover V, Crane CE, Nadle J, Petit S, Heltzel D, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Mu Y, Fridkin SK, Active Bacterial Core surveillance MRSA Investigators . 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol 47:1344–1351. doi: 10.1128/JCM.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shallcross LJ, Fragaszy E, Johnson AM, Hayward AC. 2013. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect Dis 13:43–54. doi: 10.1016/S1473-3099(12)70238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Cheung GY, Hu J, Wang D, Joo HS, DeLeo FR, Otto M. 2010. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis 202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planet PJ, LaRussa SJ, Dana A, Smith H, Xu A, Ryan C, Uhlemann AC, Boundy S, Goldberg J, Narechania A, Kulkarni R, Ratner AJ, Geoghegan JA, Kolokotronis SO, Prince A. 2013. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4:e00889-13. doi: 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez M, Hogan PG, Krauss M, Warren DK, Fritz SA. 2013. Measurement and impact of Staphylococcus aureus colonization pressure in households. J Pediatr Infect Dis Soc 2:147–154. doi: 10.1093/jpids/pit002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller LG, Eells SJ, Taylor AR, David MZ, Ortiz N, Zychowski D, Kumar N, Cruz D, Boyle-Vavra S, Daum RS. 2012. Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin Infect Dis 54:1523–1535. doi: 10.1093/cid/cis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlemann AC, Dordel J, Knox JR, Raven KE, Parkhill J, Holden MT, Peacock SJ, Lowy FD. 2014. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fricke WF, Rasko DA. 2014. Bacterial genome sequencing in the clinic: bioinformatic challenges and solutions. Nat Rev Genet 15:49–55. doi: 10.1038/nrg3624. [DOI] [PubMed] [Google Scholar]
- 19.Alam MT, Petit RA, Crispell EK, Thornton TA, Conneely KN, Jiang Y, Satola SW, Read TD. 2014. Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol Evol 6:1174–1185. doi: 10.1093/gbe/evu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laabei M, Recker M, Rudkin JK, Aldeljawi M, Gulay Z, Sloan TJ, Williams P, Endres JL, Bayles KW, Fey PD, Yajjala VK, Widhelm T, Hawkins E, Lewis K, Parfett S, Scowen L, Peacock SJ, Holden M, Wilson D, Read TD, van den Elsen J, Priest NK, Feil EJ, Hurst LD, Josefsson E, Massey RC. 2014. Predicting the virulence of MRSA from its genome sequence. Genome Res 24:839–849. doi: 10.1101/gr.165415.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inouye M, Conway TC, Zobel J, Holt KE. 2012. Short read sequence typing (SRST): multi-locus sequence types from short reads. BMC Genomics 13:338. doi: 10.1186/1471-2164-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nübel U, Dordel J, Kurt K, Strommenger B, Westh H, Shukla SK, Zemlicková H, Leblois R, Wirth T, Jombart T, Balloux F, Witte W. 2010. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog 6:e1000855. doi: 10.1371/journal.ppat.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tewhey R, Cannavino CR, Leake JA, Bansal V, Topol EJ, Torkamani A, Bradley JS, Schork NJ. 2012. Genetic structure of community acquired methicillin-resistant Staphylococcus aureus USA300. BMC Genomics 13:508. doi: 10.1186/1471-2164-13-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlicková H, Coombs G, Kearns AM, Hill RL, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramírez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nübel U. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover FC, Goering RV. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J Antimicrob Chemother 64:441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol 6:2697–2708. doi: 10.1093/gbe/evu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz FJ, Jones ME, Hofmann B, Hansen B, Scheuring S, Lückefahr M, Fluit A, Verhoef J, Hadding U, Heinz HP, Köhrer K. 1998. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob Agents Chemother 42:1249–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, Bentley SD, Parkhill J, Nejentsev S, Hoffner SE, Horstmann RD, Brown T, Drobniewski F. 2012. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res 22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. 2014. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlemann AC, Kennedy AD, Martens C, Porcella SF, DeLeo FR, Lowy FD. 2012. Toward an understanding of the evolution of Staphylococcus aureus strain USA300 during colonization in community households. Genome Biol Evol 4:1275–1285. doi: 10.1093/gbe/evs094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A 105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robicsek A, Beaumont JL, Peterson LR. 2009. Duration of colonization with methicillin-resistant Staphylococcus aureus. Clin Infect Dis 48:910–913. doi: 10.1086/597296. [DOI] [PubMed] [Google Scholar]
- 37.Jones TF, Creech CB, Erwin P, Baird SG, Woron AM, Schaffner W. 2006. Family outbreaks of invasive community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 42:e76–e78. doi: 10.1086/503265. [DOI] [PubMed] [Google Scholar]
- 38.Strommenger B, Bartels MD, Kurt K, Layer F, Rohde SM, Boye K, Westh H, Witte W, de Lencastre H, Nübel U. 2014. Evolution of methicillin-resistant Staphylococcus aureus towards increasing resistance. J Antimicrob Chemother 69:616–622. doi: 10.1093/jac/dkt413. [DOI] [PubMed] [Google Scholar]
- 39.Cheng VC, Li IW, Wu AK, Tang BS, Ng KH, To KK, Tse H, Que TL, Ho PL, Yuen KY. 2008. Effect of antibiotics on the bacterial load of meticillin-resistant Staphylococcus aureus colonisation in anterior nares. J Hosp Infect 70:27–34. doi: 10.1016/j.jhin.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 40.David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2013. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J Clin Microbiol 51:814–819. doi: 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.CLSI 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. CLSI document m07-A9 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. 2009. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Kristensen DM, Wolf YI, Mushegian AR, Koonin EV. 2011. Computational methods for gene orthology inference. Brief Bioinform 12:379–391. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Contreras-Moreira B, Vinuesa P. 2013. Get_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warnes GR, Bolker B, Lumley T. 2012. Gplots: various R programming tools for plotting data. R package version 2.6.0. The R Project for Statistical Computing, Vienna, Austria. [Google Scholar]
- 52.Grant JR, Arantes AS, Stothard P. 2012. Comparing thousands of circular genomes using the CGView comparison tool. BMC Genomics 13:202. doi: 10.1186/1471-2164-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gentry J, Long L, Gentleman R, Falcon S, Hahne F, Sarkar D, Hansen KD. 2012. Rgraphviz: provides plotting capabilities for R graph objects. R package version 2.6.0. The R Project for Statistical Computing, Vienna, Austria. [Google Scholar]
- 57.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 58.Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 60.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 61.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- 62.Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA. 2013. diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 63.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the genetic diversity and Bayesian analysis of 146 USA300 MRSA strains.
GenBank accession numbers of the S. aureus completed genomes included in this study.
Details of the USA300 MRSA isolates analyzed in this study.
Linear regression plot showing correlation between the root-to-tip genetic distance and the isolation date of S. aureus isolates inferred from the ML tree. The ML tree was constructed in RAxML under the GTRGAMMA substitution model with the core genome alignment produced by progressiveMauve. The blue and red dots represent USA300 reference strains TCH1516 and FPR3757, respectively. The remaining dots represent isolates from this study. Download
Hierarchical clustering analysis of the pangenome of 196 S. aureus strains. The rows represent 196 S. aureus genomes, and the columns represent 3,455 core (genes present in all of the strains compared) and noncore (genes present in only a subset of the strains compared) genes identified among the isolates compared. Red and blue blocks indicate the presence and absence of gene clusters, respectively. Columns with all red blocks represent core genes, whereas those with mixed colors represent noncore genes. Of the 196 isolates, 148 are USA300 (146 from this study, TCH1516 [blue dot], and FPR3757 [red dot]) and the remaining 48 are non-USA300 isolates, as listed in Table S2 in the supplemental material. As shown (designated I), the USA300 and non-USA300 isolates formed two separate clades based on the presence or absence of the gene clusters. The USA300 isolates from the same household tend to cluster together, as indicated by the color code (designated II). The color codes used to indicate households are the same as in Fig. 3. Download
Box plot showing the number of SNPs per household as determined from the core genome alignment produced by progressiveMauve. The color scheme used to indicate households is the same as in Fig. 3. Download
ML tree constructed by alignment of 1,629 core protein clusters identified in Los Angeles and Chicago USA300 S. aureus isolates. The branch tips are colored on the basis of the mutations within the grlA and gyrA genes, as indicated. (A) Rooted tree. The color scheme in column I is as follows: blue, Chicago strains; orange, Los Angeles strains; red, Newman/COL; grey, TCH1516/FPR3757. The color scheme in column II indicates households and is the same as in Fig. 3. (B) Unrooted tree. Download