ABSTRACT
Direct measurement of multiple physical properties of Geobacter sulfurreducens pili have demonstrated that they possess metallic-like conductivity, but several studies have suggested that metallic-like conductivity is unlikely based on the structures of the G. sulfurreducens pilus predicted from homology models. In order to further evaluate this discrepancy, pili were examined with synchrotron X-ray microdiffraction and rocking-curve X-ray diffraction. Both techniques revealed a periodic 3.2-Å spacing in conductive, wild-type G. sulfurreducens pili that was missing in the nonconductive pili of strain Aro5, which lack key aromatic acids required for conductivity. The intensity of the 3.2-Å peak increased 100-fold when the pH was shifted from 10.5 to 2, corresponding with a previously reported 100-fold increase in pilus conductivity with this pH change. These results suggest a clear structure-function correlation for metallic-like conductivity that can be attributed to overlapping π-orbitals of aromatic amino acids. A homology model of the G. sulfurreducens pilus was constructed with a Pseudomonas aeruginosa pilus model as a template as an alternative to previous models, which were based on a Neisseria gonorrhoeae pilus structure. This alternative model predicted that aromatic amino acids in G. sulfurreducens pili are packed within 3 to 4 Å, consistent with the experimental results. Thus, the predictions of homology modeling are highly sensitive to assumptions inherent in the model construction. The experimental results reported here further support the concept that the pili of G. sulfurreducens represent a novel class of electronically functional proteins in which aromatic amino acids promote long-distance electron transport.
IMPORTANCE
The mechanism for long-range electron transport along the conductive pili of Geobacter sulfurreducens is of interest because these “microbial nanowires” are important in biogeochemical cycling as well as applications in bioenergy and bioelectronics. Although proteins are typically insulators, G. sulfurreducens pilus proteins possess metallic-like conductivity. The studies reported here provide important structural insights into the mechanism of the metallic-like conductivity of G. sulfurreducens pili. This information is expected to be useful in the design of novel bioelectronic materials.
INTRODUCTION
It is important to understand the mechanisms for long-range electron transport along the conductive pili of Geobacter species because these “microbial nanowires” play an important role in the global geochemical cycling of metals, minerals, and carbon in the environment, bioremediation of contaminants, and microbe-electrode interactions that have potential applications in energy harvesting and the renewable production of organic commodities from carbon dioxide (1–4). Although the pili of Geobacter species are comprised of proteins, and biological proteins are typically electronic insulators (5), the pili of Geobacter sulfurreducens have the remarkable ability to transport electrons over several cell lengths, and networks of these pili can conduct electrons over centimeter distances (6). Conductive pili permit Geobacter species to transfer electrons released from the internal oxidation of organic compounds onto external electron acceptors, such as the Fe(III) minerals that are abundant in soils and sediments (7), microbial partners that use the electrons to convert carbon dioxide to methane (8, 9), or to electrodes, producing electrical current in microbial fuel cells (10–12).
Surprisingly, the pili of G. sulfurreducens exhibit organic metallic-like conductivity, similar to that observed in synthetic organic metallic nanostructures comprised of polyaniline or polyacetylene, in which charges are delocalized, free, and spread throughout the polymer (2, 6). The temperature and pH dependence of pilus conductivity were similar to those of organic metals (6). Furthermore, direct visualization of charge propagation along individual pili by electrostatic force microscopy demonstrated that charges delocalize along pili similar to metallic carbon nanotubes (13, 14). This contrasts with typical biological electron transfer, in which charges are localized within discrete sites in biomolecules and quantum-mechanically tunnel or thermally hop between molecules to effect electron transport (2, 5). The conductivity of G. sulfurreducens pili exhibits temperature, pH, and redox responses similar to those of synthetic organic metals and consistent with metallic-like conductivity (6). Lowering the temperature, gate voltage, or pH enhances the conductivity by several orders of magnitude (6). Thus, these G. sulfurreducens pili represent a model system for genetically engineering a new class of electronically functional protein nanostructures that can perform as natural conductive materials. These new materials can provide a robust alternative to synthetic conducting polymers (15) because they are nontoxic, inexpensive, and easy to synthesize as they can be mass-produced using bacteria (6).
Overlapping π-π orbitals of aromatic moieties can confer metallic-like conductivity to synthetic organic materials (16), and aromatic amino acids are thought to be responsible for the metallic-like conductivity of G. sulfurreducens pili (6, 17, 18). Genetically altering the sequence of the gene for PilA, the pilin monomer that assembles into pili, in order to substitute alanine for key aromatic amino acids, yielded a strain of G. sulfurreducens (strain Aro5) that produced nonconductive pili (17). Only the wild-type strain was effective in long-range electron transport, whereas the Aro5 mutant strain was not capable of long-range electron transport to Fe(III) oxides and electrodes in microbial fuel cells (17).
However, these previous studies did not elucidate how aromatic amino acids are organized in the pilus structure to confer metallic-like conductivity. Multiple modeling studies have suggested that aromatic amino acids are not packed tightly enough for long-range conduction along the pili via metallic-like conductivity (19–22). For example, a study that modeled the G. sulfurreducens PilA monomer structure, templated on a crystallographic structure of the Pseudomonas aeruginosa pilin monomer, noted the likely role of aromatic amino acids in electron transfer in G. sulfurreducens pili but concluded that their arrangement provided “an optimal environment for the hopping of electrons” (22). Several studies advanced beyond the study of the pilin monomer and developed models to describe the localization of aromatic amino acids in an assembled pilus (19–21). A model in which a G. sulfurreducens PilA structure (also modeled from the P. aeruginosa structure) was superimposed onto a cryo-electron microscopy-based empirical model of the Neisseria gonorrhoeae pilus (23) predicted zones rich in aromatic amino acids separated by bands devoid of aromatic amino acids (19). A similar result was obtained when the structure of the G. sulfurreducens PilA monomer determined by nuclear magnetic resonance (NMR) was superimposed onto the N. gonorrhoeae pilus model (21). The zones lacking aromatic amino acids in these models would prevent metallic-like conduction of electrons along the length of the pili. A related modeling work that docked the P. aeruginosa pilin structure into the N. gonorrhoeae structure found a lack of continuous stacking of aromatic residues (20), again inconsistent with the requirements for metallic-like conductivity.
However, the results of such homology modeling can be significantly influenced by the choice of structural template. The empirical model of the N. gonorrhoeae pilus (23), which was derived by fitting the N. gonorrhoeae PilE pilin structure into a 12.5-Å resolution cryo-electron microscopy pilus reconstruction (23), served as the template for all previous homology models of the G. sulfurreducens pilus. However, the G. sulfurreducens PilA monomer shows higher similarity to the PilA of P. aeruginosa (50%) than the PilE of N. gonorrhoeae (40%). Additional similarity between the PilA of G. sulfurreducens and P. aeruginosa over PilE of N. gonorrhoeae is due to following residues: V10, I13, I19, I21, Q23, L43, T45, E48, and A50. A cryo-electron microscopy-based empirical model of P. aeruginosa pilus is not available, but a computational model has been derived from X-ray fiber diffraction data and the X-ray crystal structure of the pilin (24). The predicted structure of the P. aeruginosa pilus has distinct helical symmetry, and the pilin subunits are packed more tightly than in the N. gonorrhoeae pilus (24). In the model for P. aeruginosa pilus, pilin subunits are assembled as a right-handed one-start helix (a helical path that connects every subunit in the filament with the smallest axial rotation) with a 41-Å pitch and 4 subunits per turn (24), whereas there are ~3.6 subunits per turn of a 37-Å-pitch one-start helix in the N. gonorrhoeae pilus (23). These key differences in the two pilus models are expected to yield different spacings between aromatic amino acids.
Here we report high-resolution synchrotron X-ray diffraction analyses that provide evidence for sufficiently tight stacking of aromatic amino acids to account for the metallic-like conductivity of G. sulfurreducens pili. A new homology model for the pilus assembly of G. sulfurreducens based on a model of the P. aeruginosa pilus as a template rather than the N. gonorrhoeae template made predictions consistent with the experimental results.
RESULTS AND DISCUSSION
Synchrotron X-ray microdiffraction reveals 3.2-Å spacing among aromatics.
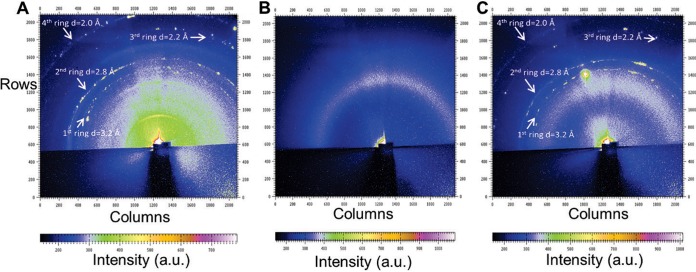
Synchrotron X-ray microdiffraction revealed 3.2-Å periodic spacing in the G. sulfurreducens pilus preparations, as well as some smaller spacings (Fig. 1A and C). The origin of the smaller spacings is not currently known, but as detailed below, only the 3.2-Å spacing was directly related to pilus conductivity. The Aro5 strain of G. sulfurreducens, which was genetically modified to produce pili without the aromatic amino acids required for conductivity (17), lacked the periodic spacing observed in the wild-type PilA (Fig. 1B), even though equal amounts of pilus protein were used for all measurements and the presence of pili in the samples was confirmed by transmission electron microscopy (see Fig. S1 in the supplemental material). Similar preparations from a strain of G. sulfurreducens that cannot produce pili because the gene for the PilA monomer was deleted also lacked the periodic spacing (see Fig. S2A in the supplemental material), as did the buffer used for making the pilus preparations (see Fig. S2B). The presence of the 3.2-Å periodic spacing only in G. sulfurreducens pili that are conductive is consistent with the spacing of 3 to 4 Å for the aromatic amino acids required for efficient orbital overlap and electron delocalization (25–27) that can confer metallic-like conductivity to the pili (16).
FIG 1 .
Synchrotron X-ray microdiffraction from a 2- by 5-µm spot focused on pili of the G. sulfurreducens wild-type strain (A), an Aro5 strain expressing modified PilA lacking aromatic amino acids required for pilus conductivity (B), and a control strain constructed in the same manner as Aro5, but with the wild-type PilA sequence, yielding conductive pili (C). Arrows in panels A and C point to the diffraction rings. a.u., arbitrary units.
Rocking-curve X-ray diffraction confirms the 3.2-Å spacing among aromatics.
The synchrotron X-ray microdiffraction studies were complemented with rocking-curve X-ray diffraction measurements, which provide more complete coverage of the diffraction space (28, 29). The rocking-curve X-ray diffraction method further confirmed the 3.2-Å periodic spacing, corresponding to a 2θ value of ≈28.4°, in the X-ray diffraction measurement of wild-type, conductive pili. The nonconductive pili of strain Aro5 lacked the 3.2-Å periodic spacing (Fig. 2), as did preparations from the G. sulfurreducens strain that could not produce pili and the buffer used to make the preparations (Fig. 3). When the samples were rotated to evaluate whether there were peaks present at any other orientation, as well as to determine sample heterogeneity, rocking-curve X-ray diffraction measurement of only wild-type pili showed the peak, further confirming that the 3.2-Å spacing is unique to wild-type, conductive, pili (Fig. 4). Therefore, these results from rocking-curve X-ray diffraction provided an independent and complementary confirmation that the 3.2-Å spacing is associated with the aromatic amino acids required for conductivity of the wild-type pili.
FIG 2 .
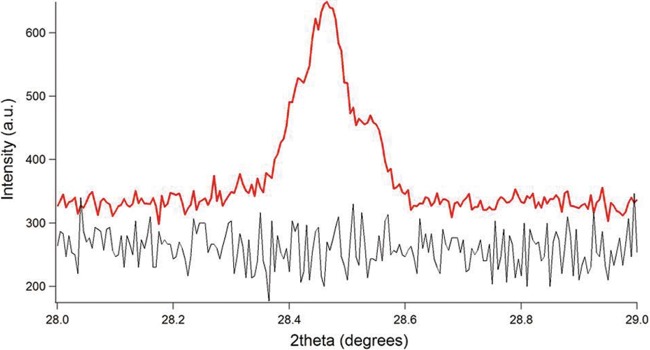
X-ray diffraction as a function of 2θ for pili of an Aro5 strain expressing modified PilA lacking aromatic amino acids required for pilus conductivity (black curve) and a positive-control strain constructed in the same manner as Aro5, but with the wild-type PilA sequence, yielding conductive pili (red curve).
FIG 3 .
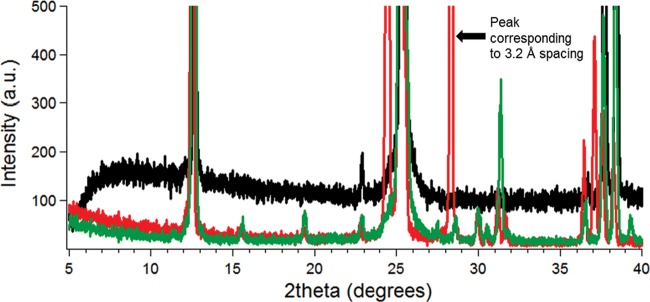
X-ray diffraction as a function of 2θ for pili of the G. sulfurreducens wild-type strain (red curve), pilus-deficient strain (black curve), and buffer (green curve).
FIG 4 .
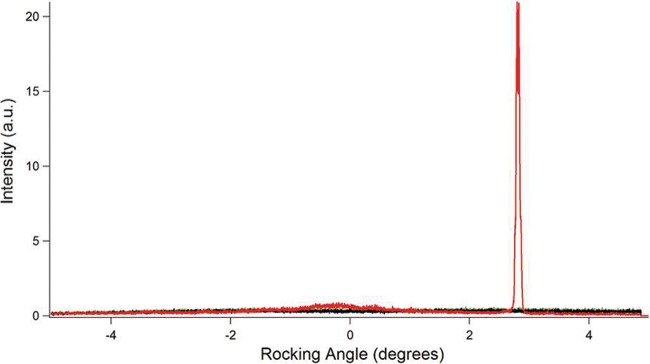
Rocking-curve X-ray diffraction, as a function of rocking angle, for pili of the G. sulfurreducens wild-type strain (red curve), pilus-deficient strain (black curve), and buffer (green curve).
Protonation induces conformational changes in aromatics that increase pilus conductivity.
In previous studies, protonation increased the conductivity of G. sulfurreducens pili by 100-fold when the pH was lowered from pH 10.5 to pH 2 (6). This result was surprising because proteins typically denature and lose functionality at low pH, whereas G. sulfurreducens pili maintained their composition at pH 2 (see Fig. S3 in the supplemental material). However, this proton-doping effect is consistent with the previous finding that the pili of G. sulfurreducens show p-type conductivity that contains positive-charge carriers (holes) (6).
In order to determine if the higher pilus conductivity at pH 2 was associated with structural changes, X-ray diffraction results from samples at pH 10.5 and pH 2 were compared (Fig. 5). There was a 100-fold increase in the intensity of the peak representing the 3.2-Å spacing at pH 2, corresponding to the 100-fold increase in conductivity at pH 2. These results demonstrate that protonation leads to conformational changes in the pili that are coincident with the dramatic increase in conductivity.
FIG 5 .
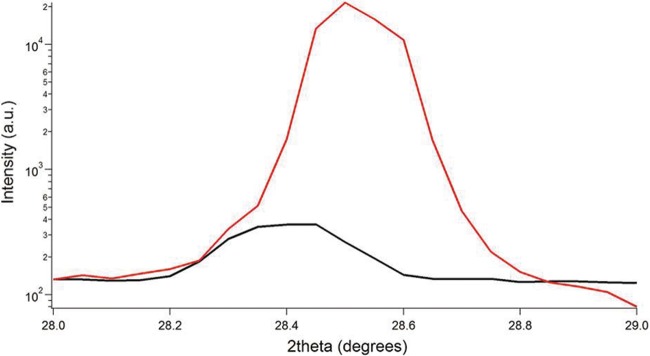
X-ray diffraction as a function of 2θ for pili of the G. sulfurreducens wild-type strain at pH 10.5 (black curve) and at pH 2 (red curve).
The mechanism underlying the pH-induced conformation change in G. sulfurreducens pilus is not yet known, but the charge transport properties of organic conductors critically depend on the packing of the individual polymer chains and the extent of the overlap between electronic wave functions of adjacent chains (30). The orientations of aromatic rings can substantially influence the conductivity of organic materials (31). For example, engineering of structural changes in polyaniline to reduce the torsion angles between the phenyl rings and the plane of the backbone resulted in more phenyl rings with a face-to-face interchain stacking distance of ~3.5 Å, increasing conductivity and improving the metallic nature (16). In a similar manner, the 100-fold increase in the intensity of the 3.2-Å peak in pili at pH 2 suggests that there is an increase of aromatics within 3.2 Å from each other. One possibility is that aromatic rotamers form a more planar structure due to α-helices interacting more tightly at lower pH, enabling more aromatics within the π-stacking distance and thus increasing the effective π conjugation length. However, additional structural studies using techniques such as circular dichroism (32, 33) will be required to evaluate this possibility. The finding that the increase in conductivity at lower pH is associated with an increase in the signal that is associated with the aromatic amino acids required for pilus conductivity further supports the hypothesis that the metallic-like conductivity of the pili can be attributed to the overlapping π-π orbitals of aromatic amino acids.
Modeling suggests aromatics are closely packed in G. sulfurreducens pili.
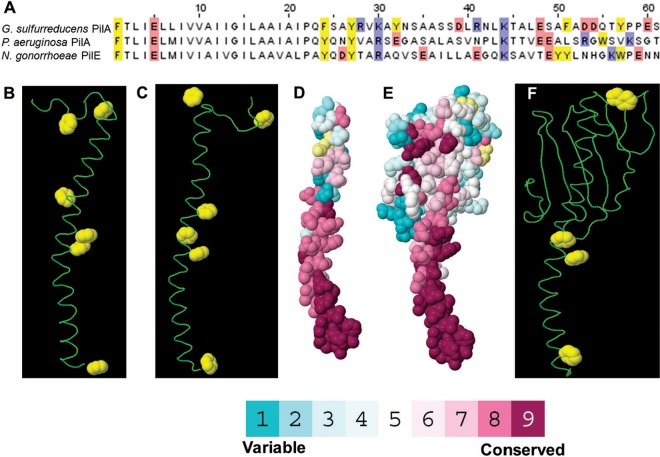
The results of previous studies (19–21) that attempted to describe the mechanisms for electron transport along G. sulfurreducens pili with homology modeling of the G. sulfurreducens pilus structure are inconsistent not only with the X-ray diffraction data but also with previously reported physical evidence (6, 13, 14) supporting the concept that G. sulfurreducens pili have metallic-like conductivity. These previous homology models used the Neisseria gonorrhoeae pilus as the structural template. However, the G. sulfurreducens pilin subunit has higher homology to the Pseudomonas aeruginosa pilin subunit (50%) than the N. gonorrhoeae pilin subunit (40%) (Fig. 6A). Therefore, a new model of the G. sulfurreducens pilus structure was constructed using the model of the P. aeruginosa strain K pilus (24) as a template (Fig. 7).
FIG 6 .
(A) Alignment of the first 61 amino acids for pilin subunits of Geobacter sulfurreducens (61 amino acids [PDB accession no. 2M7G]), Pseudomonas aeruginosa strain K (144 amino acids [PDB accession no. 1OQW]), and Neisseria gonorrhoeae (158 amino acids [PDB accession no. 2HI2]). Yellow represents aromatic amino acids, whereas blue and red represent basic and acidic residues, respectively. (B to D) Predicted structure of G. sulfurreducens pilin monomers derived from NMR (B) and homology modeling (C and D). (E and F) Structure of the Pseudomonas aeruginosa strain K pilin monomer. Colors for all-atom models in Fig. D and E represent evolutionary conservation scores from ConSurf (http://consurf.tau.ac.il). Yellow represents insufficient data to calculate conservation scores. The amino termini are at the bottom.
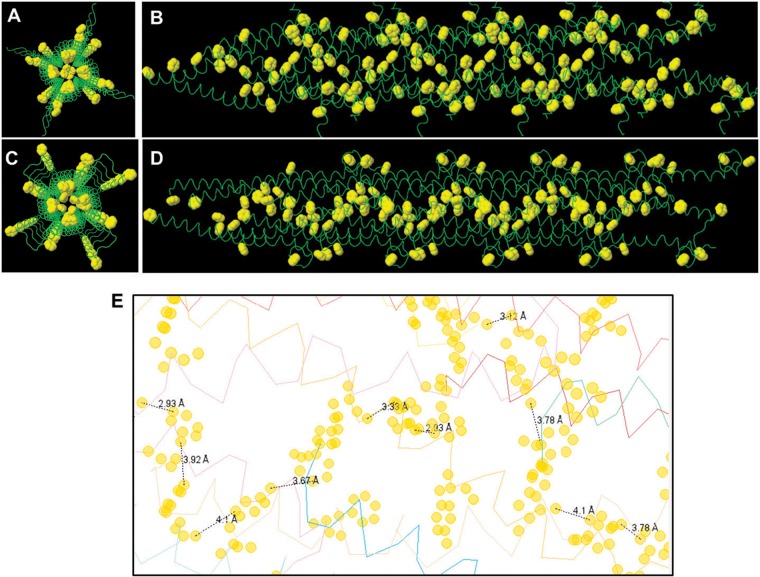
FIG 7 .
End view and side view of the predicted structure of a Geobacter sulfurreducens pilus derived using monomer pilin based on NMR structure (A and B) and homology modeling (C and D). Aromatic side-chain rings are highlighted in yellow and displayed at their van der Waals radii (carbon, 1.7 Å) (51). (E) Representative distances between different aromatic amino acids in the homology-modeled G. sulfurreducens pilus. Distances measured between proximal atoms in aromatic residues in this figure averaged 3.6 Å (range, 2.9 to 4.1 Å).
There were 18 different conformers previously reported (21) for the solution NMR analysis of the G. sulfurreducens PilA monomer (Protein Data Bank [PDB] accession no. 2M7G). In order to evaluate the best possible conformer, the clash scores and MolProbity scores (MPscores) were calculated for all 18 conformers (see Fig. S4 in the supplemental material). Conformer no. 5 (Fig. 6B) yielded the best clash and Molprobity scores and thus was the conformer utilized for modeling the pilus structure.
The predicted assembly of the G. sulfurreducens pilus using the NMR structure of the PilA monomer and the P. aeruginosa pilus template (Fig. 7A and B) yielded a better fit (root mean square deviation [RMSD] of 2.08 Å for the backbone residues) for superimposing each pilin subunit than the fit (RMSD, 2.6 Å) previously reported (21) for the model of the G. sulfurreducens pilus model based on the N. gonorrhoeae template. The model based on the P. aeruginosa pilus template predicted a pilus diameter of 3 nm, which is consistent with the diameter of G. sulfurreducens pili previously measured by atomic force microscopy (13, 34). In contrast, previous models of the G. sulfurreducens pilus based upon the N. gonorrhoeae pilus template predicted a pilus diameter of 4 to 5 nm (19–21).
The G. sulfurreducens pilus model based on the P. aeruginosa template predicted a continuous arrangement of aromatic amino acids (Fig. 7A and B). In contrast, a previous model based on the NMR structure of the G. sulfurreducens pilin subunit, but using the N. gonorrhoeae template, predicted that aromatic amino acids in the G. sulfurreducens pilus are clustered together within a sphere of radius of 15 Å, forming an aromatic-rich region separated by a region devoid of aromatic amino acids (21). This discontinuous arrangement would not facilitate metallic-like conductivity along the length of the pili.
The G. sulfurreducens pilin can adopt different conformations in solution than when crystallized (21), and the aromatic amino acids for the NMR structure have been shown to position in different orientations than a G. sulfurreducens pilin structure derived from homology modeling based on a high-resolution template structure of the P. aeruginosa pilin (22). Therefore, in order to more fully explore the possible predictions of homology models, a model of the G. sulfurreducens pilus structure based on a homology model of G. sulfurreducens pilin (Fig. 6C) was also constructed (Fig. 7C and D).
To construct a homology model of G. sulfurreducens PilA, the 61 C-terminal residues of G. sulfurreducens PilA monomer (UniProt accession no. D7AIT1) were modeled. The model was templated against the 2-Å-resolution X-ray crystal structure of the pilin of Pseudomonas aeruginosa strain K (Protein Data Bank [PDB] entry 1OQW) (35). As previously reported (19–22), the predicted structures of the G. sulfurreducens monomer using homology modeling as well as the empirical pilin structure from NMR were similar to the P. aeruginosa monomer in the N-terminal region that contains an α-helix (Fig. 6). This is expected due to the high conservation of the N-terminal sequence in the P. aeruginosa and G. sulfurreducens monomers. In the model-to-template sequence alignment, 22 of the N-terminal 23 residues are identical, whereas residues 24 to 50 align with 26% identity (Fig. 6). The P. aeruginosa template is α-helical from residues 2 to 54. The homology model of G. sulfurreducens is an α-helix spanning residues 2 to 51 (Fig. 6C). Therefore, this helical model appears reliable, based on the sequence alignment.
The P. aeruginosa PilA structure contains two bends at proline 22 and proline 42 of approximately 159 and 164°, respectively. The G. sulfurreducens model conserves proline 22 but has no other prolines in the helical portion; hence, the modest bend in the homology model at asparagine 42 is unlikely and is absent in the NMR structure (21).
The fit for superimposing each pilin subunit using the homology model of the pilin subunit (RMSD, 1.52 Å) was better than that obtained with the NMR structure of the PilA monomer. This model also predicted an abundance of aromatic amino acids positioned within 3 to 4 Å of each other (Fig. 7E), consistent with the π-stacking distance that enables π-orbital overlap and electron delocalization in synthetic organic metals (16) and synthetic conductive proteins (36).
In contrast, examination of the predicted positioning of the aromatic amino acids in the model of the P. aeruginosa pilus structure (24) indicated that the distances between the majority of the aromatic amino acids were 10 to 40 Å (see Fig. S5 in the supplemental material). This is consistent with the previous finding that the pili of P. aeruginosa are not conductive (7, 18). The pilin of P. aeruginosa contains 7 aromatics compared to 6 for G. sulfurreducens (Fig. 6). Positions 1, 24, and 27 in the α-helix are common for aromatics, whereas G. sulfurreducens pilin contains additional aromatics at positions 32, 51, and 57. P. aeruginosa contain additional aromatics at positions 55, 102, 127, and 137. Thus, G. sulfurreducens pilin possesses 2 more aromatics in its α-helix, which are absent in the α-helix of P. aeruginosa pilin. The absence of these aromatic amino acids in the P. aeruginosa α-helix might result in a gap among aromatics that prohibits charge transfer. Thus, the distribution of aromatics along the α-helix and the distance between helices may be the decisive factor in the pilus conductivity. The G. sulfurreducens pilin is much smaller (61 amino acids) than the P. aeruginosa pilin (144 amino acids), which has a globular head domain that leads to a pilus that is thicker (5 to 6 nm in diameter) (24) than the G. sulfurreducens pilus. This almost 2-fold difference in the thickness suggests that the α-helices in the pilus of P. aeruginosa are much farther apart due to the presence of the globular domain. Thus, the aromatics in the α-helix will not be able to overlap efficiently in the P. aeruginosa pilus, in contrast to the G. sulfurreducens pilus. Thus, the G. sulfurreducens pilus model was consistent with the experimental evidence suggesting that tight packing of aromatic amino acids confers metallic-like conductivity along G. sulfurreducens pili.
Increased charge carriers cannot account for increased conductivity upon protonation.
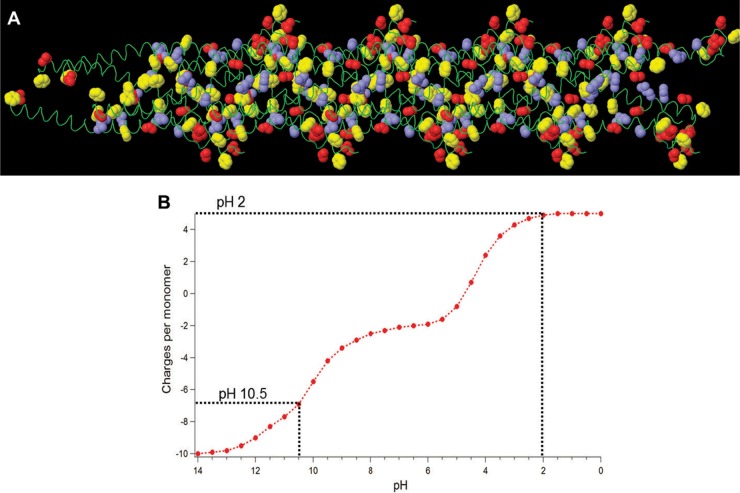
One factor potentially leading to increased conductivity of G. sulfurreducens pili at lower pH is that conductivity is proportional to the total number of positive-charge carriers and thus increasing the positive charges in the pilin monomer with greater protonation at low pH would be expected to enhance pilus conductivity. The pilin of G. sulfurreducens contains 6 acidic residues (3 aspartic acid and 3 glutamic acid) and 4 basic residues (2 arginine and 2 lysine) (Fig. 6A and Fig. 8A). Modeling the change in the charges in pilin monomer as a function of pH associated with the protonation of these charged amino acids suggested that the hole carriers in conductive pili should increase ca. 10 units (Fig. 8B). At pH 10.5, the net charge is ca. −7, whereas at pH 2, the net charge is +5 due to neutralization of 6 acidic residues and the N terminal, each losing one negative charge. This change is an order of magnitude lower than the observed 100-fold increase in conductivity, suggesting that an increase in holes due to charged residues cannot account for the disproportionately higher conductivity at pH 2. Thus, the increase in packing of aromatic amino acids at pH 2 that was observed in the X-ray diffraction studies appears to be the more likely explanation for the increased conductivity of the pili.
FIG 8 .
(A) Side view of the predicted structure of a Geobacter sulfurreducens pilus derived using homology modeling. Aromatic rings of phenylalanine and tyrosine amino acids are highlighted in yellow, whereas blue and red are applied to the side chains of basic and acidic residues, respectively. Side-chain atoms are displayed at the van der Waals radii of carbon (1.7 Å) (51). (B) Modeling of charges per monomer in Geobacter sulfurreducens pilin as a function of pH. The charges corresponding to pH 10.5 and pH 2 are represented by the dotted lines.
Implications.
The X-ray diffraction analyses suggest that the metallic-like conductivity of G. sulfurreducens pili can be attributed to the packing of aromatic amino acids with a characteristic spacing of 3.2 Å, which enables effective π-orbital overlap and electron delocalization. The 3.2-Å spacing was observed only in wild-type conductive pili, not in pili that lack the key aromatic amino acids required for conductivity. Furthermore, the abundance of the 3.2-Å spacing increased proportionally with the increase in conductivity as the pH was lowered from 10.5 to 2.
Homology modeling of the G. sulfurreducens pilus with the P. aeruginosa pilus as a template demonstrated that a pilus structure in which aromatic amino acids are packed sufficiently close for metallic-like conductivity is feasible, supporting the experimental data. However, as illustrated by the difference between the homology modeling outcome based on a P. aeruginosa template reported here and previous results with an N. gonorrhoeae template (19–21), the predictions of homology modeling are highly sensitive to the template employed. Therefore, definitive experimental determination of the G. sulfurreducens pilus structure, using techniques such as high-resolution cryo-electron microscopy (37), is required in order to unequivocally define the arrangement of aromatic amino acids within the pilus and their contribution to metallic-like conductivity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The following strains were obtained from our laboratory culture collection: Geobacter sulfurreducens strain KN400 (38); strain KN400 ΔpilA, in which the gene for pilin monomer PilA was deleted (39); strain KN400 ΔpilT, in which the gene for PilT, involved in pilus retraction, was deleted; strain Aro5, which expresses a modified PilA lacking aromatic amino acids required for pilus conductivity (17); and a control strain constructed in the same manner as Aro5, but with the wild-type PilA sequence, yielding conductive pili (17). The cultures were maintained at 30 or 25°C under strictly anaerobic conditions in growth medium supplemented with acetate (10 mM) as the electron donor and fumarate (40 mM) as the electron acceptor as described previously (7, 40).
Pilus filament preparation.
Pilus filaments were prepared as described previously (6). To remove pili from live cells of the KN400 strains, cells were washed twice in 150 mM ethanolamine buffer (pH 10.5) and vortexed for 2 min to remove pili (41). Cells were removed by centrifugation. Pili were concentrated and washed by ultracentrifugation at 100,000 × g or by ammonium sulfate precipitation and resuspended in ethanolamine buffer (41). Additional purification was performed using a sucrose gradient method (42). Protein measurements of pilus samples were carried out by the bicinchoninic acid method with bovine serum albumin as a standard as described previously (6). For all comparisons, 15 µg of pilus protein was drop-casted onto the silicon substrate for structural experiments that resulted in randomly oriented pili. The presence of pili in samples was confirmed using transmission electron microscopy as well as protein measurements. The buffer containing pili was equilibrated with aqueous HCl to perform pH experiments (43).
Synchrotron X-ray microdiffraction.
In order to experimentally evaluate the structural basis of metallic-like conductivity of the G. sulfurreducens pilus, synchrotron X-ray microdiffraction was employed. In this method, X-rays are focused to a spot size of a micrometer or less, providing very high spatial resolution (44). Due to small spot size but high intensity, the synchrotron X-ray microdiffraction method can give structural information for small samples. Furthermore, heterogeneous samples can be studied very effectively with this method as it enables measurements of local variations in the crystallographic orientations for polycrystalline samples (44). Pilus samples with a 50-µl volume were drop-casted on a diced silicon wafer prior to diffraction studies and air dried. Synchrotron X-ray microdiffraction measurements were performed at the National Synchrotron Light Source (NSLS) beamline X13B, Brookhaven National Laboratory, at an X-ray wavelength of 1.03 Å as described previously (45). X13B has microfocusing optics that can produce X-ray beam sizes of the order of 1 µm. In addition, the insertion device at X13B was used to improve the intensity of the X-ray source. The beamline uses an in vacuum undulator, described previously (45). There is a water-cooled monochromator to select the X-ray wavelength, with highly polished Si(111)-oriented surfaces, aligned in an artificial channel cut arrangement. A diamond window is the exit window from the beamline. The experiment was performed with the sample at the beam spot, which was 2 µm vertical by 5 µm horizontal focused by a Kirkpatrick-Baez (K-B) mirrors system with a working distance of 1 cm and focal lengths of 5 cm and 10 cm in the horizontal and vertical, respectively. The sample to detector distance was 150 mm. The asymmetry of the focused X-ray beam size reflected the asymmetry of the electron beam inside the NSLS synchrotron ring. Beam size was measured with fluorescence from a nano-patterned Cr knife edge. A charge-coupled device (CCD) area detector (Princeton Instruments, Trenton, NJ) was used to acquire diffraction patterns. No flood field or spatial distortion corrections were performed, due to the reasonably high fidelity of the fiber optic taper. An Alumina powder sample was used for calibration. The beam was scanned along the sample to locate pili filaments before collection of diffraction data. The FIT2D program was used to plot and analyze the diffraction data (46).
Rocking-curve X-ray diffraction.
Pili samples for rocking-curve X-ray diffraction were prepared in a manner similar to that in synchrotron studies. X-ray diffraction was measured using Cu Kα radiation (λ = 1.54 Å) on an Ultima III multipurpose diffractometer (Rigaku Corp., The Woodlands, TX) in parallel beam geometry with a 100-µm vertical beam size and a Cu-tube as the X-ray source. Prior to all measurements, samples were aligned as per the manufacturer’s instructions. This alignment is intended to ensure that the sample surface is parallel to the incident X-ray beam and that the sample surface also coincides with the center of rotation of the goniometer axes. Rocking curves are used to measure the distribution of diffraction peaks. For rocking-curve measurements, the source and the detector were fixed at the Bragg angle of interest, and the sample plane was tilted (rocked) around the position satisfying the Bragg condition (28, 29). IGOR Pro software (WaveMetrics, Inc., Portland, OR) was used for data fitting and analysis.
All of the experimental results from structural studies were confirmed by repeated measurements on at least three biological replicates.
Homology modeling of pili.
For the modeling using the NMR structure, the clash score and MolProbity score (MPscore) were calculated for all 18 conformers to determine the best conformer (47) (see Fig. S4 in the supplemental material). The clash score is defined as the number of unfavorable all-atom steric overlaps of ≥0.4 Å per 1,000 atoms (47), whereas the MolProbity score is defined as 0.426 × ln(1 + clash score) + 0.33 × ln[1 + max(0,rota_out | −1)] + 0.25 × ln(1 + max[0,rama_iffy | −2)] + 0.5, where rota_out is the percentage of side-chain conformations classed as rotamer outliers from those side chains that can be evaluated, and Rama_iffy is the percentage of backbone Ramachandran conformations outside the favored region from those residues that can be evaluated. The coefficients were derived from a log-linear fit to crystallographic resolution on a filtered set of PDB structures, so that a model’s MPscore is the resolution at which its individual scores would be the expected values (47). Thus, lower MPscores are better. The conformer no. 5 (Fig. 6B) yielded the best clash score and also the best overall MolProbity score (see Fig. S4). Therefore, this conformer was used in addition to our homology model for modeling the G. sulfurreducens pilus filament assembly.
A homology model for the full pilus filament (Fig. 7) was built using the Swiss-Pdb Viewer (DeepView) software (http://spdbv.vital-it.ch). The modeling was templated against the model of the Pseudomonas aeruginosa strain K pilus filament (24). The P. aeruginosa pilus filament model was computationally assembled using the X-ray fiber diffraction data and the X-ray crystal structure of pilin (24). In this model, pilin subunits are assembled as a right-handed one-start helix (a helical path that connects every subunit in the filament with the smallest axial rotation) with a 41-Å pitch and four subunits per turn or as a left-handed three-start helix (a set of three identical helices with the same pitch and number of subunits per turn, which together connect all subunits in the filament) with a 123-Å pitch and four submits per turn (24). The spacing between the three-start strands is 41 Å, consistent with the fiber diffraction data (24). The model does not account for side-chain flexibility that might allow tighter packing. Therefore, the structure-based model of the P. aeruginosa pilus filament is slightly thicker than the dimensions obtained from the fiber diffraction data (24).
To construct the pilus model of G. sulfurreducens using the NMR structure of the pilin monomer, 19 copies of the conformer no. 5 were superimposed, using DeepView’s “magic fit” (sequence-guided structural alignment, backbone root mean square deviation [RMSD] of 2.08 Å), onto the 19 pilin monomers of Pseudomonas aeruginosa strain K (35) to obtain a full pilus filament assembly model (Fig. 7A and B).
A homology model of the G. sulfurreducens pilin monomer was built using the Swiss-Model server (http://swissmodel.expasy.org) (48). ConSurf (http://consurf.tau.ac.il) was used for calculation of the evolutionary conservation of amino acids presented in Fig. 6D and E (49).
To construct the pilus model of G. sulfurreducens using the homology model of the pilin monomer, 19 copies of the pilin monomer homology model of G. sulfurreducens were superimposed, using DeepView’s “magic fit” (sequence-guided structural alignment, backbone RMSD of 1.52 Å), onto the 19 pilin monomers of Pseudomonas aeruginosa strain K (35) to obtain a full pilus filament assembly model (Fig. 7C and D).
FirstGlance in Jmol (http://firstglance.jmol.org) and the Jmol application (http://jmol.org) (50) were used to generate the images shown in Fig. 6 to 8. Aromatic amino acids in G. sulfurreducens PilA are highlighted in yellow in Fig. 6 to 8, and the aromatic ring atoms are displayed at their van der Waals radii (carbon, 1.7 Å) (51). Models and animations can be visualized or downloaded as described below.
The Protein Calculator tool (http://protcalc.sourceforge.net) was used to estimate protonation-induced charges in pilus filaments (Fig. 8B).
Animated models and sources for pilus assembly by NMRstructure and homology modeling.
Figures S1 to S5 are presented in the supplemental material. The atomic coordinates of the G. sulfurreducens pilus filament assembly models can be downloaded from the URLs given below in PDB format and will be visualized after publication at http://Proteopedia.Org/w/Malvankar/1. To download G. sulfurreducens pilus assembly using the NMR structure of the monomer, go to http://proteopedia.org/wiki/images/4/4a/Geobacter_Pilus_Pseuodmonas_template_homology_NMR_monomer.pdb. For G. sulfurreducens pilus assembly using homology modeling of the monomer, go to http://proteopedia.org/wiki/images/e/e4/Geobacter_Pilus_Pseuodmonas_template_homology_monomer.pdb. An animated model of G. sulfurreducens pilus assembling from 18 pilin monomers can be found at http://proteopedia.org/wiki/images/b/b5/Geobacter_pilus_assembling_animation.gif. An animated model of the final assembly of G. sulfurreducens pilus can be found at http://proteopedia.org/wiki/images/0/0f/Geobacter_pilus_assembled_rocking.gif. An animated model of the aromatic rings in the G. sulfurreducens pilus can be found at http://proteopedia.org/wiki/images/0/0a/Geobacter_pilus_aromatics_rotating.gif.
SUPPLEMENTAL MATERIAL
Transmission electron micrograph of pili of Geobacter sulfurreducens strain Aro5. Download
Synchrotron X-ray microdiffraction from a 2- by 5-µm spot focused on the (A) pilus-deficient strain. (B) Buffer. Download
Transmission electron micrographs of pili of the Geobacter sulfurreducens wild-type strain at pH 2 at scale bars 200 nm (A) and 100 nm (B). Download
The clash scores and Molprobity scores for 18 different conformers present in the NMR structure of G. sulfurreducens pilin. Conformer 5 gave the best scores and hence was used for modeling the pilus assembly. Download
Representative distances between different aromatic amino acids in the P. aeruginosa pilus. Distances measured between proximal atoms in aromatic rings in this figure averaged 19.9 Å (range, 5.2 to 37.5 Å). Download
ACKNOWLEDGMENTS
We thank Lisa Craig for providing the pilus model of P. aeruginosa strain K and for critical review of the manuscript.
This research was supported by the Office of Naval Research (grant no. N00014-12-1-0229 and N00014-13-1-0550). Research was carried out in part at the Center for Functional Nanomaterials, Brookhaven National Laboratory, which is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract no. DE-AC02-98CH10886. Nikhil S. Malvankar holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
N.S.M. performed modeling and experiments. N.S.M. and M.V. prepared pilus samples. N.S.M. and K.N. carried out strain growth. P.L.T. generated the KN400 ΔpilT strain. K.E. supervised synchrotron experiments. D.N. supervised rocking-curve experiments. E.M. supervised modeling and molecular visualization. N.S.M. and D.R.L. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
Citation Malvankar NS, Vargas M, Nevin K, Tremblay P-L, Evans-Lutterodt K, Nykypanchuk D, Martz E, Tuominen MT, Lovley DR. 2015. Structural basis for metallic-like conductivity in microbial nanowires. mBio 6(2):e00084-15. doi:10.1128/mBio.00084-15.
REFERENCES
- 1.Malvankar NS, Lovley DR. 2012. Microbial nanowires: a new paradigm for biological electron transfer and bioelectronics. ChemSusChem 5:1039–1046. doi: 10.1002/cssc.201100733. [DOI] [PubMed] [Google Scholar]
- 2.Malvankar NS, Lovley DR. 2014. Microbial nanowires for bioenergy applications. Curr Opin Biotechnol 27:88–95. doi: 10.1016/j.copbio.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru AE, Holmes DE, Franks AE, Orellana R, Risso C, Nevin KP. 2011. Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv Microb Physiol 59:1–100. doi: 10.1016/B978-0-12-387661-4.00004-5. [DOI] [PubMed] [Google Scholar]
- 4.Lovley DR, Nevin KP. 2013. Electrobiocommodities: powering microbial production of fuels and commodity chemicals from carbon dioxide with electricity. Curr Opin Biotechnol 24:385–390. doi: 10.1016/j.copbio.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Edwards PP, Gray HB, Lodge MTJ, Williams RJP. 2008. Electron transfer and electronic conduction through an intervening medium. Angew Chem Int Ed Engl 47:6758–6765. doi: 10.1002/anie.200703177. [DOI] [PubMed] [Google Scholar]
- 6.Malvankar NS, Vargas M, Nevin KP, Franks AE, Leang C, Kim BC, Inoue K, Mester T, Covalla SF, Johnson JP, Rotello VM, Tuominen MT, Lovley DR. 2011. Tunable metallic-like conductivity in microbial nanowire networks. Nat Nanotechnol 6:573–579. doi: 10.1038/nnano.2011.119. [DOI] [PubMed] [Google Scholar]
- 7.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 8.Rotaru A, Shrestha PM, Liu F, Shrestha M, Shrestha D, Embree M, Zengler K, Wardman C, Nevin KP, Lovley DR. 2014. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci 7:408–415. doi: 10.1039/c3ee42189a. [DOI] [Google Scholar]
- 9.Rotaru AE, Shrestha PM, Liu F, Markovaite B, Chen S, Nevin KP, Lovley DR. 2014. Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri. Appl Environ Microbiol 80:4599–4605. doi: 10.1128/AEM.00895-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malvankar NS, Tuominen MT, Lovley DR. 2012. Biofilm conductivity is a decisive variable for high-current-density microbial fuel cells. Energy Environ Sci 5:5790–5797. doi: 10.1039/c2ee03388g. [DOI] [Google Scholar]
- 11.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol 72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nevin KP, Kim BC, Glaven RH, Johnson JP, Woodard TL, Methé BA, DiDonato RJ Jr, Covalla SF, Franks AE, Liu A, Lovley DR. 2009. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4:e5628. doi: 10.1371/journal.pone.0005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malvankar NS, Yalcin SE, Tuominen MT, Lovley DR. 2014. Visualization of charge propagation along individual pili proteins using ambient electrostatatic force microscopy. Nat Nanotechnol 9:1012–1017. doi: 10.1038/nnano.2014.236. [DOI] [PubMed] [Google Scholar]
- 14.Lovley DR, Malvankar NS 11 November 2014. Seeing is believing: novel imaging techniques help clarify microbial nanowire structure and function. Environ Microbiol doi: 10.1111/1462-2920.12708. [DOI] [PubMed] [Google Scholar]
- 15.Qian F, Li Y. 2011. Biomaterials: a natural source of nanowires. Nat Nanotechnol 6:538–539. doi: 10.1038/nnano.2011.148. [DOI] [PubMed] [Google Scholar]
- 16.Lee K, Cho S, Park SH, Heeger AJ, Lee CW, Lee SH. 2006. Metallic transport in polyaniline. Nature 441:65–68. doi: 10.1038/nature04705. [DOI] [PubMed] [Google Scholar]
- 17.Vargas M, Malvankar NS, Tremblay PL, Leang C, Smith JA, Patel P, Snoeyenbos-West O, Nevin KP, Lovley DR. 2013. Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. mBio 4(2):e00105-13. doi: 10.1128/mBio.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Tremblay P-L, Malvankar NS, Nevin KP, Lovley DR, Vargas M. 2014. A Geobacter sulfurreducens strain expressing Pseudomonas aeruginosa type IV pili localizes OmcS on pili but is deficient in Fe(III) oxide reduction and current production. Appl Environ Microbiol 80:1219–1224. doi: 10.1128/AEM.02938-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonanni PS, Massazza D, Busalmen JP. 2013. Stepping stones in the electron transport from cells to electrodes in Geobacter sulfurreducens biofilms. Phys Chem Chem Phys 15:10300–10306. doi: 10.1039/c3cp50411e. [DOI] [PubMed] [Google Scholar]
- 20.Boesen T, Nielsen LP. 2013. Molecular dissection of bacterial nanowires. mBio 4(3):e00270-13. doi: 10.1128/mBio.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reardon PN, Mueller KT. 2013. Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J Biol Chem 288:29260–29266. doi: 10.1074/jbc.M113.498527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feliciano GT, Feliciano GT, da Silva AJ, Reguera G, Artacho E. 2012. Molecular and electronic structure of the peptide subunit of Geobacter sulfurreducens conductive pili from first principles. J Phys Chem A 116:8023–8030. doi: 10.1021/jp302232p. [DOI] [PubMed] [Google Scholar]
- 23.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Das AK, Horie M, Shaik MS, Smith AM, Luo Y, Lu X, Collins R, Liem SY, Song A, Popelier PL, Turner ML, Xiao P, Kinloch IA, Ulijn RV. 2010. An investigation of the conductivity of peptide nanotube networks prepared by enzyme-triggered self-assembly. Nanoscale 2:960–966. doi: 10.1039/b9nr00233b. [DOI] [PubMed] [Google Scholar]
- 26.Martinez CR, Iverson BL. 2012. Rethinking the term “pi-stacking”. Chem Sci 3:2191–2201. doi: 10.1039/c2sc20045g. [DOI] [Google Scholar]
- 27.Janiak C. 2000. A critical account on π-π stacking in metal complexes with aromatic nitrogen-containing ligands. J Chem Soc Dalton Trans 2000:3885–3896. doi: 10.1039/b003010o. [DOI] [Google Scholar]
- 28.Volz HM, Matyi RJ. 1999. High-resolution X-ray diffraction analyses of protein crystals. Philos Trans R Soc Lond A 357:2789–2799. doi: 10.1098/rsta.1999.0466. [DOI] [Google Scholar]
- 29.Lübbert D, Meents A, Weckert E. 2004. Accurate rocking-curve measurements on protein crystals grown in a homogeneous magnetic field of 2.4 T. Acta Crystallogr D Biol Crystallogr 60:987–998. doi: 10.1107/S0907444904005268. [DOI] [PubMed] [Google Scholar]
- 30.Brédas J-L, Beljonne D, Coropceanu V, Cornil J. 2004. Charge-transfer and energy-transfer processes in π-conjugated oligomers and polymers: a molecular picture. Chem Rev 104:4971–5004. doi: 10.1021/cr040084k. [DOI] [PubMed] [Google Scholar]
- 31.Brédas JL, Calbert JP, da Silva Filho DA, Cornil J. 2002. Organic semiconductors: a theoretical characterization of the basic parameters governing charge transport. Proc Natl Acad Sci U S A 99:5804–5809. doi: 10.1073/pnas.092143399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diegelmann SR, Gorham JM, Tovar JD. 2008. One-dimensional optoelectronic nanostructures derived from the aqueous self-assembly of pi-conjugated oligopeptides. J Am Chem Soc 130:13840–13841. doi: 10.1021/ja805491d. [DOI] [PubMed] [Google Scholar]
- 33.Vadehra GS, Wall BD, Diegelmann SR, Tovar JD. 2010. On-resin dimerization incorporates a diverse array of π-conjugated functionality within aqueous self-assembling peptide backbones. Chem Commun 46:3947–3949. doi: 10.1039/c0cc00301h. [DOI] [PubMed] [Google Scholar]
- 34.Malvankar NS, Tuominen MT, Lovley DR. 2012. Lack of cytochrome involvement in long-range electron transport through conductive biofilms and nanowires of Geobacter sulfurreducens. Energy Environ Sci 5:8651–8659. doi: 10.1039/c2ee22330a. [DOI] [Google Scholar]
- 35.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT, Tainer JA. 2003. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell 11:1139–1150. doi: 10.1016/S1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 36.Ashkenasy N, Horne WS, Ghadiri MR. 2006. Design of self-assembling peptide nanotubes with delocalized electronic states. Small 2:99–102. doi: 10.1002/smll.200500252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glaeser RM. 2013. Stroboscopic imaging of macromolecular complexes. Nat Methods 10:475–476. doi: 10.1038/nmeth.2486. [DOI] [PubMed] [Google Scholar]
- 38.Yi H, Nevin KP, Kim BC, Franks AE, Klimes A, Tender LM, Lovley DR. 2009. Selection of a variant of Geobacter sulfurreducens with enhanced capacity for current production in microbial fuel cells. Biosens Bioelectron 24:3498–3503. doi: 10.1016/j.bios.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Smith JA, Tremblay P-L, Shrestha PM, Snoeyenbos-West OL, Franks AE, Nevin KP, Lovley DR. 2014. Going wireless: Fe(III) oxide reduction without pili by Geobacter sulfurreducens strain JS-1. Appl Environ Microbiol 80:4331–4340. doi: 10.1128/AEM.01122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevin KP, Richter H, Covalla SF, Johnson JP, Woodard TL, Orloff AL, Jia H, Zhang M, Lovley DR. 2008. Power output and coulombic efficiencies from biofilms of Geobacter sulfurreducens comparable to mixed community microbial fuel cells. Environ Microbiol 10:2505–2514. [DOI] [PubMed] [Google Scholar]
- 41.Hélaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. 2005. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol 55:65–77. doi: 10.1111/j.1365-2958.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- 42.Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY. 1997. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A 94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang J, MacDiarmid AG. 1986. “Polyaniline”: protonic acid doping of the emeraldine form to the metallic regime. Synth Met 13:193–205. doi: 10.1016/0379-6779(86)90070-6. [DOI] [Google Scholar]
- 44.Cancedda R, Cedola A, Giuliani A, Komlev V, Lagomarsino S, Mastrogiacomo M, Peyrin F, Rustichelli F. 2007. Bulk and interface investigations of scaffolds and tissue-engineered bones by X-ray microtomography and X-ray microdiffraction. Biomaterials 28:2505–2524. doi: 10.1016/j.biomaterials.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 45.Gallaway JW, Gaikwad AM, Hertzberg B, Erdonmez CK, Chen-Wiegart Y-K, Sviridov LA, Evans-Lutterodt K, Wang J, Banerjee S, Steingart DA. 2013. An in situ synchrotron study of zinc anode planarization by a bismuth additive. J Electrochem Soc 161:A275–A284. doi: 10.1149/2.037403jes. [DOI] [Google Scholar]
- 46.Hammersley AP. 1997. FIT2D: an introduction and overview. ESRF, Grenoble, France. [Google Scholar]
- 47.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanson RM. 2010. Jmol—a paradigm shift in crystallographic visualization. J Appl Crystallogr 43:1250–1260. doi: 10.1107/S0021889810030256. [DOI] [Google Scholar]
- 51.Bondi A. 1964. Van der Waals volumes and radii. J Phys Chem 68:441–451. doi: 10.1021/j100785a001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transmission electron micrograph of pili of Geobacter sulfurreducens strain Aro5. Download
Synchrotron X-ray microdiffraction from a 2- by 5-µm spot focused on the (A) pilus-deficient strain. (B) Buffer. Download
Transmission electron micrographs of pili of the Geobacter sulfurreducens wild-type strain at pH 2 at scale bars 200 nm (A) and 100 nm (B). Download
The clash scores and Molprobity scores for 18 different conformers present in the NMR structure of G. sulfurreducens pilin. Conformer 5 gave the best scores and hence was used for modeling the pilus assembly. Download
Representative distances between different aromatic amino acids in the P. aeruginosa pilus. Distances measured between proximal atoms in aromatic rings in this figure averaged 19.9 Å (range, 5.2 to 37.5 Å). Download