ABSTRACT
Toxoplasma gondii is an obligate intracellular protozoan parasite that is capable of causing severe disease in immunocompromised humans. How T. gondii is able to modulate the host cell to support itself is still poorly understood. Knowledge pertaining to the host-parasite interaction could be bolstered by developing a system to specifically label parasite proteins while the parasite grows inside the host cell. For this purpose, we have created a strain of T. gondii that expresses a mutant Escherichia coli methionyl-tRNA synthetase (MetRSNLL) that allows methionine tRNA to be loaded with the azide-containing methionine analog azidonorleucine (Anl). Anl-containing proteins are susceptible to a copper-catalyzed “click” reaction to attach affinity tags for purification or fluorescent tags for visualization. The MetRSNLL-Anl system labels nascent T. gondii proteins in an orthogonal fashion, labeling proteins only in MetRSNLL-expressing parasites. This system should be useful for nonradioactive pulse-chase studies and purification of nascently translated proteins. Although this approach allows labeling of a diverse array of parasite proteins, secreted parasite proteins appear to be only minimally labeled in MetRSNLL-expressing T. gondii. The minimal labeling of secreted proteins is likely a consequence of the selective charging of the initiator tRNA (and not the elongator methionine tRNA) by the heterologously expressed bacterial MetRS.
IMPORTANCE
Studying how T. gondii modifies the host cell to permit its survival is complicated by the complex protein environment of the host cell. The approach presented in this article provides the first method for specific labeling of T. gondii proteins while the parasite grows inside the host cell. We show that this approach is useful for pulse-chase labeling of parasite proteins during in vitro growth. It should also be applicable during in vivo infections and in other apicomplexan parasites, including Plasmodium spp.
INTRODUCTION
In the past several years, the use of noncanonical amino acids (NCAAs) to label newly synthesized proteins has expanded rapidly (1–5). Numerous methionine (Met) surrogates have been developed that can be activated by wild-type (WT) methionyl-tRNA synthetases (MetRS) and incorporated into cellular proteins. Several such surrogates carry reactive functional groups that allow the attachment of affinity tags for protein purification or fluorescent probes for visualization. Among the most commonly used NCAAs are homopropargylglycine (Hpg) and azidohomoalanine (Aha) (3, 6–8), both of which are susceptible to the copper(I)-catalyzed variant of the Huisgen 1,3-dipolar cycloaddition reaction, known commonly as “click chemistry” (9–11) (Fig. 1A and B). Aha- or Hpg-containing proteins can be specifically labeled with alkyne or azide tags for visualization or purification followed by identification by mass spectroscopy.
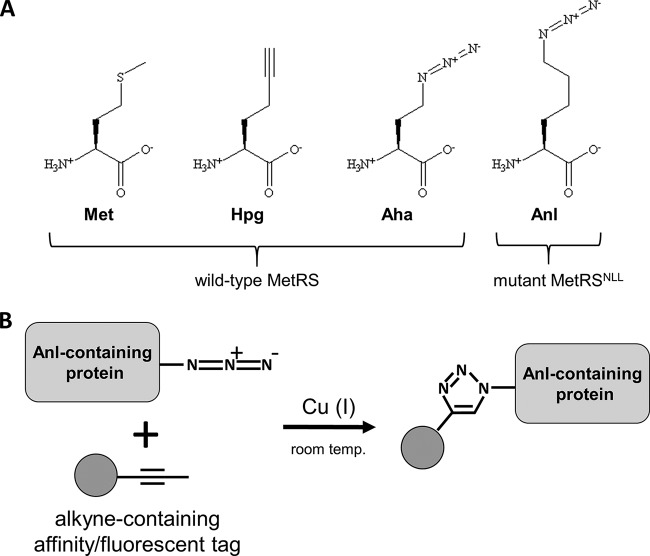
FIG 1 .
(A) The structures of methionine (Met) and methionine analogs: homopropargylglycine (Hpg), azidohomoalanine (Aha), and azidonorleucine (Anl). They can all be charged to tRNAMet by the wild-type MetRS, with the exception of Anl, which can be utilized only by a mutant synthetase like E. coli MetRSNLL. (B) Reaction scheme of a click cycloaddition between a protein that has incorporated Anl and an alkyne conjugated to an affinity/fluorescent tag.
While this is a powerful approach, it does not allow monitoring of individual cell types in mixed cell populations, as all cell types incorporate the NCAAs. This limitation prompted the development of cell-selective systems for NCAA incorporation. In 2009, Ngo and coworkers (12) used random mutagenesis on active site residues to create a mutant version of Escherichia coli MetRS (MetRSNLL) that activates the Met analog azidonorleucine (Anl), which is not utilized by the wild-type MetRS (Fig. 1A) (13). This was accomplished by mutation of three highly conserved residues in the Met-binding pocket of the enzyme (L13N, Y260L, and H301L; leading to MetRSNLL) (14, 15). E. coli expressing MetRSNLL (the Ec-MetRSNLL strain) was able to use Anl in protein synthesis, whereas wild-type E. coli was unable to do so. Furthermore, when the Ec-MetRSNLL strain was grown in coculture with other bacterial strains or with mammalian cells, Anl was detected only in Ec-MetRSNLL proteins (12). Because MetRSNLL has a higher specificity constant (kcat/Km) for Anl than for Met, the growth medium does not have to be depleted of Met for effective labeling (13).
Such a system could be extremely useful to study the interaction between the human parasite Toxoplasma gondii and its host. T. gondii is an obligate intracellular protozoan parasite of the phylum Apicomplexan (of which the causative agent of malaria, Plasmodium spp., is part) and is estimated to infect up to a third of the human population (16). For this purpose, we have developed a T. gondii strain that heterologously expresses a hemagglutinin (HA)-tagged version of E. coli MetRSNLL (the Tg-MetRSNLL strain) and show that it is fully functional.
RESULTS AND DISCUSSION
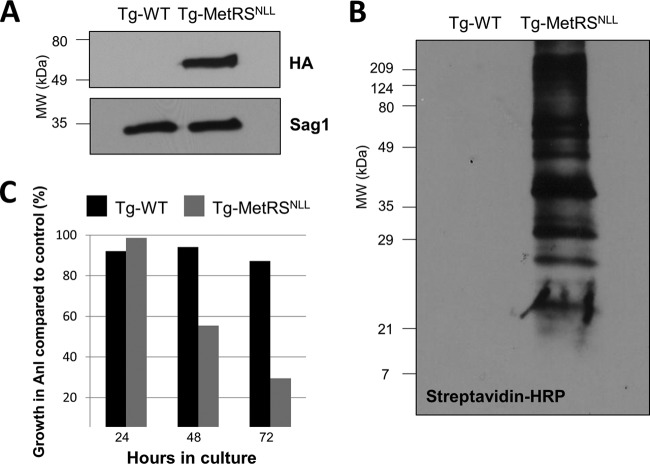
In order to implement the MetRSNLL-Anl system in T. gondii, we heterologously expressed an HA-tagged version of E. coli MetRSNLL in a type II strain of T. gondii (Me49ΔHPT:Luc strain) (17). Successful expression of the bacterial gene was confirmed by comparing lysates of human foreskin fibroblasts (HFFs) infected with either WT T. gondii or the MetRSNLL-expressing strain (Tg-MetRSNLL strain) via Western blotting (Fig. 2A). A prominent signal (at the expected 64-kDa size) is present only in the Tg-MetRSNLL strain-infected sample, confirming that E. coli MetRSNLL can be successfully expressed in T. gondii.
FIG 2 .
(A) Toxoplasma gondii is able to express the E. coli-derived MetRSNLL under control of the T. gondii GRA1 promoter. An HA-tagged version of MetRSNLL was expressed in a type II strain of T. gondii (Me49ΔHPT:Luc strain) under the control of the T. gondii GRA1 promoter. MetRSNLL expression was confirmed by anti-HA Western blotting. Anti-Sag1 antibody served as a loading control. (B) Incorporation of Anl into proteins is limited to MetRSNLL-expressing parasite strains. Lysates of Anl-treated human foreskin fibroblasts (HFFs) infected with either Tg-MetRSNLL or wild-type T. gondii parasites were subjected to click chemistry in the presence of biotin-alkyne. Successful biotinylation was assessed by probing Western blots with streptavidin-HRP. A prominent signal is seen with the Tg-MetRSNLL strain, but no signal is seen with WT T. gondii. (C) Growth of T. gondii is reduced after 48 h of Anl exposure. The effects of growing the Tg-MetRSNLL strain in 1 mM Anl over a period of 3 days were assessed by a luciferase assay. Data are represented as the percentage of growth in Anl compared to that in nonexposed controls. Growth of the Tg-MetRSNLL strain is reduced after 48 and 78 h of Anl exposure. Quantitative data are consistent with multiple qualitative observations of growth differences between the Tg-MetRSNLL and WT T. gondii strains in Anl.
The Tg-MetRSNLL strain is able to incorporate Anl into proteins.
To determine if the Tg-MetRSNLL strain is able to incorporate Anl into nascent proteins, monolayers of HFFs were infected with either WT T. gondii or the Tg-MetRSNLL strain in the presence of 1 mM Anl for 24 hours (h). It is important to note that Met-free medium was not used, such that synthesis of new proteins was not dependent on the incorporation of Anl. The growth medium was supplemented with additional Anl after 24 h, and after a total of 48 h of growth in Anl, the infected HFFs were solubilized and subjected to click chemistry using a biotin-conjugated alkyne (18, 19). Anl incorporation was confirmed via Western blotting, demonstrating that the E. coli MetRSNLL is functional in T. gondii and that it is limited to parasites that express MetRSNLL (Fig. 2B). Furthermore, a wide variety of proteins of >21 kDa are labeled only in MetRSNLL-expressing parasites. Taken together, these biotinylation data show that MetRSNLL is functional in T. gondii and is the first demonstration of the utility of this system in any protozoan pathogen. Moreover, it is likely that this system would be operational in other closely related apicomplexans, including Plasmodium spp. and Cryptosporidium spp.
It should be noted that at longer exposure times, background streptavidin-horseradish peroxidase (HRP) labeling is visible around 35, 45, 65, and 130 kDa in both the WT and MetRSNLL-expressing samples (data not shown). Biotin is a cofactor that is covalently attached to a number of carboxylase enzymes, such as the apicoplast-located acetyl coenzyme A (acetyl-CoA) carboxylase (20, 21) and the mitochondrion-located pyruvate carboxylase in T. gondii. These naturally biotinylated proteins are known to be visible on streptavidin-HRP blots (20, 22), so they should be visible regardless of Anl incorporation. The 35-kDa band is the most prominent, which may correspond to a subdomain of the multisubunit acetyl-CoA carboxylase (20), which is prone to proteolysis in rapeseed (Brassica napus) (23). The 130-kDa band likely corresponds to pyruvate carboxylase, as it agrees with its predicted size.
The effects of Anl incorporation on parasite growth and protein stability.
Incorporation of Anl into nascent proteins may lead to protein misfolding or instability, which could be toxic or lethal to replicating parasites. It is important to note that while the functional group of Anl is slightly larger than that of Met (Fig. 1A), both amino acids are nonpolar, suggesting that replacement of Met with Anl may not have a strongly disruptive effect on protein folding. Studies using a similar azide-containing methionine analog, Aha (Fig. 1A), which is capable of being charged to tRNAMet by wild-type MetRS, found that the tag was nontoxic to the growing cells and did not significantly affect protein stability or promote degradation (3, 6, 24–26). The majority of these studies were conducted using short pulse times (2 to 4 h [6, 24, 26] and up to 24 h [25]), so it remains to be seen whether prolonged exposure to Aha affects cell growth. While Anl is very similar in structure to Aha (Fig. 1A), it is slightly larger and therefore cannot be charged to tRNAMet by a wild-type MetRS. To assess the effects of Anl incorporation, we (i) examined how prolonged Anl exposure affected parasite growth and (ii) globally addressed whether Anl incorporation affects protein stability.
Examining the effect of prolonged Anl exposure on parasite growth.
The effect of Anl exposure on parasite growth was tested by growing either WT T. gondii or the Tg-MetRSNLL strain in the presence or absence of 1 mM Anl over a 72-h time period and performing luciferase assays to quantify the parasite replication rate. After 24 h of growth in 1 mM Anl, luciferase-derived signal intensities were similar between Anl-exposed and control parasite lysates (Fig. 2C). However, noticeable growth differences were observed in the MetRSNLL-expressing strain after 48 h and 72 h of Anl exposure (55% and 30% compared to wild-type controls, respectively; Fig. 2C). Based on these data, it is evident that incorporation of Anl slows parasite growth, particularly after prolonged exposures. Despite this, parasites exposed to Anl for 48 h were still able to effectively invade host cells and replicate normally after Anl removal (see Fig. 6, discussed later in the text). Parasites exposed to Anl for 72 h were also able to successfully invade host cells (data not shown). This indicates that Anl incorporation did not have long-term effects on parasite physiology and that parasites subjected to 72 h of Anl exposure could still be used in downstream experiments. While the exact mechanism of Anl-driven growth defects is not known, prolonged Anl exposure on parasite growth may result in destabilization or misfolding of T. gondii proteins, leading to a reduced parasite growth rate. Regardless, Anl-exposed parasites are fully capable of initiating new infections, indicating that key secretory proteins involved in invasion remain functional after Anl exposure.
FIG 6 .
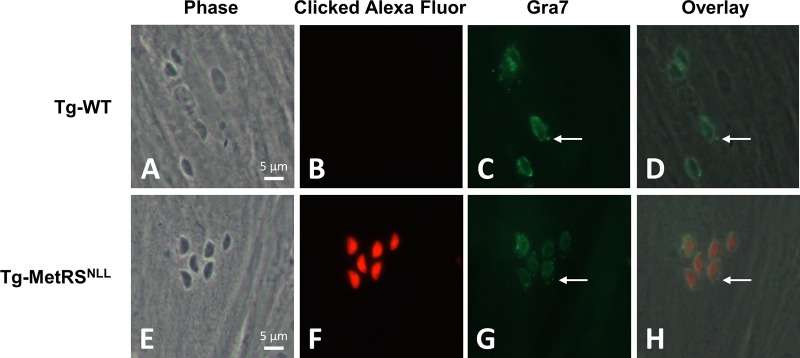
Anl-treated parasites can be labeled in situ by using click chemistry. WT T. gondii and the Tg-MetRSNLL strain were grown in the presence of Anl for 48 h, before being used to infect HFFs for 6 h. (B and F) After fixation and permeabilization, a click reaction was performed on the samples using Alexa Fluor 594-alkyne. (C and G) Additionally, the parasites were stained for the secreted protein Gra7 using a mouse Ab followed by Alexa Fluor 488 goat anti-mouse IgG. The punctate Gra7 staining emanating from around the parasites (C and G, white arrows) is not stained with the clicked Alexa fluorophore (F), suggesting secreted proteins are not labeled. The channels have been overlaid in panels D and H. Magnification at ×100 was used for all images.
The reasons for the slowed growth of T. gondii after 48 h and 72 h of Anl exposure are unclear. The Anl-exposed T. gondii parasites could be experiencing toxicity associated with aberrant folding of Anl-containing proteins, although, compared to published studies with Anl (27) and particularly Aha (6, 24, 26), the 48-h pulse time greatly exceeds typical analog pulse times. To avoid complications of the growth rate reductions, we advise using Anl pulse times less than 24 h. Importantly, if the system were to be used for pulse-chase experiments, the Anl pulse time would likely be considerably shorter than 24 h, such that significant growth defects would not be observed. Indeed, pulse-chase studies utilizing Aha as a Met analog use 2- to 4-h pulse times to achieve substantial labeling (6, 24, 26).
Assessing the stability of Anl-containing proteins.
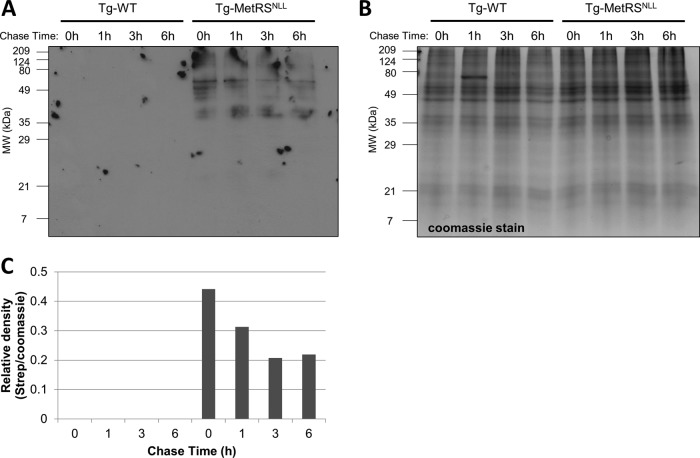
An Anl pulse-chase experiment was used to globally examine the stability of Anl-containing proteins. HFFs were infected with either WT T. gondii or the Tg-MetRSNLL strain and pulsed with Anl for 2 h. As expected, a biotinylated signal is present only in the Tg-MetRSNLL strain-infected samples (Fig. 3A, lanes 5 to 8). The signal intensity is the highest directly after the pulse, with the intensity dropping off steadily over the 6-h chase time (Fig. 3A and C). Despite the declining signal intensity, after the 6-h pulse there is still a sizeable amount of labeled protein, with the signal appearing to stabilize between 3 and 6 h. These data suggest that many Anl-labeled parasite proteins remain stable for at least 6 h. From these data, the average half-life of Anl-containing proteins is approximately 7 h. Pulse-chase studies with [35S]methionine in mammalian cells have shown that approximately 30% of newly synthesized proteins are degraded by the proteasome with a half-life of <10 min, consisting mainly of proteins that do not adopt their native fold due to translational errors (28). It remains to be determined whether a similar margin of proteins is degraded in T. gondii within this time frame. The possibility still exists that Anl incorporation has a strong destabilization effect on some proteins, leading to their rapid degradation by the proteasome, though those would be missed by the pulse-chase experiment presented here.
FIG 3 .
Anl-labeled proteins are still present 6 h after being pulsed with Anl. (A) HFFs were infected with either WT T. gondii or the Tg-MetRSNLL strain at a multiplicity of infection (MOI) of 3 and grown for 48 h before being pulsed with 1 mM Anl for 2 h. Following the 2-h pulse, Anl was washed away and the infected HFFs were harvested at different time points after the pulse (0, 1, 3, 6 h). Samples were biotinylated with click chemistry using biotin-alkyne. Successful biotinylation was assessed by Western blotting and probing with streptavidin-HRP. A biotinylation signal is visible only in the MetRSNLL-expressing strain. The signal intensity is strongest at the 0-h time point, dropping steadily over a period of 6 h. (B) Coomassie-stained SDS-PAGE gel identical to the one used to prepare the Western blot in panel A, confirming equal protein loading; (C) quantification of the data from panel B. The experiment was conducted at least 2 times with similar results.
Optimal Anl concentration for efficient incorporation into nascent proteins.
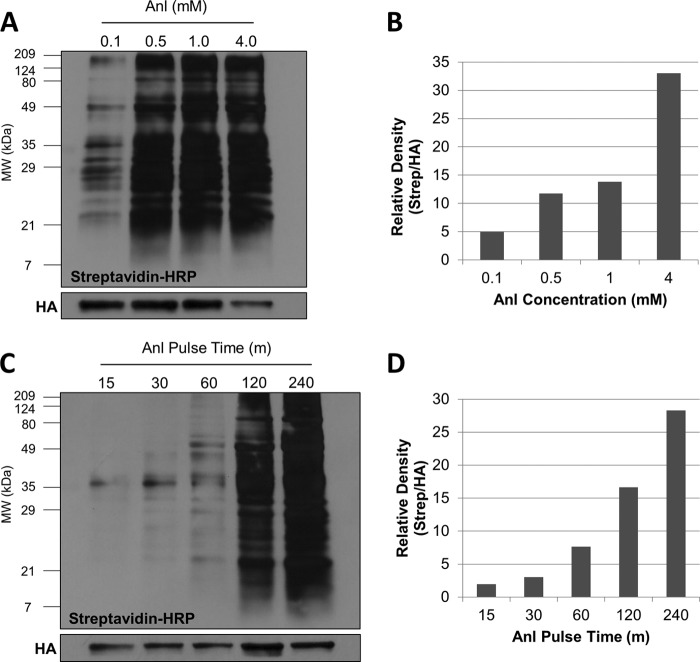
To optimize the MetRSNLL-Anl system in T. gondii, Anl incorporation was examined at concentrations ranging from 0.1 to 4 mM over a 48-h time period (Fig. 4A and B). The levels of Anl incorporation with 0.5 mM and 1.0 mM Anl were similar, though there is considerably less labeling with 0.1 mM Anl than with 0.5 mM and 1.0 mM (Fig. 4A and B). Based on visual inspection, parasite growth was similar in 0.1 mM, 0.5 mM, and 1.0 mM Anl. However, it was apparent that replication was reduced for parasites growing in 4.0 mM Anl. The reduction in parasite number was confirmed by the observed reduction in HA-tagged MetRSNLL on the Western blot (identical “cell equivalents” were loaded for each sample; Fig. 4A). While overall Anl incorporation is higher in 4.0 mM Anl than in the other tested Anl concentrations (Fig. 4B), the effect on the parasites may outweigh this benefit when large numbers of parasites are required for downstream experiments. However, this concentration may be useful in cases where parasite number is less important than overall Anl incorporation. Taken together, these data suggest that 1.0 mM Anl is a suitable concentration for maximizing Anl incorporation while keeping the negative effects on parasite growth to a minimum.
FIG 4 .
(A) Growing parasites in 1 mM Anl allows high levels of incorporation while limiting negative effects on parasite growth. HFFs were infected with the Tg-MetRSNLL strain at an MOI of 3, and either 0.1, 0.5, 1, or 4 mM Anl was added to the growth medium at the time of infection. Cells were harvested after 48 h of growth. The samples were biotinylated with click chemistry using biotin-alkyne. Successful biotinylation was assessed by Western blotting and probing with streptavidin-HRP. Anti-HA Western blotting for the HA-tagged MetRSNLL served as a loading control. Both 0.5 mM and 1.0 mM Anl result in similar levels of labeled protein, whereas 0.1 mM Anl yields considerably less labeling. Tg-MetRSNLL strain growth is greatly reduced in 4 mM Anl, as indicated by the less-intense HA-tagged MetRSNLL band. (B) Quantification of the data from panel A. The experiment was conducted at least 2 times with similar results. (C) Anl is incorporated into a substantial number of proteins just 1 h after being added to the growth medium. HFFs were infected with the Tg-MetRSNLL strain at an MOI of 3, and after 2 days of growth, 1 mM Anl was added to the growth medium. Samples were collected at time points following Anl addition: 15, 30, 60, 120, and 240 min. The samples were biotinylated with click chemistry using biotin-alkyne. Western blotting with streptavidin-HRP was used to confirm successful labeling. Anl is incorporated into proteins just 30 min after addition to the growth medium, and after 1 h of growth, a prevalent banding pattern is present, implying incorporation into a wide range of proteins. (D) Quantification of the data from panel C. The experiment was conducted at least 2 times with similar results.
Anl is incorporated into a substantial number of proteins 1 h after being added to growth medium.
Knowing that prolonged growth in Anl slows parasite growth, we wanted to determine the amount of time parasites needed to be exposed to Anl for incorporation to be detectable. To address this question, HFFs were infected with the Tg-MetRSNLL strain and allowed to grow for 48 h (such that the HFFs were well infected and would lyse the monolayer within 12 h). After 48 h of growth, the growth medium was supplemented with 1 mM Anl for various amounts of time (15, 30, 60, 120, and 240 min). Anl was then washed from the monolayers, and samples were harvested for click chemistry with biotin-alkyne. Western blotting was used to compare the levels of Anl incorporation at the different Anl pulse times (Fig. 4C and D). Anl incorporation was detected with just a 30-min Anl pulse, and after an hour of Anl exposure, a substantial number of proteins incorporated Anl, based on the extensive biotinylation banding pattern that was observed. As expected, the level of Anl incorporation increased with longer Anl pulse times, with the 4-h pulse time exhibiting the strongest signal (Fig. 4C and D).
Is Anl incorporation specific to parasite proteins?
The utility of this approach for studying an intracellular parasite like T. gondii depends on the specificity of Anl incorporation into parasite proteins. To determine if Anl incorporation is specific to T. gondii proteins compared to host cell proteins, HFFs infected with WT T. gondii or the Tg-MetRSNLL strain were treated with Anl for 48 h, solubilized, and subjected to click chemistry with biotin-alkyne. The clicked samples were then used for NeutrAvidin affinity purification. Anl-containing proteins were successfully purified (Fig. 5A) but only from the HFFs infected with the Tg-MetRSNLL strain. To assess whether host cell proteins were labeled and purified, a blot identical to the one in Fig. 5A was probed with an antibody against human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) (Fig. 5B). A distinct signal at the expected 37-kDa size is present in the input and unbound lanes of both the WT T. gondii and Tg-MetRSNLL strain samples, though it is absent from the elutions. The absence of a signal from the Tg-MetRSNLL strain elution strongly suggests that host cell proteins are not incorporating Anl and are therefore not being biotinylated and purified. To confirm the presence of parasite proteins in the Tg-MetRSNLL strain elution, a blot identical to the hGAPDH blot was probed with an anti-HA antibody to detect the parasites’ HA-tagged MetRSNLL (Fig. 5C). A distinct signal is seen for the HA-tagged MetRSNLL in both the input, unbound, and eluted samples, but only in the Tg-MetRSNLL strain samples (at the expected 64-kDa size). The signal intensity in the elution is approximately 30 to 40% of that of the input, giving a general estimation of the level of Anl incorporation. The prominent signal seen in the unbound sample likely applies to the HA-tagged MetRSNLL that did not incorporate Anl and therefore was not biotinylated and purified. These data strongly suggest that host proteins are not labeled with Anl during exposure in vitro, which is consistent with the fact that they have not been engineered to express E. coli MetRSNLL. These data also suggest that the majority, if not all, of the biotinylated proteins seen in the blot from Fig. 5A correspond to Anl-labeled parasite proteins.
FIG 5 .
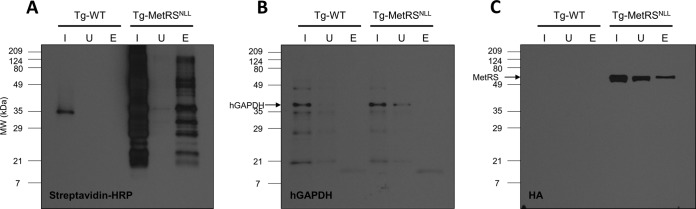
Anl incorporation is limited to parasite proteins. (A) NeutrAvidin purification of lysates of HFFs that had been infected with either WT T. gondii or the Tg-MetRSNLL strain and grown in the presence of 1 mM Anl for 2 days. The samples were biotinylated with click chemistry using biotin-alkyne and incubated with NeutrAvidin resin for 1 h. Biotinylated samples were eluted by boiling in SDS sample buffer. Labeling is present only in the Tg-MetRSNLL strain samples, where it is visible in the input (I) and elution (E), and is largely absent from the unbound (U) fraction. (B) An identical blot to the one in panel A was probed with an anti-human GAPDH antibody to see if host cell proteins were purified with NeutrAvidin. A signal for hGAPDH is present in the input and, to a lesser extent, the unbound samples but is absent from the elutions, suggesting that only parasite proteins are labeled. (C) An identical blot to the one in panel A was probed with an anti-HA antibody to identify the HA-tagged MetRSNLL. A prominent signal is seen in both the input and elution lanes of the Tg-MetRSNLL strain samples only. This suggests that the biotinylated proteins from panel A are T. gondii proteins.
Parasites can be labeled in situ using click chemistry.
The MetRSNLL-Anl system should allow for Anl-treated parasites to be labeled in situ by attaching a fluorescent tag to Anl-containing proteins using click chemistry (6, 27, 29). To test this in T. gondii, we infected HFFs with WT T. gondii or Tg-MetRSNLL parasites that had been pretreated with 1 mM Anl for 48 h. Six hours postinfection, parasites were fixed and permeabilized. Infected HFFs were then subjected to click chemistry using an Alexa Fluor 594-conjugated alkyne. Tg-MetRSNLL parasites stained brightly with Alexa Fluor 594 in the parasite cytoplasm, while WT T. gondii parasites exposed to Anl did not. This further confirms that Anl is not incorporated into host cell proteins (Fig. 6B and F). The infected HFFs were costained for the secreted parasite Dense granule protein 7 (Gra7) (Fig. 6C and G), which exhibited intense parasite staining as well as host cell cytosolic punctate staining, which is characteristic of this T. gondii protein (30). Importantly, we did not observe any such punctate staining in the Alexa Fluor 594 channel (Fig. 6F and H), suggesting that secreted proteins like Gra7 (i) are not tagged with Anl, (ii) incorporate Anl but are highly unstable and therefore undetectable, (iii) are not properly secreted from the parasite when they have incorporated Anl, or (iv) are processed in a manner that removes all Anl-containing residues. We addressed this further in the experiments described immediately below.
Anl is only minimally incorporated into secreted parasite proteins.
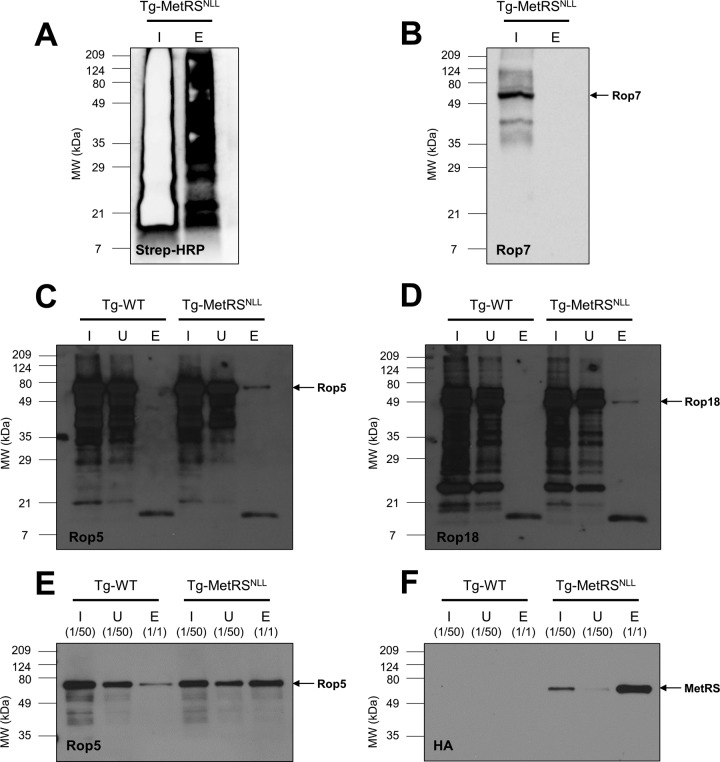
A recently published study showed that mammalian cells expressing the bacterial MetRSNLL incorporated Anl only at the N-terminal Met position in nascent proteins and not internal positions (27). This was attributed to an incompatibility between the bacterial MetRSNLL and one of the mammalian Met tRNAs, the elongator tRNA (described further below). If this were also the case in T. gondii, secretory proteins would not be labeled, since the signal peptide of secreted proteins is cleaved during protein processing, effectively removing the N-terminal Anl residue if it had been incorporated. The system would also bypass most mitochondrial proteins, which have N-terminal presequences that are removed upon successful entry into the mitochondria (31). Furthermore, most apicoplast proteins would be missed, as they have a distinct N-terminal bipartite signal consisting of a classical secretory signal peptide that is removed, followed by a transit peptide needed for import into the apicoplast (32, 33). To determine if this was a similar limitation in the T. gondii-MetRSNLL-Anl system, parasites were grown in the presence of 1 mM Anl for 2 days, solubilized, and biotinylated with click chemistry. The biotinylated samples were then purified with NeutrAvidin resin and run on a Western blot probing for the secreted T. gondii rhoptry protein 7 (Rop7) (Fig. 7B) and biotinylated proteins with streptavidin-HRP (Fig. 7A). The prevalent signal in both the input and elution samples on the streptavidin-HRP blot (Fig. 7A) implies that a sizeable number of parasite proteins incorporate Anl, allowing them to be biotinylated and purified. However, the absence of a signal in the elution on the Rop7 blot (Fig. 7B) implies that Rop7 cannot be purified, potentially because (i) it does not incorporate Anl, (ii) any incorporated Anl gets removed via processing, or (iii) incorporation destabilizes the protein, causing it to be degraded.
FIG 7 .
Proteins from T. gondii parasites grown in the presence of Anl can be biotinylated with click chemistry, though secreted rhoptry proteins are minimally labeled. (A) Western blot of a NeutrAvidin purification of biotinylated parasite proteins, probed with streptavidin-HRP. HFFs were infected with the Tg-MetRSNLL strain in the presence of 1 mM Anl and solubilized after 2 days of growth. Anl-containing proteins were biotinylated with click chemistry and purified with NeutrAvidin resin. There is a strong signal in both the input (I) and elution (E), suggesting that a sizeable number of proteins were successfully purified. Note that the large amount of biotinylated protein in each lane resulted in a rapid depletion of the chemiluminescent substrate, hence the lack of signal in the internal portion of the input lane. (B) Blot identical to the one in panel A, probed for the secreted parasite rhoptry protein 7 (Rop7). The absence of a signal for Rop7 in the elution sample suggests that it did not incorporate Anl and therefore was not biotinylated. (C) Western blot of a NeutrAvidin purification of biotinylated proteins from HFFs infected with either WT T. gondii or the Tg-MetRSNLL strain and probed with an anti-rhoptry protein 5 (Rop5) antibody. The blot was overexposed, revealing a signal for Rop5 in the elution of the Tg-MetRSNLL strain sample. (D) The blot from panel C was stripped and reprobed with anti-Rop18 antibody. As in panel C, the blot was overexposed, revealing a signal for Rop18 in the elution of the Tg-MetRSNLL strain. (E) The samples used to prepare panels C and D were used for a new blot, where the input and unbound samples were diluted 50-fold and the elution samples were undiluted. The blot was probed with an anti-Rop5 antibody, revealing Rop5 signal in both the WT T. gondii and Tg-MetRSNLL strain elutions. (F) The blot from panel E was stripped and reprobed with an anti-HA antibody to identify the HA-tagged MetRSNLL.
To investigate whether other secreted proteins could be purified, additional NeutrAvidin purifications were performed, using HFFs infected with either WT T. gondii or the Tg-MetRSNLL strain and probed for the secreted parasite proteins Rop5 (Fig. 7C) and Rop18 (Fig. 7D). Interestingly, a signal is visible in the Tg-MetRSNLL strain elution only when the blots are overexposed (Fig. 7C and D). In both cases, the clear disparity between signal intensities in the input/unbound samples and the elutions implies that both Rop5 and Rop18 are very minimally labeled. This is at odds with the purified HA-tagged MetRSNLL (Fig. 5C), which suggests around 30 to 40% of the HA-tagged MetRSNLL population is labeled and purified. If both the input and unbound samples are diluted by a factor of 50 and the elution samples remain undiluted, a Rop5 Western blot reveals that there is purified Rop5 in the elutions from both the WT T. gondii and Tg-MetRSNLL strain samples (Fig. 7E). The Rop5 signal in the WT T. gondii elution is perplexing, though it may correspond to Rop5 protein that nonspecifically adheres to the resin used in the purification. Importantly, the Rop5 signal seen in the Tg-MetRSNLL strain elution is approximately 2 times the intensity of the WT T. gondii elution, implying that there was some enrichment beyond the background level seen in the WT T. gondii sample. The Rop5 blot was stripped and reprobed with an anti-HA antibody, revealing a strong signal for the HA-tagged MetRSNLL only in the Tg-MetRSNLL strain elution sample (Fig. 7F), a signal considerably stronger than that of the eluted Rop5. These NeutrAvidin purification data are consistent with the lack of any host cell cytoplasmic labeling in the in situ tagging experiments described in Fig. 6. While it would be expected that the cytoplasmic signal from Anl-tagged (and therefore Alexa Fluor 594-labeled) proteins would be observed, we did not observe any evidence that Anl-tagged proteins could be found in multiple known locations for T. gondii-secreted proteins (primarily the host cell nucleus, as well as in cytoplasmic puncta). It has been well established that multiple rhoptry and dense granule proteins can be found in these host cell locations.
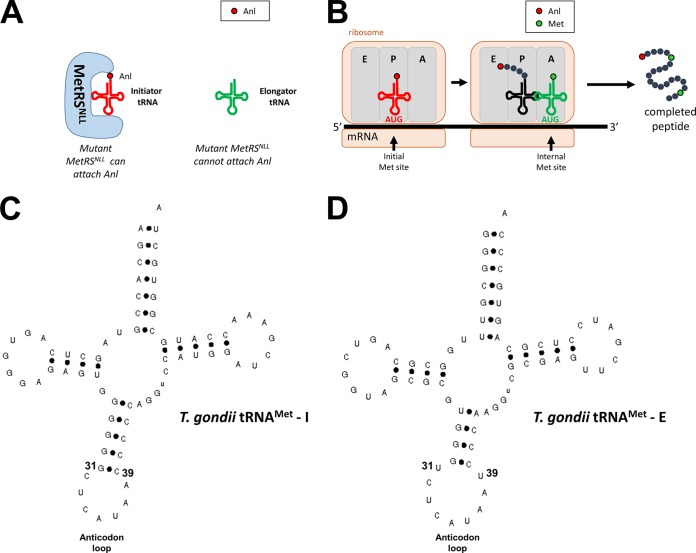
The most likely explanation for the minimal labeling of secretory proteins is that only the initiator methionine is replaced with Anl, while internal Met residues are not. This stems from the fact that there are two Met tRNAs in prokaryotes and eukaryotes, the initiator tRNA, which carries the first Met, and the elongator tRNA, which carries all of the internal Met residues (34). Normally, MetRS is responsible for attaching Met to both the initiator and elongator tRNAs. However, a bacterial MetRS cannot attach Met (or Anl) to a eukaryotic elongator Met tRNA, only to an initiator tRNA (Fig. 8A) (34–37). The reason for this inability was suggested to be due to a difference in the size of the anticodon loop of the eukaryotic elongator Met tRNA (38), which is one of the key determinants for Met tRNA binding to the MetRS (39). The initiator and elongator Met tRNAs from E. coli both have a base pair between residues 31 and 39 which forms a 7-base anticodon loop. In T. gondii (and most other eukaryotes), the anticodon loop is 7 bases in the initiator Met tRNA (Fig. 8C) but is predicted to be 9 bases in the elongator Met tRNA, due to a lack of base pairing between residues 31 and 39 (Fig. 8D). Making a single nucleotide change to introduce a Watson-Crick base pair between residues 31 and 39 in rabbit elongator Met tRNA was enough for the tRNA to be aminoacylated by a bacterial MetRS (38). Taken together, these data suggest that the E. coli MetRSNLL system is similarly incapable of replacing internal Met residues with Anl in T. gondii and therefore proteins processed via the secretory pathway would be undetectable in the Tg-MetRSNLL strain. The inability to identify Rop7 in pulldowns and the in situ data shown in Fig. 6 are consistent with this explanation. The small amount of labeled and purified Rop5 and Rop18 may correspond to newly synthesized protein that has not yet fully transitioned through the secretory pathway to the rhoptries. Another possible explanation for the low labeling of secreted proteins is that they are more susceptible to misfolding than other classes of proteins upon Anl incorporation, and are more readily degraded by quality control machinary. We will address this further in future studies using mass spectrometry to identify Anl-containing proteins and using genetics to mutate the endogenous T. gondii MetRS to incorporate Anl.
FIG 8 .
(A) Bacterial MetRS (wild type or the NLL mutant) cannot charge eukaryotic elongator Met tRNAs. (B) A diagram of the ribosome reading mRNA to synthesize a protein. Since the initiator Met tRNA can be charged with Anl, it gets incorporated into the protein. The elongator Met tRNA cannot be charged with Anl, so all of the internal Met sites stay as Met. (C) Secondary structure prediction of T. gondii initiator Met tRNA (gene models TGME49_269820, TGME49_248915, TGME49_248925). Bases 31 and 39 form a Watson-Crick base pair in the anticodon loop. (D) Secondary structure prediction of T. gondii elongator Met tRNA (gene models TGME49_212750, TGME49_202895, TGME49_257140). Bases 31 and 39 cannot form a Watson-Crick base pair, increasing the size of the anticodon loop. The models in panels C and D were generated with tRNAscan-SE 1.21 using sequences from the T. gondii Me49 strain from ToxoDB.org.
Overall summary.
Combining click chemistry and the incorporation of azide/alkyne-containing NCAAs into nascent proteins is finding increasing use in the study of protein synthesis and degradation. A combination of Aha incorporation/click chemistry and rRNA-targeted fluorescence in situ hybridization (FISH) has been used to monitor protein synthesis in diverse microbial populations (29). Zhang and colleagues used Aha incorporation to monitor the degradation of long-standing proteins via autophagy (40). These examples are all using an NCAA which can be incorporated into virtually any protein, as the NCAAs are recognized by wild-type aminoacyl-tRNA synthetases. This poses a problem when trying to use the system for studying specific cell types in a varied population, in that all the cell types can incorporate the NCAA. To surmount this problem, we have employed a system that was developed by Ngo et al. (12), using a mutant form of methionyl-tRNA synthetase in T. gondii, allowing the azide-containing Met analog, azidonorleucine, to be incorporated in nascent parasite proteins (13). This system is highly effective at labeling nascent proteins in T. gondii and is orthogonal, labeling proteins only in MetRSNLL-expressing parasites. The Tg-MetRSNLL strain is a new tool to study novel aspects of parasite protein expression while parasites are growing inside infected host cells, and strains and plasmids will be given freely upon request. However, our data also show that in T. gondii, proteins which are N-terminally processed, like those destined for secretory organelles like the dense granules and the rhoptries, are only very minimally labeled. This method could potentially be used to determine which secreted proteins, and apicoplast/mitochondrion-located proteins, are not N-terminally processed. We will address the mechanism for this labeling specificity, as well as how to label N-terminally processed parasite proteins with Anl, in future studies. A recent study in the pathogenic bacterium Yersinia enterocolitica used the MetRSNLL-Anl system to identify proteins that it secreted in HeLa cells (41), providing evidence that the system should be applicable to studying/identifying secreted proteins in apicomplexan parasites, like T. gondii, if the system can be modified to label N-terminally processed proteins.
MATERIALS AND METHODS
Host cell culture.
All assays were performed in human foreskin fibroblasts (HFFs), which were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM glutamine (Life Technologies, Rockville, MD), and 50 µg/ml each of penicillin (Life Technologies, Rockville, MD) and streptomycin (Life Technologies, Rockville, MD). The cells were maintained at 37°C in 5% CO2.
Expression of E. coli MetRSNLL in T. gondii.
The E. coli MetRSNLL gene used for creating the MetRSNLL-expressing T. gondii strain (Tg-MetRSNLL strain) was cloned into the pQE-80L bacterial expression vector. The version of the MetRS gene housed within the vector encodes a truncated form of the protein, shortened from 677 amino acids to 548. The missing C-terminal amino acids have a role in MetRS dimerization, though their absence should have minimal effects on successful incorporation of Anl into proteins, as monomeric MetRS is functionally indistinguishable from the native dimeric form (42). The MetRS gene was PCR amplified from the vector using primers with unique restriction sites (forward primer, NsiI site underlined, GTTAATGCATCCTACTATGACTCAAGTCGCGA; reverse primer, NcoI site underlined, GCTACCATGGTTTAGAGGCTTCCAGTGCTTCAACC). Using the added restriction sites, the PCR product was cloned into the pGra-HA-HPT expression plasmid (43), in frame with a C-terminal HA tag and under the control of a highly active dense granule protein promoter (GRA1) (44). The pGra-HA-HPT vector includes the parasite hypoxanthine-xanthine-guanine phosphoribosyl transferase (HPT) gene, which is used to select for stable incorporation of the vector into the genome of a ΔHPT mutant T. gondii strain when grown in the presence of mycophenolic acid (MPA) and xanthine (45, 46). The pGra-HA-HPT-MetRSNLL vector was transfected into the type II parasite strain Me49ΔHPT:Luc, and successful transformants were selected for using MPA (25 μg/ml; Sigma-Aldrich, St. Louis, MO) and xanthine (50 μg/ml; Sigma-Aldrich, St. Louis, MO) (45, 46). Drug-resistant clones were obtained by limiting dilution in 96-well plates. A MetRSNLL-expressing strain (the Tg-MetRSNLL strain) was generated with this process, along with an MPA-resistant strain which lacked the MetRSNLL gene. This strain (WT T. gondii) served as a wild-type control for the experiments, as it was prepared under the same conditions but does not express MetRSNLL.
Western blot analysis.
To probe for expression of the HA-tagged MetRSNLL, HFFs infected with either WT T. gondii or the Tg-MetRSNLL strain were lysed in 1× sodium dodecyl sulfate (SDS) sample buffer and resolved on 8% SDS-polyacrylamide gels. The samples were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). After the transfer, the membranes were blocked in Tris-buffered saline (TBS) with 0.05% Tween 20 (TBST) and 5% bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO) or 5% nonfat dry milk for 1 h. After being blocked, the membrane was incubated in anti-HA-peroxidase (Roche, Mannheim, Germany) antibody at a 1:4,000 dilution for 30 min. After incubation, the blot was washed 3 times with TBST and developed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL). To test for equal loading, the blot was stripped with Restore Western blot stripping buffer (Thermo Scientific, Rockford, IL) and reprobed with mouse anti-Tg-SAG1 (GenWay) at a 1:4,000 dilution for 1 h. After three washes with TBST, the blot was incubated with goat anti-mouse IgG-HRP (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:4,000 dilution for 1 h. After the blot was washed 3 times with TBST, it was developed by using the SuperSignal West Pico chemiluminescent substrate.
Incorporation of Anl into parasites.
A confluent monolayer of HFFs in a T25 culture flask was infected with either WT T. gondii or the Tg-MetRSNLL strain at a multiplicity of infection (MOI) of 3 and supplemented with 1 mM Anl (from a 100 mM stock in H2O). In some instances, the medium was supplemented with additional Anl 24 h after initial infection. After 48 h of growth, the infected monolayer of HFFs was washed with phosphate buffered saline (PBS; Thermo Fisher Scientific, Pittsburgh, PA) and scraped from the T25 culture flask in 3 ml of PBS. The infected HFFs were centrifuged at 800 × g for 10 min and solubilized in 100 µl of 2% SDS in PBS, followed by boiling for 5 min. The samples were diluted to 0.5% SDS with PBS, including cOmplete EDTA-free protease inhibitors (Roche Diagnostics, Indianapolis, IN).
Click cycloaddition reaction.
Cycloaddition reactions were performed on the cell lysates in a final volume of 50 µl of 50 µM biotin-alkyne (Invitrogen Molecular Probes, Eugene, OR) or 10 µM Alexa Fluor 594-alkyne (Invitrogen Molecular Probes, Eugene, OR), along with 1 mM tris(2-carboxyethyl)phosphine (TCEP; Sigma-Aldrich, St. Louis, MO), 100 mM tris(benzyltriazolylmethyl)amine ligand (TBTA; Sigma-Aldrich, St. Louis, MO), and 1 mM copper(II) sulfate (CuSO4; Sigma-Aldrich, St. Louis, MO). This mixture was incubated at room temperature for 2 h with rotation (18, 19). When using Alexa Fluor 594-alkyne, samples were obscured from light during the incubation. Samples were used directly for SDS-PAGE analysis or NeutrAvidin purification.
Determining the optimal Anl concentration for efficient incorporation into nascent proteins.
HFFs were infected with the Tg-MetRSNLL strain, and the growth medium was supplemented with various concentrations of Anl (0.1 mM, 0.5 mM, 1.0 mM, or 4.0 mM) at the time of infection. After 48 h of growth, the infected monolayer was washed to remove excess Anl, and the infected HFFs were solubilized in 2% SDS-PBS and diluted to 0.5% SDS to facilitate click chemistry. The samples were biotinylated with click chemistry using biotin-alkyne and analyzed via Western blotting, probing with streptavidin-HRP.
Luciferase growth assays.
WT T. gondii and the Tg-MetRSNLL strain were derived from the same parental parasite line, which expresses a soluble form of firefly luciferase. To determine the effects of Anl on parasite growth, 10,000 parasites were used to infect confluent HFF monolayers in a 96-well plate in triplicate. Cells were exposed to either 1 mM Anl or vehicle (water) 16 h postinfection and processed for bioluminescence at 24, 48, and 72 h after Anl exposure. Cells were lysed using 1× luciferase lysis buffer (Promega, Madison, WI) and frozen at −80°C until analysis. A total of 20 µl of each sample was added to a Greiner Cellstar 96-well plate with an opaque bottom, and 50 µl of luciferase assay reagent (Promega, Madison, WI) was added to each well immediately prior to analysis on a Centro XS3 LB 960 microplate luminometer. Luminescence was measured for 10 s in each well, with no delay between each well. For each strain (the WT T. gondii and Tg-MetRSNLL strains), data were converted to the percentage of the control by dividing the signal in the presence of Anl by the signal in its absence.
NeutrAvidin purification.
A confluent monolayer of HFFs in a T25 culture flask was infected with either WT T. gondii or the Tg-MetRSNLL strain at an MOI of 3 and supplemented with 1 mM Anl. After 48 h of growth, the infected monolayers were washed 3 times with PBS to remove unincorporated Anl. The monolayers were scraped from their culture flasks and syringe lysed by passage through 25-gauge and 27-gauge needles in 3 ml of PBS, including cOmplete EDTA-free protease inhibitors (Roche Diagnostics, Indianapolis, IN). The samples were centrifuged at 800 × g for 10 min and solubilized in 100 µl of 2% SDS in PBS, followed by boiling for 5 min. The samples were diluted to 0.5% SDS with PBS, including cOmplete EDTA-free protease inhibitors (Roche Diagnostics, Indianapolis, IN). The samples were subjected to click chemistry with biotin-alkyne. Half of the sample was saved as an input, and the rest was used for NeutrAvidin purification. Two hundred microliters of well-mixed NeutrAvidin agarose resin (Thermo Scientific, Rockford, IL) was used for each purification and was equilibrated according to the manufacturer’s instructions. The samples were incubated with the equilibrated resin for 2 h at room temperature with rotation. After incubation, the samples were centrifuged at 2,500 × g for 1 min, and the supernatant was saved as an “unbound” sample. The resin was washed 3 times with PBS, one time with PBS with 0.2% SDS, and a final time with PBS. The bound samples were eluted by boiling for 5 min in 200 µl of 1× SDS sample buffer. The eluted samples were then used directly for SDS-PAGE analysis followed by Western blotting. The mouse monoclonal anti-ROP7 1B10 antibody was generously provided by Peter Bradley (University of California, Los Angeles) (47) and used at a 1:1,000 dilution. The rabbit anti-Rop5 antibody (48) and rabbit anti-Rop18 antibody (49) were generously provided by David Sibley (Washington University School of Medicine, St. Louis, MO).
In situ labeling of parasites using click chemistry.
A confluent monolayer of HFFs in a T25 culture flask was infected with either WT T. gondii or the Tg-MetRSNLL strain at an MOI of 3 and supplemented with 1 mM Anl. After 48 h of growth, the infected monolayers were washed 2 times with PBS to remove unincorporated Anl. The monolayers were scraped from their culture flasks and syringe lysed by passage through 25-gauge and 27-gauge needles. The Anl-treated parasites were then used to infect HFF-seeded coverslips at an MOI of 5 for 6 h. The cells were washed once with PBS and then were fixed for 20 min at room temperature in 4% electron microscopy-grade paraformaldehyde (prepared from 16% stock). The coverslips were washed twice with PBS and then blocked with PBS supplemented with 5% BSA and 0.1% Triton X-100 for 1 h. After fixation and blocking, a click reaction was performed on the samples using Alexa Fluor 594-alkyne for 1 h. The coverslips were washed 2 times with PBS to remove excess Alexa Fluor 594-alkyne. Additionally, the coverslips were incubated with anti-Gra7 monoclonal mouse antibody at a dilution of 1:2,000 for 1 h, washed 3 times with PBS, and incubated with Alexa Fluor 488 goat anti-mouse IgG at a dilution of 1:2,000 for 1 h. After 3 PBS washes, the coverslips were mounted with Vectashield (Vector Laboratories, Burlingame, CA). The monoclonal antibody to Gra7 was generously provided by Peter Bradley (University of California, Los Angeles) (47).
ACKNOWLEDGMENTS
We acknowledge Frank Truong and David A. Tirrell from the Division of Chemistry and Chemical Engineering at the California Institute of Technology for generously providing the E. coli MetRSNLL gene cloned into the pQE-80L vector.
This work was funded by grants R21AI110351 and R21AI093906 (National Institutes of Health) and a Pew Scholarship from the Pew Charitable Trusts to J.P.B.
Footnotes
Citation Wier GM, McGreevy EM, Brown MJ, Boyle JP. 2015. New method for the orthogonal labeling and purification of Toxoplasma gondii proteins while inside the host cell. mBio 6(2):e01628-14. doi:10.1128/mBio.01628-14.
REFERENCES
- 1.Hohsaka T, Sisido M. 2002. Incorporation of non-natural amino acids into proteins. Curr Opin Chem Biol 6:809–815. doi: 10.1016/S1367-5931(02)00376-9. [DOI] [PubMed] [Google Scholar]
- 2.Ngo JT, Tirrell DA. 2011. Noncanonical amino acids in the interrogation of cellular protein synthesis. Acc Chem Res 44:677–685. doi: 10.1021/ar200144y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci U S A 103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagert JD, Xie YJ, Sweredoski MJ, Qi Y, Hess S, Schuman EM, Tirrell DA. 2014. Quantitative, time-resolved proteomic analysis by combining bioorthogonal noncanonical amino acid tagging and pulsed stable isotope labeling by amino acids in cell culture. Mol Cell Proteomics 13:1352–1358. doi: 10.1074/mcp.M113.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinz FI, Dieterich DC, Tirrell DA, Schuman EM. 2012. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci 3:40–49. doi: 10.1021/cn2000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. 2010. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang A, Winblade Nairn N, Johnson RS, Tirrell DA, Grabstein K. 2008. Processing of N-terminal unnatural amino acids in recombinant human interferon-beta in Escherichia coli. Chembiochem 9:324–330. doi: 10.1002/cbic.200700379. [DOI] [PubMed] [Google Scholar]
- 8.Beatty KE, Tirrell DA. 2008. Two-color labeling of temporally defined protein populations in mammalian cells. Bioorg Med Chem Lett 18:5995–5999. doi: 10.1016/j.bmcl.2008.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolb HC, Finn MG, Sharpless KB. 2001. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl 40:2004–2021. doi:. [DOI] [PubMed] [Google Scholar]
- 10.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. 2002. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl 41:2596–2599. doi:. [DOI] [PubMed] [Google Scholar]
- 11.Binder WH, Sachsenhofer R. 2007. “Click” chemistry in polymer and materials science. Macromol Rapid Commun 28:15–54. doi: 10.1002/marc.200600625. [DOI] [Google Scholar]
- 12.Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA. 2009. Cell-selective metabolic labeling of proteins. Nat Chem Biol 5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanrikulu IC, Schmitt E, Mechulam Y, Goddard WA, Tirrell DA. 2009. Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc Natl Acad Sci U S A 106:15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Link AJ, Vink MK, Agard NJ, Prescher JA, Bertozzi CR, Tirrell DA. 2006. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc Natl Acad Sci U S A 103:10180–10185. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serre L, Verdon G, Choinowski T, Hervouet N, Risler JL, Zelwer C. 2001. How methionyl-tRNA synthetase creates its amino acid recognition pocket upon l-methionine binding. J Mol Biol 306:863–876. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 16.Montoya JG, Liesenfeld O. 2004. Toxoplasmosis. Lancet 363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- 17.Tobin CM, Knoll LJ. 2012. A patatin-like protein protects Toxoplasma gondii from degradation in a nitric oxide-dependent manner. Infect Immun 80:55–61. doi: 10.1128/IAI.05543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravindran S, Lodoen MB, Verhelst SH, Bogyo M, Boothroyd JC. 2009. 4-Bromophenacyl bromide specifically inhibits rhoptry secretion during Toxoplasma invasion. PLoS One 4:e8143. doi: 10.1371/journal.pone.0008143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speers AE, Cravatt BF. 2004. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol 11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Zuther E, Johnson JJ, Haselkorn R, McLeod R, Gornicki P. 1999. Growth of Toxoplasma gondii is inhibited by aryloxyphenoxypropionate herbicides targeting acetyl-CoA carboxylase. Proc Natl Acad Sci U S A 96:13387–13392. doi: 10.1073/pnas.96.23.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jelenska J, Crawford MJ, Harb OS, Zuther E, Haselkorn R, Roos DS, Gornicki P. 2001. Subcellular localization of acetyl-CoA carboxylase in the apicomplexan parasite Toxoplasma gondii. Proc Natl Acad Sci U S A 98:2723–2728. doi: 10.1073/pnas.051629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. 2008. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci U S A 105:13574–13579. doi: 10.1073/pnas.0803862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slabas AR, Hellyer A. 1985. Rapid purification of a high molecular-weight subunit polypeptide form of rape seed acetyl CoA carboxylase. Plant Sci 39:177–182. doi: 10.1016/0168-9452(85)90171-2. [DOI] [Google Scholar]
- 24.Taskent-Sezgin H, Chung J, Banerjee PS, Nagarajan S, Dyer RB, Carrico I, Raleigh DP. 2010. Azidohomoalanine: a conformationally sensitive IR probe of protein folding, protein structure, and electrostatics. Angew Chem Int Ed Engl 49:7473–7475. doi: 10.1002/anie.201003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen LD, Zuchman R, Sorokina O, Müller A, Dieterich DC, Armstrong JD, Ziv T, Ziv NE. 2013. Metabolic turnover of synaptic proteins: kinetics, interdependencies and implications for synaptic maintenance. PLoS One 8:e63191. doi: 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirigian LS, Makareeva E, Leikin S. 2014. Pulse-chase analysis of procollagen biosynthesis by azidohomoalanine labeling. Connect Tissue Res 55:403–410. doi: 10.3109/03008207.2014.959120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngo JT, Schuman EM, Tirrell DA. 2013. Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc Natl Acad Sci U S A 110:4992–4997. doi: 10.1073/pnas.1216375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 29.Hatzenpichler R, Scheller S, Tavormina PL, Babin BM, Tirrell DA, Orphan VJ. 2014. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ Microbiol 16:2568–2590. doi: 10.1111/1462-2920.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn JD, Ravindran S, Kim SK, Boothroyd JC. 2008. The Toxoplasma gondii dense granule protein GRA7 is phosphorylated upon invasion and forms an unexpected association with the rhoptry proteins ROP2 and ROP4. Infect Immun 76:5853–5861. doi: 10.1128/IAI.01667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brydges SD, Carruthers VB. 2003. Mutation of an unusual mitochondrial targeting sequence of SODB2 produces multiple targeting fates in Toxoplasma gondii. J Cell Sci 116:4675–4685. doi: 10.1242/jcs.00750. [DOI] [PubMed] [Google Scholar]
- 32.Waller RF, Reed MB, Cowman AF, McFadden GI. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J 19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harb OS, Chatterjee B, Fraunholz MJ, Crawford MJ, Nishi M, Roos DS. 2004. Multiple functionally redundant signals mediate targeting to the apicoplast in the apicomplexan parasite Toxoplasma gondii. Eukaryot Cell 3:663–674. doi: 10.1128/EC.3.3.663-674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deniziak MA, Barciszewski J. 2001. Methionyl-tRNA synthetase. Acta Biochim Pol 48:337–350. [PubMed] [Google Scholar]
- 35.Drabkin HJ, Estrella M, Rajbhandary UL. 1998. Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol Cell Biol 18:1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence F, Blanquet S, Poiret M, Robert-Gero M, Waller JP. 1973. The mechanism of action of methionyl-tRNA synthetase. 3. Ion requirements and kinetic parameters of the ATP-PPi exchange and methionine-transfer reactions catalyzed by the native and trypsin-modified enzymes. Eur J Biochem 36:234–243. doi: 10.1111/j.1432-1033.1973.tb02905.x. [DOI] [PubMed] [Google Scholar]
- 37.Blanquet S, Petrissant G, Waller JP. 1973. The mechanism of action of methionyl-tRNA synthetase. 2. Interaction of the enzyme with specific and unspecific tRNAs. Eur J Biochem 36:227–233. doi: 10.1111/j.1432-1033.1973.tb02904.x. [DOI] [PubMed] [Google Scholar]
- 38.Meinnel T, Mechulam Y, Fayat G, Blanquet S. 1992. Involvement of the size and sequence of the anticodon loop in tRNA recognition by mammalian and E. coli methionyl-tRNA synthetases. Nucleic Acids Res 20:4741–4746. doi: 10.1093/nar/20.18.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beuning PJ, Musier-Forsyth K. 1999. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers 52:1–28. doi:. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Wang J, Ng S, Lin Q, Shen HM. 2014. Development of a novel method for quantification of autophagic protein degradation by AHA labeling. Autophagy 10:901–912. doi: 10.4161/auto.28267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahdavi A, Szychowski J, Ngo JT, Sweredoski MJ, Graham RL, Hess S, Schneewind O, Mazmanian SK, Tirrell DA. 2014. Identification of secreted bacterial proteins by noncanonical amino acid tagging. Proc Natl Acad Sci U S A 111:433–438. doi: 10.1073/pnas.1301740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellot P, Mechulam Y, Le Corre D, Blanquet S, Fayat G. 1989. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol 208:429–443. doi: 10.1016/0022-2836(89)90507-X. [DOI] [PubMed] [Google Scholar]
- 43.Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cesbron-Delauw MF, Guy B, Torpier G, Pierce RJ, Lenzen G, Cesbron JY, Charif H, Lepage P, Darcy F, Lecocq JP. 1989. Molecular characterization of a 23-kilodalton major antigen secreted by Toxoplasma gondii. Proc Natl Acad Sci U S A 86:7537–7541. doi: 10.1073/pnas.86.19.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donald RG, Carter D, Ullman B, Roos DS. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem 271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- 46.Pfefferkorn ER, Borotz SE. 1994. Toxoplasma gondii: characterization of a mutant resistant to 6-thioxanthine. Exp Parasitol 79:374–382. doi: 10.1006/expr.1994.1099. [DOI] [PubMed] [Google Scholar]
- 47.Rome ME, Beck JR, Turetzky JM, Webster P, Bradley PJ. 2008. Intervacuolar transport and unique topology of GRA14, a novel dense granule protein in Toxoplasma gondii. Infect Immun 76:4865–4875. doi: 10.1128/IAI.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A 108:9631–9636. doi: 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, Bzik DJ, Taylor GA, Turk BE, Lichti CF, Townsend RR, Qiu W, Hui R, Beatty WL, Sibley LD. 2010. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 8:484–495. doi: 10.1016/j.chom.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]