Genetic variants of maize identify genes and regions that control core carbon and nitrogen metabolism.
Abstract
Carbon (C) and nitrogen (N) metabolism are critical to plant growth and development and are at the basis of crop yield and adaptation. We performed high-throughput metabolite analyses on over 12,000 samples from the nested association mapping population to identify genetic variation in C and N metabolism in maize (Zea mays ssp. mays). All samples were grown in the same field and used to identify natural variation controlling the levels of 12 key C and N metabolites, namely chlorophyll a, chlorophyll b, fructose, fumarate, glucose, glutamate, malate, nitrate, starch, sucrose, total amino acids, and total protein, along with the first two principal components derived from them. Our genome-wide association results frequently identified hits with single-gene resolution. In addition to expected genes such as invertases, natural variation was identified in key C4 metabolism genes, including carbonic anhydrases and a malate transporter. Unlike several prior maize studies, extensive pleiotropy was found for C and N metabolites. This integration of field-derived metabolite data with powerful mapping and genomics resources allows for the dissection of key metabolic pathways, providing avenues for future genetic improvement.
Carbon (C) and nitrogen (N) metabolism are the basis for life on Earth. The production, balance, and tradeoffs of C and N metabolism are critical to all plant growth, yield, and local adaptation (Coruzzi and Bush, 2001; Coruzzi et al., 2007). In plants, there is a critical balance between the tissues that are producing energy (sources) and those using it (sinks), as the identities and locations of these vary through time and developmental stage (Smith et al., 2004). While a great deal of research has focused on the key genes and proteins involved in these processes (Wang et al., 1993; Kim et al., 2000; Takahashi et al., 2009), relatively little is known about the natural variation within a species that fine-tunes these processes in individual plants.
In addition, a key aspect of core C metabolism involves the nature of plant photosynthesis. While the majority of plants use standard C3 photosynthetic pathways, some, including maize (Zea mays) and many other grasses, use C4 photosynthesis to concentrate CO2 in bundle sheath cells to avoid wasteful photorespiration (Sage, 2004). Under some conditions (such as drought or high temperatures), C4 photosynthesis is much more efficient than C3 photosynthesis. Since these conditions are expected to become more prevalent in the near future due to climate change, various research groups are working to convert C3 crop species to C4 metabolism in order to boost crop production and food security (Sage and Zhu, 2011). Beyond this, better understanding of both C3 and C4 metabolic pathways will aid efforts to breed crops for superior yield, N-use efficiency, and other traits important for global food production.
In the last two decades, quantitative trait locus (QTL) mapping, first with linkage analysis and later with association mapping, has been used to dissect C and N metabolism in several species, including Arabidopsis (Arabidopsis thaliana; Mitchell-Olds and Pedersen, 1998; Keurentjes et al., 2008; Lisec et al., 2008; Sulpice et al., 2009), tomato (Solanum lycopersicum; Schauer et al., 2006), and maize (Hirel et al., 2001; Limami et al., 2002; Zhang et al., 2006, 2010a, 2010b). These studies identified key genetic regions underlying variation in core C and N metabolism, many of which include candidate genes known to be involved in these processes.
Previous studies of genetic variation for C and N metabolism are limited by the fact that they identified trait loci only through linkage mapping in artificial families or through association mapping across populations of unrelated individuals. Linkage mapping benefits from high statistical power due to many individuals sharing the same genotype at any given location, but it suffers from low resolution due to the limited number of generations (and hence recombination events) since the initial founders. Association mapping, in turn, enjoys high resolution due to the long recombination histories of natural populations but suffers from low power, since most genotypes occur in only a few individuals. In addition, many of these studies focused on C and N in artificial settings (e.g. greenhouses or growth chambers) instead of field conditions, running the risk that important genetic loci could be missed if the conditions do not include important (and potentially unknown) natural environmental variables.
To address these issues and improve our understanding of C and N metabolism in maize, we used a massive and diverse germplasm resource, the maize nested association mapping (NAM) population (Buckler et al., 2009; McMullen et al., 2009), to evaluate genetic variation underlying the accumulation of 12 targeted metabolites in maize leaf tissue under field conditions. This population was formed by mating 25 diverse maize lines to the reference line, B73, and creating a 200-member biparental family from each of these crosses. The entire 5,000-member NAM population thus combines the strengths of both linkage and association mapping (McMullen et al., 2009), and it has been used to identify QTLs for important traits such as flowering time (Buckler et al., 2009), disease resistance (Kump et al., 2011; Poland et al., 2011), and plant architecture (Tian et al., 2011; Peiffer et al., 2013). Most importantly, this combination of power and resolution frequently resolves associations down to the single-gene level, even when using field-based data.
The metabolites we profiled are key indicators of photosynthesis, respiration, glycolysis, and protein and sugar metabolism in the plant (Sulpice et al., 2009). By taking advantage of a robotized metabolic phenotyping platform (Gibon et al., 2004), we performed more than 100,000 assays across 12,000 samples, with two independent samples per experimental plot. Raw data and the best linear unbiased predictors (BLUPs) of these data were included as part of a study of general functional variation in maize (Wallace et al., 2014), but, to our knowledge, this is the first in-depth analysis of these metabolic data. We find strong correlations among several of the metabolites, and we also find extensive pleiotropy among the different traits. Many of the top QTLs are also near or within candidate genes relating to C and N metabolism, thus identifying targets for future breeding and selection. These results provide a powerful resource for those working with core C and N metabolism in plants and for improving maize performance in particular.
RESULTS AND DISCUSSION
Correlations among C and N Metabolites
The entire NAM population was grown in summer 2007 in Aurora, New York. Samples were collected in early August, when the largest number of lines were flowering (for details, see “Materials and Methods”). Ideally, each plant would be sampled at the exact same developmental stage relative to flowering to minimize development-related differences. Since this is not possible, especially with a population the size of NAM, we instead used the large number of data points to statistically control for development, along with other confounding factors such as daily weather, date and time of sampling, and heterogeneity across the field. We identified the levels of 12 key metabolites in the sampled tissue, namely chlorophyll a, chlorophyll b, Fru, fumarate, Glc, Glu, malate, nitrate, starch, Suc, total amino acids, and total protein. Whole-field repeatabilities for these metabolites were between 15% and 71%, and broad-sense heritabilities across the NAM founder lines were between 14% and 68% (Supplemental Table S1), indicating good prospects for accurate mapping of the genes responsible for this metabolic variation.
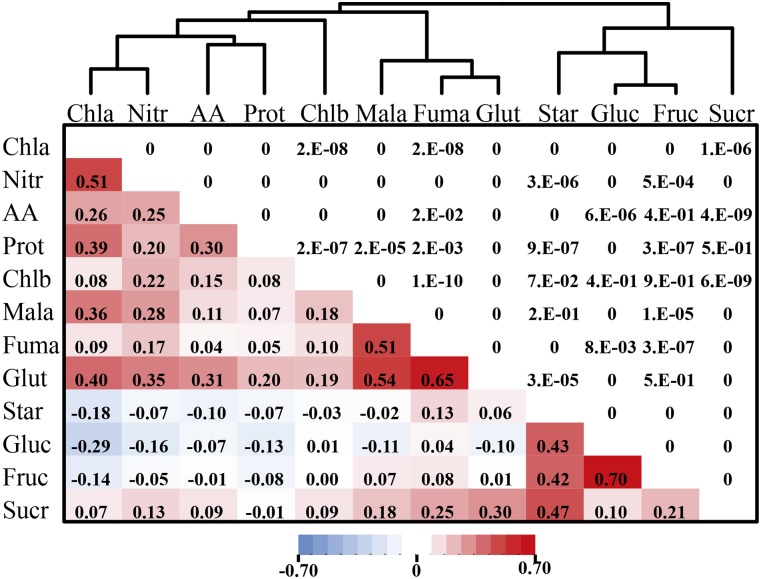
The 12 metabolites show substantial correlations across the NAM population and within each family. As expected, metabolites in either the N-related pathways or the starch-sugar pathways correlate well (Fig. 1). Some traits, such as chlorophyll b, did not substantially correlate with any other traits. We summarized across traits using both clustering and principal component analysis (Fig. 1; Supplemental Fig. S1), with the principal component analysis performed after accounting for flowering time (days to anthesis [DTA]; see “Materials and Methods”). The first two principal components (Prin1 and Prin2) explained 25% and 20% of the variation (Supplemental Fig. S1). Prin1 had its greatest contributions from N metabolites, while Prin2 was dominated by C metabolites (Supplemental Fig. S1B). These components can thus be considered as proxies for general N metabolism and general C metabolism, respectively. Both were included in later linkage and association analyses to map control points for the overall metabolic state of the plant, at least with regard to these two core pathways.
Figure 1.
Correlation matrix and clusters of the 12 C and N metabolites. The bottom left part of the matrix shows the correlation coefficients after accounting for the maturity effect, and the top right part shows the corresponding P values. P < 1 × 10−10 was rounded down to 0; P < 8 × 10−4 is significant at a Bonferroni correction at α = 0.05. Chla, Chlorophyll a; Nitr, nitrate; AA, total amino acids; Prot, protein; Chlb, chlorophyll b; Mala, malate; Fuma, fumarate; Glut, Glu; Star, starch; Gluc, Glc; Fruc, Fru; Sucr, Suc.
Linkage Analysis Shows That Most QTLs Have Modest Effects
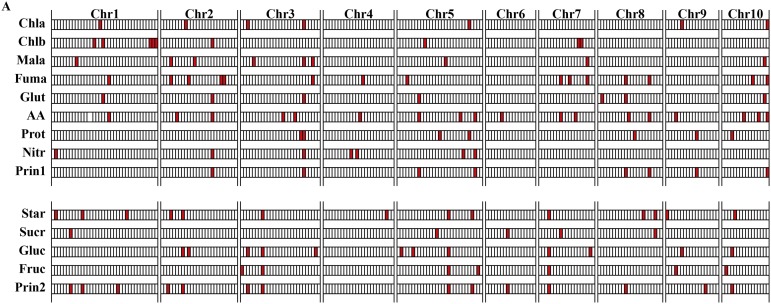
We used joint linkage analysis (Buckler et al., 2009) to identify the major QTLs segregating in the NAM population for all 12 metabolites. After accounting for flowering time, we identified five to 18 QTLs for each metabolite, Prin1, and Prin2; these QTLs explain 24% to 55% of the phenotypic variation for each trait (Fig. 2; Supplemental Tables S1 and S2), and they are generally stable even when plants are broken up by sampling date or location within the field (Supplemental Fig. S2). The effect of each QTL was determined relative to the reference line (B73), and despite the fact that each QTL is, on average, shared by six of the NAM families (Supplemental Fig. S3), the effect size and direction (positive or negative) vary based on family (Supplemental Fig. S4). Most individual QTLs explain 2% or less of the variance (Supplemental Fig. S5), and most alleles were estimated to change metabolite levels by less than 2% either way (Supplemental Fig. S6). Similar results have been observed for whole-plant traits (Laurie et al., 2004; Buckler et al., 2009; Tian et al., 2011; Peiffer et al., 2014) and disease resistance traits (Kump et al., 2011; Poland et al., 2011). We also tested for digenic (two-locus) epistatic effects among these QTLs but did not detect any significant associations.
Figure 2.
QTL distribution of the 12 C and N metabolites and the first two principal components derived from the whole field. Chla, Chlorophyll a; Chlb, chlorophyll b; Mala, malate; Fuma, fumarate; Glut, Glu; AA, total amino acids; Prot, protein; Nitr, nitrate; Prin1, first principal component; Star, starch; Sucr, Suc; Gluc, Glc; Fruc, Fru; Prin2, second principal component.
For most of the developmental and disease traits studied in NAM (Buckler et al., 2009; Kump et al., 2011; Poland et al., 2011; Tian et al., 2011; Peiffer et al., 2013, 2014), there is a strong correlation between traits across the different NAM families, but tests for pleiotropy suggest that few or no QTLs for these traits are actually shared (Poland et al., 2011; Tian et al., 2011). This implies that the phenotypic correlations are due to stacking different genes rather than variation in the same set of genes. In contrast, there is substantial overlap in QTLs between metabolic traits (Fig. 2; Supplemental Fig. S7), and the metabolites that show correlated changes across the entire NAM population (Fig. 1) also share more QTLs (Supplemental Fig. S8; r2 = 0.83, P = 2.1 × 10−26). This suggests that, in contrast to most other traits studied to date in maize, there are relatively few control points for the natural variation in N and C metabolism.
Genome-Wide Association Identifies High-Resolution Hits
To identify the genes controlling this variation, we tested the association of 1.6 million single-nucleotide polymorphisms (SNPs) from maize HapMap1 (Gore et al., 2009). Although a larger SNP data set was later available through maize HapMap2 (Chia et al., 2012), we kept the original analysis because the HapMap1 SNPs are enriched for low-methylation locations (Gore et al., 2009). Such regions are strongly enriched for genome-wide association (GWAS) results (E. Rogers-Melnick, personal communication), and adding the HapMap2 SNPs seemed to dilute signal rather than increase resolution. For reference, the HapMap2 GWAS hits (from Wallace et al., 2014) and their nearest genes are included in Supplemental Table S3.
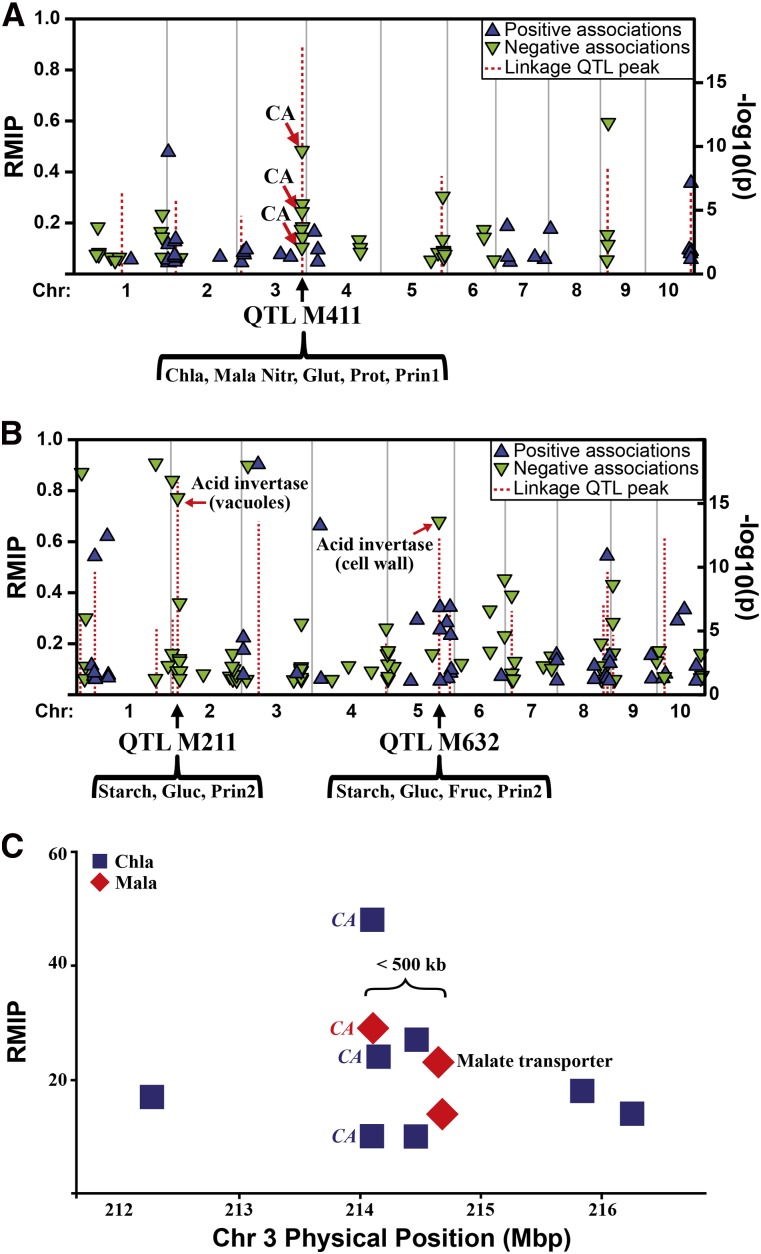
Our GWAS analysis used a method that resamples the model with random subsets of the data, as has been done previously with the NAM population (Kump et al., 2011; Poland et al., 2011; Tian et al., 2011; Peiffer et al., 2013, 2014; Wallace et al., 2014). In brief, a forward-regressing GWAS analysis is run multiple times on each chromosome after controlling for the major QTLs on other chromosomes. Each run includes only a subset of the data, which destabilizes weak or spurious associations and allows for filtering based on how robustly a given SNP is identified in the model (for details, see “Materials and Methods”). This resample model inclusion probability (RMIP) was calculated based on how often each SNP is included in each model generated from the resampled data (Valdar et al., 2006, 2009), and an RMIP of at least 0.05 (five out of 100 runs) was chosen as the cutoff. By this filter, we identified 1,394 significantly associated SNPs (Fig. 3, A and B; Supplemental Fig. S9; Supplemental Tables S4 and S5).
Figure 3.
Associations between metabolites and SNPs. All SNPs detected as significant in at least five subsamples are shown as triangles (blue with positive effect and green with negative effect) relative to their physical sequence positions. Vertical positions of triangles represent the RMIP of the SNP. QTLs are shown as red lines whose vertical positions represent their F test log(1/P) in the final joint linkage QTL model. A, Chlorophyll a and SNPs from CAs on chromosome 3. The CA region on chromosome 3 overlaps with QTL M411. Chlorophyll a (Chla), malate (Mala), nitrate (Nitr), Glu (Glut), protein (Prot), and the first principal component (Prin1) share the same QTL M411; moreover, they all have significantly associated SNPs from the CAs in this QTL. B, Starch and SNPs from invertases on chromosomes 2 and 5. The invertase region on chromosome 2 overlaps with QTL M211, which starch, Glc (Gluc), and the second principal component (Prin2) share, and that on chromosome 5 overlaps with M632, which starch, Glu, Fru (Fruc), and the second principal component share. C, Associations identified by GWAS at CA and a malate transporter region on chromosome 3. Vertical positions of diamonds represent the RMIP of the SNP.
Candidate Genes Near Major Hits
We observed significant enrichment of association around known candidate genes for C and N metabolites. Out of 514 candidate genes potentially related to C and N metabolism curated from maize pathway analyses (Supplemental Table S6), 101 are overrepresented with GWAS signals (Supplemental Fig. S10). Through Gene Ontology (GO) term enrichment analysis, most of the a priori candidates discovered in our GWAS take part in carbohydrate metabolic, alcohol metabolic, carbohydrate catabolic, and Glc metabolic processes (Supplemental Table S7). We also compared the distribution of gene classes near the 126 most significantly associated SNPs (RMIP ≥ 0.50) with that of the whole maize genome. We found that C4 and source-sink genes are overrepresented (Supplemental Fig. S11); however, most SNPs are still located in or near genes that have no known relation to C and N metabolism (Supplemental Table S5).
As for specific genes, we detected three SNPs associated with chlorophyll a variation that tag carbonic anhydrases (CAs) on chromosome 3 (Fig. 3A; Supplemental Table S4). The cytosolic isoform of CA catalyzes the first step in the C4 photosynthetic pathway in mesophyll cells, resulting in the hydration of CO2 to bicarbonate (Ludwig et al., 1998). Bicarbonate is then fixed by phosphoenolpyruvate carboxylase to produce oxaloacetate, which is reduced to malate. Malate then diffuses into the bundle sheath cells, where it is decarboxylated to generate a high internal CO2 concentration (Weiner et al., 1988). In C4 grasses, CA levels are thought to be rate limiting for photosynthesis (Hatch and Mau, 1973; Cousins et al., 2008), and in Arabidopsis, they affect water-use efficiency by acting as upstream regulators of CO2-controlled stomatal movements in guard cells (Hu et al., 2010). Our analysis identifies both CA paralogs on chromosome 3, with two SNPs in CA3.1 and one in CA3.2. This region also overlaps with QTL M411, which is stable across different subenvironments (Supplemental Fig. S2) and shared with the N metabolites malate, nitrate, Glu, total protein, and principal component 1. All of these traits also have significantly associated GWAS SNPs from the CAs within this QTL (Fig. 3A; Supplemental Table S4). The rapid decay of linkage disequilibrium here (Supplemental Fig. S12A) implies that there is single-gene resolution in this region and that genetic variation of CAs on chromosome 3 contributes to N metabolism and photosynthesis.
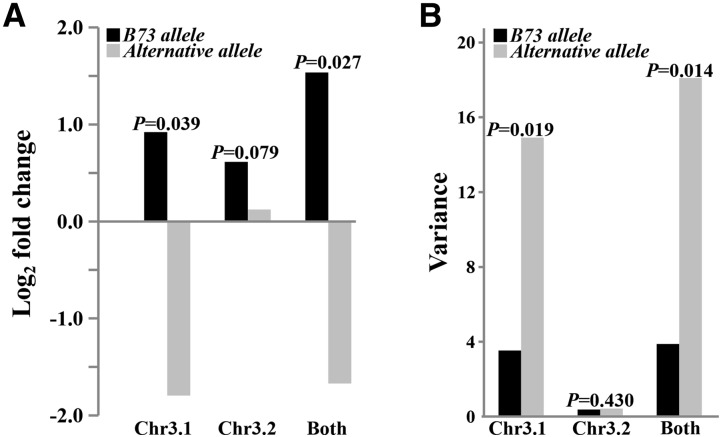
Previous data have shown that in B73 seedlings, most of the total CA expression is derived from two paralogs, CA3.1 and CA3.2 (Supplemental Fig. S13; Li et al., 2010). Since these previous analyses were performed on seedlings, we performed quantitative real-time PCR targeting the two chromosome 3 CA paralogs in the NAM founders using material sampled at flowering time, the same developmental stage as the QTL and GWAS experiment (Fig. 4A). Transcript levels are higher for the B73 allele in both cases, with a much larger difference for CA3.1. However, the variance in expression for CA3.1 is also significantly different between the two alleles (Fig. 4B). This could be for several reasons, such as these transcript levels not correlating perfectly with enzyme levels in the original sample or because we did not capture the actual causal SNP underlying the metabolite variation.
Figure 4.
Expression and corresponding variance comparison for CAs between the B73 allele and an alternative allele in NAM founder lines at flowering time. A, Expression. B, Variance.
We also identified a significantly associated SNP for malate that is approximately 2 kb away from a malate transporter gene, which in turn is approximately 500 kb away from the significantly associated CA SNP in the QTL M411 region (Fig. 3C; Supplemental Table S4). Malate transporters are required for the movement of malate between the mitochondria, cytosol, vacuoles, and chloroplasts in both mesophyll and bundle sheath cells; thus, they are integral to C4 photosynthesis (Bräutigam and Weber, 2011; Gowik and Westhoff, 2011; Nunes-Nesi et al., 2011).
In addition, we identified associations near two acid invertases that are known to regulate C metabolism (Dickinson et al., 1991; Kingston-Smith et al., 1998; Tang et al., 1999). A highly significant SNP (RMIP = 0.76) was detected for starch near a vacuolar acid invertase in the major QTL M211, and another was found near a cell wall acid invertase in the QTL M632 (Fig. 3B). Both QTL regions are also shared with Glc and Prin2, and M632 is also shared with Fru, indicating that these regions have effects across overall C metabolism.
We identified several other candidate genes with recognizable roles in C and N metabolism (Supplemental Table S4). For example, we identified a starch synthase relevant to chlorophyll a variation, a trehalose-6-phosphate synthase and a nitrate transporter relevant to Glc variation, two cellulases related to Fru and Glc, and pyruvate dehydrogenase E1 relevant to fumarate. We also detected a ribosomal protein associated with protein content, which might be of great interest, as protein synthesis is intimately linked to biomass production (Piques et al., 2009).
We found three genes annotated with functions related to cell wall synthesis and modification that are associated with protein content. This may be because protein content can be determined not only by protein synthesis per se but also by the rate of cell expansion, which depends in part on cell wall synthesis.
We also identified a chlorophyll a,b-binding protein, a Gln synthetase, an NADP-malate dehydrogenase, and a phosphoenolpyruvate carboxylase kinase associated with nitrate. NADP-malate dehydrogenase is a central component of the C4 C shuttle in mesophyll cells (reducing oxaloacetate to malate), and it is involved in the export of reducing equivalents from the chloroplast (Scheibe, 2004) to support reductive reactions in the cytosol and the formation of ATP in mitochondria. Phosphoenolpyruvate carboxylase kinase regulates the activity of phosphoenolpyruvate carboxylase, which is another component of the C4 C shuttle; it is also involved in the synthesis of malate that acts as a counterion during nitrate assimilation (Scheible et al., 1997).
In summary, we identified associations with many genes affecting C and N metabolism, including many not previously known to be involved in these pathways. These genes are good targets for improving C and N metabolism in maize and other crops, and characterization of their roles will expand the knowledge of core plant metabolism.
Prospects for Future Crop Improvement
Currently, C4 grasses are some of the most productive plants on Earth (Zhu et al., 2010). However, many of our key crops, such as rice, wheat (Triticum aestivum), and potato (Solanum tuberosum), use C3 photosynthesis, and a major international effort is under way to convert some of these crops to C4 (Sage and Zhu, 2011). This study clearly shows that fine-tuning of the expression of genes that drive the C4 C shuttle are likely key determinants of local adaptation and, hence, yield. Our results show that levels of central metabolites in C and N metabolism are determined by genetic variation in key genes involved in CO2 capture (CA) and movement (malate transporters). These proteins likely impact subprocesses in C4 photosynthesis, including the extent to which the C4 cycle draws down CO2 in the mesophyll airspaces, and the movement of malate from the mesophyll into the bundle sheath. Manipulation of these loci by breeding or transformation and then optimization in a wide range of environments could lead to even more efficient yield. For example, despite the variation present today, it is unlikely that a tropical grass like maize has evolved all the appropriate alleles to fix C well in both the cold spring and hot summer conditions prevalent in temperate latitudes. High-resolution diversity studies like this one can guide breeding efforts to identify genes that are most likely to be amenable to genetic improvement.
MATERIALS AND METHODS
Materials
For this study, we used the maize NAM population, which is composed of 5,000 recombinant inbred lines (RILs) derived from crosses between B73 and 25 highly diverse maize lines (200 RILs for each cross; McMullen et al., 2009). The NAM design integrates both linkage and association approaches to provide high resolution and statistical power for dissecting complex quantitative traits. The creation, genotyping, and map construction of the NAM population has been described previously in detail (Buckler et al., 2009; McMullen et al., 2009).
Tissue Sampling
The NAM population was planted in Aurora, New York, in May 2007. This environment coincided with the one used for the flowering time and leaf trait evaluation reported previously (Buckler et al., 2009; Tian et al., 2011). For each recombinant inbred line, two samples were taken from each row: one from the end plant and the other pooled from four middle plants. Sampling was done in the beginning of August, when most NAM lines were flowering. Tissue was punched from the base of the first leaf below the flag leaf and immediately frozen in liquid N. (The flag leaf itself was not sampled because it intercepts the most light and thus performs the most photosynthesis. We did not want to risk that wounding it would interfere with other phenotypes being measured later in the season on these same plants.) All collections occurred between 10 am and 2 pm of a given day, and all samples (approximately 12,000 total) were collected within 1 week of each other. We recorded several potential confounding variables, including the date and time of sampling, the collector, and the flowering time of the row (DTA, time between planting, and when 50% of plants are shedding pollen). Tissue was stored at −80°C until metabolite measurement.
Metabolite Measuring
Metabolites were extracted (approximately 50 mg fresh weight in a final volume of 650 µL) twice with 80% (v/v) ethanol and once with 50% (v/v) ethanol as described (Geigenberger et al., 1996). Starch and proteins were extracted from the pellet using 100 mm NaOH, as described previously (Hendriks et al., 2003). Chlorophyll content was determined immediately after the extraction according to Arnon (1949). Total free amino acids were assayed using fluorescamine as described by Bantan-Polak et al. (2001). Nitrate was determined as described by Tschoep et al. (2009). Malate and fumarate were measured as described by Nunes-Nesi et al. (2007). Glu was determined using aliquots of 10-µL extracts or standards (ranging from 0 to 20 nmol), which were pipetted onto a microplate containing 100 mm Tricine/KOH, pH 9, 3 mm NAD+, 1 mm methylthiazolyldiphenyl tetrazolium bromide, 0.4 mm phenazine ethosulfate, and 0.5% (v/v) Triton X-100. After reading the A570 for 5 min, 1 unit of Glu dehydrogenase was added, and the absorbance was read until stability. Suc, Glc, and Fru were determined in ethanolic extracts as described by Jelitto et al. (1992). Protein content was assessed as described by Bradford (1976) and starch as described by Hendriks et al. (2003). Assays were prepared on polystyrene 96-well microplates using a Janus pipetting robot (Perkin-Elmer). Absorbance was read at 340 or 570 in an ELX-800 or an ELX-808 microplate reader (Bio-Tek). Absorbance at 595, 645, or 665 nm and fluorescence (excitation at 405 nm, emission at 485 nm) were measured using a Synergy microplate reader (Bio-Tek).
Statistical Analyses
To minimize various environmental effects, the BLUPs for each trait and each line were calculated with ASREML version 2.0 software (Gilmour et al., 2005) and used for further statistical analyses, including QTL mapping. We predicted each RIL’s genetic effect on metabolite contents after accounting for spatial field effects; the amount of N, phosphorous, and potassium of the field before planting; the tissue sampling date, time, and person; the plate on which samples were put; and the metabolite measurement batch. BLUPs were also calculated for DTA by correcting the spatial field effects. These raw data and BLUPs were published as part of a previous analysis (Wallace et al., 2014) that analyzed variation across many different traits but did not look at any one of them in detail.
A correlation coefficient was calculated between end plant samples and middle plant samples across the NAM population for each metabolite using the Proc CORR procedure in SAS version 9.2, and the corresponding r2 was used to evaluate the repeatability of each metabolite. Because NAM founder lines were repeated many times in the field, we were able to estimate broad-sense heritability in them for each metabolite as described previously by Holland et al. (2003).
Correlation analyses between DTA and metabolites were conducted using the Proc CORR procedure in SAS. Since most metabolites correlate with DTA (Supplemental Table S8), we did partial correlation analyses among the 12 metabolites to account for the DTA effects using the Proc GLM procedure in SAS. To make a correction for multiple statistical tests, the sequential Bonferroni test (Holm, 1979) at α = 0.05 was conducted for the correlations of the 12 metabolites. A cluster analysis was performed based on the absolute values of the correlation matrix to detect relationships among the 12 metabolites using the Proc CLUSTER procedure in SAS and the unweighted pair group method with arithmetic mean method. The trees were constructed using the Proc TREE procedure in SAS. A principal component analysis on the 12 metabolites was performed using the Proc PRINCOMP procedure of SAS.
Joint Linkage Mapping
We used the original NAM genetic map with 1,106 SNPs (McMullen et al., 2009) to perform joint linkage mapping as described by Buckler et al. (2009). We used Proc GLMSelect to scan the genome for QTLs of the 12 metabolites plus the first two principal components at each locus of the 1,106 markers with stepwise selection at P = 1 × 10−4 . The general linear model used included a covariate of flowering time (since most metabolites correlated with development stage), a main effect for family, and marker effects nested within families. Proc GLM in SAS was used to fit selected marker effects in the general linear model, and the markers were dropped individually to confirm their significance. QTL epistasis was investigated using the same strategy described previously (Buckler et al., 2009). To examine the robustness of QTLs, this analysis was also conducted for chlorophyll a on the end plant and middle plants separately and on different sampling days (Supplemental Fig. S2).
We used the same method as Buckler et al. (2009) to examine possible pleiotropic effects among the traits. The QTL model using markers identified for one trait was fit to another and vice versa. We then correlated the allele effects across the founder lines at each locus for each trait against one another and determined the significant pleiotropy at P = 0.05. The second cluster analysis was based on the matrix of proportion of shared QTLs (Supplemental Fig. S7).
GWAS
To conduct GWAS, we used the same method reported previously by Tian et al. (2011). Briefly, 1.6 million maize HapMap1 SNPs (Gore et al., 2009) in the NAM founder lines were projected into NAM RILs using pedigree and linkage marker information. SNP positions were referenced to the B73 AGPv1 physical map (www.maizesequence.org). Each chromosome was tested separately by first fitting a model that included all QTLs on other chromosomes and performing GWAS on the residual values, with DTA included as a covariate. To obtain robust association results, 100 subsample data sets, each containing 80% of each family, were analyzed using forward regression (significance level at P < 10E-5). SNPs were scored on the number of subsamples they were identified in the RMIP (Valdar et al., 2009). SNPs detected in at least five subsamples (RMIP ≥ 0.05) were considered as significant and were examined as polymorphisms in linkage disequilibrium (LD) with potential candidate genes from the B73 filtered gene set (http://www.maizesequence.org). We used MapMan annotation to assign genes to functional categories (Thimm et al., 2004). LD analysis for all the significant SNPs and all other SNPs on the same chromosome was conducted using the same method reported previously by Kump et al. (2011)
CA Expression Study
To investigate the expression of CAs on chromosome 3, leaf samples were collected from the NAM founder lines in 2010 in Aurora, New York, using the same protocol described above. Total RNA from the leaf tissue was extracted using Trizol reagent (Invitrogen), and DNase digestion was performed with the Ambion DNA-free kit (Applied Biosystems) according to the manufacturer’s protocol. Total RNA (2 μg) treated with DNase was reverse transcribed using random hexamer. The derived first-stand complementary DNA was diluted 15 times and used as a template. The primers for CA3.1 are 5′-ACCGGATCACCCAATGCAATG-3′ (forward) and 5′-CAGTACATGAATGCTCGGCGTTAG-3′ (reverse), and those for CA3.2 are 5′-AGGAGAAGCCGTCCACAGATAC-3′ (forward) and 5′-TGCCCTTGGAGGAAGCCTTG-3′ (reverse). Ribosomal RNA (18S) was used as an internal control (forward primer, 5′-CAATGGAGATGGCTCGACTT-3′; reverse primer, 5′-GTTGCACTGCGAGCATACAT-3′). Gene Runner (http://www.generunner.net) was used for primer design, and the primers were purchased from Integrated DNA Technologies. Quantitative real-time PCR was conducted using SYBR Green (Applied Biosystems) and the ABI PRISM/Taqman 7900 Sequence Detection System (Applied Biosystems) as described previously (Li et al., 2008). Four repeats were carried out for each gene, and averaged threshold cycle numbers were used to calculate changes (log2).
Enrichment Analysis
To conduct enrichment analysis, a list of a priori candidate genes involved in maize C and N metabolism was curated using the Gramene pathway tool (Youens-Clark et al., 2011). In the curation effort, we included the pathways and superpathways in the tricarboxylic acid cycle, N metabolism, starch synthesis, and photosynthesis. The resulting list of 514 a priori candidates covers metabolic processes, regulators, transporters, electron carriers, and genes related to abiotic stimulus, enzyme, coenzyme, ion, and cation binding as well as catalytic, kinase, and transferase activities (Supplemental Table S6).
We then explored the functional implication of these maize metabolite a priori candidates by a probabilistic approach that detects the significant overrepresentation of colocalizing GWAS signals with a priori candidates. Given a linkage block size, a posterior probability of colocalization of GWAS associations and a priori candidates is thus provided; we use permutations to obtain a null hypothesis for statistical significance tests.
Local linkage block size was calculated using all 1.6 million SNPs from maize HapMap1 (Gore et al., 2009). After filtering for minor allele frequencies of 0.05 or greater, pairwise LDs of the filtered SNPs are estimated by correlation coefficients, and the linkage block size of a priori candidates is estimated by the maximum distance between the local SNP loci that are located within and flanking the gene and those that are in perfect LD (r2 = 1), from both upstream and downstream.
The conditional probability of a given a priori candidate colocalizing with GWAS associations is estimated by the empirical cumulative probability function of physical distance between GWAS associations and the gene. To assess the magnitude of GWAS signals, we performed a weighting procedure by ranking the RMIP values from each of the metabolism GWAS analyses. The RMIP cutoff was set at 0.05 as recommended; all RMIP values below the cutoff are treated at the same level. The weighted posterior probability of colocalization is by the multiplication of weighted RMIP with the conditional probability of colocalization of GWAS signals and the target candidate.
Finally, assuming that every SNP has an equal chance to be in association, we generated 1,000 randomized association sets each containing the same number of associations as the true data, sampled without replacement from all SNP loci on a chromosome. Then, we assigned each set of associations with the RMIP significance levels and computed their posterior probabilities. The adjust threshold was set to the 5% quantile of the lowest significance level from 1,000 permutations. Compared with this distribution of null hypotheses, a cutoff log of the odds value of 3 or greater indicates the significant overrepresentation of GWAS signals for a maize metabolite a priori candidate.
The maize a priori candidates that were significantly enriched in GWAS analysis then became the query list for GO analysis. The GO terms in all categories of all maize genes can be downloaded from the Gramene Ontology module (ftp://ftp.gramene.org/pub/gramene/CURRENT_RELEASE/data/ontology/). We accessed the significance level of GO Slim enrichment by Fisher’s exact test, and to correct multiple comparisons, a false discovery rate was also given (Supplemental Table S7).
The HapMap genotypes used in this article are available from the Panzea Web site: http://www.panzea.org/#!genotypes/cctl. The raw data and BLUPs used here were published previously (Wallace et al., 2014) and are available for download (http://dx.doi.org/10.1371/journal.pgen.1004845.s006).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Principal component analysis of metabolites.
Supplemental Figure S2. Sampling effects of subenvironments.
Supplemental Figure S3. Distribution of shared QTL.
Supplemental Figure S4. Proportions of QTL with both negative and positive effects.
Supplemental Figure S5. Histogram of QTL effects.
Supplemental Figure S6. Distributions of QTL allele effects.
Supplemental Figure S7. Matrix of shared QTL.
Supplemental Figure S8. Relationship between trait correlations and shared QTL.
Supplemental Figure S9. GWAS results for each trait.
Supplemental Figure S10. Overrepresentation of GWAS hits in candidate genes.
Supplemental Figure S11. Distributions of gene classes.
Supplemental Figure S12. Linkage disequilibrium of associated SNPs.
Supplemental Figure S13. Carbonic anhydrase expression.
Supplemental Table S1. Whole-field repeatability.
Supplemental Table S2. Joint-linkage results.
Supplemental Table S3. GWAS hits performed with HapMap2 SNPs.
Supplemental Table S4. Subset of SNPs associated with the metabolites.
Supplemental Table S5. GWAS results.
Supplemental Table S6. Candidate genes.
Supplemental Table S7. GO term analysis.
Supplemental Table S8. Flowering time correlation.
Supplementary Material
Acknowledgments
We thank Sara Miller in the Buckler laboratory at Cornell University for technical editing; researchers in the Buckler laboratory at Cornell University for sample collection and data analyses; researchers in the Stitt laboratory at the Max Planck Institute of Molecular Plant Physiology for assistance with the metabolite measurements; Marcela Monaco in the Ware laboratory at Cold Spring Harbor Laboratories for providing some of the candidate genes; and Eli Rodgers-Melnick in the Buckler laboratory at Cornell University for providing scripts for translating genome coordinates and identifying nearby genes.
Glossary
- C
carbon
- N
nitrogen
- QTL
quantitative trait locus
- NAM
nested association mapping
- BLUPs
best linear unbiased predictors
- DTA
days to anthesis
- SNP
single-nucleotide polymorphism
- RMIP
resample model inclusion probability
- GO
Gene Ontology
- CA
carbonic anhydrase
- RIL
recombinant inbred line
Footnotes
This work was supported by the National Science Foundation (grant nos. DBI–0501700, DBI–0321467, DBI–0820619, and IOS-1238014), the U.S. Department of Agriculture-Agricultural Research Service, and the Max Planck Society.
Articles can be viewed without a subscription.
References
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantan-Polak T, Kassai M, Grant KB (2001) A comparison of fluorescamine and naphthalene-2,3-dicarboxaldehyde fluorogenic reagents for microplate-based detection of amino acids. Anal Biochem 297: 128–136 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Weber APM (2011) Do metabolite transport processes limit photosynthesis? Plant Physiol 155: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Chia JM, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, Elshire RJ, Gaut B, Geller L, Glaubitz JC, et al. (2012) Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet 44: 803–807 [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Gutierrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins AB, Badger MR, von Caemmerer S (2008) C4 photosynthetic isotope exchange in NAD-ME- and NADP-ME-type grasses. J Exp Bot 59: 1695–1703 [DOI] [PubMed] [Google Scholar]
- Dickinson CD, Altabella T, Chrispeels MJ (1991) Slow-growth phenotype of transgenic tomato expressing apoplastic invertase. Plant Physiol 95: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Lerchl J, Stitt M, Sonnewald U (1996) Phloem-specific expression of pyrophosphatase inhibits long-distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19: 43–55 [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2005) ASReml User Guide Release 2.0. VSN International, Hemel Hempstead, UK [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Gowik U, Westhoff P (2011) The path from C3 to C4 photosynthesis. Plant Physiol 155: 56–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD, Mau SL (1973) Activity, location, and role of aspartate aminotransferase and alanine aminotransferase isoenzymes in leaves with C4 pathway photosynthesis. Arch Biochem Biophys 156: 195–206 [DOI] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Bertin P, Quilleré I, Bourdoncle W, Attagnant C, Dellay C, Gouy A, Cadiou S, Retailliau C, Falque M, et al. (2001) Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol 125: 1258–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JB, Nyquist WE, Cervantes-Martínez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22: 104 [Google Scholar]
- Holm S. (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6: 65–70 [Google Scholar]
- Hu H, Boisson-Dernier A, Israelsson-Nordström M, Böhmer M, Xue S, Ries A, Godoski J, Kuhn JM, Schroeder JI (2010) Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat Cell Biol 12: 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelitto T, Sonnewald U, Willmitzer L, Hajirezeai M, Stitt M (1992) Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188: 238–244 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Sulpice R, Gibon Y, Steinhauser MC, Fu J, Koornneef M, Stitt M, Vreugdenhil D (2008) Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol 9: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Mahé A, Brangeon J, Prioul JL (2000) A maize vacuolar invertase, IVR2, is induced by water stress: organ/tissue specificity and diurnal modulation of expression. Plant Physiol 124: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston-Smith AH, Galtier N, Pollock CJ, Foyer CH (1998) Soluble acid invertase activity in leaves is independent of species differences in leaf carbohydrates, diurnal sugar profiles and paths of phloem loading. New Phytol 139: 283–292 [Google Scholar]
- Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, et al. (2011) Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet 43: 163–168 [DOI] [PubMed] [Google Scholar]
- Laurie CC, Chasalow SD, LeDeaux JR, McCarroll R, Bush D, Hauge B, Lai C, Clark D, Rocheford TR, Dudley JW (2004) The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 168: 2141–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ainsworth EA, Leakey ADB, Ulanov A, Lozovaya V, Ort DR, Bohnert HJ (2008) Arabidopsis transcript and metabolite profiles: ecotype-specific responses to open-air elevated [CO2]. Plant Cell Environ 31: 1673–1687 [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Limami AM, Rouillon C, Glevarec G, Gallais A, Hirel B (2002) Genetic and physiological analysis of germination efficiency in maize in relation to nitrogen metabolism reveals the importance of cytosolic glutamine synthetase. Plant Physiol 130: 1860–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, von Caemmerer S, Dean Price G, Badger MR, Furbank RT (1998) Expression of tobacco carbonic anhydrase in the C4 dicot Flaveria bidentis leads to increased leakiness of the bundle sheath and a defective CO2-concentrating mechanism. Plant Physiol 117: 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li HH, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Pedersen D (1998) The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Araújo WL, Fernie AR (2011) Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol 155: 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Peiffer JA, Flint-Garcia SA, De Leon N, McMullen MD, Kaeppler SM, Buckler ES (2013) The genetic architecture of maize stalk strength. PLoS ONE 8: e67066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, Gardner CA, McMullen MD, Holland JB, Bradbury PJ, et al. (2014) The genetic architecture of maize height. Genetics 196: 1337–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Bradbury PJ, Buckler ES, Nelson RJ (2011) Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc Natl Acad Sci USA 108: 6893–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. (2004) The evolution of C4 photosynthesis. New Phytol 161: 341–370 [DOI] [PubMed] [Google Scholar]
- Sage RF, Zhu XG (2011) Exploiting the engine of C4 photosynthesis. J Exp Bot 62: 2989–3000 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Scheibe R. (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Betrán J, Runge ECA. , editors (2004) Corn: Origin, History, Technology, and Production. John Wiley & Sons, Inc., Hoboken, New Jersey [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Takahara K, Hashida SN, Hirabayashi T, Fujimori T, Kawai-Yamada M, Yamaya T, Yanagisawa S, Uchimiya H (2009) Pleiotropic modulation of carbon and nitrogen metabolism in Arabidopsis plants overexpressing the NAD kinase2 gene. Plant Physiol 151: 100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GQ, Lüscher M, Sturm A (1999) Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 11: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- Valdar W, Holmes CC, Mott R, Flint J (2009) Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J (2006) Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38: 879–887 [DOI] [PubMed] [Google Scholar]
- Wallace JG, Bradbury PJ, Zhang N, Gibon Y, Stitt M, Buckler ES (2014) Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet 10: e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Turgeon R, Carr JP, Berry JO (1993) Carbon sink-to-source transition is coordinated with establishment of cell-specific gene expression in a C4 plant. Plant Cell 5: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H, Burnell JN, Woodrow IE, Heldt HW, Hatch MD (1988) Metabolite diffusion into bundle sheath cells from C4 plants: relation to C4 photosynthesis and plasmodesmatal function. Plant Physiol 88: 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youens-Clark K, Buckler E, Casstevens T, Chen C, Declerck G, Derwent P, Dharmawardhana P, Jaiswal P, Kersey P, Karthikeyan AS, et al. (2011) Gramene database in 2010: updates and extensions. Nucleic Acids Res 39: D1085–D1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gibon Y, Gur A, Chen C, Lepak N, Höhne M, Zhang Z, Kroon D, Tschoep H, Stitt M, et al. (2010a) Fine quantitative trait loci mapping of carbon and nitrogen metabolism enzyme activities and seedling biomass in the maize IBM mapping population. Plant Physiol 154: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gur A, Gibon Y, Sulpice R, Flint-Garcia S, McMullen MD, Stitt M, Buckler ES (2010b) Genetic analysis of central carbon metabolism unveils an amino acid substitution that alters maize NAD-dependent isocitrate dehydrogenase activity. PLoS ONE 5: e9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ober JA, Kliebenstein DJ (2006) The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18: 1524–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.