The myosin-II heavy chain Myo1 and the formin Bni1 mediate the targeting of the myosin essential light chain Mlc1 to the division site before and during cytokinesis, respectively, in the budding yeast Saccharomyces cerevisiae.
Abstract
MLC1 is a haploinsufficient gene encoding the essential light chain for Myo1, the sole myosin‑II heavy chain in the budding yeast Saccharomyces cerevisiae. Mlc1 defines an essential hub that coordinates actomyosin ring function, membrane trafficking, and septum formation during cytokinesis by binding to IQGAP, myosin‑II, and myosin‑V. However, the mechanism of how Mlc1 is targeted to the division site during the cell cycle remains unsolved. By constructing a GFP‑tagged MLC1 under its own promoter control and using quantitative live‑cell imaging coupled with yeast mutants, we found that septin ring and actin filaments mediate the targeting of Mlc1 to the division site before and during cytokinesis, respectively. Both mechanisms contribute to and are collectively required for the accumulation of Mlc1 at the division site during cytokinesis. We also found that Myo1 plays a major role in the septin‑dependent Mlc1 localization before cytokinesis, whereas the formin Bni1 plays a major role in the actin filament–dependent Mlc1 localization during cytokinesis. Such a two‑tiered mechanism for Mlc1 localization is presumably required for the ordered assembly and robustness of cytokinesis machinery and is likely conserved across species.
INTRODUCTION
Cytokinesis is a fundamental process essential for the development and survival of single‑cell and multicellular organisms. In animal and fungal cells, cytokinesis requires spatiotemporal coordination of a contractile actomyosin ring (AMR), targeted vesicle fusion, and extracellular matrix (ECM) remodeling (Balasubramanian et al., 2004; Strickland and Burgess, 2004; Barr and Gruneberg, 2007; Pollard, 2010; Wloka and Bi, 2012). The AMR is believed to power the ingression of the cleavage furrow as well as guide ECM remodeling (or septum formation in yeast; Wloka and Bi, 2012). Targeted vesicle fusion is believed to increase cell surface area as well as deliver enzymatic cargoes for ECM remodeling at the division site (Wloka and Bi, 2012). Many components involved in cytokinesis have been identified in different model systems, but their mechanisms of action have yet to be fully understood.
Myosin‑II is a hexamer consisting of two heavy chains (HCs) bound to two essential light chains (ELCs) and two regulatory light chains (RLCs) via distinct IQ motifs (Trybus, 1991; Tan et al., 1992). The IQ motifs are located between the globular head (motor domain) and the long coiled‑coil tail of each HC. The two HCs interact with each other via their rod‑shaped tails to form a two‑headed structure. In animal cells, the role of RLCs in the regulation of myosin activation and filament assembly has been extensively studied in the context of different contractile processes, including cytokinesis (Trybus, 1991; Tan et al., 1992; Matsumura, 2005), whereas the function and mechanism of ELCs in the same processes have been virtually unexplored.
ELC is essential for cell viability and cytokinesis in all systems tested so far, whereas RLC is not (Pollenz et al., 1992; Chen et al., 1995; McCollum et al., 1995; Stevens and Davis, 1998; Boyne et al., 2000; Shannon and Li, 2000; D'souza et al., 2001; Wagner et al., 2002; Luo et al., 2004). It coordinates the functions of actomyosin ring, membrane trafficking, and septum formation during cytokinesis by binding to distinct targets (myosin-II, myosin-V, and IQ motif–containing GTPase-activating protein [IQGAP]; D'souza et al., 2001; Luo et al., 2004). One role of the ELC may be to regulate and mediate the localization of these proteins. Genetic evidence indicates that ELC acts at the top of an essential cytokinesis pathway (ELC to IQGAP to myosin-II) in both budding (Fang et al., 2010) and fission (Laporte et al., 2011) yeasts.
The mechanism of how ELC is localized to the division site remains unclear in any organism. During early bud growth, Mlc1, the ELC in budding yeast (Luo et al., 2004), localizes to the bud cortex, along with the rest of cell growth machinery (Shannon and Li, 2000; Luo et al., 2004). From G2/M to the end of cytokinesis, Mlc1 localizes to the bud neck to fulfill its essential role in cytokinesis (Boyne et al., 2000; Shannon and Li, 2000; Wagner et al., 2002; Luo et al., 2004). Several lines of evidence show that Mlc1 does not localize to the division site simply by binding to the IQ motifs of myosin‑II (Myo1), myosin‑Vs (Myo2 and Myo4), and IQGAP (Iqg1). First, Mlc1 targets to the division site during cytokinesis independently of its binding to the IQ motifs of myosin‑II and -Vs (Wagner et al., 2002; Luo et al., 2004). Second, Mlc1 is required for Iqg1 localization to the division site but not vice versa (Boyne et al., 2000; Shannon and Li, 2000). Finally, an Mlc1 variant (mlc1‑11) that is deficient in interactions with Iqg1, Myo2, and Myo1 still localizes to the division site robustly (Luo et al., 2004). Thus Mlc1 may be localized to the division site in some manner independently of the myosin heavy chains and IQGAP and serve to direct the activity and/or localization of these factors.
To understand the essential role of Mlc1 in cytokinesis, it is important to understand how it is targeted to the division site. Previous studies involved the use of GFP‑tagged MLC1 expressed from a heterologous promoter or of antibodies against the endogenous or an epitope‑tagged Mlc1 (Boyne et al., 2000; Shannon and Li, 2000; Wagner et al., 2002; Luo et al., 2004). Quantitative analyses of Mlc1 at the bud neck during the cell cycle in wild‑type and different mutant strains have not been performed. No conditions under which Mlc1 fails to localize at the bud neck have been reported. In this study, we found that septin ring and actin filaments mediate the targeting of Mlc1 to bud neck before and during cytokinesis, respectively. Mlc1 completely fails to localize at the bud neck when both mechanisms are inactivated. Furthermore, we show that Myo1 plays a major role in the septin‑dependent Mlc1 localization before cytokinesis, whereas Bni1 plays a major role in the actin filament–dependent Mlc1 localization during cytokinesis.
RESULTS
Disruption of actin filaments in a septin mutant abolishes Mlc1 localization to the cell division site
To determine the mechanism of Mlc1 localization during the cell cycle, we constructed an N‑terminally GFP‑tagged MLC1 under the control of its own promoter. This construct is functional, as strains carrying this construct in place of the endogenous MLC1 did not produce any obvious defects in growth and division (Supplemental Figure S1 and Supplemental Video S1). As expected, green fluorescent protein (GFP)–Mlc1 localized to the bud cortex in small‑budded cells and then to the bud neck of medium‑ and large‑budded cells (Boyne et al., 2000; Shannon and Li, 2000; Wagner et al., 2002). For the purpose of this study, a single copy of GFP‑MLC1 was integrated at the MLC1 locus in all the relevant strains. Consequently, each strain contained a copy of the endogenous MLC1 and a copy of GFP‑MLC1 (due to technical reasons, GFP‑MLC1 was not used to replace the endogenous allele in all the mutant strains used in this study). All the relevant strains also contained a single copy of mCherry‑tagged septin gene CDC3, which was integrated at the CDC3 locus. Because the septin hourglass‑to‑double-ring conversion coincides with the onset of cytokinesis (Lippincott et al., 2001) and is always accompanied by a 25–50% decrease in septin intensity (Dobbelaere et al., 2003; Wloka et al., 2011), the characteristic intensity drop was used as an objective criterion to mark the onset of cytokinesis in individual cells as well as to align the Mlc1 localization profiles for different strains or for the same strains grown under different conditions.
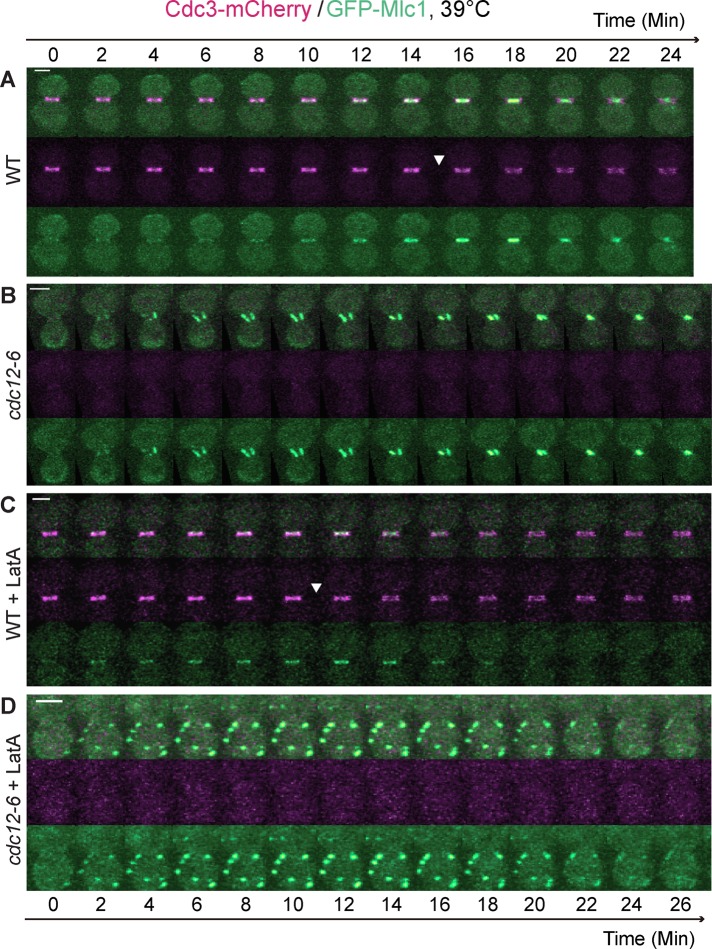
To ascertain whether the “septin ring,” which includes septin hourglass (before cytokinesis) and double ring (during cytokinesis) at the bud neck, is required for Mlc1 localization, we performed time‑lapse microscopy on wild‑type (WT) and septin mutant (cdc12‑6) cells carrying GFP‑MLC1 at the restrictive temperature (39°C). In WT cells (Figure 1A), Mlc1 accumulation at the bud neck began to increase ∼8 min before the onset of cytokinesis (Figure 1A, arrowhead) and reached its peak during cytokinesis, which was concomitant with its constriction. In mutant cells in which the septin ring was apparently absent (Figure 1B and Supplemental Video S2, left), Mlc1 also displayed efficient and cell cycle–dependent localization and constriction at the bud neck, although in an abnormal pattern. The duration of Mlc1 at the bud neck was ∼22–24 min. Thus the septin ring is dispensable for Mlc1 localization during cytokinesis, which is consistent with previous analysis of the endogenous Mlc1 localization by immunofluorescence (Shannon and Li, 2000). However, our time‑lapse analysis indicates that Mlc1 can “establish,” not just “maintain,” its localization in the absence of the septin ring. This distinction could not be drawn from the previous analysis in fixed cells (Shannon and Li, 2000).
FIGURE 1:
Septin ring and actin filaments are collectively required for the localization of Mlc1 to the bud neck during the cell cycle. (A) Time-lapse analysis of Mlc1 localization in relation to the septin ring (Cdc3-mCherry) during the cell cycle in a wild‑type (WT) strain (YEF6888; GFP‑MLC1 CDC3‑mCherry). (B) Mlc1 localizes to the bud neck during cytokinesis in the absence of the septin ring. Localization of Mlc1 in a septin mutant (YEF6884; cdc12‑6 GFP‑MLC1 CDC3‑mCherry) was analyzed by time-lapse microscopy. (C) Mlc1 localizes to the bud neck in the absence of actin filaments. Localization of Mlc1 in LatA‑treated WT cells (YEF6888) was analyzed by time-lapse microscopy. (D) Mlc1 completely fails to localize to the bud neck when both the septin ring and actin filaments are disrupted. Localization of Mlc1 in LatA‑treated septin mutant (YEF6884) was analyzed by time-lapse microscopy. All cells were grown in SC‑Leu medium to exponential phase at 39°C and then subjected to time‑lapse analysis. At least 10 cells were analyzed for each strain under each growth condition (A–D). Arrowheads indicate beginning of septin‑intensity drop during the cell cycle. Scale bars, 2 μm.
The contractile behaviors of Mlc1 (Figure 1B) and of Myo1 in the septin mutant (Dobbelaere and Barral, 2004) indicate that cells are able to assemble and maintain a functional AMR during cytokinesis in the absence of a septin ring. To determine whether the actin cytoskeleton plays any role in Mlc1 localization during cytokinesis, we treated the WT and septin mutant cells with 100 μM latrunculin A (LatA), which is known to disrupt all filamentous actin structures (actin rings, cables, and patches) in budding yeast (Ayscough et al., 1997), before time‑lapse analysis at the restrictive temperature. In WT cells, Mlc1 was still able to localize to the bud neck before and during cytokinesis (Figure 1C), although the kinetics of Mlc1 accumulation was altered by the LatA treatment (see further discussion later). Surprisingly, Mlc1 localization at the bud neck was completely abolished in the LatA‑treated septin mutant (Figure 1D and Supplemental Video S2, right); instead, Mlc1 localized as “cortical spots” in the cytoplasm with approximately the same duration as Mlc1 did at the bud neck in the absence of LatA (compare Figure 1, D to B). This result indicates that septin ring and actin filaments are collectively required for Mlc1 localization during cytokinesis.
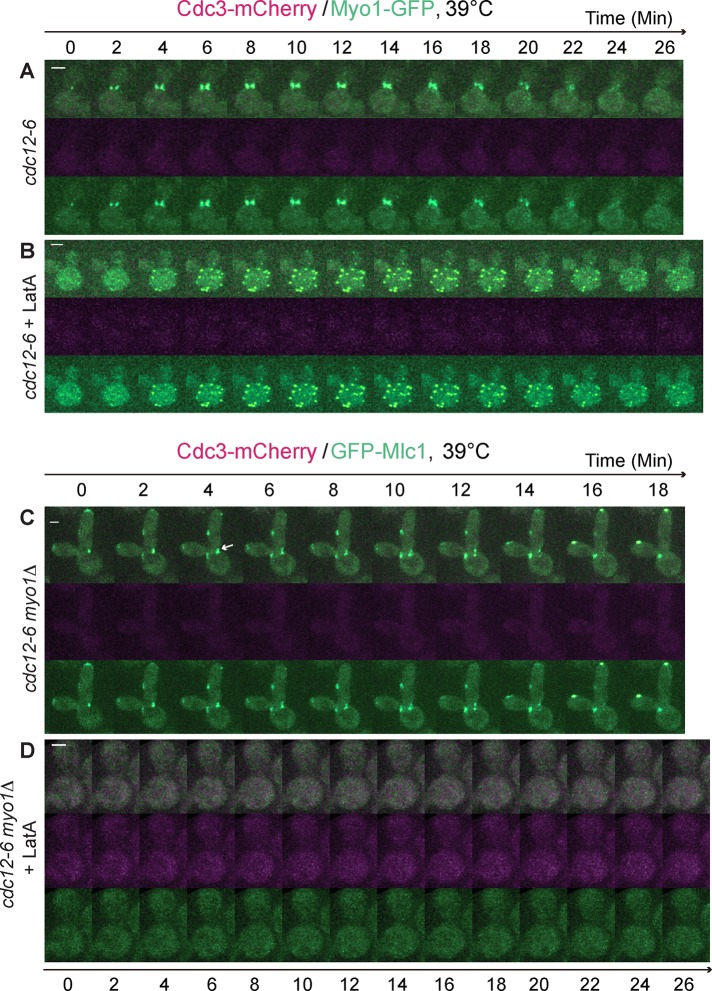
Similarly, Myo1 was able to localize and constrict at the division site in the absence of the septin ring during cytokinesis (Figure 2A and Supplemental Video S3, left; Dobbelaere and Barral, 2004). In addition, Myo1 also localized as “cortical spots” in LatA‑treated septin mutant (Figure 2B and Supplemental Video S3, right). These data suggest that Myo1 and Mlc1 might localize to the division site as a complex during cytokinesis. However, in a septin mutant carrying a myo1 deletion, Mlc1 still localized to the bud neck (Figure 2C, arrow, and Supplemental Video S4, left). These data, together with the previous observation that myo1Δ cells do not form the actin ring (Bi et al., 1998), indicate that the septin ring and the AMR are collectively dispensable for Mlc1 localization during cytokinesis. Thus other filamentous actin structures, including actin cables and actin patches, must play a role in Mlc1 localization during cytokinesis.
FIGURE 2:
Mlc1 localizes to the bud neck independently of the septin ring and Myo1, but its localization to the ectopic cortical sites formed in the absence of the septin ring and actin filaments depends on Myo1. (A, B) Myo1 displays similar localization profiles to Mlc1 in a septin mutant regardless the presence of actin filaments. Cells of the strain YEF7155 (cdc12‑6 MYO1‑GFP CDC3‑mCherry) untreated (A) or treated (B) with LatA were analyzed by time-lapse microscopy (n = 4 for each condition). (C) Mlc1 localizes to the bud neck during cytokinesis in the absence of the septin ring and Myo1. Cells of the strain YEF7081 (cdc12‑6 myo1Δ GFP‑MLC1 CDC3‑mCherry) were analyzed by time-lapse microscopy (n = 6). (D) Localization of Mlc1 to the ectopic cortical sites in LatA‑treated septin mutant depends on Myo1. LatA‑treated cells of the same strain as in C were subjected to time-lapse analysis (n = 6). Arrow indicates GFP-Mlc1 at the bud neck. All cells were grown in SC‑Leu medium at 39°C. Scale bars, 2 μm.
Strikingly, the cortical spots of Mlc1 were completely abolished in the LatA‑treated cdc12‑6 myo1Δ cells (Figure 2D and Supplemental Video S4, right). Because Myo1 is believed to undergo cell cycle–triggered higher‑order assembly (Wloka et al., 2013), the cortical spots may represent “nodes” of myosin‑II filaments containing both the heavy chain Myo1 and the ELC Mlc1, somewhat similar to the medial “nodes” observed in the fission yeast Schizosaccharomyces pombe (Wu et al., 2006). These nodes are believed to be “precursor structures” required for efficient AMR assembly (Pollard and Wu, 2010). Collectively our data demonstrate that when the septin ring is disrupted, Mlc1 localization to the bud neck depends on actin filaments but not Myo1. When the septin ring and actin filaments are both disrupted, Mlc1 is localized to the cell cortex as ectopic nodes in a Myo1‑dependent manner, suggesting that Myo1 may contribute to, although not be essential for, the targeting of Mlc1 to the bud neck during cytokinesis.
Disruption of actin filaments in a wild‑type strain prevents the increase of Mlc1 at the division site during cytokinesis
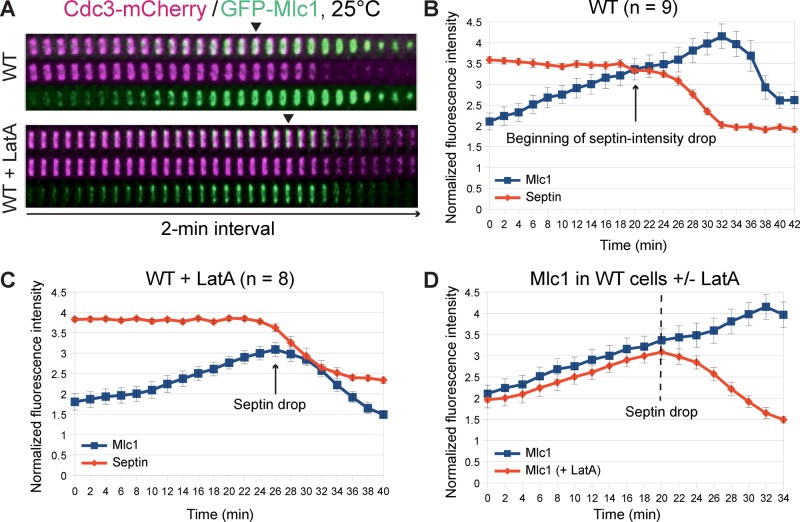
The collective requirement of the septin ring and actin filaments for the bud‑neck localization of Mlc1 suggests the involvement of multiple and distinct mechanisms in this critical event of cytokinesis. This explains why no single mutations or treatments have been found to abolish Mlc1 localization at the bud neck. It also suggests that quantitative analysis is required to reveal a specific contribution by a specific mechanism to the overall Mlc1 localization during the cell cycle. Therefore we examined the possible role of the actin cytoskeleton in Mlc1 localization during the cell cycle in WT cells by time‑lapse analysis in the presence or absence of LatA at 25°C. The ratio of the average pixel intensity of GFP‑Mlc1 at the bud neck to that in the whole cell and the ratio of the average pixel intensity of Cdc3‑mCherry at the bud neck to its maximum were plotted over time. These ratios were taken as “normalized fluorescence intensities.” To visualize clearly the septin‑intensity drop at the onset of cytokinesis, together with the Mlc1 intensity change in the same plot, we multiplied the septin ratios (≤1) by a factor of two to four.
In the absence of LatA (Figure 3, A, top, and B, and Supplemental Video S5, left), Mlc1 began to increase at the bud neck ∼20–26 min before the onset of cytokinesis (as indicated by the beginning of the septin‑intensity drop) and reached its peak during cytokinesis (as indicated by the end of the septin‑intensity drop). In the presence of LatA (Figure 3A, bottom, and C, and Supplemental Video S5, right), the overall level of Mlc1 at the bud neck decreased, especially during cytokinesis. A direct comparison of Mlc1 accumulation kinetics in the LatA‑treated versus ‑untreated cells during the same cell‑cycle period (Figure 3D) revealed that LatA caused only a slight decrease (∼7%) in the level of Mlc1 at the bud neck before cytokinesis while leaving its rate of accumulation unchanged. In contrast, LatA essentially abolished the peak of Mlc1 accumulation during cytokinesis, which represents a 26% difference between the peak levels of Mlc1 in treated versus untreated cells or is equivalent to ∼19% net increase of Mlc1 at the bud neck during cytokinesis in the untreated cells. Thus F‑actin is mainly required for Mlc1 accumulation at the division site during cytokinesis.
FIGURE 3:
Disruption of actin filaments in wild‑type cells prevents the increase of Mlc1 at the division site during cytokinesis. (A) Time-lapse analysis of Mlc1 localization with respect to the septin ring in WT cells in the presence or absence of actin filaments. Cells of the strain YEF7435 (GFP‑MLC1 CDC3‑mCherry) were grown in SC‑Leu medium at 25°C and then analyzed by time‑lapse microscopy in the absence or presence of LatA. Kymograph of a representative cell under each growth condition is shown. (B, C) “Normalized fluorescence intensity” of GFP‑Mlc1 (see Materials and Methods) and average intensity of the septin Cdc3‑mCherry at the bud neck of WT cells (YEF7435) untreated (B) or treated (C) with LatA plotted over time. (D) Direct comparison of Mlc1 localization profiles at the bud neck of WT cells in the presence or absence of LatA. Arrows indicate the beginning of the septin‑intensity drop during the cell cycle. Error bars represent SEM.
Increased accumulation of Mlc1 at the bud neck during cytokinesis depends on the formin Bni1 but not Bnr1
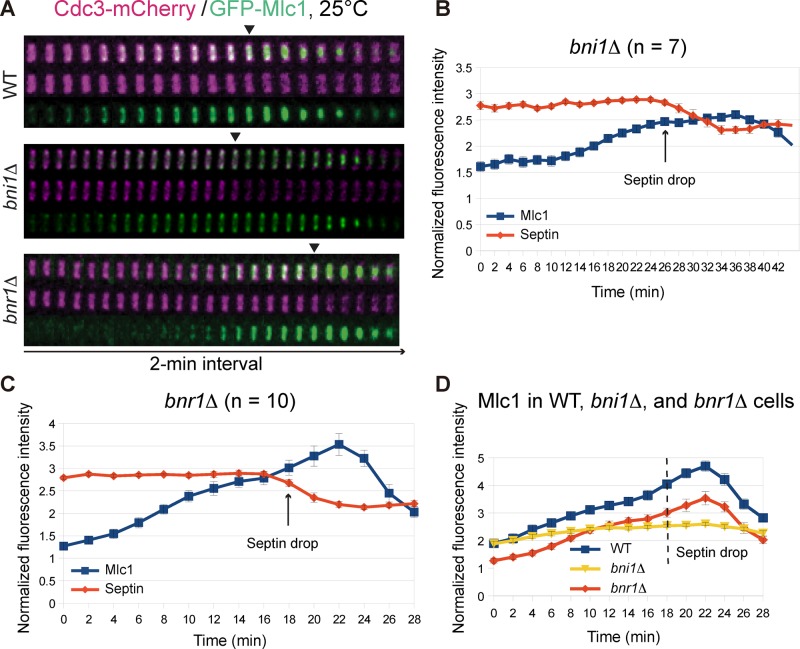
Bni1 and Bnr1, a pair of formins in Saccharomyces cerevisiae, play spatiotemporally distinct roles in the nucleation of actin filament assembly during the cell cycle and are collectively essential for cell survival (Moseley and Goode, 2006; Bi and Park, 2012). Before cytokinesis, Bni1 nucleates actin cable assembly at the bud cortex to mediate exocytosis for bud growth. During cytokinesis, Bni1 localizes to the bud neck to promote cytokinesis by stimulating actin ring and actin cable assembly (Evangelista et al., 1997; Pruyne et al., 2004; Buttery et al., 2007). In contrast, Bnr1 localizes at and promotes actin cable assembly toward the bud neck from bud emergence to the onset of cytokinesis and then disappears from the neck during cytokinesis (Pruyne et al., 2004; Buttery et al., 2007). The distinct localization profiles and other functional data suggest that Bni1 and Bnr1 play differential roles in cytokinesis, with Bni1 being the dominant player. To determine whether the formin‑nucleated actin filaments account for the role of F‑actin in Mlc1 accumulation at the bud neck during cytokinesis, we examined Mlc1 localization in bni1Δ and bnr1Δ cells during the cell cycle by time‑lapse microscopy and quantitative analysis.
In bni1Δ cells (Figure 4, A, B, and D, and Supplemental Video S6, right), Mlc1 was able to accumulate, albeit slowly, at the bud neck before cytokinesis. More strikingly, the peak of Mlc1 accumulation at the bud neck during cytokinesis was nearly abolished, which represents a ∼45% reduction compared with WT cells in the total level of Mlc1 at the bud neck during its peak time in cytokinesis (Figure 4, B and D). In contrast, the level of Mlc1 at the bud neck in bnr1Δ cells was reduced by 25–33% before cytokinesis, but the rate of Mlc1 accumulation at the neck remained essentially unchanged throughout the cell cycle (Figure 4, A, C, and D, and Supplemental Video S6, left). These data are consistent with the localization patterns of Bni1 and Bnr1 at the bud neck during the cell cycle (Pruyne et al., 2004; Buttery et al., 2007). Thus both formins play a role in Mlc1 localization, but Bnr1 is more important before cytokinesis, whereas Bni1 is more important during cytokinesis.
FIGURE 4:
Increase of Mlc1 at the bud neck during cytokinesis depends on the formin Bni1, not Bnr1. (A) Time‑lapse analysis of Mlc1 and Cdc3 at the bud neck during the cell cycle in WT (YEF7070; GFP‑MLC1 CDC3‑mCherry), bni1Δ (YEF7201; bni1Δ GFP‑MLC1 CDC3‑mCherry), and bnr1Δ (YEF7200; bnr1Δ GFP‑MLC1 CDC3‑mCherry) cells. (B, C) Fluorescence intensities of Mlc1 and Cdc3 at the bud neck in bni1Δ (YEF7201; B) and bnr1Δ (YEF7200; C) cells plotted over time. (D) Direct comparison of Mlc1 localization profiles at the bud neck of WT, bni1Δ, and bnr1Δ cells. All cells were grown in SC‑Leu medium at 25°C. Arrows (B, C) and dashed line (D) indicate beginning of the septin‑intensity drop during the cell cycle.
Myo1 plays a major role in targeting Mlc1 to the division site before cytokinesis
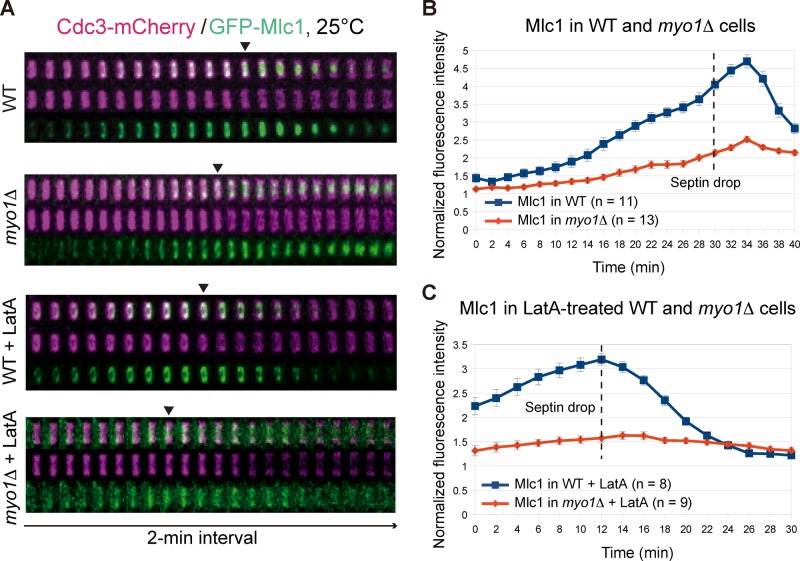
Among the Mlc1‑binding proteins, Myo1 is the only protein that localizes to the division site significantly earlier than Mlc1, which raises the possibility that Myo1 might mediate Mlc1 localization before cytokinesis. To examine this possibility, we compared Mlc1 localization in WT and myo1Δ cells during the cell cycle (Figure 5, A and B, and Supplemental Video S7, left). Mlc1 accumulation at the bud neck in myo1Δ cells was 21, 40, 47, and 46% less than in WT cells at time points 0, 18, and 30 min (septin‑intensity drop or the onset of cytokinesis) and 34 min (the Mlc1 peak during cytokinesis), respectively (Figure 5B). Thus the defect of Mlc1 localization in myo1Δ cells occurred primarily before cytokinesis. When treated with LatA (Figure 5, A and C, and Supplemental Video S7, right), Mlc1 accumulation at the bud neck in myo1Δ cells was 41 and 51% less than in WT cells at time points 0 and 12 min (septin‑intensity drop), respectively (Figure 5C). As expected, LatA treatment abolished the Mlc1 peak during cytokinesis in both strains (Figure 5, A and C). Together these data suggest that Myo1 plays a major role in Mlc1 localization before cytokinesis.
FIGURE 5:
Myo1 plays a major role in targeting Mlc1 to the division site before cytokinesis. (A) Time‑lapse analysis of Mlc1 and Cdc3 at the bud neck during the cell cycle in WT (YEF7070; GFP‑MLC1 CDC3‑mCherry) and myo1Δ (YEF7132; myo1Δ GFP‑MLC1 CDC3‑mCherry) cells in the absence or presence of LatA. (B, C) Direct comparison of Mlc1 localization profiles at the bud neck of WT (YEF7070) and myo1Δ (YEF7132) cells in the absence (B) or presence (C) of LatA. Dashed lines indicate the beginning of the septin‑intensity drop during the cell cycle. All cells were grown in SC‑Leu medium at 25°C.
The septin‑binding protein Bni5 is required for the targeting of Myo1 to the bud neck from bud emergence to the onset of anaphase (Fang et al., 2010). To determine whether Bni5 mediates the role of Myo1 in Mlc1 localization before cytokinesis, we examined GFP‑Mlc1 localization in bni5Δ cells (Supplemental Figure S2 and Supplemental Video S8) and found that Mlc1 localization was slight defective in the mutant, and this defect was restricted to a period of ∼16 min before the septin‑intensity drop (Supplemental Figure S2B). This result, together with the previous observation that Myo1 still localizes to the bud neck in bni5Δ cells during anaphase, starting at ∼14 min before the septin‑intensity drop (Fang et al., 2010), suggests that the role of Myo1 in Mlc1 localization before cytokinesis is largely independent of Bni5 (Supplemental Figure S2B).
Disruption of actin filaments in myo1Δ myo2IQ6Δ myo4Δ cells abolishes Mlc1 localization to the division site throughout the cell cycle
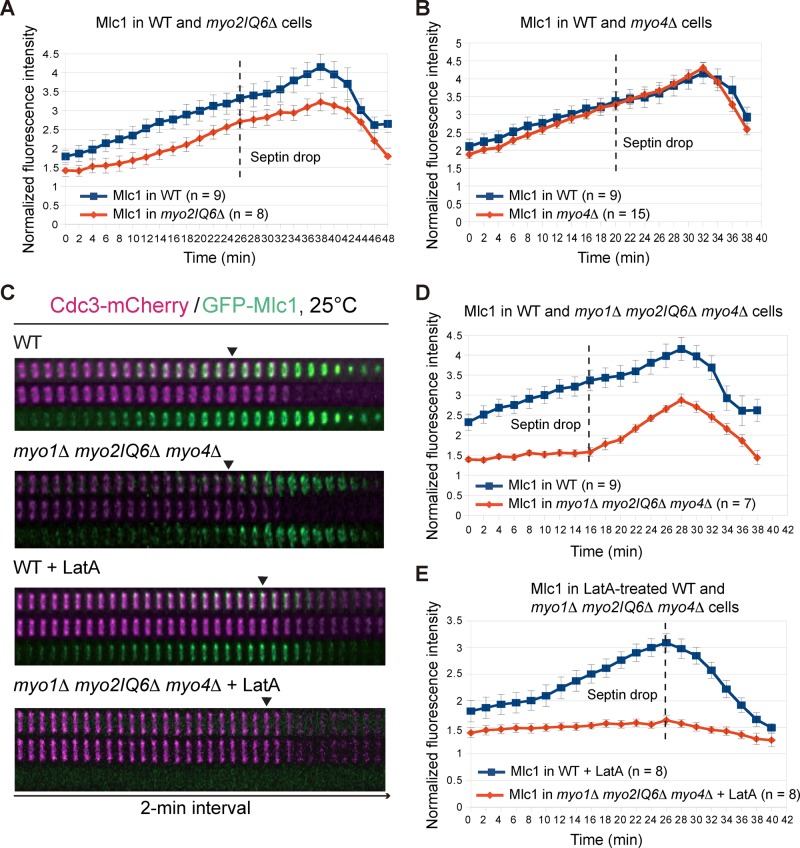
Myo2‑GFP localizes to the bud neck shortly before the septin hourglass‑to‑double ring conversion (Wloka et al., 2011), which is ∼20 min later than Mlc1 does (this study). However, this observation does not rule out the possibility that Myo2, at a subdetection level, is involved in the neck localization of Mlc1 before cytokinesis. To examine this possibility, we assessed the localization kinetics of Mlc1 in myo2IQ6Δ and myo4Δ cells. In the absence of LatA, Mlc1 localization in myo2IQ6Δ cells was remarkably similar to that in bnr1Δ cells (compare Figures 6A and 4D); in contrast, its localization in myo4Δ cells was essentially unchanged throughout the cell cycle compared with WT cells (Figure 6B). In the presence of LatA, the kinetics of Mlc1 localization was similar in both the mutants and the WT cells (unpublished data). As expected, the localization profile of Mlc1 in myo2IQ6Δ myo4Δ cells was similar to that in myo2IQ6Δ cells regardless of LatA treatment (unpublished data). Strikingly, the neck localization of Mlc1 in myo1Δ myo2IQ6Δ myo4Δ cells was nearly abolished before cytokinesis (Figure 6, C and D, and Supplemental Video S9, left). These data suggest that IQ motif–mediated binding of Mlc1 to Myo2 (and perhaps Myo4) largely accounts for the remaining Mlc1 localization in myo1Δ cells. Thus Myo1 and Myo2 play major and minor roles, respectively, in Mlc1 localization before cytokinesis.
FIGURE 6:
Myo1, Myo2, and Myo4 are collectively required for the localization of Mlc1 to the bud neck before cytokinesis. (A, B) Direct comparisons of Mlc1 localization profiles at the bud neck of WT (YEF7435) vs. myo2IQ6Δ (YEF7094; myo2IQ6Δ GFP‑MLC1 CDC3‑mCherry) cells (A) and of WT (YEF7435) vs. myo4Δ (YEF7381; myo4Δ GFP‑MLC1 CDC3‑mCherry) cells (B). (C) Time‑lapse analysis of Mlc1 and Cdc3 at the bud neck during the cell cycle in WT (YEF7435) and myo1Δ myo2IQ6Δ myo4Δ (YEF7055; myo1Δ myo2IQ6Δ myo4Δ GFP‑MLC1 CDC3‑mCherry) cells in the absence or presence of LatA. (D, E) Direct comparisons of Mlc1 localization profiles at the bud neck of WT (YEF7435) and myo1Δ myo2IQ6Δ myo4Δ (YEF7055) cells in the absence (D) or presence (E) of LatA. All cells were grown in SC‑Leu medium at 25°C. Dashed lines indicate the beginning of the septin‑intensity drop during the cell cycle.
Despite the localization defect described here, Mlc1 was able to accumulate and form a peak at the bud neck in myo1Δ myo2IQ6Δ myo4Δ cells during cytokinesis (Figure 6, C and D). However, LatA treatment essentially eliminated Mlc1 localization in the triple-mutant cells during cytokinesis (Figure 6, C and E, and Supplemental Video S9, right). Together these data indicate that Mlc1 is capable of localizing to the division site during cytokinesis independently of its binding to Myo1 as well as to the IQ motifs of Myo2, and this localization completely depends on F‑actin.
The C‑lobe of Mlc1 localizes to the division site during cytokinesis by interacting with the IQ motifs of Myo2
Structural studies indicate that Mlc1 possesses two distinct domains, the N‑ and C‑lobes, which form a cavity for the binding of IQ motifs (Terrak et al., 2003; Pennestri et al., 2007; Amata et al., 2008; Figure 7A, top). These studies further suggest that the N‑lobe binds to IQ2 and IQ3, whereas the C‑lobe binds to IQ1, IQ4, and IQ6 of Myo2. To further explore the mechanisms of Mlc1 targeting, we constructed GFP‑tagged N‑ and C‑lobes of Mlc1 (Figure 7A, bottom) and determined their localization during the cell cycle. We found that GFP‑N‑lobe (residues 1–81) did not localize to the bud neck during any point of the cell cycle (Figure 7B and Supplemental Video S10, first column from the left). In contrast, GFP‑C‑lobe (residues 82–149) localized to the bud neck only during cytokinesis in WT cells (Figure 7C and Supplemental Video S10, second column) but not in myo2IQ6Δ cells (Figure 7D and Supplemental Video S10, third column). In addition, this localization was completely abolished by LatA treatment (Figure 7E and Supplemental Video S10, last column). Together these data indicate that the C‑lobe of Mlc1 is necessary and sufficient for its interaction with the IQ motifs of Myo2, and this interaction contributes to Mlc1 localization during cytokinesis in an F‑actin–dependent manner.
FIGURE 7:
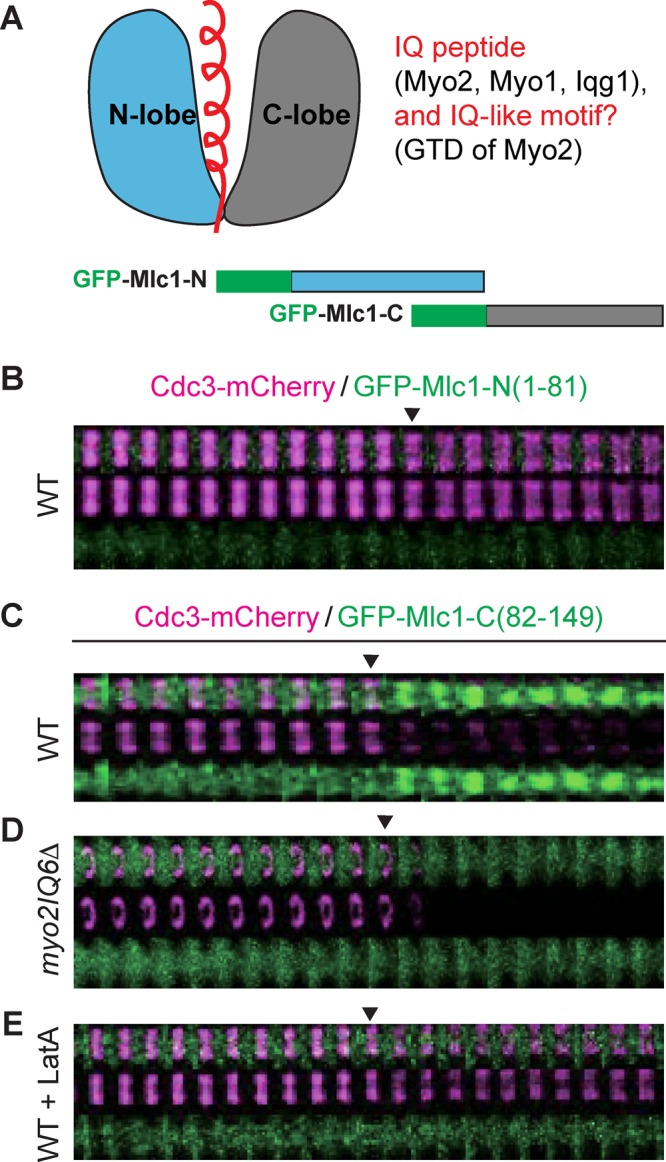
Localization of the C-lobe of Mlc1 to the bud neck depends on Myo2 and actin filaments. (A) Schematics of Mlc1 structure and its binding partners (top) and of GFP‑tagged N‑ and C‑lobes of Mlc1 (bottom). The helical IQ peptide or IQ‑like motif (top left) from the binding partners of Mlc1 (top right) are highlighted in red. (B) Time-lapse analysis of the N‑lobe of Mlc1 (amino acids 1–81) and Cdc3 during the cell cycle in WT cells (YEF7208; CDC3‑mCherry, GFP‑MLC1‑Nterm). (C–E) Time-lapse analysis of the C‑lobe of Mlc1 (amino acids 82–149) and Cdc3 during the cell cycle in WT (YEF7209; CDC3‑mCherry, GFP‑MLC1‑Cterm; C), myo2IQ6Δ (YEF7245; myo2IQ6Δ CDC3‑mCherry, GFP‑MLC1‑Cterm; (D), and LatA‑treated WT (YEF7209; E) cells.
DISCUSSION
By performing quantitative live‑cell imaging on WT cells and yeast mutants carrying multiple gene deletions and/or interaction‑specific mutations coupled with drug treatment, we determined the mechanisms underlying the targeting of ELC to the division site during the cell cycle, a critical question in cytokinesis. Here we first discuss the specific mechanisms responsible for Mlc1 targeting before and during cytokinesis and then expound on their broad significance in cytokinesis beyond budding yeast.
Myosin‑II heavy chain is chiefly responsible for septin‑dependent ELC localization before cytokinesis
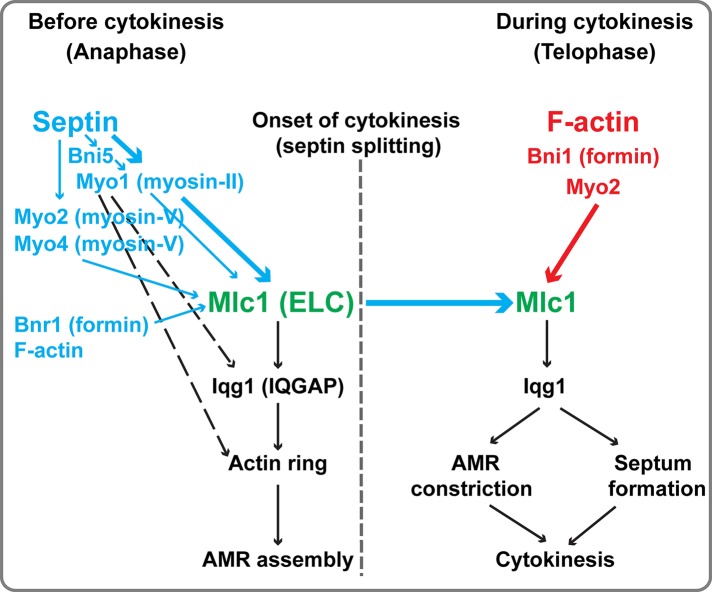
Mlc1 first localizes to the bud cortex to drive efficient bud growth before and during mitosis, and this localization depends on the interactions of Mlc1 with Myo2 and Myo4 (Stevens and Davis, 1998; Shannon and Li, 2000; Luo et al., 2004). Mlc1 then associates with the septin hourglass at the bud neck in medium‑ and large‑budded cells before the onset of cytokinesis (this study). This increase of Mlc1 at the bud neck is always accompanied by the simultaneous decrease of Mlc1 in the cell cortex, suggesting that the same pool of Mlc1 switches binding partners in a cell cycle–dependent manner. This possibility is consistent with the modeling of Mlc1 interactions during the cell cycle (Goel et al., 2011; Goel and Wilkins, 2012) and is further supported by our observation that photoactivated GFP‑Mlc1 in the cell cortex relocalized to the bud neck later in the cell cycle (unpublished data). The neck localization of Mlc1 before cytokinesis depends on the septin hourglass (Boyne et al., 2000; Shannon and Li, 2000; Luo et al., 2004). In this study, we showed that Myo1 plays a major role in the neck localization of Mlc1 before cytokinesis (Figure 8, thick blue arrow). This is consistent with the earlier arrival of Myo1 at the division site than Mlc1 during the cell cycle (Bi et al., 1998; Lippincott and Li, 1998a; Boyne et al., 2000; Shannon and Li, 2000; Wagner et al., 2002; Luo et al., 2004). Myo1 could recruit Mlc1 via direct binding between the IQ1 motif of Myo1 and Mlc1 (Luo et al., 2004) and/or an interaction involving Myo1 tail and Iqg1 (Fang et al., 2010; Figure 8, dashed black arrow), which, in turn, binds to Mlc1 via its IQ motifs (Boyne et al., 2000; Shannon and Li, 2000). Given that Myo1 localization to the bud neck also depends on the septin hourglass (Bi et al., 1998; Lippincott and Li, 1998a; Dobbelaere and Barral, 2004; Wloka et al., 2011), these observations suggest that Myo1 plays a major role in mediating the septin‑dependent Mlc1 localization before cytokinesis.
FIGURE 8:
A two‑tiered mechanism for the targeting of Mlc1 to the division site during the cell cycle. Septin ring and F‑actin (red) mediate the targeting of Mlc1 to the division site before and during cytokinesis, respectively. The septin‑dependent localization is chiefly mediated by Myo1, with some contributions from Bni5, Myo2, Myo4, Bnr1, and F‑actin (blue). In contrast, the F‑actin–dependent localization during cytokinesis is chiefly mediated by Bni1 and Myo2. See the text for more detailed description and discussion.
Besides Myo1, other factors, including Bni5, myosin‑Vs (Myo2 and Myo4), formins (Bnr1 and Bni1), and F‑actin, are also involved in Mlc1 localization before cytokinesis, although their collective role is minor in comparison to Myo1 (Figure 8, thin vs. thick arrows). Bni5 plays such a role before anaphase, presumably through its interaction with Myo1 (Fang et al., 2010). The remarkable similarity in the localization profile of Mlc1 in bnr1Δ and myo2IQ6Δ cells (this study), as well as the localization patterns of Bnr1 (at the bud neck from bud emergence to the onset of cytokinesis) versus Bni1 (at the bud cortex before anaphase and at the bud neck during cytokinesis; Pruyne et al., 2004; Buttery et al., 2007), suggest that a small pool of Mlc1 localizes to the bud neck before cytokinesis by binding to the IQ motifs of Myo2 (and perhaps Myo4), which then moves on actin cables nucleated first by Bnr1 and then by Bni1. In summary, our study demonstrates that Myo1, the IQ motifs of Myo2, and Myo4 are collectively required for Mlc1 localization to the bud neck before cytokinesis.
Formin‑nucleated actin filaments are chiefly responsible for the localization of ELC to the division site during cytokinesis
One of the major findings in this study is that Mlc1 can “establish” and “maintain” localization at the bud neck during cytokinesis in the absence of the septin ring, and this localization completely depends on F‑actin. Thus the septin ring and actin filaments mediate distinct phases of, and are collectively required for, the neck localization of Mlc1 during the cell cycle. As discussed earlier, Myo1 acts downstream of the septin ring to mediate Mlc1 localization before cytokinesis. In contrast, our study suggests that the formin Bni1‑nucleated actin filaments are essential for the further increase of Mlc1 at the division site during cytokinesis (Figure 8). This conclusion is supported by a number of observations: 1) LatA treatment of WT cells abolishes the peak of Mlc1 accumulation at the division site during cytokinesis; 2) the same peak is eliminated by the deletion of BNI1, but not BNR1; and 3) Bni1, but not Bnr1, localizes to the division site during cytokinesis (Pruyne et al., 2004; Buttery et al., 2007).
Bni1‑nucleated actin filaments are required for the assembly of actin ring (Vallen et al., 2000; Tolliday et al., 2002; Wloka et al., 2013) as well as of actin cables, which guide vesicle transport powered by myosin‑V (Myo2) during bud growth and cytokinesis (Bretscher, 2003). Because Mlc1 is still able to increase at the division site during cytokinesis in myo1Δ, as well as in cdc12‑6 myo1Δ cells, in which the actin ring is absent (Bi et al., 1998), the Bni1‑dependent actin cables must play a role in Mlc1 localization during cytokinesis. Because Mlc1 does not bind to actin filaments directly (Bruce Goode, Department of Biology, Brandeis University, personal communication), the actin cables likely mediate Mlc1 localization by guiding Myo2‑associated Mlc1, along with exocytic vesicles, to the division site during cytokinesis. Mlc1 binds to the IQ motifs of Myo2 (Stevens and Davis, 1998), and this binding contributes to Mlc1 localization during cytokinesis, as demonstrated by the localization of its C‑lobe in WT cells but not in myo2IQ6Δ or LatA‑treated WT cells (this study). Mlc1 is also a “cargo” of exocytic vesicles, as Mlc1 interacts with the globular tail domain (or cargo‑binding domain) of Myo2 (Casavola et al., 2008) and associates with secretory vesicles (Wagner et al., 2002; Bielli et al., 2006; Casavola et al., 2008). These interactions likely explain the F‑actin–dependent localization of Mlc1 in myo1Δ myo2IQ6Δ myo4Δ cells during cytokinesis (this study). Collectively our study suggests that Mlc1 is delivered to the division site during cytokinesis via polarized exocytosis, which requires Bni1‑dependent actin cables, as well as its interactions with Myo2 and secretory vesicles (Figure 8).
Why does the cell need multiple mechanisms for targeting the ELC to the division site?
Two main messages emerge from our study. First, distinct mechanisms are responsible for the targeting of Mlc1 to the division site during different phases of the cell cycle (Figure 8). The Myo1‑based mechanism acts downstream of the septin ring and plays a major role in Mlc1 localization before cytokinesis, whereas the exocytosis‑based mechanism, involving Bni1, F‑actin, and Myo2, is mainly responsible for the further increase of Mlc1 at the division site during cytokinesis. The latter mechanism also plays a minor role in Mlc1 localization before cytokinesis. Second, there is a clear reversal of localization dependence between Myo1 and Mlc1 during the cell cycle. Before cytokinesis, the neck localization of Mlc1 largely depends on Myo1, whereas during cytokinesis, Myo1 localization depends on Mlc1 and Iqg1 (Fang et al., 2010). This reversal of relationship is made possible presumably by the cell cycle–regulated switch in localization mechanisms.
What is the functional consequence of inactivating each localization mechanism? Ideally, this question should be addressed by analyzing the effects on cytokinesis of point mutations in MLC1 that are specifically defective in either the Myo1‑ or the Bni1‑mediated pathway. However, Myo1, Myo2, and Iqg1 all bind to the cavity formed by N‑ and C‑lobes of Mlc1 via their IQ motifs (Figure 7A). Thus it might be difficult to isolate mlc1 mutations that specifically affect its interaction with Myo1. According to our model, only the Bni1‑based actin cables are required for the delivery of Mlc1 to the bud neck during cytokinesis. Thus, in order to inactivate the Bni1‑based mechanism, mutations in MLC1 that specifically affect its interactions with Myo2 and secretory vesicles or mutations that specifically inactivate Bni1 during cytokinesis have to be isolated, which would be rather difficult, if not impossible, given the limited knowledge on the Mlc1 interactions and the spatiotemporal control of Bni1 activation. Despite these challenges, the observations that myo1Δ and bni1Δ cells are viable but deficient in cytokinesis (Watts et al., 1987; Rodriguez and Paterson, 1990; Bi et al., 1998; Vallen et al., 2000; Fang et al., 2010) and that the two deletions are synthetically lethal (Vallen et al., 2000) are consistent with the notion that each localization mechanism for Mlc1 plays an important role in cytokinesis.
Why does the cell evolve such elaborate mechanisms for the localization of Mlc1 to the division site? MLC1 is haploinsufficient for the survival of S. cerevisiae (Stevens and Davis, 1998) and plays an essential role in cytokinesis (Stevens and Davis, 1998; Boyne et al., 2000; Shannon and Li, 2000). Thus the level of Mlc1 in the cell and/or the ratio of Mlc1 versus its binding partners (Stevens and Davis, 1998), presumably including its local concentration at the bud neck, are critical for its function. The two‑tiered localization mechanisms for Mlc1 could provide at least two major advantages to the cell in terms of cytokinesis. First, they can ensure an ordered assembly and function of the cytokinesis machinery. As in other organisms (Pollard, 2010), the actin ring is assembled during late anaphase but before the onset of cytokinesis or mitotic exit in budding yeast (Epp and Chant, 1997; Bi et al., 1998; Lippincott and Li, 1998a; Vallen et al., 2000), and this assembly depends on Myo1 and Mlc1 (Bi et al., 1998; Boyne et al., 2000; Shannon and Li, 2000). Thus the Myo1‑dependent Mlc1 localization before cytokinesis is essential for AMR assembly. During cytokinesis, Mlc1 is required not only for the maintenance of Myo1 or AMR at the division site (Fang et al., 2010), but also for septum formation (Bi, 2001; Wagner et al., 2002; Wloka and Bi, 2012). The latter function is presumably carried out via the complex of Mlc1, Iqg1, and the F‑BAR protein Hof1 (Naylor and Morgan, 2014; Tian et al., 2014), which interacts with the C2‑domain protein Inn1 and the transglutaminase‑like protein Cyk3 to promote PS formation (Nishihama et al., 2009; Meitinger et al., 2010; Devrekanli et al., 2012). Thus distinct localization mechanisms allow Mlc1 to carry out its distinct roles in cytokinesis during different phases of the cell cycle.
The second potential advantage is to endow cytokinesis with robustness and adaptability so that cells can survive under adverse conditions. For example, when the septin- and Myo1‑dependent mechanism is compromised by a mutational event such as a septin mutation or myo1Δ, the mutant cells can survive with the second mechanism (Watts et al., 1987; Rodriguez and Paterson, 1990; Bi et al., 1998). Reciprocally, when the formin (Bni1)- and F‑actin–dependent mechanism is compromised by a mutational event such as bni1Δ, the mutant cells can survive with the first mechanism (Vallen et al., 2000). In all cases, the single mutants are viable but defective in cytokinesis and fitness. However, cells cannot survive the inactivation of both mechanisms, as demonstrated by the synthetic lethality observed between a septin mutation and a bni1 mutation (Longtine et al., 1996) or between myo1Δ and bni1Δ (Vallen et al., 2000).
Despite the fact that ELCs are evolutionarily conserved at a much higher level than their corresponding RLCs in diverse biological systems (Luo et al., 2004), the role of ELCs in cytokinesis, as opposed to that of RLCs, has been poorly studied, especially in mammalian systems. In the few cases in which functional analysis of the ELCs has been performed, such as in budding yeast (Stevens and Davis, 1998; Boyne et al., 2000; Shannon and Li, 2000; Wagner et al., 2002; Luo et al., 2004), fission yeast (Cdc4; McCollum et al., 1995), and Dictyostelium (MlcE; Pollenz et al., 1992; Chen et al., 1995), the ELCs have been proven essential for cell viability and cytokinesis. In contrast, cells lacking the RLCs in the same organisms are viable but display variable degrees of defects in cytokinesis (Chen et al., 1994; Le Goff et al., 2000; Naqvi et al., 2000; Luo et al., 2004). Thus it is critically important to understand how the ELCs perform their essential roles in cytokinesis in diverse systems. As in S. cerevisiae, the ELC in S. pombe binds to the IQ motifs of myosin‑II (Myo2 and Myp2), myosin‑V (Myo51), and IQGAP (Rng2) (D'souza et al., 2001). Moreover, ELC and IQGAP are required for maintaining myosin‑II localization at the division site during cytokinesis in both budding and fission yeast (Fang et al., 2010; Laporte et al., 2011; Padmanabhan et al., 2011; Takaine et al., 2014). Thus the mechanism of ELC targeting to the division site is likely conserved between budding yeast and other organisms.
MATERIALS AND METHODS
Strains and growth media
Yeast strains used in this study are listed in Table 1. Standard culture media and genetic techniques were used (Guthrie and Fink, 1991). Yeast strains were grown routinely at 25°C in synthetic complete (SC) minimal medium lacking specific amino acid(s) and/or uracil or in rich medium YM-1 (Lillie and Pringle, 1980) or yeast extract/peptone/dextrose (YPD). In some cases, 1 mg/ml 5-fluoroorotic acid (5‑FOA; Angus Buffers & Biochemicals, Niagara Falls, NY) was added to the media to select for the loss of URA3-containing plasmids. G418 at 200 μg/ml (Life Technologies, Gaithersburg, MD) was added to the media for the selection of kanamycin resistance (kanMX6).
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| CRY1 | a ade2-1oc can1-100 his3-11,15 leu2-3112 trp1-1 ura3-1 | Stevens and Davis (1998) |
| RSY21 | Like CRY1, except myo2IQ6Δ | Stevens and Davis (1998) |
| M-17 | a cdc12-6 leu2 ura3 | Caviston et al. (2003) |
| YEF473A | a his3 leu2 lys2 trp1 ura3 | Bi and Pringle (1996) |
| YEF1804 | Like YEF473A, except myo1Δ::KanMX6 | Bi et al. (1998) |
| YEF2692 | Like YEF473A, except bni1Δ::HIS3 | This study |
| YEF2697 | Like YEF473A, except bnr1Δ::HIS3 | This study |
| YEF3387 | Like CRY1, except myo2IQ6Δ myo1Δ::KanMX6 | This study |
| YEF6743 | Like YEF473A, except (P414‑ADH1‑MLC1) | This study |
| YEF6744 | Like YEF473A, except mlc1Δ::URA3-Kan (P414‑ADH1‑MLC1) | This study |
| YEF6757 | Like YEF473A, except GFP‑MLC1 (P414‑ADH1‑MLC1) | This study |
| YEF6800 | Like YEF473A, except GFP-MLC1 CDC3-mCherry::LEU2 | This study |
| YEF6866 | Like M‑17, except CDC12 | This study |
| YEF6870 | a cdc12-6 GFP‑MLC1::LEU2 | Derivative of M‑17 |
| YEF6884 | a cdc12-6 GFP‑MLC1::LEU2 CDC3-mCherry::URA3 | Derivative of YEF6870 |
| YEF6888 | a GFP-MLC1::LEU2 CDC3-mCherry::URA3 | Derivative of YEF6866 |
| YEF6951 | Like YEF473A, except CDC3-mCherry::URA3 | This study |
| YEF7016 | Like CRY1, except myo2IQ6Δ myo1Δ::NatMX myo4Δ::KanMX6 GFP‑MLC1::LEU2 | This study |
| YEF7055 | Like CRY1, except myo1Δ::NatMX myo2IQ6Δ myo4Δ::KanMX6 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7070 | Like YEF473A, except CDC3-mCherry::URA3 GFP-MLC1::LEU2 | This study |
| YEF7081 | Like YEF473A, except cdc12-6 myo1Δ::KanMX6 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7090 | Like CRY1, except myo2IQ6Δ CDC3-mCherry::TRP1 | This study |
| YEF7094 | Like CRY1, except myo2IQ6Δ CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7127 | Like YEF473A, except myo1Δ::KanMX6 CDC3-mCherry::TRP1 | This study |
| YEF7130 | Like CRY1, except myo2IQ6Δ myo4Δ::KanMX6 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7131 | Like CRY1, except myo2IQ6Δ myo1Δ::KanMX6 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7132 | Like YEF473A, except myo1Δ::KanMX6 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7155 | Like YEF473A, except cdc12-6 CDC3-mCherry::TRP1 MYO1-GFP-Kan | This study |
| YEF7184 | Like YEF473A, except bni1Δ::HIS3 CDC3-mCherry::TRP1 | This study |
| YEF7185 | Like YEF473A, except bnr1Δ::HIS3 CDC3-mCherry::TRP1 | This study |
| YEF7200 | Like YEF473A, except bnr1Δ::HIS3 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7201 | Like YEF473A, except bni1Δ::HIS3 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7208 | Like YEF473A, except CDC3-mCherry::URA3 [YCp111-GFP-MLC1-Nterm(1-81)] | This study |
| YEF7209 | Like YEF473A, except CDC3-mCherry::URA3 [YCp111-GFP-MLC1-Cterm(82-149)] | This study |
| YEF7245 | Like CRY1, except myo2IQ6Δ CDC3-mCherry::TRP1 [YCp111-GFP-MLC1-Cterm(82-149)] | This study |
| YEF7259 | Like YEF473A, except bni5Δ::KanMX6 CDC3-mCherry::URA3 GFP-MLC1::LEU2 | This study |
| YEF7381 | Like YEF473A, except myo4Δ::KanMX6 CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
| YEF7435 | Like CRY1, except CDC3-mCherry::TRP1 GFP-MLC1::LEU2 | This study |
Constructions of plasmids and yeast strains
All primers were purchased from Integrated DNA Technologies. Sequencing of constructs was performed at the DNA Sequencing Facility, University of Pennsylvania (Philadelphia, PA).
Plasmids YIplac128-CDC3-mCherry (integrative, LEU2; Gao et al., 2007), YIplac211-CDC3-mCherry (integrative, URA3; Fang et al., 2010), and YIplac204-CDC3-mCherry (integrative, TRP1; Wloka et al., 2011) carrying mCherry-tagged CDC3 were digested with BglII and integrated at the CDC3 locus of the recipient strains.
p414-ADH1-MLC1 (CEN, TRP1) was constructed by gap repairing PCR‑amplified MLC1 open reading frame (ORF) into PstI- and BamHI‑digested plasmid p414ADH (Mumberg et al., 1995).
Plasmid YIplac128‑GFP‑MLC1 (integrative, LEU2) and YIplac211‑GFP‑MLC1 (integrative, URA3), carrying GFP‑tagged MLC1 under its own promoter, were constructed in several steps. 1) The chromosomal MLC1 ORF in a WT strain carrying the cover plasmid p414‑ADH1‑MLC1 (YEF6743) was replaced by a PCR‑amplified URA3‑Kan fragment via homologous recombination, as described for other genes (Tong et al., 2007; Fang et al., 2010), leading to the generation of strain YEF6744 (mlc1Δ::URA3‑Kan, p414‑ADH1‑MLC1). 2) A pair of primers containing sequences immediately upstream and downstream of the MLC1 ORF, respectively, was used to PCR‑amplify GFP‑MLC1 (GFP inserted after the ATG codon of MLC1) from the plasmid pUG34‑MLC1 (GFP‑MLC1 is under the control of MET17 promoter on this plasmid; Wagner et al., 2002) and then transformed into YEF6744. The transformants were screened for the loss of URA3 and Kan on the SC‑TRP+5‑FOA and YPD+G418 plates, respectively, to generate the strain (YEF6757), in which the URA3‑Kan cassette was replaced by GFP‑MLC1. The correctness of this strain was confirmed by PCR checking and fluorescence microscopy. 3) A 2.43-kb PCR product containing GFP‑MLC1 with its putative promoter (800 base pairs upstream of the ATG codon) and terminator (424 base pairs downstream of the STOP codon) sequences were amplified from the chromosomal DNA of YEF6757. The PCR product was digested with PstI and EcoRI (both restriction sites were introduced in the PCR primers) and cloned into the corresponding sites of the plasmid YIplac128 or YIplac211 (Gietz and Sugino, 1988), thus yielding the desired plasmids. Plasmids YIplac128‑GFP‑MLC1 and YIplac211‑GFP‑MLC1 were then digested with Tth111I and integrated at the MLC1 locus of different recipient strains to generate derivatives carrying a single copy of GFP‑MLC1 under the control of its own promoter (in addition to the endogenous MLC1).
Plasmids YCp111-GFP-MLC1-Nterm(1-81) (CEN, LEU2) and YCp111-GFP-MLC1-Cterm(82-149) (CEN, LEU2) were constructed as follows. A 2.43‑kb PstI‑EcoRI fragment carrying GFP‑MLC1 was cloned from YIplac128‑GFP‑MLC1 into the corresponding sites of YCplac111 (Gietz and Sugino, 1988). The resulting plasmid, YCplac111‑GFP‑MLC1, was then digested with MscI (in the GFP ORF) and Tth111I (in MLC1 ORF) and subsequently gap repaired with a fragment containing the coding sequences for GFP and the first 81 amino acids of Mlc1 that was PCR‑amplified from pUG34‑MLC1 (Wagner et al., 2002). This led to the construction of YCp111-GFP-MLC1-Nterm(1-81). Similarly, the coding sequence for the C-lobe of Mlc1 (amino acids 82–149) was PCR amplified from pUG34‑MLC1 (Wagner et al., 2002) and then gap repaired into Tth111I‑digested YCplac111‑GFP‑MLC1, leading to the construction of YCp111-GFP-MLC1-Cterm(82-149). Correctness of the GFP‑tagged N‑ and C‑lobe constructs was confirmed by DNA sequencing.
Live-cell imaging and quantitative analysis
Cells were cultured in SC-dropout medium (a specific amino acid or uracil was omitted) to exponential phase at 25°C and then embedded in a layer of medium solidified with 1.2% low-melting-temperature agarose (Lonza, Allendale, NJ) in a polylysine-coated glass-bottom dish (MatTek, Ashland, MA; Okada et al., 2013). In some cases, cells were pretreated with 100 μM LatA or high temperature (39°C) before time-lapse analysis.
Image acquisitions were performed on a microscope (Olympus IX71, Center Valley, PA) with a spinning-disk confocal scan head (Yokogawa CSU10, Tokyo, Japan) and a 100× objective lens (1.4 numerical aperture, Plan S-Apo oil; Olympus). Acquisition and hardware were controlled by MetaMorph, version 7.7 (Molecular Devices, Downingtown, PA). An electron multiplying charge-coupled device camera (model C9100-13; ImagEM; Hamamatsu Photonics, Bridgewater, NJ) was used for acquisition. Diode lasers for excitation (488 nm for GFP and 561 nm for mCherry/red fluorescent protein) were set in a laser integrator (Spectral Applied Research, Richmond Hill, ON, Canada). Acquisition interval was 2 min. For each time point, 14 z-sections were acquired at 0.6‑μm increment. Image processing and analysis were performed using Fiji (Schindelin et al., 2012). For quantification of fluorescence intensities, image sequences generated by average projection were used. A specific polygon covering the region of interest was drawn to yield the integrated density, mean density, and area for that region. The size of the polygon was kept constant before and during AMR constriction. The ratio of the mean fluorescence intensity of Mlc1 at the bud neck versus the total cell, after background subtraction, was taken as the normalized fluorescence intensity used for all the plots in this article.
Supplementary Material
Acknowledgments
We thank T. Davis for yeast strains; A. Stout of the Imaging Core at the Perelman School of Medicine for technical assistance; J. Luo and X. Fang for strain constructions; Masayuki Onishi and Yogini Bhavsar‑Jog for critically reading the manuscript; B. Goode for sharing unpublished data; and C. Wloka, T. Wang, and other members of the Bi laboratory for valuable discussions and suggestions. This work was supported by National Institutes of Health Grant GM87365 to E.B. and a fellowship from the Chinese Scholarship Council to Z.F.
Abbreviations used:
- AMR
actomyosin ring
- ELC
essential light chain
- HC
heavy chain
- LatA
latrunculin A
- RLC
regulatory light chain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-09-1363) on January 28, 2015.
*Present address: Department of Medical Biochemistry, Kyushu University Graduate School of Medical Sciences, Fukuoka 812-8582, Japan.
References
- Amata I, Gallo M, Pennestri M, Paci M, Ragnini-Wilson A, Cicero DO. N-lobe dynamics of myosin light chain dictates its mode of interaction with myosin V IQ1. Biochemistry. 2008;47:12332–12345. doi: 10.1021/bi801178t. [DOI] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor Latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bi E. Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation. Cell Struct Funct. 2001;26:529–537. doi: 10.1247/csf.26.529. [DOI] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Park HO. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielli P, Casavola EC, Biroccio A, Urbani A, Ragnini-Wilson A. GTP drives myosin light chain 1 interaction with the class V myosin Myo2 IQ motifs via a Sec2 RabGEF-mediated pathway. Mol Microbiol. 2006;59:1576–1590. doi: 10.1111/j.1365-2958.2006.05041.x. [DOI] [PubMed] [Google Scholar]
- Boyne JR, Yosuf HM, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113:4533–4543. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J Cell Biol. 2003;160:811–816. doi: 10.1083/jcb.200301035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18:1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavola EC, Catucci A, Bielli P, Di Pentima A, Porcu G, Pennestri M, Cicero DO, Ragnini-Wilson A. Ypt32p and Mlc1p bind within the vesicle binding region of the class V myosin Myo2p globular tail domain. Mol Microbiol. 2008;67:1051–1066. doi: 10.1111/j.1365-2958.2008.06106.x. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Longtine M, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Ostrow BD, Tafuri SR, Chisholm RL. Targeted disruption of the Dictyostelium RMLC gene produces cells defective in cytokinesis and development. J Cell Biol. 1994;127:1933–1944. doi: 10.1083/jcb.127.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TL, Kowalczyk PA, Ho G, Chisholm RL. Targeted disruption of the Dictyostelium myosin essential light chain gene produces cells defective in cytokinesis and morphogenesis. J Cell Sci. 1995;108:3207–3218. doi: 10.1242/jcs.108.10.3207. [DOI] [PubMed] [Google Scholar]
- Devrekanli A, Foltman M, Roncero C, Sanchez-Diaz A, Labib K. Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J Cell Sci. 2012;125:5453–5466. doi: 10.1242/jcs.109157. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- D'souza VM, Naqvi NI, Wang H, Balasubramanian MK. Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct Funct. 2001;26:555–565. doi: 10.1247/csf.26.555. [DOI] [PubMed] [Google Scholar]
- Epp JA, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin-II. J Cell Biol. 2010;191:1333–1350. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Sperber LM, Kane SA, Tong Z, Hin Yan Tong A, Boone C, Bi E. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol Biol Cell. 2007;18:2542–2560. doi: 10.1091/mbc.E06-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goel A, Li SS, Wilkins MR. Four-dimensional visualisation and analysis of protein-protein interaction networks. Proteomics. 2011;11:2672–2682. doi: 10.1002/pmic.201000546. [DOI] [PubMed] [Google Scholar]
- Goel A, Wilkins MR. Dynamic hubs show competitive and static hubs non-competitive regulation of their interaction partners. PLoS One. 2012;7:e48209. doi: 10.1371/journal.pone.0048209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X, Motegi F, Salimova E, Mabuchi I, Simanis V. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J Cell Sci. 2000;113:4157–4163. doi: 10.1242/jcs.113.23.4157. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998a;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114:1379–1386. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Luo J, Vallen EA, Dravis C, Tcheperegine SE, Drees BL, Bi E. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae. J Cell Biol. 2004;165:843–855. doi: 10.1083/jcb.200401040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- McCollum D, Balasubramanian MK, Pelcher LE, Hemmingsen SM, Gould KL. Schizosaccharomyces pombe cdc4+ gene encodes a novel EF-hand protein essential for cytokinesis. J Cell Biol. 1995;130:651–660. doi: 10.1083/jcb.130.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Petrova B, Mancini Lombardi I, Bertazzi DT, Hub B, Zentgraf H, Pereira G. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci. 2010;123:1851–1861. doi: 10.1242/jcs.063891. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Naqvi N, Wong KCY, Tang X, Balasubramanian MK. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat Cell Biol. 2000;2:855–858. doi: 10.1038/35041107. [DOI] [PubMed] [Google Scholar]
- Naylor SG, Morgan DO. Cdk1-dependent phosphorylation of Iqg1 governs actomyosin ring assembly prior to cytokinesis. J Cell Sci. 2014;127:1128–1137. doi: 10.1242/jcs.144097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, Bi E. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J Cell Biol. 2009;185:995–1012. doi: 10.1083/jcb.200903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Leda M, Hanna J, Savage NS, Bi E, Goryachev AB. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev Cell. 2013;26:148–161. doi: 10.1016/j.devcel.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A, Bakka K, Sevugan M, Naqvi NI, D'souza V, Tang X, Mishra M, Balasubramanian MK. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr Biol. 2011;21:467–472. doi: 10.1016/j.cub.2011.01.059. [DOI] [PubMed] [Google Scholar]
- Pennestri M, Melino S, Contessa GM, Casavola EC, Paci M, Ragnini-Wilson A, Cicero DO. Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs. J Biol Chem. 2007;282:667–679. doi: 10.1074/jbc.M607016200. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollenz RS, Chen TL, Trivinos-Lagos L, Chisholm RL. The Dictyostelium essential light chain is required for myosin function. Cell. 1992;69:951–962. doi: 10.1016/0092-8674(92)90614-i. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Gao L, Bi E, Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol Biol Cell. 2004;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskeleton. 1990;17:301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon KB, Li R. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr Biol. 2000;10:727–730. doi: 10.1016/s0960-9822(00)00539-x. [DOI] [PubMed] [Google Scholar]
- Stevens RC, Davis TN. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J Cell Biol. 1998;142:711–722. doi: 10.1083/jcb.142.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland LI, Burgess DR. Pathways for membrane trafficking during cytokinesis. Trends Cell Biol. 2004;14:115–118. doi: 10.1016/j.tcb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Takaine M, Numata O, Nakano K. Fission yeast IQGAP maintains F-actin-independent localization of myosin-II in the contractile ring. Genes Cells. 2014;19:161–176. doi: 10.1111/gtc.12120. [DOI] [PubMed] [Google Scholar]
- Tan JL, Ravid S, Spudich JA. Control of nonmuscle myosins by phosphorylation. Annu Rev Biochem. 1992;61:721–759. doi: 10.1146/annurev.bi.61.070192.003445. [DOI] [PubMed] [Google Scholar]
- Terrak M, Wu G, Stafford WF, Lu RC, Dominguez R. Two distinct myosin light chain structures are induced by specific variations within the bound IQ motifs-functional implications. EMBO J. 2003;22:362–371. doi: 10.1093/emboj/cdg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wu Y, Johnsson N. Stepwise and cooperative assembly of a cytokinetic core complex in yeast Saccharomyces cerevisiae. J Cell Sci. 2014;127:3614–3624. doi: 10.1242/jcs.153429. [DOI] [PubMed] [Google Scholar]
- Tolliday N, VerPlank L, Li R. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol. 2002;12:1864–1870. doi: 10.1016/s0960-9822(02)01238-1. [DOI] [PubMed] [Google Scholar]
- Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol. 2007;179:1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus KM. Assembly of cytoplasmic and smooth muscle myosins. Curr Opin Cell Biol. 1991;3:105–111. doi: 10.1016/0955-0674(91)90172-u. [DOI] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W, Bielli P, Wacha S, Ragnini-Wilson A. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 2002;21:6397–6408. doi: 10.1093/emboj/cdf650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts FZ, Shiels G, Orr E. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 1987;6:3499–3505. doi: 10.1002/j.1460-2075.1987.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken) 2012;69:710–726. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- Wloka C, Nishihama R, Onishi M, Oh Y, Hanna J, Pringle JR, Krauss M, Bi E. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol Chem. 2011;392:813–829. doi: 10.1515/BC.2011.083. [DOI] [PubMed] [Google Scholar]
- Wloka C, Vallen EA, Thé L, Fang X, Oh Y, Bi E. Immobile myosin-II plays a scaffolding role during cytokinesis in budding yeast. J Cell Biol. 2013;200:271–286. doi: 10.1083/jcb.201208030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.