A SAXS-based structural model is described for PICK1, a key player in AMPA receptor trafficking. It is shown that the acidic C-terminal tail of PICK1 is involved in autoinhibition and motility of PICK1-associated vesicle-like structures, but, contrary to previous reports, PICK1 neither binds nor inhibits Arp2/3 complex.
Abstract
PICK1 is a modular scaffold implicated in synaptic receptor trafficking. It features a PDZ domain, a BAR domain, and an acidic C-terminal tail (ACT). Analysis by small- angle x-ray scattering suggests a structural model that places the receptor-binding site of the PDZ domain and membrane-binding surfaces of the BAR and PDZ domains adjacent to each other on the concave side of the banana-shaped PICK1 dimer. In the model, the ACT of one subunit of the dimer interacts with the PDZ and BAR domains of the other subunit, possibly accounting for autoinhibition. Consistently, full-length PICK1 shows diffuse cytoplasmic localization, but it clusters on vesicle-like structures that colocalize with the trans-Golgi network marker TGN38 upon deletion of either the ACT or PDZ domain. This localization is driven by the BAR domain. Live-cell imaging further reveals that PICK1-associated vesicles undergo fast, nondirectional motility in an F-actin–dependent manner, but deleting the ACT dramatically reduces vesicle speed. Thus the ACT links PICK1-associated vesicles to a motility factor, likely myosin, but, contrary to previous reports, PICK1 neither binds nor inhibits Arp2/3 complex.
INTRODUCTION
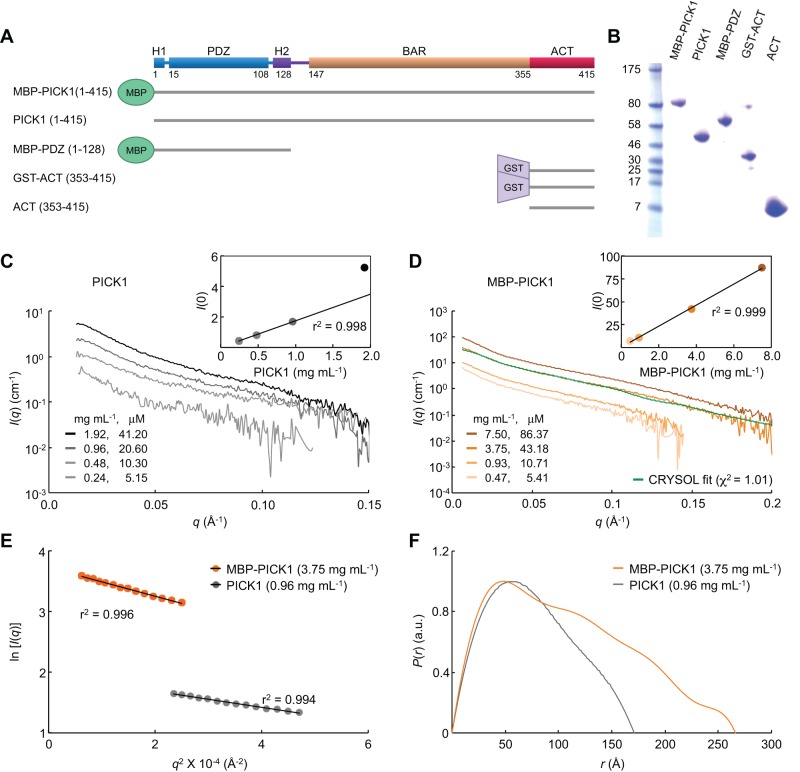
Human protein interacting with C kinase 1 (PICK1) is a 415–amino acid (aa) multidomain scaffold containing an N-terminal PSD-95/DlgA/ZO-1 (PDZ) domain, a central Bin/amphiphysin/Rvs (BAR) domain, and a 60-aa acidic C-terminal tail (ACT; Figure 1A). PICK1 is expressed in all tissues analyzed but is most abundantly expressed in brain (Xia et al., 1999), where it regulates the activity and trafficking of several important neuronal proteins (Hanley, 2008; Focant and Hermans, 2013). The interactions of PICK1 with these proteins are mediated by the PDZ domain, which selectively binds the C-termini of >30 enzymes, transporters, and receptors, including protein kinase C, the dopamine transporter (DAT), and the GluA2 subunit of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs; Erlendsson et al., 2014). Alterations in PICK1 function have been linked to several neurological disorders, including schizophrenia, Parkinson's disease, epilepsy, chronic pain, drug addiction, and amyotrophic lateral sclerosis (Focant and Hermans, 2013).
FIGURE 1:
X-ray scattering analysis of PICK1. (A) Domain diagram of PICK1 and description of constructs. (B) SDS–PAGE analysis of the PICK1 proteins used in this study. (C, D) X-ray scattering intensity plotted vs. momentum transfer for different PICK1 and MBP-PICK1 concentrations (as indicated). Insets show the scattering intensity dependence on protein concentration. A linear dependence is indicative of lack of aggregation. The green trace in D corresponds to the scattering curve calculated from the model. (E) Guinier plots of PICK1 and MBP-PICK1 calculated from the scattering data at the indicated sample concentration (color coded). (F) Normalized distance distribution functions of PICK1 and MBP-PICK1.
Most attention has been devoted to the interaction of PICK1 with AMPARs (Dev et al., 1999; Xia et al., 1999). AMPARs are glutamate-gated cation channels that mediate the vast majority of fast excitatory transmissions in the brain (Henley and Wilkinson, 2013). The number of AMPARs at the synaptic plasma membrane is a key determinant of synaptic strength and plasticity and is controlled through trafficking events involving PICK1. Thus multiple studies have shown that PICK1 plays a major role in AMPAR internalization and intracellular retention during long-term depression (Xia et al., 2000; Iwakura et al., 2001; Kim et al., 2001; Perez et al., 2001; Hanley and Henley, 2005; Steinberg et al., 2006; Citri et al., 2010), whereas other studies also have linked PICK1 to long-term potentiation (Terashima et al., 2008; Thorsen et al., 2010).
Full-length PICK1 appears to adopt an autoinhibited conformation, characterized by its uniform cytoplasmic localization, in which the membrane-binding surface is not fully exposed (Perez et al., 2001; Lu and Ziff, 2005; Madsen et al., 2008). Deletion of the PDZ domain exposes the membrane-binding capacity of the BAR domain, resulting in PICK1 relocalization to vesicle-like clusters (Perez et al., 2001; Lu and Ziff, 2005; Madsen et al., 2008). Binding to a ligand at the membrane via the PDZ domain can also trigger BAR domain–dependent membrane interaction and redistribution of PICK1 to vesicle-like structures (Madsen et al., 2008). Both the PDZ (Jin et al., 2006; Pan et al., 2007) and BAR (Jin et al., 2006; Steinberg et al., 2006; Madsen et al., 2008) domains appear to participate in membrane interactions, whereas the PDZ domain additionally binds to the C-termini of integral membrane proteins, such as AMPAR subunit GluA2 (Dev et al., 1999; Xia et al., 1999). The interaction of PICK1 with GluA2 ultimately results in AMPAR endocytosis (Chung et al., 2000; Xia et al., 2000; Kim et al., 2001; Perez et al., 2001), although the exact mechanism is poorly understood.
BAR domain proteins play important roles in endocytosis by generating or sensing membrane curvature while often also linking to the cytoskeleton for vesicle movement (Saheki and De Camilli, 2012). A link to the cytoskeleton has also been proposed for PICK1 by a study that found that PICK1 inhibits actin filament (F-actin) nucleation by Arp2/3 complex through interactions of the BAR domain with F-actin and the ACT with Arp2/3 complex (Rocca et al., 2008). These authors proposed that inhibition of Arp2/3 complex reduces membrane tension resulting from actin polymerization, which would otherwise oppose membrane invagination during endocytosis (Rocca et al., 2008; Nakamura et al., 2011). However, these conclusions contradict other studies that found that actin polymerization is used, on the contrary, to promote endocytosis under conditions of increased membrane tension (Aghamohammadzadeh and Ayscough, 2009; Boulant et al., 2011; Kaur et al., 2014). It is also unclear how PICK1 can simultaneously bind F-actin (Rocca et al., 2008) and the plasma membrane (Jin et al., 2006; Madsen et al., 2008), since the same basic residues on the BAR domain are implicated in both interactions.
Several important questions, including about the nature of autoinhibitory interactions, the relative contributions of the PDZ and BAR domains to membrane binding, the role of the ACT, and the apparent incompatibility of BAR domain interactions with F-actin and membranes, require structural information about full-length PICK1 to be addressed. However, whereas structures of the PDZ domain, including in complex with receptor tail peptides, have been determined (Elkins et al., 2007; Pan et al., 2007; Shi et al., 2010; Erlendsson et al., 2014), full-length PICK1 has so far resisted high-resolution structural analysis. Here we used small-angle x-ray scattering (SAXS) to study the structure of PICK1 in solution. We then expressed several PICK1 constructs in cells and used live-cell imaging to test the structural model of PICK1 and assess the motility of PICK1-associated vesicles. By treating cells with latrunculin B and the Arp2/3 complex inhibitor CK-666, we investigated the role of the actin cytoskeleton in the motility of PICK1-associated vesicles. By coexpressing PICK1 with organelle markers, we identified the nature of PICK1-associated vesicles. Finally, we used isothermal titration calorimetry and pyrene-actin polymerization assays to test the reported interaction and inhibition of Arp2/3 complex nucleation by PICK1. We conclude that the ACT of PICK1 is responsible for inhibition of membrane binding and is necessary for the motility of PICK1-associated organelles that colocalize with the trans-Golgi network. Contrary to previous reports, however, we found that PICK1 neither binds nor inhibits Arp2/3 complex.
RESULTS
Structural analysis of full-length PICK1 using SAXS
The structure of full-length PICK1 was investigated by SAXS, a method that yields information about the overall shape of macromolecules in solution (Blanchet and Svergun, 2013). The protein used in the experiments was highly pure (Figure 1B) and appeared soluble at the highest concentration used, ∼2 mg/ml. Yet data collected at lower concentrations through a series of dilutions showed a nonlinear dependence of the scattering intensity on protein concentration above ∼1 mg/ml (Figure 1C), which is indicative of protein aggregation (undetectable by the naked eye). Thus the highest PICK1 concentration that could be reliably used in analysis was ∼1.0 mg/ml. At this concentration, the Guinier plot was linear (Figure 1E), consistent with a pure, monodisperse species (Blanchet and Svergun, 2013).
The low protein concentration attainable with PICK1 severely limited the scattering intensity (Figure 1C). For this reason, we also studied a maltose-binding protein (MBP)–PICK1 fusion (Figures 1A). Several linker sequences between MBP and PICK1 were tested, including long, flexible linkers, but the best results in terms of solubility and stability were obtained with a rigid three-alanine linker. In this way, MBP connects directly onto the predicted N-terminal helix (H1) of PICK1 (Figure 1A), facilitating the fitting of the PDZ domain into the SAXS envelope (see next section). The data collected for MBP-PICK1 showed a linear dependence of the scattering intensity on protein concentration up to 7.5 mg/ml (Figure 1D). Yet careful scrutiny of the Guinier plot led us to choose the data collected at 3.75 mg/ml for analysis, since signs of nonlinearity (i.e., aggregation) began to appear above this concentration.
The scattering data were used to calculate the pair–distance distribution function, P(r), of PICK1 and MBP-PICK1 (Figure 1F). The P(r) function provides a measure of the mass distribution within the sample (Putnam et al., 2007). Both curves were skewed toward large r-values, indicative of elongated particles. The forward scattering, I(0), and radius of gyration, Rg, were calculated from both the Guinier plot and the P(r) function (Table 1), with the values derived from the P(r) function being considered more accurate (Blanchet and Svergun, 2013). Both PICK1 and MBP-PICK1 had large Rg values (∼59.5 and ∼86.4 Å, respectively), again indicative of elongated particles. Comparison of the Rg values of PICK1 and MBP-PICK1 further suggested that MBP adds to the overall dimensions of the molecule, and thus it is likely positioned at the ends of the antiparallel PICK1 dimer. Consistently the maximum dimension, Dmax, of PICK1 and MBP-PICK1 determined from the P(r) function was 171 and 266 Å, respectively (Figure 1F and Table 1).
TABLE 1:
Data collection and scattering-derived parameters.
| PICK1 | MBP-PICK1 | |
|---|---|---|
| Data collection | ||
| Beam line | CHESS F2 | CHESS G1 |
| Wavelength (Å) | 1.254 | 1.257 |
| q range (Å−1) | 0.0129–0.15 | 0.0066–0.2 |
| Exposure time (s) | 30 | 40 |
| Sample concentration (mg/ml) | 0.24–1.92 | 0.46–7.5 |
| Temperature (K) | 277 | 277 |
| Structural analysis | ||
| Sample concentration (mg/ml) | 0.96 | 3.75 |
| I(0) (cm−1) (from Guinier plot) | 6.94 ± 0.16 | 41.42 ± 0.68 |
| Rg (Å) (from Guinier plot) | 62.2 ± 1.39 | 83.83 ± 1.74 |
| I(0) (cm−1) [from P(r)] | 6.32 ± 0.03 | 41.38 ± 0.51 |
| Rg (Å) [from P(r)] | 59.5 ± 0.14 | 86.36 ± 1.02 |
| Dmax (Å) [from P(r)] | 171 | 266 |
| Molecular mass determination | ||
| Partial specific volume (cm3/g) | 0.746 | |
| Contrast (Δρ × 1010 cm−2) | 2.7 | |
| Molecular mass [from I(0)] (kDa) | 172.2 | |
| Theoretical mass of dimer (kDa) | 173.6 | |
| Software used | ||
| Primary data reduction | Raw | Raw |
| Data processing | Raw | Raw |
| Ab initio analysis | DAMMIF | |
| Averaging and validation | DAMAVER |
Modeling of MBP-PICK1 into the ab initio SAXS envelope
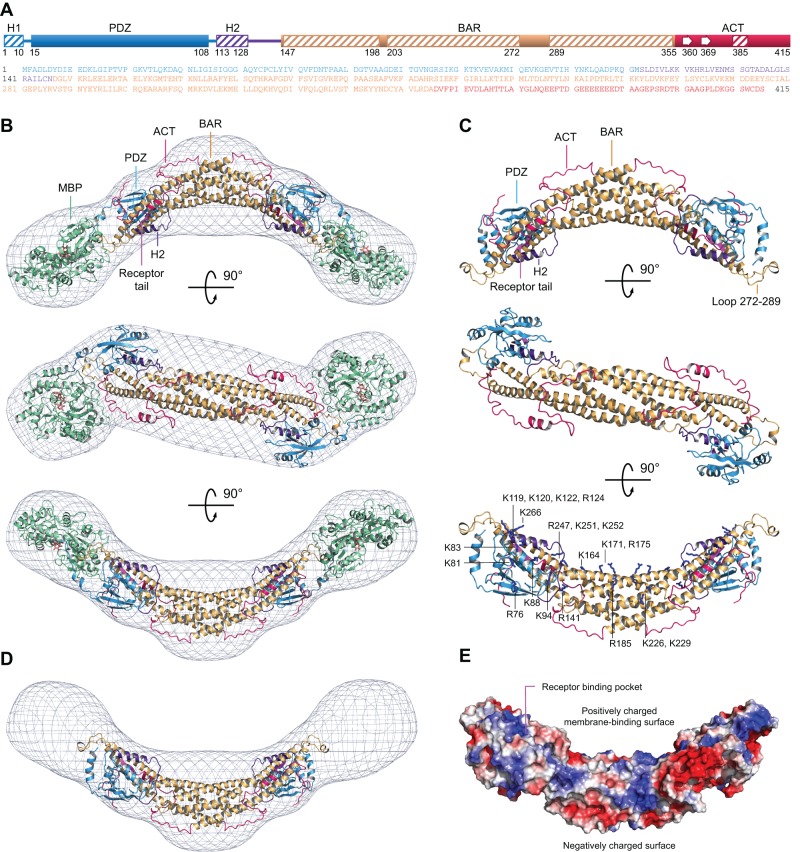
Using the higher-intensity data collected from MBP-PICK1, we calculated a low-resolution molecular envelope by averaging 20 ab initio structures. A model of MBP-PICK1 was built into the envelope (Figure 2). The atomic coordinates of MBP were from a structure determined in our laboratory (Protein Data Bank [PDB] code: 4EDQ), and those of PICK1's PDZ domain were from a recently determined structure of a complex with a GluA2 tail peptide (PDB code: 3HPK). A model of the BAR domain was built with the program SWISS-MODEL (Biasini et al., 2014), using as reference the structure of human arfaptin-2 (PDB code: 1I49), which, with 24% sequence identity, is the most closely related to PICK1 among BAR domain structures. This BAR domain consists of an antiparallel dimer of three-helix bundles, with an overall banana-like conformation.
FIGURE 2:
Atomic model of PICK1 derived from SAXS. (A) Domain diagram and sequence of PICK1 (color coded). (B) Three perpendicular views of the average SAXS envelope fit with an atomic model of the MBP-PICK1 dimer (modeling details in text). PICK1 domains are color coded as in A, and MBP is shown in green. (C) PICK1 portion of the model extracted from the images shown in B. The third orientation shows the side chains of positively charged amino acids predicted to participate in membrane binding. (D) Same as B, but subtracting the MBP portion of the model to show the contribution of PICK1 to the overall envelope. (E) Electrostatic surface representation of the PICK1 model (red, negatively charged; blue, positively charged). See also Supplemental Movies S1–S3.
The BAR domain was symmetrically positioned in the center of the envelope such that its curvature matched that of the envelope. Two MBP molecules were symmetrically fitted at the ends of the envelope, as suggested by comparison of the Rg and Dmax values of PICK1 and MBP-PICK1 (Table 1). Note that, with the BAR domain in the middle, no other volume in the envelope could fit MBP (Figure 2D), whose size (371 aa) is only marginally smaller than that of PICK1. MBP fits the envelope well (Supplemental Movie S1), and the best orientation found was one in which the C-terminus pointed toward the only remaining volume in the envelope, an area where the envelope displays a hump, which was assigned to the PDZ domain. The location of the PDZ domain in the envelope was further constrained by its position in the sequence, between MBP and the BAR domain.
Whereas the position of the PDZ domain appeared well constrained, its orientation was not. Owing to the globular shape of the PDZ domain, the receptor-binding pocket could be oriented toward either the concave or the convex side of the PICK1 dimer. We used published evidence to select the most likely orientation. The binding of the BAR domain to membranes and the PDZ domain to receptor tails can occur simultaneously through a “coincidence detection” mechanism (Madsen et al., 2008). Moreover, positively charged amino acids and a Cys-Pro-Cys motif near the receptor-binding pocket of the PDZ domain have been shown to contribute along with the BAR domain to membrane binding (Pan et al., 2007). Therefore we favor an orientation of the PDZ domain in which the receptor-binding pocket and the positively charged, membrane-binding surfaces of the PDZ and BAR domains are oriented approximately in the same direction (Figure 2, C and E).
A notable difference between PICK1 and arfaptin-2 is the presence of a 12-aa insertion in PICK1 near the junction between the second and third helices of the BAR domain (approximately corresponding to residues 272–289). This insertion could project either toward the plasma membrane, somewhat analogous to the N-terminal helix of the N-BAR domain (Gallop et al., 2006), or in the opposite direction, away from the plasma membrane and toward the PDZ domain. Because the envelope did not show any protrusions in this area, we directed the PICK1-specific insertion toward the PDZ domain (Figure 2C), but the exact role of this insertion has yet to be established.
Two additional regions remained to be modeled: the linker between the PDZ and BAR domains and the ACT. Residues 113–128 within the linker region are predicted to form an α-helix (H2). H2 contains a conserved stretch of positively charged amino acids (119KKVKHR124; Supplemental Figure S1). Remarkably, after building and connecting helix H2 C-terminally to the PDZ domain, the stretch of positively charged amino acids becomes part of the predicted membrane-binding surface on the concave side of the PICK1 dimer (Figure 2, C and E, and Supplemental Movies S2 and S3). The region between H2 and the BAR domain was built as a loop, and although its actual conformation is not resolved here, it appears likely that this loop sits at the interface between the PDZ and BAR domains.
The ACT is less well conserved than the rest of the PICK1 sequence, particularly from residue 373 onward (Supplemental Figure S1). It emerges from the C-terminus of the last helix of the BAR domain, at the top of the convex side of the dimer, coinciding with another hump in the SAXS envelope (Figure 2C). Because the C-terminal helices of the BAR domain cross each other, the ACT of one molecule in the dimer is predicted to interact almost exclusively with the other molecule. The ACT consists of 60 amino acids, and secondary structure prediction suggests that it is mostly unstructured, possibly featuring a short α-helix and two β-strands (Figure 2A). Thus the ACT likely extends along the surface of the BAR and PDZ domains. Cellular experiments described later appear to support this prediction. The final model was subjected to energy regularization with the program Phenix (Adams et al., 2010).
Cross-validation of the model against the scattering data shows that 1) the theoretical scattering curve calculated from the model fits very well the experimental data, as characterized by a discrepancy value, χ2, of 1.01 (Figure 1D), 2) the model also fits well the ab initio envelope, both visually (Supplemental Movie S1) and quantitatively, as reflected by a normalized spatial discrepancy (NSD) value of 1.44, and 3) the Rg values derived from the model for full-length PICK1 (∼57.0 Å) and MBP-PICK1 (∼81.0 Å) are in good agreement with the values obtained experimentally (Table 1). The model was then subjected to a series of tests by evaluating alternative models directly against the scattering data. Thus replacing the BAR domain with that of APPL1, a related BAR domain protein (Supplemental Figure S2), worsens the fit to the experimental scattering data, but only slightly (χ2 = 1.61). Inverting the orientation of the PDZ domain by 180° also has a relatively minor effect on the fit (χ2 = 1.5). In contrast, placing the ACT or PDZ domain outside the envelope—that is, assuming that they are separated from the BAR domain—dramatically worsens the fit to the scattering data (χ2 = 2.33 and 4.29, respectively). It thus appears that the fit to the scattering data is mostly insensitive to changes occurring within the confines of the envelope but is strongly affected by the removal of protein mass from the envelope, suggesting that the various domains of PICK1 are packed against one another. This conclusion is supported by previous evidence showing that the PDZ and BAR domains interact with each other (Lu and Ziff, 2005). Moreover, the location of the PDZ domain in the model shows striking resemblance with the location of the PH domain in the BAR-PH structure of APPL1 (Zhu et al., 2007) and the PX domain in the PX-BAR structure of sorting nexin 9 (Pylypenko et al., 2007; Supplemental Figure S2).
Testing the model—the ACT is involved in autoinhibition and vesicle motility
One important prediction of the model is that the ACT may interact with the PDZ and BAR domains, which could be the source of autoinhibition. Indeed, association of PICK1 with intracellular vesicle-like structures is observed either when the PDZ domain binds to a ligand at the membrane or when the PDZ domain is removed (Perez et al., 2001; Lu and Ziff, 2005; Madsen et al., 2008). One potential interpretation of these results is that in the inactive conformation, the PDZ domain occupies a position that interferes with membrane binding. However, it has been also suggested that the PDZ domain participates in membrane interactions (Jin et al., 2006; Pan et al., 2007), along with the BAR domain (Jin et al., 2006; Steinberg et al., 2006; Madsen et al., 2008). We thus speculated that the inhibition of membrane binding depended not on the presence of the PDZ domain or its relative position with respect to the BAR domain, but rather on the ACT, which appears to be more flexible, that is, likely to move, and has an overall negative charge that could oppose membrane binding. To test this hypothesis, we analyzed the cellular phenotype of four PICK1 constructs expressed as green fluorescent protein (GFP)–fusion proteins in HeLa (Figure 3) and B16F1 cells (see below). In contrast to most previous studies, however, we used live-cell imaging to also assess the motility of PICK1-associated organelles, leading to important new findings.
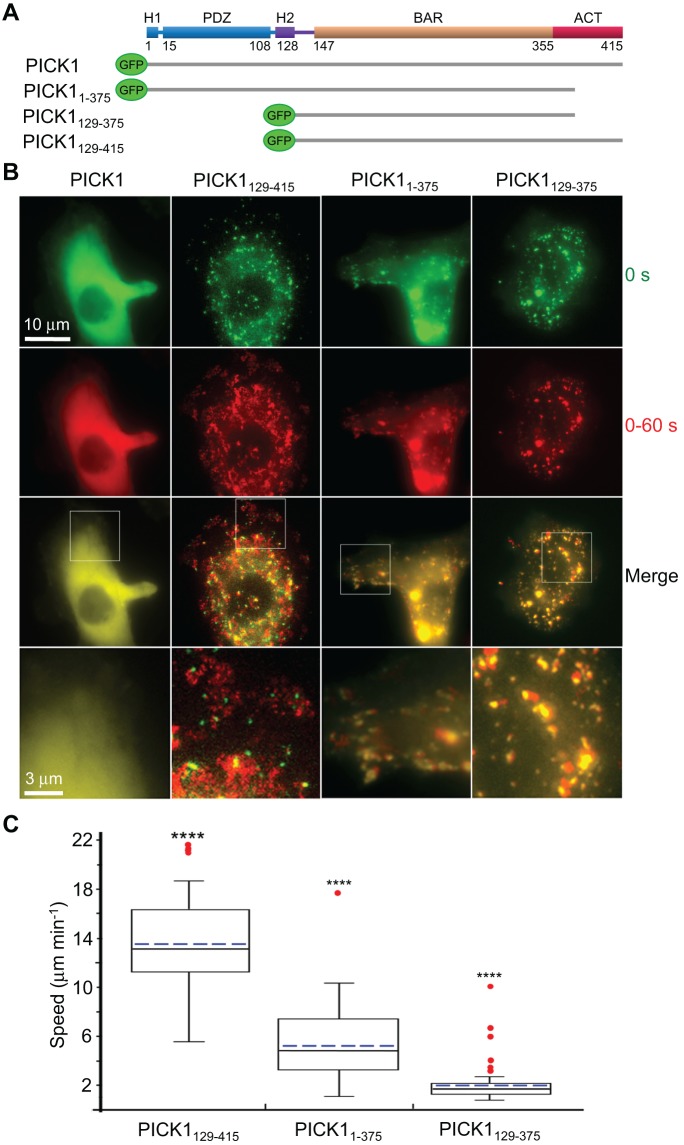
FIGURE 3:
PICK1 plays a role in intracellular organelle motility. (A) Schematic representation of the GFP-fusion constructs expressed in cells. (B) Time-lapse fluorescence microscopy analysis of organelle motility in HeLa cells expressing the indicated PICK1 constructs. First row, first frame of time-lapse sequence. Second row, maximum projection of frames 1–20 taken at 3-s intervals, showing all locations visited by the organelles during 60 s. Third row, overlay of the first two rows (see Supplemental Movie S4). Fourth row, zooms of the regions boxed in the third row. The amount of red signal (maximum projections) relative to green signal (original organelle positions) reflects the extent of organelle motility. (C) Quantification of the speeds of vesicles associated with PICK1 constructs. Data are shown as box-and-whisker plots: the boxes comprise the middle half and the whiskers the 10th–90th percentile of the data, and solid black and dashed blue lines represent median and mean values, respectively. Data outliers are indicated by red dots. All the data sets are statistically different from each other (p < 0.0001, n = 31–49 tracks from three or four cells).
As reported before (Perez et al., 2001; Lu and Ziff, 2005; Madsen et al., 2008), full-length PICK1 was uniformly distributed in the cytoplasm, whereas PICK1129-415, lacking the H1-PDZ-H2 module, localized to vesicle-like structures (Figure 3B). Next we analyzed the behavior of PICK11-375, lacking the most variable C-terminal 40-aa region of the ACT, which includes the stretch of acidic amino acids (Supplemental Figure S1). Consistent with the proposed inhibitory role of the ACT, PICK11-375 associated with punctate vesicle-like structures in cells. A similar localization was observed with PICK1129-375, lacking most of the ACT and H1-PDZ-H2.
More surprising, however, was the observation that the vesicles associated with PICK1129-415 exhibited fast nondirectional motility, with an average velocity of ∼14 μm/min (Figure 3 and Supplemental Movie S4). Deletion of the last 40 aa of the ACT dramatically reduced vesicle speed, and when both this region and H1-PDZ-H2 were deleted, the vesicles became almost static (Figure 3 and Supplemental Movie S4). The fast motility of vesicles associated with PICK1129-415 was also observed in B16F1 cells (see below), suggesting that this phenotype is not cell type specific.
These results suggested that either the ACT is directly responsible for the fast motility of PICK1-associated organelles or the PICK1 constructs localized to different organelles characterized by different intrinsic motilities. To distinguish between these two possibilities, we tested colocalization of the PICK1 constructs with several organelle markers in HeLa cells (see Materials and Methods). The Gether laboratory reported that PICK1 colocalizes with vesicles budding from the trans-Golgi apparatus (Holst et al., 2013). Consistently we found that all the PICK1 constructs, independent of their motility, colocalized and comigrated with the trans-Golgi network marker TGN38 (Supplemental Figure S3 and Supplemental Movie S5). Our results further suggested that this localization is driven by the BAR domain, since this was the smallest fragment showing such localization. We also note that whereas the PICK1 constructs generally displayed a punctate pattern, tubular structures were frequently observed with all three constructs.
PICK1 binds F-actin in vitro but does not colocalize with F-actin in cells
The finding that the ACT plays a role in the motility of PICK1-associated vesicles made us question its previously suggested role in interaction with Arp2/3 complex and inhibition of actin nucleation (Rocca et al., 2008). The same study also concluded that the BAR domain interacted with F-actin. We decided to test these observations here.
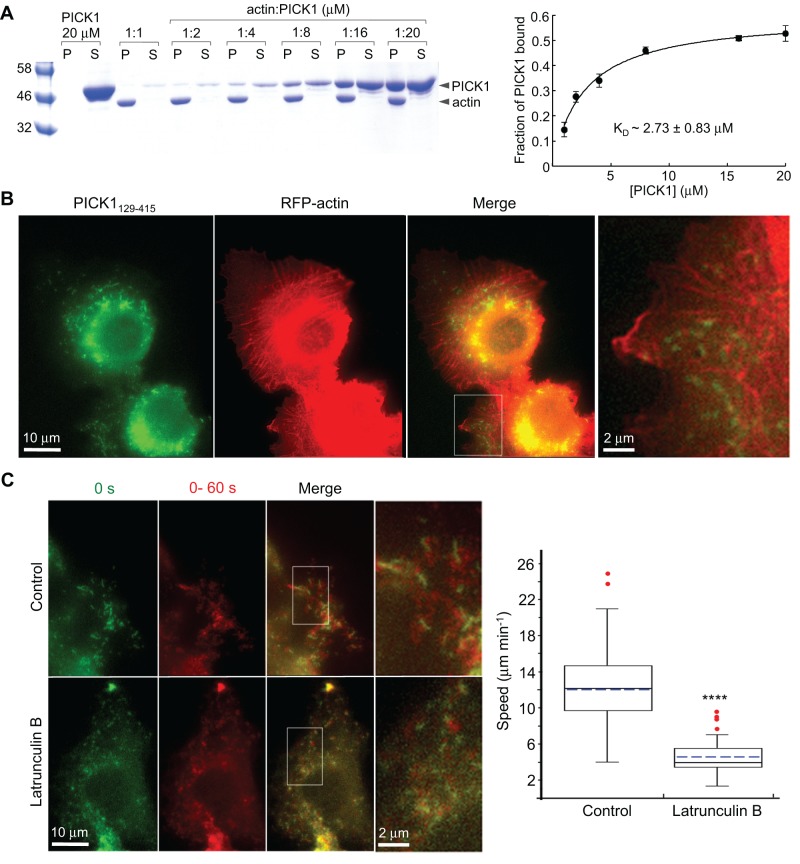
Binding of PICK1 to F-actin was confirmed using high-speed F-actin cosedimentation experiments in which the concentration of either PICK1 or F-actin was varied (Figure 4A and Supplemental Figure S4). Global fitting of three such experiments resulted in KD estimates of ∼2.0–3.0 μM for this interaction, which is somewhat lower than the value reported previously (∼0.3 μM; Rocca et al., 2008).
FIGURE 4:
PICK1 binds F-actin in vitro but not in cells. (A) SDS–PAGE analysis of the pellet (P) and supernatant (S) fractions of high-speed sedimentations performed at a fixed F-actin concentration (1 μM) and various PICK1 concentrations (1–20 μM). The graph on the right shows the fraction of F-actin–bound PICK1 as a function of PICK1 concentration, determined from densitometric analysis of the gels. The solid line represents the global fit of the data from three independent experiments. (B) Localization of GFP-PICK1129-415 (green) and RFP-actin (red) in double-transfected HeLa cells. The boxed region is enlarged on the right. Note the lack of colocalization of F-actin–rich structures with PICK1-associated vesicles. Occasional overlaps of the two markers do not persist over time (see Supplemental Movie S6). (C) Latrunculin B inhibits the motility of PICK1129-415-associated structures in B16F1 cells, shown before (control) or after treatment with 2 μM latrunculin B. First column, first frame of time-lapse sequence. Second column, maximum projection of frames 1–15 taken at 4-s intervals. Third column, overlay of the first two columns (see Supplemental Movie S7). Fourth column, zooms of the regions boxed in the third column. The plot on the right shows the quantification of vesicle speeds as described in Figure 3C. The difference between control and latrunculin B–treated cells is statistically significant (p < 0.0001, n = 56 tracks from five control cells; n = 88 tracks from five latrunculin B–treated cells).
We (Lee et al., 2007) and others (Yamagishi et al., 2004; Bompard et al., 2005; Millard et al., 2005; Mattila et al., 2007) have reported that certain BAR domains, which have a positively charged membrane-binding surface, can cosediment with F-actin, whose surface displays a periodic distribution of negative charges that is also curved because of the helical twist of the filament. A recent cryo–electron microscopy study of the F-BAR domain protein pacsin2 bound to F-actin reveals the nature of such interactions (Kostan et al., 2014). However, a functional link to such in vitro interactions has not been established; these BAR domains do not colocalize with F-actin in cells, and the interactions are believed to depend on nonspecific electrostatic contacts (Mattila et al., 2007). Consistently we found that the vesicles associated with PICK1129-415 did not colocalize with F-actin–rich structures in cells (Figure 4B), and although PICK1-positive puncta occasionally overlapped with F-actin-rich structures, time-lapse sequences showed no coordinated movement of the two markers (Supplemental Movie S6). Yet the motility of PICK1129-415-associated vesicles required a properly assembled actin cytoskeleton, since treatment with latrunculin B, a drug that inhibits actin polymerization, resulted in a dramatic reduction in organelle motility in B16F1 cells (Figure 4C and Supplemental Movie S7). Together these results suggest that PICK1 does not bind directly to F-actin–rich structures in cells, such as stress fibers and actin bundles, but may use F-actin indirectly for the movement of PICK1-associated vesicles.
PICK1 neither binds nor inhibits Arp2/3 complex
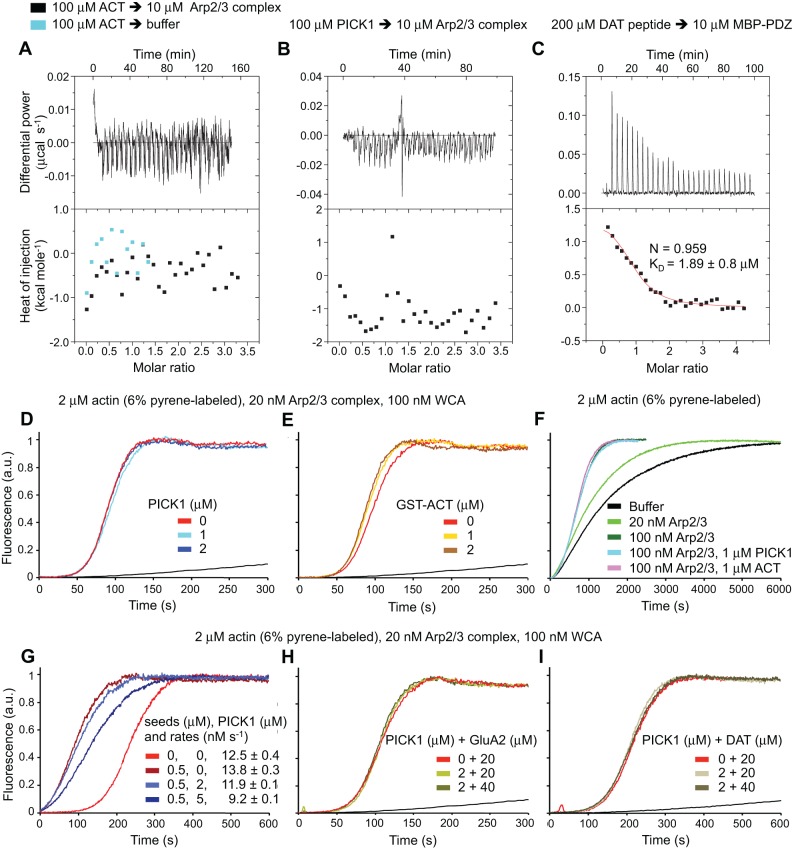
The ACT, and specifically Trp412, was implicated in the interaction of PICK1 with Arp2/3 complex (Rocca et al., 2008). To probe this interaction, we used isothermal titration calorimetry (ITC), a method we have used to analyze other interactions of Arp2/3 complex (Boczkowska et al., 2013, 2014). We tested binding of both PICK1 and the isolated ACT, which is presumably free of autoinhibitory interactions that could weaken binding to Arp2/3 complex. However, neither PICK1 nor the isolated ACT appeared to bind Arp2/3 complex (Figure 5, A and B). In contrast, binding of a C-terminal DAT peptide to the PDZ domain was confirmed by ITC (Figure 5C), with affinity similar to that obtained by fluorescence polarization (Erlendsson et al., 2014).
FIGURE 5:
PICK1 neither binds nor inhibits Arp2/3 complex. (A–C) ITC titrations of ACT (A), PICK1 (B), and a DAT peptide (C) into Arp2/3 complex or MBP-PDZ at the indicated protein concentrations. Control experiments were performed by titrating these samples into buffer (cyan data points in A). Each 10-μl injection lasted 10 s, with an interval of 300 s between injections. The lack of change in the heats of injection precluded fitting of the experiments shown in A and B to a binding isotherm. For the titration of the DAT peptide, the best fit of the data (solid red line) corresponds to a one-site binding isotherm with KD ∼1.9 μM. (D–I) Time courses of the fluorescence increase upon polymerization of 2 μM actin (6% pyrene-labeled) alone (black line) and with addition of the indicated proteins at the indicated protein concentrations. Experimental conditions are given with each experiment, and curves are color coded. For the experiments shown in G, 0.5 μM F-actin seeds were preincubated with PICK1 to test the effect of PICK1 binding to F-actin on the lag time (measured as the time to 10% polymerization) of Arp2/3 complex polymerization. Also shown are the polymerization rates at 50% polymerization. Each measurement was performed three times; one representative curve is shown. Errors are reported as SE.
Despite the lack of interaction by ITC, we conducted pyrene-actin polymerization assays to check for any effect of PICK1 on Arp2/3 complex–induced polymerization. Compared to control experiments with actin alone, polymerization was strongly stimulated in the presence of 20 nM Arp2/3 complex and 100 nM N-WASP WCA. However, the addition of increasing concentrations (0–2 μM) of PICK1 (Figure 5D) or MBP-PICK1 (Supplemental Figure S5A) had no effect on this activity. The isolated ACT also failed to inhibit nucleation by Arp2/3 complex (Supplemental Figure S5B).
Because several studies showed that Arp2/3 complex contains two WCA-binding sites (Padrick et al., 2011; Ti et al., 2011; Boczkowska et al., 2014), and the ACT was proposed to compete with WCA for binding to Arp2/3 complex (Rocca et al., 2008), we tested the ability of glutathione S-transferase (GST)–dimerized ACT to inhibit Arp2/3 complex. Again, we observed no inhibition (Figure 5E).
Because WCA binds with nanomolar affinity to Arp2/3 complex (Boczkowska et al., 2014), PICK1 may not be able to compete with WCA if it interacts very weakly with the complex. We thus tested the ability of PICK1 and the ACT to inhibit the intrinsic polymerization activity of Arp2/3 complex, that is, the very low activity observed in the absence of nucleation-promoting factors. Whereas polymerization increased somewhat with Arp2/3 complex concentration, neither PICK1 nor the ACT had any detectable effect on this activity (Figure 5F).
Because actin filaments serve as a platform for branch formation by Arp2/3 complex, their presence decreases the lag time for polymerization (Figure 5G). PICK1, which binds to F-actin (Figure 4A), could in principle inhibit this effect by blocking access of Arp2/3 complex to the mother filament. Indeed, a recent EM reconstruction shows that the F-BAR domain of pacsin2 interacts electrostatically with F-actin, with its membrane-binding positive face interacting with negative patches along the long-pitch helix of F-actin (Kostan et al., 2014). The BAR domain of PICK1 might bind similarly to F-actin and would thus be expected to overlap with the binding site for Arp2/3 complex (Rouiller et al., 2008). However, we found that even saturating concentrations of PICK1 (5 μM; Figure 4A) relative to F-actin seeds (0.5 μM) had only a minor effect on the lag phase and polymerization rate of 20 nM Arp2/3 complex (Figure 5G), suggesting that PICK1 binds nonspecifically to F-actin and is readily displaced by the Arp2/3 complex.
The binding of a GluA2 C-terminal peptide to the PDZ domain was reported to enhance the inhibitory effect of PICK1 on polymerization induced by Arp2/3 complex (Rocca et al., 2008). We tested this result using two receptor tail peptides, GluA2 and DAT. Although we confirmed binding of the DAT peptide to the PDZ domain (Figure 5C), neither peptide had a measurable effect on actin polymerization (Figure 5, H and I).
Effect of Arp2/3-complex inhibition on the motility of PICK1-associated vesicles
Although PICK1 does not directly inhibit Arp2/3 complex, nor does it colocalize with F-actin-rich structures, the motility of PICK1-associated vesicles is notably reduced by treatment with latrunculin B (Figure 4C). To explore whether Arp2/3 complex is directly involved in this motility, we treated HeLa cells expressing PICK1129-415 with the Arp2/3 complex inhibitor CK-666 (Nolen et al., 2009). Compared to control cells treated with dimethyl sulfoxide (DMSO), only a minor effect on the motility of PICK1129-415-associated vesicles was observed 30 min after treatment with 200 μM CK-666, whereas ∼50% reduction in vesicle speed was observed 60–90 min after treatment (Figure 6 and Supplemental Movie S8). For comparison, actin comet tail–driven Listeria motility is fully inhibited 60 min after treatment with CK-666, whereas lamellar motility is less sensitive to Arp2/3 complex inhibition (Nolen et al., 2009). These results suggest that the motility of PICK1-associated vesicles is not driven by a comet tail mechanism and that the reduction in vesicle speed observed after extended CK-666 treatment is likely due to overall changes in the organization of the actin cytoskeleton.
FIGURE 6:
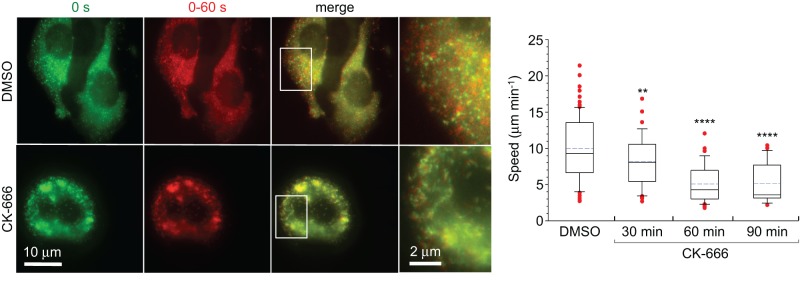
Effect of Arp2/3 complex inhibition on the motility of PICK1-associated vesicles. HeLa cells expressing GFP-PICK1129-415 treated with the Arp2/3 complex inhibitor CK-666 (200 μM) or DMSO (control). First column, first frame of time-lapse sequence. Second column, maximum projection of frames 1–20 taken at 3-s intervals. Third column, overlay of the first two columns (see Supplemental Movie S8). Fourth column, zooms of the regions boxed in the third column. The plot on the right shows the quantification of vesicle speeds 30, 60, and 90 min after treatment with CK-666 or 60 min after treatment with DMSO. The differences between control and CK-666-treated cells are statistically significant (p < 0.02 or p < 0.0001, n = 42–85 tracks from six to nine cells).
DISCUSSION
Predictions of the PICK1 model
The atomic model obtained here allows us to make several predictions about the structure–function relationship of PICK1: 1) the PDZ domain and surrounding sequences are tightly associated with the BAR domain, 2) the long loop between the second and third helices of the BAR domain likely plays a critical role by either interacting with the PDZ domain or projecting into the membrane, 3) the ACT of one subunit in the dimer likely interacts with the BAR and PDZ domains of the other subunit, 4) the ACT, which is predictably the most flexible region of the structure, could be directly responsible for inhibition of membrane binding by full-length PICK1, 5) the PDZ domain, H2, and the BAR domain form together a contiguous concave surface for membrane interaction, and 6) several positively charged amino acids on this surface are predicted to participate in membrane binding (Figure 2C). Some of these amino acids (R76, K81, K251, K252, and K266) have been mutated and shown to indeed participate in membrane interactions (Jin et al., 2006; Steinberg et al., 2006; Pan et al., 2007; Madsen et al., 2008). An additional five amino acids fall within the H2-linker region between the PDZ and BAR domains, which deletion mutants (Δ113–121 and Δ125–135) suggest is also implicated in membrane binding (Madsen et al., 2008), although the model also indicates that such deletions could have unintended global structural effects.
Our interpretation of the structural model is that most of the PICK1 structure, except for the ACT, forms a rather rigid assembly, with all the individual elements within this region being interconnected by tight interactions. This interpretation is supported by PX-BAR (Pylypenko et al., 2007) and BAR-PH (Zhu et al., 2007) structures, in which a second membrane-binding module associates tightly with the BAR domain to form a contiguous membrane-binding surface through a coincidence detection mechanism (Moravcevic et al., 2012). According to this model, it is the ACT, with its overall negative charge, that serves as a flexible on–off switch for membrane binding. Supporting this view is our finding that clustering of PICK1 on vesicle-like structures, which was previously shown to result from removal of the PDZ domain (Perez et al., 2001; Lu and Ziff, 2005; Madsen et al., 2008), can also be achieved through deletion of the ACT. The proposed inhibitory role of the ACT is in agreement with previous reports that cosedimentation of PICK1 with brain lipids increases upon removal of the ACT (Jin et al., 2006) and deletion of the acidic stretch 380–390 within the ACT enhances synaptic targeting of PICK1 (Boudin and Craig, 2001). The fact that all of the PICK1 constructs, including PICK1129-375, which lacks H1-PDZ-H2 and most of the ACT, colocalize with the trans-Golgi marker TGN38 further suggests that the BAR domain is primarily responsible for specific membrane binding. It is likely, however, that the presence of a binding partner of the PDZ domain on a different subcellular organelle could change this localization.
Role of PICK1 in synaptic receptor endocytosis
Clathrin-mediated endocytosis is the major pathway by which proteins are internalized in synapses (Saheki and De Camilli, 2012), and it is also the mechanism used for AMPAR internalization (Man et al., 2000; Wang and Linden, 2000; Lee et al., 2002; Kastning et al., 2007). BAR domain proteins, including amphiphysin, syndapin, nervous wreck, and CIP4, are central players in this process by generating or sensing membrane curvature while often linking to cytoskeletal tracks and molecular motors for vesicle movement (Saheki and De Camilli, 2012). Similarly, the BAR domain of PICK1 binds phosphoinositide-containing membranes and is believed to help stabilize the curvature of the synaptic plasma membrane during AMPAR endocytosis (Lu and Ziff, 2005; Jin et al., 2006; Steinberg et al., 2006; Hanley, 2008; Henley and Wilkinson, 2013).
Although the precise mechanism by which PICK1 regulates receptor trafficking is still under investigation, several features are now understood (Figure 7). The interaction with receptor tails puts PICK1 in close proximity to the plasma membrane and induces a conformational change that exposes its membrane-binding capacity (Madsen et al., 2008). Our results indicate that this conformational change consists of a movement of the ACT away from the positively charged, membrane-binding surface formed by the PDZ (Jin et al., 2006; Pan et al., 2007) and BAR (Jin et al., 2006; Steinberg et al., 2006; Madsen et al., 2008) domains.
FIGURE 7:
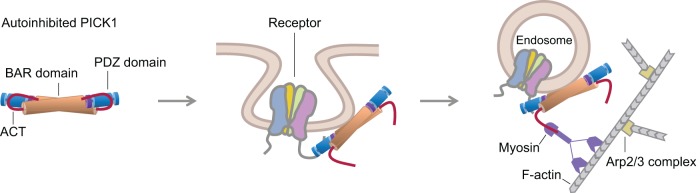
Model of PICK1 function in receptor endocytosis and trafficking. Published evidence and data presented here support a model for PICK1 involvement in receptor endocytosis. PICK1 exists in an autoinhibited state in which its membrane-binding surface is masked by interactions of the ACT with the PDZ and BAR domains. Binding of the PDZ domain to receptor tails at the membrane triggers a conformational change that exposes the membrane-binding capacity of PICK1 (Madsen et al., 2008). We propose that this conformational change consists of a movement of the ACT away from the contiguous membrane-binding surface formed by the BAR and PDZ domains. Analogous to other BAR domain proteins (Saheki and De Camilli, 2012), PICK1 may participate in the endocytic pathway by generating or sensing membrane curvature while also linking to the cytoskeleton for vesicle movement. Consistently our results suggest that the conformational change in PICK1 frees the ACT for interaction (direct or indirect) with a motility factor to propel vesicle movement. We propose that this motility factor is myosin, which is consistent with the fact that myosins are the major drivers of synaptic transport (Kneussel and Wagner, 2013).
The role of the ACT had remained obscure. It was reported that PICK1 inhibits actin filament nucleation by Arp2/3 complex through interactions of the BAR domain with F-actin and the ACT with Arp2/3 complex (Rocca et al., 2008; Nakamura et al., 2011). In subsequent studies, these authors linked this mechanism to the control of dendritic spine size, astrocyte morphology and synaptic plasticity (Nakamura et al., 2011; Murk et al., 2013; Rocca et al., 2013). According to their model, inhibition of Arp2/3 complex is required to reduce membrane tension resulting from actin polymerization, which opposes invagination and formation of the clathrin-coated pit. However, these conclusions contradict overall ideas about the role of actin polymerization in endocytosis. Actin assembly, and particularly actin nucleation and branching by Arp2/3 complex, plays essential roles in yeast and mammalian endocytosis (Saheki and De Camilli, 2012; Weinberg and Drubin, 2012). The recruitment of actin assembly factors such as the Arp2/3 complex regulator cortactin to sites of endocytosis follows a similar time course as invagination (Merrifield et al., 2005), and actin network assembly coincides precisely with the onset of invagination (Kukulski et al., 2012). Inhibition of Arp2/3 complex delays endosome formation, whereas a mutation in Arp2/3 complex that increases its intrinsic nucleation activity accelerates endosome maturation and scission (Boettner et al., 2009). Moreover, rather than opposing invagination, actin polymerization is believed to favor invagination by countering turgor pressure in yeast cells (Aghamohammadzadeh and Ayscough, 2009) or tension from osmotic swelling or mechanical stretching in mammalian cells (Boulant et al., 2011). Consistently a recent study established that increased membrane tension inhibits endocytosis in mitotic cells due to an unmet requirement for actin polymerization (Kaur et al., 2014).
Contrary to the suggested role of PICK1 in inhibition of Arp2/3 complex, we found here that PICK1 neither interacts with nor inhibits Arp2/3 complex. The proposed competition between the ACT and the acidic region of N-WASP did not materialize, which is understandable, since the two sequences are rather different; the conserved tryptophan of WASP-family proteins is embedded within the acidic patch, whereas Trp412 of PICK1 is not (Supplemental Figure S1). Moreover, whereas we confirmed the interaction of PICK1 with F-actin in vitro, PICK1 and F-actin did not colocalize in cells, suggesting that this interaction is nonspecific and likely irrelevant for PICK1's function. This is important because the interactions with F-actin and membranes involve the same positively charged surface of the BAR domain (Rocca et al., 2008) and are therefore mutually exclusive.
Our results suggest a new role for the ACT. We showed here that PICK1-associated organelles undergo fast nondirectional movement, and this motility requires the ACT. Thus the ACT is not only involved in inhibition of membrane binding by PICK1, but upon activation, it also appears to connect PICK1-associated vesicles to a motility factor. Intracellular organelle motility is driven by three fundamentally different mechanisms: microtubule-based motors, actin-based motors, or actin comet tails. Because the motility of PICK1-associated vesicles consists mostly of locally confined random trajectories (Supplemental Movie S4), microtubule-based motors that transport cargoes directionally over long distances probably contribute little, if at all, to this motility. Importantly, treatment with latrunculin B dramatically reduces the speed of PICK1-associated vesicles, suggesting that the actin cytoskeleton is involved. However, a comet tail mechanism is unlikely because PICK1-associated vesicles did not seem to colocalize with F-actin–rich structures corresponding to comet tails (Supplemental Movie S6), and, unlike Listeria, their motility was not fully inhibited by treatment with the Arp2/3 complex inhibitor CK-666 (Supplemental Movie S8). These considerations point to actin-based myosin motors as the most likely source of this motility (Figure 7), which is consistent with the fact that unconventional myosins are the major drivers of synaptic transport (Kneussel and Wagner, 2013). Testing this hypothesis and identifying the specific motors involved and the nature of the connection (direct or indirect) between PICK1 and these motors will be the subject of future studies.
MATERIALS AND METHODS
Proteins
The cDNA encoding for human PICK1 (UniProt code: Q9NRD5) was synthesized (GENEWIZ, South Plainfield, NJ) and cloned as an MBP-fusion construct between the BamHI and NotI sites of vector pRSFDuet-1 (Novagen, Madison, WI), which has a hexahistidine affinity purification tag. A TEV protease cleavage site was introduced between MBP and PICK1 for separation of full-length PICK1 after affinity purification.
For expression of the fusion protein MBP-PICK1, the PICK1 gene was cloned between the NotI and BamHI sites of a modified vector pMAL-c2X (NEB, Ipswich, MA) that carries MBP. The first modification of the vector was to introduce a hexahistidine affinity purification tag followed by a TEV protease cleavage site N-terminal to MBP. Second, to facilitate structural analysis, several surface entropy–reducing mutations (Moon et al., 2010) were introduced within the MBP moiety of the vector (DK82AA, EN172AA, K293A, E359A, KD362AA). Finally, a three-alanine insert, comprising a NotI cloning site, was introduced after MBP residue Asn-367. In this way, cloning of PICK1 at this site resulted in the incorporation of a three-alanine linker between MBP residue Asn-367 and Met-1 of PICK1. The three-alanine linker has been shown to extend the C-terminal helix of MBP (Moon et al., 2010), resulting in a rather rigid connection between MBP and PICK1 helix H1 (Figure 1A). Construct MBP-PDZ was cloned in an analogous manner. GST-ACT was cloned between the BamHI and NotI sites of vector pGEX-4T-1 (GE Healthcare, Little Chalfont, United Kingdom) to express a dimeric protein consisting of GST, a six-residue linker that includes a thrombin cleavage site, and the C-terminal 63 amino acids of PICK1.
All the proteins were expressed in BL21(DE3) cells (Invitrogen, Life Technologies, Carlsbad, CA) grown in Terrific Broth medium at 37°C until the OD600 reached a value of 1.5–2.0. Expression was induced with addition of 1 mM isopropylthio-β-d-galactoside and was carried out for 16 h at 18°C. Cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 4.0 mM benzamidine HCl, 10 mM imidazole, 1.0 mM tris(2-carboxyethyl)phosphine (TCEP), and 1 mM phenylmethylsulfonyl fluoride, and lysed using a microfluidizer apparatus (Microfluidics, Westwood, MA). All the proteins were first purified through a nickel-nitriloacetic acid (Ni-NTA) affinity column, using 30 and 250 mM imidazole during washing and elution, respectively. GST-ACT was purified on a GST affinity column. All the proteins were additionally purified through a Superdex 200 HL 26/600 gel filtration column (GE Healthcare) in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.5, 300 mM NaCl, and 2 mM dithiothreitol (DTT). The affinity histidine tag was removed by cleavage with TEV protease. In the case of GST-ACT, the tag was either kept to produce a dimeric protein or removed by cleavage with thrombin. After cleavage, the untagged proteins were separated by passage through Ni-NTA or GST affinity columns. Finally, proteins were purified through an ion exchange MonoQ 10/100 GL column (GE Healthcare) in 20 mM HEPES, pH 7.5, 2 mM DTT, and 300 mM NaCl. Under these conditions, only the contaminant proteins remain bound in the column, whereas the proteins of interest are recovered in the flowthrough. The exact boundaries of PICK1 constructs and qualitative analysis of the proteins by SDS–PAGE are shown in Figure 1, A and B, respectively.
Arp2/3 complex, actin, and the WCA of N-WASP were prepared as described (Boczkowska et al., 2014). Briefly, frozen brains were homogenized in Arp buffer (20 mM HEPES, pH 7.5, 100 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid [EGTA], and 1 mM DTT) supplemented with protease inhibitors and clarified by centrifugation at 12,000 × g for 30 min. The supernatant was loaded onto a Macro-Prep High Q column (Bio-Rad, Hercules, CA) preequilibrated with Arp buffer. The flowthrough, containing Arp2/3 complex, was purified through a WCA affinity column equilibrated with Arp buffer. Arp2/3 complex was eluted in 20 mM Tris (pH 8.0), 25 mM KCl, 400 mM MgCl2, 1 mM EGTA, and 1 mM DTT, concentrated, and further purified through an SD200HL 26/600 column in Arp buffer. Actin was purified from rabbit skeletal muscle (Pardee and Spudich, 1982). Briefly, an F-actin pellet was homogenized in G-buffer (2 mM Tris, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, and 0.01% NaN3) with the addition of 10 mM DTT to reduce actin fully. After 1 h, actin was dialyzed exhaustively against G-buffer to remove DTT and then centrifuged for 45 min at 277,000 × g to pellet any F-actin that did not depolymerize, as well as any denatured actin. The cDNA encoding for mouse N-WASP (UniProt code: Q91YD9) was obtained from the American Type Culture Collection (Manassas, VA; clone 3169027). The WCA fragment was amplified by PCR and cloned between the NdeI and EcoRI sites of vector pTYB12 (NEB), comprising a chitin-binding domain for affinity purification and an intein domain for self-cleavage of the tag after purification. This produces a fully native sequence, without any tags or extra amino acids at the ends.
The 6-aa peptides corresponding to the C-terminal tails of GluA2 (IESVKI) and DAT (RHWLKV) were synthesized.
Small-angle x-ray scattering
Collection of x-ray scattering data from samples of PICK1 and MBP-PICK1 was carried out at CHESS beam lines F2 and G1 (Table 1). For PICK1, the data collection buffer was 20 mM Tris HCl, pH 8.0, 200 mM NaCl, 2 mM DTT, and 5% (vol/vol) glycerol. For MBP-PICK1, the buffer was 50 mM Tris HCl, pH 7.5, 300 mM NaCl, 1 mM maltose, 1 mM EGTA, and 2 mM DTT. Before data collection, all the samples were centrifuged at 278,000 × g for 30 min. A 30-μl volume of each sample at the indicated concentrations (Figure 1, C and D) was loaded into a capillary flow cell using a loading robot (Nielsen et al., 2012). Data collection was carried out at 4°C. For each sample, data were collected at four different concentrations through a series of 50% dilutions. To limit radiation damage, the samples were continuously oscillated inside the capillary flow cell during data collection. The scattering profile of each sample was determined as the average of 10 (PICK1) or 20 (MBP-PICK1) independent 2-s x-ray exposures subtracted from the average of the same number of buffer exposures. To monitor for potential radiation damage, each measurement was compared with the first measurement taken from that sample. Data normalization and solvent subtraction were carried out with the program BioXTAS RAW (Nielsen et al., 2009), and the program GNOM (Svergun, 1992) was used to calculate the distance distribution function, P(r), of each sample. At high concentrations, the PICK1 sample showed a nonlinear dependence of the scattering intensity on protein concentration, such that the highest protein concentration used in analyses was 0.96 mg/ml. For the MBP-PICK1 sample, the highest concentration used in analyses was 3.75 mg/ml, based on examination of the Guinier plot, which at this concentration was linear (Figure 1E), indicative of a pure, monodisperse species (Blanchet and Svergun, 2013).
The scattering intensity was weak for PICK1, so ab initio dummy atom models were calculated only for MBP-PICK1, using the program DAMMIF (Franke and Svergun, 2009). The twofold symmetry of the MBP-PICK1 dimer was used as a restraint in these calculations, which speeds up the calculations and reduces model bias (Blanchet and Svergun, 2013). The final model of MBP-PICK1 resulted from averaging 20 ab initio dummy atom models with the program DAMAVER (Volkov and Svergun, 2003). This model was converted into a volumetric map (mesh) using the pdb2vol tool of the program SITUS (Wriggers, 2010). An atomic model of MBP-PICK1 was built into the resulting envelope as describe in the text. The program SUPCOMB (Kozin and Svergun, 2001) was used to fit the atomic model into the envelope and calculate the NSD value. A theoretical scattering curve was calculated from the atomic model and compared with the experimental curve using the program CRYSOL (Svergun et al., 1995).
Data were analyzed and reported as recommended by the Small-Angle Scattering Task Force (Table 1; Trewhella et al., 2013).
Cell culture and image analysis
GFP-fusion constructs of several PICK1 fragments (FL, 1–375, 129–375, 129–415) were cloned between the BglII and SalI sites of vector pEGFP-C1 (Clontech Laboratories, Mountain View, CA). HeLa and B16F1 cells were cultured as described (Yang et al., 2007). Cells were transiently transfected with the GFP-PICK1 constructs and red fluorescent protein (RFP)–actin using Lipofectamine LTX with Plus Reagent (Life Technologies). The transfected cells were replated into 35-mm glass-bottomed dishes 16 h after transfection and imaged 20–32 h later in phenol red–free L-15 medium supplemented with 10% fetal bovine serum at 36°C. To test the role of actin assembly in the motility of PICK1-associated vesicles, 2 μM latrunculin B was added to the medium of B16F1 cells expressing GFP-PICK1129-415 and mixed thoroughly before imaging. To test the role of Arp2/3 complex in this motility, HeLa cells expressing GFP-PICK1129-415 were treated with either 200 μM CK-666 or DMSO (control), and cells were imaged 30, 60, and 90 min after treatment. Time-lapse sequences were acquired with 3- or 4-s intervals for 1–10 min (as indicated in Figures 3, 4, and 6) using an Eclipse TE2000-U inverted microscope (Nikon, Tokyo, Japan) equipped with a Planapo 100× (1.3 numerical aperture [NA]) objective and a Cascade 512B charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) driven by MetaMorph imaging software (Molecular Devices, Sunnyvale, CA).
The trajectories of GFP-PICK1–associated organelles were analyzed using the Track Object tool of the program MetaMorph. The speed of each organelle was determined as the average of the speeds observed in 8–10 consecutive frames during a 30-s interval. Between 31 and 85 such tracks in three to nine different cells (as indicated in the figure captions) were used to determine the mean organelle velocity for each of the PICK1 constructs. The velocity distributions are represented as box-and-whisker plots. Graphic illustrations and statistical analyses were carried out using the program SigmaPlot and the Statistics Calculator of the Alcula online server (www.alcula.com/calculators/statistics/). The statistical significance of the measurements was assessed using Student's t test.
To establish the identity of the PICK1-associated organelles, we tested colocalization of PICK1 constructs with several organelle markers, including Rab5a, Rab11, Rab7, Lamp1, calnexin, GM130, and TGN38. PICK1 constructs colocalized only with the trans-Golgi marker TGN38 (Supplemental Figure S3 and Supplemental Movie S5). In this case, HeLa cells were cotransfected with mCherry-PICK1 and CFP-TGN38 and incubated for 24 h. Live-cell imaging was performed on a spinning disk confocal Olympus IX71 inverted microscope equipped with a Hamamatsu ImagEM electron-multiplying CCD camera and a 100× (1.4 NA) oil immersion objective. Frames were recorded at 2-s intervals.
Isothermal titration calorimetry
ITC measurements were carried out on a VP-ITC apparatus (MicroCal, GE Healthcare). For the titrations of PICK1 or ACT into Arp2/3 complex, protein samples were dialyzed for 2 d against 20 mM HEPES, pH 7.5, 100 mM KCl, 200 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 0.2 mM ATP. For the titrations of DAT peptide into MBP-PDZ, proteins were dialyzed against 20 mM HEPES, pH 7.5, 300 mM NaCl, and 1 mM TCEP. Titrations consisted of 10-μl injections lasting for 10 s and spaced 300 s apart. The concentration of the titrant (PICK1, ACT, or DAT peptide) was ∼10-fold higher than that of the binding partner in the 1.44-ml cell (Arp2/3 complex or MBP-PDZ). The exact concentrations are given in the caption to Figure 5. The heat of binding was corrected for the small exothermic heat of injection, determined from the titration of proteins into buffer. Data were analyzed using the program Origin (OriginLab Corporation, Northampton, MA).
Sedimentation assays
Actin (20 μM) in G-buffer (2 mM Tris, pH 7.4, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT, and 1 mM NaN3) was polymerized with addition of 100 mM KCl, 2 mM MgCl2, and 1 mM EGTA for 30 min at room temperature. PICK1 constructs were centrifuged at 278,000 × g for 30 min to remove potential aggregates. Two types of experiments were performed: either a fixed concentration of PICK1 (5 μM) was added to various concentrations of F-actin (0–25 μM), or various concentrations of PICK1 (1−20 μM) were added to a fixed concentration of F-actin (1 μM). The mixtures were then incubated for 1 h at room temperature, followed by high-speed centrifugation (278,000 × g) for 30 min. Under these conditions, PICK1 did not pellet when centrifuged in the absence of actin. Equal volumes of supernatant and resuspended pellet fractions were analyzed by SDS–PAGE. Densitometric analysis of the bands of the gels was performed using a G:Box gel scanner (Syngene, Frederick, MD) and the program ImageJ (National Institutes of Health, Bethesda, MD). The percentage of PICK1 bound to F-actin in the pellet was determined from the relative intensities of the bound and unbound fractions. SD values were determined from three independent experiments. Estimates of the dissociation constant, KD, were obtained by global fitting of the data from three independent experiments with the program IGOR Pro (WaveMetrics, Portland, OR), using a one-site binding function. Note, however, that affinity measurements from F-actin sedimentation assays are only approximate because the actual concentration of actin filaments versus monomers is not precisely determined, particularly when the concentration of F-actin is varied in the experiment.
Actin polymerization assay
Actin polymerization was measured as the time course of the fluorescence increase of pyrene-labeled actin upon its incorporation into filaments using a Cary Eclipse fluorescence spectrophotometer (Varian, Palo Alto, CA). Before data acquisition, 2 μM Mg-ATP-actin (6% pyrene-labeled) was mixed with 20 nM Arp2/3 complex alone or with 100 nM N-WASP WCA and increasing concentrations of PICK1 constructs (as indicated in Figures 5 and 6) in F-buffer (10 mM Tris, pH 7.5, 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.5 mM DTT, 0.02 mg/ml bovine serum albumin, 0.2 mM ATP). F-actin seeds were generated by polymerization of 10 μM actin in F-buffer for 1 h at room temperature. The seeds, at a final concentration of 0.5 μM, were added to the polymerization reaction either alone or after incubation with PICK1 (2 or 5 μM) for 30 min at room temperature. Data acquisition started 10 s after mixing. All the measurements were done at 25°C. Control experiments were carried out with addition of buffer alone. Polymerization rates were calculated as described (Boczkowska et al., 2013).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01 MH087950 to R.D. T.S. was supported by National Institutes of Health Grant GM095977. SAXS data collection at CHESS beam lines F2 and G1 was supported by National Science Foundation Grant DMR-0936384 and National Institutes of Health Grant GM103485.
Abbreviations used:
- ACT
acidic C-terminal tail
- BAR
Bin/amphiphysin/Rvs
- ITC
isothermal titration calorimetry
- PDZ
PSD-95/DlgA/ZO-1
- PICK1
human protein interacting with C kinase 1
- SAXS
small-angle x-ray scattering.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1448) on February 5, 2015.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet CE, Svergun DI. Small-angle X-ray scattering on biological macromolecules and nanocomposites in solution. Annu Rev Phys Chem. 2013;64:37–54. doi: 10.1146/annurev-physchem-040412-110132. [DOI] [PubMed] [Google Scholar]
- Boczkowska M, Rebowski G, Dominguez R. Glia maturation factor (GMF) interacts with Arp2/3 complex in a nucleotide state-dependent manner. J Biol Chem. 2013;288:25683–25688. doi: 10.1074/jbc.C113.493338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczkowska M, Rebowski G, Kast DJ, Dominguez R. Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat Commun. 2014;5:3308. doi: 10.1038/ncomms4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, D'Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci. 2005;118:5393–5403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- Boudin H, Craig AM. Molecular determinants for PICK1 synaptic aggregation and mGluR7a receptor coclustering: role of the PDZ, coiled-coil, and acidic domains. J Biol Chem. 2001;276:30270–30276. doi: 10.1074/jbc.M102991200. [DOI] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL. Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci. 2000;20:7258–7267. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Bhattacharyya S, Ma C, Morishita W, Fang S, Rizo J, Malenka RC. Calcium binding to PICK1 is essential for the intracellular retention of AMPA receptors underlying long-term depression. J Neurosci. 2010;30:16437–16452. doi: 10.1523/JNEUROSCI.4478-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev KK, Nishimune A, Henley JM, Nakanishi S. The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology. 1999;38:635–644. doi: 10.1016/s0028-3908(98)00230-5. [DOI] [PubMed] [Google Scholar]
- Elkins JM, Papagrigoriou E, Berridge G, Yang X, Phillips C, Gileadi C, Savitsky P, Doyle DA. Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions. Protein Sci. 2007;16:683–694. doi: 10.1110/ps.062657507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlendsson S, Rathje M, Heidarsson PO, Poulsen FM, Madsen KL, Teilum K, Gether U. PICK1 (protein interacting with C-kinase 1) binding promiscuity relies on unconventional PDZ (PSD-95/Discs-large/ZO-1 homology) binding modes for non-class II PDZ ligands. J Biol Chem. 2014;289:25327–25340. doi: 10.1074/jbc.M114.548743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focant MC, Hermans E. Protein interacting with C kinase and neurological disorders. Synapse. 2013;67:532–540. doi: 10.1002/syn.21657. [DOI] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JG. PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol Ther. 2008;118:152–160. doi: 10.1016/j.pharmthera.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Hanley JG, Henley JM. PICK1 is a calcium-sensor for NMDA-induced AMPA receptor trafficking. EMBO J. 2005;24:3266–3278. doi: 10.1038/sj.emboj.7600801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley JM, Wilkinson KA. AMPA receptor trafficking and the mechanisms underlying synaptic plasticity and cognitive aging. Dialogues Clin Neurosci. 2013;15:11–27. doi: 10.31887/DCNS.2013.15.1/jhenley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst B, Madsen KL, Jansen AM, Jin C, Rickhag M, Lund VK, Jensen M, Bhatia V, Sorensen G, Madsen AN, et al. PICK1 deficiency impairs secretory vesicle biogenesis and leads to growth retardation and decreased glucose tolerance. PLoS Biol. 2013;11:e1001542. doi: 10.1371/journal.pbio.1001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakura Y, Nagano T, Kawamura M, Horikawa H, Ibaraki K, Takei N, Nawa H. N-methyl-D-aspartate-induced alpha-amino-3-hydroxy-5-methyl-4-isoxazoleproprionic acid (AMPA) receptor down-regulation involves interaction of the carboxyl terminus of GluR2/3 with Pick1. Ligand-binding studies using Sindbis vectors carrying AMPA receptor decoys. J Biol Chem. 2001;276:40025–40032. doi: 10.1074/jbc.M103125200. [DOI] [PubMed] [Google Scholar]
- Jin W, Ge WP, Xu J, Cao M, Peng L, Yung W, Liao D, Duan S, Zhang M, Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastning K, Kukhtina V, Kittler JT, Chen G, Pechstein A, Enders S, Lee SH, Sheng M, Yan Z, Haucke V. Molecular determinants for the interaction between AMPA receptors and the clathrin adaptor complex AP-2. Proc Natl Acad Sci USA. 2007;104:2991–2996. doi: 10.1073/pnas.0611170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Fielding AB, Gassner G, Carter NJ, Royle SJ. An unmet actin requirement explains the mitotic inhibition of clathrin-mediated endocytosis. Elife. 2014;3:e00829. doi: 10.7554/eLife.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Chung HJ, Lee HK, Huganir RL. Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc Natl Acad Sci USA. 2001;98:11725–11730. doi: 10.1073/pnas.211132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel M, Wagner W. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nat Rev Neurosci. 2013;14:233–247. doi: 10.1038/nrn3445. [DOI] [PubMed] [Google Scholar]
- Kostan J, Salzer U, Orlova A, Toro I, Hodnik V, Senju Y, Zou J, Schreiner C, Steiner J, Merilainen J, et al. Direct interaction of actin filaments with F-BAR protein pacsin2. EMBO Rep. 2014;15:1154–1162. doi: 10.15252/embr.201439267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- Kukulski W, Schorb M, Kaksonen M, Briggs JA. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. Structural basis for the actin-binding function of missing-in-metastasis. Structure. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Liu L, Wang YT, Sheng M. Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron. 2002;36:661–674. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- Lu W, Ziff EB. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron. 2005;47:407–421. doi: 10.1016/j.neuron.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Madsen KL, Eriksen J, Milan-Lobo L, Han DS, Niv MY, Ammendrup-Johnsen I, Henriksen U, Bhatia VK, Stamou D, Sitte HH, et al. Membrane localization is critical for activation of the PICK1 BAR domain. Traffic. 2008;9:1327–1343. doi: 10.1111/j.1600-0854.2008.00761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man HY, Lin JW, Ju WH, Ahmadian G, Liu L, Becker LE, Sheng M, Wang YT. Regulation of AMPA receptor-mediated synaptic transmission by clathrin-dependent receptor internalization. Neuron. 2000;25:649–662. doi: 10.1016/s0896-6273(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Futterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Mueller GA, Zhong X, Pedersen LC. A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010;19:901–913. doi: 10.1002/pro.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murk K, Blanco Suarez EM, Cockbill LM, Banks P, Hanley JG. The antagonistic modulation of Arp2/3 activity by N-WASP, WAVE2 and PICK1 defines dynamic changes in astrocyte morphology. J Cell Sci. 2013;126:3873–3883. doi: 10.1242/jcs.125146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Wood CL, Patton AP, Jaafari N, Henley JM, Mellor JR, Hanley JG. PICK1 inhibition of the Arp2/3 complex controls dendritic spine size and synaptic plasticity. EMBO J. 2011;30:719–730. doi: 10.1038/emboj.2010.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SS, Moller M, Gillilan RE. High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J Appl Crystallogr. 2012;45:213–223. doi: 10.1107/S0021889812000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SS, Toft KN, Snakenborg D, Jeppesen MG, Jacobsen JK, Vestergaard B, Kutter JP, Arleth L. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Crystallogr. 2009;42:959–964. [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Wu H, Shen C, Shi Y, Jin W, Xia J, Zhang M. Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes. EMBO J. 2007;26:4576–4587. doi: 10.1038/sj.emboj.7601860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca DL, Amici M, Antoniou A, Blanco Suarez E, Halemani N, Murk K, McGarvey J, Jaafari N, Mellor JR, Collingridge GL, Hanley JG. The small GTPase Arf1 modulates Arp2/3-mediated actin polymerization via PICK1 to regulate synaptic plasticity. Neuron. 2013;79:293–307. doi: 10.1016/j.neuron.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P. Synaptic vesicle endocytosis. Cold Spring Harb Perspect Biol. 2012;4:a005645. doi: 10.1101/cshperspect.a005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yu J, Jia Y, Pan L, Shen C, Xia J, Zhang M. Redox-regulated lipid membrane binding of the PICK1 PDZ domain. Biochemistry. 2010;49:4432–4439. doi: 10.1021/bi100269t. [DOI] [PubMed] [Google Scholar]
- Steinberg JP, Takamiya K, Shen Y, Xia J, Rubio ME, Yu S, Jin W, Thomas GM, Linden DJ, Huganir RL. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron. 2006;49:845–860. doi: 10.1016/j.neuron.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun DI, Barberato C, Koch MH. CRYSOL: a program to evaluate X-ray solution scattering of biological macro-molecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Terashima A, Pelkey KA, Rah JC, Suh YH, Roche KW, Collingridge GL, McBain CJ, Isaac JT. An essential role for PICK1 in NMDA receptor-dependent bidirectional synaptic plasticity. Neuron. 2008;57:872–882. doi: 10.1016/j.neuron.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen TS, Madsen KL, Rebola N, Rathje M, Anggono V, Bach A, Moreira IS, Stuhr-Hansen N, Dyhring T, Peters D, et al. Identification of a small-molecule inhibitor of the PICK1 PDZ domain that inhibits hippocampal LTP and LTD. Proc Natl Acad Sci USA. 2010;107:413–418. doi: 10.1073/pnas.0902225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108:E463–471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewhella J, Hendrickson WA, Kleywegt GJ, Sali A, Sato M, Schwede T, Svergun DI, Tainer JA, Westbrook J, Berman HM. Report of the wwPDB Small-Angle Scattering Task Force: data requirements for biomolecular modeling and the PDB. Structure. 2013;21:875–881. doi: 10.1016/j.str.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YT, Linden DJ. Expression of cerebellar long-term depression requires postsynaptic clathrin-mediated endocytosis. Neuron. 2000;25:635–647. doi: 10.1016/s0896-6273(00)81066-1. [DOI] [PubMed] [Google Scholar]
- Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Chung HJ, Wihler C, Huganir RL, Linden DJ. Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron. 2000;28:499–510. doi: 10.1016/s0896-6273(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem. 2004;279:14929–14936. doi: 10.1074/jbc.M309408200. [DOI] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Chen J, Liu J, Brunzelle JS, Huang B, Wakeham N, Terzyan S, Li X, Rao Z, Li G, Zhang XC. Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J. 2007;26:3484–3493. doi: 10.1038/sj.emboj.7601771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.