The permeability of the baker's yeast nuclear pore complex for multidomain proteins of different sizes, both soluble and transmembrane, was measured. The permeability for soluble proteins correlated with models of the disordered phase of wild-type and mutant NPCs generated using a one bead per amino acid molecular dynamics model.
Abstract
Nuclear pore complexes (NPCs) allow selective import and export while forming a barrier for untargeted proteins. Using fluorescence microscopy, we measured in vivo the permeability of the Saccharomyces cerevisiae NPC for multidomain proteins of different sizes and found that soluble proteins of 150 kDa and membrane proteins with an extralumenal domain of 90 kDa were still partly localized in the nucleus on a time scale of hours. The NPCs thus form only a weak barrier for the majority of yeast proteins, given their monomeric size. Using FGΔ-mutant strains, we showed that specific combinations of Nups, especially with Nup100, but not the total mass of FG-nups per pore, were important for forming the barrier. Models of the disordered phase of wild-type and mutant NPCs were generated using a one bead per amino acid molecular dynamics model. The permeability measurements correlated with the density predictions from coarse-grained molecular dynamics simulations in the center of the NPC. The combined in vivo and computational approach provides a framework for elucidating the structural and functional properties of the permeability barrier of nuclear pore complexes.
INTRODUCTION
A key feature of eukaryotes is the nucleus, with the nuclear envelope (NE) forming the barrier between cytoplasm and nucleus. The compartmentalization separates DNA transcription from mRNA translation, which is critical for selective access of transcriptional regulators and control over mRNA biogenesis. Many of the key steps in cell cycle regulation require selective entry of signal molecules into the nucleus. The nuclear pore complexes (NPCs) that are embedded in the NE form the main transport route between the nucleus and cytoplasm and regulate the transport of soluble macromolecules (Aitchison and Rout, 2012). They also form the main passageway for membrane proteins to enter the inner nuclear membrane (INM) of the NE (Laba et al., 2014).
The Saccharomyces cerevisiae NPC is a 50-MDa protein complex built from ∼30 different proteins called nucleoporins (Nups), each present in multiple copies (Rout et al., 2000). A roughly cylindrical scaffold is anchored in the lipid bilayer, and ∼200 intrinsically disordered proteins extend into the interior of the NPC (Alber et al., 2007). The intrinsically disordered proteins are called FG-nups because each encodes multiple phenylalanine-glycine (FG) repeats, which are the binding sites for transport factors that shuttle specific molecules across the NPC in import or export reactions (Radu et al., 1995). The gradient of RanGTP-RanGDP (high RanGTP inside the nucleus) ensures directionality of protein transport by stimulating the dissociation or association of the transport factor–cargo complex on both sides of the NPC (Forrester et al., 1992; Corbett et al., 1995; Rexach and Blobel, 1995; Schlenstedt et al., 1995).
The main traffic route of membrane proteins between the outer nuclear membrane (ONM) and INM of the NE is also through the NPCs but is less well understood (Zuleger et al., 2012; Katta et al., 2013; Laba et al., 2014). The ONM and INM are continuous via the pore membrane at sites where NPCs are embedded, allowing transmembrane domains to diffuse between the INM and ONM (Ellenberg et al., 1997). In all membrane proteins studied, retention mechanisms are important for sorting. Import dependent on transport factors, FG-nups, and the gradient of RanGTP/RanGDP has been shown for some S. cerevisiae membrane proteins that encode nuclear localization signals (NLSs) on the extralumenal domains (King et al., 2006; Meinema et al., 2011, 2013).
The FG-nups have a dual function, as they provide a barrier for diffusion of soluble macromolecules through the NPC and slow down passive entry and exit of molecules into and out of the nucleus (influx and efflux) while selectively permitting active import and export. The influx and efflux rates depend on the size of the molecules, as well as on their surface hydrophobicity and charge (Ribbeck and Görlich, 2002; Naim et al., 2009; Colwell et al., 2010). How the FG-nups form the permeability barrier in the NPC is under debate (Adams and Wente, 2013), but cohesive Nups such as Xenopus Nup98 (Hülsmann et al., 2012) and yeast Nup100 and Nup116 (Patel et al., 2007) are important, and combining deletions of those FG domains results in lethality (Iovine and Wente, 1997). Remarkably, in S. cerevisiae, 50% of the protein mass of FG-nups could be deleted without affecting viability, and in addition no effect on permeability was seen in this strain (Strawn et al., 2004). The permeability of soluble proteins through NPCs was studied extensively in the past but recently has received renewed attention because NPCs are reported to become more permeable with chronological aging, and this may have pronounced effects on cell physiology (D'Angelo et al., 2009).
A wealth of data from diverse eukaryotes describe nuclear entry of fluorescently labeled proteins, dextrans, or gold particles in isolated nuclei or detergent-permeabilized or microinjected cells (summarized in Supplemental Table S1). These studies show that there is no clear consensus as to what size a protein would have to be to keep it from entering the nucleus on biologically relevant time scales of minutes to hours. In higher eukaryotes, the data consistently show that probes >40–60 kDa are excluded, at least on the minute time scale. However, there are also reports that proteins >>60 kDa permeate the NPC on longer time scales in vivo, both in higher eukaryotes (Chatterjee et al., 1997; Beetz et al., 2004; Lénárt and Ellenberg, 2006; Seibel et al., 2007; Wang and Brattain, 2007) and yeast (Patel et al., 2007; Gardner et al., 2011; Supplemental Table S1). A complication with the mammalian in vivo measurements is that on these longer time scales, entry into the nucleus may have happened during mitosis when the NE is broken down. In vivo studies in S. cerevisiae are easier to interpret, as the nucleus does not break down during mitosis. Direct measurements of permeability, such as fluorescence recovery after photobleaching (FRAP)–based assays, yield import, influx, and efflux rates under steady-state conditions (Wei et al., 2002; Kopito and Elbaum, 2007; Kim and Elbaum, 2013). In yeast at 30°C, efflux rate constants for green fluorescent protein (GFP)–cNLS-GFP (∼58 kDa) of 0.02 s−1 were measured (Meinema et al., 2013), indicating high passive permeability for molecules in this size range.
Compared to soluble proteins, passive entry of membrane proteins has been less studied. The lateral channels in the scaffold of the NPC have been proposed to be the sites where the soluble domains of the membrane proteins may pass (Hinshaw et al., 1992; Maimon et al., 2012), and it is unclear whether FG-nups contribute to the barrier for passive entry. The size of the lateral channels will restrict the diffusion of membrane proteins, depending on the size of the soluble domain. Proteins that use a diffusion-retention mechanism to localize to the inner nuclear membrane fail to accumulate at the INM when the cytoplasmic domain is >∼60 kDa (Soullam and Worman, 1995; Wu et al., 2002; Ohba et al., 2004; Deng and Hochstrasser, 2006; Turgay et al., 2010).
The effectiveness of a permeability barrier could be defined as the ability to effectively prevent passage of a fraction of the molecules in a biologically relevant time scale. Here we set out to provide the first systematic assessment of the permeability of the NPC for soluble and membrane proteins in live yeast cells on the time scale of hours. Our studies emphasize that although the NPCs are effective at separating transcription and translation by blocking the passage of very large structures, most membrane and soluble proteins would be small enough to pass the NPC passively based on their monomeric size when considering the time scale of hours. The in vivo permeability in strains with wild-type and mutant NPCs is compared with density predictions of a coarse-grained computational model of the NPC disordered phase. The combined in vivo and computational data highlight the importance of Nup100 for forming the barrier.
RESULTS
Permeability of NPC for soluble proteins
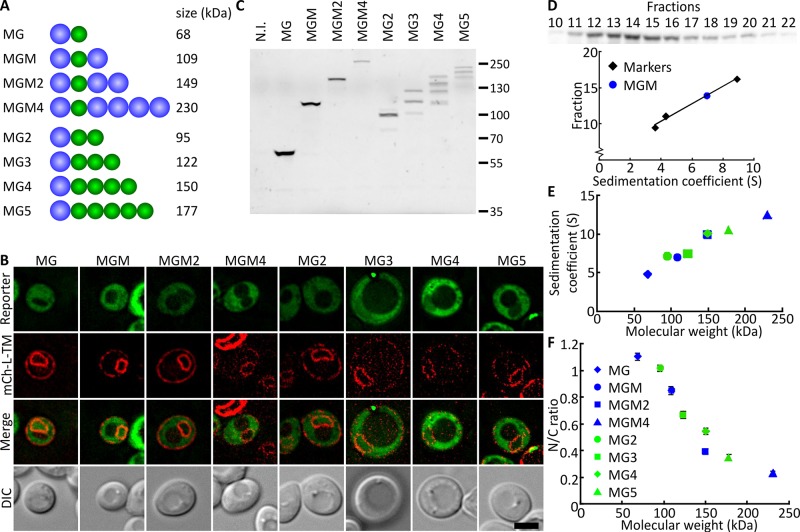
First we investigated what size of molecules was significantly excluded from the nucleus in live yeast cells. We constructed reporter proteins that consisted of one or more GFP and maltose- binding protein (MBP) domains. GFP and MBP are approximately globular, well-folded, stable proteins that have little or no interactions with native proteins in yeast (Mohr et al., 2009). We thus assumed that they freely diffuse in the crowded cytosol and the nucleus and therefore their steady-state localization reflects the permeability of the NPC. The smallest reporter is MBP-GFP (MG). In the larger ones, multiple GFP or MBP proteins were fused together to form MG2 to MG5 and MGM to MGM4, in which M refers to MBP and G to GFP (Figure 1A). We thus have two sets of proteins, and in each set the proteins are of different sizes but have comparable surface properties. As such, we accounted for potential effects of specific surface properties of MBP or GFP, or “bead size.” We expressed the reporter proteins for 3 h and stopped the expression by adding glucose. After an additional hour, to allow newly synthesized proteins to mature and equilibrate, we determined the localization of the reporters (Figure 1B). mCh-L-TM (mCherry fused to the first transmembrane helix of Heh2 with a linker in between) localizes throughout the NE–endoplasmic reticulum (ER) network and was coexpressed to indicate the position of the nucleus. As a measure of the permeability of the NPC for the reporters, we calculated the ratio of mean fluorescence in the nucleus over the cytoplasm. The smallest reporter tested (68 kDa) was not excluded from the nucleus, consistent with previous FRAP measurements (Meinema et al., 2013). The largest of the tested reporters (MGM4, 230 kDa) shows the lowest permeability, with nucleus-to-cytoplasm (N/C) ratio of 0.21 ± 0.02. We note that the 1-h equilibration time is not sufficient to reach a steady state, but this time regime was chosen to reduce the effect of protein synthesis, degradation, and inheritance (on longer time scales, cells divide) on the N/C values.
FIGURE 1:
Expression and localization of soluble reporter proteins. Permeability of the NPC correlates with sedimentation coefficient and molecular weight. (A) Cartoon representing domain composition and size of reporter proteins. G, GFP; M, MBP. (B) Deconvolved images of cells expressing reporter proteins of increasing size (green). mCh-L-TM (red) is localized in the NE-ER network and used to locate the nucleus. Cells were induced with 0.1% (wt/vol) galactose for 3 h, followed by 1-h incubation with 1% (wt/vol) glucose to block expression and allow proteins to equilibrate between cytoplasm and nucleus. Scale bar, 3 μm. (C) Lysates from cells expressing indicated reporter proteins analyzed on SDS–PAGE, in-gel fluorescence detected. N.I., not induced. A Western blot of the same samples is shown in Supplemental Figure S1A. (D) Western blot (detection with anti-GFP) of fractions from a sucrose gradient over which a whole-cell extract of cells expressing MGM (and mCh-L-TM) was separated. Three biotin-labeled marker proteins—ovalbumin (3.6 S), bovine serum albumin (4.3 S), and amylase (8.9 S)—were used as internal standard. The peak width at half-maximum for the reporter proteins is ∼6 fractions. Estimated error ± 1 S. (E) Plot of the obtained sedimentation coefficients against the molecular weight of the reporters. (F) NPC permeability, quantified by the ratio of the fluorescence intensity in the nucleus and cytosol (N/C ratio), plotted as function of the molecular weight of the reporters. N/C ratio close to 1 means that the reporter equilibrates over cytoplasm and nucleus within the time frame of the experiment. A low N/C ratio shows that the diffusion through the NPC is hindered. Plotted N/C ratios are the mean for ∼50 cells; SEM is indicated.
To resolve the relationship between nuclear entry and molecular size and shape, we confirmed full-length expression of the proteins (in-gel fluorescence in Figure 1C, Western blot in Supplemental Figure S1A) and determined their sedimentation coefficient (Figure 1D). The in-gel fluorescence shows single bands for the multi-MBP series (MGM to MGM4), indicating that the fluorescence observed with microscopy represents the full-length proteins. The multi-GFP series (MG2–MG5) also shows full-length expression—the ladder of bands reflects the different in-gel migration of folded and unfolded GFPs (Geertsma et al., 2008). The conformation of the “beads on a string” reporter proteins when entering the NPC is approximated by measurements of their sedimentation coefficient. This also allowed for comparison of the different series of reporter proteins. There is a clear correlation between the permeability of the NPC for the different MBP-GFP fusions and their sedimentation coefficients or molecular weight (Figure 1, E and F), and the MBP and GFP series behaved similarly. In contrast, the number of domains in each construct poorly correlates with the permeability; for example, MGM2 and MG3 both have four domains but differ 1.7-fold in permeability. Therefore, even though these proteins have unnatural “beads on a string” configurations, it seems that their permeability is well predicted by their sedimentation behavior. We conclude that passive diffusion through the NPC is affected by the overall size and shape of the reporters but not by the number of domains that make up the molecule. Of importance, the NPC in S. cerevisiae is highly permeable for proteins, and only the 230-kDa protein showed exclusion from the nucleus.
Permeability of NPC for membrane proteins
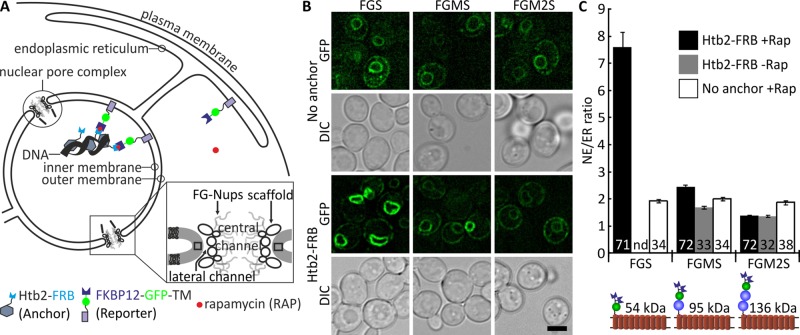
Complementing these studies on diffusion of soluble proteins through the NPC, we addressed how the passage of membrane proteins through the NPC depends on the size of their pore-facing (extralumenal) domains in a systematic way. We constructed reporter proteins with extralumenal domains of increasing size. We adapted the Anchor Away system (Haruki et al., 2008) to create an assay that reports whether a membrane protein tagged with FK506-binding protein (FKBP) has access to INM (Figure 2A). The assay relies on rapamycin-dependent retention at the INM through association with histone Htb2 tagged with FKBP12-rapamycin-binding domain (FRB). The membrane proteins consisted of the transmembrane domain of Sec61 with 10 transmembrane α-helical segments. The extralumenal domains were composed of GFP, FKBP, and no, one, or two copies of MBP (cartoons in Figure 2C). If a protein diffused to the INM, a complex would form between FRB and FKBP when rapamycin was added, trapping and accumulating the reporter at the INM. When the reporter was not able to diffuse to the INM, the ratio of fluorescence intensity in the NE to that in the ER would remain around 1, reflecting even distribution of the protein in the ER and the ONM.
FIGURE 2:
Size dependence of diffusion of membrane proteins through lateral channels of the NPC. Access to the INM depends on the size of extralumenal domains. (A) Cartoon explaining an inducible diffusion-retention assay reporting access to the INM. The reporter proteins with an extralumenal FKBP domain are mobile within the ER and the ONM and, if small enough, also the INM. Htb2-FRB is the anchor in the nucleus. On addition of rapamycin, FKBP and FRB form a complex, trapping any INM-resident reporter irreversibly in the INM. Inset, model of the NPC, indicating the lateral channel. (B) Deconvolved images of cells expressing reporter proteins with extralumenal domains increasing in size. Cells were induced with 0.1% (wt/vol) galactose for 1 h with simultaneous addition of rapamycin. Scale bar, 3 μm. (C) Permeability of NPC quantified by fluorescence intensity in NE over ER in the indicated number of cells after 1-h expression in the presence of rapamycin. High NE/ER ratio represents accumulation in the NE, showing that the reporter can diffuse to the INM and is trapped there. Error bars are SEM; numbers, cells analyzed; nd, not determined. NE/ER ratios were significantly higher for FGS and FGMS in Htb2-FRB + Rap (black bars) compared with no anchor + Rap (white bars). Cartoons of the reporters used are included; FGS, FKBP-GFP-Sec61TMs; FGMS, FKBP-GFP-MBP-Sec61TMs; FGM2S, FKBP-GFP-2xMBP-Sec61TMs. Size of the cytoplasmic domain is indicated.
The reporter with the smallest cytoplasmic domain (FGS, 54 kDa) showed a high accumulation or NE/ER ratio after 1 h of expression in the presence of rapamycin (Figure 2, B and C; NE/ER = 7.6 ± 0.6). The reporter with one MBP domain (FGMS) had an NE/ER ratio of 2.4 ± 0.1, indicating that the diffusion was hindered by the size of the soluble domain. However, the ratio was higher than what was measured without addition of rapamycin (NE/ER = 1.7 ± 0.1), showing that it could still diffuse to the INM. A cytoplasmic domain of 136 kDa stopped the influx of the reporter on the 1-h time scale, as no trapping was observed (NE/ER ratio of FGM2S was 1.4 ± 0.1 both with and without rapamycin). In control cells lacking the nuclear FRB anchor, we found NE/ER ratios for FGMS and FGM2S that were slightly higher than what we measured in the Htb2-FRB strain without addition of rapamycin, showing a small interstrain difference. FGS could not be analyzed without addition of rapamycin because a fraction of the protein did not localize in the NE/ER network but did so in spots in the cell that may represent endosomes. We conclude that membrane proteins diffuse to the inner nuclear membrane, presumably via the lateral channels, and that the size of the cytoplasmic domains determines the ability to pass. Passage of cargo with extralumenal domains of 90 kDa was already significantly hindered.
Permeability for soluble proteins in FGΔ-mutant strains
We tested which FG-nups contribute most to the permeability barrier by assessing the permeability of our differentially sized reporters in a set of strains lacking specific FG-repeat domains (Figure 3A). We expressed the reporters in yeast in which the FG domains of Nup42, Nup159, Nup1, Nup2, Nup60 (all asymmetric nups), and Nsp1 were deleted (SWY3062; Strawn et al., 2004). We saw no difference in permeability compared with the wild-type strain (Figure 3, B and C), consistent with previous results (Strawn et al., 2004). However, a leakier NPC was observed in the strain in which the NPC lacks the FG domain of Nup100 in combination with all asymmetric nups (SWY3042; Figure 3, B and C). The changed permeability was most clearly seen with the MGM2 reporter, for which the N/C ratio increased from 0.40 ± 0.02 in the wild type to 0.76 ± 0.02 in the mutant. The change in permeability is comparable to that for Nup188Δ and Nup170Δ mutants, which are known to give rise to leaky pores, as shown in Supplemental Figure S2. In the SWY3042 strain, the total mass of deleted protein is less than in SWY3062 (38 and 61%, respectively), showing that it is not the total FG domain mass that accounts for the permeability. A strain lacking the FG domains of Nup100, Nup145, and Nup57 (three symmetric GLFG-nups), SWY2950, also showed an increased permeability (Figure 3C); the N/C ratio for MGM2 increased from 0.40 ± 0.02 in wild type to 0.70 ± 0.02. These results confirm that the measured N/C ratios report NPC permeability and are consistent with previous reports indicating that specific Nups are involved in establishing the permeability barrier (Strawn et al., 2004; Patel et al., 2007; Terry and Wente, 2009; Hülsmann et al., 2012). From a comparison of the three strains, we conclude that Nup100, in combination with other Nups, is particularly critical for maintaining the permeability barrier.
FIGURE 3:
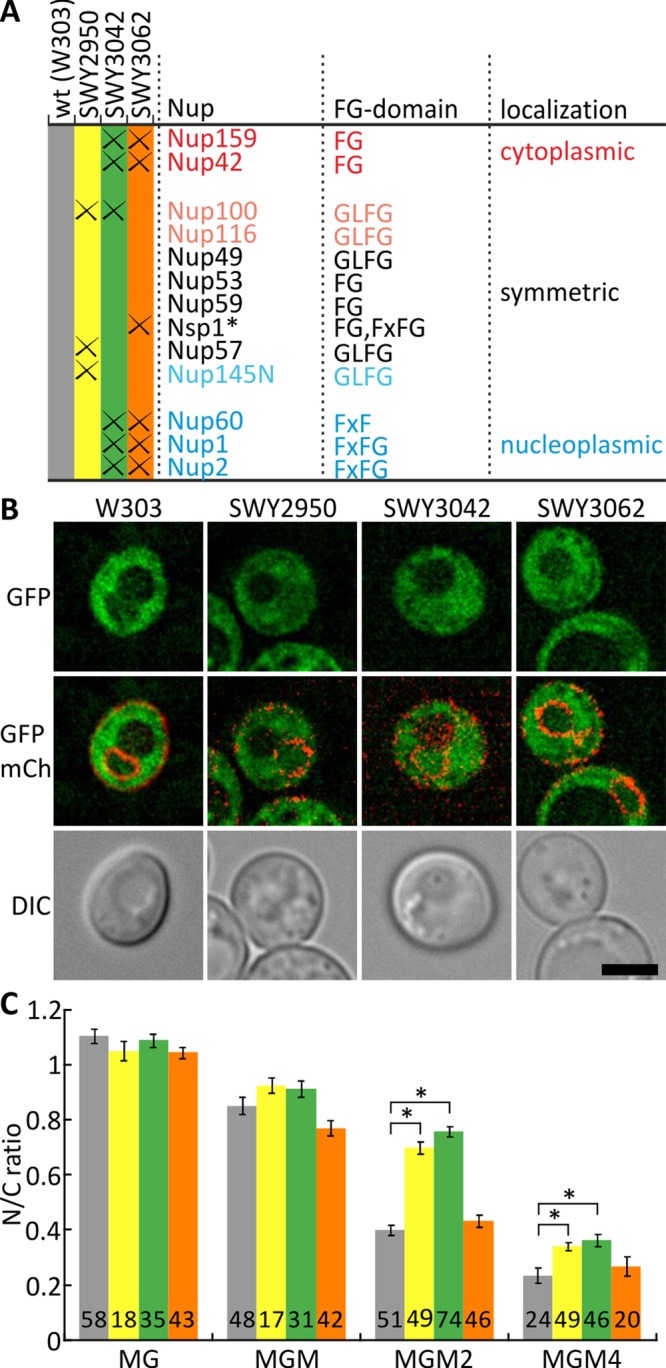
Permeability of NPC in FGΔ-mutant strains. (A) Table indicating strains used and which FG-domains are lacking. *In all strains, Nsp1 is missing amino acids 349–443; see Materials and Methods. (B) Deconvolved images of cells expressing reporter MGM2 (green), comparing wild-type W303 with three FGΔ-mutant strains. SWY3062 shows similar localization as wild type, whereas SWY2950 and SWY3042 show more nuclear localization. Scale bar, 3 μm. (C) Permeability of NPC quantified as the mean N/C ratio over the indicated numbers of cells for four different reporters—MG, MGM, MGM2, and MGM4. Error bars are SEM; numbers, cells analyzed. Significant changes are indicated with asterisks; all other pairwise comparisons between the strains are not significant.
Comparing experimental data to simulations of the disordered phase
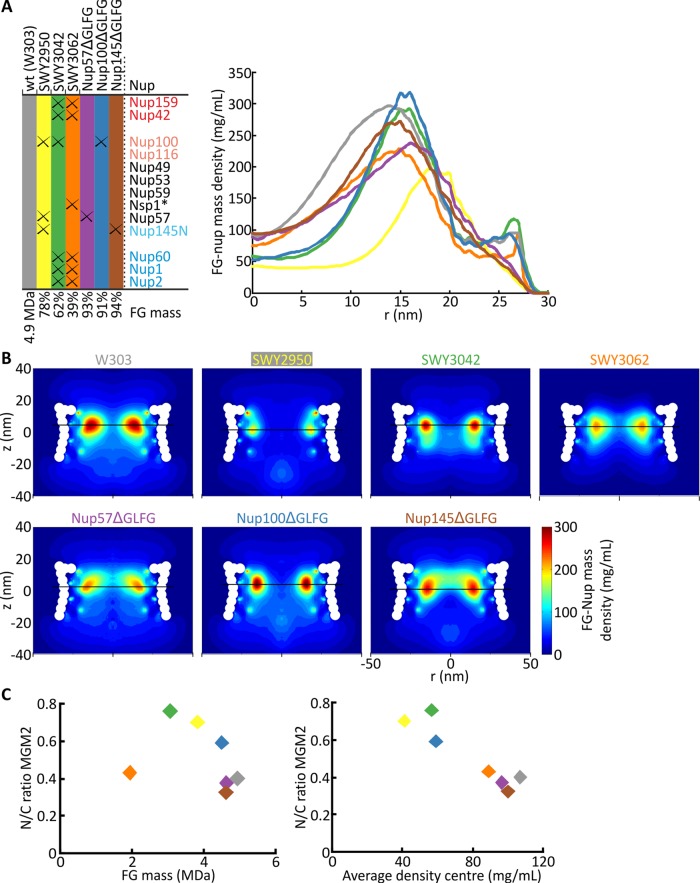
Next we compared the in vivo permeability in the different FGΔ-mutant strains with computational predictions of the density of the disordered phase of the yeast NPC (Ghavami et al., 2014). These results were generated using a one bead per amino acid molecular dynamics model, which accounts for the hydrophobic/hydrophilic and electrostatic interactions between different amino acids, polarity of the solvent, and screening of free ions. A simplified geometrical model of the NPC scaffold was constructed, onto which FG-nups were anchored at positions predicted by the architectural model of Alber et al. (2007). The modeled mutant NPCs differ slightly from the strains used for the in vivo measurements, as explained in detail in the Supplemental Methods and Supplemental Figure S3C. Figure 4A shows the radial mass density distribution at the z-location of the maximum density in different mutants averaged over the simulation time. The fluctuations of the density during the simulations are plotted in Supplemental Figure S3A for r = 0 nm (the center of the pore) and are represented by error bars over the whole r range in Supplemental Figure S3B. The density at the center of the NPC in Figure 4A was ∼35–50% lower in the two strains that had the most compromised permeability barriers in our in vivo measurements (compare SWY3042 and SWY2950 to wild type). The in vivo permeability data thus correspond to the computed protein density at the center of the NPC (r = 0–5 nm).
FIGURE 4:
Coarse-grained modeling of FGΔ-mutant strains. (A) The table shows the strains used with their respective FG-domain deletions and the remaining FG-domain mass as compared with wild-type NPCs (based on the definition in Strawn et al., 2004). The graph plots the modeled FG-nup mass density in the pore in the different FG-deletion strains, measured at z with maximum density (indicated by black horizontal lines in B). *In all strains, Nsp1 is missing amino acids 349–443; see Materials and Methods. (B) Two-dimensional FG-nup mass density plots of the mutant NPCs. A more detailed definition of the composition of the modeled NPCs and the fluctuation of the FG-nup mass density over the simulation time are given in Supplemental Methods and Supplemental Figure S3. Horizontal lines indicate z-plane with maximum density. (C) The permeability (in vivo–determined N/C ratio) of the reporter MGM2 plotted against the FG-domain protein mass (left) and against the computed average protein density in the center (r = 0–5 nm) of the NPC at z with maximum density (right).
To look more specifically at the role of single FG-nups, we used the model to create density distributions for mutants with a single FG domain deleted, namely Nup57ΔGLFG, Nup100ΔGLFG, and Nup145ΔGLFG, the three FG domains that are lacking in SWY2950. These simulations showed that when the FG domain of Nup100 is lacking, the density in the center of the NPC is comparable to the leakier FGΔ-mutant strains SWY2950 and SWY3042, whereas Nup57ΔGLFG and Nup145ΔGLFG behave comparable to wild type (Figure 4A). Indeed, also in vivo, the permeability in Nup100ΔGLFG is highest: the N/C ratio for MGM2 is 0.59 ± 0.02 in Nup100ΔGLFG compared with 0.40 ± 0.02 in wild type (significant difference), whereas Nup57ΔGLFG and Nup145ΔGLFG have an N/C ratio of 0.37 ± 0.01 and 0.33 ± 0.01, respectively, comparable to wild type. A strong correlation is observed between the density in the center of the NPC (averaged for r = 0–5 nm) and the permeability of MGM2, whereas there is no correlation with the total in vivo FG mass (Figure 4C). This strengthens our conclusion that a lower density in the center of the pore, as calculated by the model, correlates with increased permeability. It also shows that Nup100 is important for maintaining the permeability barrier, although other Nups not analyzed here, most notably Nup116, the homologue of Nup100, might show a similar effect.
DISCUSSION
Permeability for soluble proteins
The NPC is the main gateway for transport into and out of the nucleus and to the nuclear inner membrane. It selectively allows import and export of targeted soluble macromolecules while size-selecting against passive entry of larger molecules (Ribbeck and Görlich, 2002; Frey et al., 2006; Hülsmann et al., 2012). The lateral channels could size-select against membrane proteins with large extralumenal domains (Soullam and Worman, 1995; Deng and Hochstrasser, 2006). Here we measured in vivo the permeability of the NPC for multidomain proteins of different sizes using fluorescence microscopy. Our soluble reporters range from 68 to 230 kDa and are fusions of multiple globular domains. Even proteins of ∼150 kDa are still partly localized in the nucleus in a time frame of 1–4 h. Sedimentation data, reflecting the combined size and shape of the proteins in solution, show good correlation with the permeability of the NPC for the different reporters. Therefore comparisons with native, more globular proteins can be made. Indeed, a GFP fusion of Adh5, an approximately globular tetramer with a sedimentation coefficient of ∼7.5 S and molecular weight of 151 kDa, is observed inside the nucleus (Huh et al., 2003). A direct comparison with previous results obtained in yeast using the assay in which influx of GFP fused to a nuclear export signal was measured while having the cells on ice to inhibit active export (Shulga et al., 2000; Shulga and Goldfarb, 2003) is difficult, as the experimental conditions (time scale and temperature) are very different.
Overall we conclude that wild-type (young) NPCs in baker's yeast are relatively permeable for soluble proteins on the time scale relevant for cell division, and the size needed for complete exclusion from the nucleus is much larger than that of the majority of yeast proteins.
Permeability of membrane proteins
The diffusion of membrane proteins to the INM is affected by the size of the cytoplasmic domain, which has to move through the lateral channels of the NPC. We probed influx of membrane proteins as a function of the size of their extralumenal domains, using an inducible assay that mimics the diffusion-retention mechanism used by many INM proteins. We found that in yeast, a membrane protein with a 90-kDa cytoplasmic domain can still accumulate in the INM if trapped there, although much less efficiently than with a smaller domain. This result is in line with previous studies done in different cell types (Soullam and Worman, 1995; Wu et al., 2002; Ohba et al., 2004; Deng and Hochstrasser, 2006; Turgay et al., 2010). An analysis of all membrane proteins in yeast reveals that only 54 proteins have a soluble domain >80 kDa (Supplemental Table S2), of which only 26 are located in the ER. Among those, we find proteins involved in ER–plasma membrane and nucleus–vacuole junctions, ER lipid composition regulation, and ER-associated protein degradation. For example, Mga2 and Spt23, involved in regulating Ole1 transcription, should be excluded from the INM based on the size of the extralumenal domains. Indeed, activation happens only after cleavage and nuclear translocation of their cytoplasmic domains (Chellappa et al., 2001). Overall the NPC is restrictive for membrane proteins with large cytoplasmic domains, but this size selection will affect only a limited number of membrane proteins.
High NPC permeability—what does it mean?
Our results show that NPCs are significantly permeable to soluble and membrane proteins in a size range that comprises >90% of the—monomeric—soluble and membrane proteins in baker's yeast (based on analysis of all open reading frames from the Saccharomyces genome database at yeastgenome.org). Many proteins may thus roam the nuclear compartment at some point in time. Only for some proteins may nuclear exclusion be regulated, in which case the proteins associate into macromolecular complexes that are large enough to be excluded or are retained at cytosolic membranes. In addition, retention at the INM is a mechanism that renders transcription factors inactive when leaked into the nucleus (Heessen and Fornerod, 2007). Active export mechanisms further ensure low nuclear concentrations, but these are known only for soluble macromolecules and not for membrane proteins.
The barrier function of the NPC may be seen more as a by-product of the desired separation of transcription and translation than as a primary task. Indeed, the NPCs effectively separate transcription and translation by blocking the passive crossing of very large structures, such as chromatin, messenger ribonucleoprotein particles, and ribosomes. In addition to the separation of transcription and translation, an important function of the NPC is to maintain gradients across the NE and adjust these in response to changing conditions. For both, the important parameter is the difference in the rates of transport and leak rather than their absolute rates.
FG-nups contributing to permeability
FG-nups in the center of the pore are important for the permeability of the NPC (Strawn et al., 2004; Patel et al., 2007; Terry and Wente, 2009). We tested which FG-nups contribute most to the permeability barrier by assessing the permeability of our differentially sized reporters in a set of strains lacking specific FG-repeat domains. We show that the total mass of FG-nups per pore does not correlate with permeability. Comparing pores lacking the FG-repeat domains of Nsp1 or Nup100 in combination with the asymmetric nups, we find that the latter strain is more permeable, whereas the former strain has greater reduction in FG-repeat mass (39 and 62% of FG-domain protein mass left, respectively). In addition, when NPCs lack only a combination of the FG domains of GLFG-nups Nup100, Nup57, and Nup145N (78% of wild-type protein mass left), the NPCs show increased permeability. Clearly, specific Nups are important for forming the barrier for passive diffusion. This is shown even more strikingly by the single-FGΔ-mutant strains, for which deletion of the FG domain of Nup100 alone results in a more permeable NPC, whereas the single deletion of Nup57-GLFG or Nup145N-GLFG does not change the permeability. Nup100 is one of the cohesive Nups, interacting with the other cohesive nups, namely Nup116, Nup57, Nup49, Nup145N, and Nup42 (Patel et al., 2007). Nup98 is the mammalian analogue of Nup100, Nup116, and Nup145N from yeast and is essential for forming a permeability barrier in reconstituted pores from Xenopus extracts (Hülsmann et al., 2012). This all points toward a role of cohesive GLFG-nups—in yeast in particular, Nup100—in forming the permeability barrier, consistent with earlier data (Patel et al., 2007; Hülsmann et al., 2012).
We compared our data to the results of a recently developed coarse-grained molecular dynamics model of the disordered phase of the yeast NPC (Ghavami et al., 2014). Consistent with the measurements of permeability, the simulations show that the density distribution of the mutant NPC lacking Nsp1 and the asymmetric Nups is overall lower than wild type, but the shape is similar and, of importance, the density in the center is comparable. With Nup100, Nup57, and Nup145N deleted, the density in the center of the pore is lower and the radius of the low-density region is larger. Consistently, NPCs lacking Nup100 and the asymmetric Nups or Nup100 alone also show a reduced density in the center of the pore. An area of lower density in the center of the pore may thus result in a more permeable NPC. Single-molecule studies indeed support passive diffusion of Alexa Fluor–labeled GFP, tandem-GFP, and dextrans through the center of the NPC (Ma et al., 2012; Yang, 2013), but electron microscopy studies show that GFP diffuses throughout the entire NPC (Fiserova et al., 2010).
When comparing different FGΔ-mutant strains, the presence of a high-density region correlates with viability (Ghavami et al., 2014). In addition, when comparing the strains used in this study, we see this correlation: the maximum density is lowest for the temperature-sensitive strains SWY2950 and SWY3062 (Strawn et al., 2004), whereas the strains with an impaired permeability barrier, and a corresponding reduced density in the center of the pore, grow fine. Further, on the basis of our conclusion that the NPCs are significantly permeable even in wild-type young cells, we speculate that the impairment in maintenance of gradients across the NE poses a larger problem for the cell than that of entry of detrimental protein components. Future studies are required to further test and validate these hypotheses and potentially relate them to changes in the NPC with aging (D'Angelo et al., 2009), for which the computational tools and in vivo assays reported here can help to dissect the structure–function relationships.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions
Strains used in this study were all in the W303 background. The specific strains are listed in Supplemental Table S3. We noticed that our W303 strain, as well as W303-derived strains from other laboratories (e.g., Strawn et al., 2004; Haruki et al., 2008) and the wild-type W303 strain distributed by the European Saccharomyces cerevisiae Archive for Functional Analysis (Frankfurt, Germany), have a mutation in Nsp1, missing amino acids 349–443. This deletion was present in all strains used. Plasmids used are listed in Supplemental Table S4. Cells were grown at 30°C in selective drop-out medium (Sigma-Aldrich, St. Louis, MO), supplemented with 2% (wt/vol) d-raffinose. Soluble reporter proteins were expressed under control of the GAL1 promoter by 3-h induction with 0.1% (wt/vol) d-galactose, followed by 1-h incubation with 1% (wt/vol) d-glucose to stop expression. Some aggregates were observed when expressing the multidomain proteins, but these were not taken along for analysis. Full-length expression was confirmed by in-gel fluorescence and Western blot (Figure 1B and Supplemental Figure S1). Membrane reporter proteins were expressed by 1-h induction with 0.1% (wt/vol) d-galactose in the presence of 5 μg/ml rapamycin.
Microscopy
Imaging was done on a DeltaVision Deconvolution Microscope (Applied Precision), using InsightSSITM Solid State Illumination of 488 and 594 nm and an Olympus UPLS Apo 100× oil objective with 1.4 numerical aperture. Detection was done with a CoolSNAP HQ2 camera. Image stacks were deconvolved using standard settings. Data were analyzed with open source software Fiji (Schindelin et al., 2012). Two-tailed Student's t test was used to determine whether changes were significant, using cut-off p < 0.05.
Gradients
Sedimentation coefficients of the reporter proteins were determined essentially as described (Alber et al., 2007) from fractionation of whole cell cryolysis extracts over 5–20% sucrose gradients. For details see the Supplemental Methods.
Modeling
Simulations were performed using a one bead per amino acid coarse-grained molecular dynamics model (Ghavami et al., 2014). The model distinguishes between 20 amino acids and takes into account the hydrophobic and hydrophilic as well as the electrostatic interactions between residues. To perform NPC simulations, a simplified geometrical model for the scaffold of the yeast nuclear pore complex was built based on published data (Alber et al., 2007). The FG-nups were anchored at the estimated position inside the central channel of the NPC with an initial conformation taken from single FG-nup simulations. The domains used are detailed in the Supplemental Methods. For each simulation, the system was first energy minimized. The subsequent molecular dynamics simulation was carried out for at least 3.5 × 107 steps, with the first 5 × 106 steps neglected in generating the results. Simulations were run until the density at r = 0 converged to a stable average. To obtain density maps, the NPC was centered in a box of 100 nm by 100 nm along the sides and 140 nm along the vertical axis of the NPC, which is discretized using (0.5 nm)3 cells. The number of residues in each cell was counted over the total simulation time, and a three-dimensional density profile was generated, which was averaged in the circumferential direction to obtain two-dimensional (2D) density plots. Finally, the radial density distribution was obtained by averaging these 2D density maps in the vertical direction.
Supplementary Material
Acknowledgments
This work was financed by research programs from the Zernike Institute for Advanced Materials and the Netherlands Organization for Scientific Research (VIDI and ECHO grant to L.M.V.). We acknowledge the support of NWO Physical Sciences, SURFsara (www.surfsara.nl), and the Centre for High Performance Computing and Visualization of the University of Groningen in providing supercomputer facilities. We thank members of the Veenhoff, Chang, and Poolman laboratories for valuable discussions. We are grateful to Ben Timney and Michael Rout (Rockefeller University), and Erik van der Giessen (University of Groningen, The Netherlands) for critical reading of the manuscript. We thank the reviewers for their valuable suggestions. We thank S.R. Wente from Vanderbilt University School of Medicine for the generous gift of the NPC mutant strains.
Abbreviations used:
- ER
endoplasmic reticulum
- FG
phenylalanine glycine
- FKBP
FK506-binding protein
- FRAP
fluorescence recovery after photobleaching
- FRB
FKBP12-rapamycin–binding domain
- GFP
green fluorescent protein
- INM
inner nuclear membrane
- MBP
maltose-binding protein
- NE
nuclear envelope
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- Nup
nucleoporin
- ONM
outer nuclear membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1175) on January 28, 2015.
REFERENCES
- Adams R, Wente S. Uncovering nuclear pore complexity with innovation. Cell. 2013;152:1218–1221. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J, Rout M. The yeast nuclear pore complex and transport through it. Genetics. 2012;190:855–883. doi: 10.1534/genetics.111.127803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Beetz C, Brodhun M, Moutzouris K, Kiehntopf M, Berndt A, Lehnert D, Deufel T, Bastmeyer M, Schickel J. Identification of nuclear localisation sequences in spastin (SPG4) using a novel Tetra-GFP reporter system. Biochem Biophys Res Commun. 2004;318:1079–1084. doi: 10.1016/j.bbrc.2004.03.195. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Javier M, Stochaj U. In vivo analysis of nuclear protein traffic in mammalian cells. Exp Cell Res. 1997;236:346–350. doi: 10.1006/excr.1997.3746. [DOI] [PubMed] [Google Scholar]
- Chellappa R, Kandasamy P, Oh C, Jiang Y, Vemula M, Martin C. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J Biol Chem. 2001;276:43548–43556. doi: 10.1074/jbc.M107845200. [DOI] [PubMed] [Google Scholar]
- Colwell L, Brenner M, Ribbeck K. Charge as a selection criterion for translocation through the nuclear pore complex. PLoS Comput Biol. 2010;6:e1000747. doi: 10.1371/journal.pcbi.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett A, Koepp D, Schlenstedt G, Lee M, Hopper A, Silver P. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo M, Raices M, Panowski S, Hetzer M. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- Ellenberg J, Siggia E, Moreira J, Smith C, Presley J, Worman H, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J, Richards S, Wente S, Goldberg M. Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J Cell Sci. 2010;123:2773–2780. doi: 10.1242/jcs.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3'-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- Frey S, Richter R, Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Gardner J, Smoyer C, Stensrud E, Alexander R, Gogol M, Wiegraebe W, Jaspersen S. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J Cell Biol. 2011;193:489–507. doi: 10.1083/jcb.201011017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertsma ER, Groeneveld M, Slotboom D, Poolman B. Quality control of overexpressed membrane proteins. Proc Natl Acad Sci USA. 2008;105:5722–5727. doi: 10.1073/pnas.0802190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavami A, Veenhoff L, van der Giessen E, Onck P. Probing the disordered domain of the nuclear pore complex through coarse-grained molecular dynamics simulations. Biophys J. 2014;107:1393–1402. doi: 10.1016/j.bpj.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli U. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 2007;8:914–919. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J, Carragher B, Milligan R. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Huh W, Falvo J, Gerke L, Carroll A, Howson R, Weissman J, O'shea E. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hülsmann B, Labokha A, Görlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Iovine M, Wente S. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta S, Smoyer C, Jaspersen S. Destination: inner nuclear membrane. Trends Cell Biol. 2013;24:221–229. doi: 10.1016/j.tcb.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Kim S, Elbaum M. A simple kinetic model with explicit predictions for nuclear transport. Biophys J. 2013;105:565–569. doi: 10.1016/j.bpj.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M, Lusk C, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kopito RB, Elbaum M. Reversibility in nucleocytoplasmic transport. Proc Natl Acad Sci USA. 2007;104:12743–12748. doi: 10.1073/pnas.0702690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laba J, Steen A, Veenhoff L. Traffic to the inner membrane of the nuclear envelope. Curr Opin Cell Biol. 2014;28:36–45. doi: 10.1016/j.ceb.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Lénárt P, Ellenberg J. Monitoring the permeability of the nuclear envelope during the cell cycle. Methods. 2006;38:17–24. doi: 10.1016/j.ymeth.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Ma J, Goryaynov A, Sarma A, Yang W. Self-regulated viscous channel in the nuclear pore complex. Proc Natl Acad Sci USA. 2012;109:7326–7331. doi: 10.1073/pnas.1201724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Meinema A, Laba J, Hapsari R, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk C, Poolman B, Veenhoff L. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- Meinema A, Poolman B, Veenhoff L. Quantitative analysis of membrane protein transport across the nuclear pore complex. Traffic. 2013;14:487–501. doi: 10.1111/tra.12048. [DOI] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Güttler T, Görlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim B, Zbaida D, Dagan S, Kapon R, Reich Z. Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier. EMBO J. 2009;28:2697–2705. doi: 10.1038/emboj.2009.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Schirmer E, Nishimoto T, Gerace L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Belmont B, Sante J, Rexach M. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Radu A, Moore M, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Görlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M, Aitchison J, Suprapto A, Hjertaas K, Zhao Y, Chait B. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Saavedra C, Loeb J, Cole C, Silver P. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of poly(A)+ RNA in the cytoplasm. Proc Natl Acad Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel N, Eljouni J, Nalaskowski M, Hampe W. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal Biochem. 2007;368:95–99. doi: 10.1016/j.ab.2007.05.025. [DOI] [PubMed] [Google Scholar]
- Shulga N, Goldfarb D. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol Cell Biol. 2003;23:534–542. doi: 10.1128/MCB.23.2.534-542.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Mosammaparast N, Wozniak R, Goldfarb DS. Yeast nucleoporins involved in passive nuclear envelope permeability. J Cell Biol. 2000;149:1027–1038. doi: 10.1083/jcb.149.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman H. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L, Shen T, Shulga N, Goldfarb D, Wente S. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Terry L, Wente S. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryotic Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Brattain M. The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 2007;581:3164–3170. doi: 10.1016/j.febslet.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Henke VG, Strübing C, Brown EB, Clapham DE. Real-time imaging of nuclear permeation by EGFP in single intact cells. Biophys J. 2002;84:1317–1327. doi: 10.1016/S0006-3495(03)74947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Lin F, Worman H. Intracellular trafficking of MAN1, an integral protein of the nuclear envelope inner membrane. J Cell Sci. 2002;115:1361–1371. doi: 10.1242/jcs.115.7.1361. [DOI] [PubMed] [Google Scholar]
- Yang W. Distinct, but not completely separate spatial transport routes in the nuclear pore complex. Nucleus. 2013;4:166–175. doi: 10.4161/nucl.24874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N, Kerr A, Schirmer E. Many mechanisms, one entrance: membrane protein translocation into the nucleus. Cell Mol Life Sci. 2012;69:2205–2216. doi: 10.1007/s00018-012-0929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.