Abstract
Loss of the mitochondrial protease activity of Omi causes mitochondrial dysfunction, neurodegeneration with parkinsonian features and premature death in mnd2 (motor neuron degeneration 2) mice. However, the detailed mechanisms underlying this pathology remain largely unknown. Here, we report that Omi participates in the process of mitochondrial biogenesis, which has been linked to several neurodegenerative diseases. The mitochondrial biogenesis is deficit in mnd2 mice, evidenced by severe decreases of mitochondrial components, mitochondrial DNA and mitochondrial density. Omi cleaves glycogen synthase kinase 3β (GSK3β), a kinase promoting PPARγ coactivator-1α (PGC-1α) degradation, to regulate PGC-1α, a factor important for the mitochondrial biogenesis. In mnd2 mice, GSK3β abundance is increased and PGC-1α abundance is decreased significantly. Inhibition of GSK3β by SB216763 or overexpression of PGC-1α can restore mitochondrial biogenesis in mnd2 mice or Omi-knockdown N2a cells. Furthermore, there is a significant improvement of the movement ability of mnd2 mice after SB216763 treatment. Thus, our study identified Omi as a novel regulator of mitochondrial biogenesis, involving in Omi protease-deficient-induced neurodegeneration.
Mitochondria have a vital role in neuronal death and survival.1 As critical cellular organelles, mitochondria have highly dynamic properties, including mitochondrial fission, fusion, transport, biogenesis and degradation. The changes of those properties affect mitochondrial functions, leading to the occurrence of diseases.2, 3 Growing lines of evidence suggest that the mitochondrial dysfunction is involved in aging and neurodegenerative diseases, such as Alzheimer's disease (AD), Huntington's disease (HD) and Parkinson's disease (PD).4, 5 Similar to other neurodegenerative diseases, PD is a progressive neurological disorder, which is characterized by the development of cytoplasmic aggregates known as Lewy bodies and degeneration of dopaminergic (DA) neurons in the substantia nigra of midbrain and other brain regions.6 In PD, dysfunction of mitochondria has been documented to be associated with disease pathogenesis in PD brains and both genetic- and toxin-induced PD animal models. In PD brains, mutations in mitochondrial DNA (mtDNA) occur more frequently than those in age-matched control; and mutations in the nuclear-encoded mtDNA polymerase-γ gene, which impair mtDNA replication and result in multiple mtDNA deletions, cause PD-like symptoms.5 Meanwhile, several PD-associated gene products, including α-synuclein, parkin, DJ-1, PINK1 (PTEN-induced putative kinase 1), leucine-rich repeat kinase 2, ubiquitin carboxy-terminal hydrolase L1 and Omi, have been identified to be associated with PD, and lead to mitochondrial dysfunction with changes in mitochondrial morphology, biogenesis and mitophagy in vivo and in vitro.5, 7, 8, 9 Besides, mitochondrial toxins, such as MPTP (1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine) and rotenone that inhibit complex I of the mitochondrial respiratory chain, cause clinically parkinsonian phenotype.10, 11
The serine protease Omi (also known as HtrA2) belongs to the high-temperature requirement factor A (HtrA) family, and was originally identified as a mammalian homolog of the Escherichia coli heat-shock-induced serine protease HtrA/DegP and DegS.12 Omi is mainly localized in mitochondria, although a fraction of it is also found in nucleus.13 Omi is released from the mitochondria into the cytosol to mediate cell death by caspase-dependent or -independent pathways in response to apoptotic stimuli.14, 15 However, the notion that Omi is an apoptosis inducer in the central nervous system was challenged by studies of Omi-overexpressing or -deficient mice. Omi-overexpressing mice show normal development without any sign of apoptotic cell death.16 On the other hand, mnd2 (motor neuron degeneration 2) mice that harbor protease-deficient Omi S276C mutants, and Omi-knockout mice both suffer from progressive neurodegeneration, especially in striatum, and motor abnormalities similar to PD. Both mice fail to gain weight and die before postnatal day 40 due to neurodegeneration with progressive mitochondrial damage.17, 18, 19 Besides, mutations in the Omi gene have also been identified in PD patients.20, 21 Previous studies have shown that Omi has a vital role in the mitochondrial integrity, and the loss of protease activity leads to mitochondrial dysfunction, such as abnormal mitochondrial morphology and increased mtDNA mutation and deletions, increased susceptibility of mitochondrial membrane permeabilization, decreased mitochondrial membrane potential, and reduced mitochondrial density in mnd2 mice and Omi-knockout mice.17, 18, 22 Omi has been found to act downstream of PINK1, but parallel to parkin, in a mitochondrial stress sensing pathway to sense the different stresses, which may be defective in PD.23 These findings suggest that the primary function of Omi is involved in neuroprotection, especially in the maintenance of mitochondrial homeostasis.23, 24
In this article, we identified that Omi cleaves glycogen synthase kinase 3β (GSK3β) to regulate PPARγ coactivator-1α (PGC-1α) abundance and to ensure mitochondrial biogenesis.
Results
Omi regulates mitochondrial components
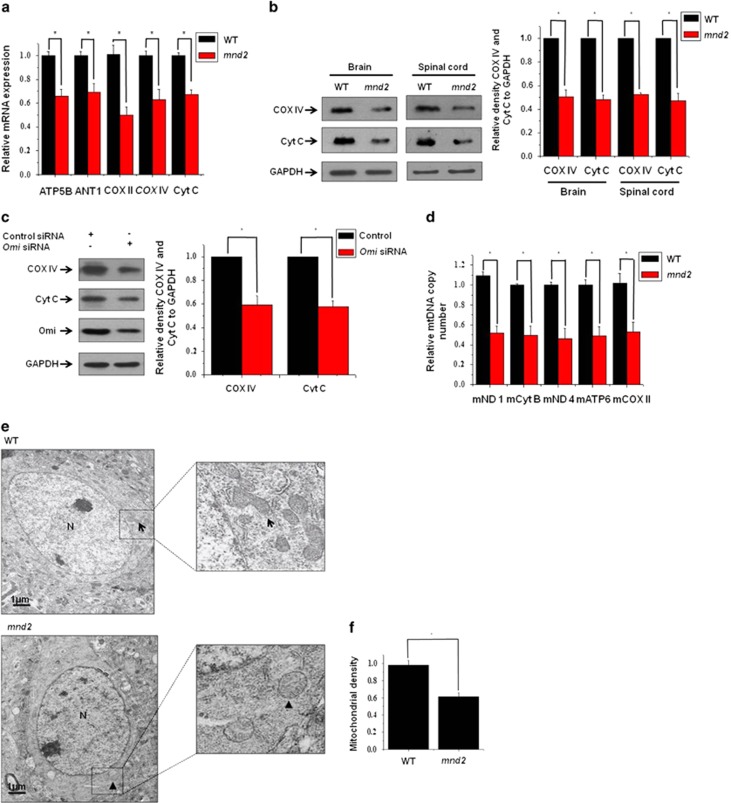
Recent findings show that mitochondrial density is likely to be reduced in tissues derived from Omi-knockout mice,19 and that the number of mitochondria, displaying reduced electron density and open cristae, is increased in Omi-deletion mutant fly.25 To determine whether mitochondrial components are altered when Omi protease activity is deficient, we examined the expressions of genes involved in mitochondrial respiratory chain function and oxidant metabolism, from the brains of 25-day-old mnd2 mice and age-matched wide-type mice using real-time quantitative PCR (RT-qPCR). The mRNA levels of the essential components of the respiration chain, including the nuclearly encoded mitochondrial ATP synthase 5 β subunit (ATP5B) and cytochorome c oxidase subunit IV (COX IV), the mitochondrially encoded cytochrome c oxidase subunit II (COX II) and cytochrome c (Cyt C), and the component of mitochondrial oxidant metabolism, adenine nucleotide translocase-1 (ANT1), were all decreased significantly in mnd2 mice compared with those in wide-type mice (Figure 1a). We next performed western blotting to analyze the abundance of COX IV and Cyt C at protein levels. Consistence with RT-qPCR analyses, the protein levels of COX IV and Cyt C were also decreased (Figure 1b). Meanwhile, in Omi-knockdown SH-SY5Y cells (Figure 1c), a human neuroblastoma cell line, and N2a cells (Supplementary Figure S1), a mouse neuroblastoma cell line, the abundance of COX IV and Cyt C was also reduced, similar to the alterations in mnd2 mice. Further, we detected the amount of mtDNA copy number in brains of mnd2 mice using RT-qPCR. The β-globin gene was used as the target template for the determination of nuclear DNA (nDNA); five different mitochondrial-derived genes, that is, mitochondrial NADH dehydrogenase 1 (mNADH1), mitochondrial cytochrome B, mitochondrial NADH dehydrogenase 4 (mNADH4), mitochondrial ATP synthase 6 (mATP6) and mitochondrial cytochrome oxidase subunit II (mCOX II) were used in the determination of mtDNA quantities. By calculating the ratio of the mitochondrial genes to the nDNA (mtDNA/nDNA), we found that mtDNA copy number was decreased dramatically in mnd2 mice compared with that in wide-type mice (Figure 1d). Moreover, to further identify whether mitochondrial number is certainly reduced in mnd2 mice, we performed transmission electron microscopic analyses to examine mitochondria in striatum from 25-day-old mnd2 mice, where neurodegeneration is most prominent. The mnd2 mice showed an obviously decreased number of mitochondria when compared with wide-type mice (Figure 1e). Quantitative morphometry of transmission electron micrographs confirmed that the mitochondrial density (mitochondrial number/cytoplasmic area) was significantly lower in mnd2 mice than that in wide-type mice (Figure 1f). Thus, our data suggest that Omi affects mitochondrial components and density.
Figure 1.
Omi regulates mitochondrial components. (a) Real-time RT-qPCR assays were performed showing that the mRNA levels of the components of the mitochondrial respiration chain and oxidant metabolism, including ATP5B, COX IV, COX II, CytC and ANT1 are decreased in the mnd2 mice of 25 days compared with those in wide-type mice of the same age. n=3–4 per group, *P<0.05; one-way ANOVA. (b) Western blotting analysis was performed showing that COX IV and Cyt C protein abundances were decreased in the brains and spinal cord of mnd2 mice of 25 days compared with those in wild-type mice of the same age. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (c) Western blotting analysis was performed showing that the knockdown of Omi decreases COX IV and Cyt C protein abundance in SH-SY5Y cells. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (d) Real-time RT-qPCR assays were performed showing that the relative mtDNA copy number was decreased in the mnd2 mice of 25 days compared with those wide-type mice of the same age. n=3–4 per group, *P<0.05; one-way ANOVA. (e) Transmission electron microscopy was performed showing an obviously decreased proliferation of mitochondria in striatum from 25-day-old mnd2 mice when compared with that in the wide-type mice. The arrow indicates normal mitochondria, arrowhead indicates damage mitochondria and N indicates nucleus. (f) Quantitative morphometric measurements of the mitochondrial density were based on the analysis of Electron micrographs (five sections from every animal and three animals per group). The magnification is × 5000. Densitometric analyses were quantified by one-way ANOVA, *P<0.05
Omi regulates PGC-1α levels
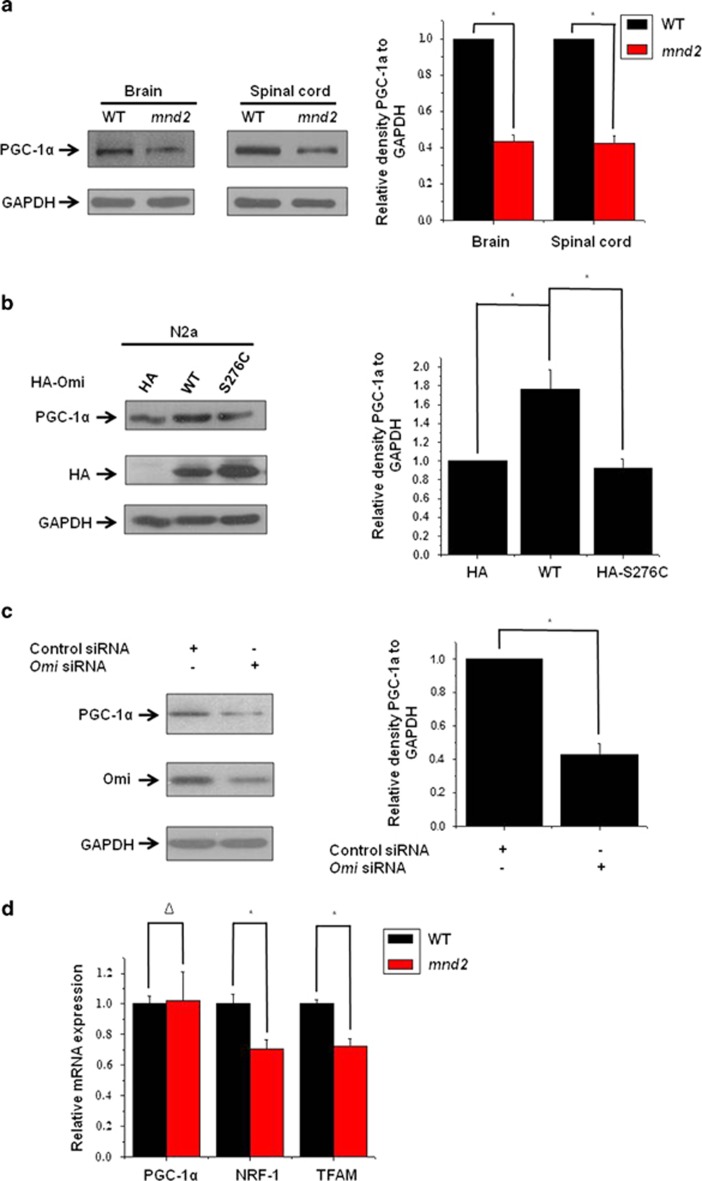
As mitochondrial components and density were decreased in mnd2 mice compared with those in wide-type mice, we wonder that whether mitochondrial biogenesis is decreased in mnd2 mice. It was reported that PGC-1α has a key role in the mitochondrial biogenesis.26, 27 We, therefore, examined the levels of PGC-1α in mnd2 mice. The abundance of PGC-1α was significantly decreased in mnd2 mouse brain and spinal cord (Figure 2a), suggesting that the loss of protease activity of Omi may be associated with the reduction of PGC-1α. We next transfected N2a cells with hemagglutinin (HA)-tagged full-length wild-type or S276C mutant Omi. The overexpression of wide-type Omi resulted in an increased abundance of PGC-1α, whereas the overexpression of S276C Omi did not change PGC-1α abundance (Figure 2b). Moreover, in Omi-knockdown SH-SY5Y cells (Figure 2c) and N2a cells (Supplementary Figure S1), the abundance of PGC-1α was also decreased, consistent with the data from mnd2 mice. Besides, immunoprecipitation assays showed that endogenous Omi did not interact with endogenous PGC-1α in the brain of mice (Supplementary Figure S2), suggesting no direct interaction between Omi and PGC-1α. We further examined two PGC-1α target genes, nuclear respiratory factor-1 (NRF-1) and mitochondrial transcription factor A (TFAM), which are important factors for mitochondrial biogenesis. We found that the levels of NRF-1 and TFAM mRNA were reduced in mnd2 mouse brain compared with those in wide-type mouse brain (Figure 2d). In addition, we found no alteration in the levels of PGC-1α mRNA in mnd2 mouse brain versus wide-type mouse brain (Figure 2d), suggesting that Omi may regulate PGC-1α abundance at protein levels, and that the regulation depends on Omi protease activity. The above data suggest that Omi regulates mitochondrial biogenesis.
Figure 2.
Omi protease activity regulates PGC-1α levels. (a) Western blotting analysis was performed showing that PGC-1α protein abundances were lower in the brains and spinal cord of mnd2 mice of 25 days compared with those in wild-type mice of the same age. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (b) Western blotting analysis was performed showing that HA-tag full-length Omi, but not HA alone or HA-tagged full-length S276C mutant Omi, increased PGC-1α protein abundance. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (c) Western blotting analysis was performed, showing that knockdown of Omi decreases PGC-1α protein abundance in SH-SY5Y cells. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (d) Real-time RT-qPCR assays were performed showing that the mRNA levels of NRF-1 and TFAM were decreased and that PGC-1α is not altered in the mnd2 mice of 25 days compared with wide-type mice of the same age. n=3–4 per group, *P<0.05; Δ, no significance; one-way ANOVA
Omi represses GSK3β expression
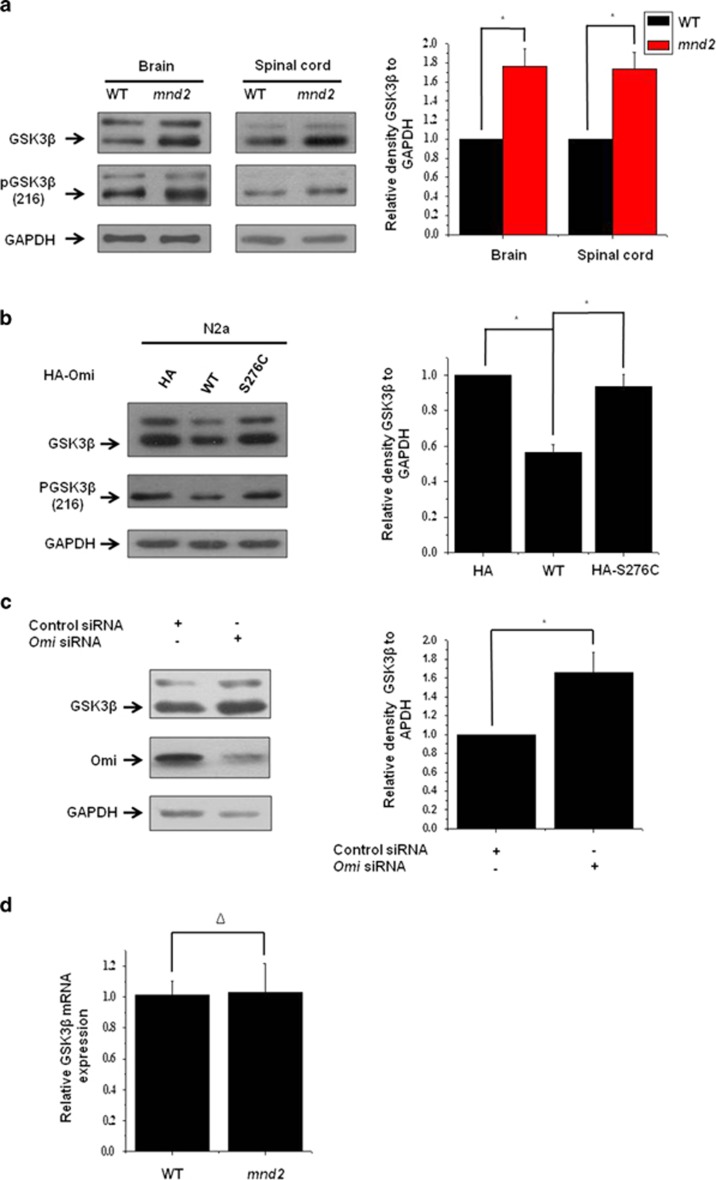
As we found that PGC-1α is decreased in mnd2 mice, we wonder whether some factors that regulate PGC-1α are changed in mnd2 mice. We therefore examined regulators that influence PCG-1α abundance or activity, including p38, GSK3β, p70s6k (ribosomal protein S6 kinase), AKT, pAMPK (AMP-activated protein kinase), Sirtuin-1 (SIRT-1) and Cdc 4 (Supplementary Figures S3A and B). Within them, GSK3β, which can phosphorylate PGC-1α and promote its ubiquitin-mediated degradation (Supplementary Figures S4A and B),28, 29 was upregulated significantly in mnd2 mouse brain and spinal cord compared with that in wide-type mice (Figure 3a). We next investigated whether Omi downregulates GSK3β in vitro. In N2a cells, the overexpression of wide-type full-length Omi, but not S276C mutant Omi, reduced the endogenous GSK3β abundance (Figure 3b), as well as enhanced green fluorescent protein (EGFP)-, Flag- and HA-tagged GSK3β (Supplementary Figures S5A, B and C), whereas knockdown of Omi increased the abundances of GSK3β in SH-SY5Y cells (Figure 3c) and N2a cells (Supplementary Figure S1). In addition, we examined the mRNA levels of GSK3β in brains of mnd2 mice and found that there was no difference between mnd2 mice and control mice (Figure 3d).
Figure 3.
Omi represses GSK3β expression. (a) Western blotting analysis was performed, showing that GSK3β protein abundances were increased in the brains and spinal cord of mnd2 mice of 25 days compared with those in wild-type mice of the same asge. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (b) Western blotting analysis was performed showing that HA-tag full-length Omi, but not HA alone or HA-tagged full-length S276C mutant Omi, decreased GSK3β protein abundance. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (c) Western blotting analysis was performed showing that the knockdown of Omi increased GSK3β protein abundance in SH-SY5Y cells. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (d) Real-time RT-qPCR assays were performed showing that the mRNA levels of GSK3β were not altered in the mnd2 mice of 25 days compared with those in wide-type mice of the same age. n=3–4 per group, Δ, no significance; one-way ANOVA
GSK3β is the substrate of Omi
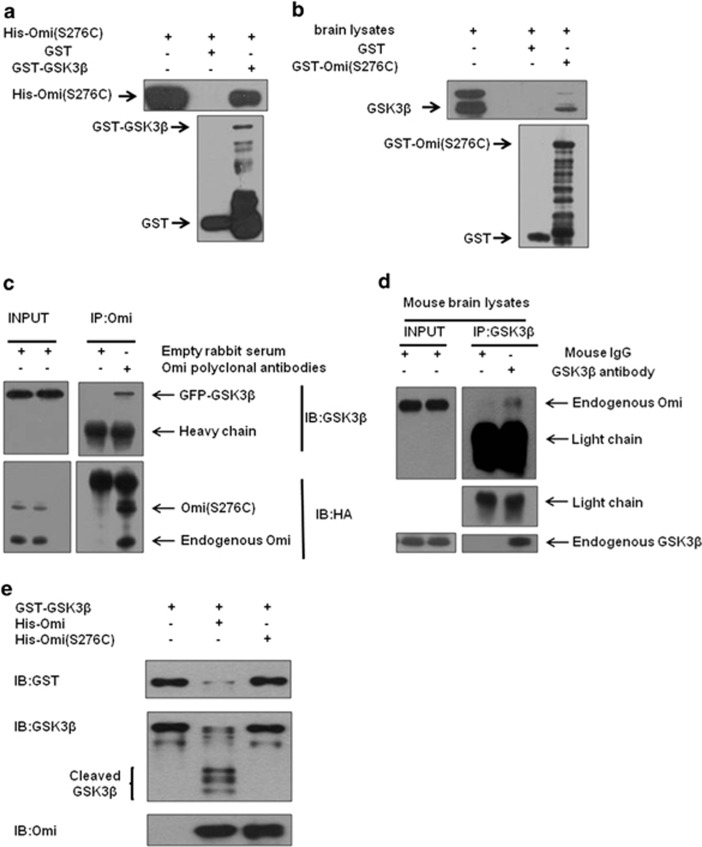
As the increase of GSK3β abundace in both mnd2 mouse brain and Omi-knockdown cells is associated with Omi protease activity and the mRNA levels of GSK3β is not changed in mnd2 mice, we wonder whether GSK3β is a substrate of Omi. First, we examine the subcellular localization of GSK3β and Omi in SH-SY5Y and N2a cells. Subcellular fractionation assay showed that GSK3β was localized in both cytosolic and mitochondrial fractions, and Omi was localized in mitochondrial fraction (Supplementary Figure S6), suggesting that GSK3β and Omi may interact in mitochondria physiologically. Next, we examined the interactions between Omi and GSK3β. Glutathione S-transferase (GST) pulldown assays showed that GST-GSK3β pulled His-tagged Omi (His-Omi) down, whereas GST alone did not (Figure 4a). Besides, GST-S276C mutant Omi pulled down endogenous GSK3β from brain lysates, whereas GST alone did not (Figure 4b). We further performed immunoprecipitation assays using N2a cells, in which HA-tagged full-length S276C mutant Omi and EGFP-GSK3β were co-transfected, to further confirm the interactions between Omi and GSK3β. As shown in Figure 4c, EGFP-GSK3β was coimmunoprecipitated with Omi when endogenous Omi or HA-Omi was precipitated with Omi-specific antibodies. These data indicate that Omi interacts with GSK3β directly in vitro. Moreover, endogenous Omi was coimmunoprecipitated with endogenous GSK3β after endogenous GSK3β was immunoprecipitated from mnd2 mouse brain lysates with a GSK3β-specific antibody (Figure 4d), further suggesting that Omi interacts with GSK3β under normal physiological conditions. Last, in vitro cleavage assay was performed to identify whether GSK3β is a substrate of Omi directly. The results showed that GST-GSK3β was cleaved by wide-type Omi but not by S276C Omi (Figure 4e), whereas GST alone was not cleaved by Omi (Supplementary Figure S7A). Meanwhile, the Omi substrate β-casein was cleaved by the wild-type Omi, but not S276C Omi, which demonstrated a protease activity of Omi (Supplementary Figure S7B). Taken together, these data indicate that GSK3β is a substrate of Omi.
Figure 4.
GSK3β is a substrate of Omi. (a) In vitro pulldown assays were performed showing that GST-GSK3β but not GST alone, interacted with His-Omi. Three independent experiments were performed. (b) The semi in vivo pulldown assays were performed showing that GST-S276C mutant Omi but not GST alone, pulled down endogenous GSK3β from brain lysates. Three independent experiments were performed. (c) Immunoprecipitation assays were performed showing that Omi interacted with GSK3β in N2a cells. Three independent experiments were performed. (d) Immunoprecipitation assays showed that endogenous Omi interacted with endogenous GSK3β in the brain of mice. Three independent experiments were performed. (e) In vitro cleavage assays showed that GST-GSK3β was cleaved by His-Omi, but not by His-Omi S276C. In vitro purified GST-GSK3β was incubated with WT or protease-inactive S276C Omi for 60 min in protease buffer at 37 °C. The incubated mixtures were subjected to western blotting analysis with an anti-GST or anti-GSK3β antibody. The brace indicates proteolytic fragments. Three independent experiments were performed
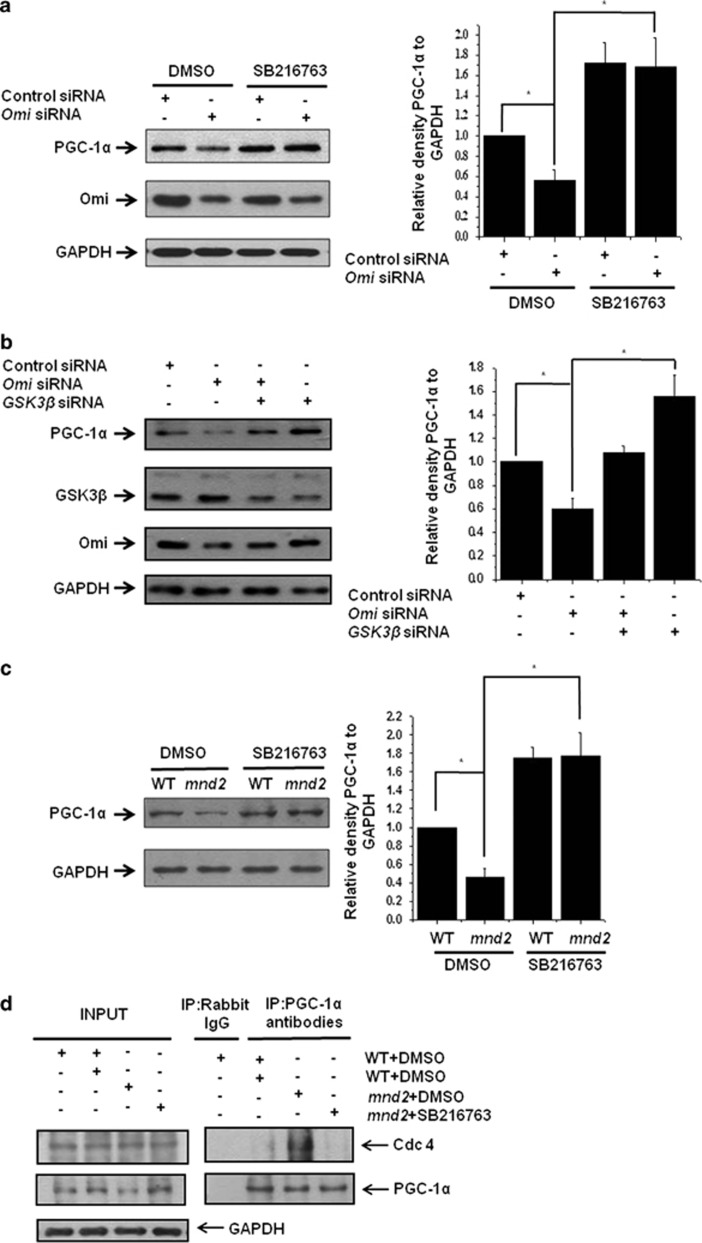
Regulation of PGC-1α by Omi is mediated by GSK3β
We already found that Omi decreases PGC-1α and cleaves GSK3β. As GSK3β is a kinase involved in PGC-1α degradation, we wonder whether GSK3β mediates the downregulation of PGC-1α by Omi. We therefore examined PGC-1α levels, in the presence or absence of a GSK3β specific inhibitor SB216763. In SH-SY5Y cells in which Omi was knocked down, a significant decrease in PGC-1α abundance was observed; however, the effect of Omi knockdown on PGC-1α abundance was blocked by SB216763 (Figure 5a). Similar results were observed in mnd2 mice after SB216763 treatment (Figure 5b). Besides, the decreased abundance of PGC-1α caused by Omi knockdown can also be blocked by the knockdown of GSK3β (Figure 5c). Moreover, immunoprecipitation assays showed that the interaction of PGC-1α and Cdc4, a component of SCF-Cdc4 E3 ligase that regulates PGC-1α for the ubiquitin-mediated degradation, was significantly increased in dimethylsulfoxide (DMSO)-treated mnd2 mice as compared with wild-type mice; however, the interaction of PGC-1α and Cdc4 was decreased in mnd2 mice after SB216763 treatment (Figure 5d), suggesting that Omi protease activity influences the interaction between PGC-1α and its associated SCF-Cdc4 E3 ligase complex in UPS pathway and that GSK3β affects the interaction, evidenced by the data obtained from the usage of GSK3β inhibitor. Thus, our data suggest that GSK3β mediates Omi-regulated PGC-1α abundance.
Figure 5.
Omi regulates PGC-1α through GSK3β. (a) Western blotting analysis was performed showing that the knockdown of Omi decreased PGC-1α protein abundance; however, the effects of Omi knockdown were blocked by treatment with SB216763 (5 μM) for 24 h. SHSY-5Y cells were transfected with si-NC or si-Omi, and 24 h after transfection, cells were treated with DMSO or 5 μM SB216763 for another 24 h. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (b) Western blotting analysis was performed showing that the knockdown of Omi decreased PGC-1α protein abundance; however, PGC-1α protein abundances were restored when GSK3β was also knockdown. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (c) Western blotting analysis was performed showing that PGC-1α protein abundances were decreased in the brains in mnd2 mice; however, PGC-1α protein abundances were increased significantly in SB216763-treated mnd2 mice of 25 days compared with DMSO-treated mnd2 mice. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (d) Immunoprecipitation assays showed that the interaction of PGC-1α and Cdc4 was significantly increased in DMSO-treated mnd2 mice as compared with wild-type mice; however, the interaction of PGC-1α and Cdc4 was decreased in mnd2 mice after SB216763 treatment
Inhibition of GSK3β and overexpression of PGC-1α restore mitochondrial biogenesis influenced by Omi
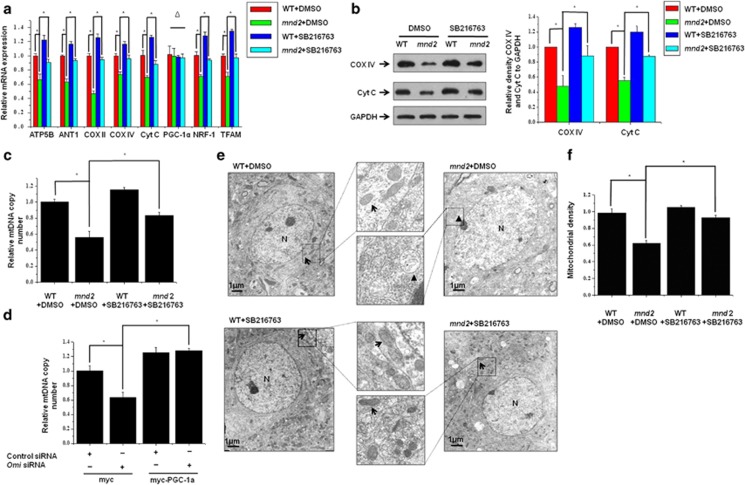
As GSK3β is effective on PGC-1α abundance at the downstream of Omi, we wonder whether the inhibition of GSK3β is able to restore mitochondrial biogenesis in vivo and in vitro. We therefore treated mnd2 mice with GSK3β inhibitor SB216763 and detected the state of mitochondrial biogenesis. We found that the mRNA levels of ATP5B, ANT1, COX II, COX IV, CytC, NRF-1α and TFAM were significantly increased in mnd2 mice (Figure 6a). Consistence with RT-qPCR analyses, the protein levels of COX IV and Cyt C were also restored significantly (Figure 6b). Further, the inhibition of GSK3β significantly increased mtDNA in mnd2 mice (Figure 6c). Similar results were obtained in PGC-1α-overexpressing N2a cells, in which Omi was knocked down (Figure 6d). Moreover, transmission electron microscopic analyses showed that the mitochondria number was increased significantly in striatum in mnd2 mice after a treatment with SB216763 as compared with that in mnd2 mice without SB216763 treatment (Figure 6e). Quantitative morphometry of transmission electron micrographs confirmed that the mitochondrial density was significantly recovered in mnd2 mice treatment with SB216763 than that in mnd2 mice without SB216763 treatment (Figure 6f). Thus, our data suggest that inhibition of GSK3β and overexpression of PGC-1α significantly restore mitochondrial biogenesis that is influenced by Omi deficiency.
Figure 6.
Inhibition of GSK3β and overexpression of PGC-1α restore mitochondrial biogenesis influenced by Omi. (a) Real-time RT-qPCR assays were performed, showing that the mRNA levels of ATP5B, ANT1, COX II, COX IV, CytC, NRF-1 and TFAM were decreased; however, the mRNA was increased significantly in SB216763-treated mnd2 mice of 25 days compared with DMSO-treated mnd2 mice. The mRNA levels of PGC-1α was not changed in SB216763 or DMSO-treated mnd2 mice. n=3–4 per group, *P<0.05; Δ means no significance; one-way ANOVA. (b) Western blotting analysis was performed showing that COX IV and Cyt C protein abundances were decreased in the brains in mnd2 mice; however, COX IV and Cyt C protein abundances were increased significantly in SB216763-treated mnd2 mice of 25 days compared with DMSO-treated mnd2 mice. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (c) Real-time RT-qPCR assays show that mtDNA copy number was decreased in mnd2 mice; however, the mtDNA copy number was increased significantly in SB216763-treated mnd2 mice of 25 days compared with DMSO-treated mnd2 mice. n=4 per group,*P<0.05; one-way ANOVA. (d) Real-time RT-qPCR assays were performed, showing that knockdown of Omi reduced expression of mtDNA copy number; however, the effect of Omi knockdown was rescued by overexpressing PGC-1α in N2a cell. Densitometric analyses from three independent experiments were quantified by one-way ANOVA, *P<0.05. (e) Transmission electron microscopy was performed showing an obviously decreased proliferation of mitochondria in striatum from mnd2 mice; however, the effects were rescued in SB216763-treated mnd2 mice of 25 days compared with DMSO-treated mnd2 mice. The arrow indicates normal mitochondria, arrowhead indicates damage mitochondria and N indicates nucleus. (f) Quantitative morphometric measurements of the mitochondrial density were based on analysis of electron micrographs (five sections from every animal and three animals per group). The magnification is × 5000. Densitometric analyses were quantified by one-way ANOVA, *P<0.05
Inhibition of GSK3β improves movement ability of mnd2 mice
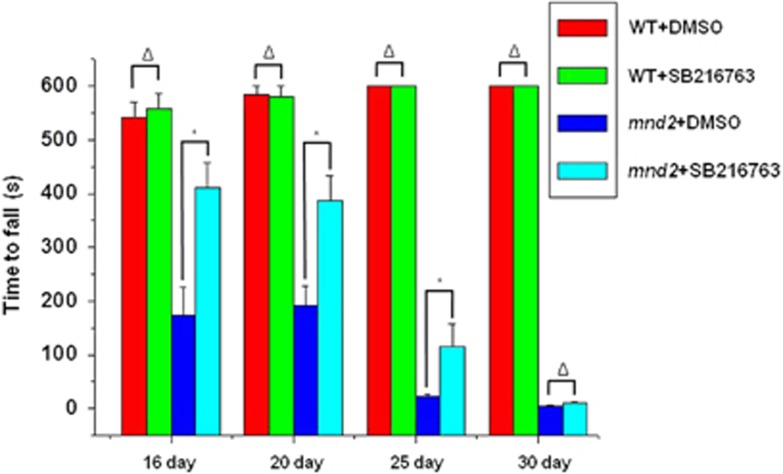
As inhibition of GSK3β is able to restore mitochondrial biogenesis influenced by Omi in vivo and in vitro, we wonder whether there is a functional improvement in mnd2 mice after SB216763 treatment. We therefore performed the rotarod test, which is widely used to evaluate the overall motor deficits in rodent models of PD and others.30 We found that the performance of mnd2 mice was severely deteriorated compared with wide-type mice; however, the performance was significantly improved when the mnd2 mice was administrated with SB216763 (Figure 7).
Figure 7.
Inhibition of GSK3β improves movement ability of mnd2 mice. The rotarod test showed that the performance of mnd2 mice was severely deteriorated compared with wide-type mice treated with DMSO; however, the performance was significantly improved in SB216763-treated mnd2 mice of 16, 20, 25 and 30 days old compared with DMSO-treated mnd2 mice. There was no significant difference of wide-type mice treated with DMSO or SB216763. n=10 per group, *P<0.05; Δ, no significance; one-way ANOVA
Discussion
In this study, we demonstrated that Omi regulates mitochondrial biogenesis, which may involve in Omi protease-deficient-induced neurodegeneration. Omi protease-deficient mnd2 mice present decreases of mtDNA and mitochondrial density in brain, with a reduced amount of the essential components COX IV and Cyt C of the mitochondrial respiratory chain, as well as other components. The mitochondrial biogenesis factor PGC-1α is downregulated and the kinase GSK3β that promotes PGC-1α degradation is increased in mnd2 mice. Beisdes, GSK3β inhibitor SB216763 significantly improved the movement ability of mnd2 mice. Thus, our findings suggest that Omi protease activity is involved in the regulation of mitochondrial biogenesis and the deficiency of mitochondria biogenesis may be associated with Omi protease-deficient-induced neurodegeneration.
Mitochondrial biogenesis is a complex process that requires the synthesis, import and incorporation of proteins, lipids and mtDNA to the existing mitochondrial reticulum, and it involves the coordinated expression of mitochondrial and nuclear genes.31 A growing body of evidence supports the hypothesis that the abnormality of mitochondrial biogenesis is tightly associated with neurodegenerative diseases, including PD.2, 32, 33 mtDNA depletion may occur in neurons in sporadic PD brains, which reflects an impairment of mitochondrial biogenesis.34 The Tfam-knockout mice show reduced mtDNA level and respiratory chain deficiency in the midbrain DA neurons, with adult onset of slowly progressive typical parkinsonian features, also supporting that the disturbance of mtDNA integrity or mitochondrial function may underlie neurodegenerative disorders.35 Normally, mitochondrial biogenesis and function are dynamically regulated to adapt to the energetic and metabolic demands in response to physiological signals.32 In many neurodegenerative diseases, brain mitochondrial function is impaired, and mitochondrial biogenesis is increased to compensate for functional declines. Neurodegeneration will occur in the case of insufficient mitochondrial biogenesis to balance the functional declines.
In our study, we identified that Omi regulates mitochondrial biogenesis. We found that: first, the expression of the components of the mitochondrial respiration chain and oxidant metabolism is downregulated from transcriptional level; second, the amount of mtDNA is also decreased; third, the mitochondrial density is significantly reduced in mnd2 mouse brain; last, the abundance of PGC-1α is downregulated in the brain and spinal cord of mnd2 mice.
PGC-1α, as the central regulator of mitochondrial biogenesis and function, integrates diverse physiological signals that impact mitochondrial biogenesis and coordinates multiple transcription factors, such as nuclear receptors such as NRF-1, NRF-2 and the orphan nuclear hormone receptors peroxisome proliferator–activated receptor α, PPARδ, estrogen-related receptors and thyroid hormone receptor, to induce broad sets of mitochondrial genes.26, 36 PGC-1α is highly expressed in tissues with high-energy demands and abundant mitochondria, such as brown adipose tissue, heart muscle, slow-twitch skeletal muscle and brain.36, 37 The association of PGC-1α with neurodegenerative disorders has been addressed in many studies: on one hand, the increased risk of the development of both PD and HD is associated with single nucleotide polymorphisms in the gene encoding PGC-1;37, 38 on the other hand, the abundance of PGC-1α was decreased in AD, HD and PD patients.37, 39, 40 Recently, several studies have suggested that an impairment of PGC-1α function is involved in PD pathogenesis. For example, a gene set enrichment analysis shows a coordinate downregulation of 425 PGC-1α regulated genes in PD patients.41 Furthermore, an activation of PGC-1α can protect DA neuron from the damage of mutant α-synuclein or the pesticide rotenone or MPTP in primary neuron models.42 In addition, PD-related gene product parkin regulates the levels of PGC-1α and enables PGC-1α-dependent gene expression.40
Interestingly, PGC-1α-knockout mice display impaired mitochondrial function such as diminished mitochondrial number and respiratory capacity in muscle and brown fat, with movement disorder, including exaggerated startle responses, dystonic posturing, stimulus-induced myoclonus and frequent limb clasping, accompanied by neurodegeneration, particularly in the striatum, which show some similarities to the mnd2 mice.18, 19, 43, 44 As PGC-1α abundance is decreased in mnd2 mice, it is possible that the Omi protease-deficient-induced neurodegeneration may be associated with the decreased abundance of PGC-1α. Besides, it was reported that PGC-1α is a potent suppressor of reactive oxygen species (ROS) and induces the production of ROS-scavenging enzymes. Thus, the decreased expression of PGC-1α caused by Omi deficiency may also result in the insufficiency to defend ROS and neurodegeneration.42 On the other hand, as a protease, Omi involves many pathways and regulates many critical cellular processes, such as autophagy and neuroinflammation;45, 46 however, most functions regulated by PGC-1α are not associated with cell death directly. It is possible that besides PGC-1α, other factors and pathways may be involved in the pathogenesis of mnd2 mice, so that the phenotype by loss of Omi activity is much dramatic and the life span of PGC-1α-knockout mice is longer than mnd2 mice.
GSK3β is an important serine/threonine kinase to regulate PGC-1α degradation.28, 29 GSK3β reduces PGC-1α levels by phosphorylating PGC-1α and subsequently stimulating PGC-1α degradation by the ubiquitin–proteasomal system. An inhibition of GSK3β promotes mitochondrial biogenesis during ischemia cerebral injury.47 In our study, we found that the interaction of PGC-1α and the component of E3 ubiquitin ligase Cdc4 was significantly increased in mnd2 mice and the interaction of PGC-1α and Cdc4 was decreased in mnd2 mice after SB216763 treatment. We also demonstrated that the regulation of PGC-1α by Omi is mediated by GSK3β. Omi cleaves GSK3β to reduce PGC-1α degradation and maintain mitochondrial biogenesis. Increases of GSK3β and decreases of PGC-1α abundance are presented in Omi knockdown SH-SY5Y cells and mnd2 mice, whereas a restoration of PGC-1α abundance and mitochondrial biogenesis occurs after a treatment with the GSK3β inhibitor SB216763.
The function of GSK3β in PD has been documented in several studies. The polymorphisms in GSK3β, which affect its transcription and splicing, are associated with disease risk in PD.48 Besides, GSK3β activity is elevated in the striatum in postmortem tissue from PD patients and mouse models of PD,49, 50 and GSK3β levels are increased in peripheral blood lymphocytes in PD patients.50, 51 In addition, the abnormal activation of GSK3β is associated with several psychiatric disorders and AD, and the overexpression of GSK3β results in neurodegeneration in mice.52, 53 Thus, abnormal increases of GSK3β activity or abundance are able to induce PGC-1α degradation, subsequently leading to neurodegeneration. These studies suggest that it is essential for neurons to ensure GSK3β activity in a reasonable level for maintenance of cellular homeostasis. GSK3β has been reported to be located in cytosol, nucleus and mitochondria (Supplementary Figure S4),54, 55 and GSK3β activity is mainly regulated through phosphorylation by kinases, such as Akt, PKC and P70S6K, or interacted with other proteins, such as and DISC 1.56 In our study, we identified that the kinase GSK3β is a novel substrate of Omi, and Omi cleaves GSK3β to repress its levels. The abundance of GSK3β and the phosphorylation on tyrosine (Tyr) residues (Tyr 216) are upregulated in the brain and spinal cord of mnd2 mice, as well as in Omi-knockdown SH-SY5Y and N2a cells. Overexpression of wide-type Omi, but not of S276C Omi, substantially represses the total level of GSK3β. In addition, in vitro cleavage assays show that Omi cleaves GSK3β directly. Furthermore, GSK3β inhibitor SB216763 significantly improves the movement ability of mnd2 mice indicated by rotarod test. Thus, as a substrate of Omi, GSK3β that is the abnormally high expressed due to Omi protease deficiency may be associated with the pathogenesis in mnd2 mice.
In summary, our study identified that GSK3β is a novel substrate of Omi. Loss of Omi protease activity results in an abnormal increase of GSK3β, leading to the degradation of PGC-1α, which causes an impairment of mitochondrial biogenesis and induces neurodegeneration.
Materials and Methods
Animal experiments
The mnd2 mice and age-matched wild-type mice were obtained from crossing paired heterozygotes (HtrA2mnd2, C57BL/6J) that were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Identifications of the genotypes of the offspring mice were performed according to the protocol provided by JAX mice (The Jackson Laboratory). Mice were maintained in a specific pathogen-free environment in compliance with national regulations. All animal experiments were approved by the animal welfare advisory committee of the University of Science and Technology of China. The mice were given an intraperitoneal injection of vehicle or SB216763 (2 mg/kg), suspended in PBS every 2 days for 15 or 20 days, from the time they were 10 days old.
Cell culture, transfections and drug treatments
SH-SY5Y cells, mouse neuroblastoma (N2a) cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY, USA) F12 or DMEM containing 10% fetal bovine serum (Gibco) with penicillin (100 mg/ml) and streptomycin (100 mg/ml). Transfections were performed using Lipofectamine 2000 reagent (Invitrogen, La Jolla, CA, USA) according to the manufacturer's instructions. The cells were lysated and subjected to western blotting analysis 48 h after transfection. For the SB216763 treatment (Sigma-Aldrich, St. Louis, MO, USA), cells were treated with 5 mM SB216763, and after 24 h incubation, cell lysates were subjected to western blotting analysis.
Plasmids
The myc-CMV-PGC-1α was kindly provided by Dr Chang Liu (Nanjing Normal University, China). The Omi-related plasmids were previously described.45 Full-length human GSK3β complementary DNA (cDNA) was amplified by PCR from an adult human brain cDNA library with the primers 5′-AACTGCAGACCATGTCAGGGCGGCCCAGAA-3′ and 5′-TGCTCTAGAGGTGGAGTTGGAAGCTGATGC-3′ and inserted in-frame into the pGEX-5x-1 vector at the Bgl II/ECOR I sites or into the pEGFP-C2 vector at the BamH I/ECOR I sites. The C-terminal HA-tagged and Flag-tagged GSK3β was created by subcloning the PCR product amplified with primers 5′-AACTGCAGACCATGTCAGGGCGGCCCAGAA-3′ and 5′-TGCTCTAGAGGTGGAGTTGGAAGCTGATGC-3′ or 5′-TAAAGATCTGATGTCAGGGCGGCCCAGAAC-3′and 5′-TAAGTCGACGGTGGAGTTGGAAGCTGATGC-3′ into pKH3-HA vector or p3x-Flag-CMV vector at sites Pst I/Xba I or Bgl II/Sal I.
RNA interference
The oligonucleotides were transfected with Oligofectamine (Invitrogen) according to the manufacturer's instructions. The oligonucleotide sequences targeting to Omi (si-Omi) and the negative control small interfering RNA (si-NC) were previously described,45 and the oligonucleotide sequences targeting to GSK3β are shown as follows: sense 1, 5′-CTGCATTTATCGTTAACCTAATTdTdT-3′ and antisense 1, 5′-AATTAGGTTAACGATAAATGCAGdTdT-3′ sense 2, 5′-CAGGGCAGTGATTCTGTTCAATTdTdT-3′ and antisense 2, 5′-AATTGAACAGAATCACACTGCCCTGdTdT-3′.
Western blotting analysis and antibodies
Tissue homogenate and cell extracts were lysed in the 1 × RIPA lysis buffer (25 mM tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40 and 1% sodium deoxycholate) in the presence of a protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Nearly 20 μg of cell lysate was resolved by SDS–PAGE and transferred onto a PVDF membrane (Millipore, Bedford, MA, USA). Western blotting analysis was carried out with the following primary antibodies: anti-α-Tubulin (CP06, Calbiochem, San Diego, CA, USA), anti-AKT1 (sc-1618, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-AMPKα (23A3, Cell Signaling, Beverly, MA, USA), anti-Cdc4 (ab171961, Abcam, Cambridge, MA, USA), anti-COX IV (S0553, Epitomics, Burlingame, CA, USA), anti-CytC (3895-1, Epitomics), anti-Flag (F3165, Sigma-Aldrich), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (MAB374, CHEMICON, Temecula, CA, USA), anti-GFP (sc-9996, Santa Cruz Biotechnology), anti-GSK-3β (NBP1-47470, Novus, Littleton, CO, USA or sc-7291, Santa Cruz Biotechnology), anti-GST (sc-138, Santa Cruz Biotechnology), anti-p38 (sc-7972, Santa Cruz Biotechnology), anti-HA (sc-7392, Santa Cruz Biotechnology), anti-His (M30111, Abmart, Shanghai, China), anti-Omi (AF1458, R&D Systems, Minneapolis, MN, USA or prepared by our laboratory), anti-p70s6k (#3204-1, Epitomics), anti-PGC-1α (ST1202, Calbiochem, or sc-13067, Santa Cruz Biotechnology), anti-Phospho-AMPKα (Thr 172; 40H9, Cell Signaling), anti-Phospho-AKT1 (#2118-1, Epitomics), anti-Phospho-GSK3α/β (pY279/pY261) (#2309-1-1, Epitomics), anti-Phospho-GSK3β (Ser9; AG753, Biyuntian, Shanghai, China), anti-Phospho-p38 MAPK (pT180/pY182; 1229-1, Epitomics), anti-Phospho-p70s6k (#1494-1, Epitomics), anti-SIRT-1 (13161-AP, Proteintech, Chicago, IL, USA), anti-TOM70 (14528-1-AP, Proteintech), anti-Ub (sc-8017, Santa Cruz Biotechnology). The anti-mouse and anti-rabbit secondary antibodies coupled to horseradish peroxidase were from Amersham Pharmacia Biotech (Peapack, NJ, USA). The proteins were viewed with an ECL detection kit (Amersham Pharmacia Biotech, Arlington Heights, IL, USA).
GST pulldown assays
For in vitro pulldown assay, an aliquot containing 20 mg/μg of GST or GST- GSK3β that was expressed in E. coli strain JM109 was incubated with 30 ml of Glutathione Sepharose 4B (Amersham Biosciences, Tokyo, Japan) for 30 min at 4 °C. GST or GST-GSK-3β bound to Sepharose was incubated with 50 mg of His-Omi protein from the supernatants of E. coli crude extracts containing His-tagged proteins in 0.25 ml of HNTG buffer (20 mM Hepes-KOH, pH 7.5, 0.1% Triton X-100, 100 mM NaCl and 10% glycerol) for 1 h at 4 °C. For semi in vivo pulldown assay, an aliquot containing 20 μg of GST or GST-S276C mutant Omi was incubated with 30 ml of Glutathione Sepharose 4B for 30 min at 4 °C. GST or GST-Omi bound to Sepharose was incubated with mouse brain tissue homogenates in 0.25 ml of HNTG buffer for 1 h at 4 °C. After incubation, the beads were washed six times with 1 ml of HNTG buffer each time to remove unbound protein. Bound protein was eluted by boiling in SDS–PAGE sample buffer, and then subjected to western blotting analysis.
Immunoprecipitation
Homogenates from mnd2 mouse brains and cells that were transfected with the indicated plasmids were lysed in TSPI buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM sodium chloride, 1 mM EDTA and 1% NP-40 supplemented with a complete mini protease inhibitor cocktail (Roche).
The lysate was incubated with anti-Omi antibodies or normal rabbit immunoglobulin G overnight at 4 °C. After incubation, protein G-Sepharose (Roche) was used for precipitations. The beads were washed with TSPI buffer six times and were eluted with SDS sample buffer for western blotting analysis.
In vitro proteolytic cleavage assay
GST, GST-GSK3β or β-casein (Sigma-Aldrich) was incubated with 3 μg of 6 × His-tagged mature Omi in protease cleavage buffer (50 mM tris-HCl, pH 7.6, 1 mM dithiothreitol) for 60 min at 37 °C. The reactions were stopped by the addition of 2 × SDS loading buffer and boiled for 10 min. The samples were subjected to western blotting analyses with an anti-GST or anti-GSK3β antibody.
RNA isolation and real-time PCR
Total RNA was extracted from brain using Trizol (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized using a TransScript First-Strand cDNA Synthesis Kit (Takara, Shiga, Japan). Real-time PCR was performed with SYBR Green Real-Time PCR Master Mix (Takara) reagents using a BioRad MiniOpticon System. Relative gene expressions were calculated by the 2−ΔΔCt method. Primers for RT-qPCR used here are shown as follows: PGC-1α forward 5′-AGCCGTGACCACTGACAACGAG-3′, reverse 5′-GCTGCATGGTTCTGAGTGCTAAG-3′ PGC-1β forward 5′-GGCAGGTTCAACCCCGA-3′, reverse 5′-CTTGCTAACATCACAGAGGATATCTTG-3′ GSK3β forward 5′-TAAGGAAGGAAAAGGTGAtTCGA-3′, reverse 5′-CCAAAAGCTGAAGGCTGCTG-3′ NRF-1 forward 5′-GTTGGTACAGGGGCAACAGT-3′, reverse 5′-TCGTCTGGATGGTCATTTCA-3′ TFAM forward 5′-CTGATGGGTATGGAGAAGGAGG-3′, reverse 5′-CCAACTTCAGCCATCTGCTCTTC-3′ UCP2 forward 5′-ACTTTCCCTCTGGATACCGC-3′, reverse 5′-ACGGAGGCAAAGCTCATCTG-3′ ANT1 forward 5′-GTCTCTGTCCAGGGCATCAT-3′, reverse 5′-ACGACGAACAGTGTCAAACG-3′ ATP5B forward 5′-GAGGGATTACCACCCATCCT-3′, reverse 5′-CATGATTCTGCCCAAGGTCT-3′ CYTC forward 5′-CCAAATCTCCACGGTCTGTTC-3′, reverse 5′-ATCAGGGTATCCTCTCCCCAG-3′ COX IV forward 5′-CGAGAGCTTCGCCGAGATGAAC-3′, reverse 5;-GCAGCTTCCAGCATGCCGAGG-3′ COX II forward 5′-ACGAAATCAACAACCCCGTA-3′, reverse 5′-GGCAGAACGACTCGGTTATC-3′ β-actin forward 5′-TGTCCACCTTCCAGCAGATGT-3′, reverse 5′-AGCTCAGTAACAGTCCGCCTAGA-3′.
Analysis of mtDNA
Total genomic DNA was extracted using a GenElute mammalian genomic DNA kit (Sigma-Aldrich) according to the manufacturer's instructions. mNADH1, mCYTB, mNADH4, mATP6 and mCOX II were used as mtDNA markers, and nuclear intron of β-globin was used as a nDNA marker. mTND1, mCYTB, mMTND4, mATP6 and mCOX II primers were designed from regions of mtDNA that are not found in nuclear-encoded mitochondrial pseudogenes, with the primers which were described elsewhere as following:57, 58, 59 mNADH1 forward 5′-TCTGCCAGCCTGACCCATAG-3′, reverse 5′-CCGGCTGCGTATTCTACGTT-3′ mCYTB forward 5′-TATTCCTTCATGTCGGACGA-3′, reverse 5′-AAATGCTGTGGCTATGACTG-3′ mNADH4 forward 5′-ATTATTATTACCCGATGAGGGAACC-3′, reverse 5′-ATTAAGATGAGGGCAATTAGCAGT-3′ mATP6 forward 5′-AATTACAGGCTTCCGACACAAAC-3′, reverse 5′-TGGAATTAGTGAAATTGGAGTTCCT-3′ mCOX2 forward 5′-GCCGACTAAATCAAGCAACA-3′, reverse 5′- CAATGGGCATAAAGCTATGG-3′ β-globin forward 5′-GAAGCGATTCTAGGGAGCAG-3′, reverse 5′-GGAGCAGCGATTCTGAGTAGA-3′. The quantification of abovementioned product was analyzed after being normalized to β-globin using one-way ANOVA.
Subcellular Fractionation Assay
The cytosolic and mitochondrial fractions from SH-SY5Y and N2a cells were isolated according to the instructions of the Mitochondria Isolation kit for cultured Cells (Beyotime, China). The lysates of fractions were subjected to western blotting analyses. GAPDH was used as the cytosolic marker and TOM70 was used as the mitochondrial marker.
Rotarod test
Mice were detected for their ability to run on a 3-cm diameter rotarod at speed of 10 revolutions per minute (r.p.m.) as previously described with modification.30 To reach a stable performance, mice were trained at speed of 5–10 r.p.m. for 2 days before the first-time test. The tests were performed using mice that were 16, 20, 25 and 30 days old, with a treatment using SB216763 or DMSO. The time that the mice spent on the rotarod before falling was recorded for each trial, and the trial was repeated thrice. To reduce stress and fatigue, there was at least 10 min of rest between trials, and a cut-off time point was set at 600 s.
Electron microscopy
For electron microscopic analysis of the brain, the tissue was prepared as previously described.18 Mnd2 mice that were 25 days old and age-matched wild-type mice were transcardially perfused with 0.1 M phosphate buffer for 6–8 min, followed by 2% formaldehyde/4% glutaraldehyde fixative in 0.1 M phosphate buffer for 6–8 min. Dissected striatum were further fixed with the same fixative for 2–4 h at 4 °C and postfixed with 2% OsO4 in buffer and embedded in Araldite. Ultrathin sections of striatum were cut onto formvar-coated slot grids stained with uranyl acetate and lead citrate and viewed with a JEM-1230 electron microscope. To calculate mitochondrial density, six different fields at the magnification of 5000 for each animal were quantified as described previously.60
Statistical analysis
Densitometric analysis of western blots from three independent experiments was implemented with Adobe Photoshop 7.0 and the data were analyzed with Origin 6.0 (OriginLab, Northampton, MA, USA). Quantitative data are presented as means±S.E.M. Statistical significance was assessed via one-way ANOVA and the criterion of significance was set at P<0.05.
Acknowledgments
We are grateful to Dr. Hyangshuk Rhim (the Catholic University of Korea) for full-length Omi cDNA. This work was supported in part by the National High-tech Research and Development program of China 973-projects (2011CB504102), the National Natural Sciences Foundation of China (Nos. 31330030 and 81371393), National Natural Sciences Foundation of Jiangsu Province (No. BK2013) and Jiangsu Key Laboratory of Translational Research and Therapy for Neuro-Psycho-Diseases (BM2013003).
Glossary
- AD
Alzheimer's disease
- AMPK
the AMP-activated protein kinase
- ANT1
adenine nucleotide translocase-1
- ATP5B
ATP synthase 5 β subunit
- ATP6
ATP synthase 6
- COX II
cytochrome c oxidase subunit II
- COX IV
cytochorome c oxidase subunit IV
- Cyt B
cytochorome B
- Cyt C
cytochrome C
- DA
dopaminergic
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
Dimethylsulfoxide
- EGFP
enhanced green fluorescent protein
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GSK3β
glycogen synthase kinase 3β
- GST
glutathione S-transferase
- HA
hemagglutinin
- HD
Huntington's disease
- HtrA
high-temperature requirement factor A
- mnd2
motor neuron degeneration 2
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- mtDNA
mitochondrial DNA
- NADH1
NADH dehydrogenase 1
- NADH4
NADH dehydrogenase 4
- nDNA
nuclear DNA
- NRF-1
nuclear respiratory factor-1
- p70s6k
ribosomal protein S6 kinase
- PD
Parkinson's disease
- PGC-1α
PPARγ coactivator-1α
- PINK1
PTEN-induced putative kinase 1
- ROS
reactive oxygen species
- RT-qPCR
real-time quantitative PCR
- SIRT-1
Sirtuin-1
- TFAM
mitochondrial transcription factor A
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschellà
Supplementary Material
References
- Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- Van Laar VS, Berman SB. The interplay of neuronal mitochondrial dynamics and bioenergetics: implications for Parkinson's disease. Neurobiol Dis. 2013;51:43–55. doi: 10.1016/j.nbd.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne Stetler R, Leak RK, Gao Y, Chen J. The dynamics of the mitochondrial organelle as a potential therapeutic target. J Cereb Blood Flow Metab. 2013;33:22–32. doi: 10.1038/jcbfm.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat Rev Neurosci. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- Wu Y, Le W, Jankovic J. Preclinical biomarkers of Parkinson disease. Arch Neurol. 2011;68:22–30. doi: 10.1001/archneurol.2010.321. [DOI] [PubMed] [Google Scholar]
- Wang X, Yan MH, Fujioka H, Liu J, Wilson-Delfosse A, Chen SG, et al. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum Mol Genet. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H, et al. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornai F, Schluter OM, Lenzi P, Gesi M, Ruffoli R, Ferrucci M, et al. Parkinson-like syndrome induced by continuous MPTP infusion: convergent roles of the ubiquitin-proteasome system and alpha-synuclein. Proc Natl Acad Sci USA. 2005;102:3413–3418. doi: 10.1073/pnas.0409713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, et al. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem. 2000;267:5699–5710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- Hartkamp J, Carpenter B, Roberts SG. The Wilms' tumor suppressor protein WT1 is processed by the serine protease HtrA2/Omi. Mol Cell. 2010;37:159–171. doi: 10.1016/j.molcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Blink E, Maianski NA, Alnemri ES, Zervos AS, Roos D, Kuijpers TW. Intramitochondrial serine protease activity of Omi/HtrA2 is required for caspase-independent cell death of human neutrophils. Cell Death Differ. 2004;11:937–939. doi: 10.1038/sj.cdd.4401409. [DOI] [PubMed] [Google Scholar]
- Liu MJ, Liu ML, Shen YF, Kim JM, Lee BH, Lee YS, et al. Transgenic mice with neuron-specific overexpression of HtrA2/Omi suggest a neuroprotective role for HtrA2/Omi. Biochem Biophys Res Commun. 2007;362:295–300. doi: 10.1016/j.bbrc.2007.07.118. [DOI] [PubMed] [Google Scholar]
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- Rathke-Hartlieb S, Schlomann U, Heimann P, Meisler MH, Jockusch H, Bartsch JW. Progressive loss of striatal neurons causes motor dysfunction in MND2 mutant mice and is not prevented by Bcl-2. Exp Neurol. 2002;175:87–97. doi: 10.1006/exnr.2002.7868. [DOI] [PubMed] [Google Scholar]
- Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, et al. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum Mol Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- Bogaerts V, Nuytemans K, Reumers J, Pals P, Engelborghs S, Pickut B, et al. Genetic variability in the mitochondrial serine protease HTRA2 contributes to risk for Parkinson disease. Hum Mutat. 2008;29:832–840. doi: 10.1002/humu.20713. [DOI] [PubMed] [Google Scholar]
- Kang S, Louboutin JP, Datta P, Landel CP, Martinez D, Zervos AS, et al. Loss of HtrA2/Omi activity in non-neuronal tissues of adult mice causes premature aging. Cell Death Differ. 2013;20:259–269. doi: 10.1038/cdd.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, et al. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat Cell Biol. 2007;9:1243–1252. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- Inagaki R, Tagawa K, Qi ML, Enokido Y, Ito H, Tamura T, et al. Omi / HtrA2 is relevant to the selective vulnerability of striatal neurons in Huntington's disease. Eur J Neurosci. 2008;28:30–40. doi: 10.1111/j.1460-9568.2008.06323.x. [DOI] [PubMed] [Google Scholar]
- Tain LS, Chowdhury RB, Tao RN, Plun-Favreau H, Moisoi N, Martins LM, et al. Drosophila HtrA2 is dispensable for apoptosis but acts downstream of PINK1 independently from Parkin. Cell Death Differ. 2009;16:1118–1125. doi: 10.1038/cdd.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, et al. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Barger JL, Edwards MG, Braun KH, O'Connor CE, Prolla TA, et al. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen J, Wang L, Zhang L, Lu M, Chopp M. Intracerebral transplantation of bone marrow stromal cells in a 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson's disease. Neurosci Lett. 2001;316:67–70. doi: 10.1016/s0304-3940(01)02384-9. [DOI] [PubMed] [Google Scholar]
- Diaz F, Moraes CT. Mitochondrial biogenesis and turnover. Cell Calcium. 2008;44:24–35. doi: 10.1016/j.ceca.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango IG, Lu J, Rodova M, Lezi E, Crafter AB, Swerdlow RH. Regulation of neuron mitochondrial biogenesis and relevance to brain health. Biochim Biophys Acta. 2010;1802:228–234. doi: 10.1016/j.bbadis.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Lin TK, Liou CW, Chen SD, Chuang YC, Tiao MM, Wang PW, et al. Mitochondrial dysfunction and biogenesis in the pathogenesis of Parkinson's disease. Chang Gung Med J. 2009;32:589–599. [PubMed] [Google Scholar]
- Keeney PM, Dunham LD, Quigley CK, Morton SL, Bergquist KE, Bennett JP., Jr Cybrid models of Parkinson's disease show variable mitochondrial biogenesis and genotype-respiration relationships. Exp Neurol. 2009;220:374–382. doi: 10.1016/j.expneurol.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, et al. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc Natl Acad Sci USA. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C. The biology of PGC-1alpha and its therapeutic potential. Trends Pharmacol Sci. 2009;30:322–329. doi: 10.1016/j.tips.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Jones AW, Yao Z, Vicencio JM, Karkucinska-Wieckowska A, Szabadkai G. PGC-1 family coactivators and cell fate: roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion. 2012;12:86–99. doi: 10.1016/j.mito.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Clark J, Reddy S, Zheng K, Betensky RA, Simon DK. Association of PGC-1alpha polymorphisms with age of onset and risk of Parkinson's disease. BMC Med Genet. 2011;12:69. doi: 10.1186/1471-2350-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Hu Q, Wang H, Man N, Ren H, Wen L, et al. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ. 2010;17:1773–1784. doi: 10.1038/cdd.2010.55. [DOI] [PubMed] [Google Scholar]
- Hu Q, Li B, Xu R, Chen D, Mu C, Fei E, et al. The protease Omi cleaves the mitogen-activated protein kinase kinase MEK1 to inhibit microglial activation. Sci Signal. 2012;5:ra61. doi: 10.1126/scisignal.2002946. [DOI] [PubMed] [Google Scholar]
- Valerio A, Bertolotti P, Delbarba A, Perego C, Dossena M, Ragni M, et al. Glycogen synthase kinase-3 inhibition reduces ischemic cerebral damage, restores impaired mitochondrial biogenesis and prevents ROS production. J Neurochem. 2011;116:1148–1159. doi: 10.1111/j.1471-4159.2011.07171.x. [DOI] [PubMed] [Google Scholar]
- Kwok JB, Hallupp M, Loy CT, Chan DK, Woo J, Mellick GD, et al. GSK3B polymorphisms alter transcription and splicing in Parkinson's disease. Ann Neurol. 2005;58:829–839. doi: 10.1002/ana.20691. [DOI] [PubMed] [Google Scholar]
- Wills J, Jones J, Haggerty T, Duka V, Joyce JN, Sidhu A. Elevated tauopathy and alpha-synuclein pathology in postmortem Parkinson's disease brains with and without dementia. Exp Neurol. 2010;225:210–218. doi: 10.1016/j.expneurol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei P, Ayton S, Bush AI, Adlard PA. GSK-3 in neurodegenerative diseases. Int J Alzheimers Dis. 2011;2011:189246. doi: 10.4061/2011/189246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armentero MT, Sinforiani E, Ghezzi C, Bazzini E, Levandis G, Ambrosi G, et al. Peripheral expression of key regulatory kinases in Alzheimer's disease and Parkinson's disease. Neurobiol Aging. 2011;32:2142–2151. doi: 10.1016/j.neurobiolaging.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Pei JJ, Braak E, Braak H, Grundke-Iqbal I, Iqbal K, Winblad B, et al. Distribution of active glycogen synthase kinase 3beta (GSK-3beta) in brains staged for Alzheimer disease neurofibrillary changes. J Neuropathol Exp Neurol. 1999;58:1010–1019. doi: 10.1097/00005072-199909000-00011. [DOI] [PubMed] [Google Scholar]
- Lucas JJ, Hernandez F, Gomez-Ramos P, Moran MA, Hen R, Avila J. Decreased nuclear beta-catenin, tau hyperphosphorylation and neurodegeneration in GSK-3beta conditional transgenic mice. EMBO J. 2001;20:27–39. doi: 10.1093/emboj/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit-Paitel A, Brau F, Cazareth J, Chabry J. Involvment of cytosolic and mitochondrial GSK-3beta in mitochondrial dysfunction and neuronal cell death of MPTP/MPP-treated neurons. PLoS One. 2009;4:e5491. doi: 10.1371/journal.pone.0005491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Allouche M, Esposti DD, Fanelli E, Boursier C, Henry C, et al. Glycogen synthase kinase 3-mediated voltage-dependent anion channel phosphorylation controls outer mitochondrial membrane permeability during lipid accumulation. Hepatology. 2013;57:93–102. doi: 10.1002/hep.25967. [DOI] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimovsky EV, Ushakova TE, Kudriavtcev AA, Gaziev AI. Alteration of mtDNA copy number, mitochondrial gene expression and extracellular DNA content in mice after irradiation at lethal dose. Radiat Environ Biophys. 2011;50:181–188. doi: 10.1007/s00411-010-0329-6. [DOI] [PubMed] [Google Scholar]
- Strum JC, Shehee R, Virley D, Richardson J, Mattie M, Selley P, et al. Rosiglitazone induces mitochondrial biogenesis in mouse brain. J Alzheimers Dis. 2007;11:45–51. doi: 10.3233/jad-2007-11108. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.