Abstract
The fitness landscape is a powerful metaphor for describing the relationship between genotype and phenotype for a population under selection. However, empirical data as to the topography of fitness landscapes are limited, owing to difficulties in measuring fitness for large numbers of genotypes under any condition. We previously reported a case of reciprocal sign epistasis (RSE), where two mutations individually increased yeast fitness in a glucose-limited environment, but reduced fitness when combined, suggesting the existence of two peaks on the fitness landscape. We sought to determine whether a ridge connected these peaks so that populations founded by one mutant could reach the peak created by the other, avoiding the low-fitness “Valley-of-Death” between them. Sequencing clones after 250 generations of further evolution provided no evidence for such a ridge, but did reveal many presumptive beneficial mutations, adding to a growing body of evidence that clonal interference pervades evolving microbial populations.
Keywords: Evolution, epistasis, yeast, genomics
Introduction
Natural Historians are fond of citing the many ways in which organisms are well-suited to their environments so much so that the adaptationist paradigm holds a seductive power over the scientific imagination [1]. In its most extreme form, that paradigm leads to the conclusion that an organism can be atomized into a set of traits, each of which is optimally adapted to serve a particular function. But every trait, be it the ratio of a beak’s length to its depth or a protein’s kcat/Km, arises from interactions among its component elements, subject to past phylogenetic and current biophysical constraints. Moreover, every trait itself interacts with myriad elements to generate other traits. Thus, the study of the adaptive process is fundamentally the study of how these nested interactions respond over time to mutation and selection. Proving that a trait is “adaptive” is non-trivial, as it requires showing not only that the trait confers a fitness benefit but that it also has a heritable basis [2]. Demonstrating how an adaptive trait arose can prove even more daunting because detailed knowledge is also required of the phylogenetic history of the population in which the trait arose. For those many species that have a long generation time and a sparse fossil record, the adaptive process can only be studied retrospectively using comparative methods. By contrast, because many microbes have short generation times and can be cryopreserved, the adaptive process can be studied in these species prospectively using experimental methods. Empowered by advances in high-throughput sequencing and phenotyping, experimental microbial evolution now offers the possibility of mapping the adaptive process as it unfolds at the genomic scale in real time.
A powerful metaphor for envisioning the adaptive process is the fitness landscape, first introduced by Sewall Wright in the 1930s [3]. This concept enables us to represent in multiple dimensions the relationship between genotype and fitness for a population under selection, and thereby identify the various mutational paths by which lineages can evolve from genotypes that lie in valleys of low fitness to genotypes that occupy peaks of high fitness [4]. The fitness landscape has fine-scale topography: when it is smooth, all adaptive mutational paths will eventually converge on one genotype, the global optimum, whose fitness is greatest for the selection regime under consideration. When the topography is rugged, there exist multiple peaks and adaptation is constrained, as the mutations available to increase the population’s fitness depend on the founder’s genotype [5]. Different genotypes may evolve to reach different peaks in such a landscape (and indeed, a single genotype may not always reach the same peak when evolved under the same condition). For example, E. coli’s TEM-1 can be co-opted to confer a novel phenotype, cefotaxime resistance; however, biophysical pleiotropy in this protein limits the number of mutational trajectories leading to the novel phenotype [6]. In vitro selection experiments have shown that cefotaxime resistance evolves with a high degree of parallelism: three mutations usually occur in fixed order [7]. Rarely, different initial substitutions create alternative paths that lead to cefotaxime resistance, albeit at lower levels than that achieved via the most common trajectory. All else being equal, the rate of adaptive evolution in a “rugged” landscape having multiple adaptive peaks is slower than in a smooth landscape having one global optimum: “rugged” landscapes are more functionally constrained [8], and some fraction of the population can become trapped on peaks of different height.
Although a plethora of theoretical and empirical studies have explored the causes and consequences of different fitness landscapes, it is still an outstanding question as to whether most are simple, essentially being Mount Fuji-like [9], containing a single global fitness optimum, or whether they are complex, with selective constraints on differing mutational trajectories creating multiple local fitness optima [5, 10–13]. In part, this uncertainty arises from the perspective of what is the trait under consideration and what aspect of that trait is being optimized by selection in the framework of a fitness landscape? Is it a protein’s activity [14] or its cofactor binding affinity [15]? Is it the regulatory response of a repressor-operator pair [16] or is it the rate of a process such as CO2 assimilation [9]? And if it actually is true Darwinian fitness, is fitness estimated in terms of differential reproduction [17] or in terms of differential survivorship [7], or as is possible in microbial studies, both?
Regardless of one’s perspective, genetic constraints on the adaptive landscape stem from a common source, epistasis, where a mutation’s effect on fitness depends on the genetic background in which it arises [12]. Theory has shown that sign epistasis, wherein a mutation is beneficial within the context of some genetic backgrounds, but detrimental within others, can constrain mutational trajectories on fitness landscapes [18]. In particular, the creation of rugged fitness landscapes with their local peaks and valleys depends critically on “reciprocal sign epistasis” (RSE), wherein the mutational path between two genotypes becomes inaccessible to selection due to an intermediate low-fitness genotype [13, 18, 19]. Such valleys of low fitness are less likely to be “crossed” by the action of natural selection, mutation and drift [20]. Under RSE, genotypes that reside at local fitness optima may become evolutionary dead-ends. Even if a higher fitness peak exists elsewhere on the landscape, “valleys-of-death” adjacent to these local optima create barriers to adaptive evolutionary change.
A variety of experimental approaches have been used to discover how mutational constraints shape fitness landscapes, each with its own measure of ‘fitness.’ At the level of single proteins, differences in catalytic activity have been assayed by substituting different residues at an enzyme active site(s) [14, 21, 22], and by constructing evolutionary intermediates between the ancestral and evolved states [6, 23, 24]. When the phenotypic consequences of mutations in different genes have been studied sign epistasis appears to be prevalent ([17, 25–28] but see [25]), which may explain why adaptation takes surprisingly few mutational paths to optimal genotypes. Some genotype-phenotype mapping studies using molecular data have inferred a multi-peaked landscape using proxies for fitness, but the extent of the role played by local optima during adaptation was either unknown [28] or limited [21, 29].
For clonal populations evolving in a constant environment, theory suggests that RSE is required to establish a rugged adaptive landscape [19]. But how often do empirical fitness landscapes exhibit such topography? Analysis of in vivo data on repressor-operator pairs in the lac system of Escherichia coli reveals 19 distinct peaks [16]. Multiple, deep fitness valleys have recently been demonstrated in adaptation of HIV-1 to its secondary cell-surface chemokine co-receptor, CXCR4 [30, 31]. In the plant RNA virus TEV, RSE was the most common type of epistasis, accounting for 55% of significant gene interactions [32]. In addition, a considerable body of clinical data points to the existence of inaccessible evolutionary trajectories in diseases such as colorectal cancer [33, 34], blastic plasmacytoid dendritic cell neoplasm [35], secondary glioblastomas [36] and lung adenocarcinoma [37]. That so many alleles linked to cancers are mutually exclusive, suggests that reciprocal sign epistasis may be a pervasive feature of their adaptive landscapes.
A clonal population may traverse an adaptive “valley-of-death” stemming from RSE when environmental change alters the topography of its fitness landscape [38], as well as when recombination (e.g., [39]) and/or gene flow creates a genetic overpass. Alternatively, a neutral or adaptive ridge might provide an alternative path connecting one peak to another. The likelihood that such a route can be successfully followed depends on mutational path length and the ease with which the population can explore that ridge, subject to its size and mutation rate. For example, in the TEM β-lactamase system, although the landscape leading to cefotaxime resistance appears locally rugged, recent analyses indicate the existence of multiple adaptive pathways that lead to a global optimum of maximal resistance [40]. Clearly, predictive treatment of infectious disease and cancer will benefit from maps of empirically determined fitness landscapes that describe not only the height of local and global optima, but also the topography of adaptive ridges that may provide routes for therapeutic escape.
We have previously described a rugged fitness landscape that we discovered during the experimental evolution of yeast in a constant, glucose-limited environment [41]. We identified two mutually exclusive mutations, a nonsense mutation in mth1-1 and an amplification involving the HXT7/6 locus. Each mutation arose multiple times in independent lineages; individually each mutation was strongly adaptive (with the HXT7/6 amplification resulting in higher fitness than the mth1-1 mutant), but in combination, within the same individual, the resulting strain was not only less fit than the two single mutants, but also less fit than their wild-type ancestor, a clear case of reciprocal sign epistasis. HXT7 and HXT6 are almost identical, tandemly-arrayed genes encoding high-affinity hexose transporters that have been repeatedly observed to undergo fusion and amplification in yeast evolving under glucose limitation [42, 43]. MTH1 encodes a negative regulator of the glucose-sensing pathway, and its inactivation has been shown to elevate HXT expression [44]). Here, we set out to determine whether there exists either a neutral or an adaptive ridge between the two fitness peaks defined by these mutually exclusive mutations. We founded replicate experimental evolution experiments wherein one of the mutations, mth1-1, was fixed in the population, and then sought to determine whether the other mutation arose. Using PCR, we screened for hxt7/6 amplifications in populations that had evolved for ~250 generations under conditions identical to those described in [41]. In addition, we also performed whole genome resequencing on multiple clones drawn from each of three replicate experiments. We did not uncover any evidence for the occurrence of the other mutation, suggesting the existence of an adaptive ‘valley-of-death’ in which hxt7/6 amplifications cannot rise to appreciable frequency. However, we did discover a myriad of other mutations distributed amongst the clones we sequenced, many of which are plausibly beneficial. The discovery of multiple adaptive lineages coexisting in the same population adds to a growing body of evidence that clonal interference is a pervasive feature of microbial evolution, and that at any given time there may exist a plethora of highly fit lineages, each of which prevents the others from going to fixation.
Results
Evolution of the mth1-1 mutant under glucose limitation provides evidence for clonal interference
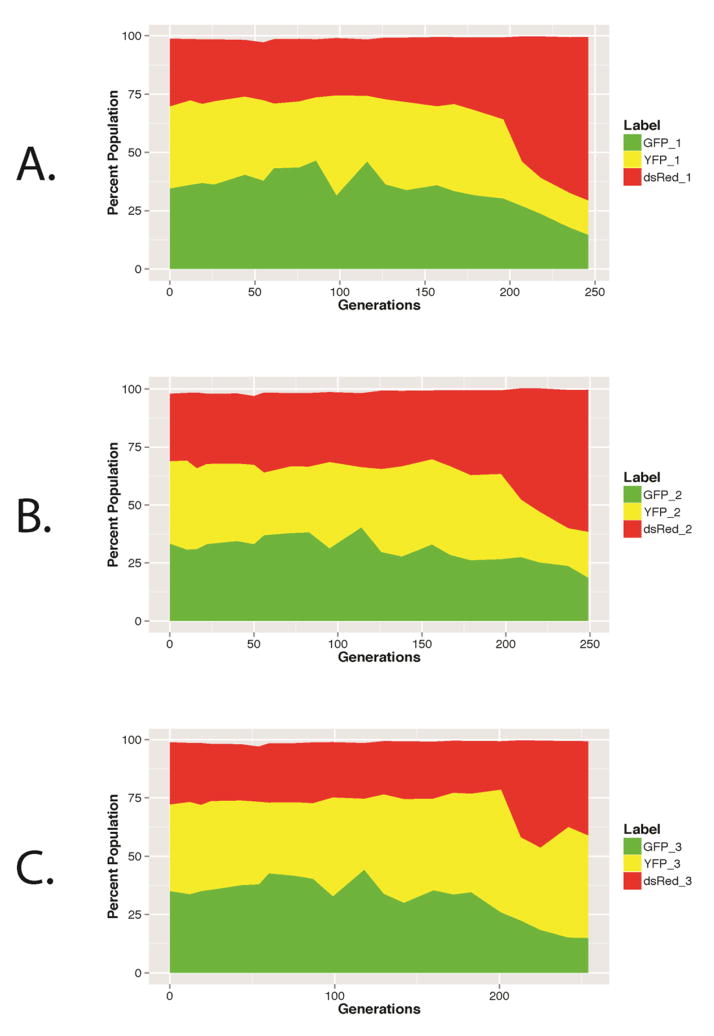
The mth1-1 allele, originally isolated and described in [44], contains a C to T transition that results in a Gln to Stop nonsense mutation at amino acid 338 of Mth1, removing the terminal 96 amino acids. We introduced mth1-1 into three otherwise isogenic Mat-α strains that carried one of three different fluorescent markers: GFP, DsRed, YFP (see Materials and Methods). Our previous work [44] had demonstrated that the growth rates of these marked strains did not significantly differ from one another. Overnight cultures of fluorescently tagged mth1-1 strains were pooled at approximately equal frequencies, and used to inoculate three independent bioreactors, each of which contained 300 mL 0.08% glucose Delft minimal media [45]. Following overnight incubation in batch, cultures were switched to continuous mode and grown for ~25 generations at D≈0.1 h−1 then for 225 generations at D≈0.17 h−1 under continuous glucose limitation. In no case did we observe a single fluorescently-tagged lineage go to fixation following 250 generations of evolution (Fig. 1), suggesting that clonal interference maintained a complex population genetic structure in evolving populations originating from an mth1-1 ancestor, as we had seen previously in experiments founded by a wild-type clone [44].
Figure 1.
Dynamics of fluorescent tags indicate pervasive clonal interference: (A) Population 1, (B) Population 3, (C) Population 3.
A PCR screen reveals no increase in HXT copy number among independent populations founded by mth1-1 nonsense mutants
To determine whether the HXT6/7 amplification arose in any of the three independent populations founded by haploid yeast carrying the mth1-1 allele, we performed PCR on genomic DNA from population samples, using primers that specifically detect the tandem duplication of the locus. We determined that we can detect the amplification when it is present in a frequency of ≥0.10 from reconstruction experiments. Thus, in mixing experiments, containing genomic DNA from a wild-type strain mixed with genomic DNA from a mutant strain carrying the tandem duplication, we were consistently able to PCR amplify a band when the DNA of the mutant strain was in a frequency of 0.10 or greater of the DNA mix, but not when the DNA was present at a frequency of ≤0.01 (data not shown), To determine whether the amplification had arisen in any of the three evolved populations, we performed PCR as described above on DNA extracted from generation ~250 on samples for each of the three evolved populations. In no case did we observe the diagnostic band that is produced by the tandem duplication of the HXT6/7 locus, observed in positive controls (data not shown). To further determine whether the amplification might be present at low frequency, we performed the diagnostic PCR on 48 individual clones randomly selected from the ~250 generation population for each of the 3 evolved populations. Again, in no instance did we observe amplification of the diagnostic band, which was present in positive controls, suggesting that if the amplification is present with the 250 generation population, then it is present in ~2% or less of the population. We therefore conclude that, while amplifications of the locus may occur, they do not rise to high frequency in our experimental evolutions, likely due to the reciprocal sign epistasis between them and the pre-existing mth1-1 allele.
Clonal sequencing also provides additional evidence that HXT6/7 amplification mutations do not reach high frequency in an mth1-1 background evolving under glucose limitation
To investigate further whether the HXT6/7 amplification occurs in an mth1-1 background, and to also determine if other mutations arose during this evolution, we selected a total of 17 clones from the terminal experimental evolution populations (four in population 1, six in population 2, seven in population 3) and performed whole genome re-sequencing. Significantly, all of these clones retained the mth1-1 allele; consistent with our PCR results, while none showed (hxt6/7)n amplification. Thus, while the hxt6/7 amplification can be crossed into an mth1-1 background, resulting in a strain with fitness lower than wild-type [41], we uncovered no evolutionary trajectory that allowed this combination of alleles to arise.
Clone sequencing uncovers other mutations arising in an mth1-1 background, some of which may have adaptive value under glucose limitation
A total of 81 Single Nucleotide Polymorphisms (SNPs) and indels were discovered among the seventeen sequenced mth1-1 clones relative to the founder (Table 1–3). 43 of these resulted in non-synonymous changes to coding sequences, while 4 resulted in synonymous changes; 6 introduced premature STOP codons, and 2 resulted in frameshifts. 1 SNP was in an intron, 8 SNPs occurred upstream of coding sequences, 8 occurred immediately downstream and 8 were classified as intergenic. All of the evolved clones retained the ancestral mth1-1 nonsense mutation. Only one other SNP was observed in three populations: fourteen of seventeen clones shared a T→A transversion at position 371865 on Chr. II. This SNP occurs downstream of TIP1, which encodes a major cell wall mannoprotein. Other than mth1 and tip1 mutations, fifteen SNPs were observed in more than one clone; as expected for independently evolving populations, all fifteen were shared only among a subset of clones isolated from any one population (Tables 1–3).
Table 1.
Summary of SNPs detected in sequenced clones from population 1, where yellow indicates the presence of the SNP.
| #CHROM POS | GENE(S) | Effect | Codon | AA | REF | ALT | GSY2737 | GSY2736 | GSY2734 | GSY2735 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chrII | 371865 | TIP1 | DOWNSTREAM | T | A | ||||||
|
|
|||||||||||
| chrII | 275399 | GAL7 | STOP_GAINED | taC/taA | Y/* | G | T | ||||
|
|
|||||||||||
| chrIV | 727219 | YCF1 | NON_SYNONYMOUS | atG/atT | M/I | C | A | ||||
|
|
|||||||||||
| chrVI | 102407 | LPD1 | NON_SYNONYMOUS | Gca/Cca | A/P | C | G | ||||
|
|
|||||||||||
| chrVII | 348047 | MAD1 | NON_SYNONYMOUS | gAt/gTt | D/V | A | T | ||||
|
|
|||||||||||
| chrVII | 994190 | INTERGENIC | C | T | |||||||
|
|
|||||||||||
| chrVIII | 81842 | ETP1, PRS3 | INTERGENIC | G | T | ||||||
|
|
|||||||||||
| chrX | 172326 | URA2 | NON_SYNONYMOUS | atG/atT | M/I | C | A | ||||
|
|
|||||||||||
| chrXII | 221252 | YLR036C | DOWNSTREAM | G | T | ||||||
|
|
|||||||||||
| chrXIII | 891857 | PSE1 | NON_SYNONYMOUS | cTa/cCa | L/P | A | G | ||||
|
|
|||||||||||
| chrXV | 593444 | LSC1 | STOP_GAINED | Caa/Taa | Q/* | C | T | ||||
|
|
|||||||||||
| chrVIII | 58100 | RIM4 | NON_SYNONYMOUS | atG/atT | M/I | G | T | ||||
|
|
|||||||||||
| chrVIII | 85558 | LTR | G | A | |||||||
|
|
|||||||||||
| chrXII | 893967 | VAC14 | NON_SYNONYMOUS | Gca/Tca | A/S | G | T | ||||
|
|
|||||||||||
| chrXIV | 674942 | BUD17 | NON_SYNONYMOUS | aCg/aAg | T/K | C | A | ||||
|
|
|||||||||||
| chrII | 237120 | ECM15, HTB2 | DOWNSTREAM | G | T | ||||||
|
|
|||||||||||
| chrXI | 609417 | PXL1 | NON_SYNONYMOUS | Cat/Aat | H/N | C | A | ||||
|
|
|||||||||||
Table 3.
Summary of SNPs detected in sequenced clones from population 3, where yellow indicates the presence of the SNP. Gene names in red, bold font are genes which were either mutated recurrently in our experiments, or in which mutations have been observed previously in other glucose limited chemostat evolutions.
| #CHROM | POS | GENE(S) | Effect | Codon | AA | REF | ALT | GSY2750 | GSY2754 | GSY2756 | GSY2751 | GSY2755 | GSY2757 | GSY2753 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chrII | 371865 | TIP1 | DOWNSTREAM | T | A | |||||||||
|
|
||||||||||||||
| chrI | 128481 | SYN8 | FRAMESHIFT | S/Frameshift | G | - | ||||||||
|
|
||||||||||||||
| chrV | 364256 | SSA4 | UPSTREAM | A | G | |||||||||
|
|
||||||||||||||
| chrVIII | 519892 | CRG1 | SYNONYMOUS | ccC/ccT | P/P | C | T | |||||||
|
|
||||||||||||||
| chrXV | 178574 | IRA2 | NON_SYNONYMOUS | aCa/aAa | T/K | C | A | |||||||
|
|
||||||||||||||
| chrMito | 80513 | COX3 | DOWNSTREAM | C | G | |||||||||
|
|
||||||||||||||
| chrIV | 1289911 | SIZ1 | NON_SYNONYMOUS | tCt/tAt | S/Y | C | A | |||||||
|
|
||||||||||||||
| chrX | 666710 | STR2 | NON_SYNONYMOUS | tCt/tTt | S/F | G | A | |||||||
|
|
||||||||||||||
| chrXII | 147914 | DNM1 | NON_SYNONYMOUS | Cct/Act | P/T | C | A | |||||||
|
|
||||||||||||||
| chrXV | 606620 | MDM32 | NON_SYNONYMOUS | cGt/cCt | R/P | G | C | |||||||
|
|
||||||||||||||
| chrIV | 67810 | TIM22, YDL218W | DOWNSTREAM | C | A | |||||||||
|
|
||||||||||||||
| chrIV | 183707 | RPC53 | NON_SYNONYMOUS_CODING | tCt/tTt | S/F | C | T | |||||||
|
|
||||||||||||||
| chrVIII | 167568 | YHR028W-A | NON_SYNONYMOUS | YHR028W-A:c.217T>G,218C>G,219C>A | TCC | GGA | ||||||||
|
|
||||||||||||||
| chrX | 304208 | JEM1 | NON_SYNONYMOUS | tCg/tGg | S/W | C | G | |||||||
|
|
||||||||||||||
| chrXI | 158736 | RPS27A | INTRON | A | G | |||||||||
|
|
||||||||||||||
| chrXI | 647912 | FLO10 | SYNONYMOUS | tcT/tcA | S | T | A | |||||||
|
|
||||||||||||||
| chrXVI | 416459 | YTA6 | NON_SYNONYMOUS | Caa/Aaa | Q/K | C | A | |||||||
|
|
||||||||||||||
| chrII | 452501 | PHO88 | UPSTREAM | G | T | |||||||||
|
|
||||||||||||||
| chrXV | 557840 | UBP2 | NON_SYNONYMOUS | aGa/aCa | R/T | C | G | |||||||
|
|
||||||||||||||
| chrIII | 283793 | CDC39 | NON_SYNONYMOUS | tAt/tGt | Y/C | A | G | |||||||
|
|
||||||||||||||
| chrVII | 321148 | INTERGENIC | T | C | ||||||||||
|
|
||||||||||||||
| chrVIII | 524447 | INTERGENIC | C | G | ||||||||||
|
|
||||||||||||||
| chrIV | 675571 | YDR109C | NON_SYNONYMOUS | Caa/Aaa | Q/K | G | T | |||||||
|
|
||||||||||||||
| chrIV | 785856 | TRM82 | NON_SYNONYMOUS | aCa/aTa | T/I | C | T | |||||||
|
|
||||||||||||||
| chrVIII | 418602 | TDA11 | STOP_GAINED | Gag/Tag | E/* | G | T | |||||||
|
|
||||||||||||||
| chrIX | 89756 | TMA108, TPM2 | INTERGENIC | G | C | |||||||||
|
|
||||||||||||||
| chrXII | 838026 | ILV5 | DOWNSTREAM | C | A | |||||||||
|
|
||||||||||||||
| chrXV | 179275 | IRA2 | STOP_GAINED | Gag/Tag | E/* | G | T | |||||||
|
|
||||||||||||||
| chrXVI | 95218 | CIN2, IQG1 | INTERGENIC | G | A | |||||||||
|
|
||||||||||||||
| chrXVI | 421772 | MUK1 | STOP_GAINED | taC/taA | Y/* | C | A | |||||||
|
|
||||||||||||||
| chrV | 143507 | MIT1 | NON_SYNONYMOUS | gAa/gCa | E/A | A | C | |||||||
|
|
||||||||||||||
| chrXII | 921722 | BDF1, YLR400W | UPSTREAM | C | G | |||||||||
|
|
||||||||||||||
| chrXV | 213387 | PRS5 | NON_SYNONYMOUS | Acc/Gcc | T/A | A | G | |||||||
|
|
||||||||||||||
| chrIX | 69868 | SLN1 | NON_SYNONYMOUS | Cca/Gca | P/A | G | C | |||||||
|
|
||||||||||||||
| chrXIV | 119311 | BOR1 | NON_SYNONYMOUS | aCa/aGa | T/R | C | G | |||||||
|
|
||||||||||||||
| chrXVI | 898617 | SEC23 | NON_SYNONYMOUS | Gat/Cat | D/H | C | G | |||||||
|
|
||||||||||||||
| chrMito | 10368 | INTERGENIC | T | C | ||||||||||
|
|
||||||||||||||
| chrMito | 84816 | RPM1, tM(CAU)Q2 | UPSTREAM | A | T | |||||||||
|
|
||||||||||||||
A number of these mutations occurred in genes in which mutations have been observed in other yeast evolution experiments carried out under continuous glucose limitation. For example, three of six clones sequenced in population 2, carry a nonsense mutation in GIN4 (G→C, S1035*) that encodes a protein kinase involved in bud growth and assembly. A GIN4 missense mutation was reported by Gresham and colleagues [43] to attain >20% frequency in populations founded by haploid S288C after 200 generations of evolution under glucose-limitation. As these same three clones in population 2 also share a missense mutation in PBS2 (G→C, A56G) and an intergenic mitochondrial T→A transversion, they are likely members of the same lineage. Notably, mutations in PBS2 have also been observed to arise multiple times in populations evolving under glucose limitations, in each case those mutations predicted to be disruptive [46]. Four of seven clones sequenced in population 3 carry a missense mutation (T2502K) in IRA2, a GTPase-activating protein that negatively regulates RAS. Because these four ira2 mutants share SNPs not only at MTH1, but also at SSA4, CRG1, and COX3, they are almost certainly members of the same lineage (though they can be distinguished from one another by additional unique SNPs). A fifth clone in population 3 bears a nonsense mutation at a different position in IRA2 (E2736*). Presumptive loss of function mutations in IRA2, as well as in its paralog IRA1, have previously been observed in adaptive clones evolving under conditions where glucose is either periodically [47, 48] or continuously limiting [44, 46, 49]. Indeed, loss of nutrient signaling via any one of several possible mechanisms appears to be selected for in populations evolving in continuous culture under glucose limitation. Significantly, we have shown previously that IRA1/2 and MTH1 mutations can arise in the same background, and attain high frequency [46]. It is noteworthy that allelic variation at IRA1 and IRA2, which are homologs of the hypermutable human neurofibromatosis gene NF1, results in diverse morphological and growth phenotypes in Saccharomyces paradoxus [50].
Other mutational targets in our experiments have previously been reported as being selected under constant or periodic glucose limitation. These include ACE2 (Y594S), a transcription factor required for septum destruction after cytokinesis, observed in two clones from Population 2 and also by [46]. Interestingly, mutations in ACE2 have recently been shown to underlie rapid evolution of multicellular, fast-sedimenting phenotypes in yeast [51]. We also observed mutations in the sphingolipid biosynthetic gene LCB1 (D320E) in two different clones from Population 2; mutants in the same pathway (LCB3) were previously observed by [46] in evolving populations under glucose limitation. As these pairs of clones share, respectively, mutations in PBS2 (G→C, A56G) and in PET10 (G→T, S65R), we conclude that repeated observation of ACE2 and LCB1 alleles in population 2 is not due to their having arisen independently but rather to their being shared by descent. We also observed a nonsense mutation in MUK1 (C→A, Y275*), which truncates the predicted Muk1 protein at less than half its length, removing a candidate VPS9 domain. We have previously observed MUK1 mutations in other populations evolving under glucose limitation [44, 49]. Recently, MUK1 has been shown to encode a guanosine nucleotide exchange factor (GEF) for proteins involved in Golgi trafficking and endocytosis [52, 53]. A link between these functions and increased fitness in populations evolving under glucose-limitation is unclear; indeed, we previously showed [41] that alone, MUK1 has no fitness effect, suggesting that either it requires another mutation in the background in an epistatic fashion, or possibly that MUK1 simply has a higher mutation rate than many genes.
Finally, in addition to observing two IRA2 mutations, we also observed independent mutations in MIT1, in populations one and three. Little is know about MIT1, though it appears to be a transcriptional regulator of biofilm development [54].
Discussion
The spectrum of mutations arising in an mth1-1 background is conspicuously lacking in other variants that would degrade nutrient signaling pathways
Amplification of hxt6/7 chimeras has been repeatedly observed to occur in yeast as an adaptive response to glucose limitation, but this path to increased fitness appears to be closed to mth1-1 strains. Other mutations that might be expected to increase the capacity of cells to scavenge limiting glucose include those that alter nutrient signaling via the cAMP/PKA [43, 46, 49] and HOG pathways [46]. As noted, five of seven sequenced clones in population 3 had IRA2 mutations that likely to diminish Ras activity, and thus cAMP/PKA signaling, while three of six clones in population 2 had mutations in PBS2, and one of seven clones in population 3 has a mutation in SLN1, both of which are predicted to affect signaling via the HOG pathway, as has been observed in [46, 49]. However, a number of other targets, whose impact on nutrient signaling is known to confer a fitness advantage under glucose limitation, were not mutated these targets include: RIM15 and RGT2 [43, 46], and CYR1 [49]. Failure to recover mutants at these loci may be due to their diminishing returns on fitness in an mth1-1 background, to the limited number of clones we sampled, the relatively short duration of the experiment, or possibly, to negative sign epistasis preventing (htx6/7)n mutants from attaining appreciable frequency. Clearly, population sequencing or similar approaches, with the ability to identify mutations that reach a 0.01 allele frequency or more, are needed to gain deep insight into how the beneficial mutational spectra might differ between replicate populations evolving under the same conditions, but with different founding genotypes.
Does introgression of mth1-1 elevate wild-type yeast onto a fitness peak?
Theory suggests that there are generally two mechanisms by which higher fitness can be achieved in a continuous, resource-limited environment [55]. A genotype may become so efficient at scavenging the limiting substrate that the substrate effectively becomes inaccessible to potential competitors, or a genotype may simply become more proficient at using the limiting substrate so that it produces more progeny per unit resource than potential competitors. In yeast, one or both strategies may come play, depending on evolutionary trajectory and on the availability of oxygen as the terminal electron acceptor for respiratory metabolism. Ferea et al. [56] demonstrated that evolution of an S288C-derivative under glucose limitation resulted in genotypes that exhibited, relative to their common ancestor, superior glucose uptake kinetics and lower residual substrate concentrations. Under aerobic conditions, evolved diploids showed greater than 3-fold yield than their common ancestor. When cultured under anaerobic conditions, these yield differences largely disappeared, yet evolved strains nevertheless were nevertheless more fit than their common ancestor, owing to their superior glucose uptake capacities [49]. We know that the mth1-1 allele confers a fitness advantage over wild type in pairwise competition [41], but we do not know if this genotype occupies a fitness peak that restricts further improvements in performance. To test this we measured residual glucose (s), cell number, and dry weight biomass in populations founded by mth1-1 mutants before and after 250 generations of evolution. We found that while the scope for improvement in physiological performance did not approach that observed in previously reported populations evolving under glucose-limitation, cells did become significantly more proficient in their uptake of the limiting substrate (s=0.043±0.013 mg mL−1 vs. 0.028±0.044 mg mL−1, t-test, P=0.0321) and in their yield of cells per mg glucose consumed (4.33±0.29 × 107 vs. 5.72±0.31 × 107, t-test, P=0.0313). We therefore conclude that under glucose limitation the scope for selection for (hxt6/hxt7)n and other beneficial mutations appears to be diminished in an mth1-1 background. While this genotype may not occupy the summit of a fitness peak, it very likely resides at a higher elevation than wild type strains used to initiate previous populations evolving under glucose-limitation.
The nature of reciprocal sign epistasis
The predicted product of the wild-type MTH1 gene is a 433 amino acid protein that interacts with Snf3 and Rgt1 and Rgt2, and serves as a negative regulator of the glucose-sensing pathway. Mth1 inhibits protein kinase A-dependent phosphorylation of Rgt1 [57], while inactivation of Mth1 and its paralog Std1 leads to hyperphosphorylation of Rgt1 and its dissociation from hexose transporter promoters (HXT) in the absence of glucose [58], bypassing the requirement of Grr1for HXT induction. We showed previously that this nonsense mutation in MTH1 increases HXT expression, especially of HXT1 and HXT4 (see Supp. Fig 1 in [44]), providing a clear fitness advantage to yeast evolving in constant low glucose conditions. Further analysis of yeast populations evolved under glucose limitation (see [44]) revealed additional, independently derived mth1 alleles that also contain nonsense mutations [41]. Because both mth1-1 and (HXT6/7) amplification mutations are predicted to increase the abundance of hexose transporters in the plasma membrane, their apparent mutual exclusivity suggests that there may be either an upper limit to, or and optimal number of these proteins under the glucose limiting conditions. In the case of the double mutant, this limit or optimum is overshot, leading to decreased fitness. Whether this is a generalizable principle for mutually exclusive mutations (that is, when a pathway has an optimum under one condition, that optimum may shift under another condition) will require significantly more investigation. However, in the particular case of hexose transporter expression, it may be relatively straightforward to derive a function mapping fitness to hexose transporter transcription under various experimental conditions then to determine if the resulting surface has a simple maximum.
Alternative approaches to find a neutral or beneficial ridge
In this investigation, we attempted to determine if amplification of the HXT6/7 locus could occur during experimental evolution on an mth1-1 background. While we did not observe this outcome, there are many reasons that we may not have observed it, chief among them that an insufficient number of generations had elapsed by the termination of the populations. Alternatively, it is possible that we could have observed the HXT6/7 amplification on the mth1-1 background, but that the other mutations required to make that observation might have placed the evolved individuals on yet a new peak, rather than at a new point on our imagined HXT6/7 peak. Distinguishing between such possibilities would be challenging, in the absence of a mapping of the fitness landscape at high resolution across many related genotypes. An alternative approach might be to start evolution experiments with the double mth1-1 (hxt6/7)n mutant, and to select for compensatory mutations, which relieve the reciprocal sign epistasis. If clones were isolated with mutations that are neutral in the wild-type background, it would suggest that they might provide such a neutral ridge as we originally postulated.
Materials and methods
Strains
The ancestral strains used in these experiments are derived from GSY1171 (strain M1 in Kao and Sherlock 2008) [44]. The ancestral population consisted of equal numbers of GSY3087, GSY2674, and GSY2672, which were constructed so as to contain the mth1-1 allele (Gln338*) previously described by [44] and [41] as well as a chromosomally integrated gene encoding GFP, YFP or DsRed. Founder strains were created by crossing GSY1171 (Mat-α ura3-52 GAL2+ YBR209W::Act1p-GFP-Act1t-URA3 mth1 in an S288c background) to one of three FP-marked Mat-a strains derived from FY2 [59]: GSY1221 (ura3-52 GFP), GSY1222 (ura3-52 YFP) and GSY1223 (ura3-52 DsRed). The diploids produced by these matings were sporulated, and Mat-alpha haploids isolated to create the ancestral population.
Media and culture conditions
Yeast strains were routinely maintained on either liquid or solid YEP medium containing 1% (w/v) dextrose). The ancestral population was created by culturing single colony isolates of GSY3087, GSY2674, and GSY2672 overnight in YEPD at 30°C, then apportioning equal numbers of the differently labeled mth1-1 strains into each of three 300 mL working volume bioreactors (ATR SixFors fermentation apparatus, ATR Biotechnologies). Following overnight batch culture, reactors were initially run in continuous mode at a dilution rate, D, of 0.1 h−1 for ~25 generations, then for an additional 225 generations at D=0.17 h−1. Evolution experiments were carried out at 30°C using the Delft minimal medium described by [45] amended with 0.08% (w/v) glucose. Aerobic conditions were maintained by sparging cultures with sterile humidified air (25 L h-1). Glucose-limited chemostats were sampled were sampled every day to determine absorbance at 660 nm, to create an archive of −80°C glycerol stocks (20% v/v), and to estimate the number of colony forming units (CFU) per mL on YEPD agar. At 50-generation intervals cells were withdrawn and processed for Fluorescence Activated Cell Sorting (FACS), as described below.
Fluorescence Activated Cell Sorting (FACS)
Quantitative cell sorting by fluorimetry was performed using methods previously described [44].
PCR detection of HXT6/7 amplification
PCR to identify the amplification of was carried out using primers and PCR conditions as described in [41].
Whole genome sequencing and mutation determination
Clones from each of the 3 replicate chemostat populations were paired-end sequenced (2×100bp) using the Illumina HiSeq 2000. Sequencing libraries were constructed for each clone using the Illumina Genomic DNA sample prep kit from 5 μg of genomic DNA and each libraries were multiplexed on a single flow cell lane. Sequence analysis was performed as follows, using default parameters unless otherwise noted. Fastq files were first trimmed to remove adaptor sequences using cutadapt [60], and reads were mapped to a modified S288c reference genome that we generated previously [41] using bwa-short in BWA v0.5.9-r16 [61]. PCR duplicates in the resulting bam files were marked using Picard MarkDuplicates v1.45, and then indel realignment and base quality score recalibration performed with GATK v1.0.5777 [62, 63]. SNPs and indels were then called with the GATK UnifiedGenotyper using default parameters. SNPs were then hard filtered using filters: “QD < 2.0” “MQ < 40.0” “FS > 60.0” “HaplotypeScore > 13.0” “MQRankSum < −12.5” “ReadPosRankSum < −8.0”, while indels were hard filtered using: “FS > 200.0” “QD < 2.0” “ReadPosRankSum < −20.0”. A list of all SNPs and indels were then compiled, and annotated to indicate the affected gene, and likely outcome of that change.
Table 2.
Summary of SNPs detected in sequenced clones from population 2, where yellow indicates the presence of the SNP. Gene names in red, bold font are genes which were either mutated recurrently in our experiments, or in which mutations have been observed previously in other glucose limited chemostat evolutions.
| #CHROM POS | GENE(S) | Effect | Codon | AA | REF | ALT | GSY4318 | GSY4322 | GSY4319 | GSY4323 | GSY4321 | GSY4324 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chrII | 371865 | TIP1 | DOWNSTREAM | T | A | ||||||||
|
|
|||||||||||||
| chrIV | 329069 | YDL073W | SYNONYMOUS | ctG/ctC | L/A | G | C | ||||||
|
|
|||||||||||||
| chrV | 143645 | MIT1 | NON_SYNONYMOUS | YEL007W:c.1754T>G, YEL007W:p.Met585Arg | T | G | |||||||
|
|
|||||||||||||
| chrV | 143646^143647 | MIT1 | FRAMESHIFT | YEL007W:c.1755_1756insG, YEL007W:p.Tyr586fs | - | G | |||||||
|
|
|||||||||||||
| chrXI | 524880 | PET10 | NON_SYNONYMOUS | agC/agA | S/R | G | T | ||||||
|
|
|||||||||||||
| chrXIII | 859932 | LCB1 | NON_SYNONYMOUS | gaC/gaA | D/E | G | T | ||||||
|
|
|||||||||||||
| chrXV | 988368 | PUT4 | NON_SYNONYMOUS | Tgc/Cgc | C/R | A | G | ||||||
|
|
|||||||||||||
| chrII | 600545 | PCH2 | UPSTREAM | A | G | ||||||||
|
|
|||||||||||||
| chrVI | 108690 | IES1 | NON_SYNONYMOUS | gAg/gGg | E/G | T | C | ||||||
|
|
|||||||||||||
| chrXIII | 420692 | tV(AAC)M2 | UPSTREAM | A | G | ||||||||
|
|
|||||||||||||
| chrXI | 313785 | tH(GUG)K | UPSTREAM | C | T | ||||||||
|
|
|||||||||||||
| chrXII | 770185 | BUD6 | NON_SYNONYMOUS | aaC/aaA | N/K | G | T | ||||||
|
|
|||||||||||||
| chrXIV | 368081 | SRV2 | NON_SYNONYMOUS | ttT/ttA | F/L | T | A | ||||||
|
|
|||||||||||||
| chrIV | 1462683 | GIN4 | STOP_GAINED | tCa/tGa | S/* | G | C | ||||||
|
|
|||||||||||||
| chrX | 179937 | PBS2 | NON_SYNONYMOUS | gCt/gGt | A/G | G | C | ||||||
|
|
|||||||||||||
| chrMito | 57268 | INTERGENIC | T | A | |||||||||
|
|
|||||||||||||
| chrXII | 405042 | ACE2 | NON_SYNONYMOUS | tAc/tCc | Y/S | T | G | ||||||
|
|
|||||||||||||
| chrII | 85054 | SSA3 | SYNONYMOUS | gcT/gcG | A/A | A | C | ||||||
|
|
|||||||||||||
| chrX | 237313 | LSB6 | NON_SYNONYMOUS | aaG/aaC | K/N | G | C | ||||||
|
|
|||||||||||||
| chrX | 364859 | MHP1 | NON_SYNONYMOUS | gCa/gTa | A/V | C | T | ||||||
|
|
|||||||||||||
| chrXIV | 144125 | IST1, PIK1 | DOWNSTREAM | C | G | ||||||||
|
|
|||||||||||||
| chrXVI | 926957 | OPT2 | UPSTREAM | G | T | ||||||||
|
|
|||||||||||||
| chrVII | 44172 | TAD1 | NON_SYNONYMOUS | cCa/cGa | P/R | C | G | ||||||
|
|
|||||||||||||
| chrXV | 902077 | SPS4 | NON_SYNONYMOUS | atT/atG | I/M | A | C | ||||||
|
|
|||||||||||||
| chrV | 476850 | BEM2 | NON_SYNONYMOUS | gGa/gCa | G/A | C | G | ||||||
|
|
|||||||||||||
| chrVIII | 197604 | INM1 | NON_SYNONYMOUS | Tat/Cat | Y/H | A | G | ||||||
|
|
|||||||||||||
| chrXII | 953417 | PUN1 | NON_SYNONYMOUS | aGc/aAc | S/N | C | T | ||||||
|
|
|||||||||||||
| chrXIV | 736085 | BIO3 | DOWNSTREAM | C | T | ||||||||
|
|
|||||||||||||
Acknowledgments
The authors gratefully acknowledge funding from NIH (R01-HG003328) to GS and NASA to FR (NNX07AJ28G), which supported our research and preparation of this manuscript. The manuscript was significantly improved by helpful discussions with Margie Kinnersley, Evgueny Kroll, and we thank Julian Adams and Michael Desai for additional comments. Analysis of fluorescently labeled yeast populations was made possible by the expertise of Pam Shaw, staff scientist at the University of Montana Fluorescence Cytometry Core Facility, which has been in part supported by National Institutes of Health grant P20RR017670.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gould SJ, Lewontin RC. Proceedings of the Royal Society of London. Series B, Containing papers of a Biological character. Vol. 205. Royal Society; 1979. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme; pp. 581–598. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen R. Adaptionism-30 years after Gould and Lewontin. Evolution; international journal of organic evolution. 2009;63:2487–2490. doi: 10.1111/j.1558-5646.2009.00799.x. [DOI] [PubMed] [Google Scholar]
- 3.Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proceedings of The Sixth Congress on Genetics; Ithaca, New York. 1932; pp. 355–366. [Google Scholar]
- 4.Wright S. Surfaces of Selective Value Revisited. The American Naturalist. 1988;131:115–123. [Google Scholar]
- 5.Whitlock MC, Phillips PC, Moore FBG, Tonsor SJ. Multiple Fitness Peaks and Epistasis. Annu Rev Ecol Syst. 1995;26:601–629. [Google Scholar]
- 6.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 7.Salverda ML, Dellus E, Gorter FA, Debets AJ, van der Oost J, Hoekstra RF, Tawfik DS, de Visser JA. Initial mutations direct alternative pathways of protein evolution. PLoS genetics. 2011;7:e1001321. doi: 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povolotskaya IS, Kondrashov FA. Sequence space and the ongoing expansion of the protein universe. Nature. 2010;465:922–926. doi: 10.1038/nature09105. [DOI] [PubMed] [Google Scholar]
- 9.Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber AP, Lercher MJ. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell. 2013;153:1579–1588. doi: 10.1016/j.cell.2013.04.058. [DOI] [PubMed] [Google Scholar]
- 10.Brodie ED. Why Evolutionary Genetics Does Not Always Add Up. In: Wolf JB, Brodie ED, Wade MJ, editors. Epistasis and the Evolutionary Process. Oxford University Press; New York: 2000. [Google Scholar]
- 11.Gavrilets S. Fitness Landscapes and the Origin of Species. Princeton University Press; 2004. [Google Scholar]
- 12.Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nature reviews Genetics. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- 14.Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science. 2005;310:499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- 15.Zhu G, Golding GB, Dean AM. The selective cause of an ancient adaptation. Science. 2005;307:1279–1282. doi: 10.1126/science.1106974. [DOI] [PubMed] [Google Scholar]
- 16.Dawid A, Kiviet DJ, Kogenaru M, de Vos M, Tans SJ. Multiple peaks and reciprocal sign epistasis in an empirically determined genotype-phenotype landscape. Chaos. 2010;20:026105. doi: 10.1063/1.3453602. [DOI] [PubMed] [Google Scholar]
- 17.Chou HH, Chiu HC, Delaney NF, Segre D, Marx CJ. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science. 2011;332:1190–1192. doi: 10.1126/science.1203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution; international journal of organic evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- 19.Poelwijk FJ, Tanase-Nicola S, Kiviet DJ, Tans SJ. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. Journal of theoretical biology. 2011;272:141–144. doi: 10.1016/j.jtbi.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Weissman DB, Desai MM, Fisher DS, Feldman MW. The rate at which asexual populations cross fitness valleys. Theoretical population biology. 2009;75:286–300. doi: 10.1016/j.tpb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozovsky ER, Chookajorn T, Brown KM, Imwong M, Shaw PJ, Kamchonwongpaisan S, Neafsey DE, Weinreich DM, Hartl DL. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellner A, Raitses Gurevich M, Tawfik DS. Mechanisms of protein sequence divergence and incompatibility. PLoS genetics. 2013;9:e1003665. doi: 10.1371/journal.pgen.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lunzer M, Golding GB, Dean AM. Pervasive cryptic epistasis in molecular evolution. PLoS genetics. 2010;6:e1001162. doi: 10.1371/journal.pgen.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332:1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 26.Hinkley T, Martins J, Chappey C, Haddad M, Stawiski E, Whitcomb JM, Petropoulos CJ, Bonhoeffer S. A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nature genetics. 2011;43:487–489. doi: 10.1038/ng.795. [DOI] [PubMed] [Google Scholar]
- 27.Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I. Positive epistasis drives the acquisition of multidrug resistance. PLoS genetics. 2009;5:e1000578. doi: 10.1371/journal.pgen.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Visser JA, Park SC, Krug J. Exploring the effect of sex on empirical fitness landscapes. Am Nat. 2009;174(Suppl 1):S15–30. doi: 10.1086/599081. [DOI] [PubMed] [Google Scholar]
- 29.Poelwijk FJ, Kiviet DJ, Tans SJ. Evolutionary potential of a duplicated repressor-operator pair: simulating pathways using mutation data. PLoS computational biology. 2006;2:e58. doi: 10.1371/journal.pcbi.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da Silva J, Wyatt SK. Fitness valleys constrain HIV-1’s adaptation to its secondary chemokine coreceptor. Journal of evolutionary biology. 2014 doi: 10.1111/jeb.12329. [DOI] [PubMed] [Google Scholar]
- 31.da Silva J, Coetzer M, Nedellec R, Pastore C, Mosier DE. Fitness epistasis and constraints on adaptation in a human immunodeficiency virus type 1 protein region. Genetics. 2010;185:293–303. doi: 10.1534/genetics.109.112458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lalic J, Elena SF. Magnitude and sign epistasis among deleterious mutations in a positive-sense plant RNA virus. Heredity. 2012;109:71–77. doi: 10.1038/hdy.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, Diaz LA, Jr, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature medicine. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 35.Stenzinger A, Endris V, Pfarr N, Andrulis M, Johrens K, Klauschen F, Siebolts U, Wolf T, Koch PS, Schulz M, Hartschuh W, Goerdt S, Lennerz JK, Wickenhauser C, Klapper W, Anagnostopoulos I, Weichert W. Targeted ultra-deep sequencing reveals recurrent and mutually exclusive mutations of cancer genes in blastic plasmacytoid dendritic cell neoplasm. Oncotarget. 2014 doi: 10.18632/oncotarget.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, Hu BQ, Yan W, Zhang W, Akers J, Ramakrishnan V, Li J, Carter B, Liu YW, Hu HM, Wang Z, Li MY, Yao K, Qiu XG, Kang CS, You YP, Fan XL, Song WS, Li RQ, Su XD, Chen C, Jiang T. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome research. 2014 doi: 10.1101/gr.165126.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshizawa A, Sumiyoshi S, Sonobe M, Kobayashi M, Uehara T, Fujimoto M, Tsuruyama T, Date H, Haga H. HER2 status in lung adenocarcinoma: A comparison of immunohistochemistry, fluorescence in situ hybridization (FISH), dual-ISH, and gene mutations. Lung cancer. 2014 doi: 10.1016/j.lungcan.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Flynn KM, Cooper TF, Moore FB, Cooper VS. The environment affects epistatic interactions to alter the topology of an empirical fitness landscape. PLoS genetics. 2013;9:e1003426. doi: 10.1371/journal.pgen.1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moradigaravand D, Kouyos R, Hinkley T, Haddad M, Petropoulos CJ, Engelstadter J, Bonhoeffer S. Recombination accelerates adaptation on a large-scale empirical fitness landscape in HIV-1. PLoS genetics. 2014;10:e1004439. doi: 10.1371/journal.pgen.1004439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schenk MF, Szendro IG, Salverda ML, Krug J, de Visser JA. Patterns of Epistasis between beneficial mutations in an antibiotic resistance gene. Molecular biology and evolution. 2013;30:1779–1787. doi: 10.1093/molbev/mst096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kvitek DJ, Sherlock G. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS genetics. 2011;7:e1002056. doi: 10.1371/journal.pgen.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Molecular biology and evolution. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 43.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS genetics. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nature genetics. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 46.Kvitek DJ, Sherlock G. Whole genome whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLoS genetics. 2013;9:e1003972. doi: 10.1371/journal.pgen.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, Desai MM. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 2014;344:1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS genetics. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roop JI, Brem RB. Rare variants in hypermutable genes underlie common morphology and growth traits in wild Saccharomyces paradoxus. Genetics. 2013;195:513–525. doi: 10.1534/genetics.113.155341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oud B, Guadalupe-Medina V, Nijkamp JF, de Ridder D, Pronk JT, van Maris AJ, Daran JM. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4223–4231. doi: 10.1073/pnas.1305949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paulsel AL, Merz AJ, Nickerson DP. Vps9 family protein Muk1 is the second Rab5 guanosine nucleotide exchange factor in budding yeast. The Journal of biological chemistry. 2013;288:18162–18171. doi: 10.1074/jbc.M113.457069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabrera M, Arlt H, Epp N, Lachmann J, Griffith J, Perz A, Reggiori F, Ungermann C. Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. The Journal of biological chemistry. 2013;288:5166–5175. doi: 10.1074/jbc.M112.431536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayhew D, Mitra RD. Transcription factor regulation and chromosome dynamics during pseudohyphal growth. Molecular biology of the cell. 2014;25:2669–2676. doi: 10.1091/mbc.E14-04-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dykhuizen DE, Dean AM. Experimental evolution from the bottom up. In: Garland T Jr, Rose MR, editors. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments. University of California Press; 2009. pp. 67–87. [Google Scholar]
- 56.Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy A, Jouandot D, 2nd, Cho KH, Kim JH. Understanding the mechanism of glucose-induced relief of Rgt1-mediated repression in yeast. FEBS open bio. 2014;4:105–111. doi: 10.1016/j.fob.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, Wittenberg C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Molecular biology of the cell. 2003;14:3230–3241. doi: 10.1091/mbc.E03-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 60.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 61.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]