Abstract
The increased longevity of the C. elegans electron transport chain mutants isp-1 and nuo-6 is mediated by mitochondrial ROS (mtROS) signaling. Here we show that the mtROS signal is relayed by the conserved, mitochondria-associated, intrinsic apoptosis signaling pathway (CED-9/Bcl2, CED-4/Apaf1 and CED-3/Casp9) triggered by CED-13, an alternative BH3-only protein. Activation of the pathway by an elevation of mtROS does not affect apoptosis but protects from the consequences of mitochondrial dysfunction by triggering a unique pattern of gene expression that modulates stress sensitivity and promotes survival. In vertebrates, mtROS induce apoptosis through the intrinsic pathway to protect from severely damaged cells. Our observations in nematodes demonstrate that sensing of mtROS by the apoptotic pathway can, independently of apoptosis, elicit protective mechanisms that keep the organism alive under stressful conditions. This results in extended longevity when mtROS generation is inappropriately elevated. These findings clarify the relationships between mitochondria, ROS, apoptosis, and aging.
INTRODUCTION
The observed association of the aging process with the biology of reactive oxygen species (ROS), in particular ROS originating from mitochondria (mtROS), has led to the formulation of the oxidative stress theory of aging. Recently, however, more nuanced interpretations have been proposed to explain the basic observations that led to the formulation of the theory (Lapointe and Hekimi, 2010; Sena and Chandel, 2012). One possibility is that ROS damage is not causally involved in the aging process but that ROS levels are correlated with the aged phenotype because they modulate signal transduction pathways that respond to cellular stresses brought about by aging (Hekimi et al., 2011). In other words, ROS generation may be enhanced by the aging process because, in their role as signaling molecules, ROS help to alleviate the cellular stresses caused by aging. This hypothesis is supported by findings in a variety of organisms, in particular in C. elegans where changes in ROS generation or detoxification can be uncoupled from any effect on lifespan (Doonan et al., 2008; Van Raamsdonk and Hekimi, 2009, 2010; Yang et al., 2007). Most strikingly, moderate mitochondrial dysfunction (Felkai et al., 1999; Feng et al., 2001; Yang and Hekimi, 2010b), severe loss of mtROS detoxification (Van Raamsdonk and Hekimi, 2009), elevated mtROS generation (Yang and Hekimi, 2010a), as well as treatments with pro-oxidants (Heidler et al., 2010; Lee et al., 2010; Van Raamsdonk and Hekimi, 2012; Yang and Hekimi, 2010a), can all lengthen rather than shorten lifespan. In addition, the pro-longevity effects of both dietary restriction (Schulz et al., 2007), and reduced insulin signaling in C. elegans (Zarse et al., 2012), appear to involve an increase in ROS levels. Such observations are not limited to C. elegans. For example, mtROS signalling can act to extend chronological lifespan of the yeast S. cerevisiae (Pan et al., 2011).
The longevity phenotype of isp-1(qm150) (Feng et al., 2001) and nuo-6(qm200) (Yang and Hekimi, 2010b) mutants is most unequivocally connected to mtROS generation (Yang and Hekimi, 2010a). isp-1 encodes the ‘Rieske’ iron sulfur protein, one of the major catalytic subunits of mitochondrial complex III, and nuo-6 encodes the mitochondrial complex I subunit NDUFB4. The qm150 and qm200 mutations are missense mutations that do not lead to a full loss of protein function. Mitochondria isolated from both mutants show elevated superoxide generation, as measured by fluorescence sorting of purified mitochondria incubated with the dye MitoSox (Yang and Hekimi, 2010a). This is a very specific phenotype that is not accompanied by an increase in overall mitochondrial oxidative stress, nor by a measurable increase in overall oxidative damage. The long-lived phenotype can also be phenocopied by treatment of the wild type with a very low level (0.1mM) of the superoxide generator paraquat (PQ). In contrast, treatment of the mitochondrial mutants with PQ has no effect, suggesting that treatment with PQ extends lifespan by the same mechanisms as the mitochondrial mutations (Yang and Hekimi, 2010a).
Increased longevity can also result from induction of the mitochondrial unfolded protein stress response (mtUPR) which can be triggered by RNA interference knock-down of mitochondrial components (Dillin et al., 2002; Durieux et al., 2011; Lee et al., 2003). This response is however distinct from the response to elevated mtROS as the lifespan increases produced by the elevated mtROS in the mutants and by the activated mtUPR are fully additive (Yang and Hekimi, 2010b).
How might elevated mtROS promote longevity? ROS are well known to act as modulators in signal transduction pathways, and it is as such that they might be enhancing longevity. One candidate signaling pathway that could include potential mtROS sensors as well as a mechanism of downstream signalling is the intrinsic apoptosis pathway. Apoptosis is a highly controlled process that in mammals is sensitive to mitochondrial function, including mtROS, via the intrinsic apoptosis signal pathway (Wang and Youle, 2009). In C. elegans the intrinsic apoptotic machinery consists of the BH3-only protein EGL-1, CED-9 (Bcl2-like), CED-4 (Apaf1-like) and CED-3 (Casp9-like). CED-9 is tethered to the outer mitochondrial membrane and binds CED-4. However, in contrast to vertebrates, there is no evidence for any role for mtROS in regulating apoptosis in C. elegans.
Interestingly, the individual proteins or pairs of interacting proteins of the apoptotic signalling machinery appear to be able to carry out apoptosis-independent functions. For example, EGL-1 and CED-9 affect mitochondrial dynamics (Lu et al., 2011), CED-4 and CED-3 promote neuronal regeneration (Pinan-Lucarre et al., 2012), CED-4 appears to be involved in hypoxic pre-conditioning (Dasgupta et al., 2007) and S-phase checkpoint regulation (Zermati et al., 2007). These and similar findings in other organisms (Galluzzi et al., 2008) suggest that the proteins of the intrinsic apoptotic pathway have bona fide signal transduction activities in other processes. However, in no case to date has the full pathway, from a BH3-only protein to a caspase, been found to be involved in a process distinct from apoptosis.
Here we show that the isp-1 and nuo-6 mutations and 0.1mM PQ treatment induce a unique pattern of changes in gene expression. Strikingly, we found that mutations in the conserved intrinsic apoptosis signaling pathway (ced-9, ced-4 and ced-3) suppress the longevity of isp-1 and nuo-6 mutants, independently of inhibition of apoptosis. Moreover, unlike apoptosis, which requires the BH3-only protein EGL-1, the suppression of isp-1 and nuo-6 requires the BH3-only protein CED-13, which is not required for apoptosis. Treatment with PQ can bypass the need for CED-13, suggesting that mitochondrial ROS acts directly on activation of the pathway, possibly by acting on CED-9 and CED-4, which are associated with mitochondria. Loss of apoptotic signaling also suppresses most of the other phenotypes of isp-1 and nuo-6, such as slow development, slow behaviors, altered gene expression, and sensitivity to heat, but not the primary defects of low oxygen consumption and low ATP levels. The finding that the hypo-metabolic and hyper-sensitive phenotypes can be suppressed at the same time as longevity, without suppression of the low oxygen consumption and ATP concentration, indicates that these phenotypes are actively induced by mtROS to protect from mitochondrial dysfunction.
RESULTS
Pro-longevity mtROS signalling induces a unique pattern of gene expression
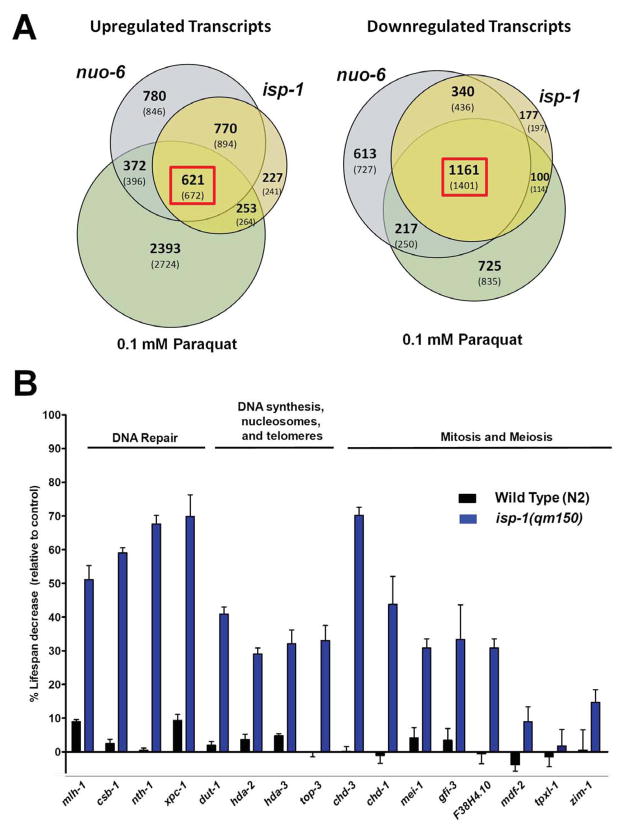
The effects of isp-1(qm150) and nuo-6(qm200) mutations on longevity are not additive, and a low dose of PQ (0.1mM) increases the lifespan of the wild type, but not of either of the mutants, (Yang and Hekimi, 2010a, b). These observations suggest that a common mechanism underlies the increased longevity in all three conditions. To test this further we used Affymetrix C. elegans microarrays to characterize patterns of gene expression in the two mutants and in wild type PQ-treated worms. The lists of genes with significantly altered expression overlap considerably (Table S1). 621 genes were up-regulated and 1161 were down-regulated in all three conditions (Figure 1A). This confirms that PQ treatment and the two mutations induce common changes in worm physiology. However, the only feature revealed by GO-term analysis was a down-regulation of large families of kinases and phosphatases whose expression is linked to the production of sperm (Reinke et al., 2000). GO-term analysis did not reveal any obvious pattern pointing to a specific biological process that could be responsible for the slow aging phenotype (Table S2). In particular, the list of genes up-regulated in all three conditions is not significantly enriched in ROS-detoxifying activities or ROS-damage repair activities (Table S3). This strongly suggest that the longevity increase under the three conditions is not the result of an over-compensatory increase in oxidative defences in response to increased levels of mtROS.
Figure 1.
Whole genome expression profiling of isp-1 and nuo-6 mutants, and the wild type treated with 0.1mM paraquat. A) Venn diagrams illustrating the number of significantly up-regulated or down-regulated transcripts found in each condition tested when compared to untreated wild type. Bolded numbers represent the actual number of probes whose expression was significantly changed relative to wild type expression, while numbers in brackets represent the maximum number of different transcripts that could be detected as a result of high homology. B) Lifespan changes resulting from treatment of wild type and isp-1 mutants with RNAi against genes that are up-regulated in all three conditions and whose activities are expected to be involved in genome stability. The majority of genes had large effects on the mutant but no, or very little, effect on the wild type. Bars represent the degree of lifespan shortening relative to control and error bars represent SEM. See also Figures S1, S2, S3; Tables S1, S2, S3, S4 and S6. Numerical values and statistical analyses for all lifespan experiments are presented in Table S5. The GEO ascension number for all gene array data in this paper is GSE54024.
We compared the lists of gene with similar lists obtained by genome-wide expression studies of other mutants and treatments related to the aging process (Table S4). The overlaps were from 1 to 20% for up-regulated genes and from 0 to 33% for down-regulated genes. The highest overlap was with the short-lived gas-1(fc21) mutant. Although gas-1 is short-lived it encodes a subunit of a mitochondrial complex I, which is likely the source of the similarities. However, the comparison did not identify a particular process by GO-term analysis (Table S2). Finally, in contrast to a recent study in yeast where mitochondrial ROS signaling ultimately resulted in the specific silencing at sub-telomeric loci (Schroeder et al., 2013), we found a uniform distribution of the down-regulated loci across all chromosomes (Figure S1). Taken together our findings suggest that the pattern of gene expression induced by elevated mtROS is unique, which is consistent with the observation that PQ treatment is fully or partially additive to the pro-longevity effects of mutations in daf-2, eat-2 and clk-1, and is not fully suppressed by mutations in daf-16, aak-2, wwp-1, hif-1, skn-1, or hsf-1 (Yang and Hekimi, 2010a).
The changes in gene expression are necessary for the pro-longevity effect of mtROS
To provide a proof-of-principle demonstration of the relevance for lifespan of the observed changes in gene expression, we focused on a small group of genes. The possibility that genome instability is important in determining lifespan remains a strong hypothesis in biogerontology. Furthermore, there are strong links between DNA damage and general stress responses (Ermolaeva et al., 2013). We tested 16 up-regulated genes belonging to this group (Figure 1B, S2 and S3). All numerical values and statistics for survival data presented in the paper are provided in Table S5. The knockdown of 3 of the genes in isp-1 mutants only had a very small effect on lifespan. The knockdown of the 13 other genes had a much larger effect on the mutant than on the wild type and the knockdown of 7 of these had virtually no effect on the wild type but strongly suppressed the longevity of the mutants (Figure 1B, S2 and S3). The fact that the knockdown of at least some of the up-regulated genes limits the longevity of isp-1 without affecting the wild type strongly suggest that at least some and probably many of the changes in gene expression we observed are necessary for the longevity resulting from mtROS signaling.
The longevity response of isp-1 and nuo-6, but not that of other longevity mutants, requires the conserved intrinsic apoptotic signaling pathway
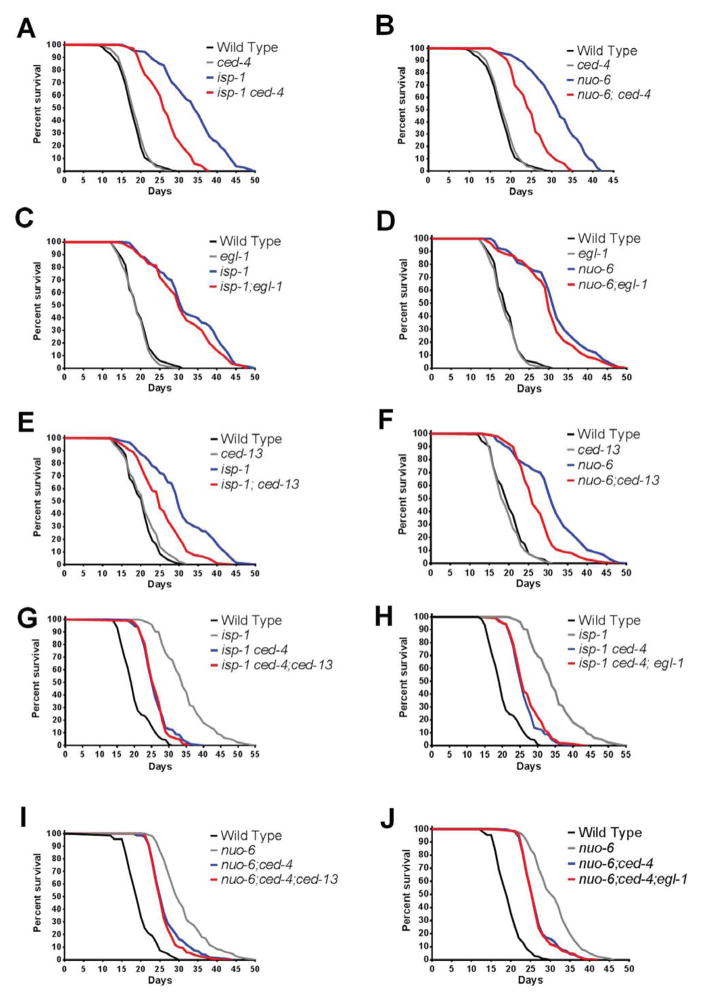
The intrinsic pathway of apoptosis, which uses conserved signaling proteins, is physically associated with mitochondria, and in vertebrates is sensitive to mitochondrial oxidative stress. We therefore tested whether this pathway was involved in the pro-longevity signal in worms by scoring the lifespan of double mutants of isp-1 and nuo-6 with ced-9gf, ced-4, and ced-3 mutations (Figure 2A–B, S4A–D). The mutations in all three ced genes significantly suppressed the longevity of both isp-1 and nuo-6, without having any significant effects on lifespan by themselves. The suppression by ced-4(n1162) was consistently the most robust. The suppression by ced-9(n1950) was somewhat less effective, possibly because it is a gain-of-function allele and might therefore not be fully equivalent to a loss of ced-4(n1162). The somewhat lesser suppression by ced-3(n717) suggests that CED-4 recruits other effectors as well.
Figure 2.
Genetic interactions between longevity and cell death genes. A) and B): effect of ced-4(n1162) on the survival of isp-1(qm150) and nuo-6(qm200). C) and D): effect of egl-1(n1084n3082) on the survival of isp-1 and nuo-6. E) and F): effect of ced-13(sv32) on the survival of isp-1 and nuo-6. G) Effects of ced-4 and ced-13 on isp-1 survival in the triple mutant combination. H) Effects of ced-4 and egl-1 on isp-1 survival in the triple mutant combination. I) Effects of ced-4 and ced-13 on nuo-6 survival in the triple mutant combination. J) Effects of egl-1 and ced-4 on nuo-6 survival in the triple mutant combination. See also Table S7; Figures S4 and S5. Numerical values and statistical analyses for all lifespan experiments are presented in Table S5.
We tested possible effects of ced-4 on other lifespan mutants that had previously been shown to be genetically distinct from isp-1/nuo-6, including eat-2, clk-1, daf-2 and glp-1. For this, we tested lifespan in double mutant combination with ced-4, but no effects on the lifespan of these mutants were detected (Figure S4E–H). In addition, previous findings had suggested that RNAi against subunits of the ETC prolong lifespan by a mechanism that is distinct from that of the genomic mutants isp-1 and nuo-6 (Yang and Hekimi, 2010b). We therefore tested RNAi against isp-1 and nuo-6 on ced-4 mutants and, as predicted, ced-4 did not suppress the longevity induced by the RNAi treatments (Figure S4I,J). We conclude that the intrinsic apoptotic signaling machinery uniquely mediate the longevity of isp-1 and nuo-6.
The longevity response is independent of apoptosis per se
As ced-9gf, ced-4 and ced-3 affect apoptosis, we scored embryonic and pharyngeal apoptosis in isp-1 and nuo-6 mutants as well as in ced-4, isp-1;ced-4 and nuo-6;ced-4 double mutants (Table S6). The pattern of apoptosis in the mitochondrial mutants was indistinguishable from the wild type, and the pattern of apoptosis in the double mutants with ced-4 was indistinguishable from that produced by the ced-4 mutation alone, that is, most cell deaths were eliminated. These findings indicated that isp-1 and nuo-6 do not affect normal or mutant apoptosis, but they cannot establish whether the normal pattern of apoptosis is necessary for the mutants’ increased longevity. For this we turned to the BH3-only protein EGL-1, which is required for all apoptosis in C. elegans. We scored both apoptosis and lifespan in egl-1 mutants as well as in egl-1;isp-1 and egl-1;nuo-6 double mutants. As expected, the egl-1 mutation, like the ced-4 mutation, abolished apoptosis in all three genotypes (Table S6). However, in contrast to ced-4, ced-9 and ced-3, egl-1 had no effect at all on lifespan (Figure 2C,D). Thus it is not the absence of apoptosis in the intrinsic pathway mutants that suppresses the lifespan of the mitochondrial mutants.
The activity of the intrinsic apoptotic signaling pathway on longevity requires CED-13, an alternative BH3-only protein
For canonical apoptotic signaling, the intrinsic pathway requires stimulation by a BH3-only protein. CED-13 is the only other protein in C. elegans to possess a BH3 domain (Schumacher et al., 2005). CED-13 has been shown to be able to have some effect on somatic apoptosis when overexpressed, and is also able to interact with CED-9 in vitro in a way that is similar to that of EGL-1 (Fairlie et al., 2006). However, loss of CED-13 has very limited effects and only on DNA damage-induced germline apoptosis. We found however that the ced-13(sv32) mutation suppressed the longevity of isp-1 and nuo-6 mutants as efficiently as the mutations in the genes of the core pathway (Figure 2E,F). We verified whether CED-13 acted indeed in the same pathway as the other CED proteins by testing whether the effects of ced-13(sv32) were additive to those of ced-4(n1162) for suppression of the lifespan of isp-1. We found that the lifespan of the triple mutants isp-1; ced-4; ced-13 and nuo-6; ced-4; ced-13 were indistinguishable from those of the double mutants isp-1; ced-4 and nuo-6; ced-4, respectively (Figure 2G,I), indicating that ced-13 acts in the same pathway as ced-4. As expected, egl-1 had no effect either in triple combinations (Figure 2H,J). Thus, rather than EGL-1, CED-13 is the BH3-only protein that is required for pro-longevity signaling through the intrinsic pathway.
mtROS act downstream of CED-13 for longevity
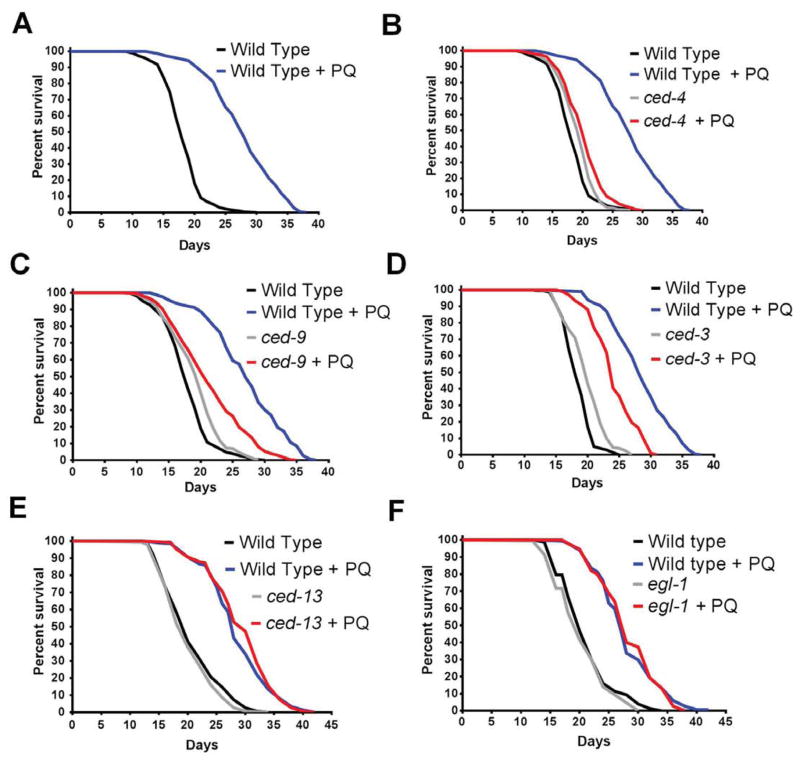
We first determined if treatment with 0.1mM PQ (which does lengthen wild type lifespan) or with 0.5mM PQ (which is too toxic to lengthen wild type lifespan) had any effect on apoptosis (Table S6). No effect was found at either concentration, which is consistent with mtROS being capable of regulating the CED-13-dependent activation of the pathway and not apoptosis. We then treated mutants of all four genes (ced-13, ced-9gf, ced-4 and ced-3) with 0.1mM PQ. The effect of PQ on lifespan was almost completely suppressed by ced-4 and ced-9, partially by ced-3 but not at all by ced-13 and egl-1 (Figure 3). This suggested that PQ (and thus mtROS) act downstream of CED-13. As the ced-13 mutation is capable of suppressing the lifespan of isp-1 and nuo-6 mutants, its inability to suppress the longevity induced by PQ suggests that the level of mtROS is insufficient in the isp-1 and nuo-6 mutants to trigger the pathway in the absence of stimulation by CED-13 but that the level of mtROS induced by PQ treatment is sufficient to directly activate the mitochondria-associated CED-9 and/or CED-4. Although ced-4 does not suppress the longevity induced by RNAi against ETC subunits the position of CED-13 upstream of CED-4 and of mtROS could in principle allow it to regulate RNAi-dependent longevity through a parallel pathway. However, no suppression of isp-1(RNAi) by ced-13 or egl-1 was observed (Figure S5). All further analyses of the pathway described below were conducted with ced-4 for part of the pathway downstream of ROS activity, with ced-13 for the part of the pathway upstream of ROS activity, and with egl-1 as control for apoptosis per se.
Figure 3.
Lifespan extension by 0.1mM paraquat (PQ) requires the intrinsic apoptosis pathway. A) Effect of 0.1mM PQ treatment on the wild type. Effects of 0.1mM PQ treatment on: B) ced-4(n1162), C) ced-9(n1950gf), D) ced-3(n717), E) ced-13(sv32) and F) egl-1(n1084n3082). See also Table S5. Numerical values and statistical analyses for all lifespan experiments are presented in Table S5.
Loss of the intrinsic pathway signaling does not suppress low oxygen consumption and ATP levels
isp-1 and nuo-6 encode subunits of mitochondrial respiratory complexes and the mutations lead to reduced oxygen consumption (Figure S6A and Table S5). This is likely a primary phenotype directly resulting from altered function of the electron transport chain. Lower electron transport chain function is expected to lead to ATP depletion. We found that ATP levels were low in both mutants and particularly severely in isp-1 mutants (Figure S6B and Table S5). Neither oxygen consumption nor ATP levels were affected in ced-4, ced-13, or egl-1. To test whether suppression by the ced mutations was achieved by restoration of electron transport or ATP levels, we measured oxygen consumption and ATP levels in suppressed double mutants (Figure S6C,D and Table S5). No effect on oxygen consumption or ATP levels was observed, indicating that this is not the mechanism by which phenotypic suppression is achieved.
Loss of the intrinsic pathway suppresses the hypo-metabolic and gene expression phenotypes of isp-1 and nuo-6 mutants
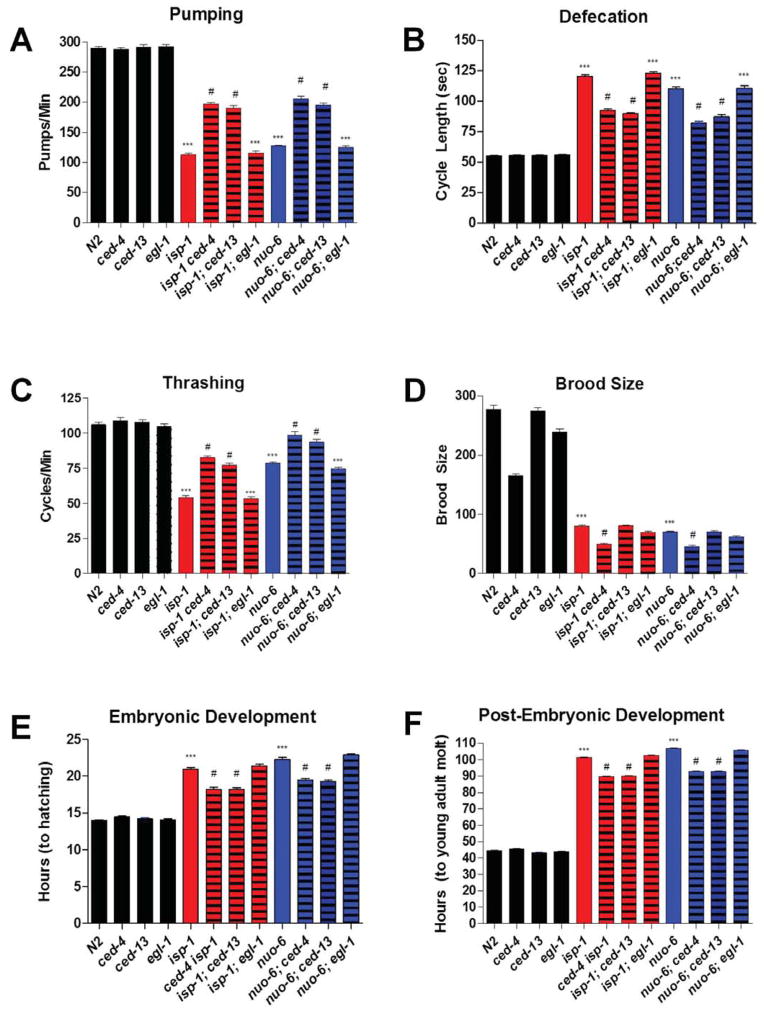
The isp-1 and nuo-6 mutations induce other phenotypes in addition to an increase in lifespan, including slow embryonic and post-embryonic development, as well as slow behaviors such as pumping, defecation and thrashing. Mutations in ced-13 and ced-4 but not egl-1 partially suppressed all these phenotypes of both isp-1 and nuo-6, (Figure 4 and Table S5). The fact that the egl-1 mutation, which abolishes cell death but has no effect on lifespan, had no effect on any of the phenotypes implies, as for longevity, that these phenotypes do not depend on changes in apoptosis. One phenotype that is not rescued and is in fact worsened by ced-4 is brood size (Figure 4D). This suggest that the germline phenotype due to mitochondrial dysfunction does not involve the longevity pathway we have uncovered. Recent findings suggest that apoptosis in the germline is necessary for oocyte quality (Andux and Ellis, 2008), which might be the cause of the reduction in brood size.
Figure 4.
The behavioural and growth defects of isp-1(qm150) and nuo-6(qm200) mutants are partially suppressed by ced-4(n1162) and ced-13(sv32) but not egl-1(n1084n3082). A) Pharyngeal pumping rate. isp-1 and nuo-6 pump at a significantly slower rate than the wild type. Loss of ced-4 or ced-13 but not egl-1 partially rescues the slow pumping rates of isp-1 and nuo-6. None of the cell death genes affects the pumping rate of the wild type. B) Defecation cycle length. isp-1 and nuo-6 mutants have a significantly lengthened defecation cycle length. Loss of ced-4 or ced-13 but not egl-1 partially rescues the slow defecation phenotype of isp-1 and nuo-6. None of the cell death genes affect the defecation cycle length of the wild type. C) Thrashing rate. isp-1 and nuo-6 mutants have a significantly decreased rate of thrashing. Loss of ced-4 or ced-13 but not egl-1 partially rescues the slow thrashing phenotype of isp-1 and nuo-6. None of the cell death genes affect the thrashing rate of the wild type. D) Brood size (the number of progeny produce by self-fertilization of a single hermaphrodite). Both isp-1 and nuo-6 have significantly reduced brood sizes. The reduction in brood size was enhanced by loss of ced-4 but not ced-13 or egl-1. Loss of ced-4, and to a lesser degree egl-1, also significantly reduced brood size the wild-type background. E) Length of Embryonic Development. The time taken for a 2-cell stage embryo to reach hatching is significantly increased in isp-1 and nuo-6 mutants. Loss of ced-4 and ced-13 but not egl-1 partially rescues this phenotype of isp-1 and nuo-6. None of the cell death genes affect the rate of embryonic development of the wild type. F) Length of post-embryonic development. The time taken for freshly hatched L1-stage larva to reach the young adult stage is significantly increased in isp-1 and nuo-6 mutants. Loss of ced-4 and ced-13 but not egl-1 partially rescues this phenotype of isp-1 and nuo-6. None of the cell death genes affect the rate of post-embryonic development of the wild type. Bars represent the mean value of 25 animals. Error bars represent standard error of the mean. Significance was determined using a Student’s t-test (*** denotes P < 0.0001 as compared to the wild type; # denotes P < 0.0001 as compared to either isp-1 or nuo-6 single mutants). Complete numerical values and statistics are provided in Table S5.
As described above, the isp-1 and nuo-6 mutations result in many changes in gene expression relative to wild type. We determined whether ced-4(n1162), which suppresses the increased lifespan of the mutants as well as most other phenotypes also suppressed the gene expression changes. Using Affymetrix C. elegans microarrays as before we compared the changes in gene expression in isp-1;ced-4 and nuo-6;ced-4 double mutants relative to wild type to those in the single mutants relative to wild type (Table S1). The ced-4 mutation partially suppressed both up-regulated and down-regulated changes in both isp-1 and nuo-6. 57% of the genes up-regulated in nuo-6 were back to wild type levels in nuo-6;ced-4, and 36% of the genes up-regulated in isp-1 were back to wild type level in isp-1;ced-4. Similarly ced-4 suppressed the down-regulation of 62% of the genes in the case of nuo-6 but only 18% in the case of isp-1. GO-term analysis of the list of genes affected by ced-4 in both mutants showed a meaningful enrichment only in the kinases and phosphatases linked to sperm production that were down-regulated in the mutants (Table S2). However, as the low brood size of the mutants was not suppressed by ced-4 the significance of this observation is unclear.
Constitutive activation of the CED pathway leads to heat-stress hyper-sensitivity
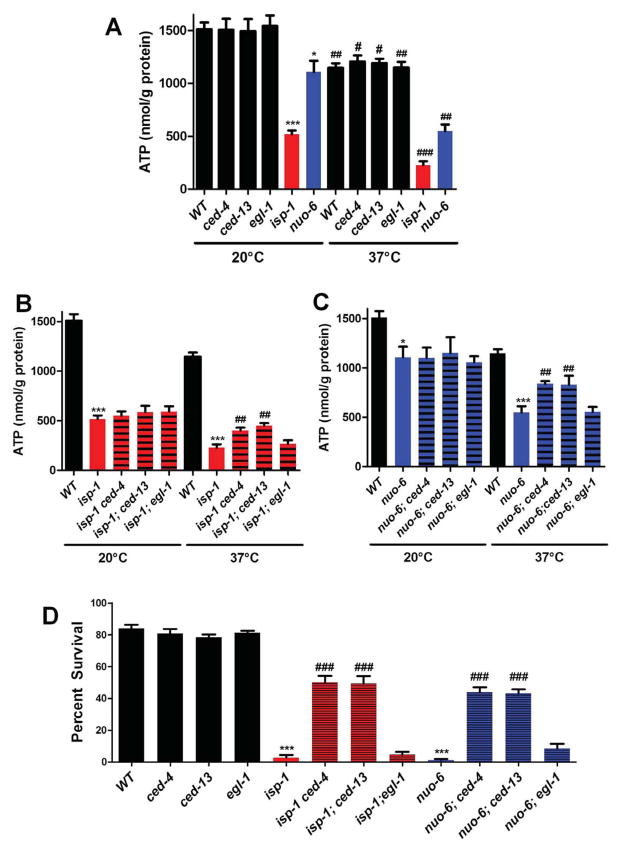
To further investigate the hypothesis that the CED pathway is a stress pathway that responds to mitochondrial dysfunction, we examined the effects of a severe heat stress. To establish the level of stress on mitochondrial function produced by this treatment we measured ATP levels after the animals had experienced 37°C for 1.5 hours. The stress led to a severe ATP depletion in all genotypes (Figure 5A and Table S5). However, the depletion was substantially more severe for the two mitochondrial mutants. While the wild type, ced-13, ced-4 and egl-1 experienced a ~30% drop, isp-1 and nuo-6 lost >50% of their already low ATP levels. Surprisingly, but consistent with our other findings, ced-13 and ced-4 but not egl-1 suppressed the severity of this effect (Figure 5B and 5C, and Table S5). To explore this further we treated young adult for 4 hours at 37°C and scored survival (Treinin et al., 2003). Treatment of all genotypes with this longer heat stress decreased survival, but much more severely in the mitochondrial mutants. Treatment of the wild type, ced-13, ced-4 and egl-1 resulted in ~80% survival but the treatment killed virtually all isp-1 or nuo-6 mutants (Figure 5D and Table S5). Again, ced-13 or ced-4 but not egl-1 suppressed the mitochondrial mutants such that the double mutants had much higher survival rates (40–50%). Taken together these observations suggest that resources required for acute survival are not available in animals in which the CED pathway is strongly and constitutively activated by mitochondrial dysfunction because they have been diverted to processes involved in long term survival.
Figure 5.
Effects of isp-1, nuo-6 and cell death genes on ATP levels and survival under heat stress. A) isp-1 and nuo-6 mutants, but not ced-4, ced-13 or egl-1 mutants exhibit reduced ATP levels when grown under standard conditions (20°C). Acute exposure (1.5h) to heat (37°C) reduces the ATP levels of all genotypes. B) Loss of ced-4, ced-13 or egl-1 does not affect ATP levels in isp-1 mutants at 20°C. However, the reduction in ATP levels after heat stress is significantly reduced in ced-4;isp-1 and isp-1;ced-13 but not egl-1;isp-1 double mutants compared to isp-1(qm150). C) Mutations in ced-4, ced-13 and egl-1 do not affect ATP levels in nuo-6 mutants at 20°C. However, the reduction in ATP levels after heat stress is significantly less in ced-4;nuo-6 and nuo-6;ced-13 but not egl-1;isp-1 double mutants compared to nuo-6(qm200). D) Exposure to heat stress for 4h significantly decreases the survival of all genotypes, but much more severely for isp-1(qm150) and nuo-6(qm200) mutants. However, loss of ced-4 or ced-13 but not egl-1 strongly rescues the survival of isp-1 and nuo-6 mutants. Significance was determined using a Student’s t-test (a) * denotes P < 0.05, *** denotes P < 0.0001 as compared to the wild type. # denotes P < 0.05 compared to the control at 20°C, ## denotes P < 0.05 compared to the wild type at 37°C. ### denotes P < 0.005 compared to the wild type at 37°C. (b) *** denotes P < 0.0005 relative to the wild type control, ## denotes P < 0.05 relative to isp-1(qm150) at 37°C. (c) * denotes P < 0.05 as compared to the wild type control at 20°C, *** denotes P < 0.001 as compared to the wild type control at 37°C, ## denotes P < 0.005 as compared to the nuo-6(qm200) at 37°C. Complete numerical values and statistics are provided in Table S5.
CED-13 acts upstream of mtROS for all phenotypes
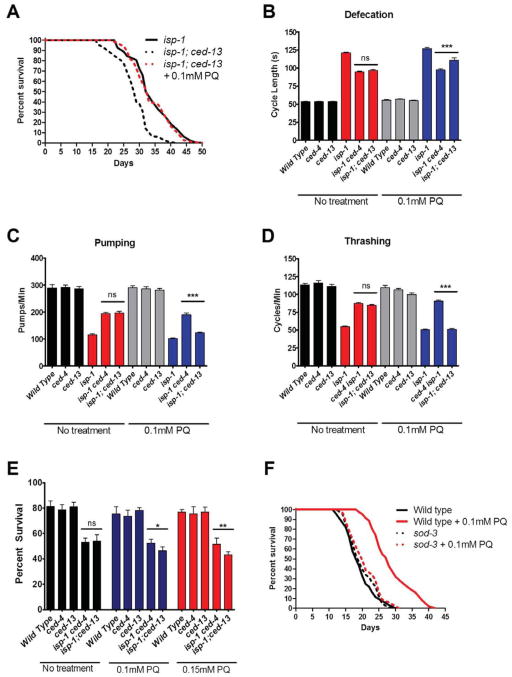
We focused on isp-1 to explore further the epistatic relationships in the ced-13-dependent pathway. Previous observations indicated that the longevity effect of PQ is not additive to isp-1 (Yang and Hekimi, 2010a), which we have confirmed (Table S5). On the other hand, the observation that ced-13 does not suppress the longevity induced by PQ treatment (Figure 3), places its action upstream of that of mtROS. This suggests that PQ should suppress the suppressed longevity of isp-1;ced-13 double mutants, which is what we observed (Figure 6A). Similarly, the slow defecation, pumping and thrashing of isp-1 mutants are partially suppressed by ced-13 and by ced-4 (Figure 4A–C). If mtROS act downstream of ced-13 but upstream of ced-4, PQ treatment should suppress the suppressive effect of ced-13 but not that of ced-4, which is what we observed (Figure 6B–D). Finally, ced-13 and ced-4 partially suppress the lethality induced by heat treatment (Figure 5D). Thus treatment with PQ should partially suppress the lethality suppression of ced-13 but not that of ced-4, which is what we observed (Figure 6E).
Figure 6.
Epistatic relationships between genotypes and treatments. A) Treatment of isp-1;ced-13 with 0.1mM PQ rescues lifespan to the isp-1 level (n>50, P < 0.0001 for the difference between treated and untreated double mutants). B) Treatment with 0.1mM PQ does not affect the defecation of isp-1 ced-4 but partially restores the defecation of isp-1;ced-13 toward the isp-1 level (n=25). C) Treatment with 0.1mM PQ does not affect the pumping rate of isp-1 ced-4 but partially restores isp-1;ced-13 pumping toward the isp-1 level (n=10). D) Treatment with 0.1mM PQ does not affect the trashing rate of isp-1 ced-4 but partially restores isp-1;ced-13 thrashing toward the isp-1 level (n =15). F) Treatment with 0.1mM and 0.15mM PQ decreases the acute survival of isp-1;ced-13 worms but not of isp-1 ced-4 at 37°C (for 4 hours). Significance for all experiments was determined using the Student’s t-test (* denotes P < 0.05, ** denotes P < 0.01, *** denotes P < 0.001. G) Treatment with 0.1mM PQ increases wild type lifespan but not sod-3(tm783) lifespan (n =150, P <0.0001 for the difference between the wild type and sod-3 treated with PQ). Error bars represent mean + SEM. See also Figure S6. Complete numerical values and statistics are provided in Table S5.
SOD-3 is involved in generating the pro-longevity mtROS signal
Treatment with PQ and altered ETC function in the mitochondrial mutants are believed to generate superoxide (Yang and Hekimi, 2010a). However, only peroxide is believed to cross membranes readily, which might be necessary to affect the CED pathway proteins, which are associated with the outer mitochondrial membrane. The main mitochondrial superoxide dismutase SOD-2 is not required to generate the pro-longevity ROS signal, as PQ treatment can further lengthen the already long lifespan of sod-2 mutants (Van Raamsdonk and Hekimi, 2009), which we have confirmed (Table S5). sod-3, which encodes a minor, inducible, mitochondrial superoxide dismutase very similar to SOD-2 in structure was the only ROS handling enzyme whose expression was found to be increased by microarray analysis (Table S3). Interestingly, we found that sod-3 was absolutely required for the pro-longevity signal induced by PQ treatment as this treatment was without effect on longevity in the sod-3(tm760) knockout background. (Figure 6F). This suggests that peroxide is the necessary intermediate for pro-longevity signaling. The specificity of the action of SOD-3 could be achieved by specific sub-mitochondrial localization in relation to the outer membrane localization of CED-4/CED-9 complexes. Interestingly, both SODs have recently been found to be closely associated with the ETC (Suthammarak et al., 2013).
DISCUSSION
A model for lifespan determination by mitochondrial dysfunction and mtROS signaling
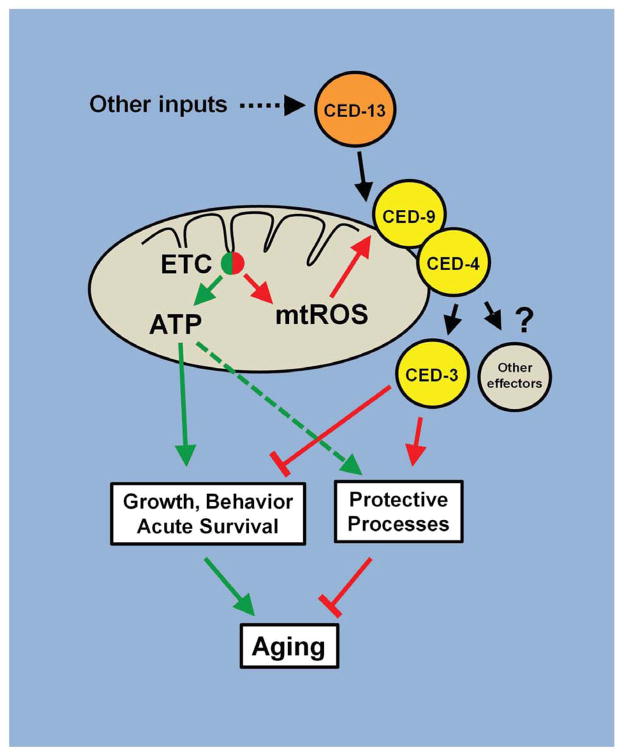
Previous studies have suggested that the mechanism of lifespan extension operating in the long-lived isp-1 and nuo-6 mutants is based on increased mtROS generation due to mitochondrial dysfunction. Here we provide further evidence for this by showing that PQ treatment and the mutations induce a common pattern of changes in gene expression which is at least in part required for longevity. Most importantly, we show that the mtROS signal requires the activity of the intrinsic apoptosis signaling pathway (including CED-9, CED-4 and CED-3), activated by a dedicated BH3-only protein, CED-13 (Figure 7). However, the recruitment of this pathway by mtROS and the consequences on longevity are fully independent of apoptosis per se. The known association of CED-9 and CED-4 with mitochondria, their involvement in sensing mtROS in vertebrates, and our findings from epistasis analysis that PQ, and therefore ROS, acts immediately downstream of CED-13, suggest that this pathway is the most immediately affected by mtROS and likely functions upstream of other pathways that might also be engaged (Lee et al., 2010; Walter et al., 2011). We found that loss of CED-3 suppresses less efficiently than loss of CED-4, suggesting that CED-4 could have other effectors in addition to CED-3, which is consistent with the existence of CED-3-independent activities of CED-4. ROS-independent activation by CED-13 provides the opportunity for input from upstream signals to modulate the sensitivity of the pathway to mtROS. For example, ced-13 expression appears to be regulated by cep-1, the C. elegans homologue of p53 (Schumacher et al., 2005). Furthermore, cep-1 appears to affect lifespan modulation by mitochondrial dysfunction in a complex manner (Baruah, 2014; Ventura et al., 2009). Thus the action of CED-13 might have affinities with that of BH3-only proteins such as PUMA and NOXA, which are regulated by p53 (Nakano and Vousden, 2001; Oda et al., 2000).
Figure 7.
A model for the regulation of lifespan by mtROS signaling through the intrinsic apoptosis pathway. The intrinsic apoptosis pathway (composed of CED-9, CED-4 and CED-3) is sensitive to mtROS from the ETC when it is activated by the alternative BH3-only protein CED-13. Mitochondrial dysfunction leads to an increase in mtROS which activates the CED signalling pathway to reduce ATP usage and redistribute it to protective rather than active functions. We propose that the mitochondrial dysfunction in isp-1(qm150) and nuo-6(qm200) mutants induces the mutant phenotypes, including longevity, both by directly lowering ATP generation and by stimulating mtROS signaling to alter ATP usage. In the wild type this mechanism could provide a protective role in case of transient mitochondrial dysfunction or nutrient shortage. In the mutants its continuous action leads to the mutant phenotypes, including longevity.
Loss of the CED pathway cannot rescue the low oxygen consumption and the low ATP levels. This is expected as isp-1 and nuo-6 mutations are point mutations in subunits of the mitochondrial respiratory chain and the ATP and oxygen phenotypes are likely primary defects that cannot be fixed. However, the loss of the CED pathway abolishes a large part of the increased longevity and several other phenotypes of the mutants, such as slow growth and behavior, a large sub-set of the changes in gene expression, and the hypersensitivity to heat stress. Taken together this suggests that mtROS acts in the mutants through the CED pathway to trigger phenotypic changes that alleviate the consequences of the primary defects, including protective changes that ultimately result in increased lifespan. Interestingly, the slow growth and behavioral phenotypes are the type of effects expected from mitochondrial dysfunction and the resulting low ATP production. Even slow aging can be postulated to result from low energy production, based on the observation that cold temperature and low metabolic rates are associated with longer lifespans. Thus it appears that the mtROS/CED pathway amplifies phenotypes that would be produced to a lesser extent by the immediate effects of mitochondrial dysfunction on energy metabolism alone. This could be a protective mechanism that, in the wild type, allows the mitochondria to recover their function when the dysfunction is only transient, sparing ATP and re-routing its use to protective mechanisms (Figure 7). In this model, longevity in the mutants is the result of both a slowing down of ATP-dependent processes that limit lifespan and by an abnormally intense activation of the protective pathway induced by elevated mtROS acting through the CED cascade. This model is consistent with the finding that, in contrast to the mutants which are only partially suppressed by ced-4 and ced-9gf, the longevity induced by PQ, which does not affect oxygen consumption nor ATP levels (Yang and Hekimi, 2010a), can be almost completely suppressed by ced-4 and ced-9 (Figure 3).
Activation of the apoptotic pathway by mtROS
In vertebrates mtROS are involved in the regulation of apoptosis by the intrinsic mitochondrial pathway. However, no clear role for mtROS in C. elegans apoptosis has yet been discovered. We confirmed this by showing that neither mtROS-generating mitochondrial mutations nor PQ treatment affect the extent of somatic apoptosis (Table S5). Two biological roles for apoptosis have been proposed: a role in shaping the development of multicellular organisms by eliminating cells that are not needed, and a protective role by eliminating cells that are damaged. In the somatic lineage of worms, apoptosis appears to have a developmental role but in the germline it might have a protective role for fertility by eliminating damaged gamete precursors (Gartner et al., 2008) and reallocating resources to produce high quality gametes (Andux and Ellis, 2008). In vertebrates, the mtROS-sensitive intrinsic pathway is part of a protective program and participates in the elimination of defective cells, including cells with defective mitochondria. Our findings suggest that in C. elegans, the intrinsic apoptotic machinery, including CED-9, CED-4 and CED-3, is also sensitive to mtROS when stimulated by the BH3-only protein CED-13. Stimulation by CED-13 leads to the activation of a protective program but not to apoptosis. How stimulation of the same pathway by CED-13 and EGL-1 results in different outcomes is unknown at the present time but likely involves cell type-specific differences. A program of protective apoptosis similar to that in vertebrates is probably not possible in C. elegans because of its very small number of post-mitotic cells. Losing damaged cells is not an option without losing important functions and bodily integrity. However, stimulating protective and repair mechanisms in the face of injury remains useful. Thus it appears that what is conserved from nematode to vertebrates is the use of the proteins of the intrinsic pathway to transduce a mtROS signal that stimulates a protective response to mitochondrial dysfunction. It is interesting to speculate whether a non-apoptotic protective function of the intrinsic pathway is also acting in vertebrate post-mitotic cells such as neurons and could have a role in protecting from neurodegeneration.
EXPERIMENTAL PROCEDURES
Strains and Genetics
All strains were maintained by standard methods, at 20°C, on solid agar (NGM plates), and fed E. coli OP50. The following genotypes were used: Bristol N2 (wild type); LGI: nuo-6(qm200), sod-2(ok1030); LGII: eat-2(ad1116); LGIII: daf-2(e1370), clk-1(qm30), ced-4(n1162), ced-9(n1950); glp-1 (e2141ts); LGIV: isp-1(qm150), ced-3(n717); LGV: egl-1(n1084n3082); LGX: ced-13(sv32), sod-3(tm783).
Lifespan Analysis
All lifespan measurements were performed at 20°C and set up using a 4 hour limited lay. An experimental pool of 50 animals was used for each genotype in any given experiment, and lost or animals that died prematurely were replaced from a backup pool. Statistical analysis was performed using GraphPad Prism (v5.0) and Student’s t-tests in Microsoft Excel.
Paraquat (PQ) Treatment
Paraquat (Sigma-Aldrich, St. Louis, USA) was added to NGM plates at a final concentration of 0.1mM, 0.15mM or 0.5mM. OP50 grown on regular NGM plates was transferred onto NGM-PQ plates using a platinum pick instead of seeding directly onto the NGM-PQ plates. Control NGM plates containing no PQ were treated in a similar fashion.
Gene Expression Studies
2000 synchronized young adults grown at 20°C on NGM plates were collected, frozen in liquid nitrogen and total RNA was extracted using a Qiagen RNeasy Tissue Microarray Mini kit. Total RNA samples were analyzed for concentration and dissolution spectrophotometrically using a Nanodrop ND-100 Spectrophotometer. RNA samples were processed by Génome Québec (Montreal) and hybridized onto Affymetrix C. elegans GeneChips. Raw expression data was analyzed using FlexArray v1.6.1 (Génome Québec) and normalized using the GC-RMA method. Comparisons of each genotype were compared to the wild type using the Empirical Base (Wright & Simon) algorithm and fold changes were represented on a log2 scale. A threshold of p<0.05 and a fold change of 1.3 (log2) was set to determine differentially expressed targets.
Comparisons of Gene Expression Patterns
Comparisons made to other published data sets were done using raw Affymetrix data sets wherever possible (obtained from NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). Raw data was imported to FlexArray and handled identically to the data that was generated in this study. For studies that did not deposit their data to GEO or used technologies other than Affymetrix, comparisons of gene lists (upregulated and downregulated transcripts) were conducted using Microsoft Excel.
Gene Ontology (GO) Term Analysis
Gene ontology (GO) term analysis was performed using Cytoscape (v2.8.3) and the BiNGO plugin (v2.44). A hypergeometric test using the Benjamini & Hochberg false discovery rate (FDR) correction was implemented at a significance level of 0.05.
Measurement of Apoptosis
Quantification of corpses or cells was performed as previously described (Lu et al., 2009; Schwartz, 2007).
Whole worm phenotypes
All phenotypes were measured as before (Yang and Hekimi, 2010b).
Oxygen Consumption
Mixed populations of worms were collected and washed 3x in M9 to a final volume of 50μL of packed worms. 25μL of worms was then resuspended to a final volume of 50μL using M9 buffer and loaded into a chamber of an Oroboros Oxygraph-2K. The remaining 25μL of worms were freeze-thawed 3x in liquid nitrogen and resuspended in lysis buffer for immediate determination of protein concentration by a BCA Protein Assay kit (Thermo Scientific, Rockford, USA).
ATP Measurements
Young adult populations were collected using a 4 hour limited lay. Worms were picked and washed 3 times in M9. Worm pellets were subjected to 3 cycles of freeze-thaw using liquid nitrogen and subsequently spun down for 15 minutes at top speed. The resulting supernatant was assayed using an ATP Determination kit (Life Technologies, Carlsbad, USA). Protein concentrations were determined as described above.
Heat Stress Assays
Young adults were picked onto NGM plates that were pre-heated to 37°C and incubated for 4 hours at 37°C. Animals were allowed to recover for 30 minutes and scored for viability. For ATP measurements after heat stress, mixed populations were transferred onto pre-heated NGM plates and incubated for 1.5 hours at 37°C. Animals were then collected and washed 3 times with M9 and flash frozen and stored in liquid nitrogen. ATP measurements were performed as described above. For experiments performed using paraquat, paraquat plates were made as described and worms were grown on paraquat for one generation. Young animals that were grown on paraquat were subsequently assayed on pre-heated paraquat plates.
Supplementary Material
Acknowledgments
We thank Robyn Branicky for thoughtfully commenting on the manuscript, Eve Bigras for technical assistance, and Steve Hodgkinson for technical advice. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The work was funded by grants from the Canadian Institutes of Health Research to SH: MOP-114891, MOP-123295, MOP-97869 and INE-117889 (through the CoEN initiative (www.coen.org), as well as by McGill University. SH is Strathcona Chair of Zoology and Campbell Chair of Developmental Biology.
References
- Andux S, Ellis RE. Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS genetics. 2008;4:e1000295. doi: 10.1371/journal.pgen.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah HC, Hall Mathew, Yuan Jie, Gordon Sarah, Johnson Erik, Shtessel Ludmila L, Yee Callista, Hekimi Siegfried, Brent Derry W, Lee Siu Sylvia. CEP-1, the Caenorhabditis elegans p53 homolog, mediates opposing longevity outcomes in mitochondrial electron transport chain mutants. PLoS genetics. 2014 doi: 10.1371/journal.pgen.1004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N, Patel AM, Scott BA, Crowder CM. Hypoxic preconditioning requires the apoptosis protein CED-4 in C. elegans. Current biology : CB. 2007;17:1954–1959. doi: 10.1016/j.cub.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes & development. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Segref A, Dakhovnik A, Ou HL, Schneider JI, Utermohlen O, Hoppe T, Schumacher B. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501:416–420. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairlie WD, Perugini MA, Kvansakul M, Chen L, Huang DC, Colman PM. CED-4 forms a 2 : 2 heterotetrameric complex with CED-9 until specifically displaced by EGL-1 or CED-13. Cell death and differentiation. 2006;13:426–434. doi: 10.1038/sj.cdd.4401762. [DOI] [PubMed] [Google Scholar]
- Felkai S, Ewbank JJ, Lemieux J, Labbe JC, Brown GG, Hekimi S. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. The EMBO journal. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Joza N, Tasdemir E, Maiuri MC, Hengartner M, Abrams JM, Tavernarakis N, Penninger J, Madeo F, Kroemer G. No death without life: vital functions of apoptotic effectors. Cell death and differentiation. 2008;15:1113–1123. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK. Germline survival and apoptosis. WormBook. 2008:1–20. doi: 10.1895/wormbook.1.145.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidler T, Hartwig K, Daniel H, Wenzel U. Caenorhabditis elegans lifespan extension caused by treatment with an orally active ROS-generator is dependent on DAF-16 and SIR-2.1. Biogerontology. 2010;11:183–195. doi: 10.1007/s10522-009-9239-x. [DOI] [PubMed] [Google Scholar]
- Hekimi S, Lapointe J, Wen Y. Taking a “good” look at free radicals in the aging process. Trends in cell biology. 2011;21:569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. When a theory of aging ages badly. Cellular and molecular life sciences : CMLS. 2010;67:1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Current biology : CB. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature genetics. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lu N, Yu X, He X, Zhou Z. Detecting apoptotic cells and monitoring their clearance in the nematode Caenorhabditis elegans. Methods in molecular biology. 2009;559:357–370. doi: 10.1007/978-1-60327-017-5_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Rolland SG, Conradt B. A molecular switch that governs mitochondrial fusion and fission mediated by the BCL2-like protein CED-9 of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E813–822. doi: 10.1073/pnas.1103218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Molecular cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell metabolism. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinan-Lucarre B, Gabel CV, Reina CP, Hulme SE, Shevkoplyas SS, Slone RD, Xue J, Qiao Y, Weisberg S, Roodhouse K, et al. The core apoptotic executioner proteins CED-3 and CED-4 promote initiation of neuronal regeneration in Caenorhabditis elegans. PLoS biology. 2012;10:e1001331. doi: 10.1371/journal.pbio.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, et al. A global profile of germline gene expression in C. elegans. Molecular cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell metabolism. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell metabolism. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A, Conradt B, Shaham S. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell death and differentiation. 2005;12:153–161. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- Schwartz HT. A protocol describing pharynx counts and a review of other assays of apoptotic cell death in the nematode worm Caenorhabditis elegans. Nature protocols. 2007;2:705–714. doi: 10.1038/nprot.2007.93. [DOI] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Molecular cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthammarak W, Somerlot BH, Opheim E, Sedensky M, Morgan PG. Novel interactions between mitochondrial superoxide dismutases and the electron transport chain. Aging cell. 2013;12:1132–1140. doi: 10.1111/acel.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treinin M, Shliar J, Jiang H, Powell-Coffman JA, Bromberg Z, Horowitz M. HIF-1 is required for heat acclimation in the nematode Caenorhabditis elegans. Physiological genomics. 2003;14:17–24. doi: 10.1152/physiolgenomics.00179.2002. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS genetics. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Reactive Oxygen Species and Aging in Caenorhabditis elegans: Causal or Casual Relationship? Antioxidants & redox signaling. 2010;13:1911–1953. doi: 10.1089/ars.2010.3215. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, Johnson TE. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging cell. 2009;8:380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS biology. 2011;9:e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Youle RJ. The role of mitochondria in apoptosis*. Annual review of genetics. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS biology. 2010a;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging cell. 2010b;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell metabolism. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zermati Y, Mouhamad S, Stergiou L, Besse B, Galluzzi L, Boehrer S, Pauleau AL, Rosselli F, D’Amelio M, Amendola R, et al. Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Molecular cell. 2007;28:624–637. doi: 10.1016/j.molcel.2007.09.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.