Abstract

Influenza A is an RNA virus with a genome of eight negative sense segments. Segment 7 mRNA contains a 3′ splice site for alternative splicing to encode the essential M2 protein. On the basis of sequence alignment and chemical mapping experiments, the secondary structure surrounding the 3′ splice site has an internal loop, adenine bulge, and hairpin loop when it is in the hairpin conformation that exposes the 3′ splice site. We report structural features of a three-dimensional model of the hairpin derived from nuclear magnetic resonance spectra and simulated annealing with restrained molecular dynamics. Additional insight was provided by modeling based on 1H chemical shifts. The internal loop containing the 3′ splice site has a dynamic guanosine and a stable imino (cis Watson–Crick/Watson–Crick) GA pair. The adenine bulge also appears to be dynamic with the A either stacked in the stem or forming a base triple with a Watson–Crick GC pair. The hairpin loop is a GAAA tetraloop closed by an AC pair.
Influenza virus infections annually contribute to 3300–49000 deaths1 and more than 200000 hospitalizations in the United States.2 The largest influenza pandemic, known as the Spanish flu (H1N1, 1918–1919), killed as many as 40–50 million people worldwide.3 Lesser pandemics consist of the Asian (H2N2, 1957), Hong Kong (H3N2, 1968), and Russian (H1N1, 1977) flus.3 Available drugs are neuraminidase inhibitors and M2 ion channel blockers (adamantanes).4 However, the emergence of influenza strains with resistance to both classes of drugs, especially neuraminidase inhibitors,5,6 has led to interest in identifying new antiviral therapeutics.7 Antiviral agents may selectively target viral RNA structure with small molecules,8−10 oligonucleotides,11 or synthetic peptides.12
The influenza A genome consists of eight segments of negative sense vRNA, which encode at least 11 proteins.13 A pandemic of influenza occurs when RNA segments of human and animal viruses reassort to give rise to new strains to which humans have no immunity.13 The extreme ends of each segment are highly conserved and base pair to form a promoter for RNA synthesis.13
With bioinformatics approaches, Moss et al.14 identified conserved and stably folded secondary structures of influenza A mRNAs. Three of the secondary structures have been confirmed by chemical mapping,15,16 and a fourth was found to fold into a hairpin rather than a predicted multibranch loop.17 Two of the conserved secondary structures contain the 3′ splice site of segment 7 mRNA. Segment 7 encodes the essential M1 and M2 and/or M42 proteins.18,19 An equilibrium between a two-hairpin folding and a pseudoknot folding may regulate expression of M1 and M2 and/or M4215,19 (Figure 1). For example, the equilibrium populations of these conformations may depend on factors in the cellular environment such as pH, protein binding, or the presence of metabolites. In chemical mapping experiments on the two-hairpin model of the 3′ splice site of segment 7 mRNA (Figure 1), the smaller hairpin, 14 nucleotides (nt), is dynamic on the basis of high reactivity to enzymes and small molecules.15 Herein, we report the NMR structure of the consensus sequence of the larger hairpin, 37 nt, containing the 3′ splice site (Figure 1).
Figure 1.
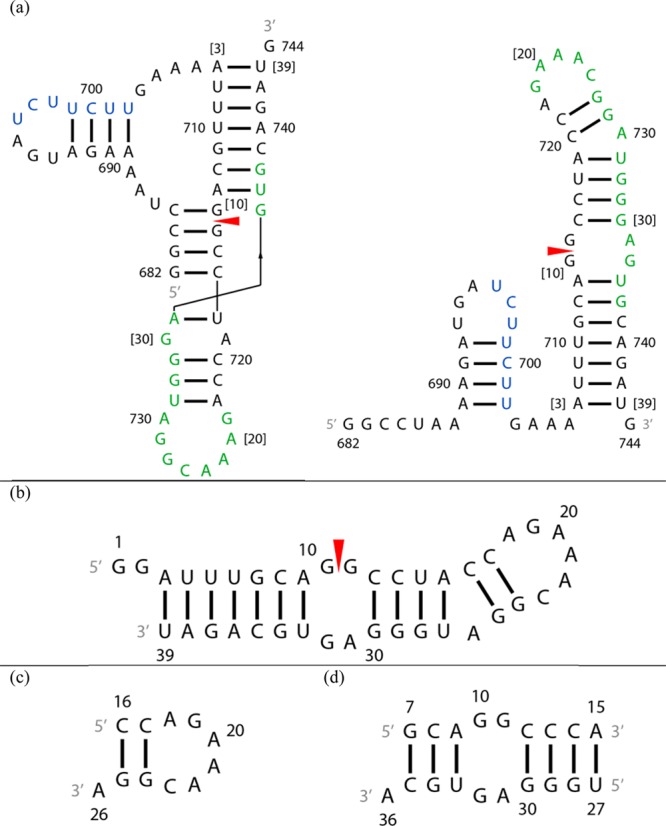
Secondary structures of constructs of the 3′ splice site region of segment 7 mRNA. A red arrowhead denotes the splice site. (a) Pseudoknot and hairpin conformations from ref (15). The SF2/ASF exonic splicing enhancer binding site is colored green and a polypyrimidine tract blue.15 Numbers in brackets correspond to numbering of residues of the 39 nt hairpin studied with NMR. (b) The 39 nt hairpin studied with NMR. (c) The 11 nt hairpin mimic. (d) The 19 nt duplex model containing the 2 nt × 2 nt internal loop. The U14 residue in the 39-mer was substituted with a cytidine to stabilize formation of the target heterodimer over a homodimer.
Experimental Methods
Preparation of 39 nt Hairpin Samples
Milligram quantities of a 39 nt construct containing the segment 7 hairpin (Figure 1) were prepared with in vitro transcription by T7 RNA polymerase.20 T7 RNA polymerase was synthesized from a plasmid supplied by B. S. Tolbert (Case Western Reserve University, Cleveland, OH) and purified via nickel column affinity chromatography.21 A pUC18 plasmid containing an insert for the RNA sequence was constructed and purified from Escherichia coli competent cells with standard plasmid preparation protocols (see the Supporting Information for the plasmid construct). The 5′ tail of the hairpin was replaced with a 5′ GG dinucleotide to enhance the efficiency of transcription initiation.22 The plasmid was linearized with EcoRV-HF restriction endonuclease (New England BioLabs) at 37 °C prior to in vitro transcription. Transcription mixtures typically consisted of 25 mM Mg2+, 1 mg/mL DNA template, rNTPs (12–13 mM each), 40 mM DTT, and 0.65–0.70 mg/mL T7 RNA polymerase at pH 7.5–8.0. After transcription mixtures had been incubated for 2 h at 37 °C, 2.5 μL of 0.5 M EDTA and 6 μL of 50% glycerol were added for every 30 μL of transcription mixture to stop reactions. Transcription mixtures were purified via FPLC using three 5 mL HiTrap DEAE Sepharose FF columns (GE Healthcare) connected in series.23 FPLC fractions with purified RNA were concentrated and then exchanged with an Amicon Ultra-15 Centrifugal Filter Unit (EMD Millipore) into NMR buffer [80 mM KCl, 20 mM KH2PO4/K2HPO4, and 0.02 mM Na2EDTA (pH 6.0)] to yield 3.8 mg of RNA. The final NMR sample had 1.1 mM RNA in 300 μL, including 15 μL of D2O to provide a lock signal.
A second sample of the 39 nt hairpin was synthesized from the linearized plasmid template with T7 High Yield RNA Synthesis Kits (New England BioLabs). This sample was initially purified via FPLC and concentrated, as described above. The second sample was further purified via denaturing polyacrylamide gel electrophoresis, extracted from gels via electroelution, concentrated, and exchanged into NMR buffer.24 The final NMR sample had 1.4 mM RNA in 400 μL of NMR buffer, including 15 μL of D2O. NMR spectra for the two samples were essentially identical.
Design of 11 nt Hairpin and 19 nt Duplex Model Mimics
The relatively large size of the 39 nt hairpin resulted in spectral overlap of resonances that made resonance assignments difficult. To aid resonance assignments, two smaller model constructs were assembled: (1) a 19 nt duplex RNA corresponding to the 2 nt × 2 nt internal loop region and (2) an 11 nt RNA corresponding to the hairpin loop region (Figure 1). In the 19-mer duplex, the residue that corresponds to U14 of the 39-mer was substituted with cytidine to stabilize formation of the intended heterodimer over a homodimer (Figure S1 of the Supporting Information). The 11 nt hairpin and the bottom strand of the 19 nt duplex each have a 3′ dangling nucleotide to stabilize their helices.25,26
Preparation of Model Mimic Samples
Oligoribonucleotides were purchased from Integrated DNA Technologies, Inc. (IDT), and dissolved in 315 μL of NMR buffer, including 15 μL of D2O. The highest concentration of r(5′CCAGAAACGGA) was 4.0 mM. The final concentration of the duplex, r(5′GCAGGCCCA) + r(5′UGGGAGUGCA), was 1.0 mM. MgCl2 was added to each sample to a final concentration of 5 mM.
NMR Spectroscopy
NMR spectra of samples in Shigemi NMR tubes (Shigemi, Inc.) were acquired on Varian Inova 500 and 600 MHz spectrometers. For samples in H2O, one-dimensional spectra were recorded for all constructs at a series of temperatures, with a 1–1–echo pulse to suppress the water signal.27 For 2D spectra, a WATERGATE pulse with flipback28,29 or an S-pulse30 was applied during acquisition to suppress the water signal. 2D NOESY spectra with different mixing times were acquired at −2 and 20 °C for all constructs and at additional temperatures for the two smaller constructs. 2D TOCSY spectra were acquired with mixing times between 30 and 50 ms for all constructs. Imino chemical exchange peaks of the 39-mer were detected with a 2D ROESY experiment, where their sign is the opposite of that of peaks arising from direct cross-relaxation.31 2D NOESY spectra32 were acquired on the smaller constructs in D2O at a series of temperatures to overcome ambiguities due to overlaps.
Proton chemical shifts were referenced internally to the frequency of water, with 2,2-dimethylsilapentane-5-sulfonic acid (DSS) as the external reference standard, and carbon chemical shifts were referenced indirectly to DSS on the basis of the absolute proton frequency according to Biological Magnetic Resonance Data Bank (BMRB) standards.31,33 2D NMR spectra were processed with NMRPipe.34
Modeling Methods
NMR Spectra for Obtaining Restraints
Resonances were assigned with standard procedures using 1H–1H NOESY, 1H–1H TOCSY, 1H–13C HSQC, and 1H–31P HETCOR spectra and SPARKY.35 Distance restraints were generated from spectra with mixing times between 50 and 150 ms, to minimize contributions from spin diffusion.31,36
Methods for Obtaining NOE Restraints
Most distance restraints for pairs of hydrogen atoms were obtained by integrating NOE volumes with SPARKY.35 Some that were difficult to integrate were manually assigned to a range of distances based on the relative size of their NOEs. NOE volumes were converted to distance restraints by referencing to volumes from fixed distances: H2′–H1′ (2.75 Å), H4′–H1′ (3.35 Å), pyrimidine H5–H6 (2.45 Å), cytosine H42–H41 (1.75 Å), cytosine H41–guanine H1 in a CG pair (2.70 Å), and adenine H2–uracil H3 in an AU pair (2.85 Å). Hydrogen bonds between bases were restrained to 2.1 ± 0.3 Å for all canonical base pairs. H1′–H2′ scalar coupling information was used to restrain all canonically base paired residues to the C3′-endo conformation. The χ dihedral angle was held between 170° and 340° (anti) for all residues except G10, where NMR evidence indicates that it is flexible.
In general, if an NOE in equivalent chemical environments is present in spectra of the 39 nt hairpin and either smaller construct, a distance restraint was obtained from the smaller construct to reduce complications of peak overlap. Because of different chemical environments, NOEs from the 39 nt hairpin were used for distance restraints rather than terminal residues G7, A15, U27, and A36 in the 19 nt duplex and C16 and A26 in the 11 nt hairpin. Intraresidue NOE volumes from those residues, however, were used to calculate reference distances for the 19 nt duplex and 11 nt hairpin. Restraints were also not obtained from C14 in the 19-mer duplex because the C14-G28 pair is structurally different from the U14-G28 pair in the 39-mer. Restraints for the 11-mer and 19-mer duplex constructs were obtained from spectra acquired with Mg2+ present because they generally had narrower and better-resolved cross-peaks that could be more accurately integrated.
Structure Calculation
Structures were refined with a simulated annealing37,38 protocol on a starting structure built with NUCGEN.39 Solvent was simulated with the generalized Born implicit solvent model and 0.1 M NaCl.40 The system was heated from 0 to 3000 K in 5000 steps for 5 ps and cooled to 100 K in 93000 steps for 93 ps and then to 0 K in 2000 steps for 2 ps. Force constants were 12 kcal mol–1 Å–2 for NOE restraints and 12 kcal mol–1 rad–2 for dihedral angle restraints. The weight of the restraints was increased from 0.1 to 1 during the first 3000 steps, i.e., during heating, and held at 1 for the remainder of the simulation. These restrained molecular dynamics calculations were conducted with AMBER 1441 using the parm99χ_YIL force field.42 The simulated annealing procedure was repeated with different initial velocities to generate an ensemble of 200 structures. The 22 structures without violations along with the eight structures with the lowest distance restraint violation energies and violations between 0.1 and 0.2 Å were refined with the same simulated annealing protocol except that they were heated to 600 K. Force constants for refinement were 30 kcal mol–1 Å–2 for NOE restraints and 30 kcal mol–1 rad–2 for dihedral angle restraints. The 20 structures with the lowest distance restraint violation energies that also agreed with NMR experimental restraints were selected as a final ensemble of structures. Similar structure minimization and refinement protocols were followed for the 19 nt duplex and 11 nt hairpin. Rmsds of the ensemble of structures were calculated with VMD.43 Images of 3D models of the RNA were generated with PyMOL.
Relationship between Chemical Shifts and Structure
Chemical shifts can provide structural information about RNA.44 Programs have been developed to predict 3D structure from chemical shifts45,46 or chemical shifts from a model structure.47,48 For RNA, nonexchangeable 1H chemical shifts calculated with programs such as NUCHEMICS47 and RNAShifts49 agreed well with experiments.49,50
The ROSETTA software suite46,51 on the ROSIE server52 was used to model separately two fragments of the 39 nt hairpin: a 19 nt hairpin containing residues 12–30 with the hairpin loop and A26 bulge and an 18 nt duplex containing residues 7–15 and 27–35 with the internal loop. Input files for each computation consisted of the RNA sequence, a predicted secondary structure, and a set of assigned chemical shifts (Table S1 and Figure S2 of the Supporting Information). Chemical shifts of nonexchangeable protons of the 39 and 11 nt hairpins and the 19 nt duplex were also calculated with NUCHEMICS for ensembles of 20 minimized 3D structures restrained by NOEs. For comparison with experiment, the chemical shift of each hydrogen was averaged among the 20 minimized structures for each construct.
Results
The Secondary Structure from NMR Is That Predicted from Bioinformatics
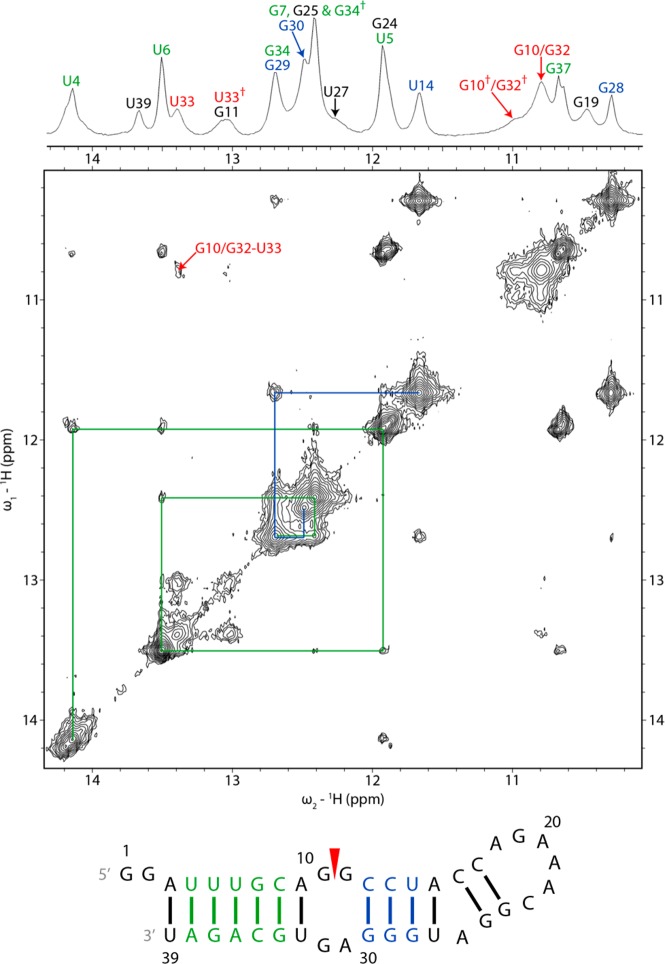
NMR spectra of the 39 nt hairpin confirm the helices predicted by bioinformatics and supported by chemical mapping (Figures 2 and 3).14,15 Resonance assignments of exchangeable protons in the helices of the 39-mer commenced with identifying signature GH1–UH3 cross-peaks of GU pairs in the 10–12 ppm region of NOESY spectra at −2 °C (Figure 2).53 Imino resonances corresponding to helices U4-A38 to C8-G34 and C12-G30 to U14-G28 were assigned from these cross-peaks according to the imino proton chemical shifts of AU and GC pairs (Figure 2 and Table S2 of the Supporting Information).53 A table of all assigned chemical shifts is given in the Supporting Information.
Figure 2.
Imino proton region of 1D and 2D proton NMR spectra of the 39 nt construct showing sequential proton walks with blue and green lines. The water signal was suppressed with a 1–1–echo pulse in the 1D spectrum and an S-pulse in the 2D NOESY spectrum. The daggers in the 1D spectrum mark chemical exchange peaks. The spectra were acquired at −2 °C with a mixing time of 125 ms for the 2D spectrum. Addition of 5 and 10 mM Mg2+ caused minor shifts and sharpening of the imino resonances, including U4, U5, U6, G11, G19, G24, and U33 (Figure S3 of the Supporting Information).
Figure 3.

Schematic of the secondary structure of the 39 nt hairpin with assigned interresidue NOEs. Blue lines denote NOEs identified in the 39-mer and the 19 nt duplex, green lines NOEs identified in the 39-mer and the 11 nt hairpin, and red lines NOEs identified only in the 39-mer.
A cross-peak between resonances at 12.41 and 12.69 ppm in a NOESY spectrum at −2 °C was determined from a ROESY spectrum to be a chemical exchange peak for G34H1. The primary G34H1 peak lies at 12.69 ppm. The secondary peak overlaps with G7H1 (Figure 2). The A15H2–U27H3 cross-peak is weak because of solvent exchange (Figure S4 of the Supporting Information). The G24H1 peak was assigned to 11.92 ppm in the −2 °C NOESY spectrum based on cross-peaks to C17 amino protons (Figure S4 of the Supporting Information) but is missing from the 20 °C NOESY spectrum. The G25H1 peak was assigned to 12.41 ppm in the 20 °C NOESY spectrum on the basis of cross-peaks to C16 amino protons but could not be identified in the −2 °C NOESY spectrum. These assignments indicate that C16 and G25, in addition to C17 and G24, form Watson–Crick base pairs, though an NOE between G24H1 and G25H1 could not be identified.
The U5H3 resonance is overlapped with the G24H1 peak. U27H3 appears as a shoulder to the G7/G34 peak. Resonances corresponding to G11H1 and G19H1 were identified at 13.0–13.1 and 10.4–10.5 ppm, respectively (vide infra). The resonance at 10.8 ppm was assigned to G10H1 and/or G32H1, and their presence indicates that they are at least partially protected from solvent exchange.54 Only the solvent-exposed G1 and G2 could not be located in any imino proton spectrum.
The imino proton NOEs (Figure 2) are consistent with the predicted secondary structure (Figure 1). Thus, hydrogen bonding restraints for each type of canonical base pair predicted in the secondary structure were applied during structure modeling.
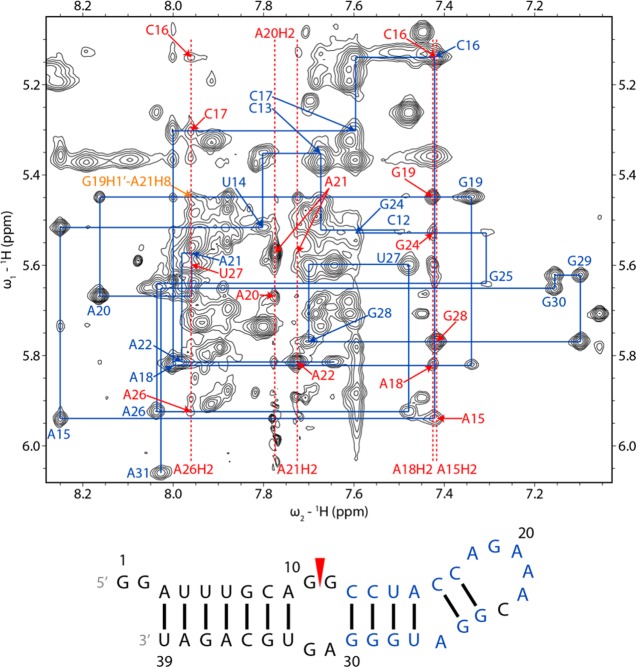
Assignments of nonexchangeable protons in the 39-mer (Table S2 of the Supporting Information) were facilitated by spectra of the smaller constructs (Figure 3), and there was a high degree of correlation between the final chemical shifts (Figure 4 and Tables S3 and S4 of the Supporting Information). Assignments were initiated by identifying pyrimidine H5–H6 cross-peaks from TOCSY spectra. Cytosine H5 and H6 peaks were assigned by intraresidue NOE cross-peaks to amino protons and from those to associated guanosine imino protons. Intraresidue and interresidue H1′–H6/H8 NOEs from G2 to A9 and from U33 to U39 confirm the A-form geometry of the stem below the internal loop in the constructs (Figure S5 of the Supporting Information).53 The same types of H1′–H6/H8 NOEs from C12 to C17, from G24 to G25, and from U27 to G30 confirm the A-form geometry of the stem above the internal loop. Cross-peaks from an adenine H2 to the 3′ H1′ on the same strand and the 3′ H1′ on the opposite strand also identify A-form helical regions of the constructs.55 Specifically, NOEs are present from A3H2 to U4H1′, A9H2 to G34H1′, A15H2 to C16H1′ and U27H1′, A36H2 to G7H1′ and G37H1′, and A38H2 to U5H1′ and U39H1′. In RNA helices, a similar pattern of NOEs from guanine H1 to the 3′ H1′ on the same strand and the 3′ H1′ on the opposite strand is present (Figure S4 of the Supporting Information)56 and was used to confirm H1′ assignments of residues such as U6, A38, U14, and G29, near GC and GU pairs in the 39-mer.
Figure 4.
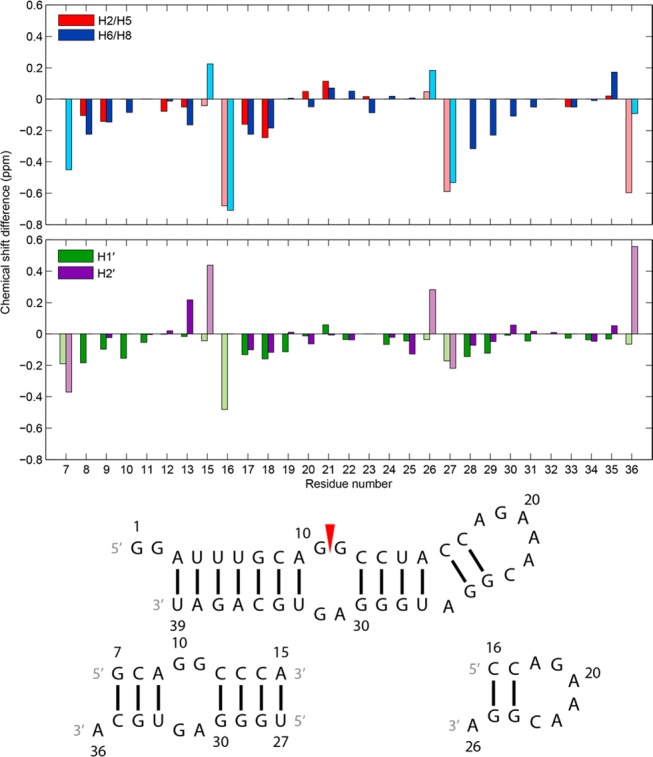
Differences in chemical shifts between the 39 nt hairpin and 19 nt duplex (residues 7–15 and 27–36) and between the 39 nt hairpin and 11 nt hairpin (residues 16–26) for select nonexchangeable aromatic and sugar protons. Chemical shift data were obtained from spectra at 20 °C for the 39-mer and 11-mer and at 25 °C for the 19-mer. Spectra for the 11-mer and 19-mer were acquired with 5 mM Mg2+. Bars colored with light shades belong to terminal helix residues of the 11-mer (residues 16 and 26) and 19-mer (residues 7, 15, 27, and 36) that are not at the termini of any helices of the 39-mer and thus are in structurally inequivalent regions among the constructs. Residue numbers on the x-axis align with the middle of each set of two bars in each plot. Residue 14 is not included because of a U to C substitution.
The Hairpin Loop Is a GAAA Tetraloop Closed with an AC Pair
Thermodynamic calculations with the nearest neighbor model57,58 predicted that the 11 nt construct would form a hairpin rather than a duplex (Figure S6 of the Supporting Information). To check the prediction, 1D imino proton NMR spectra were recorded at 0.2 and 4 mM at 2 °C (Figure S7 of the Supporting Information).59 The number of chemical shifts and resonances and their relative intensities were similar. Therefore, the sequence forms a hairpin at a strand concentration of 4 mM used to measure NOEs for modeling.
In the 39 and 11 nt hairpins, G19H1 was assigned to 10.46 and 10.43 ppm, respectively, consistent with a sheared GA (trans Hoogsteen/sugar edge) pair.60−62 Moreover, NOEs from G19 imino and amino protons to A22H8 in the 39-mer and 11-mer are consistent with G19N2 and A22N7 being close in a sheared GA-like conformation in a GNRA tetraloop.63
A sequential walk consisting of H1′–H6/H8 correlations was completed from C17 to C23 in the 39-mer (Figures 3 and 5) and 11-mer (Figure 6). Intraresidue H1′–H6/H8 NOEs for C17 to A22 were of typical intensity for bases in the anti orientation. An NOE between G19H2′ and A21H8 indicates that A21 lies inside the loop. Cross-peaks from A21H1′ to A20H2, from A22H1′ to A21H2, and from C23H1′ to A22H2 in the 11-mer agree with the expected H2i–H1′i+1 pattern of NOE interactions for stacked bases.55 In spectra of the 11-mer, H1′ of C23 (4.85 ppm at 20 °C) is relatively upfield compared to typical A-form values, and the large line width indicates that it is even further upfield some of the time.64,65 However, the chemical shifts of H1′ of a 3′ uridine in a loop-closing AU pair and of a 3′ cytidine in a loop-closing GC pair of two different GAAA tetraloops are 3.82 and 3.28 ppm, respectively.66,67 The different structure of an A-C pair relative to a Watson–Crick pair may result in a weaker effect of ring current on C23H1′ from the A22 base. On the basis of its large line width, however, the H1′ chemical shift of C23 in some conformations may be near the expected upfield range of H1′ chemical shifts of a 3′ tetraloop-closing residue. H3′ of A22 could not be identified in NMR spectra of the 39-mer because of overlap but was assigned to 4.62 ppm in a 20 °C spectrum of the 11-mer (4.58 ppm at −2 °C) in D2O without Mg2+, relatively upfield of the chemical shift (∼5.0 ppm) expected for the last adenosine of a GAAA tetraloop closed by a canonical base pair.65,67−70
Figure 5.
H1′–H6/H8 region of a 2D proton NOESY spectrum of the 39 nt hairpin showing sequential proton walks for residues C12–C23 and G24–A31. The C23H1′–H6 and C23H1′–G24H8 NOEs are missing from the walk because the C23H1′ and H2O resonances are close to each other. H1′–H6/H8 walk NOEs are labeled in blue. Adenine H2 signals are labeled with red dashed lines. H1′–adenine H2 NOEs are labeled in red with only the label of the residue for H1′. A G19H1′ (5.45 ppm)–A21H8 (7.96 ppm) NOE is labeled in orange. The spectrum was acquired at 20 °C and a mixing time of 350 ms with a WATERGATE pulse to suppress the water signal. In the secondary structure of the 39 nt hairpin, residues whose intraresidue H1′–H6/H8 NOEs were identified in the NOESY walks are labeled in blue. Spectrum and walks for residues G2−G10 and U33−U39 are in Figure S5 of the Supporting Information.
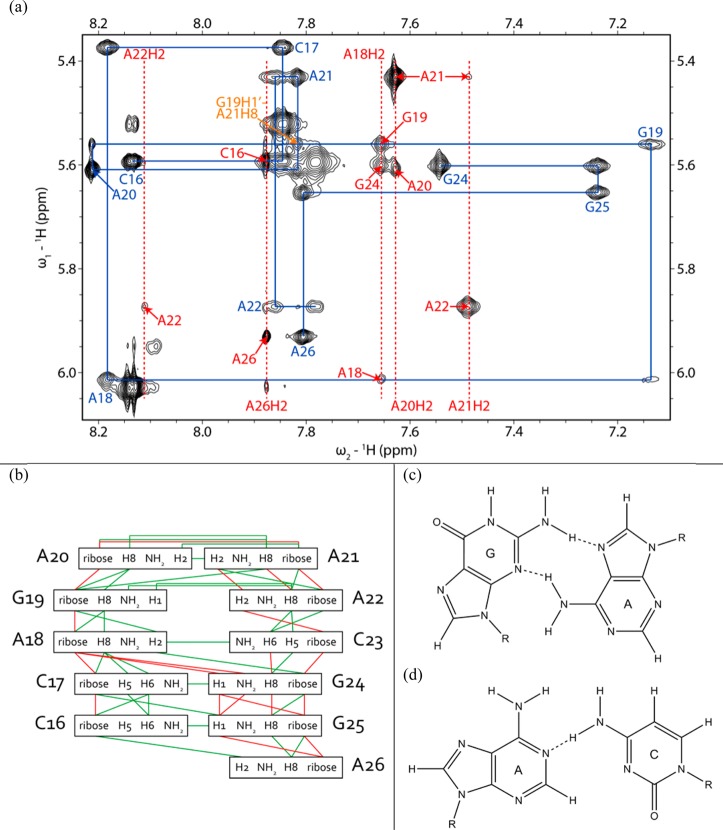
Figure 6.
(a) H1′–H6/H8 region of a 2D proton NOESY spectrum of the 11 nt hairpin showing a sequential proton walk with blue lines. H1′–H6/H8 walk NOEs are labeled in blue. Adenine H2 signals are labeled with red dashed lines. H1′–adenine H2 NOEs are labeled in red with only the label of the residue for H1′. The G19H1′ (5.56 ppm)–A21H8 (7.89 ppm) NOE is labeled in orange and is consistent with the formation of a GNRA-like U-turn. The spectrum was recorded at −2 °C in D2O and 5 mM Mg2+ with a mixing time of 400 ms. Cross-peaks from C23H1′ to A22H2 and C23H6 were not observed in this spectrum, but in a spectrum acquired at 20 °C with a mixing time of 400 ms. (b) Secondary structure of the 11 nt hairpin with assigned interresidue NOEs. Green lines denote NOEs identified in the 11-mer and the 39 nt hairpin and red lines NOEs identified only in the 11-mer. (c) Geometry of the G19-A22 sheared GA pair observed in the AMBER-refined structures. (d) Geometry of the A18-C23 pair observed in most of the AMBER-refined structures.
The possible occurrence of a protonated A+C pair at the base of the hairpin loop was investigated with homonuclear NOESY and 13C–1H HSQC experiments. A+C pairs can have a pKa as high as 6.5 for protonation of adenine N1.71−73 Formation of A+C pairs is accompanied by an upfield shift of adenine C2 by ∼7 ppm (to ∼145 ppm) relative to other adenine C2 resonances, a downfield shift of adenine H2 to above 8 ppm, and an adenine H1 shift of ∼14.5 ppm.71,73 The A18H2 chemical shifts of 7.43 and 7.67 ppm in the 39-mer and 11-mer (20 °C), respectively, the absence of imino proton signals at ≥14.5 ppm, and the absence of adenine C2 resonances below 150 ppm are inconsistent with an A+C pair. Furthermore, the presence of an NOE from A18H2 to C23 amino proton(s) in a NOESY spectrum74 of the 11-mer is structurally inconsistent with the orientation of the adenine H2 and cytosine amino protons in opposite grooves of an A+C pair (signal overlap prevented this NOE from being identified in NOESY spectra of the 39-mer). This NOE, however, is consistent with a cis Watson–Crick bifuricated AC pair (Figure 6).62 In summary, NMR spectra are consistent with a GAAA tetraloop closed by an AC pair.
The Internal Loop Has an Imino GA Pair and Is Dynamic
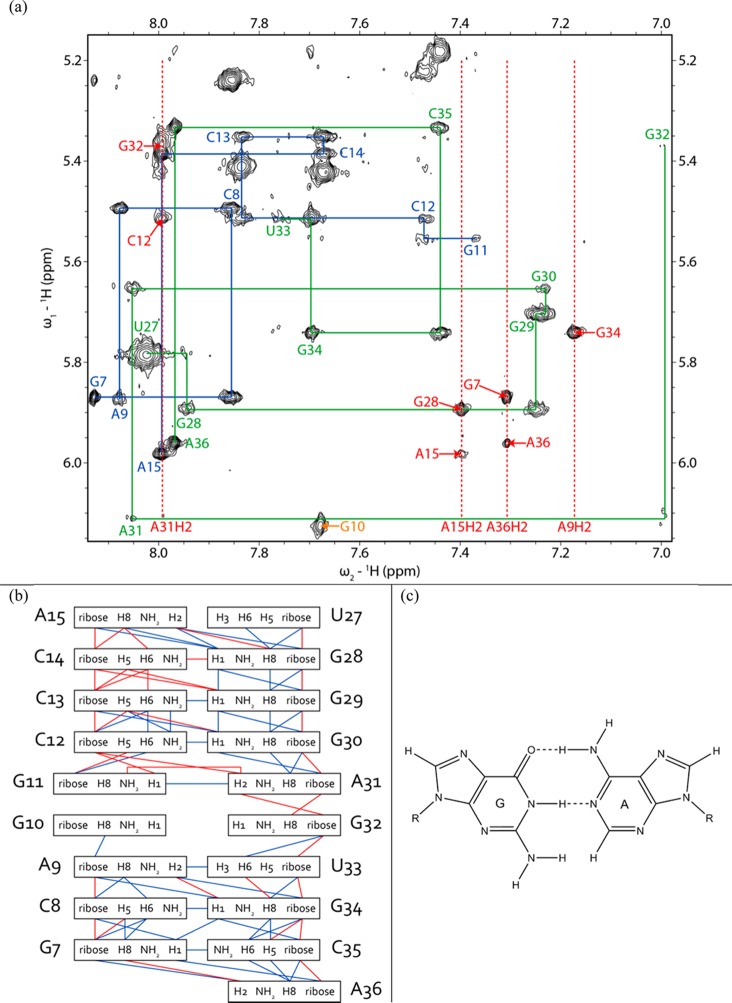
Fewer cross-peaks were observed among residues within the internal loop than within the hairpin loop (Figure 7). The G11H1 peak of the 39 nt hairpin and 19 nt duplex at 13.08 ppm has a cross-peak to A31H2 at 7.96 and 7.99 ppm, respectively, consistent with an imino (cis Watson–Crick/Watson–Crick) GA pair (Figure 7).61,62,65,75 Cross-peaks from H1′ and H2′ of G30 to A31H8 in spectra of the 39-mer, in addition to a cross-peak from C12H1′ to A31H2 in spectra of the 19-mer duplex, suggest that A31 is stacked below G30. In both the 39-mer hairpin and 19-mer duplex, there are weak NOEs from G11H1 and U33H3 to G10H1 and/or G32H1, in addition to an NOE from G11H1 to C12H1′.
Figure 7.
(a) H1′–H6/H8 region of a 2D proton NOESY spectrum of the 19 nt duplex showing a sequential proton walk with blue lines for residues 7–15 and green lines for residues 27–36. H1′–H6/H8 walk NOEs are labeled with the same respective colors. The G32H1′–H8 cross-peak is small because G32 is dynamic. Adenine H2 signals are labeled with red dashed lines. H1′–adenine H2 NOEs are labeled in red with only the label of the residue for H1′. The U27H1′–H6 NOE overlaps with the U27H5–H6 NOE. The G10H1′ (6.13 ppm)–H8 (7.68 ppm) NOE is labeled in orange. Additional G10 NOEs are absent because G10 is dynamic. The spectrum was acquired at −2 °C in D2O and 5 mM Mg2+ with a mixing time of 400 ms. (b) Secondary structure of the 19 nt duplex with assigned interresidue NOEs. Blue lines denote NOEs identified in the 19-mer and the 39 nt hairpin and red lines NOEs identified in only the 19-mer duplex. (c) Geometry of the G11-A31 imino GA pair.
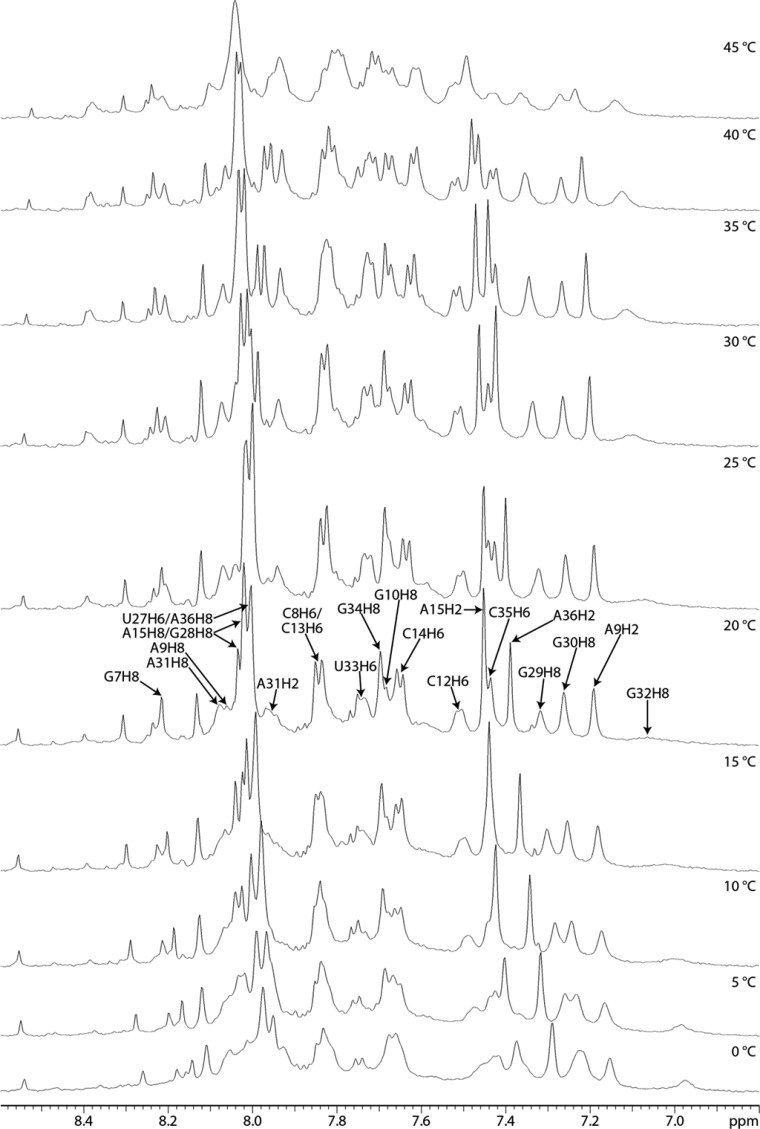
A strong G10H1′–H8 NOE and weak A9H1′– and A9H2′–G10H8 NOEs of the 39-mer and 19-mer duplex indicate that G10 has a syn conformation,36,55,76,77 or an equilibrium of syn and anti conformations (Figure 7 and Figure S4 of the Supporting Information). An A9H2–G10H1′ NOE of the 39-mer is weaker than other H2i–H1′i+1 cross-peaks in A-form helices. The G10H8 chemical shift (7.60 ppm in the 39-mer at 20 °C) is upfield of its unshielded reference value (8.10 ppm),47 consistent with populations in which it is stacked in the helix. On the other hand, few NOEs were detected for G10, consistent with an extrahelical conformation. Apparently, G10 is in an equilibrium of conformations in which G10 is stacked in or extruded from the helix (Figure 7 and Figure S4 of the Supporting Information). Exchange cross-peaks from 13.4 to 13.0 ppm and from 10.8 to 11.0 ppm in a ROESY spectrum correspond to U33 and G10 and/or G32, respectively, consistent with dynamics at the start of the loop.
Spectra of the 19-mer duplex acquired in D2O have a broad G32H8 resonance. The expected intraresidue H1′–H8 NOEs of G11 and G32 are present in a 400 ms spectrum of the 19-mer duplex in D2O, but absent in H2O and D2O spectra of the 39-mer and 19-mer duplex taken with shorter mixing times. In 1D spectra of the 19-mer duplex acquired between 0 and 40 °C in D2O, a broad peak for G32H8 is observed at 0 °C, which further broadened initially, almost disappearing as the temperature was increased to 20 °C, and then sharpened above 30 °C (Figure 8). This observation suggests interconversion of G32 or an adjacent residue between two conformations in an intermediate time range at low temperatures, changing to fast exchange at 30 °C, resulting in a single peak.78 Evidently, G32 is dynamic. Taken together, NMR spectral properties of the internal loop region demonstrate that the GA pair is relatively fixed but the GG pair is dynamic.
Figure 8.
Aromatic region of 1D proton NMR spectra of the 19 nt duplex acquired from 0 to 45 °C in D2O and 5 mM Mg2+.
Bulge Loop
NOEs from A15H1′ and A15H2′ to C16H6 and from A15H2 to C16H1′ indicate that A15 and C16 are close, so the A26 bulge does not prevent their stacking (Figure 5). In contrast, there is no evidence of stacking between G25 and U27 as there are no NOEs between those residues. NOE cross-peaks from G25H1′ and G25H3′ to A26H8 and from A26H1′ to U27H6 have normal A-form intensities. Cross-peaks from G25H2′ to A26H8 and from A26H2′ to U27H6 could be present but are overlapped by intraresidue H2′–H6/H8 cross-peaks of A26 and U27, respectively. The seven steps of a sequential H1′–H6/H8 NOE walk were observed from nucleotides G24 to A31, inclusive, and the A26H8 chemical shift (8.04 ppm) is within the range of the H8 chemical shifts observed for stacked adenines. These data indicate that A26 is not completely bulged out of the helix.79
RNA FRABASE80 was searched for 3D structures with an adenine bulge flanked by canonical base pairs as for A26. One such structure has the bulged A excluded from the helix81 on the basis of evidence for sequential NOE connectivity between residues on each side of the A and the AH8 chemical shift of 8.48 ppm, close to the reference value of 8.64 ppm for AH8 when the chemical shift is not affected by neighboring ring currents.47,81 In contrast, another structure has a bulged A stacked in the helix on the basis of cross-peaks of nearly equal intensity from AH2 to H1′ of consecutive cross-strand 3′ C’s and an AH8 chemical shift of 8.18 ppm.82 Similarly, a bulged A stacked in the helix of a duplex was revealed by interresidue, intrastrand, and interstrand H2–H1′ cross-peaks, sequential H1′–H6/H8 cross-peaks through the bulged A, and an AH8 chemical shift of 7.88 ppm.79 The 60 ms NOESY spectrum of the 39-mer has a cross-peak from A26H2 to C17H1′ that is stronger than that from A26H2 to C16H1′ even though A26 is closer to C16 than C17 in the secondary structure. This spectral feature can be explained, however, by formation of a (C16-G25)A26 base triple of the type seen in crystal structures.83,84
Effect of Magnesium on the Structure of the GAAA Tetraloop and 2 nt × 2 nt Internal Loop
Solvated Mg2+ ions bind to RNA by diffuse, nonspecific interactions with the backbone or by specific interactions with the RNA.85 The former predominate due to the energetic cost of dehydrating Mg2+ for site-specific interactions.86 A sheared GA pair can directly coordinate Mg2+, while an imino GA pair binds fully hydrated Mg2+.61,87,88 To study the effect of Mg2+ on the structures of the GAAA tetraloop and 2 nt × 2 nt internal loop, the short RNA constructs were studied in the presence of 5 mM MgCl2.
The 5 mM Mg2+ caused some minor shifts and sharpening of imino resonances of the 11 nt hairpin (Figure S8 of the Supporting Information) and 19 nt duplex (Figure S9 of the Supporting Information). Nevertheless, exchange cross-peaks of imino resonances for G10/G32 and U33 remained. The chemical shifts of G11H1 and A31H2 in the 19-mer duplex changed little, but resonances for G11H1 in the 39-mer were easier to observe in the presence of Mg2+ (Figure S3 of the Supporting Information). These observations indicate that addition of Mg2+ did not significantly stabilize the helical or internal loop regions or induce a conformational change in the flexible G10 and/or G32. No new imino proton resonances were observed in the presence of Mg2+. Thus, Mg2+ did not introduce new elements of secondary or tertiary structure.65,66,89
Mg2+ sharpened some of the nonexchangeable resonances of the 11-mer hairpin and 19-mer duplex. Minor shifts (typically <0.1 ppm) of nonexchangeable resonances occurred in these constructs (Figures S10 and S11 of the Supporting Information). On the basis of its H1′–H8 cross-peak, G10 of the 19-mer duplex was in a syn–anti equilibrium in the presence and absence of Mg2+.
The similarities of chemical shifts obtained on the model mimics with and without 5 mM Mg2+ indicate that Mg2+ did not significantly impact the structures of the 11-mer hairpin or 19-mer duplex.90 The data are also consistent with the expected small effect of counterion charge on 1H chemical shift.45,91
Modeling Helices and the Hairpin Loop
The simulated annealing protocol provided structures for the 39 nt hairpin that are consistent with the NMR details described above (Table 1 and Figures S12 and S13 of the Supporting Information). The helices formed as expected,14,15 including two wobble GU and Watson–Crick A15-U27 and C16-G25 pairs.
Table 1. Structural Refinement Statistics for the 39 nt Hairpin, 19 nt Duplex, and 11 nt Hairpin for the Average of 20 Structures of Each RNA Construct.
| 39 nt hairpin | 19 nt duplex | 11 nt hairpin | |
|---|---|---|---|
| no. of restraints | |||
| all distance restraints, including hydrogen bonds | 224 | 106 | 68 |
| all NOE restraints | 192 | 87 | 59 |
| intraresidue | 89 | 50 | 33 |
| sequential residues | 65 | 26 | 20 |
| long range | 38 | 11 | 6 |
| hydrogen bond | 32 | 19 | 9 |
| dihedral restraints | 176 | 93 | 24 |
| rmsd of experimental restraints | |||
| distances (Å) | 6.4 × 10–4 | 6.0 × 10–4 | 7.7 × 10–4 |
| dihedral angles (deg) | 1.5 | 1.1 | 0.0 |
| rmsd of structures for heavy atoms (Å) | |||
| all residues (except 1 and 2 in the 39-mer) | 2.90 ± 0.57 | 1.23 ± 0.43 | 0.75 ± 0.21 |
| internal loop (residues 10, 11, 31, and 32) | 1.65 ± 0.28 | 0.62 ± 0.23 | – |
| base triple (residues 16, 25, and 26) | 1.14 ± 0.18 | – | – |
| hairpin loop and AC pair (residues 18–23) | 0.83 ± 0.16 | – | 0.67 ± 0.24 |
| helix 12/30–15/27 (excluding C16-G25 and C17-G24 base pairs) | 0.47 ± 0.13 | 0.39 ± 0.20 | – |
In the 39 and 11 nt hairpins, the modeled AGAAAC loop has properties of GNRA tetraloops (Figure 9 and Figure S13 of the Supporting Information). A sharp U-turn exists between G19 and A20 with the following atoms within hydrogen bonding distance as expected (Figure 9c):64,92 (1) G19H2 and A22OP, (2) G19 2′OH and A21N7, (3) G19H2 and A22N7, and (4) G19H1 and A22OP. Stacking of G19 on the 5′ side of the loop and A20–A22 on the 3′ side of the loop is also consistent with models of GNRA tetraloops.64 The distance between G19N3 and A22 amino protons, however, is too long (>3.4 Å) for a hydrogen bond.93 This is consistent with observations for some GNRA tetraloops,94 including several in crystal structures of rRNAs.83,84,95 Jucker et al.64 reported that the GA pair in a GAAA loop has a GN3–AN6 hydrogen bonding distance ranging from 3.4 to 5.1 Å with an average of 4.28 Å; the long length may indicate a water-mediated interaction. The unrestrained sugar puckers of A20–A22 are primarily C3′-endo (δ near 84°),96 consistent with relatively weak or unobservable H2′–H1′ cross-peaks in a 1H–1H TOCSY spectrum acquired at 20 °C (Figure S15 of the Supporting Information), even though the second to fourth residues of a GNRA loop may experience C2′-endo states (δ near 147°).64,96 In the modeled structures, a C23 amino proton forms a hydrogen bond with A18N1 or A18N3, consistent with the previously mentioned cross-peak between a C23 amino proton and A18H2 in a water spectrum of the 11-mer74,97,98 and the absence of a protonated A+C pair (Figure 9 and Figure S13 of the Supporting Information).
Figure 9.

(a) Model of the GAAA loop of the 39 nt hairpin construct calculated with AMBER, showing the 3′ A3 stack and an AC pair with a hydrogen bond from the C23 amino group to A18N1. (b) Space-filling model of the A18-C23 pair. (c) Space-filling model of the G19-A22 pair.
Modeling the Internal Loop
Except for terminal residues in the 19 nt duplex model, all of the residues primarily have a C3′-endo sugar pucker (Figure S16 of the Supporting Information). The modeled 2 nt × 2 nt internal loop contains a G11-A31 imino (cis Watson–Crick/Watson–Crick) pair in the 39 nt hairpin and 19 nt duplex (Figure 10 and Figure S16 of the Supporting Information). If the force field allowed the exocyclic amine of G11 to be out of plane, then it could have a favorable interaction with the carbonyl of C12.99−102 While the G11 and U33 imino peaks are broadened because of solvent exchange, local conformational dynamics, or both (Figure 2), cross-peaks from these resonances to A31H2 and A9H2 (Figure S4 of the Supporting Information), respectively, and chemical shifts typical of hydrogen-bonded imino protons indicate that formation of G11-A31 and A9-U33 (cis Watson–Crick/Watson–Crick) pairs is dominant.
Figure 10.

Calculated model of the internal loop of the 39 nt hairpin construct. (a) G10 stacked in the helix with a syn conformation. (b) G10 flipped out of the helix with an anti conformation. G10 was also observed in syn and anti conformations flipped out of and stacked in the helix, respectively. The averages of G10 chemical shifts calculated with NUCHEMICS47 for the 20 structures with the lowest distance restraint violation energies generated by simulated annealing are consistent with the structural ensemble. (c) Space-filling model of the G11-A31 pair.
Consistent with the NMR characteristics, some structural models have G10 extruded from the helix while others have G10 positioned within the helix, in a syn conformation or an anti conformation (Figure 10 and Figure S16 of the Supporting Information). On the basis of the G10H1′–H8 distance of 3.02 Å derived from NOE volumes, G10 is estimated to be in the syn and anti conformations in 26 and 74% of the populations of the 39 nt hairpin, respectively. G10 forms no hydrogen bonds with G32 in two of the 20 lowest-violation energy structures. G32 is also dynamic but is always within the helix in the ensemble of generated structures.
The A26 Bulge Can Form a Base Triple
The A26 bulge can form a (C16-G25)A26 cWW/cSH base triple (Figure 11),103 as observed in the 30S and 70S ribosomal subunits of Thermus thermophilus [PDB entries 2UXC for (C1260-G1274)A1275 and 2B9N for (C965-G952)A2267].83,84,104 A characteristic feature of this type of base triple is that the adenine is in the minor groove of a canonical CG pair.103,105 The structures of the base triple in the models of the segment 7 hairpin generally agree with expected structures of the cWW/cSH CGA base triple.103,106 In most of the modeled structures, A26N7 and at least one, if not both, of the A26 amino hydrogens are within hydrogen bonding distance of one of the G25 amino hydrogens and C16O2, respectively. One of the A26 amino hydrogens is also within hydrogen bonding distance of C16O2′. The location of A26 in the minor groove of the C16-G25 pair is consistent with cross-peaks, including those from A26H2 to C16H1′, C17H1′, and U27H1′, and A26H8 to G25H3′.106 The distance between A26H2 and C16H1′ is greater than that between A26H2 and C17H1′, which agrees with the slightly weaker NOE from A26H2 to C16H1′ compared to that of A26H2 to C17H1′. A similar arrangement of a C, a G, and an A was observed in a hairpin from Caenorhabditis elegans, but the adenine appears to be stabilized by a hydrogen bond between its amino group and the ribose of a 3′ cross-strand residue rather than the ribose of the C.107 In chemical modification experiments,15 A26 was modified by DEPC, which carbethoxylates an exposed adenine N7, such as one not buried in the major groove of an RNA helix.108 This is inconsistent with the formation of a hydrogen bond between A26N7 and the amino group of G25, suggesting that the base triple is dynamic. In short, the NMR-guided models suitably explain the observed NOE data for residues around A26, but not the DEPC mapping data. Both types of data can be rationalized, however, by a dynamic model for A26 (vide infra).
Figure 11.
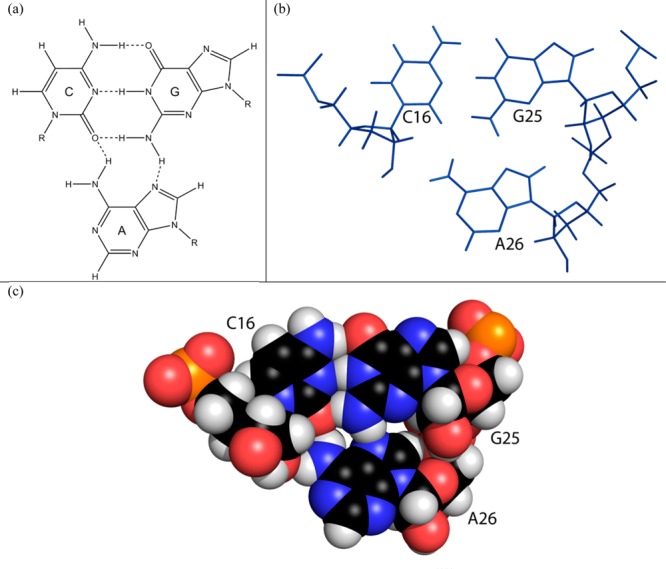
(a) Schematic of a CGA base triple of the cWW/cSH family103 with expected hydrogen bonds from the adenine to cytosine and guanine: A-N7 to G-H22 and A-H61 to C-O2. Not shown is a hydrogen bond from A-H62 to C-O2′. (b) Model of the (C16-G25)A26 base triple of the 39 nt hairpin construct refined by AMBER. (c) Space-filling model of the (C16-G25)A26 base triple.
Prediction of Structure from Chemical Shifts
Predicted structures for the loops were also generated with CS-ROSETTA-RNA46 by applying chemical shift restraints from the NMR spectra to secondary structures containing the same loops. Canonical base pairs in helical regions of the secondary structure for each RNA fragment were present in 3D models of the 20 lowest-energy structures. All 20 lowest-energy structures of a 19 nt hairpin mimic, r(5′CCUACCAGAAACGGAUGGG3′) (Figure S2 of the Supporting Information), containing the hairpin loop and A26 bulge, have a 3′ stack of bases A20–A22 and a sheared-like G19-A22 base pair. A cWC/WC A18-C23 pair forms in all 20 structures. A26 stacks below G25 in all 20 structures without forming a (C16-G25)A26 base triple, and the distance from A26H2 to C17H1′ is much longer than that to C16H1′, contrary to the larger C17H1′–A26H2 NOE compared to the C16H1′–A26H2 NOE. This again suggests that A26 is dynamic so that one structure does not satisfy all the data.
The G11-A31 imino pair was present in all 20 lowest-energy CS-ROSETTA-RNA structures of an 18 nt duplex containing the 2 nt × 2 nt internal loop (Figure S2 of the Supporting Information). Sixteen structures have G10-G32 in a base pair that resembles a trans Hoogsteen/sugar edge pair stabilized by a G10O6 to G32H1 hydrogen bond in 10 structures and a G10O6 to G32H2 hydrogen bond in six structures. The four remaining structures have a G10-G32 base pair resembling a cis Watson–Crick/Hoogsteen pair stabilized by a G10H1 and/or G10H2 to G32N7 hydrogen bond. In all 20 structures, G10 is in an anti conformation, contrary to the syn character revealed by its relatively large H1′–H8 NOE. Taken together, ROSETTA provides reasonable 3D models of the RNA from sequence, secondary structure, and assigned chemical shifts. Comparisons between structures based on chemical shifts and distance restraints, however, reveal dynamics.
Prediction of Chemical Shifts from Structure
The program NUCHEMICS47 was used to predict chemical shifts for the ensembles of 20 structures of the RNA constructs generated with distance restraints. Average calculated chemical shifts of H1′, H2′, H2, H5, and H6/H8 of the 39 nt hairpin agree for most residues within 0.4 ppm of those assigned in NMR spectra at 20 °C (Figure 12). Chemical shifts of sugar resonances of the internal loop are within ∼0.2 ppm of experiment despite the flexibility of the loop (Figure 12 and Figure S18 of the Supporting Information). For the AC closed GAAA loop, larger differences between predicted and experimental chemical shifts are observed. The A18H2 proton was predicted to be at 7.03 ppm in the 39-mer and 7.11 ppm in the 11 nt hairpin, which are ∼0.4 and ∼0.6 ppm smaller than experimental chemical shifts for their respective structures (Figure 12 and Figure S19 of the Supporting Information). The C23H1′ chemical shift for the 11-mer was 4.85 ppm but was predicted to be 3.77 ppm. To explore whether the discrepancy is seen with other RNAs, differences were analyzed between chemical shifts predicted with NUCHEMICS for 3D structures of three GAAA hairpins65,67,70 from the PDB104 and their assigned chemical shifts from the BMRB.33 Each of the three types of canonical base pairs is represented among these structures as a closing pair of the GAAA hairpin loop. Indeed, for a given RNA, experimental and predicted chemical shifts for each of these hairpins differ most (up to 2.3 ppm) within the GAAA hairpin loop and closing base pair (Figures S20–S22 of the Supporting Information), confirming that either NUCHEMICS inconsistently predicts true chemical shifts of these residues or the structures are inaccurate or dynamic.
Figure 12.
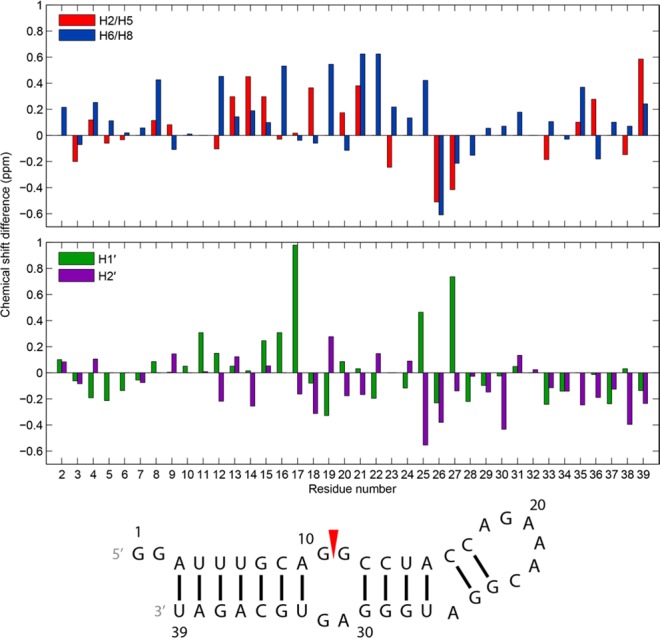
Chemical shift differences of the 39 nt hairpin between experiment, assigned at 20 °C, and those predicted by NUCHEMICS for H2/H5, H6/H8, H1′, and H2′ in an ensemble of 20 structures generated with NMR restraints. Residue numbers on the x-axis align with the middle of each set of two bars in each plot.
Chemical shifts were predicted and averaged for the 20 lowest-energy structures generated with CS-ROSETTA for the 19 nt hairpin and 18 nt duplex (Figure S2 of the Supporting Information). Except for those of terminal residues in each construct, most H1′, H2′, H2, H5, and H6/H8 chemical shifts of the CS-ROSETTA structures, including those of GAAA loop residues, were predicted to be within 0.3 ppm of those assigned for NMR spectra of the 39 nt hairpin at 20 °C (Figures S23 and S24 of the Supporting Information). The average predicted chemical shift of A26H2 in the CS-ROSETTA structures of the 19 nt hairpin is within 0.1 ppm of the experimental chemical shift. Thus, the stacking of A26 below G25 in the CS-ROSETTA structures may occur in the 39 nt hairpin, consistent with a dynamic base triple.
Discussion
Influenza is a public health problem that is incompletely controlled by yearly vaccination.109 Available therapeutics target neuraminidase and M2 proteins but are not particularly efficacious. While most therapeutics used clinically target proteins, RNA research is revealing a wealth of potential RNA targets, including splice sites. For example, beta thalassemia and Duchenne muscular dystrophy have been treated or reversed with oligonucleotides that block a splice site.110−112
If a splice site has a stable structure, it should also be possible to affect splicing with small molecules that bind specifically to a loop.8,113,114 Most splice sites, though, are thought not to have stable structures.115−118 One exception is the 3′ splice site of segment 7 mRNA from influenza A.14,15 Splicing at this site determines the relative abundance of two essential proteins, M1 and M2.119 The amount of splicing may be determined by an equilibrium between a pseudoknot and two-hairpin structure around the splice site (Figure 1). The results reported here reveal several interesting characteristics of the hairpin containing the 3′ splice site.
The splice site is between G10 and G11 in a conformationally flexible internal loop where G10 appears to be in equilibria between intrahelical and extrahelical conformations as well as syn–anti conformations for the base relative to the ribose (Figure 10). Local flexibility may be an important characteristic for a 3′ splice site.115 In contrast to G10 and G32, the adjacent G11-A31 and A9-U33 pairs are relatively stable. Alternative conformations of the splice site region are also consistent with exchange peaks for imino protons of residues G10 and/or G32, U33, and G34 and two-site exchange observed for G32H8. The flexibility of the internal loop and its functional importance to the viral life cycle as a splice site make it an attractive target for binding and inhibition by therapeutic agents.120,121
Specificity of binding to RNA can be improved by targeting two loops by coupling together two small molecules.8,122 Presumably, the bulge A loop, the GAAA tetraloop capping the hairpin, or both could be targeted along with the internal loop. The GAAA tetraloop is closed by an AC pair, which is an unusual combination. The (C16-G25)A26 base triple formed by the bulged A is known to occur in other RNAs83,84 but is rare.
Internal loops with adjacent GG and imino GA pairs were observed in loop E of E. coli 5S rRNA, r(5′CGAUGGUAG79/3′GAUGAGAGC97),123 the HIV Rev responsive element (RRE), r(5′GGGC49/3′GGUAC74),120,121 and the 2 nt × 2 nt internal loop, (5′AGGU271/3′UGAA282), from a hairpin of a group II intron of Oceanobacillus iheyensis (Figure S25 of the Supporting Information).88,124,125 The first two loops participate in protein recognition but are larger than the 2 nt × 2 nt loop containing the influenza 3′ splice site.
The sequence of the r(5′AGGU271/3′UGAA282) loop is similar to that of the r(5′AGGC/3′UGAG) influenza splice site loop. To structurally compare the loops, hydrogen atoms were added to Protein Data Bank (PDB) X-ray structures of the group II intron (3EOH, 4E8M, 4E8Q, 4FAR, 4FAW, and 4FAX) using Reduce with the NOFLIP option.126 In each of the crystal structures, G270 and A283 form an imino GA pair, the same as G11-A31 in the segment 7 hairpin. G269 and G284, however, form a trans Hoogsteen/sugar edge pair, perhaps as a result of the noncanonical A268-U285 pair below it. In contrast to the Watson Crick A9-U33 pair closing the splice site loop, the A268-U285 pair in the group II intron is a trans Watson–Crick/Watson–Crick pair in the 2008 structure124 and a trans Watson–Crick/Hoogsteen pair in the 2012 structures.125 In all six structures, the A268 amino proton that does not contact U285 points away from the major groove of its helix and lies within hydrogen bonding proximity of O4′ of an extrahelical residue, G321. There is also a hydrogen bond from the amino of G269 to O2′ of G321. Similar extrahelical hydrogen bonds were found in X-ray crystal structures of ribosomes.127−129 The terminal base pair of the group II intron motif consists of a canonical GC pair with G267 forming an extrahelical trans sugar edge/sugar edge pair with G320 to form a cWW/tSS (C286-G267)G320 base triple.62,103
Nucleotide details of the X-ray structures of the r(5′AGGU271/3′UGAA282) internal loop region of the group II intron hairpin differ from those of the NMR solution structure of the segment 7 hairpin. G269 of the group II intron has an anti conformation, in contrast to the mix of syn and anti conformations of G10 of the segment 7 hairpin. A268 of the 2008 X-ray structure124 has a syn conformation, compared to the anti conformation of A9 in the segment 7 hairpin. In the A268-U285 pair of the 2012 X-ray structures, the orientation of A268H2 away from U285H3 is inconsistent with the presence of an NOE from A9H2 to U33H3 in NMR spectra of the segment 7 hairpin. Evidently, tertiary interactions from the amino protons of A268 and G269 to G321 stabilize G269-G284 and noncanonical A268-U285 base pairs in the group II intron. Similar large differences have been observed between an NMR structure of an isolated internal loop and the same sequence loop in crystals of ribosomes.130 The results suggest certain internal loops may be poised for molecular recognition by induced fit, structure capture,131 adaptive recognition,132,133 or all of them. Independent of mechanism, the results also suggest that a variety of small molecules could bind tightly to the influenza internal loop and serve as therapeutics.
Ultimately, it should be possible to predict the structure and dynamics of RNAs and of molecules to bind them. The results presented here provide a useful benchmark for testing such predictions. The minimal effect of Mg2+ on the 2 nt × 2 nt internal loop and AGAAAC tetraloop implies that it will not be necessary to include Mg2+ in such calculations.
Acknowledgments
We thank the following individuals for their contributions to this work: Dr. Blanton Tolbert for providing a plasmid that expresses T7 RNA polymerase and a protocol for in vitro transcription of RNA by T7 RNA polymerase; Dr. Jesse Kleingardner, Dr. Kara Bren, and Hiram Lyon for assistance with purifying RNA transcripts; Dr. Brendan Mort for assistance with computations; and Dr. Anton Petrov for providing information about base triples from literature. Minimizations by simulated annealing were performed with computational resources provided by the Center for Integrated Research Computing (CIRC).
Glossary
Abbreviations
- 1D
one-dimensional
- 2D
two-dimensional
- 3D
three-dimensional
- FPLC
fast protein liquid chromatography
- HETCOR
heteronuclear correlation
- HIV-1
human immunodeficiency virus-1
- HSQC
heteronuclear single-quantum coherence
- NMR
nuclear magnetic resonance
- NOESY
nuclear Overhauser effect spectroscopy
- rmsd
root-mean-square deviation
- ROESY
rotating frame Overhauser spectroscopy
- TOCSY
total correlation spectroscopy.
Supporting Information Available
(I) Equilibria of 19 nt duplex mimics of a portion of the 39 nt hairpin with homodimers of the top strands of their duplexes. (II) Fragments of the 39 nt hairpin used for modeling with CS-ROSETTA. (III) 1D and 2D proton spectra of the constructs studied. (IV) Thermodynamic calculation for formation of the 11 nt hairpin. (V) Differences in chemical shifts of select nonexchangeable hydrogens of the 11 nt hairpin and 19 nt duplex with and without 5 mM Mg2+ at −2 °C. (VI) Ensembles of the 10 structures of the 11 nt hairpin and 19 nt duplex modeled in AMBER with NMR restraints. (VII) Lowest distance restraint violation energy models of the GAAA loop and closing AC pair of the 11 nt hairpin and of the internal loop of the 19 nt duplex calculated in AMBER with NMR restraints. (VIII) Chemical shift differences of the 19 nt duplex and 11 nt hairpin between experiment, assigned at 25 and 20 °C, respectively, in the presence of 5 mM Mg2+, and those predicted by NUCHEMICS for H2/H5, H6/H8, H1′, and H2′ in an ensemble of 20 structures generated with NMR restraints. (IX) Differences between experimental and NUCHEMICS-predicted chemical shifts of GAAA hairpins closed by a GU, GC, or AU pair. (X) Differences between experimental chemical shifts of the 39 nt hairpin, assigned at 20 °C, and those predicted by NUCHEMICS for H2/H5, H6/H8, H1′, and H2′ for an ensemble of 20 structures each of the 19 nt hairpin and 18 nt duplex as generated by CS-ROSETTA with chemical shift constraints. (XI) Secondary structure of a hairpin with a 2 nt × 2 nt internal loop from a group II intron of O. iheyensis. (XII) Chemical shifts used to model fragments of the segment 7 hairpin with CS-ROSETTA. (XIII) Assigned chemical shifts of the 39 and 11 nt hairpins and 19 nt duplex. (XIV) Plasmid insert design for in vitro transcription of the 39 nt 3′ segment 7 mRNA hairpin by T7 RNA polymerase. (XV) Distance and dihedral restraints for AMBER modeling of the 39 and 11 nt hairpins and the 19 nt duplex. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00012.
Accession Codes
NMR chemical shifts for the 11 nt hairpin, 19 nt duplex, and 39 nt hairpin were deposited in the Biological Magnetic Resonance Data Bank as entries 25414, 25415, and 25416, respectively. Structural coordinates for these constructs were deposited in the RCSB Protein Data Bank as entries 2MXJ, 2MXK, and 2MXL, respectively.
This work was supported by National Institutes of Health Grant GM22939.
The authors declare no competing financial interest.
Supplementary Material
References
- Centers for Disease Control and Prevention (2010) Estimates of deaths associated with seasonal influenza: United States, 1976–2007. In Morbidity and Mortality Weekly Report, pp 1057–1062, Centers for Disease Control and Prevention, Atlanta. [PubMed] [Google Scholar]
- Thompson W. W.; Shay D. K.; Weintraub E.; Brammer L.; Bridges C. B.; Cox N. J.; Fukuda K. F. (2004) Influenza-associated hospitalizations in the United States. JAMA, J. Am. Med. Assoc. 292, 1333–1340. [DOI] [PubMed] [Google Scholar]
- Baigent S. J.; McCauley J. W. (2003) Influenza type A in humans, mammals and birds: Determinants of virus virulence, host-range and interspecies transmission. BioEssays 25, 657–671. [DOI] [PubMed] [Google Scholar]
- Hsu J.; Santesso N.; Mustafa R.; Brozek J.; Chen Y. L.; Hopkins J. P.; Cheung A.; Hovhannisyan G.; Ivanova L.; Flottorp S. A.; Sæterdal I.; Wong A. D.; Tian J.; Uyeki T. M.; Akl E. A.; Alonso-Coello P.; Smaill F.; Schünemann H. J. (2012) Antivirals for treatment of influenza: A systematic review and meta-analysis of observational studies. Ann. Int. Med. 156, 512–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharan N. J.; Gubareva L. V.; Meyer J. J.; Okomo-Adhiambo M.; McClinton R. C.; Marshall S. A.; St. George K.; Epperson S.; Brammer L.; Klimov A. I.; Bresee J. S.; Fry A. M. (2009) Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA, J. Am. Med. Assoc. 301, 1034–1041. [DOI] [PubMed] [Google Scholar]
- Sheu T. G.; Fry A. M.; Garten R. J.; Deyde V. M.; Shwe T.; Bullion L.; Peebles P. J.; Li Y.; Klimov A. I.; Gubareva L. V. (2011) Dual resistance to adamantanes and oseltamivir among seasonal influenza A(H1N1) viruses: 2008–2010. J. Infect. Dis. 203, 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukol A.; Hughes D. J. (2014) Large-scale analysis of influenza A virus nucleoprotein sequence conservation reveals potential drug-target sites. Virology 454–455, 40–47. [DOI] [PubMed] [Google Scholar]
- Guan L.; Disney M. D. (2013) Small-molecule-mediated cleavage of RNA in living cells. Angew. Chem., Int. Ed. 52, 1462–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer A. C.; Frank A. T.; Kratz J. D.; Swanson M. D.; Gonzalez-Hernandez M. J.; Lee J. H.; Andricioaei I.; Markovitz D. M.; Al-Hashimi H. M. (2011) Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 7, 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-K.; Bottini A.; Kim M.; Bardaro M. F. Jr.; Zhang Z.; Pellecchia M.; Choi B.-S.; Varani G. (2014) A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chem. Commun. 50, 368–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiusto D. L.; Krishnan A.; Li L.; Li H.; Li S.; Rao A.; Mi S.; Yam P.; Stinson S.; Kalos M.; Alvarnas J.; Lacey S. F.; Yee J.-K.; Li M.; Couture L.; Hsu D.; Forman S. J.; Rossi J. J.; Zaia J. A. (2010) RNA-based gene therapy for HIV with lentiviral vector-modified CD34+ cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2, 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeley B. D.; McNaughton B. R. (2014) Synthetic RNA recognition motifs that selectively recognize HIV-1 trans-activation response element hairpin RNA. ACS Chem. Biol. 9, 1320–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier N. M.; Palese P. (2008) The biology of influenza viruses. Vaccine 26S, D49–D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss W. N.; Priore S. F.; Turner D. H. (2011) Identification of potential conserved RNA secondary structure throughout influenza A coding regions. RNA 17, 991–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss W. N.; Dela-Moss L. I.; Kierzek E.; Kierzek R.; Priore S. F.; Turner D. H. (2012) The 3′ splice site of influenza A segment 7 mRNA can exist in two conformations: A pseudoknot and a hairpin. PLoS One 7, e38323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T.; Kennedy S. D.; Moss W. N.; Kierzek E.; Turner D. H. (2014) Secondary structure of a conserved domain in an intron of influenza A M1 mRNA. Biochemistry 53, 5236–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priore S. F.; Kierzek E.; Kierzek R.; Baman J. R.; Moss W. N.; Dela-Moss L. I.; Turner D. H. (2013) Secondary structure of a conserved domain in the intron of influenza A NS1 mRNA. PLoS One 8, e70615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A.; Lai C.-J.; Choppin P. W. (1981) Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: Colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. U.S.A. 78, 4170–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise H. M.; Hutchinson E. C.; Jagger B. W.; Stuart A. D.; Kang Z. H.; Robb N.; Schwartzman L. M.; Kash J. C.; Fodor E.; Firth A. E.; Gog J. R.; Taubenberger J. K.; Digard P. (2012) Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 8, e1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle J. L., and Green R. (2013) In vitro transcription from plasmid or PCR-amplified DNA. Methods Enzymol. 530, 101–114. [DOI] [PubMed] [Google Scholar]
- Kashlev M.; Martin E.; Polyakov A.; Severinov K.; Nikiforov V.; Goldfarb A. (1993) Histidine-tagged RNA polymerase: Dissection of the transcription cycle using immobilized enzyme. Gene 130, 9–14. [DOI] [PubMed] [Google Scholar]
- Milligan J. F.; Groebe D. R.; Witherell G. W.; Uhlenbeck O. C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton L. E.; Shibata Y.; Lukavsky P. J. (2010) Rapid, nondenaturing RNA purification using weak anion-exchange fast performance liquid chromatography. RNA 16, 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A., Wu T., Puglisi E. V., and Puglisi J. D. (2013) RNA purification by preparative polyacrylamide gel electrophoresis. Methods Enzymol. 530, 315–330. [DOI] [PubMed] [Google Scholar]
- Freier S. M.; Burger B. J.; Alkema D.; Neilson T.; Turner D. H. (1983) Effects of 3′ dangling end stacking on the stability of GGCC and CCGG double helixes. Biochemistry 22, 6198–6206. [Google Scholar]
- Sugimoto N.; Kierzek R.; Turner D. H. (1987) Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry 26, 4554–4558. [DOI] [PubMed] [Google Scholar]
- Sklenář V.; Bax A. (1987) Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra. J. Magn. Reson. 74, 469–479. [Google Scholar]
- Piotto M.; Saudek V.; Sklenář V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665. [DOI] [PubMed] [Google Scholar]
- Grzesiek S.; Bax A. (1993) The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115, 12593–12594. [Google Scholar]
- Smallcombe S. H. (1993) Solvent suppression with symmetrically-shifted pulses. J. Am. Chem. Soc. 115, 4776–4785. [Google Scholar]
- Cavanagh J., Fairbrother W. J., Palmer A. G. I., and Skelton N. J. (1996) Protein NMR Spectroscopy: Principles and Practice, pp 160–180 and 384–402, Academic Press, San Diego. [Google Scholar]
- Kumar A.; Ernst R. R.; Wüthrich K. (1980) A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95, 1–6. [DOI] [PubMed] [Google Scholar]
- Ulrich E. L.; Akutsu H.; Doreleijers J. F.; Harano Y.; Ioannidis Y. E.; Lin J.; Livny M.; Mading S.; Maziuk D.; Miller Z.; Nakatani E.; Schulte C. F.; Tolmie D. E.; Kent Wenger R.; Yao H.; Markley J. L. (2008) BioMagResBank. Nucleic Acids Res. 36, D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Goddard T. D., and Kneller D. G. (2004) SPARKY, NMR Assignment and Integration Software, version 3, University of California, San Francisco. [Google Scholar]
- Ulyanov N. B.; Mujeeb A.; Du Z. H.; Tonelli M.; Parslow T. G.; James T. L. (2006) NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J. Biol. Chem. 281, 16168–16177. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick S.; Gelatt C. D.; Vecchi M. P. (1983) Optimization by simulated annealing. Science 220, 671–680. [DOI] [PubMed] [Google Scholar]
- Schmitz M.; Steger G. (1996) Description of RNA folding by “Simulated Annealing”. J. Mol. Biol. 255, 254–266. [DOI] [PubMed] [Google Scholar]
- Bansal M.; Bhattacharyya D.; Ravi B. (1995) NUPARM and NUCGEN: Software for analysis and generation of sequence dependent nucleic acid structures. Comput. Appl. Biosci. 11, 281–287. [DOI] [PubMed] [Google Scholar]
- Still W. C.; Tempczyk A.; Hawley R. C.; Hendrickson T. (1990) Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 112, 6127–6129. [Google Scholar]
- Case D. A., Babin V., Berryman J. T., Betz R. M., Cai Q., Cerutti D. S., Cheatham T. E. I., Darden T. A., Duke R. E., Gohlke H., Goetz A. W., Gusarov S., Homeyer N., Janowski P., Kaus J., Kolossváry I., Kovalenko A., Lee T. S., LeGrand S., Luchko T., Luo R., Madej B., Merz K. M., Paesani F., Roe D. R., Roitberg A., Sagui C., Salomon-Ferrer R., Seabra G., Simmerling C. L., Smith W., Swails J., Walker R. C., Wang J., Wolf R. M., Wu X., and Kollman P. A. (2014) AMBER 14, University of California, San Francisco. [Google Scholar]
- Yildirim I.; Stern H. A.; Kennedy S. D.; Tubbs J. D.; Turner D. H. (2010) Reparameterization of RNA χ torsion parameters for the AMBER force field and comparison to NMR spectra for cytidine and uridine. J. Chem. Theory Comput. 6, 1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Lam S. L.; Chi L. M. (2010) Use of chemical shifts for structural studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 56, 289–310. [DOI] [PubMed] [Google Scholar]
- van der Werf R. M.; Tessari M.; Wijmenga S. S. (2013) Nucleic acid helix structure determination from NMR proton chemical shifts. J. Biomol. NMR 56, 95–112. [DOI] [PubMed] [Google Scholar]
- Sripakdeevong P.; Cevec M.; Chang A. T.; Erat M. C.; Ziegeler M.; Zhao Q.; Fox G. E.; Gao X.; Kennedy S. D.; Kierzek R.; Nikonowicz E. P.; Schwalbe H.; Sigel R. K. O.; Turner D. H.; Das R. (2014) Structure determination of noncanonical RNA motifs guided by 1H NMR chemical shifts. Nat. Methods 11, 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromsigt J. A. M. T. C.; Hilbers C. W.; Wijmenga S. S. (2001) Prediction of proton chemical shifts in RNA: Their use in structure refinement and validation. J. Biomol. NMR 21, 11–29. [DOI] [PubMed] [Google Scholar]
- Frank A. T.; Bae S.-H.; Stelzer A. C. (2013) Prediction of RNA 1H and 13C chemical shifts: A structure based approach. J. Phys. Chem. B 117, 13497–13506. [DOI] [PubMed] [Google Scholar]
- Barton S.; Heng X.; Johnson B. A.; Summers M. F. (2013) Database proton NMR chemical shifts for RNA signal assignment and validation. J. Biomol. NMR 55, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A. T.; Horowitz S.; Andricioaei I.; Al-Hashimi H. M. (2013) Utility of 1H NMR chemical shifts in determining RNA structure and dynamics. J. Phys. Chem. B 117, 2045–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Lange O.; Delaglio F.; Rossi P.; Aramini J. M.; Liu G.; Eletsky A.; Wu Y.; Singarapu K. K.; Lemak A.; Ignatchenko A.; Arrowsmith C. H.; Szyperski T.; Montelione G. T.; Baker D.; Bax A. (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S.; Chou F.-C.; Conchúir S. Ó.; Der B. S.; Drew K.; Kuroda D.; Xu J.; Weitzner B. D.; Renfrew P. D.; Sripakdeevong P.; Borgo B.; Havranek J. J.; Kuhlman B.; Kortemme T.; Bonneau R.; Gray J. J.; Das R. (2013) Serverification of molecular modeling applications: The Rosetta Online Server That Includes Everyone (ROSIE). PLoS One 8, e63906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürtig B.; Richter C.; Wohnert J.; Schwalbe H. (2003) NMR spectroscopy of RNA. ChemBioChem 4, 936–962. [DOI] [PubMed] [Google Scholar]
- Burkard M. E.; Xia T.; Turner D. H. (2001) Thermodynamics of RNA internal loops with a guanosine-guanosine pair adjacent to another noncanonical pair. Biochemistry 40, 2478–2483. [DOI] [PubMed] [Google Scholar]
- Varani G.; Aboulela F.; Allain F. H. T. (1996) NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc. 29, 51–127. [Google Scholar]
- Heus H. A.; Pardi A. (1991) Novel 1H nucleic magnetic resonance assignment procedure for RNA duplexes. J. Am. Chem. Soc. 113, 4360–4361. [Google Scholar]
- Xia T. B.; SantaLucia J.; Burkard M. E.; Kierzek R.; Schroeder S. J.; Jiao X. Q.; Cox C.; Turner D. H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- Turner D. H.; Mathews D. H. (2010) NNDB: The nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 38, D280–D282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszczyk M. M.; Zanier K.; Sattler M. (2008) A NMR strategy to unambiguously distinguish nucleic acid hairpin and duplex conformations applied to a Xist RNA A-repeat. Nucleic Acids Res. 36, 7068–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J.; Turner D. H. (1993) Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry 32, 12612–12623. [DOI] [PubMed] [Google Scholar]
- Rüdisser S.; Tinoco I. Jr. (2000) Solution structure of cobalt(III) hexammine complexed to the GAAA tetraloop, and metal-ion binding to G·A mismatches. J. Mol. Biol. 295, 1211–1223. [DOI] [PubMed] [Google Scholar]
- Leontis N. B.; Stombaugh J.; Westhof E. (2002) The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 30, 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman M.; Eldho N. V.; Padgett R. A.; Dayie K. T. (2006) Structure of a self-splicing group II intron catalytic effector domain 5: Parallels with spliceosomal U6 RNA. RNA 12, 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker F. M.; Heus H. A.; Yip P. F.; Moors E. H. M.; Pardi A. (1996) A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264, 968–980. [DOI] [PubMed] [Google Scholar]
- Desjardins G.; Bonneau E.; Girard N.; Boisbouvier J.; Legault P. (2011) NMR structure of the A730 loop of the Neurospora VS ribozyme: Insights into the formation of the active site. Nucleic Acids Res. 39, 4427–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. H.; Tonelli M.; Scott L. G.; Jaeger L.; Williamson J. R.; Butcher S. E. (2005) RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 351, 371–382. [DOI] [PubMed] [Google Scholar]
- Korth M. M. T.; Sigel R. K. O. (2012) Unusually high-affinity Mg2+ binding at the AU-rich sequence within the antiterminator hairpin of a Mg2+ riboswitch. Chem. Biodiversity 9, 2035–2049. [DOI] [PubMed] [Google Scholar]
- Cevec M.; Thibaudeau C.; Plavec J. (2008) Solution structure of a let-7 miRNA:lin-41 mRNA complex from C. elegans. Nucleic Acids Res. 36, 2330–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevec M.; Thibaudeau C.; Plavec J. (2010) NMR structure of the let-7 miRNA interacting with the site LCS1 of lin-41 mRNA from Caenorhabditis elegans. Nucleic Acids Res. 38, 7814–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruschel D.; Skilandat M.; Sigel R. K. O. (2014) NMR structure of the 5′ splice site in the group IIB intron Sc.ai5γ-conformational requirements for exon-intron recognition. RNA 20, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D.; Wyatt J. R.; Tinoco I. Jr. (1990) Solution conformation of an RNA hairpin loop. Biochemistry 29, 4215–4226. [DOI] [PubMed] [Google Scholar]
- Legault P.; Pardi A. (1997) Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 119, 6621–6628. [Google Scholar]
- Flinders J.; Dieckmann T. (2001) A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J. Mol. Biol. 308, 665–679. [DOI] [PubMed] [Google Scholar]
- Vallurupalli P.; Moore P. B. (2003) The solution structure of the loop E region of the 5 S rRNA from spinach chloroplasts. J. Mol. Biol. 325, 843–856. [DOI] [PubMed] [Google Scholar]
- Wu M.; Turner D. H. (1996) Solution structure of (rGCGGACGC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry 35, 9677–9689. [DOI] [PubMed] [Google Scholar]
- Kolk M. H.; van der Graaf M.; Fransen C. T. M.; Wijmenga S. S.; Pleij C. W. A.; Heus H. A.; Hilbers C. W. (1998) Structure of the 3′-hairpin of the TYMV pseudoknot: Preformation in RNA folding. EMBO J. 17, 7498–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A.; Jardine P. J.; Anderson D. L.; Grimes S.; Matsuo H. (2008) Analysis of intermolecular base pair formation of prohead RNA of the phage circle divide 29 DNA packaging motor using NMR spectroscopy. Nucleic Acids Res. 36, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner I. R.; Foster M. P. (2011) An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta 1814, 942–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda L.; Adamiak R. W.; Gdaniec Z. (2008) Bulged adenosine influence on the RNA duplex conformation in solution. Biochemistry 47, 5059–5067. [DOI] [PubMed] [Google Scholar]
- Popenda M.; Blazewicz M.; Szachniuk M.; Adamiak R. W. (2008) RNA FRABASE version 1.0: An engine with a database to search for the three-dimensional fragments within RNA structures. Nucleic Acids Res. 36, D386–D391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov J.; Gaudin M.; Podbevšek P.; Olsthoorn R. C. L.; Petersen M. (2012) The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter. RNA 18, 992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital D. G.; Allmann A. M.; Van Doren S. R.; Butcher S. E. (2003) Structural basis for a lethal mutation in U6 RNA. Biochemistry 42, 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S.; Brodersen D. E.; Murphy F. V. IV; Dunham C. M.; Selmer M.; Tarry M. J.; Kelley A. C.; Ramakrishnan V. (2005) Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Dunham C. M.; Selmer M.; Phelps S. S.; Kelley A. C.; Suzuki T.; Joseph S.; Ramakrishnan V. (2007) Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA 13, 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V. K.; Draper D. E. (1998) On the role of magnesium ions in RNA stability. Biopolymers 48, 113–135. [DOI] [PubMed] [Google Scholar]
- Misra V. K.; Shiman R.; Draper D. E. (2003) A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers 69, 118–136. [DOI] [PubMed] [Google Scholar]
- Maderia M.; Horton T. E.; DeRose V. J. (2000) Metal interactions with a GAAA RNA tetraloop characterized by 31P NMR and phosphorothioate substitutions. Biochemistry 39, 8193–8200. [DOI] [PubMed] [Google Scholar]
- Marcia M.; Pyle A. M. (2014) Principles of ion recognition in RNA: Insights from the group II intron structures. RNA 20, 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.; Tinoco I. Jr. (1998) RNA folding causes secondary structure rearrangement. Proc. Natl. Acad. Sci. U.S.A. 95, 11555–11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z.; Tinoco I. Jr. (1996) Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35, 6026–6036. [DOI] [PubMed] [Google Scholar]
- Wijmenga S. S.; Kruithof M.; Hilbers C. W. (1997) Analysis of 1H chemical shifts in DNA: Assessment of the reliability of 1H chemical shift calculations for use in structure refinement. J. Biomol. NMR 10, 337–350. [DOI] [PubMed] [Google Scholar]
- Jucker F. M.; Pardi A. (1995) GNRA tetraloops make a U-turn. RNA 1, 219–222. [PMC free article] [PubMed] [Google Scholar]
- Bloomfield V. A., Crothers D. M., and Tinoco J. I. (2000) Nucleic Acids: Structures, Properties, and Functions, pp 25–27, University Science Books, Herndon, VA. [Google Scholar]
- Correll C. C.; Swinger K. (2003) Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 Å resolution. RNA 9, 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D. J.; Schmeing T. M.; Moore P. B.; Steitz T. A. (2001) The kink-turn: A new RNA secondary structure motif. EMBO J. 20, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray L. J. W.; Arendall W. B. I.; Richardson D. C.; Richardson J. S. (2003) RNA backbone is rotameric. Proc. Natl. Acad. Sci. U.S.A. 100, 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman Y. V.; Kennedy S. D.; Shankar N.; Parisien M.; Major F.; Turner D. H. (2011) NMR structure of a 4 × 4 nucleotide RNA internal loop from an R2 retrotransposon: Identification of a three purine-purine sheared pair motif and comparison to MC-SYM predictions. RNA 17, 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S. H.; Cheong H. K.; Lee J. H.; Cheong C.; Kainosho M.; Choi B. S. (2001) Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 98, 10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza P.; Šponer J. (1999) Structure, energetics, and dynamics of the nucleic acid base pairs: Nonempirical ab initio calculations. Chem. Rev. 99, 3247–3276. [DOI] [PubMed] [Google Scholar]
- Disney M. D.; Turner D. H. (2002) Molecular recognition by the Candida albicans group I intron: Tertiary interactions with an imino G·A pair facilitate binding of the 5′ exon and lower the KM for guanosine. Biochemistry 41, 8113–8119. [DOI] [PubMed] [Google Scholar]
- Šponer J.; Mokdad A.; Šponer J. E.; Špačková N.; Leszczynski J.; Leontis N. B. (2003) Unique tertiary and neighbor interactions determine conservation patterns of cis Watson-Crick A/G base-pairs. J. Mol. Biol. 330, 967–978. [DOI] [PubMed] [Google Scholar]
- Yildirim I.; Stern H. A.; Sponer J.; Spackova N.; Turner D. H. (2009) Effects of restrained sampling space and nonplanar amino groups on free-energy predictions for RNA with imino and sheared tandem GA base pairs flanked by GC, CG, iGiC or iCiG base pairs. J. Chem. Theory Comput. 5, 2088–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Almakarem A. S.; Petrov A. I.; Stombaugh J.; Zirbel C. L.; Leontis N. B. (2012) Comprehensive survey and geometric classification of base triples in RNA structures. Nucleic Acids Res. 40, 1407–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish P. V.; Hennig M.; Giedroc D. P. (2005) A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 102, 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda L.; Bielecki L.; Gdaniec Z.; Adamiak R. W. (2009) Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple. Arkivoc 3, 130–144. [Google Scholar]
- Greenbaum N. L.; Radhakrishnan I.; Patel D. J.; Hirsh D. (1996) Solution structure of the donor site of a trans-splicing RNA. Structure 4, 725–733. [DOI] [PubMed] [Google Scholar]
- Weeks K. M.; Crothers D. M. (1991) RNA recognition by Tat-derived peptides: Interaction in the major groove?. Cell 66, 577–588. [DOI] [PubMed] [Google Scholar]
- Osterholm M. T.; Kelley N. S.; Sommer A.; Belongia E. A. (2012) Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect. Dis. 12, 36–44. [DOI] [PubMed] [Google Scholar]
- Dominski Z.; Kole R. (1993) Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 90, 8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemans N. M.; Tulinius M.; van den Akker J. T.; Burm B. E.; Ekhart P. F.; Heuvelmans N.; Holling T.; Janson A. A.; Platenburg G. J.; Sipkens J. A.; Sitsen J. M. A.; Aartsma-Rus A.; van Ommen G.-J. B.; Buyse G.; Darin N.; Verschuuren J. J.; Campion G. V.; de Kimpe S. J.; van Deutekom J. C. (2011) Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 364, 1513–1522. [DOI] [PubMed] [Google Scholar]
- Voit T.; Topaloglu H.; Straub V.; Muntoni F.; Deconinck N.; Campion G.; De Kimpe S. J.; Eagle M.; Guglieri M.; Hood S.; Liefaard L.; Lourbakos A.; Morgan A.; Nakielny J.; Quarcoo N.; Ricotti V.; Rolfe K.; Servais L.; Wardell C.; Wilson R.; Wright P.; Kraus J. E. (2014) Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): An exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 13, 987–996. [DOI] [PubMed] [Google Scholar]
- Disney M. D. (2013) Rational design of chemical genetic probes of RNA function and lead therapeutics targeting repeating transcripts. Drug Discovery Today 18, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D.; Zaninovic N.; Cohen M. S.; Rosenwaks Z.; Yang W.-Y.; Gerhardt J.; Disney M. D.; Jaffrey S. R. (2014) Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343, 1002–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K. B.; Green M. R.; Redfield A. G. (1988) Structure of a pre-mRNA branch point/3′ splice site region. Proc. Natl. Acad. Sci. U.S.A. 85, 704–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbink T. E.; Berkhout B. (2008) RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J. Virol. 82, 3090–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf M. B.; Diegel J. V.; von Hippel P. H.; Berglund J. A. (2009) The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl. Acad. Sci. U.S.A. 106, 9203–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf M. B.; Berglund J. A. (2010) Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 35, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J.; Portela A.; Ortín J. (1991) Regulated M1 mRNA splicing in influenza virus-infected cells. J. Gen. Virol. 72, 1301–1308. [DOI] [PubMed] [Google Scholar]
- Battiste J. L.; Mao H. Y.; Rao N. S.; Tan R.; Muhandiram D. R.; Kay L. E.; Frankel A. D.; Williamson J. R. (1996) α helix-RNA major groove recognition in an HIV-1 Rev peptide-RRE RNA complex. Science 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Hung L.-W.; Holbrook E. L.; Holbrook S. R. (2000) The crystal structure of the Rev binding element of HIV-1 reveals novel base pairing and conformational variability. Proc. Natl. Acad. Sci. U.S.A. 97, 5107–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzuczek S. G.; Park H.; Disney M. D. (2014) A toxic RNA catalyzes the in cellulo synthesis of its own inhibitor. Angew. Chem., Int. Ed. 53, 10956–10959. [DOI] [PubMed] [Google Scholar]
- Dallas A.; Moore P. B. (1997) The loop E loop D region of Escherichia coli 5S rRNA: The solution structure reveals an unusual loop that may be important for binding ribosomal proteins. Structure 5, 1639–1653. [DOI] [PubMed] [Google Scholar]
- Toor N.; Rajashankar K.; Keating K. S.; Pyle A. M. (2008) Structural basis for exon recognition by a group II intron. Nat. Struct. Mol. Biol. 15, 1221–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M.; Pyle A. M. (2012) Visualizing group II intron catalysis through the stages of splicing. Cell 151, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Word J. M.; Lovell S. C.; Richardson J. S.; Richardson D. C. (1999) Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 285, 1735–1747. [DOI] [PubMed] [Google Scholar]
- Schmeing T. M.; Huang K. S.; Kitchen D. E.; Strobel S. A.; Steitz T. A. (2005) Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20, 437–448. [DOI] [PubMed] [Google Scholar]
- Kurata S.; Weixlbaumer A.; Ohtsuki T.; Shimazaki T.; Wada T.; Kirino Y.; Takai K.; Watanabe K.; Ramakrishnan V.; Suzuki T. (2008) Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U·G wobble pairing during decoding. J. Biol. Chem. 283, 18801–18811. [DOI] [PubMed] [Google Scholar]
- Ulyanov N. B.; James T. L. (2010) RNA structural motifs that entail hydrogen bonds involving sugar-phosphate backbone atoms of RNA. New J. Chem. 34, 910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar N.; Kennedy S. D.; Chen G.; Krugh T. R.; Turner D. H. (2006) The NMR structure of an internal loop from 23S ribosomal RNA differs from its structure in crystals of 50S ribosomal subunits. Biochemistry 45, 11776–11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. M.; Cech T. R. (1996) Assembly of a ribonucleoprotein catalyst by tertiary structure capture. Science 271, 345–348. [DOI] [PubMed] [Google Scholar]
- Hermann T.; Patel D. J. (2000) Adaptive recognition by nucleic acid aptamers. Science 287, 820–825. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Sun X.; Watt E. D.; Al-Hashimi H. M. (2006) Resolving the motional modes that code for RNA adaptation. Science 311, 653–656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.