Abstract
Background
The gibberellin (GA) pathway plays a central role in the regulation of plant development, with the 2-oxoglutarate-dependent dioxygenases (2-ODDs: GA20ox, GA3ox, GA2ox) that catalyse the later steps in the biosynthetic pathway of particularly importance in regulating bioactive GA levels. Although GA has important impacts on crop yield and quality, our understanding of the regulation of GA biosynthesis during wheat and barley development remains limited. In this study we identified or assembled genes encoding the GA 2-ODDs of wheat, barley and Brachypodium distachyon and characterised the wheat genes by heterologous expression and transcript analysis.
Results
The wheat, barley and Brachypodium genomes each contain orthologous copies of the GA20ox, GA3ox and GA2ox genes identified in rice, with the exception of OsGA3ox1 and OsGA2ox5 which are absent in these species. Some additional paralogs of 2-ODD genes were identified: notably, a novel gene in the wheat B genome related to GA3ox2 was shown to encode a GA 1-oxidase, named as TaGA1ox-B1. This enzyme is likely to be responsible for the abundant 1β-hydroxylated GAs present in developing wheat grains. We also identified a related gene in barley, located in a syntenic position to TaGA1ox-B1, that encodes a GA 3,18-dihydroxylase which similarly accounts for the accumulation of unusual GAs in barley grains. Transcript analysis showed that some paralogs of the different classes of 2-ODD were expressed mainly in a single tissue or at specific developmental stages. In particular, TaGA20ox3, TaGA1ox1, TaGA3ox3 and TaGA2ox7 were predominantly expressed in developing grain. More detailed analysis of grain-specific gene expression showed that while the transcripts of biosynthetic genes were most abundant in the endosperm, genes encoding inactivation and signalling components were more highly expressed in the seed coat and pericarp.
Conclusions
The comprehensive expression and functional characterisation of the multigene families encoding the 2-ODD enzymes of the GA pathway in wheat and barley will provide the basis for a better understanding of GA-regulated development in these species. This analysis revealed the existence of a novel, endosperm-specific GA 1-oxidase in wheat and a related GA 3,18-dihydroxylase enzyme in barley that may play important roles during grain expansion and development.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0520-7) contains supplementary material, which is available to authorized users.
Keywords: Gibberellin, Wheat, Biosynthesis, Signalling, Gene sequences, De novo assembly, Transcriptomics, Heterologous expression, GA 1-oxidase
Background
Gibberellins (GAs) are a group of plant secondary products based on the diterpenoid ent-gibberellane skeleton; a small subset of bioactive GAs such as GA4 and GA1 act as plant hormones and participate in a wide range of developmental processes. Although classically involved in the promotion of growth processes such as germination and stem elongation, GA signalling has also been shown to be important in root elongation [1], lateral root formation [2], skotomorphogenesis [3], cambial activity [4], leaf expansion [5], trichome development [6], floral induction [7], anther and pollen development (reviewed in Plackett et al.[8]), fruit growth [9] and seed development [10]. Furthermore, GAs mediate environmental effects on growth and development through modulation of both biosynthetic and signalling components [11]. The central components of GA signalling, GRAS-domain proteins containing an N-terminal “DELLA” motif that repress growth, also act as nodes in the interactions with several other plant hormones, including jasmonate [12], brassinosteroids [13] and strigolactones [14]. In short, GAs play a central role in plant development and environmental responses, with impacts on crop yield and quality.
The importance of GA signalling in determining plant stature is clear from evidence in both wild and crop species showing phenotypic effects of genetic variation in GA biosynthetic and signalling genes. In wheat (Triticum aestivum L.), semi-dwarfing alleles of the Rht DELLA genes were key to increasing yield during the Green Revolution as not only did the shorter stature protect against lodging under high fertiliser application rates, but also enhanced harvest index by reducing straw biomass and, in many genetic backgrounds, increasing grain numbers per ear [15]. In rice (Oryza sativa L.), a similar height phenotype was conferred by loss-of-function mutations in a key GA biosynthetic gene, OsGA20ox2 [16], and there is evidence that semi-dwarfing of barley (Hordeum vulgare L.) by the sdw1/denso gene is associated with reduced expression of the orthologous HvGA20ox2 gene [17].
Although Rht semi-dwarfing alleles are widespread in modern wheat varieties, the involvement of Rht in all GA responses results in pleiotropic effects on many other traits. For example, even mild alleles such as Rht-B1b and Rht-D1b impart reduced leaf area [18]. These alleles also have a strong effect on coleoptile elongation which prevents deeper sowing under dry conditions [19]. Better targeting of the dwarfing effect to stem tissues might therefore confer significant advantages. In contrast to Rht, the enzymes acting during the latter stages of GA biosynthesis are encoded by multiple paralogs with overlapping domains of expression and, in Arabidopsis, mutations in individual genes have more localised effects [20-22]. This suggests that the characterisation of the GA biosynthetic genes of wheat has the potential to identify targets for the development of novel semi-dwarfing alleles with fewer undesirable pleiotropic effects than the current Rht alleles.
GAs are also thought to play a critical role in wheat grain development: endogenous GA levels are very high in developing grain and increase during grain expansion [10] and wheat lines containing Rht alleles have smaller grains [15]. Despite grain size being an important component of wheat yield and quality, little is known regarding the spatial or temporal regulation of GA biosynthesis and signalling in the grain. A fuller understanding of the role of GA during grain development is required to engineer improvements in this trait in modern wheat varieties.
The GA biosynthetic pathway has been extensively characterized in both rice and Arabidopsis (reviewed by Yamaguchi, 2008 [23]), and the early genes in the pathway, from copalyl diphosphate synthase (CPS) to ent-kaurenoic acid oxidase (KAO) that produce the GA precursors GA12 and GA53 have also been identified and characterised in wheat [24-26]. The final steps in GA biosynthesis and inactivation are catalysed by soluble 2-oxoglutarate-dependent dioxygenases (2-ODDs) (Fig. 1). GA 20-oxidase catalyses the multi-step oxidation of GA12 and GA53 to form the C19 skeleton, while GA 3-oxidase produces the final bioactive products, GA4 and GA1. A third class of 2-ODD, GA 2-oxidase, is involved in inactivation, with two sub-classes of enzyme that act against either bioactive GAs (GA4, GA1) and their immediate C19 precursors (GA9, GA20) [27] or against C20-GAs earlier in the pathway (e.g., GA12, GA53) [28]. However, our knowledge of the size, structure and function of the gene families encoding these enzymes in wheat is limited.
Figure 1.
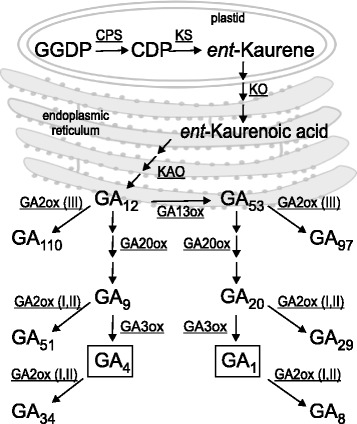
Principal reactions of the GA biosynthetic pathway in plants. Enzymes are underlined; numerals after GA2ox genes indicate the class of enzyme as defined by Lee & Zeevaart (2005). The bioactive GAs are boxed
Most of the evidence points to dynamic regulation of GA biosynthesis through regulation of the 2-ODD genes which, in contrast to the terpene cyclase and cytochrome P450 genes earlier in the pathway [24], are encoded by small multigene families [23]. Although a small number of the 2-ODDs involved in the GA pathway of wheat have been identified previously [24, 29], this study represents the first comprehensive analysis of the paralogous and homoeologous genes encoding the enzymes in this crop species. In this report we identify and characterise the biochemical function of GA-biosynthetic 2-ODD genes in wheat and demonstrate novel biochemical functions. We also catalogue the orthologous genes, where data is available, in durum wheat, barley and Brachypodium distachyon. Finally, we identify tissue-specific patterns of expression in wheat that suggest specialized roles in plant development for the different paralogs.
Results
Identification of wheat genes involved in GA metabolism
Our strategy to identify the wheat complement of genes of the 2-ODD class from the GA biosynthetic pathway, previously catalogued in rice [30, 31] is outlined in Additional file 1 (Figure S1) and involved first identifying orthologous genes from Brachypodium by BLASTP search at www.phytozome.org (Table 1 and Additional file 2) using the rice peptide sequences as queries. Coding sequences from the Brachypodium genes were then used to BLAST partially-assembled genomic survey sequence from the International Wheat Genome Sequencing Consortium (IWGSC), generated by Illumina sequencing of DNA from individual wheat chromosome arms [32]. We were thus able to identify high-quality contigs containing wheat orthologs of many of the rice and Brachypodium GA 2-ODD genes.
Table 1.
Rice, Brachypodium and bread wheat genes encoding 2-ODDs from the GA pathway
| Target | Rice gene | Brachypodium gene | T. aestivum chromosome arm assembly | ||
|---|---|---|---|---|---|
| A | B | D | |||
| GA20ox1 | Os03g63970 | Bradi1g00950 | FL (4AL) | FL (5BL) | FL (5DL) |
| GA20ox2 | Os01g66100 | Bradi2g57030 | Partiala (3A) | FLa(3B) | FLa (3D) |
| GA20ox3 | Os07g07420 | Bradi1g56200 | Partiala (3A) | FL (3B) | FLa (3D) |
| GA20ox4 | Os05g34854 | Bradi2g24980 | FLa (1AL) | FLa,b (1BL) | FLa,b (1DL) |
| GA3ox1 | Os05g08540 | - | - | - | - |
| GA3ox2 | Os01g08220 | Bradi2g04840(a) Bradi4g23570(b) | Partiala,c (3A) | FL (3B) | FLa (3D) |
| GA3ox3 | - | - | FLa (2AL) | FLa (2BL) | FLa,d (2DL) |
| GA1ox1 | - | - | - | FL (2BL) | - |
| GA2ox1 | Os05g06670 | Bradi2g34840 | FLa (1AS) | FL (1BS) | FL (1DS) |
| GA2ox2 | Os01g22910 | Bradi2g12440 | - | Partial (7BL) | FLa (7D) |
| GA2ox3 | Os01g55240 | Bradi2g50280 | Partiale (3AL) | FL (3B) | FLa (3D) |
| GA2ox4 | Os05g43880 | Bradi2g19900 | FLa (1AL) | FLa (1B) | FLa (5BL) |
| GA2ox5 | Os07g01340 | Bradi1g59570 | - | - | - |
| GA2ox6 | Os04g44150 | Bradi5g16040 | FLa (2AL) | FL (2BL) | FLa (2DL) |
| GA2ox7 | Os01g11150 | Bradi2g06670 | Partial (3AS) | FLa (3B) | FLa (3DS) |
| GA2ox8 | Os05g48700 | Bradi2g16730(a) Bradi2g16750(b) | FLa (1AL) | Partial (1BL) | FLa,b (5BL) |
| GA2ox9 | Os02g41954 | Bradi3g49390 | FL (6AL) | Partial (6BL) | FLa,b (6DL) |
| GA2ox10 | Os05g11810 | Bradi2g32580 | FLa (1AS) | FL (1BS) | FL (1DS) |
| GA2ox11 | - | - | FL (4AS) | FL (4BL) | FLa (4DL) |
| GA2ox12 | - | - | - | FL (4BL) | - |
| GA2ox13 | - | - | - | FL (4BL) | - |
Genes absent from, or incomplete in, the wheat genomic survey data were initially assembled from shotgun genomic reads of wheat cv. Chinese Spring [33] located at http://www.cerealsdb.uk.net. The raw 454 reads were identified by BLASTN with the Brachypodium CDS sequences and assembled at high stringency; for most targets this resulted in the identification of genomic contigs covering the bulk of the coding region of the gene; however, the low genome coverage of the raw data, approximately 5x, [33] coupled with the relatively high error rate of 454 sequencing and the hexaploid genome of wheat resulted in contigs that contained ambiguous bases and, in many cases, chimeric sequences derived from more than one homoeolog. Hence, these consensus sequences were used in BLAST searches of the unassembled Illumina data from individual chromosome arms [32], followed by mapping the Illumina reads to the 454 assemblies to generate homoeolog-specific sequences, as annotated in Table 1 and, in greater detail, in Additional file 2. Thus, across the GA20ox, GA3ox and GA2ox gene families, we were able to identify or assemble at least one complete homoeolog, and often all three, for most of the target 2-ODD genes, as described in detail below. In addition, we identified or assembled wheat sequences encoding the GA 13-hydroxylase (GA13ox) biosynthetic enzymes and for the GID1 and GID2 signalling components.
The genomic survey data from bread wheat cv. Chinese Spring was complemented with assembled RNA-seq data from tetraploid durum wheat (T. turgidum L. subsp. durum Desf.) cv. Kronos and the diploid progenitor T. urartu [34]. In addition, we identified, where possible, likely orthologs of each gene in the genomic survey sequences of T. urartu [35] and Aegilops tauschii [36], the diploid progenitors of the A and D genomes, respectively, of bread wheat (Additional file 2). Homoeolog-specific sequences from the tetraploid species were assigned to the A or B genomes by BLAST to the bread wheat chromosome arm-specific genomic survey data above (Table 1). Finally, we identified likely orthologs of each of the wheat genes in barley (Additional file 2), within the recent draft genome sequence of this species [37].
Structure and biochemical function of wheat GA 2-ODD genes
We identified likely Brachypodium orthologs for each of the four GA20ox genes, two GA3ox genes and ten GA2ox genes previously described in rice [30, 31]. The only exception was GA3ox1, which appears to be absent from Brachypodium, as shown in Table 1 and in the phylogenetic analysis presented in Fig. 2B; however, this species contains two genes related to GA3ox2, as discussed below. Brachypodium also contains a tandem duplication of GA2ox8. As neither hexaploid bread wheat, nor its tetraploid or diploid progenitors, possesses a fully-sequenced genome, we cannot be unequivocal about the number of GA 2-ODD genes present in wheat. However, with the exception of GA2ox5, we identified in bread wheat at least one homolog of each Brachypodium gene, and usually complete or partial sequence evidence of homoeologs on each of the three chromosomes. Thus we identified in bread wheat four homoeologous sets of GA20ox genes, at least two sets of GA3ox genes and at least nine sets of GA2ox genes, as detailed below.
Figure 2.
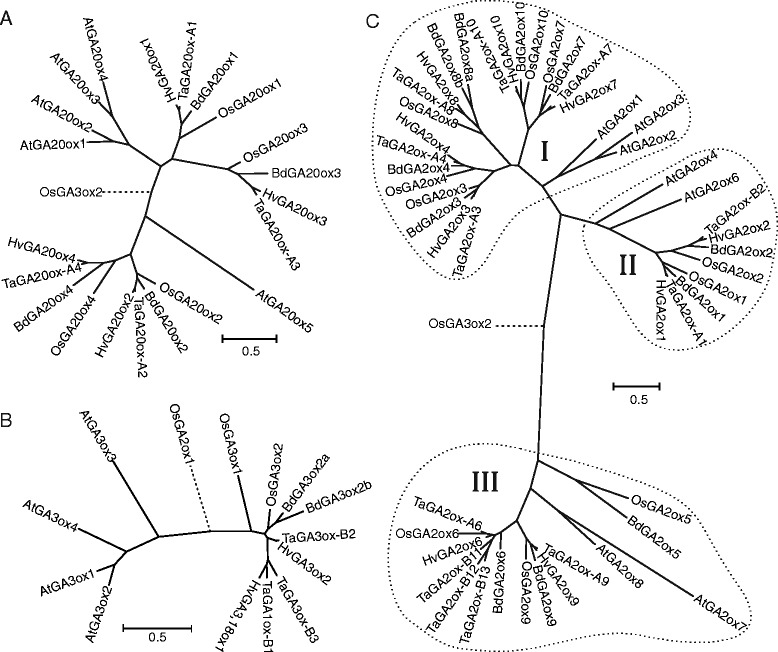
Phylogenetic relationships between gibberellin 2-ODD amino acid sequences from bread wheat (Ta), barley (Hv), rice (Os), Brachypodium (Bd) and Arabidopsis thaliana (At). Dotted lines indicate sequences used as outgroups for the rooted tree. Scale bars indicate number of amino acid substitutions per site. A: GA20ox; B: GA3ox (including TaGA1ox1 and HvGA3,18ox1); C: GA2ox. Clades labelled I, II and III in panel C reflect classes of GA2ox as defined by Lee and Zeevaart (2005)
GA 20-oxidases
Phylogenetic analysis of the 2-ODD genes showed that the grass GA20ox genes fall into four paralogous clades (Fig. 2A) each including one of the four rice GA20ox genes, OsGA20ox1 through OsGA20ox4. The assignment of the wheat GA20ox1, GA20ox2 and GA20ox4 genes to the corresponding rice groups is supported by syntenic relationships between rice and wheat chromosomes [38]. In contrast, GA20ox3 which was expected to be found on wheat chromosome group 2 based on its position in rice and Brachypodium, was found on the three homoeologs of chromosome group 3 [39]. We also identified full-length or partial sequences for all GA20ox paralogs in the tetraploid and diploid wheat species and full-length coding sequences from barley (Additional file 2). As previously reported [20], phylogenetic analysis indicates that the four paralogs of GA20ox in grass species are not directly orthologous to any of the five paralogs identified in Arabidopsis: four of the five Arabidopsis genes lie in a single clade of the tree (Fig. 2A), suggesting that the expansion in paralogs of GA20ox occurred after the separation of the monocot and eudicot lineages.
We have previously reported the biochemical function of all three homoeologs of TaGA20ox1 by heterologous expression in the pET3d vector [29]. In this study, we present a similar characterization for TaGA20ox2, TaGA20ox3 and TaGA20ox4. We expressed the coding regions of one representative homoeolog of each of these three wheat paralogs as fusion proteins in E. coli (see Methods) and demonstrated their ability to carry out the series of sequential oxidations of GA12 to GA9 (Additional file 1: Figure S2). These results demonstrate that all four wheat paralogs encode fully active GA 20-oxidases (Fig. 1).
GA 3-oxidases
Phylogenetic relationships between rice, Brachypodium, barley and wheat are less clear in the GA3ox gene family. Neither Brachypodium, barley nor either of the wheat polyploid species or their progenitors appear to possess a true ortholog of OsGA3ox1 (Fig. 2B). However, Brachypodium and wheat possess likely orthologs of OsGA3ox2, with three homoeologs in bread wheat as described previously [29]. Similar sequences were also found for the A genome in T. durum (Table 1), in T. urartu, Ae. tauschii (as a partial genomic sequence), and barley (Additional file 2). Brachypodium contains a second sequence, Bradi4g23570, related to OsGA3ox2, but the predicted coding regions of the genes from several accessions of this species at www.brachypodium.org contain a frame shift in exon 2 due to the apparent insertion of a G residue at nucleotide 765. However, PCR amplification and sequencing of this region from B. distachyon Bd21 genomic DNA clearly showed the inserted base present in the database sequence to be an artefact. Removal of G765 from the database sequence resulted in a complete open reading frame most closely related to BdGA3ox2a (Bradi2g04840) (Fig. 2B) and Bradi4g23570 was therefore assigned as BdGA3ox2b. Heterologous expression of synthetic coding sequences of BdGA3ox2a and BdGA3ox2b in E. coli followed by incubation of bacterial lysates of these cultures with radiolabelled substrates showed that both Brachypodium genes encode GA 3-oxidase enzymes, converting GA9 to GA4 (Fig. 3G,H).
Figure 3.
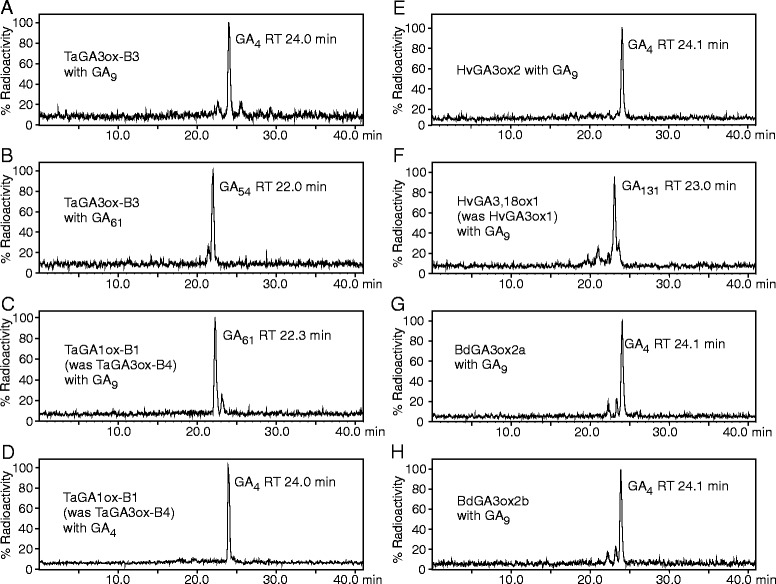
HPLC separation of incubations of GA3ox-like enzymes from wheat, barley and Brachypodium with [1-14C]-GA substrates. A,B: TaGA3ox-B3; C,D: TaGA1ox-B1 (was TaGA3ox-B4); E: HvGA3ox2; F: HvGA3,18ox1 (was HvGA3ox1); G: BdGA3ox2a; H: BdGA3ox2b. X-axis shows HPLC retention time in while the Y-axis is scaled such that the height of the largest peak of radioactivity is 100 %.
Syntenic relationships and phylogenetic analysis (Fig. 2B) support the assignment of wheat GA3ox2 genes on the group 3 chromosomes as orthologous to rice GA3ox2. However, in bread wheat we identified four further related sequences encoding potential GA 3-oxidases, all located on the long arms of the group 2 chromosomes, some of which were also found in the tetraploid and diploid species (Additional file 2). Phylogenetic analysis and the location on chromosomes 2AL, 2BL and 2DL suggested that the three most closely-related of these four novel sequences probably form a homoeologous group (Additional file 1: Figure S12) and therefore were named as TaGA3ox-A3, TaGA3ox-B3 and TaGA3ox-D3 (Table 1). Near-identical sequences to the A and B homoeologs were identified in the durum wheat transcript assembly [34] and a partial sequence from T. urartu was also identified; however, TaGA3ox3 was not found in the Ae. tauschii assembly. The fourth novel gene, located on bread wheat chromosome 2BL and also identified in the durum wheat transcript assembly, was provisionally named TaGA3ox-B4; no homoeologs of this sequence were identified in the A or D genomes of wheat or in the A and D diploid progenitor species, although a similar sequence (79.3 % amino acid identity) had been previously identified in barley and annotated as HvGA3ox1 [39].
We have previously determined the biochemical function of the bread wheat GA3ox2 genes by heterologous expression of the cDNAs in E. coli [29]: products from all three homoeologs converted GA9 to GA4 and GA20 to GA1, demonstrating GA 3β-hydroxylase (GA 3-oxidase) activity. In this study we present the functional characterization of TaGA3ox3 and TaGA3ox4 through expression in E. coli of synthetic cDNAs. When lysates from induced bacterial cells containing synthetic cDNA constructs were incubated with [1-14C]GA9, TaGA3ox-B3 was shown to encode a functional GA 3-oxidase, converting the substrate to [1-14C]GA4 (Fig. 3A), while expression products of TaGA3ox-A3 did not have any detectable catalytic activity; TaGA3ox-D3 was not tested as the Chinese Spring sequence contains a 7 bp insertion in exon 2, indicating that this gene is unlikely to be functionally active.
An unexpected result was observed for TaGA3ox-B4, which converted [1-14C]GA9 to a product with an HPLC retention time different to that of [1-14C]GA4 (Fig. 3C). This product was analysed by combined gas chromatography-mass spectroscopy (GC-MS) and had a mass spectrum consistent with [1-14C]GA61 (1β-hydroxy-GA9) [40], identifying TaGA3ox-B4 as a GA 1β-hydroxylase (GA 1-oxidase; Fig. 4), the first time an enzyme with such a catalytic activity has been described. Based on this result we propose to rename this wheat gene as TaGA1ox-B1.
Figure 4.
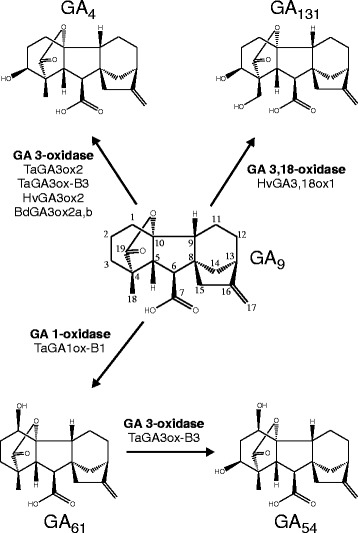
Reactions catalysed in vitro by homologs of GA3ox from bread wheat (Ta), barley (Hv) and Brachypodium (Bd)
GA61 was originally identified in the endosperm of developing grains of bread wheat [41] along with the more abundant 1β,3β-dihydroxylated form, GA54 (1-hydroxy-GA4; Fig. 4) [42]. To determine the likely sequence of reactions leading to the production of GA54 in grain, expression products of TaGA3ox-B3 and TaGA1ox-B1 were separately incubated with 14C-labelled GA9, GA4 and GA61 in the presence of co-substrates and cofactors and the products analysed by HPLC. The 3-oxidase enzyme, TaGA3ox-B3, was active against both GA9 (producing GA4) and GA61 (producing GA54, Fig. 3A,B) whereas the GA 1-oxidase, TaGA1ox-B1 acted only upon GA9 (producing GA61), and not upon GA4 (Fig. 3C,D). This suggests that the order of reactions in developing wheat grains is GA9 → GA61 → GA54, catalysed by TaGA1ox-B1 and TaGA3ox-B3, respectively (Fig. 4).
As reported above, we identified a published sequence in barley, annotated as HvGA3ox1 [39] but most closely related to TaGA1ox1 and TaGA3ox3 (Fig. 2B) and located on the syntenic barley chromosome arm, 2HL. We investigated the catalytic activity of HvGA3ox1 and HvGA3ox2 by heterologous expression of synthetic cDNAs in E. coli as above. When incubated with [1-14C]GA9, HvGA3ox2 yielded GA4 (Fig. 3E), as expected for a GA 3-oxidase, whereas HvGA3ox1 generated a product with an HPLC retention time different from both GA4 and GA61. GC-MS analysis of this novel product revealed that it was GA131 (3β,18-dihydroxy-GA9; Fig. 3F) [43], thus identifying HvGA3ox1 as a bifunctional enzyme, a GA 3β,18-dihydroxylase (GA 3,18-oxidase). We therefore propose to rename HvGA3ox1 as HvGA3,18ox1. It has been previously shown that whereas developing grains of wheat accumulate 1-hydroxy-GAs, grains from barley accumulate 18-hydroxy-GAs including GA131 (18-hydroxy-GA4) and GA132 (18-hydroxy-GA1) [43-45]. It therefore seems highly likely that HvGA3,18ox1 is the only enzyme required for the production of GA131 and GA132 from GA9 and GA20, respectively, in developing barley grains, while biosynthesis of bioactive GA4 and GA1 from these substrates in the rest of the plant is catalysed by HvGA3ox2 (Fig. 4).
As phylogenetic analysis (Fig. 2B) suggested a close relationship between TaGA3ox3, TaGA1ox-B1 and HvGA3,18ox1, we investigated their chromosomal locations. All these novel genes are located on the long arms of the group 2 chromosomes of wheat and barley, respectively. To further refine the syntenic relationships, the POPSEQ mapping data of wheat [32] was interrogated and showed the contig containing TaGA3ox-A3 to be located on chromosome 2AL at 120.3 cM while TaGA1ox-B1 was on chromosome 2BL at 134.03 cM; the contigs containing TaGA3ox-B3 and TaGA3ox-D3 were absent from the POPSEQ data. Predicted genes from the wheat contigs mapped to the same location as TaGA3ox-A3 and TaGA1ox-B1 were screened by BLASTN against the pseudomolecule of barley chromosome 2H [37], on which HvGA3,18ox1 is located at 608.9 Mbp; 95 % of the wheat genes in the same mapping bin as TaGA3ox-A3 had a top BLAST hit on barley chromosome 2H within 3 cM of HvGA3,18ox1, while 90 % of the genes co-locating with TaGA1ox-B1 also had a top BLAST hit within the same window. This suggests that TaGA3ox3, TaGA1ox-B1 and HvGA3,18ox1 are in orthologous positions in the wheat and barley genomes and are likely to be derived from a common ancestral gene, as suggested by the phylogenetic analysis (Fig. 2B). Similar BLAST searches of the rice and Brachypodium genomes with the wheat genes flanking TaGA3ox-A3 and TaGA1ox-B1 did not identify any linkage to OsGA3ox1, OsGA3ox2, BdGA3ox2a or BdGA3ox2b.
GA 2-oxidases
Ten GA2ox genes, OsGA2ox1 through OsGA2ox10, have been described in rice (Table 1); although the biochemical function of some of the rice genes has been demonstrated by heterologous expression in E. coli (e.g., OsGA2ox1, [46]; OsGA2ox5 [47]), by transactivation in rice by T-DNA insertion (e.g., OsGA2ox3, [46]) or by ectopic expression in transgenic plants (e.g., OsGA2ox5, [46]), most have not been fully characterised. Based on phylogenetic analysis of protein sequences from a number of dicot species, Lee and Zeevaart [48] proposed three structural classes of GA2ox enzymes. A phylogenetic analysis of GA2ox sequences from Arabidopsis, rice, and Brachypodium suggested that the grass enzymes can each be assigned to one of these classes (Fig. 2C). Class I, exemplified by AtGA2ox1, -2 and -3, includes the rice and Brachypodium paralogs GA2ox3, -4, -7, -8 and -10; Class II contains AtGA2ox4 and -6, and the grass paralogs GA2ox1 and -2; Class III is represented by AtGA2ox7 and -8 and the grass paralogs GA2ox5, -6 and -9. Previous data suggest that most GA2ox enzymes in Classes I and II almost exclusively use C19-GAs as substrates, while class III enzymes reportedly metabolize only C20-GAs.
In wheat, bioinformatic analysis of assembled and raw chromosome arm data revealed likely orthologs for each of the rice and Brachypodium GA2ox genes (Table 1) with the sole exception of OsGA2ox5, which was not detected in any wheat species, or in barley (Additional file 2). In bread wheat, T. urartu and Ae. tauschii we identified an additional group of GA2ox genes on the homoeologous group 4 chromosomes, that were most similar to TaGA2ox6; the bread wheat genes were named as TaGA2ox-A11, -B11 and -D11 and we identified two further related paralogs in the bread wheat genome assembly, TaGA2ox-B12 and TaGA2ox-B13, both on chromosome 4BL (Table 1) and not detected in the A and D genomes.
In general, the chromosomal locations of the wheat GA2ox genes as indicated by the chromosome arm survey data was as predicted by synteny with rice [38]. However, GA2ox2 is located on rice chromosome 1 and the orthologous genes in wheat would be expected to be found on the group 3 chromosomes but instead are on the group 7 chromosomes (Table 1). Also, the A and B homoeologs of TaGA2ox4 and TaGA2ox8 were found on the long arms of the wheat group 1 chromosomes, as predicted from synteny with rice, but in both cases no homoeolog was found on chromosome 1D, although partial sequences with high nucleotide sequence identity (94–98 %) were identified in the Ae. tauschii assembly (Additional file 2). However, genes very closely related to TaGA2ox4 and TaGA2ox8 and to the candidate orthologous sequences in Ae. tauschii were identified in the chromosome arm assembly for 5BL, and these genes were tentatively named TaGA2ox-D4(5BL) and TaGA2ox-D8(5BL).
To confirm the biochemical activities of wheat GA2ox genes we expressed one representative of each paralogous group as a fusion protein in E. coli and tested for activity against C20 (GA12) and C19 (GA9) substrates. The activity detected in each bacterial lysate was as predicted by the phylogenetic analysis (Fig. 2C): TaGA2ox-D1, -D2, -B3, -D4, -D7, -D8 and -D10 were all active against the C19 substrate, [1-14C]GA9, while TaGA2ox-D6 and TaGA2ox-D9 were active against the C20 substrate, [1-14C]GA12, (Additional file 1: Figure S3); no activity against either substrate was detected for any of the three homoeologs of TaGA2ox11; TaGA2ox-B12 and TaGA2ox-B13 were not tested. TaGA2ox-D2, -B3, -D4 and -D10 also further oxidised the GA51 product of GA9 to its catabolite, which is almost certainly derived by rearrangement of the ketone, 2-oxo-GA9, formed by a second round of oxidation at C-2. In contrast to most species, however, we found that some of the wheat enzymes showed markedly reduced substrate specificity towards C20- or C19-GAs. Notably, TaGA2ox-B3, TaGA2ox-D4 and TaGA2ox-D10, all from GA2ox Class I by phylogeny (Fig. 2C), efficiently converted GA12 to GA110 (2β-hydroxy-GA12) while TaGA2ox-D6, a Class III enzyme, converted GA9 to GA51 (2β-hydroxy-GA9) (Additional file 1: Figure S3). TaGA2ox9, also in Class III, showed partial activity against GA9, producing an unidentified product with a retention time different to both GA51 and its catabolite.
Transcript levels for GA biosynthetic and signalling gene expression in wheat tissues by RNA-seq
To determine the relative expression levels of the wheat GA genes across the life cycle of wheat, we exploited a dataset of RNA-seq samples derived from five different organs (root, leaf, stem, spike, and grain) each at three developmental stages of bread wheat cv. Chinese Spring, generated as part of the analysis of chromosome 3B [49]. The paired-end RNA-seq reads were mapped to a transcriptome reference consisting of all the wheat coding sequences identified above together with non-redundant cDNA sequences from the IWGSC wheat chromosome arm survey (see Methods). Mean fragments per kb per million mapped reads (FPKM) values (from biological duplicates) for each homoeolog of each gene are presented in Additional file 3 and histograms of expression levels of each gene family, summing the FPKM values from each homoeolog, are shown in Fig. 5.
Figure 5.
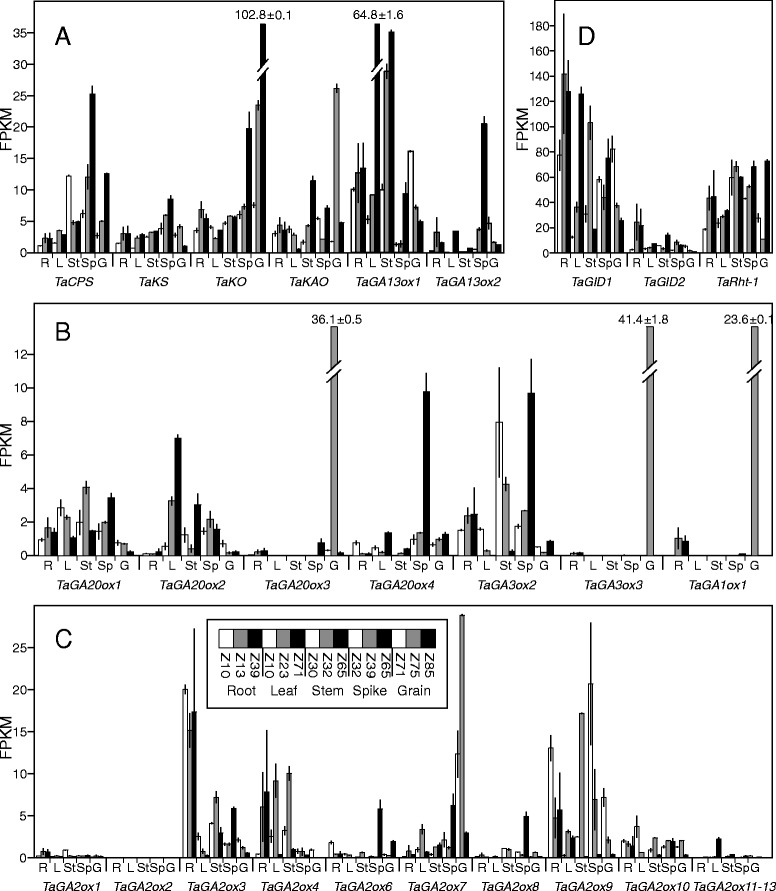
Expression of GA biosynthetic and signalling genes in five bread wheat tissues each at three developmental stages. Expression levels from each homolog were summed and then averaged across replicates, ± standard errors. A: Early GA pathway genes; B: GA-biosynthetic genes; C: GA-inactivating genes; D: GA signalling genes. GA2ox11-13 represents the sum of all additional paralogs of TaGA2ox6: GA2ox11,-12, and -13. FPKM: Fragments per kb per million mapped reads; Z: Zadoks developmental stage
The genes encoding the early enzymes in GA biosynthesis, catalysing the steps from GGDP to GA53, were found to be expressed in all tissues and stages (Fig. 5A and Additional file 3), although the homoeologous gene sets for TaCPS, TaKS, TaKO and TaKAO were more highly expressed in the spike at anthesis than in most other tissues. The TaKO genes, particularly TaKO-D1, also appear to be very highly expressed late in developing grain at Zadoks stage 85 (Additional file 3), although the physiological basis for this is unclear. GA13ox, which catalyses the 13-hydroxylation of GA12 to form GA53, is encoded by two paralogs as in rice [50]. The homoeologues of TaGA13ox1 are more highly expressed than those of TaGA13ox2 in all tissues except mature spikes (Fig. 5A and Additional file 3) and in most organs the GA13ox genes have their highest levels of expression in the most mature developmental stage sampled.
The biosynthetic 2-ODD gene families, GA20ox and GA3ox, showed tissue specific expression profiles (Fig. 5B). TaGA20ox1 and TaGA20ox2 were the most highly expressed GA20ox genes in vegetative tissues, while TaGA20ox4 was highest in the spike at anthesis and TaGA20ox3 expression was very high in and almost completely restricted to expanding grain. TaGA20ox1 was the most highly expressed GA20ox in roots, whereas in leaves and stems TaGA20ox1 and TaGA20ox2 showed a contrasting pattern, with the former having higher expression early in development of the organ and the latter being higher towards tissue maturity. The tissue specificity of GA3ox paralogs was even more striking: TaGA3ox2 appeared to be responsible for GA biosynthesis in vegetative and floral organs, while TaGA1ox-B1 and TaGA3ox3 were expressed at a very high level and almost exclusively at the mid-way stage of grain development, although TaGA1ox-B1also showed some expression in roots (Fig. 5B).
The GA2ox gene family, responsible for GA inactivation, showed variation in expression between both genes and tissue/time points (Fig. 5C). Several genes had very low or undetectable expression: no transcripts from TaGA2ox2 homoeologs were found in any tissue and TaGA2ox1 was expressed at very low levels. The only transcripts detected from the novel wheat paralogs related to TaGA2ox6 were found in mature leaves, from TaGA2ox-B12 (Fig. 5C and Additional file 3). TaGA2ox3, TaGA2ox4 and TaGA2ox9 were the most highly expressed GA2ox genes overall, contributing most to GA2ox levels in roots, leaves and stems, while TaGA2ox9 was the most highly expressed GA2ox in the developing spike. In addition, several other GA2ox genes, namely TaGA2ox3, TaGA2ox6, TaGA2ox7 and TaGA2ox8, also contributed to GA2ox transcript levels in the spike at anthesis (Fig. 5C and Additional file 3). TaGA2ox7 was by far the most highly expressed GA2ox in developing grain, with some contribution from TaGA2ox9 at the earliest stage of grain development.
We also investigated expression of the GA signalling components TaRht, TaGID1 and TaGID2: these genes were expressed in all tissues studied (Fig. 5D). The genes encoding the GA receptor, TaGID1, were most highly expressed in roots, mature leaves and elongating stems while TaGID2 expression was highest in roots. TaRht showed relatively little variation in expression except for a lower level in expanding grain.
There were some noticeable differences between expression levels of homoeologs for each of the studied genes, as shown in Additional file 3. In general, where one homoeolog was expressed at a significantly higher or lower level than the other two homoeologs, this was reflected across most of the 15 tissue/time samples, as has recently been described for homoeologous gene expression in grain tissues [51]. However, there were clear exceptions to this rule: for example, TaKO-A1 was most highly expressed in nearly all vegetative, spike and grain samples, but TaKO-D1 dominated in late grain development. Similarly, TaCPS-A1 showed the lowest level of expression in most vegetative tissues, but was higher in most reproductive tissues.
qRT-PCR of GA genes in tissues of durum wheat
To support the expression patterns determined by RNA-seq analysis in bread wheat we developed qRT-PCR assays for all the biosynthetic and signalling genes (Additional file 4). As we did not have access to the tissue samples used for the RNA-seq analysis, we assessed transcript levels in existing samples of RNA from shoot, root, grain and spike tissues in the tetraploid (durum) wheat cv. Kronos [34]. Redundant primers were designed to amplify both A and B homoeologous copies of each gene where both sequences were known. Additional file 1 (Figure S4) shows the distribution of expression of each paralog between tissues. The results of this analysis broadly reflect the RNA-seq analysis above: in particular, TaGA20ox3, TaGA1ox1 and TaGA3ox3 transcripts were detected almost exclusively in grain tissues. TaGA20ox1 and TaGA20ox2 were expressed in vegetative tissues, mainly the shoot, although appreciable levels of TaGA20ox4 were also found in this tissue as well as in the spike. As with the RNA-seq analysis TaGA3ox2 was expressed in root, shoot and spike tissues but at a much lower level in grain. There was also reasonably good agreement in the expression profiles of the GA2ox genes: the qRT-PCR showed TaGA2ox3 expression to be highest in roots, TaGA2ox4 highest in shoots, TaGA2ox7 highest in grain, while TaGA2ox8 and TaGA2ox9 expression was highest in the spike.
Expression patterns of Brachypodium GA genes
To gain insight into conservation of expression patterns between members of the grass family, we accessed RNA-seq reads from Brachypodium distachyon Bd21 [52] and mapped them to the Brachypodium reference transcriptome from Ensembl Plants (http://plants.ensembl.org/). This revealed a high degree of conservation of expression pattern between this species and wheat (Additional file 1: Figure S5): as in wheat, BdGA20ox1 and BdGA20ox2 were most highly expressed in vegetative tissues, while BdGA20ox3 expression was highest in developing seed. Notably, BdGA20ox4 expression was highly expressed in anthers, suggesting that the high level of TaGA20ox4 expression observed in wheat spikes close to anthesis (Fig. 5B) may also have been in this organ. Also, while BdGA3ox2a expression was confined to vegetative tissues, anthers and developing embryos, BdGA3ox2b was expressed predominantly in developing seed tissues, specifically the endosperm (Additional file 1: Figure S5B). The expression profiles of the GA2ox gene family were somewhat less well conserved between Brachypodium and wheat, although GA2ox3 and GA2ox7 were major contributors in both species. The highest level of GA2ox expression was in the pistil (BdGA2ox3 and BdGA2ox7), anther (BdGA2ox3 and BdGA2ox8) and in the embryo (BdGA2ox8 and BdGA2ox10) and endosperm (BdGA2ox7) of developing seeds (Additional file 1: Figure S5C).
GA gene expression patterns in developing wheat grain
The above results demonstrated that the developing wheat grain exhibits high expression levels of GA biosynthetic and signalling genes, including the novel GA 1-oxidase TaGA1ox-B1. To investigate further these patterns of expression within the expanding grain, we generated a set of RNA-seq reads from tissue layers of developing grains of bread wheat cv. Holdfast at 12 days post-anthesis: the endosperm, the inner seed coat/pericarp layer (consisting of the aleurone, nucellar epidermis, integuments, any remaining tube cells, and cross cells) and the outer pericarp layer of immature grain, consisting of the mesocarp parenchyma, hypodermis and epidermis (Additional file 1: Figure S6A). These tissue distributions were confirmed using known cell-specific gene expression patterns (Additional file 1: Figure S6B), although it is likely that there is some cross-contamination of the samples from adjacent layers. Single-end RNA-seq reads were mapped to the wheat transcriptome reference described above (see Methods), and average RPKM (reads per kilobase per million mapped reads) values for each gene from three biological replicates were calculated. The results in Fig. 6 represent combined counts from A, B and D homoeologs, while expression values for individual genes are presented in Additional file 5.
Figure 6.
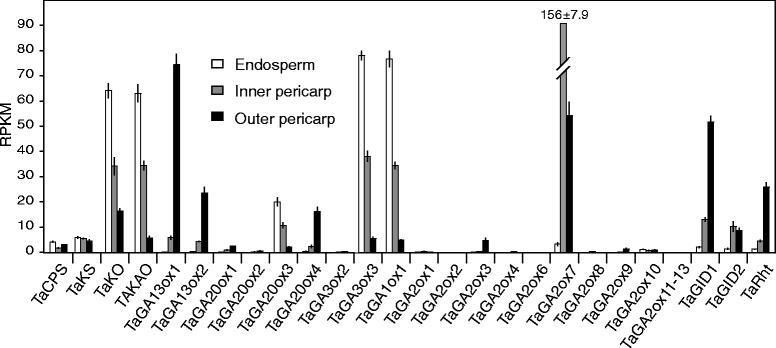
Expression of GA biosynthetic and signalling genes in different layers of the developing bread wheat grain at 10 days post anthesis. Expression levels from each homolog were summed and averaged across replicates, ± standard error. Tissue layers are as defined in Additional File 1 (Figure S6). GA2ox11-13 represents the sum of all additional paralogs of TaGA2ox6: GA2ox11, -12, and -13. RPKM: Reads per kb per million mapped reads
Taken together, RPKM expression values validated those of the whole grain sample from the previous experiments, showing that the early GA biosynthetic genes TaKO and TaKAO were highly expressed in the grain and that TaGA20ox3, TaGA20ox4, TaGA3ox3, and TaGA2ox7 account for the majority of GA 20-oxidase, GA 3-oxidase, and GA 2-oxidase expression, respectively. TaGA1ox-B1 was also highly expressed in grain. The three GA signalling genes assayed, TaGID1, TaGID2 and TaRht, were all expressed in the grain. However, there were very clear distinctions in expression levels of these genes between different grain tissue layers.
While the cyclase genes, TaCPS and TaKS, were expressed in all three grain layers, TaKO and TaKAO were more highly expressed in the endosperm and inner pericarp than in the outer layer. In stark contrast, transcripts of both paralogs of TaGA13ox were absent from the endosperm and concentrated in the outer pericarp. The later GA biosynthetic genes TaGA20ox3 and TaGA3ox3, and also the novel TaGA1ox gene, were found to be more highly expressed in the endosperm than in the seed coat/pericarp layers and, within the seed coat/pericarp, more highly in the inner layer than the outer layer (Fig. 6). TaGA20ox4 was an exception to this trend, showing highest levels of expression in the outer pericarp layers. A contrasting trend was observed for the GA catabolic gene TaGA2ox7, which was expressed at negligible levels in the endosperm, but at higher levels in the seed coat/pericarp tissues and highest in the inner layer (Fig. 6). The GA signalling genes TaGID1, TaGID2 and TaRht were expressed almost exclusively in the seed coat/pericarp layers, almost no expression being detected in the endosperm. TaGID1 and TaRht were also predominantly expressed in the outer of the two seed coat/pericarp layers.
Discussion
2-Oxoglutarate-dependent dioxygenases in the GA pathway of wheat
Since GAs contribute to nearly every stage of plant development, modifying their biosynthesis, perception and turnover has great potential to engineer improved varieties of agriculturally important crops. A prerequisite for such an approach is a more complete description and characterization of the components of the GA biosynthesis and signalling pathways. In the current study, we describe the sequence and biochemical activity of the GA20ox, GA3ox and GA2ox gene families in wheat, building upon a previous study which described the genes encoding the enzymes catalysing the earlier stages of GA biosynthesis in wheat [24]. While the absence of a fully-sequenced wheat genome precluded a precise determination of the final number of genes in each family, the transcript and genome assemblies used for this study cover a large proportion of the wheat genes [32, 34]. For missing or incomplete genes, we also successfully reassembled chromosome arm-specific sequence reads onto consensus sequences derived from assembly of 454 reads, allowing us to further extend the coverage, and also illustrating the potential for improving the wheat genome assembly using existing short read sequence data. Moreover, the number of 2-ODD paralogs found in wheat are very similar to those described for both rice and Brachypodium that have completely sequenced genomes, and in most cases we identified full-length or partial sequences for all three homoeologs, suggesting that the great majority of the genes from the GA pathway were identified in this study.
We identified or assembled full-length or partial sequences for all three homoeologs of four GA20ox genes in wheat (Table 1), corresponding to the four characterized members of this family in Brachypodium and rice (Fig. 2A). We confirmed that all four paralogs encode fully functional GA 20-oxidase enzymes, each capable of catalysing the complete series of reactions from GA12 to GA9. We similarly identified wheat homologs of all but one of the ten characterized rice GA2ox genes and confirmed that representative homoeologs of each of the enzymes encoded by these genes have GA 2-oxidase activity against either C19- or C20-GAs as substrates. However, these assays revealed that the substrate specificities of some of the wheat GA2ox enzymes do not conform to the current observations in other species where enzymes in Classes I and II, structurally-related to Arabidopsis AtGA2ox1 through AtGA2ox6, are active against C19-GAs whereas enzymes related to AtGA2ox7 and AtGA2ox8 (i.e., from Class III; Fig. 2C) are active only against C20-GAs [48]. Previously, the only identified exception to this pattern was the Class I enzyme CsGA2ox4 from cucumber, which showed weak activity against the C20-GA GA12 [53]. However, this study shows that several of the wheat enzymes have broader substrate specificities: TaGA2ox-B3, TaGA2ox-D4 and TaGA2ox-D10, all from GA2ox Class I by sequence similarity (Fig. 2C), were also active against C20-GAs, while TaGA2ox-D6, a Class III enzyme, was active against C19-GAs (Additional file 1: Figure S3). The significance of this wider substrate specificity is unclear, particularly since there do not appear to be distinct biological roles for the three structural classes of GA2ox enzymes or distinct class-specific expression profiles during development [21].
In addition to the GA2ox genes in rice, Brachypodium and barley, we identified five additional GA2ox gene sequences in wheat, comprising one homoeologous group on the long arm of the group 4 chromosomes, TaGA2ox11, plus two more individual paralogs on chromosome 4B, TaGA2ox-B12 and TaGA2ox-B13. All of these additional genes are closely related to TaGA2ox6, suggestive of gene duplication events in the ancestors of wheat. The importance of these GA 2-oxidase genes to wheat development remains unclear, however, since expression of all three homoeologs of TaGA2ox11 was negligible in all tissues studied and none of the encoded enzymes could be shown to exhibit GA 2-oxidase activity. Expression of the TaGA2ox-B12 and TaGA2ox-B13 paralogs was similarly very low in the assayed tissues.
Of the three 2-ODD families, the largest divergence from rice was observed in the GA 3-oxidases. The rice genome contains two GA3ox genes [54]; the wheat genes orthologous to OsGA3ox2 were identified and characterized in a previous study and shown to act principally as GA 3-oxidases, but also with additional minor activities including GA 2-oxidase, 2,3-desaturation and even 13-hydroxylation [29]. We did not identify a clear wheat ortholog of OsGA3ox1, either in sequence homology or in syntenic position, and this gene was also absent from the Brachypodium genome. Instead, during the current study, we found novel GA3ox gene sequences in bread wheat, three of which were closely related to one another and most likely represent a homoeologous group (TaGA3ox3) while a fourth gene, TaGA1ox-B1 (initially named TaGA3ox-B4) was identified only in the B genome (Fig. 2B); orthologs of these genes were found in durum wheat but not in rice or Brachypodium. Of the GA3ox3 group, only TaGA3ox-B3 could be shown to encode an active GA 3-oxidase, converting GA9 to GA4; TaGA3ox-D3 contains a 7 bp insertion in the second exon and while TaGA3ox-A4 appeared to encode a full-length protein, no enzyme activity could be detected in the E. coli expression products. This might be explained by polymorphisms observed in this paralog that result in mis-sense changes in amino acid residues conserved between all grass GA3ox sequences (Additional file 1: Figure S9), but technical problems with heterologous expression cannot be ruled out.
In contrast, expression products of TaGA3ox-B4 were found to catalyse 1β-hydroxylation of GA9 to yield GA61 (Figs. 3B and 4), rather than the 3β-hydroxylation suggested by its similarity to other GA 3-oxidases. This is the first description of an enzyme with GA 1-oxidase activity, and the gene was therefore renamed as TaGA1ox-B1. We found a single ortholog of TaGA1ox-B1 in durum wheat but not in the A genome or the A and D diploid progenitors, and little sequence data is available for Ae. speltoides, the closest living relative of the B genome progenitor. However, a search of the draft barley genome [37] identified two GA3ox-like genes: HvGA3ox2 was shown to encode a ‘normal’ GA 3-oxidase (Fig. 3D) and had 95-96 % nucleotide sequence identity with the three wheat TaGA3ox2 homoeologs. In contrast, HvGA3ox1 [39] is most closely related to TaGA1ox-B1 (87.6 % nucleotide sequence identity; Fig. 2B) and is located on the orthologous chromosome arm in barley, 2HL. We showed that HvGA3ox1 encodes a GA-3β,18-dihydroxylase, converting GA9 to GA131 (Figs. 3D and 4) and was therefore renamed as HvGA3,18ox1.
Analysis of the POPSEQ mapping data [32] showed that both TaGA3ox-A3 and TaGA1ox1-B1 were in orthologous positions on the group 2 chromosomes of wheat to HvGA3,18ox1 on barley chromosome 2H. However, these genes are not orthologous to either OsGA3ox1 or OsGA3ox2, which are located on rice chromosomes 5 and 1, respectively, or to the two GA3ox2 variants in Brachypodium, where are located on chromosomes 2 and 4 from that species. It seems highly likely, therefore, that the three Triticeae-specific genes evolved from an unidentified common ancestor but have subsequently diverged, acquiring different catalytic activities. Amino acid sequence identity ranged from 66 % between HvGA3,18ox1 and TaGA3ox-A3 to 79 % between HvGA3,18ox1 and TaGA1ox1-B1, whereas amino acid sequence identity between the wheat and barley GA3ox2 enzymes responsible for bioactive GA production in vegetative tissues is 95–96 %. This suggests the existence of a strong purifying selection of mutations in the GA3ox2 genes compared with the genes of novel function identified here.
Investigation of the expression patterns of these novel GA3ox-like genes showed that TaGA1ox-B1 is only expressed in the developing grain of both bread and durum wheat (Fig. 5 and Additional file 1: Figure S4), predominantly in the endosperm (Fig. 6) and that TaGA3ox-B3 shows a similar expression profile, although with some expression detected in vegetative tissues; the other homoeologues of TaGA3ox3 are also expressed in developing endosperm, but at a much lower level (Additional file 5). Similarly, inspection of the barley Gene Expression Atlas at http://www.plexdb.org revealed that HvGA3,18ox1 (Affymetrix probeset contig9888_at) is only expressed in the endosperm of developing barley grains. Supporting the expression and functional analyses described above, developing wheat grains accumulate 1β-hydroxylated GAs, including GA54 (1β-hydroxy-GA4) and GA55 (1β-hydroxy-GA1) [42]. Given that we showed that TaGA1ox-B1 is active against GA9 but not GA4, whereas TaGA3ox-B3 is active against both GA9 and GA61 (1β-hydroxy-GA9), the order of reaction in planta is likely to be GA9 → GA61 → GA54, catalysed by TaGA1ox-B1 and TaGA3ox-B3, respectively (Fig. 4), and it seems likely that these two enzymes also produce the 13-hydroxylated equivalent, GA55, from GA20 in developing grain via the same sequence of reactions. In contrast, 1-hydroxy-GAs have not been identified in developing barley grains, which accumulate a number of 18-hydroxylated GAs, including GA131 (18-hydroxy-GA4) [44], presumably produced through the action of HvGA3,18ox1 on GA9 (Fig. 4).
Tissue specificity of GA biosynthesis, signalling and turnover in wheat
Our survey of GA gene expression in five tissues of bread wheat each at three stages of development (Fig. 5) and also different tissues of durum wheat (Additional file 1: Figure S4) and Brachypodium (Additional file 1: Figure S5) indicated that all organs and time points were competent in GA biosynthesis, perception and inactivation, exhibiting measurable transcript levels for at least one paralog of each component assayed. One caveat to consider when drawing conclusions from GA biosynthetic expression data is the existence of feedback and feed-forward mechanisms which act upon the transcription of 2-ODD genes. As part of a homeostatic mechanism, increased concentrations of bioactive GA inhibit GA20ox and GA3ox expression and promote GA2ox expression [23]. Therefore, high expression of GA biosynthetic genes is not necessarily indicative of higher endogenous bioactive GA levels and may instead result from their up-regulation under conditions of low GA. The early biosynthetic genes, TaCPS, TaKS, TaKO and TaKAO, and the signalling genes TaRht, TaGID1 and TaGID2, are expressed at all stages of development, albeit with marked variation in transcript levels between tissues and stages. However, it is clear that the multiple paralogs of the 2-ODD genes encoding GA20ox, GA3ox and GA2ox have more specific patterns of expression. In particular, the expression of TaGA20ox3, TaGA3ox3 and TaGA1ox1 is very high in the grain, specifically at Zadoks 75 developmental stage, but is much lower in the other tissues assayed. This is similar to the expression patterns observed for OsGA20ox3 and OsGA3ox1 in rice [55], although no wheat ortholog of OsGA3ox1 was identified; OsGA20ox3 and TaGA20ox3 are similarly not orthologous by chromosome location, although the phylogenetic relationship appears clear (Fig. 2A).
In the leaf and stem, TaGA20ox1 is expressed more highly in early, rapidly-elongating stages of development while TaGA20ox2 appears to be highest in more mature tissue, after the period of maximal elongation growth (Fig. 5B), implying that TaGA20ox1 might be more important in determining growth rate and therefore final organ size. However, both paralogs of these two genes contribute to plant height in rice [16,56], although only OsGA20ox2 (SD-1) has been exploited in plant breeding [57]. TaGA20ox1, -2 and -3 are also expressed to some extent at all stages of spike development, a result consistent with a recent study which showed that these three GA biosynthetic genes are up-regulated by long days in the shoot apical meristem of wheat, where bioactive GA is required for the up-regulation of floral meristem identity genes SOC1-1 and LFY in spike development [7]. Relatively few genes from the GA2ox family show high levels of expression in the tissues assayed, and some have narrow expression domains. Thus TaGA2ox3, TaGA2ox4 and TaGA2ox9 account for most of the vegetative GA2ox expression, while TaGA2ox6 and TaGA2ox8 predominate in the flowering spike and TaGA2ox7 in developing grain.
Analysis of GA gene expression in separate tissue layers of the developing grain revealed that the GA biosynthetic genes, notably TaKO, TaGA20ox3, TaGA3ox3 and TaGA1ox1, were predominantly expressed in the endosperm. In contrast, genes involved in GA perception, signalling and turnover were expressed at only low levels in the endosperm and were instead expressed at their highest levels in the inner and outer seed coat/pericarp layers. Indeed, expression levels of the GA receptor, TaGID1, and the central signalling component, TaRht, were 26- and 22-fold higher, respectively, in the outer pericarp than in the endosperm (Fig. 6). These results suggest that the main site of GA biosynthesis in the young developing grain is the endosperm tissue while GA signalling occurs predominantly in the outer layers of the wheat grain, possibly implying movement of bioactive GAs between the tissues. As this period of grain development, Zadoks 73, is one of rapid radial expansion growth, it is tempting to speculate that bioactive GA produced by the endosperm promotes cell expansion in the outer grain layers which otherwise would constrain growth of the endosperm. In support of this, dwarfing mutations in TaRht genes that reduce sensitivity to bioactive GAs also reduce grain size [15], implying a role for GAs and GA signalling in grain expansion.
However, no defined role for GA in seed development has been demonstrated in any species. Although developing seeds of plants often accumulate high levels of GAs, in many cases these are inactive or partially active forms. Thus, in rice the OsGA3ox1 gene that is expressed in developing grain encodes an enzyme with both 3β-hydroxylase and 2β-hydroxylase activities [54], although the latter activity is a minor component, at least when assayed in vitro. Unlike the other sequenced grass species (maize, sorghum and Setaria italica), wheat, barley and Brachypodium lack orthologs of GA3ox1 but in each case have evolved novel paralogs of the GA3ox2 gene responsible for GA biosynthesis in vegetative tissues (Fig. 2B and Additional file 1: Figure S7) that have high expression in developing grain. In the case of bread wheat, we show above that two novel paralogs, TaGA3ox3 and TaGA1ox1, combine to produce the unusual gibberellin GA54 (1β-hydroxy-GA4) in grain. Similarly, in barley HvGA3,18ox1 encodes a bifunctional GA 3β,18-dihydroxylase; this gene is closely related to TaGA3ox3 and TaGA1ox1 and its expression is confined to developing grain. It is presumably, therefore, responsible for the accumulation of 18-hydroxy-GAs, such as GA131 (18-hydroxy-GA4), in seeds of this species. Brachypodium also contains a paralogous gene related to GA3ox2, Bradi4g23570, that we have named BdGA3ox2b. This gene is similarly expressed exclusively in seed tissues (Additional file 1: Figure S5) and encodes an enzyme with only GA 3-oxidase activity. Thus, rice, barley, wheat, and Brachypodium have independently evolved paralogs of GA3ox that produce high levels of GAs in developing grain. However, the GAs synthesized in wheat and barley grain exhibit reduced biological activity: GA131 is ~4-fold less active than GA4 [58] and the biological activity of 1-hydroxy GAs is similarly lower than of their non-1-hydroxylated equivalents (personal communication from the late Dr. John Lenton). The biological relevance of these unusual GA-modifying activities is therefore unclear and merits further investigation.
Target genes for novel dwarfing effects in wheat
The introduction of the Rht dwarfing alleles during the Green Revolution had a major impact on global wheat productivity because of a reduced incidence of lodging and improved assimilate partitioning [59]. However, because Rht is a negative regulator of GA signalling in all tissues, these alleles are also associated with negative pleiotropic effects, such as the requirement for planting closer to the surface and a slight reduction in grain size [15, 19]. Results from our study confirm that Rht was expressed in all tissues assayed, including the outer layers of expanding grain and suggests that alternative dwarfing alleles, more specific and targeted to the stem, may limit these negative pleiotropic effects. Several genetic studies to identify plant height QTLs are being carried out in order to identify alternative dwarfing alleles for wheat, although to date no specific gene has been identified [60].
One solution may lie in the selection of loss-of-function mutations in GA biosynthetic genes using currently available TILLING populations [61, 62]. In rice, the most widely used dwarfing gene is GA20ox2 [16, 17], where multiple loss-of-function alleles have been identified which reduce height. These mutations have only limited effects on reproductive development, presumably because other GA 20-oxidase genes regulate GA biosynthesis in these tissues. In wheat, TaGA20ox1, TaGA20ox2 and TaGA20ox4 are expressed in the leaf and stem, suggesting that these are promising candidates in which to search for disruptive mutations. However, since all three homoeologs of both TaGA20ox1 and TaGA20ox2 are expressed in stem tissues (Additional file 3) multiple mutations may necessary to obtain economically relevant height reductions. Current efforts to sequence the exomes of wheat TILLING lines [63] is accelerating the discovery of mutations for most of the genes presented in this study, which will facilitate the experimental validation of their function and the testing of their effects on plant height and productivity.
Conclusions
In this study we identified the genes encoding 2-ODD enzymes from the GA biosynthetic pathway of bread wheat, through a combination of interrogation of publicly available, partially-assembled genome sequences and de novo assembly of shotgun reads from individual chromosome arms. We also catalogued the GA 2-ODD genes from barley and Brachypodium. With a few exceptions, hexaploid bread wheat contains three homoeologs of each paralog of the genes identified in rice. In each case the function of the enzymes encoded by the genes was demonstrated by heterologous expression and their expression was analysed across a range of tissues and developmental stages. Certain paralogs of the GA20ox, GA3ox and GA2ox gene families were shown to be exclusively expressed in developing grain, including a novel, highly-expressed GA 1-oxidase gene in wheat endosperm (and a related GA 3,18-dihydroxylase gene in barley). The presence of these genes likely explains the high levels of 1β-hydroxylated GAs in wheat grain, and 18-hydroxylated GAs in barley, in both cases the activities resulting in a reduction in the biological activity of the GAs produced. The pattern of transcript accumulation of the GA genes in early developing grain suggested that, while most GA biosynthesis is carried out in the endosperm, GA inactivation, perception and signalling are confined to the seed coat and pericarp, consistent with a role for GA in grain expansion. This comprehensive identification and characterisation of the GA 2-ODD genes in wheat will provide the basis for a better understanding of GA-regulated development in this species, including the involvement of GA biosynthesis and signalling in grain development.
Methods
Identification and phylogeny of wheat genes for components of the GA pathway
To identify wheat (Triticum aestivum L.) genes encoding components of the GA biosynthetic and signalling pathway, sequences from Brachypodium (Brachypodium distachyon L.), identified by BLASTP at www.phytozome.org using rice (Oryza sativa L.) peptide sequences [31] as queries, were used to search the IWGSC chromosome arm survey assembly at urgi.versailles.inra.fr. In many cases it was not possible to identify all three wheat homoeologs and, therefore, guided assembly of homologous genomic sequence reads from chromosome-arm-specific shotgun sequencing assembled to 454 contigs [32, 33] was carried out using the Geneious software (Biomatters Ltd) as outlined in Additional file 1 (Figure S1). Accession numbers for all full-length 2-ODD sequences identified from wheat and other grasses are shown in Additional file 2. In addition, full-length genomic, coding sequence and polypeptide sequences of all the wheat 2-ODD genes, plus coding sequences of other genes in the pathway that were used for transcript analysis, are provided as FASTA files in Additional file 6.
For phylogenetic analysis, peptide sequences were aligned using MUSCLE [64] and the resulting alignments (Additional file 1: Figures S8-S10) were edited to remove unaligned sequences in the N- and C-terminal regions. For GA20ox the N-terminal 82 and C-terminal 31 columns of the alignment were removed; similarly for GA3ox the 39 N-terminal and C-terminal 45 columns and for GA2ox the N-terminal 69 and C-terminal 49 columns were deleted. Phylogenetic analysis was carried out on the resulting alignments using the PhyML algorithm within TOPALi v2.5 [65], including model optimisation and bootstrapping (100 repeats); an outgroup was used for rooting and phylogenetic trees were drawn using MEGA5 [66]. The trees obtained using Neighbor Joining methods were not significantly different. To simplify presentation, only one homoeolog of each paralogous bread wheat gene was included in the phylogenetic trees presented in Figure 2, but complete versions of each tree with bootstrap values are shown in Additional file 1 (Figures S11–S13).
Heterologous expression of wheat, barley and Brachypodium GA 2-ODDs
Coding sequences for a single representative homoeolog of each wheat 2-ODD, and of barley (Hordeum vulgare L.) and Brachypodium (Brachypodium distachyon L.) GA3ox genes, from the GA pathway were synthesised (Genscript, Piscataway, USA) and inserted in-frame as a fusion with thioredoxin in the expression vector pET32b. The constructs were expressed in E. coli strain Rosetta2 (DE3) pLysS (Merck Millipore Ltd) by induction of log phase cultures with 0.5 mM IPTG followed by growth at 25 °C for 5–6 h. Cells were harvested by centrifugation, suspended in 100 mM Tris-Cl pH7.5, 5 mM DTT, 10 u.ml-1 DNAseI (Sigma), lysed by freeze-thaw and sonication and assayed using radiolabelled GA substrates in the presence of co-substrates and co-factors as described previously [67]. Control assays were also run to demonstrate that neither E. coli proteins nor products encoded by the pET32b vector were active against the GA12 and GA9 substrates. All products were identified by comparison of HPLC retention times with those of standards, or, in the case of 14C-labelled GA54, GA61 and GA131, by comparison of their mass spectra with published spectra [40] after combined gas chromatography–mass spectrometry of methyl esters trimethylsilyl ethers as described previously [68] using a MAT95XP mass spectrometer coupled to a Trace GC (ThermoElectron).
Mapping RNA-seq reads to the wheat reference transcriptome
Paired-end RNA-seq reads from root, leaf, stem, spike and grain tissues of wheat cv. Chinese Spring [49] were obtained from http://urgi.versailles.inra.fr/. Paired-end RNA-seq reads from Brachypodium tissues were downloaded from the Short Read Archive at Genbank (accession number SRA046377). For developing grain samples, bread wheat cv. Holdfast seedlings were vernalized for 8 weeks at 8 °C and transplanted to pots of Rothamsted Prescription Mix including slow-release fertilizer. The plants were grown in a glasshouse with supplemental lighting under a 16 h day (18 °C), 8 h (14 °C) night and tagged at anthesis (anther emergence). Immature grain at 12 days post-anthesis were dissected into endosperm, inner and outer seed coat/pericarp tissues and RNA extracted as described by Wan et al. [69]. Illumina RNA-seq libraries from three biological replicates of each tissue were prepared using the TruSeq mRNA sample kit (Illumina TruSeq RNA sample preparation guide Part 15008136 Rev A, Nov 2010) using 4 μg total RNA. Single end reads, 110 bp, were generated on an Illumina GAIIx sequencer and RNA-seq analysis was carried out within the Galaxy environment [70]. For all sets of RNA-seq data, sequencing reads were trimmed for quality using Trimmomatic [71] and reads from each library were mapped using BWA-mem using default parameters [72] to a wheat reference transcriptome consisting of the predicted cDNA sequences from the IWGSC assembly (v21) at www.plants.ensembl.org/Triticum_aestivum. Where genes of interest were absent from this reference, full-length coding sequences developed in this work were added to replace either partial or missing sequences as detailed in Additional file 7. Gene sequences from the early part of the GA pathway were obtained from online databases: TaCPS [EMBL:GU980886, EMBL:GU980887, EMBL:GU980888); TaKS [GenBank:FR719731, EMBL: GU980889, EMBL: GU980890], TaKO [EMBL:GU980893, EMBL: GU980894, EMBL: GU980895]; TaKAO [EMBL:GU980891, EMBL:GU980892, EMBL:GU143912]. FPKM (for paired-end reads) and RPKM (for single-end reads) values were generated using the eXpress tool [73].
Availability of supporting data
The RNA-seq data from tissue layers of developing grain described in this work are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3103. The phylogenetic input data and analysis for the GA20ox, GA3ox and GA2ox trees are available from Dryas (doi:10.5061/dryad.sk2nd).
Additional files
Strategy for identification and assembly of wheat GA 2-ODD sequences. Figure S2: HPLC traces of incubations of heterologous expression products of GA20ox genes with [1-14C]-GA12. Figure S3: HPLC traces of incubations of heterologous expression products of GA2ox genes with [1-14C]-GA12 and [1-14C]-GA9. Figure S4: Relative expression of GA biosynthetic and signalling genes in durum wheat tissues determined by qRT-PCR. Figure S5: Transcript analysis of GA biosynthetic and signalling genes in Brachypodium distachyon by RNA-seq. Figure S6: Dissection of tissue layers from developing grain of bread wheat. Figure S7: Phylogenetic tree of grass GA3ox-like protein sequences. Figures S8–10: GA20ox, GA3ox and GA2ox protein alignments, respectively. Figures S11–13: Phylogenetic trees of all GA20ox, GA3ox and GA2ox protein sequences, respectively, with bootstrap support values.
Rice, Brachypodium, barley and wheat genes encoding GA 2-ODDs including accession numbers.
Homoeolog-specific expression of GA biosynthetic and signalling genes across five tissues of wheat at three developmental stages.
qRT-PCR primers used in this study and qRT-PCR methods.
Homoeolog-specific expression of GA biosynthetic and signalling genes in different layers of the wheat grain.
FASTA files containing genomic, coding sequence and polypeptide sequences of all full-length wheat genes described in this work.
Sequences in the IWGSC reference used for RNA-seq mapping substituted with full-length CDS sequences from this work.
Acknowledgments
We thank Rothamsted glasshouse staff for care of plant material, the Rothamsted CSYS Bioinformatics Support Group for advice and provision of Galaxy tools and Stephen Powers for statistical advice. We also thank Prof. Jizeng Jia for prepublication access to assembled genomic sequences from Triticum urartu and Triticum tauschii. Financial support: work by AKH, IMP, SPV, BG, AP, PH and ALP was carried out under the 20:20 Wheat® Institute Strategic Programme at Rothamsted Research, funded by the Biotechnology and Biological Sciences Research Council of the UK; YL was funded by the OPTICHINA network within the Seventh Framework Programme of the EU. SP and JD acknowledge support from the Howard Hughes Medical Institute, the Gordon and Betty Moore Foundation and from the U.S. Department of Agriculture National Institute of Food and Agriculture grants 2011–67013–30077 and 2011–68002–30029.
Abbreviations
- 2-ODD
2-oxoglutarate-dependent dioxygenases
- FPKM
Fragments per kilobase per million mapped reads
- GA
Gibberellin
- GA1ox
GA 1-oxidase (GA 1β-hydroxylase)
- GA13ox
GA 13-hydroxylase
- GA20ox
GA 20-oxidase
- GA2ox
GA 2-oxidase (GA 2β-hydroxylase)
- GA3ox
GA 3-oxidase (GA 3β-hydroxylase)
- GC-MS
Coupled gas chromatography–mass spectrometry
- HPLC
High performance liquid chromatography
- IWGSC
International Wheat Genome Sequencing Consortium
- RPKM
Reads per kilobase per million mapped reads
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
- TILLING
Targeting induced local lesions in genomes
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SP, PH, AH and ALP conceived and designed the experiments; SP, AH, IP, BG, JD and ALP assembled sequences of wheat and barley 2-ODD genes and performed phylogenetic analyses; AH and SV performed dissection and RNA isolation from developing wheat grains; JC carried out RNA-seq library construction and sequencing; SP carried out qRT-PCR on durum wheat samples; YL, AH, AP and ALP performed PCR, DNA sequencing and heterologous expression; PH executed enzyme assays and HPLC and GC-MS analysis of products; SP and ALP analysed RNA-seq data; and SP, PH, AH, JD and ALP drafted the manuscript and generated the figures and tables. All authors read and approved the final manuscript.
Contributor Information
Stephen Pearce, Email: sppearce@ucdavis.edu.
Alison K Huttly, Email: alison.huttly@rothamsted.ac.uk.
Ian M Prosser, Email: ian.prosser@gmx.com.
Yi-dan Li, Email: liyidan@foxmail.com.
Simon P Vaughan, Email: simon.vaughan@rothamsted.ac.uk.
Barbora Gallova, Email: bylynka14@hotmail.com.
Archana Patil, Email: archana.patil@rothamsted.ac.uk.
Jane A Coghill, Email: jane.coghill@bristol.ac.uk.
Jorge Dubcovsky, Email: jdubcovsky@ucdavis.edu.
Peter Hedden, Email: peter.hedden@rothamsted.ac.uk.
Andrew L Phillips, Email: andy.phillips@rothamsted.ac.uk.
References
- 1.Ubeda-Tomás S, Federici F, Casimiro I, Beemster GTS, Bhalerao R, Swarup R, et al. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 2009;19:1194–9. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Gou JQ, Strauss SH, Tsai CJ, Fang K, Chen YR, Jiang XN, et al. Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell. 2010;22:623–39. doi: 10.1105/tpc.109.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alabadí D, Gil J, Blázquez MA, García-Martínez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004;134:1050–7. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mauriat M, Moritz T. Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 2009;58:989–1003. doi: 10.1111/j.1365-313X.2009.03836.x. [DOI] [PubMed] [Google Scholar]
- 5.Appleford NEJ, Lenton JR. Gibberellins and leaf expansion in near-sogenic wheat lines containing Rht1 and Rht3 dwarfing alleles. Planta. 1991;183:229–36. doi: 10.1007/BF00197793. [DOI] [PubMed] [Google Scholar]
- 6.Perazza D, Vachon G, Herzog M. Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 1998;117:375–83. doi: 10.1104/pp.117.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013;163:1433–45. doi: 10.1104/pp.113.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plackett ARG, Thomas SG, Wilson ZA, Hedden P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011;16:568–78. doi: 10.1016/j.tplants.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Ozga JA, van Huizen R, Reinecke DM. Hormone and seed-specific regulation of pea fruit growth. Plant Physiol. 2002;128:1379–89. doi: 10.1104/pp.010800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radley M. The development of wheat grain in relation to endogenous growth substances. J Exp Bot. 1976;27:1009–21. doi: 10.1093/jxb/27.5.1009. [DOI] [Google Scholar]
- 11.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 12.Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A. 2012;109:E1192–200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li QF, Wang CM, Jiang L, Li S, Sun SSM, He JX. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal. 2012;5:ra72. doi: 10.1126/scisignal.2002908. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura H, Xue Y-L, Miyakawa T, Hou F, Qin H-M, Fukui K, et al. Molecular mechanism of strigolactone perception by DWARF14. Nat Commun. 2013;4:2613. doi: 10.1038/ncomms3613. [DOI] [PubMed] [Google Scholar]
- 15.Flintham JE, Börner A, Worland AJ, Gale MD. Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J Agric Sci. 1997;128:11–25. doi: 10.1017/S0021859696003942. [DOI] [Google Scholar]
- 16.Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A. 2002;99:9043–8. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia QJ, Zhang JJ, Westcott S, Zhang XQ, Bellgard M, Lance R, et al. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct Integr Genomics. 2009;9:255–62. doi: 10.1007/s10142-009-0120-4. [DOI] [PubMed] [Google Scholar]
- 18.Calderini DF, Miralles DJ, Sadras VO. Appearance and growth of individual leaves as affected by semidwarfism in isogenic lines of wheat. Ann Bot. 1996;77:583–9. doi: 10.1093/aob/77.6.583. [DOI] [Google Scholar]
- 19.Ellis MH, Rebetzke GJ, Chandler P, Bonnett D, Spielmeyer W, Richards RA. The effect of different height reducing genes on the early growth of wheat. Funct Plant Biol. 2004;31:583–9. doi: 10.1071/FP03207. [DOI] [PubMed] [Google Scholar]
- 20.Plackett ARG, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–60. doi: 10.1105/tpc.111.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rieu I, Eriksson S, Powers SJ, Gong F, Griffiths J, Woolley L, et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 2008;20:2420–36. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, et al. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–18. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–51. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Yang W, Pei Z, Guo X, Liu D, Sun J, et al. The genes for gibberellin biosynthesis in wheat. Funct Integr Genomics. 2012;12:199–206. doi: 10.1007/s10142-011-0243-2. [DOI] [PubMed] [Google Scholar]
- 25.Wu YS, Zhou K, Toyomasu T, Sugawara C, Oku M, Abe S, et al. Functional characterization of wheat copalyl diphosphate synthases sheds light on the early evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry. 2012;84:40–6. doi: 10.1016/j.phytochem.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou K, Xu MM, Tiernan M, Xie Q, Toyomasu T, Sugawara C, et al. Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry. 2012;84:47–55. doi: 10.1016/j.phytochem.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci U S A. 1999;96:4698–703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–63. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appleford NEJ, Evans DJ, Lenton JR, Gaskin P, Croker SJ, Devos KM, et al. Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta. 2006;223:568–82. doi: 10.1007/s00425-005-0104-0. [DOI] [PubMed] [Google Scholar]
- 30.Lo SF, Yang SY, Chen KT, Hsing YL, Zeevaart JAD, Chen LJ, et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20:2603–18. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, et al. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008;49:1429–50. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.IWGSC A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 33.Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D’Amore R, Allen AM, et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 2012;491:705–10. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krasileva KV, Buffalo V, Bailey P, Pearce S, Ayling S, Tabbita F, et al. Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol. 2013;14:R66. doi: 10.1186/gb-2013-14-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling HQ, Zhao SC, Liu DC, Wang JY, Sun H, Zhang C, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496:87–90. doi: 10.1038/nature11997. [DOI] [PubMed] [Google Scholar]
- 36.Jia JZ, Zhao SC, Kong XY, Li YR, Zhao GY, He WM, et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496:91–5. doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- 37.Mayer KFX, Waugh R, Langridge P, Close TJ, Wise RP, Graner A, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–7. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 38.Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, et al. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 2003;13:1818–27. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spielmeyer W, Ellis M, Robertson M, Ali S, Lenton JR, Chandler PM. Isolation of gibberellin and metabolic pathway genes from barley and comparative mapping in barley, wheat and rice. Theor Appl Genet. 2004;109:847–55. doi: 10.1007/s00122-004-1689-6. [DOI] [PubMed] [Google Scholar]
- 40.Gaskin P, MacMillan J. GC-MS of the gibberellins and related compounds: Methodology and a library of spectra. Bristol, U.K.: Cantock’s Enterprises; 1992. [Google Scholar]
- 41.Kirkwood PS, MacMillan J. Gibberellins A60, A61, and A62 : partial syntheses and natural occurrence. J Chem Soc Perkin Trans. 1982;1:689–97. doi: 10.1039/p19820000689. [DOI] [Google Scholar]
- 42.Gaskin P, Kirkwood PS, Lenton JR, Macmillan J, Radley ME. Identification of gibberellins in developing wheat grain. Agric Biol Chem. 1980;44:1589–93. doi: 10.1271/bbb1961.44.1589. [DOI] [Google Scholar]
- 43.Crow JR, Thomson RJ, Mander LN. Synthesis and confirmation of structure for the gibberellin GA131 (18-hydroxy-GA4) Org Biomol Chem. 2006;4:2532–44. doi: 10.1039/b603570a. [DOI] [PubMed] [Google Scholar]
- 44.Gaskin P, Gilmour SJ, Lenton JR, Macmillan J, Sponsel VM. Endogenous gibberellins and kauranoids identified from developing and germinating barley grain. J Plant Growth Reg. 1984;2:229–42. doi: 10.1007/BF02042252. [DOI] [Google Scholar]
- 45.Green LS, Faergestad EM, Poole A, Chandler PM. Grain development mutants of barley - α-amylase production during grain maturation and its relation to endogenous gibberellic acid content. Plant Physiol. 1997;114:203–12. doi: 10.1104/pp.114.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, et al. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001;125:1508–16. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shan C, Mei ZL, Duan JL, Chen HY, Feng HF, Cai WM. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One. 2014;9:e87110. doi: 10.1371/journal.pone.0087110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee DJ, Zeevaart JAD. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 2005;138:243–54. doi: 10.1104/pp.104.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345:1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- 50.Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, et al. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci U S A. 2013;110:1947–52. doi: 10.1073/pnas.1215788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, Mayer KFX, Olsen O-A, IWGSC Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014;345:1250091. doi: 10.1126/science.1250091. [DOI] [PubMed] [Google Scholar]
- 52.Davidson RM, Gowda M, Moghe G, Lin HN, Vaillancourt B, Shiu SH, et al. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 2012;71:492–502. doi: 10.1111/j.1365-313X.2012.05005.x. [DOI] [PubMed] [Google Scholar]
- 53.Lange MJP, Liebrandt A, Arnold L, Chmielewska SM, Felsberger A, Freier E, et al. Functional characterization of gibberellin oxidases from cucumber, Cucumis sativus L. Phytochemistry. 2013;90:62–9. doi: 10.1016/j.phytochem.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci U S A. 2001;98:8909–14. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–53. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol. 2004;55:687–700. doi: 10.1007/s11103-004-1692-y. [DOI] [PubMed] [Google Scholar]
- 57.Rutger JN. Applications of induced and spontaneous mutation in rice breeding and genetics. Adv Agron. 1983;36:383–413. [Google Scholar]
- 58.Morrison E, Chandler PM, Thomson RJ, Mander LN. Synthesis and bioactivity of the gibberellin, 18-hydroxy-GA1 (GA132) Org Biomol Chem. 2008;6:1416–24. doi: 10.1039/b800728d. [DOI] [PubMed] [Google Scholar]
- 59.Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/S0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 60.Griffiths S, Simmonds J, Leverington M, Wang YK, Fish L, Sayers L, et al. Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mol Breed. 2012;29:159–71. doi: 10.1007/s11032-010-9534-x. [DOI] [PubMed] [Google Scholar]
- 61.Botticella E, Sestili F, Hernandez-Lopez A, Phillips A, Lafiandra D. High resolution melting analysis for the detection of EMS induced mutations in wheat SbeIIa genes. BMC Plant Biol. 2011;11:156. doi: 10.1186/1471-2229-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uauy C, Paraiso F, Colasuonno P, Tran R, Tsai H, Berardi S, et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115–28. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henry IM, Nagalakshmi U, Lieberman MC, Ngo KJ, Krasileva KV, Vasquez-Gross H, et al. Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell. 2014;26:1382–97. doi: 10.1105/tpc.113.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, et al. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 2009;25:126–7. doi: 10.1093/bioinformatics/btn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ward DA, MacMillan J, Gong F, Phillips AL, Hedden P. Gibberellin 3-oxidases in developing embryos of the southern wild cucumber, Marah macrocarpus. Phytochemistry. 2010;71:2010–8. doi: 10.1016/j.phytochem.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 68.MacMillan J, Ward DA, Phillips AL, Sánchez-Beltrán MJ, Gaskin P, Lange T, et al. Gibberellin biosynthesis from gibberellin A12-aldehyde in endosperm and embryos of Marah macrocarpus. Plant Physiol. 1997;113:1369–77. doi: 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan Y, Underwood C, Toole G, Skeggs P, Zhu T, Leverington M, et al. A novel transcriptomic approach to identify candidate genes for grain quality traits in wheat. Plant Biotechnol J. 2009;7:401–10. doi: 10.1111/j.1467-7652.2009.00405.x. [DOI] [PubMed] [Google Scholar]
- 70.Goecks J, Nekrutenko A, Taylor J, The Galaxy Team: Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 2010;11:R86. [DOI] [PMC free article] [PubMed]
- 71.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forster SC, Finkel AM, Gould JA, Hertzog PJ. RNA-eXpress annotates novel transcript features in RNA-seq data. Bioinformatics. 2013;29:810–2. doi: 10.1093/bioinformatics/btt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strategy for identification and assembly of wheat GA 2-ODD sequences. Figure S2: HPLC traces of incubations of heterologous expression products of GA20ox genes with [1-14C]-GA12. Figure S3: HPLC traces of incubations of heterologous expression products of GA2ox genes with [1-14C]-GA12 and [1-14C]-GA9. Figure S4: Relative expression of GA biosynthetic and signalling genes in durum wheat tissues determined by qRT-PCR. Figure S5: Transcript analysis of GA biosynthetic and signalling genes in Brachypodium distachyon by RNA-seq. Figure S6: Dissection of tissue layers from developing grain of bread wheat. Figure S7: Phylogenetic tree of grass GA3ox-like protein sequences. Figures S8–10: GA20ox, GA3ox and GA2ox protein alignments, respectively. Figures S11–13: Phylogenetic trees of all GA20ox, GA3ox and GA2ox protein sequences, respectively, with bootstrap support values.
Rice, Brachypodium, barley and wheat genes encoding GA 2-ODDs including accession numbers.
Homoeolog-specific expression of GA biosynthetic and signalling genes across five tissues of wheat at three developmental stages.
qRT-PCR primers used in this study and qRT-PCR methods.
Homoeolog-specific expression of GA biosynthetic and signalling genes in different layers of the wheat grain.
FASTA files containing genomic, coding sequence and polypeptide sequences of all full-length wheat genes described in this work.
Sequences in the IWGSC reference used for RNA-seq mapping substituted with full-length CDS sequences from this work.


