Abstract
An extraordinary diversity of amino acid sequences in the peptide-binding region (PBR) of human leukocyte antigen [HLA; human major histocompatibility complex (MHC)] molecules has been maintained by balancing selection. The process of accumulation of amino acid diversity in the PBR for six HLA genes (HLA-A, B, C, DRB1, DQB1, and DPB1) shows that the number of amino acid substitutions in the PBR among alleles does not linearly correlate with the divergence time of alleles at the six HLA loci. At these loci, some pairs of alleles show significantly less nonsynonymous substitutions at the PBR than expected from the divergence time. The same phenomenon was observed not only in the HLA but also in the rat MHC. To identify the cause for this, DRB1 sequences, a representative case of a typical nonlinear pattern of substitutions, were examined. When the amino acid substitutions in the PBR were placed with maximum parsimony on a maximum likelihood tree based on the non-PBR substitutions, heterogeneous rates of nonsynonymous substitutions in the PBR were observed on several branches. A computer simulation supported the hypothesis that allelic pairs with low PBR substitution rates were responsible for the stagnation of accumulation of PBR nonsynonymous substitutions. From these observations, we conclude that the nonsynonymous substitution rate at the PBR sites is not constant among the allelic lineages. The deceleration of the rate may be caused by the coexistence of certain pathogens for a substantially long time during HLA evolution.
Keywords: allelic lineage, balancing selection, HLA, pathogen, peptide-binding region, genetics of immunity, innate immunity, complex genetics, tolerance, complex immunity, infection, resistance
The human leukocyte antigen (HLA) system, also known as the major histocompatibility complex (MHC) in nonhumans, codes for molecules that bind to both self and nonself peptides and presents them to T lymphocytes. In humans, six classical HLA molecules, three of class I (HLA-A, B, and C) and three of class II (HLA-DR, DQ, and DP), have been identified as important for the initiation of T cell–mediated immune response.
The peptide-binding region (PBR) for each HLA is the most polymorphic coding region in the human genome, and this variation is thought to be primarily maintained by balancing selection (Hughes and Nei 1988, 1989; Hughes and Yeager 1998; Klein et al. 2007). There are three major (nonmutually exclusive) mechanisms of balancing selection: heterozygote advantage (overdominant selection) (Doherty and Zinkernagel 1975); rare allele advantage (Slade and McCallum 1992); and fluctuating selection (Hill 1991) [see Spurgin and Richardson (2010) for an in-depth review of these hypotheses].
Of the different mechanisms proposed for balancing selection, there are several studies that have identified an important role of the heterozygote advantage in maintaining genetic variation of the HLA. Heterozygote advantage acts when individuals who are heterozygous at a given locus have a higher fitness than individuals who are homozygous because they can bind a larger suite of antigens and confer resistance to a broader range of pathogens (Doherty and Zinkernagel 1975). The first study to provide molecular evidence for overdominant selection was conducted by Hughes and Nei (1989). They found that the rate of nonsynonymous substitutions in the PBR exceeded that of synonymous substitutions in the entire gene. Studies examining human resistance to specific pathogens have also indicated a role for heterozygote advantage. For example, heterozygotes for HLA class II loci were more resistant to the hepatitis B virus than homozygotes (Thursz et al. 1997). Similarly, later onset and slower progression of acquired immunodeficiency syndrome (AIDS) have been observed in heterozygotes for HLA class I loci, rather than in homozygotes (Carrington et al. 1999). In addition, a review of MHC studies in free-ranging animal populations highlighted the functional importance of MHC variability in parasite resistance (Sommer 2005). Similar studies have also shown associations between MHC polymorphism and parasite resistance, although some studies did not support the heterozygote advantage hypothesis (Penn et al. 2002; Musolf et al. 2004; Oliver et al. 2009).
In addition to balancing selection via amino acid substitutions in the PBR, evolutionary forces to the selection seem to be in operation, purifying selection due to functional (or structural) constraints. Consequently, nonsynonymous substitutions in the PBR show several characteristics. One is a frequent flip-flop substitution of Val and Gly at site 81 in the HLA-DRB1 gene. This specific amino acid switching is believed to be due to functional or structural constraints for the peptide-binding groove (Ong et al. 1991; Demotz et al. 1993; Verreck et al. 1996). In addition, the extent of MHC diversity at the PBR appears to be limited, as too much diversity limits T lymphocyte repertoires as a result of extensive negative selection during the T cell development process in the thymus (Vidović and Matzinger 1988; Nowak et al. 1992; Woelfing et al. 2009).
Despite the unusually large amount of variation found at the MHC PBR, the theory (allelic genealogy) of allelic lineage under symmetric balancing selection (Takahata 1990; Takahata et al. 1992) predicts that nucleotide substitutions at the PBR sites are accumulated with the divergence time of the alleles. This is because symmetric balancing selection assumes that large numbers of alleles are equivalent to each other in their fitness, and thus the probability for producing descendants is also equal. Therefore, the genealogical characteristic is quite similar to the neutral case, except for its time scale (Takahata 1990). This time scale is determined by a “scaling factor” that is a function of the mutation rate, selection coefficient, and effective population size. For the molecular clock to operate for nonsynonymous substitutions in the PBR, the selection intensity for all the lineages must be constant. However, despite many studies that examined selective mechanisms of HLA diversity, no previous studies have elucidated the temporal aspects of nucleotide substitution patterns at the PBR nonsynonymous sites. To determine the temporal evolutionary trajectory of HLA diversity, amino acid substitution patterns at the PBR nonsynonymous sites were examined at six HLA loci (HLA-A, B, C, DRB1, DQB1, and DPB1), with special focus on HLA-DRB1. Here, we provide evidence for episodic amino acid substitutions in the PBR of humans, in primate evolution, suggesting significant fluctuation of selection intensity of the allelic lineage in HLA-DRB1. The extent of amino acid diversity in the PBR is directly linked to how many kinds of pathogens can be recognized by HLA molecules. Hence, tracing the transitional change of HLA diversity may allow us to describe the relationships between humans and pathogens in particular time periods.
Materials and Methods
Collection of nucleotide sequence data for six HLA loci
Nucleotide sequences for the six HLA loci (HLA-A, B, C, DRB1, DQB1, and DPB1) were obtained from the HLA/IMGT database (http://www.ebi.ac.uk/imgt/hla/) (Robinson et al. 2011). The sequences of HLA-A, B, C, and DRB1 containing the entire coding region (HLA-A/-B/-C, approximately 1100 bp; HLA-DRB1, approximately 800 bp) were used in this analysis. Because the number of alleles covering the whole sequence of the coding region in the HLA-DQB1 and HLA-DPB1 was limited, slightly shorter sequences were included in the analysis (HLA-DQB1, approximately 690 bp; HLA-DPB1, approximately 540 bp). Possible recombinant alleles were excluded by using the method described by Satta (1992). This method assumes that the relationship between the number of substitutions in a particular region and that in the entire region is binomially distributed. Additionally, the ratio of the number of these substitutions is proportional to the size in the corresponding regions (see Kusaba et al. 1997 for more details). Consequently, the dataset used in this analysis included 50 HLA-A alleles, 143 HLA-B, 129 HLA-C, 56 HLA-DRB1, 55 HLA-DQB1, and 38 HLA-DPB1. HLA-DRB1 alleles observed in a Japanese population were investigated using the Allele Frequency Net Database (http://www.allelefrequencies.net/) (Gonzalez-Galarza et al. 2011).
Phylogenetic analyses
Multiple sequence alignments of MHC-DRB1 nucleotide sequences and amino acid translations were performed using MEGA v5.10 (Tamura et al. 2011). A phylogenetic tree of nucleotide sequences in DRB1 non-PBR was constructed based on the maximum-likelihood (ML) method using the Hasegawa-Kishino-Yano (HKY) substitution model (Hasegawa et al. 1985) implemented in MEGA. Nearest-neighbor-interchange (NNI) was applied as the ML heuristic method. Bootstrap values were obtained from 1000 replications. Amino acid distances at the PBR between alleles were calculated using the Jones-Taylor-Thornton (JTT) model (Jones et al. 1992) as per the ML tree topology for non-PBR using software in the PHYLIP 3.69 package (Felsenstein 2009). In the phylogenetic analysis of 56 HLA-DRB1 alleles (described above), 6 HLA-DRB3, 4 HLA-DRB4, 2 HLA-DRB5, 11 chimpanzee (Pan troglodytes) Patr-DRB1, 22 rhesus monkey (Macaca mulatta) Mamu-DRB1, and 3 crab-eating macaque (Macaca fascicularis) Mafa-DRB1 sequences were also included after possible recombinant alleles were removed using the method by Satta (1992) (see above). Two HLA-DQB1 sequences (HLA-DQB1*02:01:01 and HLA-DQB1*06:02:01) were used for determining the root of the tree. The dataset of HLA-DRB3, HLA-DRB4, HLA-DRB5, and HLA-DQB1 allele sequences was obtained from the HLA/IMGT database, whereas the nonhuman primate DRB1 sequences were obtained from the IPD-MHC NHP database (http://www.ebi.ac.uk/ipd/mhc/nhp/) (Robinson et al. 2005).
Examination for temporal aspect of amino acid substitution pattern in the PBR
Sites 57, 67, and 90 were not identified as the PBR sites of HLA-DRB1 molecule by Brown et al. (1993). However, Brown and collaborators have subsequently shown that these sites are involved in peptide-binding pockets that are crucial for peptide capture (Stern et al. 1994). In addition, recent assays for the peptide-binding prediction of HLA molecules have re-identified these three sites as peptide-binding residues (Reche and Reinherz 2003). Therefore, they were included as PBR in this analysis, and 27 amino acids in total were regarded as PBR. To examine the nucleotide substitution rate, the mean number of nonsynonymous substitutions in PBR [KB(m)] among allelic pairs that had the particular number of substitutions (m = KS + KN, where KS is the number of synonymous substitutions in the entire region and KN is the number of nonsynonymous substitutions in non-PBR) at putative neutral sites (synonymous + non-PBR nonsynonymous sites) was examined. The molecular clock for KN was tested using the ML method in MEGA.
The expected KB(m) value was calculated on the basis of equation 12 described in Takahata et al. (1992), and its SE was estimated by maximum variance (Takahata and Tajima 1991). The variables necessary for the equation were selected such that the first several observations showed good agreement with expectations.
Estimation of the divergence time of HLA-DRB1 alleles and amino acid substitution rates in the PBR
We calculated the divergence time of allelic pairs on the basis of neutral divergence, d = (KS + KN/f)/(LS + LN), where LS and LN are the number of synonymous and non-PBR nonsynonymous sites, respectively. The KN/f value is the number of converted neutral substitutions obtained by dividing by the functional constraint f, which is the mean ratio of nonsynonymous to synonymous substitution rates at the non-PBR of all allelic pairs. The divergence time (T) of each allele pair is given by the formula T = d/2μ, where μ is the neutral substitution rate of 10−9 per site per year at the primate MHC loci (Satta et al. 1994).
To evaluate the amino acid substitution rate in the PBR for each allelic lineage, the number of PBR amino acid substitutions on each branch of the non-PBR ML tree was compared with the ML estimate of the corresponding branch length. Here, the linear correlation between these two values was normally expected if the molecular clock worked in both PBR and non-PBR. Significant outliers were identified based on 95% confidence interval of the regression line. These outliers were further examined for whether the ML estimate of a branch length was significantly different from zero, based on a confidence interval of the estimates. In addition, internodal branches supported by more than 80% bootstrap values were selected. Finally, we identified allelic lineages or branches with a fast or slow PBR substitution rate compared with the average.
Computer simulation and prediction of peptide-binding specificity for HLA-DRB1 alleles
The assumptions underlying the computer simulation were as follows. We supposed that there is a PBR phylogenetic relationship that shows the same topology of the non-PBR tree based on nucleotide sequences of 56 HLA-DRB1 alleles. According to the expected time length (Ti) for a branch i, the number of PBR substitutions (Ki) on the branch follows a Poisson distribution with mean λ = μTi, where μ is the substitution rate (=1/unit time). The Ti value is the number of neutral nucleotide substitutions corresponding to each branch length on the non-PBR tree. The Ki value on a slow-rate branch was determined by dividing the original λ value by 10, whereas that on a fast branch was calculated by multiplying the original λ value by 10. In this manner, Ki values for every branch were determined and the number of substitutions in a pair of alleles was obtained by summing corresponding branches.
The dataset of epitopes or source organisms bound to HLA-DRB1 molecules was obtained from the Immune Epitope Database (IEDB) (http://www.immuneepitope.org) (Vita et al. 2010).
Results
Variation in amino acid diversity at the PBR
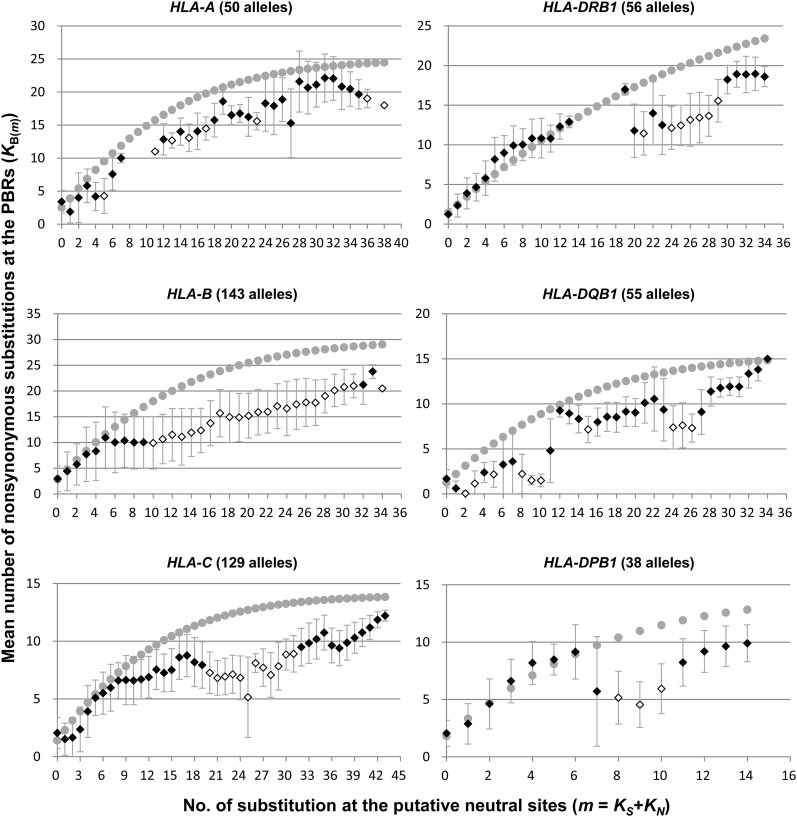
For each of the six HLA loci, the mean number of nonsynonymous substitutions in the PBR [KB(m)] among particular allele pairs with putative neutral substitutions of m (m = KS + KN) was calculated (Figure 1). Since the KN is the number of nonsynonymous substitutions at the non-PBR, we performed molecular clock tests for the substitutions. As a result, the homogeneous evolutionary rate of KN in each HLA locus was statistically supported (P < 0.05), with the exception of HLA-B and HLA-C loci. According to the theory previously described (Takahata et al. 1992), the value of KB(m) is expected to increase in proportion to m, the coalescent time (or divergence time) between alleles, and the value is then saturated with m of distantly related allele pairs (Supporting Information, Figure S1). However, the observed KB(m) value did not show the linear relationship with m for most loci (Figure 1). For the intermediate values of m, it appeared to decrease and cease the accumulation of the substitutions. After this plateau, the KB(m) value increased again. Although most of the HLA genes showed a generally similar pattern of KB(m) distribution, their detailed patterns were somewhat different for each locus. For instance, KB(m) values in HLA-DQB1 decreased until m = 2, whereas those in HLA-DPB1 decreased from m = 6 to m = 9. Interestingly, our preliminary research indicated that the rat (Rattus norvegicus) DRB1 (RT1-Db1) also showed a similar pattern of KB(m) saturation (Figure S2).
Figure 1.
The level of amino acid diversity at the peptide-binding region among HLA allele pairs that share the same coalescence time. The ordinate axis indicates the mean number of nonsynonymous substitutions at the peptide-binding region (PBR) among allele pairs [KB(m)]. The abscissa axis indicates the number of substitutions at putative neutral sites (m = KS + KN). The gray dot represents the expected KB(m) value. The diamond represents the observed KB(m) value. The open diamond indicates the statistically significant difference between the observed and expected KB(m) values (P < 0.05).
The comparison between the observed and expected KB(m) values showed that the observed KB(m) was quite lower than expected in all HLA loci, and the difference was statistically significant (Z test, P < 0.05) (Figure 1). This result indicates that KB(m) fluctuation was not likely caused by stochastic error. Instead, nonsynonymous substitutions at the PBR may have been unexpectedly suppressed due to an unknown biological phenomenon.
An acceleration of KB(m) was expected in order to cope with the rapid evolutionary change of pathogens; however, we observed a decline of KB(m), and the reason was unexplained. It was intriguing to understand why amino acid substitutions at the PBR were suppressed despite balancing selection. Therefore, we investigated why KB(m) did not increase in proportion to the divergence time of alleles (m). We explored HLA-DRB1 alleles, which showed a typical graphical pattern and provided relatively rich experimental data for binding peptides. Depending on the extent of the KB(m) increment, we categorized the graph into four phases, I through IV (Figure 2). In phases I and III, KB(m) increase was proportional to m, the divergence time between alleles, whereas in phases II and IV, KB(m) values were more or less constant, irrespective of the m values. In phase IV, the accumulation of substitutions might have reached a ceiling because of an enhanced PBR nonsynonymous nucleotide substitution rate due to balancing selection. In phase II, the KB(m) values [m = 20 to 28, mean KB(m) = 12.7] appeared to be extensively decreased as compared to that [m = 19, KB(m) = 17.0] in phase I, suggesting that PBR amino acid substitutions of alleles in phase II were limited. Table S1 shows the allelic pairs that constituted phase II. When we approximately estimated the divergence time of allele pairs in phase II (see Materials and Methods), the divergence time ranged from approximately 18 MYA to 24 MYA, suggesting that those allele pairs have diverged at least before speciation events in the subfamily Homininae (or Hominidae). The KB(m) decline was not observed in smaller m, suggesting that the decline had not occurred within the human lineage only.
Figure 2.
The mean number of nonsynonymous substitutions at the PBR among HLA-DRB1 allele pairs that share the same coalescence time. The ordinate axis represents the mean number of nonsynonymous substitutions at the PBR among allele pairs [KB(m)]. The abscissa axis represents the number of substitutions at putative neutral sites (m = KS + KN). The Roman numerals indicate the four phases that are classified as per variation in patterns of KB(m) values (see text). Error bars indicate the SD from the mean.
Phylogenetic analysis for the DRB1 locus
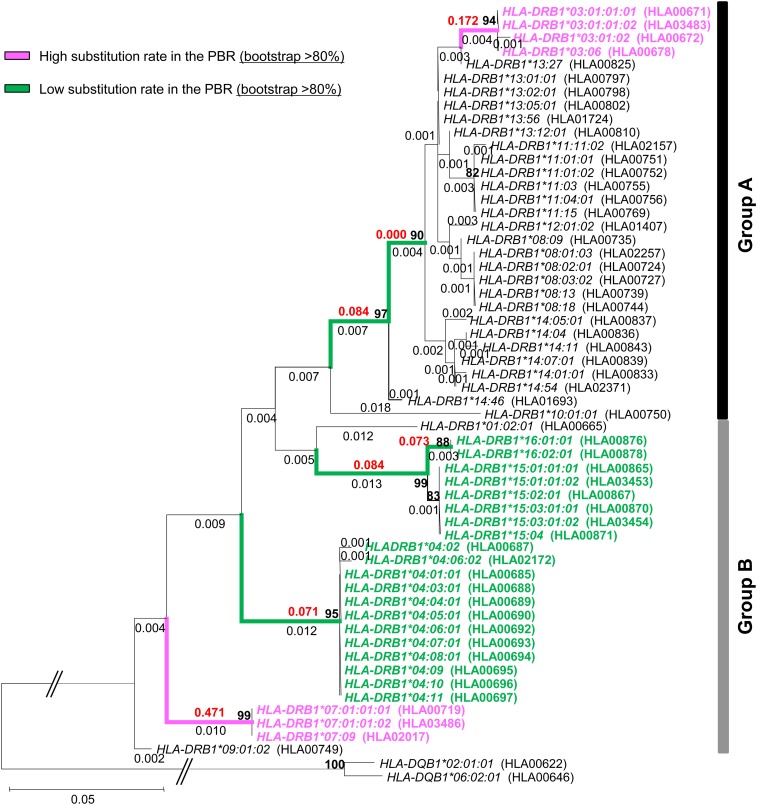
We constructed an ML tree for DRB alleles, including HLA-DRB1/3/4/5, Patr-DRB1, Mamu-DRB1, and Mafa-DRB1, based on total substitutions in only non-PBR nucleotide sequences. In the tree, some HLA-DRB1 alleles formed a monophyletic group, whereas other alleles were polyphyletic with DRB alleles from nonhuman primates (Figure 3). This topology was also supported by the neighbor-joining (NJ) tree (Saitou and Nei 1987) method based on both nucleotide and amino acid sequences in the region, although bootstrap values of their groups were not high (Figure S3). Here, we refer to these two groups as group A and group B, respectively (Figure 3). Among the 14 known HLA allelic lineages, group A consisted of seven lineages, namely DRB1*03, *08, *10, *11, *12, *13, and *14. The remaining seven lineages, DRB1*01, *02, *04, *07, *09, *15, and *16, were in group B. The allelic lineages HLA-DRB1*01 and *10 of the same DR1 haplotype were included in different groups, whereas other alleles of the same haplotype belonged to either group A or group B.
Figure 3.
Phylogenetic relationships between MHC DRB alleles (including those of humans, chimpanzees, and macaques) as determined by the maximum likelihood (ML) method on the basis of nucleotide sequences (690 bp) in the non-PBR. Only bootstrap values more than 80% are shown here. Two HLA-DQB1 sequences are used as the out-group. Evolutionary distances were computed using the Hasegawa-Kishino-Yano (HKY) model. Groups A and B indicate two major phylogenetic groups of HLA-DRB1 alleles. HLA, humans; Patr, chimpanzees; Mamu, rhesus monkeys; Mafa, crab-eating macaques. IMGT/HLA and IPD accession numbers are in parentheses.
When the non-PBR phylogeny (Figure 3) based on total nucleotide substitutions was compared with the tree based on PBR amino acid substitutions, the phylogenetic position of allelic lineages in the non-PBR tree did not correspond to that in the PBR amino acids tree (Figure S4). This means that when members of an allele pair are distantly related to each other in their non-PBR sequences, the amino acids in the PBR are often more closely related (and vice versa). When the allele pairs in phase II were placed on the tip of non-PBR tree, each allele for a particular pair was frequently found (73% of allele pairs in phase II) from either group A or group B.
The constant accumulation of PBR nonsynonymous substitutions appears to be violated in certain allele pairs. The reasons for the observed nonlinearity of KB(m) includes one of the following possibilities: (1) saturation of transitions in nucleotide substitution precedes that of transversion, and this difference in saturation timing causes the phenomenon; (2) parallel substitutions in the PBR mask the linear relationship between KB(m) and m; or (3) slow-down of the PBR nonsynonymous substitution rate in particular allelic lineages skews the constant KB(m) accumulation.
Biased rate of transitions and transversions and parallel substitutions in the PBR among HLA-DRB1 allele pairs
It is well-known that transitions (Ts) occur at a higher frequency than transversions (Tv) in both animal mitochondrial and nuclear DNA sequences. Because of this bias and the lower number of Ts than Tv states, Ts is likely to reach a plateau earlier than Tv. Thus, there is a possibility that excess of Ts in the KB(m) explains the observed accumulation pattern. To examine this possibility, we tested the relationship between numbers of Ts and Tv in the PBR (Figure S5). In phase I, Ts values of 0.7 to 6.5 were observed with Tv of 0.6 to 11.0; in the phase II, Ts of 4.5 to 6.7 were seen with Tv of 7.7 to 11.0; in phase III, Ts of 5.5 to 7.3 were associated with Tv of 10.6 to 14.5; and in phase IV, Ts of 4.6 to 7.1 were found with Tv of 14.0 to 16.6. Although Ts values seem to be saturated when Tv = 14, the corresponding allele pairs were all in phase III or phase IV. Thus, the timing of Ts saturation did not explain the cessation of KB(m) accumulation in phase II.
The non-PBR phylogenetic relationship among HLA-DRB1 sequences was not consistent with the PBR phylogenetic relationship. This is likely due to parallel substitutions or recombination between alleles, and these events probably mask the linear relationship between KB(m) and m. Because we excluded the possibility of recombinants when filtering alleles used in this analysis (see Materials and Methods), the effect of recombination on the KB(m) accumulation pattern is quite small. Next, parallel substitutions in the PBR were investigated. An example of a parallel substitution is the substitution between Val and Gly at the site 81 (Gregersen et al. 1986), which was frequently observed on several branches in the non-PBR tree. Dimorphism (Val and Gly) at the site has been shown to affect the conformation of the peptide-binding pocket (Ong et al. 1991) and allo-recognition of peptides (Demotz et al. 1993). This dimorphism appears to be associated with the selection of binding-peptide groups with different motifs (Verreck et al. 1996). However, this parallel substitution at site 81 does not explain the phase II substitution pattern in KB(m) (see below).
We performed an extensive search for parallel substitutions at the PBR amino acid sites by manually placing the PBR amino acid substitutions on branches in the non-PBR tree (Figure S4). The number of PBR substitutions was estimated by the maximum parsimonious method. Here, the same amino acid substitution on different branches was defined as parallel substitution. The number of such parallel substitutions was counted in each allele pair, not only in phase II but also in other phases. Consequently, it is interesting that a large number of parallel substitutions are observed even in closely related pairs of alleles as frequently as in distantly related allele pairs. Parallel substitutions at not only site 81 but also other PBR sites were widely observed across all phases (I–IV) (Table S2). However, KB(m) values in phase I and phase III still increased with m, even though parallel substitutions were observed in these phases. Therefore, KB(m) saturation in phase II was unlikely to be affected by parallel substitutions.
Heterogeneity in the PBR substitution rate among allelic lineages
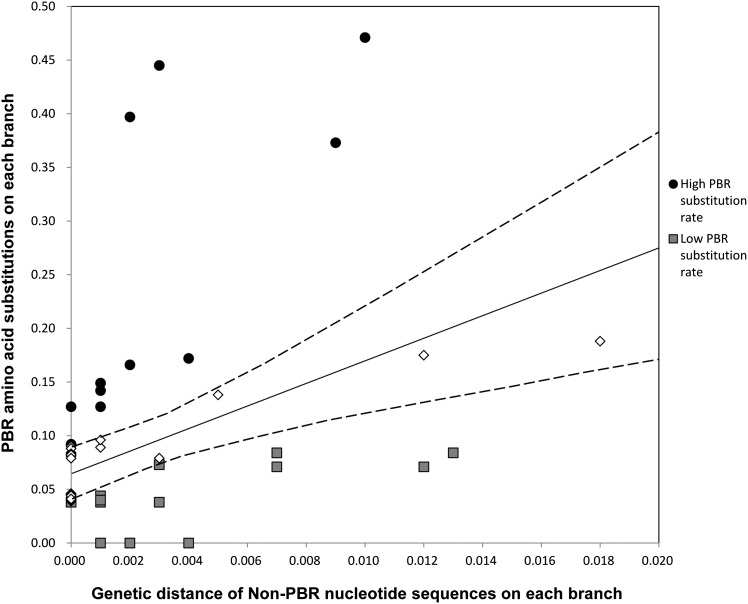
To evaluate the possibility of heterogeneity in the substitution rate, we tested whether the PBR amino acid substitution rate for each allelic lineage was constant by means of the comparison between the number of nonsynonymous substitutions at non-PBR and PBR sites on each branch of the non-PBR tree (see Materials and Methods). This analysis identified five and two branches with significantly slow and fast PBR substitution rates, respectively, as compared with the average (Figure 4 and Figure 5). Allelic lineages of HLA-DRB1*04, *15, and *16 had a slow PBR substitution rate, and those of *03, and *07 had a fast rate. As expected, alleles with slow PBR substitution rates were observed frequently (44%) in phase II, whereas in phases I, III, and IV the frequencies of such alleles were 1–6% (Table 1).
Figure 4.
The relationship between the PBR and non-PBR genetic distances on each branch of the phylogenetic tree. The ordinate axis represents amino acid substitutions at the PBR among allele pairs on each branch of the non-PBR ML tree. The substitutions were estimated by the JTT model. The abscissa axis represents nucleotide substitutions at the non-PBR among allele pairs on each branch of the ML tree. The substitutions were estimated by the HKY model. The solid line represents a linear regression. The broken line represents the 95% confidential interval for the regression coefficient.
Figure 5.
Phylogenetic relationships between HLA-DRB alleles as determined by the ML method on the basis of nucleotide sequences (690 bp) in the non-PBR. Only bootstrap values more than 80% are shown here. The values are shown by a bold character. Two HLA-DQB1 sequences are used as the out-group. Evolutionary distances were computed using the HKY model. Group A and group B indicate two major phylogenetic groups of HLA-DRB1 alleles. IMGT/HLA accession numbers are in parentheses. The numeric character shown in black on each branch of the tree represents nucleotide substitutions at the non-PBR among allele pairs. The numeric character shown in red on each branch of the tree represents amino acid substitutions estimated by the JTT model at the PBR among allele pairs. Branches and alleles with slow PBR amino acid substitutions are shown in green, whereas those with fast PBR substitutions are shown in pink.
Table 1. The frequency of alleles with fast or slow PBR substitution rates in phase I to IV.
| Phase I | Phase II | Phase III | Phase IV | |||||
|---|---|---|---|---|---|---|---|---|
| Fast | Slow | Fast | Slow | Fast | Slow | Fast | Slow | |
| Frequency of countsa, b | 0.10 | 0.01 | 0.01 | 0.44 | 0.15 | 0.06 | 0.47 | 0.04 |
| Countsa | 107 | 8 | 10 | 757 | 16 | 6 | 94 | 7 |
| No. of allele pairsa (×2)b | 537 (1074) | 851 (1702) | 53 (106) | 99 (198) | ||||
When both alleles in an allelic pair include the allelic lineage with fast or slow PBR substitution rates, the frequency or number is counted twice. When an allelic pair includes both allelic lineages with fast and slow PBR substitution rates, the frequency or number is not counted.
Denominator is twice the total number of allele pairs in each phase.
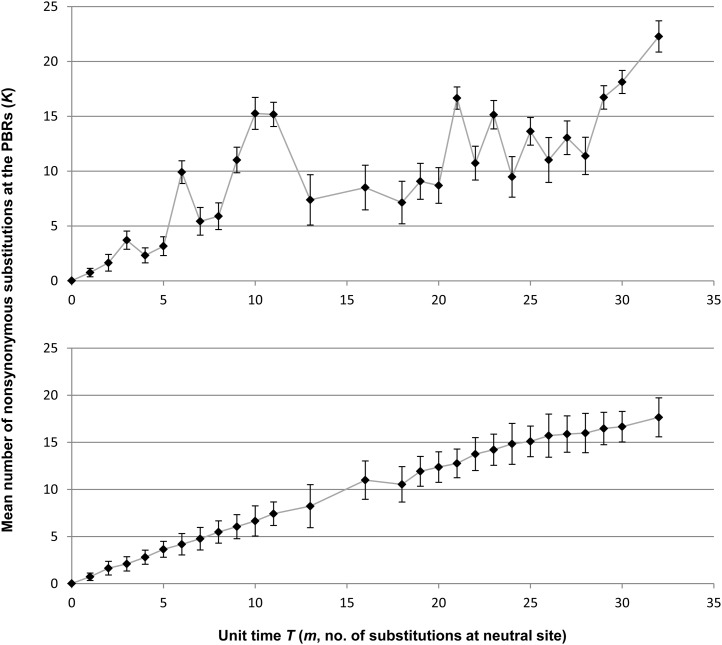
A computer simulation was performed to confirm whether the presence of such lineages with slow and fast substitution rates led to the nonlinearity of KB(m) accumulation using 56 fictitious alleles that had the same phylogenetic relationship as the non-PBR tree (for details of the simulation methods, see Materials and Methods). After the simulation was repeated 200 times, we examined the relationship between the divergence time of alleles and the number of substitutions in the 56 allele pairs (Figure 6). As expected, KB(m) values estimated in the computer simulation exhibited a similar pattern as the observation shown in Figure 2. We also performed a similar computer simulation (100 replications) without a reduction or enhancement of the PBR substitution rate. The simulation showed that KB(m) values proportionally increased with the divergence time among the alleles (m). These results suggest that PBR nonsynonymous substitutions of HLA-DRB1 alleles were decelerated or enhanced because of the heterogeneity of the PBR substitution rates among the allelic lineages.
Figure 6.
The K values of allele pairs that share the same coalescent time T. The ordinate axis represents the K value. The abscissa axis represents unit time T corresponding to the number of substitutions at neutral sites (m). The upper graph represents the accumulation of the mean number of nonsynonymous substitutions in the PBR among allele pairs (K) when PBR substitution rates of particular branches in a phylogenetic tree are slowed down or enhanced. The bottom graph represents the accumulation of K values without reduction or enhancement of PBR substitution rates. Error bars indicate the SD from the mean. The simulations for upper and bottom graphs were repeated 200 times and 100 times, respectively.
Discussion
The cessation of accumulation in amino acid variation at the PBR
Contrary to expectations, HLA loci showed a KB(m) decline in intermediate values of m. We estimated the m value on the basis of KS and KN in the analysis. It is possible that the KB(m) decline was caused by the heterogeneous evolutionary rate of KN. Although constant non-PBR nonsynonymous substitution rates in the HLA-B and HLA-C were not supported by the molecular clock test, class II loci showed molecular clock in non-PBR nonsynonymous substitutions. Therefore, the unequal substitution rate of KN values does not necessarily affect the KB(m) decline.
The symmetric balancing selection model assumes that all HLA alleles are sampled from a panmictic population. To investigate the possibility that the biased sampling of sequences may result in the KB(m) accumulation pattern, we examined the relationship between KB(m) and m using HLA-DRB1 alleles from a Japanese population, which is a putative panmictic population. The result showed that even if the DRB1 alleles from a putative panmictic population were used, the pattern of KB(m) decline in intermediate values of m was still observed (Figure S6). Therefore, KB(m) reduction was unlikely to be caused due to the inappropriate sampling of HLA alleles in the present study.
Although possible recombinant alleles were removed from the analyses of the present study by Satta’s method (Satta 1992), we further investigated for reciprocal recombination or gene conversion using the GENECONV program (Sawyer 1989) to dismiss the possibility that recombination caused KB(m) deceleration. Permutation test for the sequences used in this analysis indicated intragenic recombination between some pairs of alleles. However, the Bonferroni-corrected BLAST-like test showed no significant values. Even though possible recombinants (19 alleles) estimated by both of the permutation tests in GENECONV and Satta’s method were excluded from the examination of the relationship between KB(m) and m, KB(m) deceleration was still observed in 45 nonrecombinant alleles (Figure S7). Therefore, filtering of recombinants in the present study was unlikely to skew the KB(m) distribution.
Because KB(m) decline was also observed in the KN and KS comparison (Figure S8), it was interesting to know whether such KN saturation affected the KB(m) accumulation in proportion to KS + KN. This question was addressed in the following manner. We assumed that KB(m) values increase proportionately with KS, but that the KN value is somewhat saturated for intermediate KS values (Figure S9, A–C). Next, the KB(m) accumulation along with KS + KN was examined. We observed that even if KN did not increase constantly with KS, a linear relationship was observed between KB(m) and KS + KN (Figure S9D). This result suggests that KN saturation did not affect our results, although the reason for KN saturation against KS remains unknown.
Figure 2 shows that allele pairs with m = 14 to 18 were not observed. This was due to a lack of allelic pairs with intermediate values of KN (KN = 7 and 8; Figure S10A), whereas KS values are continuously observed (Figure S10B). The absence of certain allele pairs may be due to gaps in the database information. Many sequences obtained from the database provided only exon 2 sequences, and there is a clear need for submitting complete HLA-DRB1 coding sequences to the database in the future.
Phylogenetic relationships among HLA-DRB1 alleles
The phylogenetic tree for DRB alleles of primates suggested that the group A DRB1 alleles in humans formed a monophyletic group from other DRB alleles. However, the remaining HLA-DRB1 alleles were polyphyletic. HLA-DRB1*01 and *10 belonged to the same DR1 haplotype, but only those alleles were assigned to different groups (group A and group B in Figure 3). Andersson (1998) showed that the DR1 haplotype with HLA-DRB1*01 or HLA-DRB1*10 is likely derived from the DR51 haplotype (HLA-DRB1*15 and HLA-DRB1*16) in group B during recent primate evolution. The exon 2 sequence in HLA-DRB1*10 appears to be similar to that of HLA-DRB1*01 (Gongora et al. 1997). However, the configuration of repetitive elements in a segment encompassing the promoter region to upstream of exon 2 (including exon 1) showed higher similarity of HLA-DRB1*10 with that of HLA-DRB1*03 in DR52 haplotype in group A than with HLA-DRB1*01. The assignment of HLA-DRB1*10 to group A in the non-PBR tree may be affected by conversion of exon 1 and surrounding sequences from HLA-DRB1*03 to HLA-DRB1*10, although the exon 1 sequence was quite short. Because the bootstrap value on the node leading to the group A branch was not high (33%), the clustering of group A still remains a matter of debate.
The cause of stagnation of KB(m) accumulation
A comparison of the relationship between non-PBR and PBR genetic distances on each branch of the non-PBR ML tree showed three allelic lineages (HLA-DRB1*04, *15, and *16) with a slow PBR substitution rate and two lineages (HLA-DRB1*03 and *07) with a fast rate. The allelic lineages with the slow substitution rate were frequently included in phase II. This result suggests that a decline of the PBR substitution rate likely caused the stagnation of KB(m) accumulation. Interestingly, although the PBR substitution rate was reduced, most dN/dS values of those branches were higher than unity, suggesting that balancing selection is still active.
The computer simulation supported the finding that the slow-down of amino acid substitution rate in the PBR caused the stagnation of KB(m) accumulation. The duration of the tentative plateaus was slightly different between the simulation and that observed. The extent of rate acceleration or deceleration was arbitrary in this simulation and branches with fast or slow rates may have been underestimated due to the strict criteria for heterogeneity identification (see Materials and Methods). This probably caused the difference in the timing of plateaus.
In the symmetric balancing selection model, nonsynonymous substitutions at the PBR sites proportionally increased with divergence time and eventually reached a plateau (Takahata et al. 1992) (Figure S1). However, in this study KB(m) values did not constantly increase with the divergence time of alleles. The theory of asymmetric balancing selection may explain this phenomenon because the fitness of heterozygotes or homozygotes is not equivalent as per this theory. However, asymmetric balancing selection does not fit the model of polymorphism for the actual data in the simulation (Takahata and Nei 1990). Thus, asymmetric balancing selection could not explain the skewed KB(m) distribution in the present study.
We used PBR sites identified by Brown et al. (1993) to estimate KB(m) values. When PBR sites identified by recent assays for higher-order structures of the HLA molecule (S. Kusano and S. Yokoyama, personal communication) were used, the KB saturation pattern was also observed in phase II. In addition, allele pairs with low PBR substitution rates were also assigned to phase II. The PBR definitions did not affect our results.
Biological significance of substitution rate heterogeneity
Slow nucleotide substitution rates, in particular allelic lineages (HLA-DRB1*04, *15, and *16), may be necessary to retain the binding affinity for specific peptides. In general, evolutionary rates of hosts and pathogens have been accelerated by virtue of their arms race. However, HLA molecules preferentially target evolutionarily conserved regions that are functionally important sites for pathogens. Among HLA alleles there appears to be a difference of correlation between the peptide-binding affinity and conservation of the targeted regions (Hertz et al. 2011). If this is true, then these slower lineages must be highly specific to certain pathogens, which are the source of these specific peptides.
Using the IEDB database, we examined peptides bound by DRB1 allelic lineage with the slow PBR substitution rate (HLA-DRB1*04, *15, and *16) and pathogens from which the peptides were derived (Table S3). The prediction from the database showed that one pathogen was specifically recognized by the HLA molecule with a fast PBR substitutions rate, whereas 12 pathogens were specifically recognized by HLA molecules with a slow PBR substitution rate. These more slowly evolving HLA molecules recognized viruses and bacteria that cause disease specific to humans. In addition, HLA-DRB1*04:01 and HLA-DRB1*04:07 bound peptides from lactic bacteria, which are also specific to humans. To cope with these specific pathogens, the nucleotide substitution rate at particular DRB1 alleles had possibly slowed down relative to those at other alleles. One may hypothesize that evolutionary rates of amino acids at sites 9, 11, 13, and 37 in the HLA-DRB1*04 lineage and at sites 11 and 13 in the DRB1*15/*16 lineages were decelerated, respectively, to retain the affinity of the conserved proteomic region in pathogens (green branches in Figure 5 and Figure S4). According to our estimate, the divergence time of allelic pairs in phase II is 18 MYA to 24 MYA. Humans and the pathogens described above might have coexisted for long periods of time, at least before speciation events of the subfamily Homininae.
In summary, we observed the episodic fluctuations of the amino acid substitution rate in the PBR over the course of HLA evolution. This observation was not anticipated because we believed that nonsynonymous substitutions in the PBR increased with the divergence time of the alleles until the substitutions reached saturation. In the case of the DRB1 gene, such fluctuation is probably due to a decrease of amino acid substitution rates at the PBR on the stem of particular allelic lineages. This suggests that a part of the peptide-binding repertoire at the HLA-DRB1 locus has been limited over long periods of time due to the recognition of certain pathogens.
Supplementary Material
Acknowledgments
We give special thanks to Dr. Naoyuki Takahata for providing valuable comments, and to Dr. Seisuke Kusano, Dr. Shigeyuki Yokoyama, and the Institute of Physical and Chemical Research (RIKEN). We thank Dr. John A. Eimes for the critical checking of the English language of this manuscript. This work was supported by a grant-in-aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (22133007).
Footnotes
Communicating editor: S. W. Scherer
Literature Cited
- Andersson G., 1998. Evolution of the human HLA-DR region. Front. Biosci. 3: d739–d745. [DOI] [PubMed] [Google Scholar]
- Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., et al. , 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364: 33–39. [DOI] [PubMed] [Google Scholar]
- Carrington M., Nelson G. W., Martin M. P., Kissner T., Vlahov D., et al. , 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283: 1748–1752. [DOI] [PubMed] [Google Scholar]
- Demotz S., Barbey C., Corradin G., Amoroso A., Lanzavecchia A., 1993. The set of naturally processed peptides displayed by DR molecules is tuned by polymorphism of residue 86. Eur. J. Immunol. 23: 425–432. [DOI] [PubMed] [Google Scholar]
- Doherty P. C., Zinkernagel R. M., 1975. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256: 50–52. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 2009. PHYLIP (Phylogeny Inference Package) ver.3.69. Distributed by the author, Department of Genome Sciences, University of Washington, Seattle, USA. [Google Scholar]
- Gongora R., Figueroa F., Klein J., 1997. Complex origin of the HLA-DR10 haplotype. J. Immunol. 159: 6044–6051. [PubMed] [Google Scholar]
- Gonzalez-Galarza F. F., Christmas S., Middleton D., Jones A. R., 2011. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 39: D913–D919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen P. K., Shen M., Song Q. L., Merryman P., Degar S., et al. , 1986. Molecular diversity of HLA-DR4 haplotypes. Proc. Natl. Acad. Sci. USA 83: 2642–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T., 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hertz T., Nolan D., James I., John M., Gaudieri S., et al. , 2011. Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J. Virol. 85: 1310–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A. V. S., 1991. HLA associations with malaria in Africa: some implications for MHC evolution, pp. 403–419 in Molecular evolution of the major histocompatibility complex, edited by Klein J., Klein D. Springer, Berlin, Heidelberg. [Google Scholar]
- Hughes A. L., Nei M., 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335: 167–170. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M., 1989. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc. Natl. Acad. Sci. USA 86: 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Yeager M., 1998. Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 32: 415–435. [DOI] [PubMed] [Google Scholar]
- Jones D., Taylor W., Thornton J., 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Klein J., Sato A., Nikolaidis N., 2007. MHC, TSP, and the origin of species: from immunogenetics to evolutionary genetics. Annu. Rev. Genet. 41: 281–304. [DOI] [PubMed] [Google Scholar]
- Kusaba M., Nishio T., Satta Y., Hinata K., Ockendon D., 1997. Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: implications for the evolution and recognition mechanism. Proc. Natl. Acad. Sci. USA 94: 7673–7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musolf K., Meyer-Lucht Y., Sommer S., 2004. Evolution of MHC-DRB class II polymorphism in the genus Apodemus and a comparison of DRB sequences within the family Muridae (Mammalia: Rodentia). Immunogenetics 56: 420–426. [DOI] [PubMed] [Google Scholar]
- Nowak M., Tarczy-Hornoch K., Austyn J., 1992. The optimal number of major histocompatibility complex molecules in an individual. Proc. Natl. Acad. Sci. USA 89: 10896–10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver M. K., Telfer S., Piertney S. B., 2009. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc. Biol. Sci. 276: 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong B., Willcox N., Wordsworth P., Beeson D., Vincent A., et al. , 1991. Critical role for the Val/Gly86 HLA-DRβ dimorphism in autoantigen presentation to human T cells. Proc. Natl. Acad. Sci. USA 88: 7343–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D. J., Damjanovich K., Potts W. K., 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl. Acad. Sci. USA 99: 11260–11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche P. A., Reinherz E. L., 2003. Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 331: 623–641. [DOI] [PubMed] [Google Scholar]
- Robinson J., Waller M. J., Stoehr P., Marsh S. G. E., 2005. IPD-the Immuno Polymorphism Database. Nucleic Acids Res. 33: D523–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J., Mistry K., McWilliam H., Lopez R., Parham P., et al. , 2011. The IMGT/HLA database. Nucleic Acids Res. 39: D1171–D1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Satta, Y., 1992 Balancing selection at HLA loci, pp. 111–131 in The Proceedings of the 17th Taniguchi Symposium, edited by N. Takahata. Japan Science Society Press, Tokyo.
- Satta Y., C. O’hUigin, N. Takahata, and J. Klein., 1994. Intensity of natural selection at the major histocompatibility complex loci. Proc. Natl. Acad. Sci. USA 91: 7184–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526–538. [DOI] [PubMed] [Google Scholar]
- Slade R. W., McCallum H. I., 1992. Overdominant vs. frequency-dependent selection at MHC loci. Genetics 132: 861–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S., 2005. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin L. G., Richardson D. S., 2010. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. Biol. Sci. 277: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L. J., Brown J. H., Jardetzky T. S., Gorga J. C., Urban R. G., et al. , 1994. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368: 215–221. [DOI] [PubMed] [Google Scholar]
- Takahata N., 1990. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl. Acad. Sci. USA 87: 2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Nei M., 1990. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Tajima F., 1991. Sampling Errors in Phylogeny. 8: 494–502. [Google Scholar]
- Takahata N., Satta Y., Klein J., 1992. Polymorphism and balancing selection at major histocompatibility complex loci. Genetics 130: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursz M. R., Thomas H. C., Greenwood B. M., Hill A. V., 1997. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat. Genet. 17: 11–12. [DOI] [PubMed] [Google Scholar]
- Verreck F. A., van de Poel A., Drijfhout J. W., Amons R., Coligan J. E., et al. , 1996. Natural peptides isolated from Gly86/Val86-containing variants of HLA-DR1, -DR11, -DR13, and -DR52. Immunogenetics 43: 392–397. [DOI] [PubMed] [Google Scholar]
- Vidović D., Matzinger P., 1988. Unresponsiveness to a foreign antigen can be caused by self-tolerance. Nature 336: 222–225. [DOI] [PubMed] [Google Scholar]
- Vita R., Zarebski L., Greenbaum J. A., Emami H., Hoof I., et al. , 2010. The immune epitope database 2.0. Nucleic Acids Res. 38: D854–D862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfing B., Traulsen A., Milinski M., Boehm T., 2009. Does intra-individual major histocompatibility complex diversity keep a golden mean? Philos. Trans. R. Soc. Lond. B Biol. Sci. 364: 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.