Abstract
Mini-chromosome maintenance (MCM) proteins form complexes that are required for DNA replication and are highly conserved throughout evolution. The replicative helicase of eukaryotic organisms is composed of the six paralogs MCM2-7, which form a heterohexameric ring structure. In contrast, the structure of the archaean replicative MCM helicase is a single Mcm protein that forms a homohexameric complex. Atomic structures of archaeal MCMs have identified multiple beta-finger structures in Mcm proteins whose in vivo function is unknown. In the present study, we have investigated the physiological role of the pre-sensor 1 beta-hairpin (PS1-hp) beta-fingers of Saccharomyces cerevisiae Mcm4p and Mcm5p in DNA replication initiation and elongation in vivo. The PS1-hp beta-finger mutant of Mcm5p (mcm5-HAT K506A::URA3) has a growth defect at both 18° and 37°. Mutation of the Mcm4p PS1-hp beta-finger (mcm4-HA K658A::TRP1) does not have a growth defect, indicating different functional contributions of the PS1-hp beta-finger structures of different MCM helicase subunits. Both Mcm4p and Mcm5p PS1-hp beta-finger mutants can coimmunoprecipitate Mcm2p, indicating the formation of the hexameric MCM helicase complex. Both PS1-hp beta-finger mutants have a plasmid loss phenotype that is suppressible by origin dosage, indicating a defective replication initiation. Surprisingly, a defect in the binding of PS1-hp MCM mutants to origins of DNA replication was not found by chromatin immunoprecipitation, suggesting a novel interpretation in which the defect is in a subsequent step of DNA strand separation by the MCM helicase. The double mutant mcm4-HA K658A::TRP1mcm5-HAT K506A::URA3 is lethal, displaying a terminal MCM mutant phenotype of large budded cells.
Keywords: replication, helicase, S phase, cell cycle, yeast
DNA replication in eukaryotes is a tightly controlled process. Failure of DNA replication control mechanisms leads to chromosomal instability, mutations, and aneuploidy, which are hallmarks of human disease, including birth defects, aging, and cancer (Jackson et al. 2014). The exact duplication of the genome is an essential cellular process. DNA replication control mechanisms ensure that genomic information is replicated exactly once per cell cycle (Sclafani and Holzen 2007). DNA replication begins at DNA sequences termed “origins.” In higher eukaryotes, origins have no defined DNA sequence; however, in Saccharomyces cerevisiae, origins of DNA replication have a 14-bp consensus site termed the autonomous replicating sequence (ARS) (Leonard and Mechali 2013; Sclafani and Holzen 2007). Origins are continually bound throughout the cell cycle by the origin recognition complex (ORC), which is a six-subunit ATP-dependent protein complex encoded in yeast by ORC1-6 (Bell and Dutta 2002; Leonard and Mechali 2013; Sclafani and Holzen 2007).
A two-step process regulates DNA replication. The first step is termed “licensing of the origin,” characterized by the formation of the prereplicative complex (Labib 2010). Licensing consists of loading the mini-chromosome maintenance (MCM) helicase during the G1 phase of the cell cycle onto double-stranded DNA (dsDNA) by Cdt1p and Cdc6p in yeast (Frigola et al. 2013; Labib 2010). The MCM helicase is composed of six paralogs MCM2-7 (Forsburg 2004) and is loaded as a double hexamer (Bell and Botchan 2013; Frigola et al. 2013; Gambus et al. 2011). Mcm2-7p is thought to be the replicative helicase responsible for DNA stand separation (Bell and Botchan 2013; Forsburg 2004; Tye 1999). The inactive MCM helicase loaded onto the origin of replication along with ORC forms the prereplicative complex. In the second step, the origin of DNA replication is activated in S phase by cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK) (Sclafani and Holzen 2007).
DDK, which is composed of the catalytic Cdc7p and regulatory Dbf4p subunits, phosphorylates the N-terminal tails of Mcm 2/4/6. Phosphorylations are thought to induce structural changes that activate the MCM helicase (Hoang et al. 2007; Sclafani and Holzen 2007). It has also been shown that S-phase CDK (S-CDK) phosphorylates the MCM helicase; however, the essential targets of S-CDK are Sld2p and Sld3p (Randell et al. 2010; Tanaka et al. 2007; Zegerman and Diffley 2007) Phosphorylation of Sld2p and Sld3p allows for the binding of Dpb11p, which leads to the stable association of Cdc45p and the recruitment of the GINs complex and DNA polymerases, activating the origin of DNA replication (Labib 2010). The formation of Cdc45p, GINs, and Mcm2-7p complex is known as the CMG complex and is thought to be the active form of the replicative helicase (Bell and Botchan 2013). The DNA replication origins are activated in a temporal fashion during S phase, with some origins activating (firing) early in S phase, whereas others fire in the middle and later parts of S phase (McCune et al. 2008). Temporal control of origin activation is thought to be controlled by DNA replication factor binding mediated by the kinase activities of CDK and DDK.
MCM Helicase Structure and Function
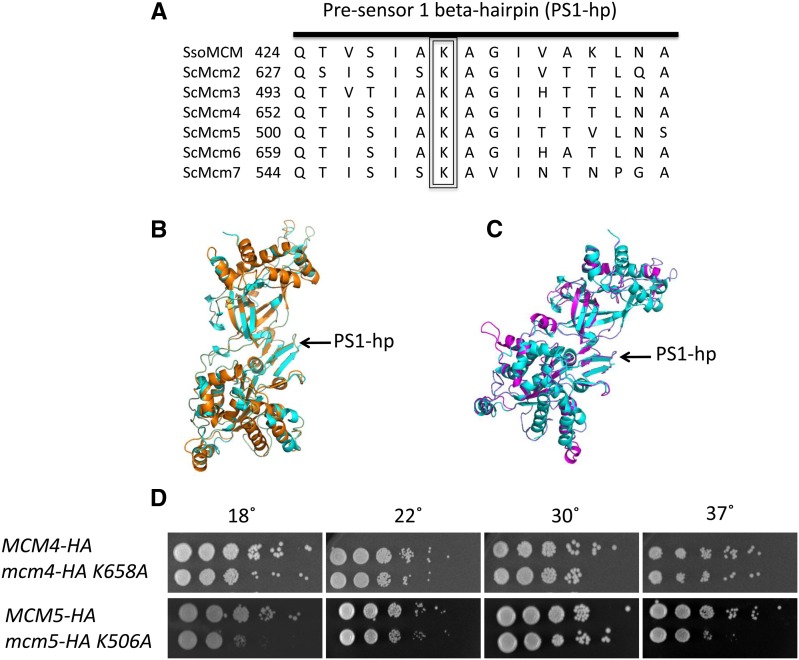
MCM genes are found in both eukaryotes and archaea (Bochman and Schwacha 2009). The eukaryotic MCM helicase is composed of six paralogs MCM 2-7 that belong to the highly conserved AAA+ ATPase family (Bochman and Schwacha 2009). Mcm2-7p are thought to have diverged from a common ancestor and form a hexameric ring structure with a 1:1:1:1:1:1 stoichiometry (Brewster et al. 2008). All Mcm2-7 proteins are required for DNA replication and are essential for viability (Bochman and Schwacha 2009). A single gene encodes the archeal MCMs in Sulfolobus solfataricus (ssoMCM) and Methanothermobacter thermautotrophicus (mtMCM) (Brewster et al. 2008; Fletcher et al. 2003). Analysis of the structures of the N-terminal domain of mtMCM protein and the near full-length structure of ssoMCM protein has revealed interesting beta-hairpin structures of each subunit that are located in the central channel or side channels in the MCM hexamer (Brewster et al. 2008; Fletcher et al. 2003). Previous work from our laboratory has demonstrated the importance and functional conservation of the N-terminal beta finger in Saccharomyces cerevisiae. The N-terminal beta finger of Mcm5p is important for the MCM complex to bind to origins of DNA replication (Leon et al. 2008). The pre-sensor 1 beta-hairpin (PS1-hp) of MCM complexes is highly conserved (Figure 1) and a mutation in the PS1-hp of ssoMCMp greatly decreases helicase activity on both a 3′-tailed substrate and a Y-shaped substrate in vitro (McGeoch et al. 2005). The PS1-hp is thought to have nucleotide dependent movement that promotes helicase activity (Bell and Botchan 2013; Slaymaker and Chen 2012). The PS1-hp is conserved in the distantly related Superfamily 3 helicases, which includes SV40 T antigen, and has been shown to undergo significant movement during nucleotide binding, hydrolysis, and release cycle of the helicase (Bell and Botchan 2013; Gai et al. 2004; Slaymaker and Chen 2012). In this current study, we present a molecular genetic analysis of mutations of the highly conserved lysine residue of the PS1-hp in Saccharomyces cerevisiae Mcm4p and Mcm5p (Figure 1).
Figure 1.
Pre-sensor 1 beta-hairpin structure and growth of mutants. (A) Conservation of the PS1-hp amino acids between Sulfolobus solfataricus Mcm and Saccharomyces cerevisiae Mcm2-7p. (B) Structure prediction of scMcm4p using Phyre software using the ssoMcm structure as a template. Alignment of the ssoMcm(cyan) and scMcm4p (orange) using pymol software, PS1-hp is indicated with an arrow. (C) Structure prediction of scMcm5p using Phyre software using the ssoMcm structure as a template. Alignment of the ssoMcm(cyan) and scMcm5p (magenta) using pymol software, PS1-hp indicated with an arrow. (D) Serial dilution analysis of growth indicates no growth defect of the Mcm4p PS1-hp mutant compared with wild type (Mcm4p-HA). Yeast with the PS1-hp mutation in Mcm5p (mcm5-K506A::URA3) has a cold sensitivity phenotype at 18° and a temperature sensitivity phenotype at 37° as indicated by reduced growth compared with the wild type (Mcm5p-HA).
Materials and Methods
Yeast strains, media, and plasmids
Yeast strains are listed in Table 1 and plasmids are listed in Table 2. Yeast strains were grown using standard procedures as well as methods published previously (Leon et al. 2008). To produce integrating plasmids carrying mcm4-HA K506A and mcm5-HA K506A, Quickchange mutagenesis (Stratagene) was used on plasmid pRS663 (MCM4-HA) and pRPL104 (MCM5-HAT) and mutations were marked by the addition of restriction sites PstI (MCM4-HA) and PvuII (MCM5-HAT). All mutations were verified by polymerase chain reaction (PCR), restriction digest, and by sequencing. Yeast strains were constructed using a plasmid shuffle strategy using a covering wild-type (WT) gene on a plasmid. Plasmids carrying either MCM4-HA (pRS663) or mcm4-HA K658A (pCJR101) were digested with NruI and integrated at the endogenous MCM4 locus, creating a partial duplication with the active copy containing a hemagglutinin (HA) tag or HA tag and the PS1-hp mutation. MCM5-HA or mcm5-HA K506A was constructed using pRPL104 (MCM5-HAT) or pRS734 (mcm5-K506A-HAT) digested with PpuMI to integrate at the ura3-1 locus. MCM5 at the endogenous locus was deleted with KanMX4 and covered with a WT MCM5 plasmid (pRS660) that was shuffled with the tagged version and the tagged PS1-hp mutant version to test for viability.
Table 1. S. cerevisiae strains used.
| Yeast Strain | Genotype | Source |
|---|---|---|
| RSY311 | MATa trp1 leu2 ura3 can1 his6 bar1 | (Leon et al. 2008) |
| CRY106 | MATa trp1 leu2 ura3 can1 his6 bar1 MCM4-HA::TRP1 | This study |
| CRY107 | MATa trp1 leu2 ura3 can1 his6 bar1 mcm4-HA K658A::TRP1 | This study |
| CRY109 | MATα trp1 leu2 ura3 can1 his6 bar1 mcm4-HA K658A::TRP1 | This study |
| CRY111 | MATa trp1 leu2 ura3 can1 his6 bar1 MCM4-HA::TRP1[ pDK243] | This study |
| CRY112 | MATa trp1 leu2 ura3 can1 his6 bar1 MCM4-HA::TRP1 [pDK368-7] | This study |
| CRY113 | MATa trp1 leu2 ura3 can1 his6 bar1 mcm4-HA K658A::TRP1[ pDK243] | This study |
| CRY114 | MATa trp1 leu2 ura3 can1 his6 bar1 mcm4-HA K658A::TRP1[ pDK368-7] | This study |
| CRY119 | MATα trp1 leu2 ura3 can1 his6 bar1 mcm4-HA K658A::TRP1 | This study |
| CRY175 | MATa ura3::MCM5-HAT URA3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 [pDK243] | This study |
| CRY176 | MATa ura3::MCM5-HAT URA3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 [pDK368-7] | This study |
| CRY177 | MATa ura3::mcm5-K506A-HAT URA3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 [pDK243] | This study |
| CRY178 | MATa ura3::mcm5-K506A-HAT URA3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 [pDK368-7] | This study |
| CRY210 | MATa ura3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 [pRS660 MCM5::LEU2] | This study |
| CRY211 | MATa ura3::mcm5-K506A-HAT URA3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 [pRS660 MCM5::LEU2] | This study |
| CRY212 | MATa ura3::mcm5-K506A-HAT URA3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 mcm4-K658A-HA::TRP1 [pRS660 MCM5::LEU2] | This study |
| RSY1259 | MATa ura3 tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 [ pRS414 MCM5::TRP1] | (Leon et al. 2008) |
| RSY1336 | MATa ura3::URA3 MCM5-HAT tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 bar1::LEU2 | (Leon et al. 2008) |
| RSY1345 | MATa ura3::URA3 mcm5-K506A-HAT tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 bar1::LEU2 | (Leon et al. 2008) |
| RSY1148 | MATa trp1 leu2 ura3 can1 his6 bar1 mcm5-461 | (Leon et al. 2008) |
| CRY205 | MATa trp1 leu2 ura3::URA3 MCM5-HAT can1 his6 bar1 mcm5-461 | This study |
| CRY206 | MATa trp1 leu2 ura3::URA3 mcm5-K506A-HAT can1 his6 bar1 mcm5-461 | This study |
| CRY207 | MATa trp1 leu2 ura3::URA3 MCM5-HAT can1 his6 bar1 mcm5-461 mcm4-K658A-HA::TRP1 | This study |
| CRY208 | MATa trp1 leu2 ura3::URA3 mcm5-K506A-HAT can1 his6 bar1 mcm5-461 MCM4-HA::TRP1 | This study |
| CRY209 | MATa trp1 leu2 ura3::URA3 mcm5-K506A-HAT can1 his6 bar1 mcm5-461 mcm4-K658A-HA::TRP1 | This study |
| CRY213 | MATa ura3::URA3 MCM5-HAT tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 | This study |
| CRY214 | MATa ura3::URA3 mcm5-K506A-HAT tyr1 leu2 ade1 ade2 mcm5∆::kanMX4 trp1 cyh2 | This study |
Table 2. Recombinant plasmids used.
| Plasmid Name | Genotype | Source |
|---|---|---|
| pRAS660 | pRS305 ARS CEN LEU2 MCM5 | (Leon et al. 2008) |
| pRAS662 | pRS305 ARS CEN LEU2 MCM4 | (Leon et al. 2008) |
| pRPL104 | pRS306 ARS CEN URA3 MCM5-HAT | (Leon et al. 2008) |
| pDK243 | ARS(1x) CEN LEU2 | (Hogan and Koshland 1992) |
| pDK368-7 | ARS(8X) CEN LEU2 | (Hogan and Koshland 1992) |
| pRAS663 | pRS404 mcm4-HA | (Aparicio et al. 1999) |
| pCJR101 | pRS404 mcm4-K658A-HA | This study |
| pRAS734 | pRS306 mcm5-K506A-HAT | This study |
Plasmid stability assay
To generate the strains used for plasmid loss assays, CRY106, CRY107, CRY213, and CRY214 were transformed with either pDK243 LEU2 (1X-ARS site) or pDK368-7 LEU2 (8X-ARS sites), and Leu+ transformants were selected generating strains. WT and PS1-hp mutant strains containing either pDK243 LEU2 (1X-ARS site) or pDK368-7 (8X-ARS sites) with a LEU2 marker were grown in synthetic complete media–leucine to stationary phase, then plated onto YEPD and synthetic complete media–leucine to determine plasmid stability under selection. Stationary cultures were then diluted to 2 × 103 cells/mL and grown in synthetic complete media + leucine (relaxed conditions) and allowed to grow to stationary phase. Stationary cultures were then plated onto yeast extract peptone dextrose and synthetic complete media – leucine to determine plasmid stability under relaxed selection. The plasmid loss rate was determined by the observed loss of the plasmid on synthetic complete–leucine plates compared with yeast extract peptone dextrose plates under relaxed selection as described previously (Hogan and Koshland 1992; Leon et al. 2008). At least three independent loss rates were calculated for each condition.
Co-immunoprecipitation (IP) and western blot analysis:
Protein extracts were prepared as described previously (Leon et al. 2008). In summary, 2 mg of protein extract were incubated with 30 μL of protein G-Sepharose beads (Sigma-Aldrich) blocked in lysis buffer supplemented with complete protease inhibitor tablets from Roche (cat. no. 05892970001) for 1 hr at 4°. IP was performed using 50 μg of anti-HA antibody (Roche). Negative controls were performed without antibody, containing only protein G-sepharose beads, and all reactions were incubated with end-over-end rotation at 4° for 3 hr. Samples were then centrifuged at 240g at 4° and the supernatant was collected and mixed with 5X sodium dodecyl sulfate (SDS) buffer to a final of 1X and boiled for 5 min. Pellets were washed three times in 500 μL of lysis buffer containing complete protease inhibitors tablets from Roche (cat. no. 05892970001) and were then resuspended in 50 μL of 2.5X SDS buffer. Western blot analysis used 10% of each fraction, whole-cell extract, supernatant, and pellet. Samples of IPs were resolved on a 7% SDS-polyacrylamide gel electrophoresis gel, transferred to a nitrocellulose membrane, and probed with anti-HA antibody (Roche) at a 1:2500 dilution, anti-Mcm2p antibody (Santa Cruz) at a 1:1000 dilution, and anti-Cdt1p (UM185) at a 1:15000 dilution. For HA blots, a secondary antimouse antibody conjugated to horseradish peroxidase (HRP; Jackson ImmunoResearch) was used at 1:3000 dilution. For Mcm2p blots, a secondary antigoat antibody conjugated to HRP (Santa Cruz Biotechnology) was used at a 1:3000 dilution. For Cdt1p blots, a secondary antirabbit antibody conjugated to HRP (Bio-Rad) was used at a 1:3000 dilution. Immunoblots were visualized on film or using a ChemiDoc MP scanner (Bio-Rad) employing an ECL chemiluminescence kit (Millipore).
Chromatin immunoprecipitation (ChIP) analysis:
ChIP analysis was performed as previously described (Adkins et al. 2004) with the following modifications. Forty-five milliliters of alpha factor G1 arrested yeast cultures (2 × 107 cells/mL) were fixed with formaldehyde for 1 hr at room temperature. Cells were lysed using a mini bead buster (Biospec) for 3X 2-min intervals in ChIP lysis buffer 1 [50 mM HEPES, pH 7.5; 140 mM NaCl; 1 mM ethylenediaminetetraacetic acid (EDTA); 1% Triton X-100; 0.1% deoxychloic acid; complete protease inhibitor tablet (Roche cat nol. 05892970001)]. Cell lysis was sonicated using a Bioruptor (Diagenode) for 30 cycles of 30 sec on/off at 4°. Cell debris was removed with two 20,000g spins for 5 min. The chromatin fraction was precleared with the addition of 10 μL of Protein G sepharose beads (Sigma-Aldrich) with end-over-end rotation for 1 hr followed by centrifugation at 880g for 1 min. Then, 50 μL of cleared lysate was taken as an input sample with the addition of 30 μL of ChIP elution buffer (50 mM Tris-HCl, pH 7.5; 10mM EDTA; 1% SDS). IPs were set up with 350 μL of chromatin solution per IP reaction. For HA IPs, the chromatin fraction was incubated with 25 μg of ant-HA antibody (Roche). All IPs were incubated for 15 hr at 4°. Protein G sepharose beads (30 μL) were added to each reaction for 1 hr at 4°. Beads were washed 2X with ChIP lysis buffer 1 (50 mM HEPES, pH 7.5; 140mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% deoxycholic acid), twice with lysis buffer 2 (50 mM HEPES, pH 7.5; 500 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% deoxycholic acid), twice with lysis buffer 3 (10 mM Tris, pH 7.9; 250 mM LiCl; 1 mM EDTA; 0.5% NP-50; 0.5% deoxycholic acid), and once with TE. Beads were resuspended in 80 μL of ChIP elution buffer with 20 μL of pronase at 20 mg/mL (Calbiochem), incubated at 42° for 2 hr, followed by 8 hr at 65° to reverse the crosslinks. Chromatin-associated DNA isolated by ChIP was subjected to quantitative PCR, using 2X Sybergreen master mix from (Roche) and a Roche Lightcycler 480 thermocycler. Chromatin associated DNA was subjected to PCR analysis with primers to the following DNA replication origins: ARS1, ARS305, ARS306, ARS603, ARS607, and ARS1412.
Determining whether Mcm4p Mcm5p PS1-hp double mutant requires a WT Mcm5p encoding plasmid for viability
Yeast strain CRY214 (mcm5-HA K506A::URA3) was transformed with pRS660 (WT MCM5). Next, mcm4-HA K658A::TRP was created by integration and partial duplication at the MCM4 locus. To test whether the viability of the Mcm4p Mcm5p PS1-hp double mutants requires the WT Mcm5p encoding plasmid, the loss rate of the WT MCM5 plasmid under relaxed conditions was compared between the double PS1-hp mutant and the single mutant mcm5-HA K658A::URA3.
Lethality of the Mcm4p Mcm5p PS1-hp double mutant by tetrad analysis:
CRY119 (MATα mcm4-HA K658A::TRP1) was mated to RSY1345 (MATa mcm5-HA K506A::URA3 integrated at ura3-1 mcm5::KanMX4) (Table 1). Diploids were selected on synthetic complete –TRP –URA plates and sporulated in 3% potassium acetate. Ninety-one tetrads were dissected, and spores were germinated on YPD plates at 30°. Spores that were Ura+ G418R were tested for the Trp+ phenotype, indicating double mutant (mcm4-HA K658A::TRP1mcm5-HA K658A::URA3mcm5::KanMX4) viability. The terminal phenotype was determined by photographing spores know to be a double PS1-hp mutants.
Results
PS1-hp mutations of Mcm4 and Mcm5 are viable
The PS1-hp beta-finger structure is conserved in both Mcm4 and Mcm5 proteins of yeast by both primary sequence and structural alignment (Figure 1, A−C). Importantly, a conserved lysine is present at the tip of the beta-finger in both proteins. Yeast strains with integrated copies of the lysine (K) to alanine (A) PS1-hp mutations in Mcm5p and Mcm4p were constructed to produce mcm4 K658A::TRP1 and mcm5 K506A::URA3 strains CRY107 and RSY1345, respectively, as described in the Materials and Methods. These strains have a PS1-hp mutant mcm4 or mcm5 gene as the only copy of the gene in the cell, under the control of its native promoter. Because both temperature-sensitive and cold-sensitive mcm mutants have been described previously (Tye 1999), we tested for growth at a range of temperatures. Ten-fold serial-dilution analysis of the mcm5 K506A::URA3 strain indicates both a temperature-sensitive phenotype and a cold-sensitive phenotype, with reduced growth compared with the MCM4 or MCM5 wt strain at 37° and 18° (Figure 1D). In contrast, the corresponding PS1-hp mutation on Mcm4p (mcm4 K658A::TRP1) does not display a temperature-sensitive or cold-sensitive phenotype (Figure 1D). To test for sensitivity of PS1-hp mutants to replication stress, 10-fold serial-dilution analysis of growth was performed on plates containing different amounts of hydroxyurea, but no hydroxyurea sensitivity was found (Supporting Information, Figure S2) indicating that the checkpoint is intact in PS1-hp mutants. Introduction of the PS1-hp mutation into Mcm4p or Mcm5p does not have an effect on protein stability and both mutant proteins are expressed from their own promoters (Figure S1).
The Mcm4p and Mcm5p PS1-hp mutant proteins are incorporated into the Mcm2-7p complex
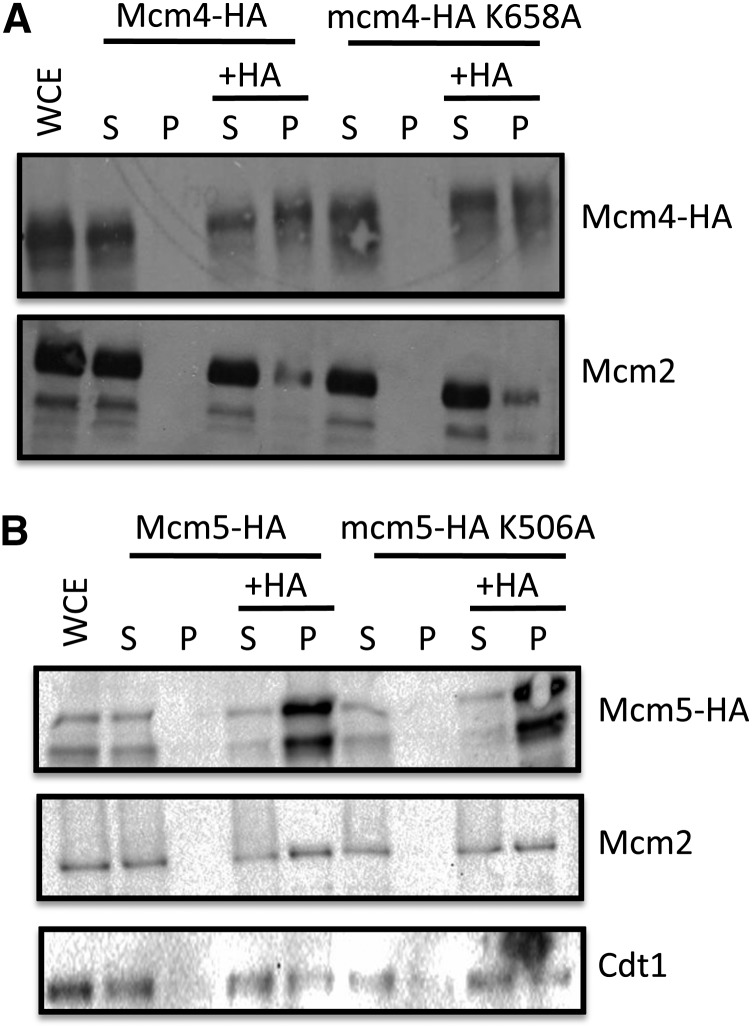
To demonstrate that the PS1-hp mutants get incorporated into the Mcm2-7p hexamer, both WT and PS1-hp mutants of both Mcm4p and Mcm5p were tagged with an HA epitope. An anti-HA antibody was used to precipitate each of the following: Mcm4p-HA, mcm4p-HA K658A, Mcm5p-HA, and mcm5p-HA K658A. Immunoprecipitates were run on 7.5% SDS-polyacrylamide gel electrophoresis gels, transferred to nitrocellulose, and probed for Mcm2p using an anti-Mcm2p antibody to demonstrate Mcm2-7p complex formation. Mcm2p binds directly to Mcm5p and Mcm6p and only binds indirectly with Mcm4p in the hexameric MCM ring. Immunoprecipitates from both Mcm4p-HA and mcm4p-HA K658A strains contain similar amounts of Mcm2p, indicating that the PS1-hp of Mcm4p forms the hexameric MCM complex in vivo (Figure 2A). Similar to Mcm4p, both Mcm5p-HA and mcm5p-HA K506A immunoprecipitates contain equal amounts of Mcm2p, demonstrating Mcm5p binds Mcm2p in the Mcm5p PS1-hp mutant (Figure 2B.). In addition to Mcm2p, mcm5p-HA K506A co-immunoprecipitates with Cdt1p at WT levels (Figure 2B), indicating that Mcm5p PS1-hp can interact with Cdt1p, which is important for MCM helicase loading at origins of DNA replication (Tanaka and Diffley 2002). Because Cdt1p is known to interact with the hexameric MCM helicase, these data indicate Mcm5p PS1-hp mutant can form the MCM complex. These results are consistent with results found in vitro when analyzing the MCM complex formation of the PS1-hp mutants of SsoMCM (McGeoch et al. 2005).
Figure 2.
Co-immunoprecipitation of the Mcm4p and Mcm5p PS1-hp mutants. (A) Immunoprecipitations of Mcm4p-HA, mcm4p-HA K658A, Mcm5p-HA, and mcm5p-HA K506A were performed using an anti-HA antibody or a no antibody negative control. Western blots were probed with anti-HA antibody for Mcm5p, an anti-Mcm2p antibody, and an anti-Ctd1p antibody. Supernatant and pellet fractions are represented by “S” and “P,” respectively. HA, hemagglutinin.
Plasmid loss phenotype of Mcm4p and Mcm5p PS1-hp mutants
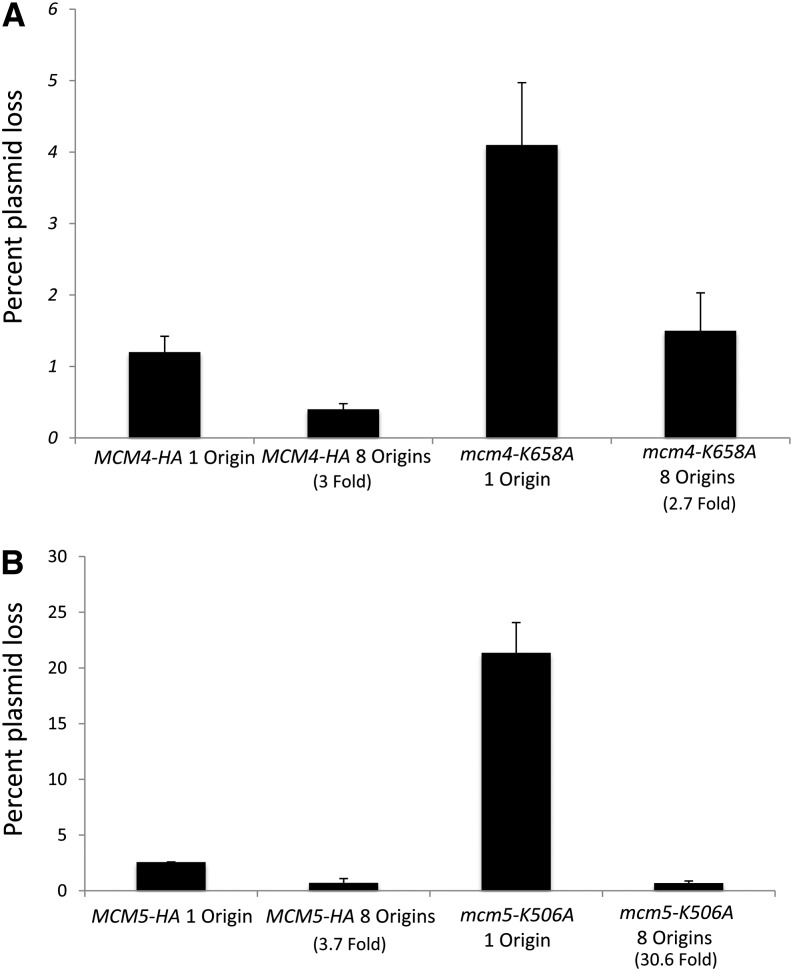
Yeast strains containing the mcm4-K658A PS1-hp mutation have a plasmid loss rate per generation of 4.1%, which is threefold higher than the loss rate of 1.2% per generation in wt cells (Figure 3A). To test whether the plasmid loss was due to initiation of DNA replication or elongation, we examined the plasmid loss rate with a plasmid carrying eight origins (Hogan and Koshland 1992). The plasmid loss rate was decreased 2.5-fold in a Mcm4p-PS1-hp mutant compared to a 3-fold reduction in wt with the addition of seven ARS elements, consistent with a defect in replication initiation (Figure 3A). This result was unexpected and inconsistent with the published elongation defect seen for the ssoMCM PS1-hp mutant in vitro (McGeoch et al. 2005). Next, we examined the plasmid loss rate of the PS1-hp mutant of Mcm5p. The mcm5-HA K506A::URA3 PS1-hp mutant has an increased plasmid loss rate of 21.4%, which is 8-fold greater than the loss rate of 2.6% in wt cells. With the addition of seven origins, the loss rate was reduced about 30-fold for mcm5-k506A::URA3 compared with 3.5-fold in wt cells(Figure 3B). Thus, both mcm4 and mcm5 PS1-hp mutants had an increase in plasmid loss with a clear MCM phenotype that was partially suppressible by increased origin dosage. To further investigate this phenotype, we used ChIP analysis to determine Mcm2-7p helicase binding to chromatin at cellular origins of DNA replication.
Figure 3.
Plasmid stability of Mcm4p and Mcm5p PS1-hp mutants. (A) Wild type or mcm4-HA K658A::TRP1 or (B) wild type or mcm5-HAT K506A::URA3 yeast strains were transformed with either pDK243 (1 origin of replication) or pDK368-7 (eight origins of replication), and plasmid stability was analyzed. Plasmid loss rates are shown in % loss per generation. The increase in stability with the addition of seven origins of replication is presented as fold increase in stability compared with one origin of replication.
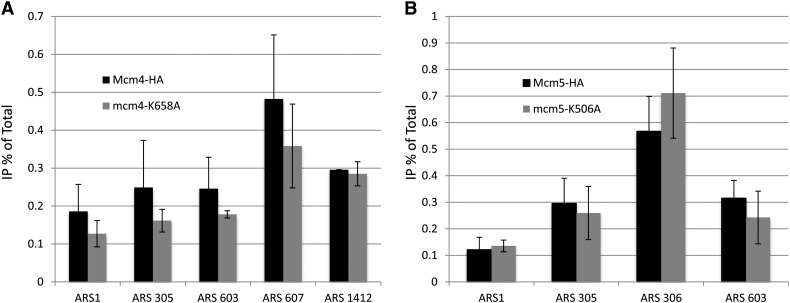
We used cells arrested in G1 phase of the cell cycle where Mcm2-7 protein binding to origin chromatin is maximal (Aparicio et al. 1999). Different origins were examined using ChIP to gain a broad understanding of the affects of the PS1-hp mutation in MCM complex binding to origins of DNA replication. We found no significant defect in origin binding by ChIP of MCM complexes containing the Mcm4p PS1-hp mutation at all 5 origins examined (Figure 4A), which includes early (ARS305, ARS607), mid (ARS1 = ARS416), and late (ARS603, 1412) origins (Raghuraman et al. 2001). Analysis of Mcm5p PS1-hp mutants binding to four origins of DNA replication also showed no binding defects detectable by ChIP analysis in G1 arrested cultures (Figure 4B).
Figure 4.
Chromatin immunoprecipitation (ChIP) analysis of Mcm4p and Mcm5p PS1-hp mutant proteins at origins of replication. (A) ChIP analysis of wild-type (Mcm4-HA) and PS1-hp mutant of Mcm4p (mcm4-K658A-HA) from G1-arrested cultures. Log-phase cultures were arrested in G1 with the addition of alpha factor for 3 hr and an anti-HA antibody was used for ChIP. Chromatin associated DNA was analyzed using quantitative polymerase chain reaction using primers specific to origins of DNA replication. (B) Similar ChIP analysis of wild type (Mcm5-HA) and PS1-hp mutant Mcm5p (mcm5-K506A-HA) strains.
To gain further insight into the plasmid loss defect of PS1-hp mutants, a conditional Mcm4p PS1-hp and a Mcm5p PS1-hp double mutant was constructed in a background containing the temperature-sensitive allele mcm5-461 (Dalton and Hopwood 1997; Leon et al. 2008). At 37° the Mcm5-461 protein is unable to integrate into the Mcm2-7p complex (Dalton and Hopwood 1997; Leon et al. 2008), creating a double mcm4p mcm5p PS1-hp mutant cell. The following strains were used to observe S-phase progression by flow cytometry of the PS1-hp double mutant: RSY1148 (mcm5-461) and CRY209 (mcm5-461 mcm5-HAT K506A::URA3mcm4-HA K658A::TRP1). Yeast strains were arrested in alpha factor for 3 hr at 22° then raised to 37° in alpha factor for 1 hr, then released into S-phase at 37° by the addition of pronase. The resolution of this flow cytometry experimental approach was unable to determine if PS1-hp double mutants progressed more slowly through S-phase when compared to either single PS1-hp mutant alone (Figure S3). The mcm5-461 temperature sensitive mutant is leaky because eventually cells progress through the cell cycle and will form colonies at 37° after several days (Leon et al. 2008). Therefore, we produced double mcm4-HA K658A::TRP1mcm5-HAT K506A::URA3 mutants using either plasmid shuffling or yeast meiotic recombination techniques.
Mutation of the PS1-hp in both mcm4 and mcm5 is lethal
We tested the viability of a mcm4-HA K658A::TRP1mcm5-HA K506A::URA3 double mutant in two different ways. Yeast strains with an integrated mcm5-HA K506A::URA3 mutation and a WT 2μ LEU2 MCM5 plasmid can easily lose the plasmid in that >99% of the cells had lost the plasmid (n = 1836 colonies) after growth in nonselective media for >10 doublings. In contrast, the double mcm4-HA K658A::TRP1mcm5-HA K506A::URA3 mutant maintained the plasmid 100% of the time in that 0% of the cells (n = 2039 colonies; P < 0.0025) had lost the plasmid under similar conditions, indicating a requirement of the plasmid MCM5 gene for viability.
Because the MCM5 gene was on plasmid pRAS690, it is possible that only the plasmid with a single ARS is unable to replicate as was found for the mcm5-bob1 mutation in the absence of Cdc7p function (Hoang et al. 2007). This creates an artificial situation in which the cells do not divide only because the essential plasmid, but not the genome, is unable to replicate. To confirm the synthetic lethality of the two PS1-hp mutants when both are integrated in the genome, CRY119 (mcm4-HA K658A::TRP1 integrated as a partial duplication at the mcm4 locus) was mated to RSY1345 (mcm5-HA K658A::URA3 at the ura3-1 locus) that also had a mcm5::KanMX4 deletion to form diploids, which were then sporulated. Tetrads were dissected and spores that were Ura+ G418R, which contain both the mcm5::KanMX4 and mcm5-HA K658A::URA3 mutations, were tested for the presence of the mcm4-HA K658A::TRP1 allele. As mcm5 and mcm4 are unlinked, we expected to find 50% Trp+ colonies that carry the mcm4 PS1-hp mutation. With mcm5-HA K658A::URA3, no Trp+ colonies were found (n = 44, P < 0.005) (Figure S4A). Thus, the double mcm4mcm5 PS1-hp mutant is inviable in cells with the normal complement of cellular origins. The terminal phenotype of the germinated spore of a double mcm4mcm5 PS1-hp mutant is two large budded cells indicative of a helicase defect as originally described for mcm2∆ and mcm3∆ mutants (Figure S4B)(Yan et al. 1991).
Discussion
Using the archaeal structure of MCM as a model, we have confirmed the role of the SsoMCM PS1-hp structure that had been examined in in vitro experiments (McGeoch et al. 2005) as also being important for downstream events after binding of DNA origins in vivo using the model system Saccharomyces cerevisiae. This is further in vivo evidence that structural models of the homohexameric structures of archaeal MCMs are a valuable tool in understanding the function of eukaryotic heterohexameric MCM complexes (Brewster et al. 2008; Fletcher et al. 2003; Leon et al. 2008). Our results demonstrate that in vivo yeast PS1-hp mutants can form the heterohexameric MCM complex and bind origins of DNA replication, but fail to maintain plasmid stability, indicating a defect in the transition to an active MCM complex, or an MCM complex that is defective in efficient strand separation during replication elongation.
The MCM helicase has been clearly shown to load onto dsDNA in the G1 phase of the cell cycle (Leon et al. 2008; Remus et al. 2009). Our laboratory has previously shown the importance of N-terminal beta-fingers for the MCM complex to bind dsDNA (Leon et al. 2008). The circular MCM complex is loaded onto dsDNA using a break in the MCM ring between Mcm5p and Mcm2p (Bochman and Schwacha 2007) by the actions of Ctd1 and Cdc6p (Randell et al. 2006; Remus et al. 2009). One model of DNA strand separation by the MCM helicase is that of strand exclusion, in which the leading strand DNA remains in the central cavity of the complex bound to the β-hairpins, and the lagging strand is extruded and bound to the PS1 hairpin (Bell and Botchan 2013; Bochman and Schwacha 2009; Fu et al. 2011; Slaymaker and Chen 2012). One possible defect of Mcm PS1-hp mutants is the inability to undergo the transformation of binding dsDNA in the inactive form to excluding one strand of DNA in the active form, which would be consistent with our plasmid loss and ChIP results. During the review of this paper, we became aware of a new publication that demonstrates a PS1-hp mutation in yeast Mcm3p is lethal, although the MCM complex with the mutant Mcm3p is properly formed as interpreted from gel filtration experiments (Lam et al. 2013)
We found that MCM helicase PS1-hp mutants bind origins of DNA replication at normal occupancy levels and propose PS1-hp mutants are defective in a transition step, which is recovered by the addition of multiple origins by mass action. In plasmid loss experiments, only mutants in initiation such as in cdc6 (Hogan and Koshland 1992) or orc2 (Loo et al. 1995) are suppressed by increased origin gene dosage. Similarly, mcm5 mutants in the N-terminal β-hairpins have a similar phenotype and displayed reduced binding to origins by ChIP (Leon et al. 2008). In contrast, plasmid loss rates are not suppressed in pol1 mutants, which are defective in elongation by DNA polymerase α (Hogan and Koshland 1992). We propose that the Mcm PS1-hp mutant proteins can bind to origins normally but have increased dissociation due to a reduced ability to bind the extruded lagging strand. In support of this idea, the DNA helicase activity and the single-stranded DNA (ssDNA) binding stability of recombinant MCM helicases in an electromobility shift assay in vitro was lower for MCM2-7 complexes containing a PS1-hp mutation in Mcm3p, possibly because the ssDNA-MCM complexes formed are unstable (Lam et al. 2013)
In this scenario, this dissociation defect is suppressed by having more Mcm2-7p complexes at more origins with only one active helicase still bound to complete replication. Our study suggests a different interpretation of the plasmid stability assay when considering mutants of the Mcm helicase or of other proteins that load in G1 phase and are activated in S phase. This leads to a new understanding of the plasmid stability assay, such that multiple origins not only recover binding defects but also defects in transition from an inactive helicase form to an active replicative form.
We found that the mcm5 and mcm4 PS1-hp mutants behave differently in that the mcm5 mutant was more deleterious with a higher plasmid loss defect (Figure 3) and displays both temperature-sensitive and cold-sensitive conditional phenotypes (Figure 1). This is similar to our previous analysis with the β-hairpin mutants (Leon et al. 2008). Perhaps, the Mcm5 protein is more important in stabilizing the channel in ssDNA strand exclusion.
Also similar to our previous studies (Leon et al. 2008), lethality required mutations in two different MCM genes, which we propose reflects the fact that the six PS1-hp structures in the heterohexamer must cooperate for function and do not act independently, as also has been found for mutations in the ATPase site (Schwacha and Bell 2001). It is also possible that lethality results because the Mcm4 and Mcm5 proteins lie opposite each other in the hexameric ring (Bell and Botchan 2013). In the recent study cited previously (Lam et al. 2013) all combinations of the other MCM helicase PS1-hp mutations (MCM2, 4, 5, 6, and 7) were lethal except for the mcm4-K568A and mcm5-K506A combination, which had a slow growth phenotype. The later result contradicts our findings that the mcm4-K568A and mcm5-K506A combination is lethal. Furthermore, our mcm5-K506A mutant has a conditional phenotype (Figure 1), while theirs had no phenotype. One possible explanation is we used a different yeast strain background (A364a) compared to them (S288C). Nonetheless, our proposal in which important structures in MCM subunits cooperate and have similar functions is supported by both studies.
Supplementary Material
Acknowledgments
We thank Karen Helm of the University of Colorado Comprehensive Cancer Center (P30CA046934) Flow Cytometry Shared Resources. We thank for Marisa Ruehle assisting with the structure-based alignments of yeast and archaeal Mcm proteins. We thank John Diller and Rebecca Ferguson for their critical reading of this manuscript. We also thank Stephanie Williams for the initial characterization of the mcm5 mutant. This work was supported by a Public Health Service award to RAS (GM35078) from the NIH.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Adkins M. W., Howar S. R., Tyler J. K., 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14: 657–666. [DOI] [PubMed] [Google Scholar]
- Aparicio O. M., Stout A. M., Bell S. P., 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. USA 96: 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. D., Botchan M. R., 2013. The Minichromosome Maintenance Replicative Helicase. Cold Spring Harb. Perspect. Biol. 5: a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Dutta A., 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333–374. [DOI] [PubMed] [Google Scholar]
- Bochman M. L., Schwacha A., 2007. Differences in the single-stranded DNA binding activities of MCM2–7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J. Biol. Chem. 282: 33795–33804. [DOI] [PubMed] [Google Scholar]
- Bochman M. L., Schwacha A., 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 73: 652–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster A. S., Wang G., Yu X., Greenleaf W. B., Carazo J. M., et al. , 2008. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. USA 105: 20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Hopwood B., 1997. Characterization of Cdc47p-minichromosome maintenance complexes in Saccharomyces cerevisiae: identification of Cdc45p as a subunit. Mol. Cell. Biol. 17: 5867–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher R. J., Bishop B. E., Leon R. P., Sclafani R. A., Ogata C. M., et al. , 2003. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 10: 160–167. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., 2004. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 68: 109–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J., Remus D., Mehanna A., Diffley J. F., 2013. ATPase-dependent quality control of DNA replication origin licensing. Nature 495: 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. V., Yardimci H., Long D. T., Ho T. V., Guainazzi A., et al. , 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai D., Zhao R., Li D., Finkielstein C. V., Chen X. S., 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119: 47–60. [DOI] [PubMed] [Google Scholar]
- Gambus A., Khoudoli G. A., Jones R. C., Blow J. J., 2011. MCM2–7 form double hexamers at licensed origins in Xenopus egg extract. J. Biol. Chem. 286: 11855–11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang M. L., Leon R. P., Pessoa-Brandao L., Hunt S., Raghuraman M. K., et al. , 2007. Structural changes in Mcm5 protein bypass Cdc7-Dbf4 function and reduce replication origin efficiency in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 7594–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan E., Koshland D., 1992. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89: 3098–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Laskey R. A., Coleman N., 2014. Replication proteins and human disease. Cold Spring Harb. Perspect. Biol. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. K., Ma X., Sing T. L., Shilton B. H., Brandl C. J., et al. , 2013. The PS1 hairpin of Mcm3 is essential for viability and for DNA unwinding in vitro. PLoS ONE 8: e82177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon R. P., Tecklenburg M., Sclafani R. A., 2008. Functional conservation of beta-hairpin DNA binding domains in the Mcm protein of Methanobacterium thermoautotrophicum and the Mcm5 protein of Saccharomyces cerevisiae. Genetics 179: 1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. C., Mechali M., 2013. DNA replication origins. Cold Spring Harb. Perspect. Biol. 5: a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S., Fox C. A., Rine J., Kobayashi R., Stillman B., et al. , 1995. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol. Biol. Cell 6: 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune H. J., Danielson L. S., Alvino G. M., Collingwood D., Delrow J. J., et al. , 2008. The temporal program of chromosome replication: genomewide replication in clb5Δ Saccharomyces cerevisiae. Genetics 180: 1833–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch A. T., Trakselis M. A., Laskey R. A., Bell S. D., 2005. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 12: 756–762. [DOI] [PubMed] [Google Scholar]
- Raghuraman M. K., Winzeler E. A., Collingwood D., Hunt S., Wodicka L., et al. , 2001. Replication dynamics of the yeast genome. Science 294: 115–121. [DOI] [PubMed] [Google Scholar]
- Randell J. C., Bowers J. L., Rodriguez H. K., Bell S. P., 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol. Cell 21: 29–39. [DOI] [PubMed] [Google Scholar]
- Randell J. C., Fan A., Chan C., Francis L. I., Heller R. C., et al. , 2010. Mec1 Is one of multiple kinases that prime the Mcm2–7 helicase for phosphorylation by Cdc7. Mol. Cell 40: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., et al. , 2009. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Bell S. P., 2001. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell 8: 1093–1104. [DOI] [PubMed] [Google Scholar]
- Sclafani R. A., Holzen T. M., 2007. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 41: 237–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M., Chen X. S., 2012. MCM structure and mechanics: what we have learned from archaeal MCM. Subcell. Biochem. 62: 89–111. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Diffley J. F., 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2–7 during G1 phase. Nat. Cell Biol. 4: 198–207. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., et al. , 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332. [DOI] [PubMed] [Google Scholar]
- Tye B. K., 1999. MCM proteins in DNA replication. Annu. Rev. Biochem. 68: 649–686. [DOI] [PubMed] [Google Scholar]
- Yan H., Gibson S., Tye B. K., 1991. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 5: 944–957. [DOI] [PubMed] [Google Scholar]
- Zegerman P., Diffley J. F., 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.