Abstract
Transgenic techniques offer a valuable tool for determining gene functions. Although various promoters are available for use in gene overexpression, gene knockdown, and identification of transgenic individuals, there is nevertheless a lack of versatile promoters for such studies, and this dearth acts as a bottleneck, especially with regard to nonmodel organisms. Here, we succeeded in identifying a novel strong and ubiquitous promoter/enhancer in the silkworm. We identified a unique silkworm strain whose reporter gene showed strong and ubiquitous expression during the establishment of enhancer trap strains. In this strain, the transposon was inserted into the 5′UTR of hsp90, a housekeeping gene that is abundantly expressed in a range of tissues. To determine whether the promoter/enhancer of hsp90 could be used to induce strong gene expression, a 2.9-kb upstream genomic fragment of hsp90 was isolated (hsp90P2.9k), and its transcriptional activation activity was examined. Strikingly, hsp90P2.9k induced strong gene expression in silkworm cell cultures and also strongly induced gene expression in various tissues and developmental stages of the silkworm. hsp90P2.9k also exhibited significant promoter/enhancer activity in Sf9, a cell culture from the armyworm, suggesting that this fragment might possibly be used as a gene expression tool in other Lepidoptera. We further found that 2.0 kb of hsp90P2.9k is sufficient for the induction of strong gene expression. We believe that this element will be of value for a range of studies such as targeted gene overexpression, gene knockdown and marker gene expression, not only in the silkworm but also in other insect species.
Keywords: promoter/enhancer, hsp90, silkworm, enhancer trap, transgenic
Transgenesis is an important research technique in both basic and applied biological sciences. With respect to basic research, transgenic manipulations offer the opportunity to carry out in vivo functional analysis of genes or to identify their cis regulatory elements. The use of transgenesis methodologies has contributed greatly to advances in a wide range of biological research, including molecular biology, genetics, physiology, and others. In applied research, transgenic organisms have been used as a bioreactor for production of medically important compounds or materials or for pest control (Robinson et al. 2004; Tatematsu et al. 2012). For both basic and applied research, it is important that expression of a transgene is regulated very precisely, both in target tissues and at the appropriate time. Because this regulation usually is mediated by the activity of the promoter connected to the transgene, it is clear that promoter activity is very critical for transgenic analyses. However, the lack of a suitable promoter is still a bottleneck for some studies, especially those using nonmodel organisms.
The silkworm, Bombyx mori, has a long history of use in research. B. mori belongs to the order Lepidoptera and was the first species of this order to have its genome sequenced (The International Silkworm Genome Consortium 2008). Transgenic techniques also have been developed for use in this species, enabling studies on gene overexpression, gene knockdown, and knockout, among many others (Tamura et al. 2000; Imamura et al. 2003; Dai et al. 2008; Takasu et al. 2010; Ma et al. 2012; Sajwan et al. 2013; Takasu et al. 2013). In Bombyx, several different promoters have been exploited in these studies: the actin A3 promoter, which can induce widespread gene expression (Tamura et al. 2000; Imamura et al. 2003); the 3xP3 artificial promoter, which has three binding sites for Pax-6 and functions in the stemmata or complex eyes (Berghammer et al. 1999; Thomas et al. 2002; Imamura et al. 2003); the hsp70 promoter, which can induce a significant level of gene expression after heat shock treatment (Uhlířová et al. 2002); and the sericin or fibroin promoter, which acts specifically in the middle or posterior silk gland (Imamura et al. 2003; Sezutsu et al. 2009; Tatematsu et al. 2010). In addition, we recently have succeeded in identifying a novel hemocyte oenocytoid promoter (Tsubota et al. 2014). These promoters have been used not only for gene function analyses or recombinant protein production but also for identification of transgenic organisms (Thomas et al. 2002). Much of the recent progress in transgenic silkworm research can be attributed to the exploitation of such promoters. Unfortunately, however, these promoters nevertheless have limitations regarding their application. The A3 promoter, for example, is not ubiquitous but shows little to no activity in several larval tissues and in the embryonic stage (Tamura et al. 2000; Uchino et al. 2008). It is thus not practical to use this promoter for functional analyses in these tissues and stage. Another recently described baculovirus immediate-early promoter, ie1, also exhibits limited activity in the larvae (Masumoto et al. 2012), demonstrating the difficulty of identifying a bona fide ubiquitous promoter. The 3xP3 promoter is widely used for transgenic marker gene expression; however, because the promoter functions just in a very limited region, screening is difficult and time-consuming (Thomas et al. 2002). As a consequence of the problems associated with use of many of the known promoters, identification of more versatile promoters is crucial for exploitation of transgenic methods to promote further progress in silkworm research.
The enhancer trap system provides one of the most powerful tools in functional genomic research. In this system, a transposon is induced to insert randomly into the host genome using transposase activity. Usually the transposon contains a reporter (e.g., green fluorescent protein [GFP], cyan fluorescent protein [CFP], or lacZ) or a driver gene (e.g., GAL4). If the transposon is inserted within the promoter/enhancer region of a gene, its activity can be detected by monitoring reporter gene expression. This method was first exploited in the fruit fly Drosophila melanogaster (O’Kane and Gehring 1987; Cooley et al. 1988) and now applied to various organisms, including insects such as red flour beetle (Tribolium castaneum) and mosquito (Anopheles stephensi), vertebrates such as zebrafish (Danio rerio) and medaka (Oryzias latipes), and plants such as rice (Oryza sativa) and thale cress (Arabidopsis thaliana) (Lorenzen et al. 2003; Ito et al. 2004; Parinov et al. 2004; Ellingsen et al. 2005; Liu et al. 2005; Lorenzen et al. 2007; O’Brochta et al. 2011). A significant number of genes or their promoter/enhancer elements have been identified using this system (Bier et al. 1989; Wilson et al. 1989; Ito et al. 2004; Parinov et al. 2004; Ellingsen et al. 2005; Liu et al. 2005; Vijaybhaskar et al. 2008). Our laboratory has previously established more than 300 silkworm enhancer trap strains using the piggyBac transposon and released descriptions of reporter gene expression into public databases (Uchino et al. 2008; Shimomura et al. 2009). The detection of highly variable expression profiles strongly indicates that the silkworm enhancer trap system can contribute to the identification of a wide variety of novel genes and/or their promoter/enhancer elements (Uchino et al. 2008). These strains are also valuable resources for tissue-specific driver strains since the transposon contains a GAL4 gene (Ito et al. 2008; Daimon et al. 2012).
In this study, we identified a unique transgenic silkworm strain whose reporter gene showed strong and widespread expression pattern during the process of enhancer trap strain establishment. Analysis of the insertion site revealed that the transposon was inserted into Bombyx hsp90, a housekeeping gene that is expressed abundantly in various tissues. We examined the promoter/enhancer activity of an hsp90 2.9 kb upstream sequence (hsp90P2.9k) and found that it induced strong gene expression in silkworm cell cultures as well as in a range of tissues and developmental stages. hsp90P2.9k also drove strong transcription in an armyworm cell line, Sf9, suggesting that the fragment could be used as a gene expression tool in other lepidopteran species. We further found that just 2.0 kb would be sufficient for the induction of strong gene expression. We believe that the identified fragment will be of value in various types of investigation, such as widespread gene expression or efficient transgenic individual screening, and will thereby contribute to further advances in research in silkworms and other insects.
Materials and Methods
Silkworm strains
Silkworms, B. mori, were reared on an artificial diet (Nihon Nosan Kogyo, Yokohama, Japan) at 25° under a 12-hr light/dark photoperiod. A silkworm enhancer trap strain, AyFib-042, was used as a mutator strain (Sezutsu et al. 2009). This strain was crossed with a jumpstarter strain, Js15 (Uchino et al. 2008), for the mobilization of the piggyBac transposon. The established strains were crossed with the UAS-GFP homozygous strain (Uchino et al. 2006) and the mobilization of the transposon was confirmed by the lack of the stripe-like GFP expression of the original AyFib-042 strain (Sezutsu et al. 2009). The AyFib-431a strain was obtained in this way.
Detection of GFP and DsRed fluorescence
The expression of the fluorescent proteins GFP and DsRed was detected using an MZFLIII (Leica, Solms, Germany), MZ16FA (Leica), SZX16 (Olympus, Tokyo, Japan), or VB-S20 fluorescence microscope (Keyence, Osaka, Japan) with a GFP band pass filter (excitation: 440/70 nm, emission: 525/50 nm or excitation: 440/70 nm, emission: 535/550 nm or excitation: 460/80 nm, emission: 495/540 nm), GFP long-pass filter (excitation: 440/70 nm, emission: 510 nm) or DsRed filter (excitation: 525/40 nm, emission: 572 nm or excitation: 530/45 nm, emission: 620/60 nm). Images were captured using a Leica DC200 (Leica), Leica DFC300FX (Leica), DP71 (Olympus), or VB-7000 (Keyence) system.
Inverse polymerase chain reaction (PCR)
Genomic DNA was isolated from adult moths by sodium dodecyl sulfate−phenol extraction (Ohshima and Suzuki 1977). The DNA was digested with NcoI and heated at 70° for 15 min to inactivate the enzymes. The digested DNA was purified using a QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany) or QIAquick Gel Extraction Kit (QIAGEN) and ligated using T4 DNA ligase (New England Biolabs, Ipswich, MA) at 4°overnight. The DNA was purified using a QIAquick PCR Purification Kit (QIAGEN) and was used as the template for PCR. PCR was carried out using the ExTaq polymerase (TaKaRa, Otsu, Japan) as follows: 95° for 2 min followed by five cycles of 95° for 30 sec, 65° for 30 sec and 72° for 2 min, 5 cycles of 95° for 30 sec, 60° for 30 sec and 72° for 2 min, 35 cycles of 95° for 30 sec, 55° for 30 sec and 72° for 2 min, and 72° for 10 min. The amplified fragments were sequenced using Applied Biosystems 3130xl (Life Technologies, Carlsbad, CA) after cycle sequencing with BigDye Terminator V3.1 (Life Technologies). The obtained sequence was subjected to a BLAST search in the silkworm genome database, KAIKObase (http://sgp.dna.affrc.go.jp/KAIKObase/) for the identification of the insertion site.
Southern hybridization analysis
Southern hybridization analysis was performed as described in Sezutsu et al. (2009). In brief, 2 µg of the genomic DNA was digested with NcoI or SacI and subjected to electrophoresis, blotted onto a Hybond-N+ nylon membrane (GE Healthcare, Chalfont St. Giles, UK), hybridized with a GAL4 probe and detected using an Alkphos Direct Labeling and Detection System (GE Healthcare).
Probe synthesis for in situ hybridization
5′-ATGCCGGAAGAAATGGAGAC-3′ and 5′-TGCTCGGAACTCTAACTGAC-3′ primers with T7 sequence was used for PCR amplification of the hsp90 gene. The PCR product was purified and used as a template for a probe labeling reaction. For probe labeling, DIG RNA labeling kit (SP6/T7) was used (Roche Diagnostics, Basel, Switzerland).
In situ hybridization
Embryos (ca. stage 20) were dissected and fixed in 4% paraformaldehyde at room temperature overnight. They were stored in 100% methanol at −20° until use. After replacement of the methanol with phosphate-buffered saline (PBS), the embryos were placed in 0.2 M HCl for 10 min and washed with PBS. They were then treated with 10 µg/mL proteinase K (Roche diagnostics) at 37° for 30 min, washed with PBS, and acetylated using 0.1 M triethanolamine-HCl (pH 8.0) and 0.25% acetic anhydride for 10 min. This solution was gradually replaced by hybridization buffer, and the embryos were hybridized with a labeled probe (100 ng/ml concentration) at 60° overnight. After hybridization, they were washed in PBS, and unhybridized probe was degraded using 50 µg/mL RNaseA. For antibody reaction, the blocking reaction was carried out using 10% Goat Normal Serum, and the anti-Dig antibody (Roche Diagnostics) was applied at a 1:2000 dilution. The embryos were then stained with NBT/BCIP (Roche Diagnostics), mounted in 100% glycerol and examined using an Axioplan microscope (Carl Zeiss, Oberkochen, Germany).
Reverse-transcription PCR of hsp90
Total RNAs were extracted from each tissue of w-c spinning stage larvae using ISOGEN (Nippongene, Tokyo, Japan) and SV Total RNA Isolation System (Promega, Madison, WI). The RNA concentration was measured using a NanoDrop ND2000 UV spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). The RNA was used for reverse-transcription using Superscript III (Life Technologies). The KOD-FX polymerase (Toyobo, Osaka, Japan) was used for PCR, with the following primer sequences: 5′-ATGCCGGAAGAAATGGAGAC-3′ and 5′-TGCTCGGAACTCTAACTGAC-3′ for hsp90 and 5′-CAGGCGGTTCAAGGGTCAATAC-3′ and 5′-TGCTGGGCTCTTTCCACGA-3′ for rp49.
Plasmid construction for the luciferase reporter assay and for transgenic silkworm generation
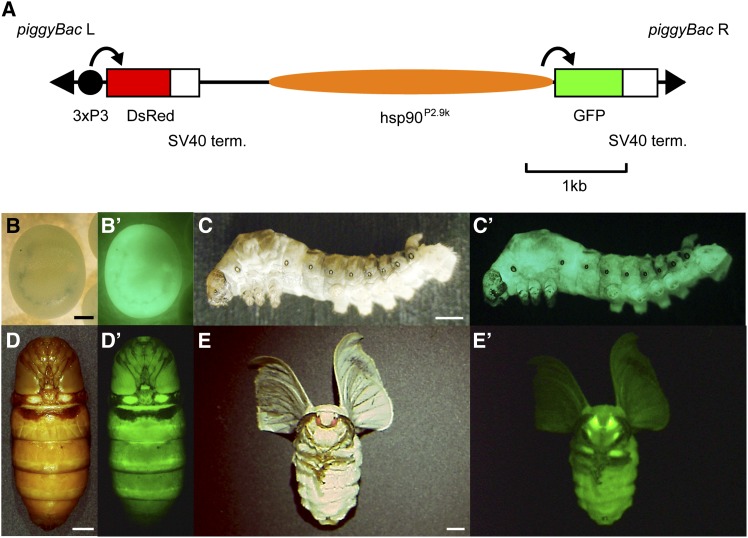
The pGL3-A3 plasmid was generated for the luciferase reporter assay of the A3 promoter. First, the 682 bp A3 promoter fragment was amplified from pHA3PIG (Tamura et al. 2000) using primers 5′-CTAGCTAGCCCGGGCTCAAGCTTGATGCG-3′ and 5′-CCGCTCGAGTGAATTAGTCTGCAAGAA-3′. The amplified fragment was inserted into the Nhe I/Xho I site of the pGL3 vector (Promega) to generate pGL3-A3. A pGL3- hsp90P2.9k plasmid was generated for the hsp90P2.9k luciferase assay. The hsp90P2.9k genomic sequence was first determined by inverse PCR because the genome sequence upstream of hsp90 was not available in the KAIKObase. After sequence determination, the hsp90P2.9k genomic fragment was amplified from the w-c strain using primers 5′-TTAATTCACACAAAATGACTAGAGGG-3′ and 5′-CCATGGCTCAGTTCGCTTTAAATA-3′ and was inserted into pCR-BluntII-TOPO vector (Life Technologies). We verified the sequence, and the hsp90P2.9k fragment was inserted into the Nhe I/Xho I site of the pGL3 vector to generate pGL3-hsp90P2.9k. pGL3-hsp90P2.0k or pGL3-hsp90P1.6k plasmid was generated for the luciferase reporter assay of hsp90P2.9k partial fragment. The 916- to 2033-bp (hsp90P2.0k) or 1330- to 2033-bp (hsp90P1.6k) fragment of hsp90P2.9k was first amplified from pGL3-hsp90P2.9k using primers 5′-TCGCGTTTCCTTCACTCGCG-3′ and 5′-TCTAGAAAAGCATCGAAATTTTAACATTACAGA-3′ or 5′-GGCGCTTCTCAAATGGAACTTTCG-3′ and 5′- TCTAGAAAAGCATCGAAATTTTAACATTACAGA-3′, respectively. The amplified fragment was inserted into the pCR-BluntII-TOPO vector and the sequence was verified. Next, the plasmid was digested with XbaI, and the nucleotides downstream of hsp90P2.9k 2034 bp was connected by inserting the fragment obtained by XbaI digestion of pGL3-hsp90P2.9k. The plasmid (pCR-BluntII-hsp90P2.0k or pCR-BluntII-hsp90P1.6k) was digested with NheI/XhoI, and the hsp90P2.0k or hsp90P1.6k fragment was inserted into the NheI/XhoI site of pGL3 to generate pGL3-hsp90P2.0k or pGL3-hsp90P1.6k. The hsp90-GFP transgenic strain was generated using the pBachsp90GFP-3xP3DsRed plasmid. To construct this vector, the pBacA3dGAL4/3xP3-DsRed(ANB) vector (Tsubota et al. 2014) was digested with AscI/XbaI and the A3dGAL4 fragment was removed. Then, GFP and SV40 terminator sequences were inserted to generate pBacGFP-3xP3DsRed. Next, the hsp90P2.9k fragment was inserted into the Xba I site located upstream of GFP to generate pBachsp90GFP-3xP3DsRed.
Luciferase reporter assay
The luciferase reporter assay using NIAS-Bm-oyanagi2 was conducted as described in Tanaka et al. (2009). In summary, the reporter plasmid (pGL3-A3, pGL3-hsp90P2.9k, pGL3-hsp90P2.0k or pGL3-hsp90P1.6k) and pOpIE2-core-Rluc (Tanaka et al. 2012) were transfected into NIAS-Bm-oyanagi2 cells using FuGENE HD (Promega). Firefly luciferase activity in cell extracts was measured 72 hr later and normalized against Renilla luciferase activity. Both luciferase activities were measured using a lumicounter (Nition, Funabashi, Japan) with the Dual-luciferase assay system (Promega).
For the reporter assay of the cell lines NIAS-Bm-aff3 (Kayukawa et al. 2012), BmN (Katakura Industries Co., Tokyo, Japan), Sf9 (Tsubota et al. 2010), S2 (Kayukawa et al. 2012), or Tc81 (Kayukawa et al. 2013), cells were seeded at a density of 1.5 × 105 cells per well in 200 µL of medium in a 96-well plate 1 d before transfection. The reporter plasmid was transfected with pIZT_RLuc vector using FuGENE HD Transfast transfection Reagent (Promega); 72 hr later, luciferase activity was measured using a luminometer ARVO (PerkinElmer, Waltham, MA) with the Dual-luciferase assay system (Promega).
Generation of transgenic silkworms
The transgenic silkworm strain was produced by germline transformation as described elsewhere (Tamura et al. 2000, 2007). The plasmid DNA for the injection was purified using a QIAGEN Plasmid Midi Kit (QIAGEN). The DNA was injected into w1-pnd embryos at the preblastodermal stage. Screening of G1 embryos were performed around stage 25 (6 or 7 d after oviposition). The transgenic strains were maintained by crossing with diapausing w-c strain.
Results
Isolation of a novel transgenic silkworm strain with strong and ubiquitous DsRed expression
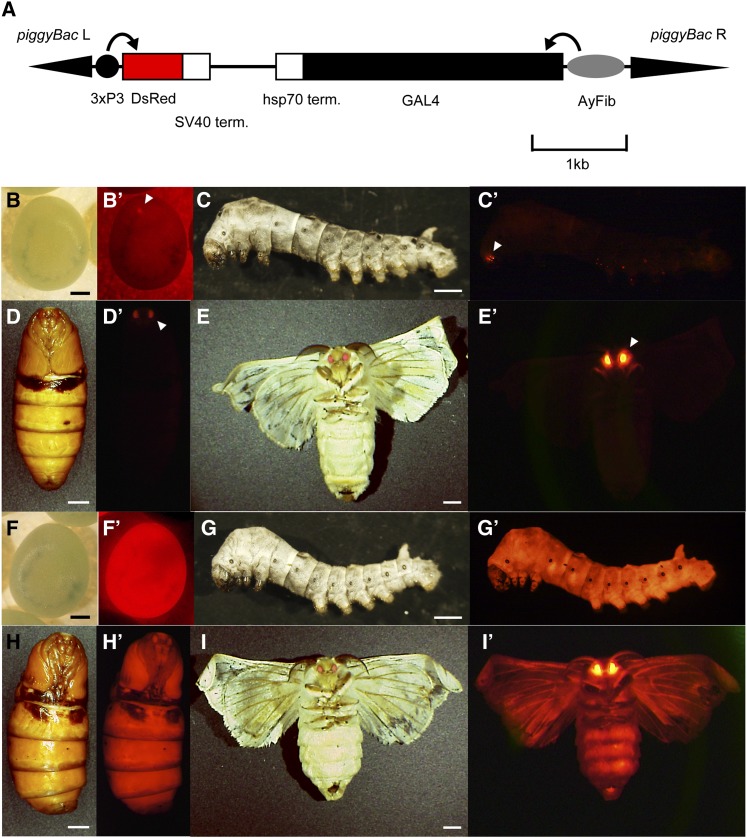
We previously constructed a large number of silkworm enhancer trap strains for use in Bombyx functional genomics (Uchino et al. 2008). We also have proceeded to establish the novel enhancer trap strains using the AyFib-042 strain as the mutator (see the section Materials and Methods), and during this process, we could obtain a unique strain designated AyFib-431a. AyFib strains possess an Antheraea yamamai fibroin promoter-GAL4 cassette and a 3xP3 promoter-DsRed transgenic marker (Figure 1A). The eye-specific activity of the 3xP3 promoter drives DsRed expression only in the stemmata and complex eyes (Figure 1, B−E′). In AyFib-431a, however, DsRed expression was strong and widespread in embryonic, larval, pupal, and adult stages (Figure 1, F−I′). The results indicated that the 3xP3 had trapped a promoter/enhancer with strong and ubiquitous activity.
Figure 1.
DsRed expression in AyFib-042 and AyFib-431a strains. (A) Structure of the piggyBac transposon vector used for AyFib enhancer trap strains. L, left arm; R, right arm; 3xP3, artificial 3xP3 promoter (Berghammer et al. 1999); SV40 term., SV40 terminator; hsp70 term., hsp70 terminator; AyFib, Antheraea yamamai fibroin promoter (Sezutsu et al. 2009). (B−E′) Expression of DsRed protein in the AyFib-042 strain. (B, C, D, E) Bright-field images. (B′, C′, D′, E′) DsRed-fluorescence images. (B, B′) Embryo 7 d after oviposition. (C, C′) Third instar larva. (D, D′) Pupa. (E, E′) Adult. Arrowheads in (B′), (C′), (D′), and (E′) indicate the eye-specific DsRed expression in AyFib-042. (F−I′) Expression of DsRed protein in AyFib-431a strain. (F, G, H, I) Bright-field images. (F′, G′, H′, I′) DsRed fluorescence images. (F, F′) Embryo 7 d after oviposition. (G, G′) Third instar larva. (H, H′) Pupa. (I, I′) Adult. Bars in (B, F) are 0.3 mm, (C, G) are 1 mm, and (D, E, H, I) are 3 mm.
To confirm that the transposon had altered its position, a southern hybridization analysis was carried out. Genomic DNAs were extracted from the AyFib-042 and AyFib-431a strains and used for hybridization with a probe for the GAL4 fragment. The two strains showed different band patterns on gels (Supporting Information, Figure S1). Therefore, we conclude that the piggyBac transposon had indeed been mobilized and inserted into a novel genomic locus in the AyFib-431a strain. The southern hybridization analysis also showed that both AyFib-042 and AyFib-431a have a single copy of the piggyBac transposon (Figure S1). Our findings suggested that the strong DsRed expression in AyFib-431a was mediated by the activity of a strong promoter/enhancer located at a single genomic locus.
Detailed expression analysis of DsRed in AyFib-431a
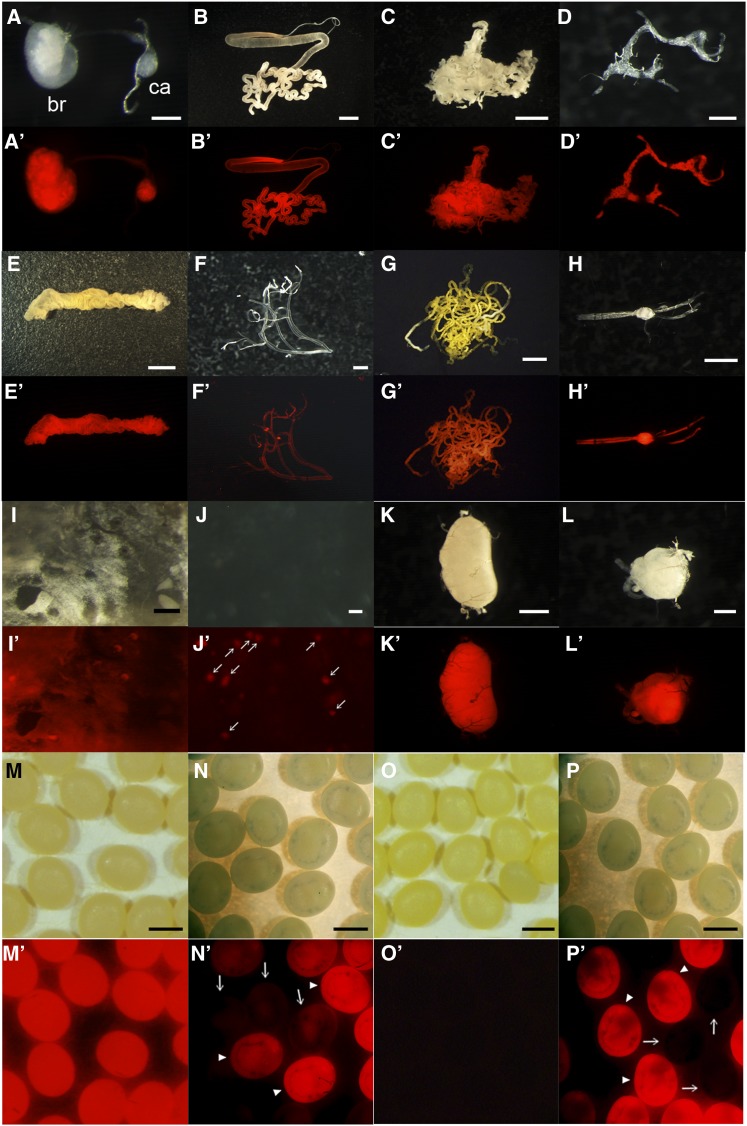
Next, we examined DsRed expression in more detail. The expression was investigated in each larval tissues and interestingly, all of the investigated tissues, namely, brain, corpora allata, silk gland, fat body, prothoracic gland, gut, trachea, Malpighian tubules, ventral nerve, epidermis, hemocyte, testis, and ovary, exhibited strong level of DsRed expression (Figure 2, A−L′). Thus, it is suggested that the genomic region in which the piggyBac is inserted in AyFib-431a is under the regulation of a highly ubiquitous promoter/enhancer. We found that the DsRed expression levels were particularly high in the brain, corpora allata, fat body, ventral nerve, testis and ovary (Figure 2, A−L′).
Figure 2.
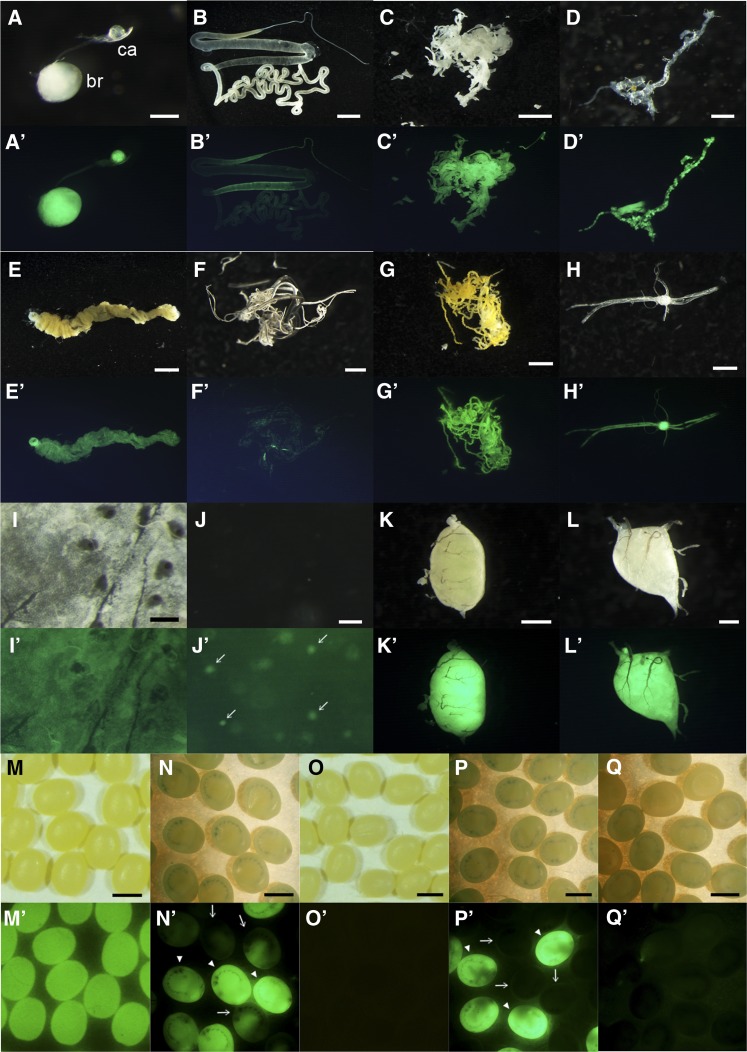
DsRed expression of AyFib-431a in each tissue of final instar larvae (A−L′) or in the embryo (M−P′). (A, A′) Brain and corpora allata. (B, B′) Silk gland. (C, C′) Fat body. (D, D′) Prothoracic gland. (E, E′) Gut. (F, F′) Trachea. (G, G′) Malpighian tubules. (H, H′) Ventral nerves. (I, I′) Epidermis. (J, J′) Hemolymph. (K, K′) Testis. (L, L′) Ovary. (A, B, C, D, E, F, G, H, I, J, K, L) Bright-field images. (A′, B′, C′, D′, E′, F′, G′, H′, I′, J′, K′, L′) DsRed-fluorescent images. br, brain; ca, corpora allata. Arrows in (J′) indicate hemocyte cells. (M, M′, N, N′) Eggs laid by AyFib-431a heterozygous female crossed with w-c male. (M, M′) Just after oviposition. (N, N′) 7 days after oviposition. Arrowheads indicate the putative DsRed+ (AyFib-431a/+) and arrows indicate DsRed− (+/+) individuals. Residual maternal DsRed protein was detected in the putative +/+ embryos. (O, O′, P, P′) Eggs laid by w-c female crossed with AyFib-431a heterozygous male. (O, O′) Just after oviposition. (P, P′) 7 days after oviposition. Arrowheads indicate the putative DsRed+ (AyFib-431a/+) and arrows indicate DsRed− (+/+) individuals. Bars represent 0.25 mm in (A, I), 5 mm in (B, E), 2.5 mm in (C, G), 0.5 mm in (D, L), 1 mm in (F, H, K, M−P), and 0.05 mm in (J).
The strong DsRed expression in the ovary was of interest as it suggested the possibility of maternal inheritance. To investigate this possibility, we crossed a female of AyFib-431a heterozygote with a male of the nontransgenic strain w-c and analyzed DsRed expression in the progeny. If the DsRed shows Mendelian inheritance, then the ratio of DsRed positive to negative eggs will be 1:1. Contrary to this expectation, however, all of the eggs were found to express DsRed very strongly just after oviposition (Figure 2, M and M′), suggesting that maternal inheritance has occurred. The level of DsRed expression diminished in about half of the embryos by the late embryonic stage, i.e., around seven days after egg laying (Figure 2, N and N′). On the other hand, none of the progenies of the cross between w-c females and AyFib-431a heterozygous males showed DsRed immediately after oviposition, but approximately half of them showed expression of DsRed by seven days after oviposition (Figure 2, O−P′, data not shown).
A. yamamai fibroin promoter activity in AyFib-431a
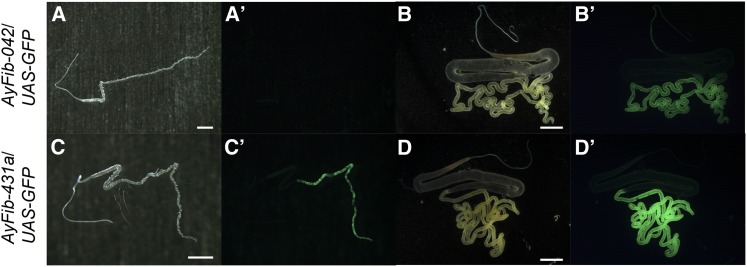
Since AyFib-431a possesses not only the 3xP3 promoter but also the A. yamamai fibroin promoter (Figure 1A, Sezutsu et al. 2009), it was possible that the fibroin promoter also was activated ubiquitously, similarly to 3xP3. The A. yamamai fibroin promoter has been shown to be active just in the posterior silk gland (Figure 3, B and B′, Sezutsu et al. 2009). We examined fibroin promoter activity of AyFib-431a by observing GAL4 expression and found that the activity was still confined to the posterior silk gland (Figure 3, D and D′). Therefore, the A. yamamai fibroin did not appear to have trapped the ubiquitous promoter/enhancer that was trapped by 3xP3. Interestingly, we found a slight modification of GAL4 expression pattern. In AyFib-042, GAL4 expression can be detected only after the middle or late stage of fifth instar larvae (Figure 3, A and B′, Sezutsu et al. 2009), whereas in AyFib-431a its expression was detected at an earlier stage, namely, in the third instar larvae (Figure 3, C and C′) and, occasionally, even in the first instar larvae (data not shown).
Figure 3.
GAL4 expression in AyFib-042 (A−B′) and AyFib-431a (C−D′). AyFib strains were crossed with the UAS-GFP homozygous strain (Uchino et al. 2006) and green fluorescent protein in the F1 hybrid was observed to monitor GAL4 expression. (A, A′, C, C′) First day of third instar larva. (B, B′, D, D′) Spinning stage of final instar larva. (A, B, C, D) Bright-field images. (A′, B′, C′, D′) GFP-fluorescent images. Bars represent 1 mm in (A, C) and 5 mm in (B, D).
Identification of the AyFib-431a piggyBac insertion site
The aforementioned results indicate that the 3xP3 but not the A. yamamai fibroin of AyFib-431a trapped a strong and ubiquitous silkworm promoter/enhancer. To identify the genomic region responsible for this strong and ubiquitous promoter/enhancer activity, we analyzed the piggyBac insertion site (see the Materials and Methods). This analysis indicated that the transposon was inserted into the 5′UTR of the Bombyx hsp90 gene (Figure 4, Landais et al. 2001).
Figure 4.
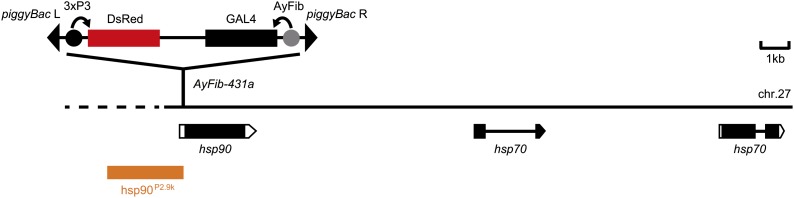
Insertion site of the piggyBac transposon in the AyFib-431a strain. The transposon is inserted into the 5′UTR of hsp90 gene. The dashed line indicates that the genome sequence of this region is not available in the KAIKOBase.
hsp90 is a molecular chaperone gene that is conserved from bacteria to eukaryotes (Lindquist and Craig 1988). The gene is expressed strongly and ubiquitously in various organisms, including lepidopteran insects (Sonoda et al. 2006a,b, 2007; Tungjitwitayakul et al. 2008; Gkouvitsas et al. 2009; Zhang and Denlinger 2010; Xu et al. 2011). A previous analysis showed that silkworm Hsp90 protein is present in all larval stages (Sosalegowda et al. 2010). We carried out a detailed expression analysis and identified strong and widespread expression in the embryonic stage and in final instar larvae (Figure S2).
The insertion of piggyBac into the 5′UTR of hsp90 suggested that the AyFib-431a strain might lack its function. To determine whether this was the case, we crossed AyFib-431a heterozygotes and examined their F1 progeny. Approximately one quarter of the progeny showed lethality at the embryonic stage (data not shown), supporting this hypothesis.
hsp90 promoter/enhancer activity in the silkworm
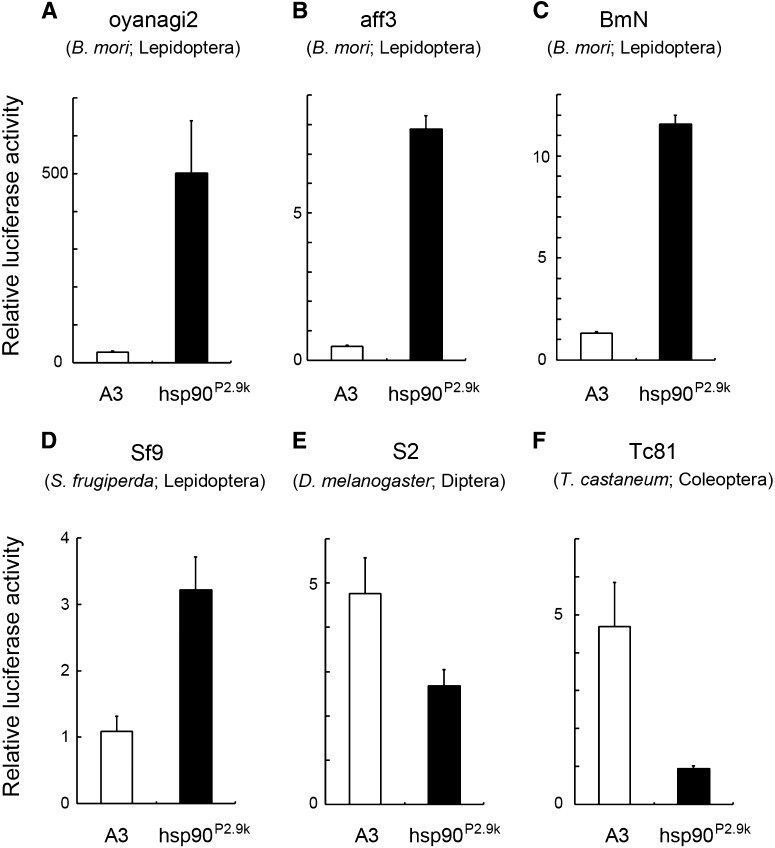
As piggyBac was inserted into the hsp90, a gene with a significant level of expression in various developmental stages and/or tissues (Figure S2, Sosalegowda et al. 2010), we hypothesized that the 3xP3 of AyFib-431a had trapped the hsp90 promoter/enhancer. We therefore attempted to identify the genomic region responsible for the strong expression of hsp90. In mammals and flies, hsp90 expression is regulated by a promoter/enhancer located in an upstream and/or intronic region (Xiao and Lis 1989; Shen et al. 1997; Zhang et al. 1999; Theodoraki et al. 2008). Because the silkworm hsp90 lacks intron (Figure 4, Landais et al. 2001), we focused on its 5′ flanking region. A 2.9-kb upstream genomic fragment of hsp90 (hsp90P2.9k) was amplified by PCR (Figure 4, see the section Materials and Methods) and its promoter/enhancer activity was examined via a luciferase reporter assay using various silkworm cell cultures. Strikingly, this fragment induced a high level of reporter gene expression in all of the Bombyx cells examined (Figure 5, A−C). In comparison to the A3 promoter, hsp90P2.9k showed 10 times or greater activity in various cultures (Figure 5, A−C).
Figure 5.
Transcriptional activation activity of Bombyx A3 and hsp90P2.9k in cell culture. The promoter was connected to the firefly luciferase gene, and the plasmid was transfected into the following cell lines to measure activity: (A) oyanagi2, (B) aff3, (C) BmN, (D) Sf9, (E) S2, and (F) Tc81.
We next examined the in vivo activity of hsp90P2.9k. For this assay, a transgenic silkworm expressing GFP under hsp90P2.9k was generated (hsp90-GFP strain, Figure 6A). We found that hsp90P2.9k-mediated GFP expression was present in all developmental stages (Figure 6, B−E′) and also in the various larval tissues in which we identified DsRed expression in AyFib-431a (Figure 7, A−L′). Interestingly, tissues with strong DsRed expression in AyFib-431a also expressed a high level of GFP in the hsp90-GFP strain (Figure 7, A−L′). The strong GFP expression in the ovary resulted in maternal inheritance similar to that seen for DsRed in the AyFib-431a strain (Figure 7, M−P′, compare with Figure 2, M−P′). Strong GFP expression in embryos was confirmed in four other independent hsp90-GFP strains (Figure S3), suggesting that the GFP expression was not due to a positional effect but was the result of hsp90P2.9k activity. We also compared the levels of GFP expression in hsp90-GFP and A3-GFP strains and found a considerably higher level of expression in the former (Figure 7, Q and Q′). Taken together, our findings indicate that the hsp90P2.9k fragment is responsible for the high level of expression of the Bombyx hsp90 gene in vivo and also suggest that the strong and ubiquitous DsRed expression of AyFib-431a was, in part at least, due to the activity of hsp90P2.9k.
Figure 6.
(A) Schematics of the vector used for transgenesis of hsp90-GFP. (B-E′) GFP expression in the hsp90-GFP strain. (B, C, D, E) Bright-field images. (B′, C′, D′, E′) GFP-fluorescent images. (B, B′) Embryo 7 d after oviposition. (C, C′) Third instar larva. (D, D′) Pupa. (E, E′) Adult. Bars represent 0.3 mm in (B), 1 mm in (C), and 3 mm in (D, E).
Figure 7.
GFP expression of hsp90-GFP in each tissue of final instar larvae (A−L′) or in the embryo (M−P′). (A, A′) Brain and corpora allata. (B, B′) Silk gland. (C, C′) Fat body. (D, D′) Prothoracic gland. (E, E′) Gut. (F, F′) Trachea. (G, G′) Malpighian tubules. (H, H′) Ventral nerves. (I, I′) Epidermis. (J, J′) Hemolymph. (K, K′) Testis. (L, L′) Ovary. (A, B, C, D, E, F, G, H, I, J, K, L) Bright-field images. (A′, B′, C′, D′, E′, F′, G′, H′, I′, J′, K′, L′) Green fluorescent protein (GFP) images. br, brain; ca, corpora allata. Arrows in (J′) indicate hemocyte cells. (M−N′) Eggs laid by hsp90-GFP heterozygous female crossed with w-c male. (M, M′) Just after oviposition. (N, N′’) 7 d after oviposition. Arrowheads indicate the putative GFP+ (hsp90-GFP/+), and arrows indicate GFP- (+/+) individuals. Residual maternal GFP protein can be detected in the putative +/+ embryos. (O−P′) Eggs laid by w-c female crossed with hsp90-GFP heterozygous male. (O, O′) Just after oviposition. (P, P′) 7 days after oviposition. Arrowheads indicate the putative GFP+ (hsp90-GFP/+) and arrows indicate GFP− (+/+) individuals. (Q, Q′) GFP expression in the day seven embryo of A3-GFP/+ individuals. (Q) Bright-field image. (Q′) GFP-fluorescent image. Note that (N′), (P′), and (Q′) are imaged under the same exposure conditions. Bars represent 0.25 mm in (A, I), 5 mm in (B, E), 2.5 mm in (C, G), 0.5 mm in (D, L), 1 mm in (F, H, K, M−Q), and 0.05 mm in (J).
hsp90P2.9k activity in cells of other insect species
We investigated whether the hsp90P2.9k fragment could also be used as a gene expression tool for other insect species. In Sf9 cells, which are derived from the lepidopteran Spodoptera frugiperda, hsp90P2.9k induced stronger gene expression than Bombyx A3 by a factor of more than three (Figure 5D). A previous analysis showed that Bombyx A3 exhibited substantial promoter activity in the larvae of the swallowtail butterfly Papilio xuthus (Ando and Fujiwara 2013). Therefore, we conclude that hsp90P2.9k can act as a strong and/or ubiquitous promoter/enhancer in Lepidoptera. However, hsp90P2.9k showed weaker activity than Bombyx A3 in the fruit fly and red flour beetle cells (Figure 5, E and F). It would therefore be necessary to improve its function in order to use this fragment as a significant gene expression tool in non-Lepidopteran insects.
The promoter/enhancer activity of hsp90P2.9k partial fragment
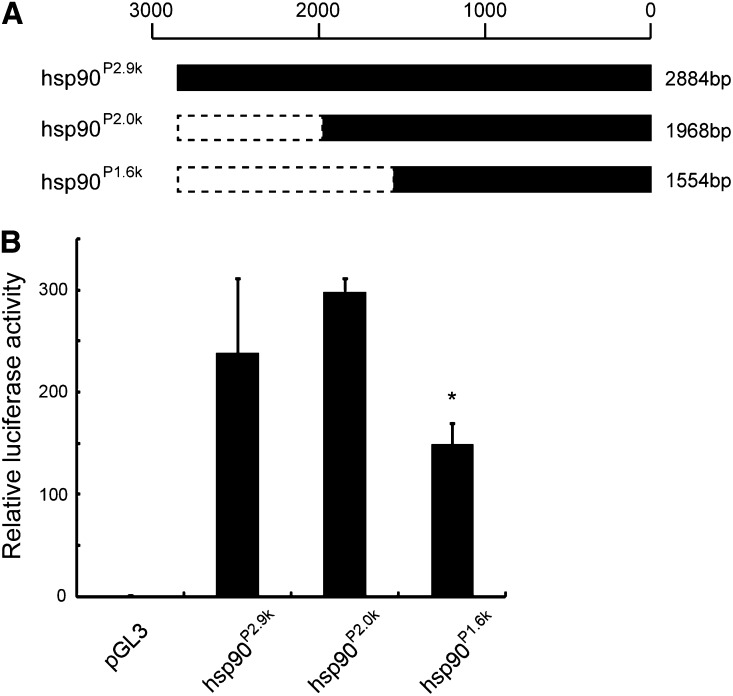
As shown above, we revealed that hsp90P2.9k fragment can be used as a valuable tool for various research fields in the silkworm and other related species. However, this fragment is 2.9 kb in length; it would be more preferable to minimize this fragment for use. Thus, we deleted the fragment and investigated whether the strong promoter/enhancer activity is still present. In case that the 5′ ∼900 bp was removed (hsp90P2.0k), its transcription activation activity was almost comparable to that of the full-length (Figure 8). Therefore, this 2.0-kb fragment can potentially be used as a tool for strong gene expression instead of hsp90P2.9k. However, the removal of additional ∼400-bp sequence (hsp90P1.6k) resulted in the significant reduction of the activity (Figure 8). We conclude that the hsp90P2.0k would be the current minimum fragment that is applicable to the induction of strong gene expression.
Figure 8.
Promoter activity of hsp90P2.9k and its partial fragment. (A) Schematics of the promoter fragment used in this analysis. The name of each fragment is shown to the left, and the nucleotide length is shown to the right. (B) The transcription activation activity of each fragment. The promoter was connected to the firefly luciferase gene, and the plasmid was transfected into the oyanagi2 cells to measure activity. For pGL3, just the empty vector was transfected. *P < 0.05 vs. hsp90P2.9k (Welch’s t-test).
Discussion
Recent progress in transgenic techniques has provided the means to perform a wide range of gene functional analyses. These techniques have now been applied to a range of insect species, such as D. melanogaster (Diptera), Ceratitis capitata (Diptera), Anopheles gambiae (Diptera), A. stephensi (Diptera), B. mori (Lepidoptera), Plutella xylostella (Lepidoptera), Cydia pomonella (Lepidoptera), T. castaneum (Coleoptera), Athalia rosae (Hymenoptera), and Gryllus bimaculatus (Orthoptera) (Rubin and Spradling 1982; Miller et al. 1987; Catteruccia et al. 2000; Tamura et al. 2000; Lorenzen et al. 2003; Sumitani et al. 2003; Nakamura et al. 2010; Ferguson et al. 2011; Martins et al. 2012). However, at present, a restricted set of promoters are available for functional studies except for D. melanogaster. In the silkworm, for example, the A3 and ie1 promoters show limited activity in some tissues and/or developmental stages (Uchino et al. 2008; Masumoto et al. 2012). This restricts the use of these promoters as a tool for strong and widespread gene expression. The A3 promoter has another drawback in that it is very susceptible to positional effects (Uchino et al. 2006, 2008). The promoter identified here, hsp90P2.9k, offers the possibility of strong gene expression across many cell and tissue types and in different developmental stages. Its ability to induce gene expression in a wide variety of larval tissues should be of especial value because this will provide greater opportunity for in vivo gene function analyses. Use of hsp90P2.9k will enable functional studies in tissues such as the ovary and testis in which no active promoters have been available to date. Its activity in the ovary and the maternal transmission of transcripts and/or proteins will be of value to the analysis of early embryogenesis. Similar maternal promoters were previously not available in insects except for D. melanogaster (Rørth 1998). We also showed that hsp90P2.9k is less sensitive to the positional effects (Figure S3). In addition to functional analyses, hsp90P2.9k could be used for transgenic marker gene expression. Because this fragment induced very strong and ubiquitous gene expression in embryos (Figure 6B′), its employment would make screening much more efficient. It should also be possible to use hsp90P2.9k for more effective production of the recombinant proteins, such as drugs or other materials, through transgene expression. Taken together, exploitation of this novel promoter/enhancer might be expected to benefit research across both basic and practical sciences. In an attempt to minimize this fragment, we found that ∼2.0 kb of hsp90P2.9k would be sufficient for the induction of strong gene expression (Figure 8). We are now in the process for further determining the core element of this fragment.
The hsp90P2.9k promoter/enhancer identified here is also of relevance to studies in species other than the silkworm. As described previously, the in vivo functional analysis is now possible in a number of insect species. In addition, such analysis should also be feasible even in nonmodel organisms because of the recent development of a method for rapid and efficient gene introduction, namely, electroporation-mediated somatic transgenesis, (Ando and Fujiwara 2013). The development and exploitation of versatile and/or universal promoters is thus a topic of considerable interest. In this context, our result that a novel strong and ubiquitous promoter was identified in the silkworm, a non-Drosophilid insect, should have a significant implication. This promoter/enhancer can be applicable to multiple purposes in the silkworm research, as described. Moreover, we consider that hsp90P2.9k showed clear evidence of being use in studies of other lepidopteran insects because: 1) it exhibited more activity than A3 promoter in both silkworm and armyworm cell cultures (Figure 5, A−D); 2) Bombyx A3 has been shown to be active in the swallowtail butterfly (Ando and Fujiwara 2013); and 3) hsp90 is expressed strongly and ubiquitously in various Lepidoptera (Sonoda et al. 2006a,b, 2007; Tungjitwitayakul et al. 2008; Gkouvitsas et al. 2009; Zhang and Denlinger 2010; Xu et al. 2011). The widespread activity of hsp90P2.9k in the silkworm indicates that this fragment is also likely to be ubiquitously active in other lepidopteran species. However, use of Bombyx hsp90P2.9k in non-Lepidopteran insects might be more problematic because it showed weak activity in Drosophila and Tribolium cells (Figure 5, E and F). Weak expression driven by a Bombyx enhancer in distantly related insect species could be due to evolutionary divergence of the core transcription machinery, as has been suggested recently for T. castaneum (Schinko et al. 2010). To more generally test the capacity of hsp90 promoters to drive ubiquitous expression, it might be therefore necessary to isolate the respective endogenous promoters in the species of interest.
The regulation of hsp90 gene transcription has been studied previously in the yeast, fungus, fruit fly, medfly, mouse, and humans (Xiao and Lis 1989; Giardina and Lis 1995; Dale et al. 1997; Shen et al. 1997; Erkine et al. 1999; Zhang et al. 1999; Theodoraki et al. 2008; Lamoth et al. 2014). These studies have reported that transcription of the gene is activated by a transcription factor, heat shock factor (HSF), that binds to the cis-element of hsp90 [heat shock element (HSE)] (Giardina and Lis 1995; Shen et al. 1997; Erkine et al. 1999). HSF also regulates the expression of other genes, such as hsp70, via HSEs whose consensus sequences are identical to that of hsp90 (Wu 1984). Studies in the silkworm have shown that HSF can bind to the HSE of hsp70 or samui genes, suggesting the possible conservation of HSF function in this species (Kihara et al. 2011). We analyzed Bombyx hsp90P2.9k or hsp90P2.0k and found several HSE-like sequences (Figure S4). Thus, it is very likely that these elements play critical roles for hsp90 transcription. Previously, we found that a 108-bp upstream sequence of Bombyx hsp90 that does not contain any HSEs shows no promoter activity (Figure S4, Uchino et al. 2006); this observation is suggestive of a possible role for HSE-like sequences. Interestingly, a search of lepidopteran genomes showed that these elements are also present upstream of hsp90 gene in species such as Manduca sexta, Danaus plex, and Heliconius melpomene (data not shown). Apart from HSEs, very little is known about cis regulatory elements for hsp90. In D. melanogaster it has been suggested that a 7 bp module (CGTTTTG) might play a role in expression of hsp90 in the ovary (Xiao and Lis 1989), but this element does not appear to be present in the medfly (Theodoraki et al. 2008). The silkworm likewise does not have this element despite the fact that Bombyx hsp90P2.9k can induce strong gene expression in the ovary (Figure 7L′, data not shown). One possibility is that the strong hsp90 expression in the ovary might be mediated by species-specific mechanisms or strategies.
This study has shown that a Bombyx hsp90 upstream fragment can be used as an effective gene expression tool in both Bombyx and closely related species. Our analyses also demonstrated that use of this fragment was effectively limited to Lepidoptera. We are currently attempting to identify further versatile promoters that can be used in a wide range of organisms. Exploitation of emerging genomic information and/or gene expression data, in combination with a variety of in vivo analytical tools, should allow us to identify further useful promoters.
Supplementary Material
Acknowledgments
We thank Mr. Kaoru Nakamura, Mr. Koji Hashimoto, and Mr. Toshihiko Misawa for maintaining the silkworms and Ms. Yoshiko Mise and Ms. Satoko Kawamoto for their technical assistance. This work was supported by grants-in-aid from the Ministry of Agriculture, Forestry, and Fisheries.
Footnotes
Communicating editor: M. Boutros
Literature Cited
- Ando T., Fujiwara H., 2013. Electroporation-mediated somatic transgenesis for rapid functional analysis in insects. Development 140: 454–458. [DOI] [PubMed] [Google Scholar]
- Berghammer A. J., Klingler M., Wimmer E. A., 1999. A universal marker for transgenic insects. Nature 402: 370–371. [DOI] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Shepherd S., Lee K., McCall K., et al. , 1989. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 3: 1273–1287. [DOI] [PubMed] [Google Scholar]
- Catteruccia F., Nolan T., Loukeris T. G., Blass C., Savakis C., et al. , 2000. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature 405: 959–962. [DOI] [PubMed] [Google Scholar]
- Cooley L., Kelley R., Spradling A., 1988. Insertional mutagenesis of the Drosophila genome with single P elements. Science 239: 1121–1128. [DOI] [PubMed] [Google Scholar]
- Dai H., Ma L., Wang J., Jiang R., Wang Z., et al. , 2008. Knockdown of ecdysis-triggering hormone gene with a binary UAS/GAL4 RNA interference system leads to lethal ecdysis deficiency in silkworm. Acta Biochim. Biophys. Sin. (Shanghai) 40: 790–795. [PubMed] [Google Scholar]
- Daimon T., Kozaki T., Niwa R., Kobayashi I., Furuta K., et al. , 2012. Precocious metamorphosis in the juvenile hormone-deficient mutant of the silkworm, Bombyx mori. PLoS Genet. 8: e1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale E. C., Yang X., Moore S. K., Shyamala G., 1997. Murine 86-kDa heat shock protein gene and promoter. Cell Stress Chaperones 2: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingsen S., Laplante M. A., König M., Kikuta H., Furmanek T., et al. , 2005. Large-scale enhancer detection in the zebrafish genome. Development 132: 3799–3811. [DOI] [PubMed] [Google Scholar]
- Erkine A. M., Magrogan S. F., Sekinger E. A., Gross D. S., 1999. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol. Cell. Biol. 19: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson H. J., Neven L. G., Thibault S. T., Mohammed A., Fraser M., 2011. Genetic transformation of the codling moth, Cydia pomonella L., with piggyBac EGFP. Transgenic Res. 20: 201–214. [DOI] [PubMed] [Google Scholar]
- Giardina C., Lis J. T., 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 15: 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkouvitsas T., Kontogiannatos D., Kourti A., 2009. Expression of the Hsp83 gene in response to diapause and thermal stress in the moth Sesamia nonagrioides. Insect Mol. Biol. 18: 759–768. [DOI] [PubMed] [Google Scholar]
- Imamura M., Nakai J., Inoue S., Quan G. X., Kanda T., et al. , 2003. Targeted gene expression using the GAL4/UAS system in the silkworm Bombyx mori. Genetics 165: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Kidokoro K., Sezutsu H., Nohata J., Yamamoto K., et al. , 2008. Deletion of a gene encoding an amino acid transporter in the midgut membrane causes resistance to a Bombyx parvo-like virus. Proc. Natl. Acad. Sci. USA 105: 7523–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Eiguchi M., Kurata N., 2004. Establishment of an enhancer trap system with Ds and GUS for functional genomics rice. Mol. Genet. Genomics 271: 639–650. [DOI] [PubMed] [Google Scholar]
- Kayukawa T., Minakuchi C., Namiki T., Togawa T., Yoshiyama M., et al. , 2012. Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. USA 109: 11729–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayukawa T., Tateishi K., Shinoda T., 2013. Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Sci. Rep. 3: 1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara F., Niimi T., Yamashita O., Yaginuma T., 2011. Heat shock factor binds to heat shock elements upstream of heat shock protein 70a and Samui genes to confer transcriptional activity in Bombyx mori diapause eggs exposed to 5 °C. Insect Biochem. Mol. Biol. 41: 843–851. [DOI] [PubMed] [Google Scholar]
- Lamoth F., Juvvadi P. R., Gehrke C., Asfaw Y. G., Steinbach W. J., 2014. Transcriptional activation of heat shock protein 90 mediated via a proximal promoter region as trigger of caspofungin resistance in Aspergillus fumigatus. J. Infect. Dis. 209: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais I., Pommet J.-M., Mita K., Nohata J., Gimenez S., et al. , 2001. Characterization of the cDNA encoding the 90 kDa heat-shock protein in the Lepidoptera Bombyx mori and Spodoptera frugiperda. Gene 271: 223–231. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Craig E. A., 1988. The heat shock proteins. Annu. Rev. Genet. 22: 631–677. [DOI] [PubMed] [Google Scholar]
- Liu P.-P., Koizuka N., Homrichhausen T. M., Hewitt J. R., Martin R. C., et al. , 2005. Large-scale screening of Arabidopsis enhancer-trap lines for seed germination-associated genes. Plant J. 41: 936–944. [DOI] [PubMed] [Google Scholar]
- Lorenzen M. D., Berghammer A. J., Brown S. J., Denell R. E., Klinger M., et al. , 2003. piggyBac-mediated germline transformation in the beetle Tribolium castaneum. Insect Mol. Biol. 12: 433–440. [DOI] [PubMed] [Google Scholar]
- Lorenzen M. D., Kimzey T., Shippy T. D., Brown S. J., Denell R. E., et al. , 2007. piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol. Biol. 16: 265–275. [DOI] [PubMed] [Google Scholar]
- Ma S., Zhang S., Wang F., Liu Y., Liu Y., et al. , 2012. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS ONE 7: e45035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S., Naish N., Walker A. S., Morrison N. I., Scaife S., et al. , 2012. Germline transformation of the diamondback moth, Plutella xylostella L., using the piggyBac transposable element. Insect Mol. Biol. 21: 414–421. [DOI] [PubMed] [Google Scholar]
- Masumoto M., Ohde T., Shiomi K., Yaginuma T., Niimi T., 2012. A baculovirus immediate-early gene, ie1, promoter drives efficient expression of a transgene in both Drosophila melanogaster and Bombyx mori. PLoS One 7: e49323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. H., Sakai R. K., Romans P., Gwadz R. W., Kantoff P., et al. , 1987. Stable integration and expression of a bacterial gene in the mosquito, Anopheles gambiae. Science 237: 779–781. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yoshizaki M., Ogawa S., Okamoto H., Shinmyo Y., et al. , 2010. Imaging of transgenic cricket embryos reveals cell movements consistent with a syncytial patterning mechanism. Curr. Biol. 20: 1641–1647. [DOI] [PubMed] [Google Scholar]
- O’Brochta D. A., Alford R. T., Pilitt K. L., Aluvihare C. U., Harrell R. A., 2011. piggyBac transposon remobilization and enhancer detection in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 108: 16339–16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Suzuki Y., 1977. Cloning of the silk fibroin gene and its flanking sequences. Proc. Natl. Acad. Sci. USA 74: 5363–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane C. J., Gehring W. J., 1987. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 84: 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S., Kondrichin I., Korzh V., Emelyanov A., 2004. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231: 449–459. [DOI] [PubMed] [Google Scholar]
- Robinson A. S., Franz G., Atkinson P. W., 2004. Insect transgenesis and its potential role in agriculture and human health. Insect Biochem. Mol. Biol. 34: 113–120. [DOI] [PubMed] [Google Scholar]
- Rørth P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Sajwan S., Takasu Y., Tamura T., Uchino K., Sezutsu H., et al. , 2013. Effieicnt disruption of endogenous Bombyx gene by TAL effector nucleases. Insect Biochem. Mol. Biol. 43: 17–23. [DOI] [PubMed] [Google Scholar]
- Schinko J. B., Weber M., Viktorinova I., Kiupakis A., Averof M., et al. , 2010. Functionality of the GAL4/UAS system in Tribolium requires the use of endogenous core promoters. BMC Dev. Biol. 10: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezutsu H., Uchino K., Kobayashi I., Tatematsu K. I., Iizuka T., et al. , 2009. Conservation of fibroin gene promoter function between the domesticated silkworm Bombyx mori and the wild silkmoth Antheraea yamamai. J. Insect Biotechnol. Sericology 78: 1–10. [Google Scholar]
- Shen Y., Liu J., Wang X., Cheng X., Wang Y., et al. , 1997. Essential role of the first intron in the transcription of hsp90β gene. FEBS Lett. 413: 92–98. [DOI] [PubMed] [Google Scholar]
- Shimomura M., Minami H., Suetsugu Y., Ohyanagi H., Satoh C., et al. , 2009. KAIKObase: Anintegrated silkworm genome database and data mining tool. BMC Genomics 10: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda S., Ashfaq M., Tsumuki H., 2006a Cloning and nucleotide sequencing of three heat shock protein genes (hsp90, hsc70, and hsp19.5) from the damondback moth, Plutella xylostella (L.) and their expression in relation to developmental stage and temperature. Arch. Insect Biochem. Physiol. 62: 80–90. [DOI] [PubMed] [Google Scholar]
- Sonoda S., Fukumoto K., Izumi Y., Yoshida H., Tsumuki H., 2006b Cloning of heat shock protein genes (hsp90 and hsc70) and their expression during larval diapause and cold tolerance acquisition in the rice stem borer, Chilo suppressalis walker. Arch. Insect Biochem. Physiol. 63: 36–47. [DOI] [PubMed] [Google Scholar]
- Sonoda S., Ashfaq M., Tsumuki H., 2007. A comparison of heat shock protein genes from cultured cells of the cabbage armyworm, Mamestra brassicae, in response to heavy metals. Arch. Insect Biochem. Physiol. 65: 210–222. [DOI] [PubMed] [Google Scholar]
- Sosalegowda A. H., Kundapur R. R., Boregowda M. H., 2010. Molecular characterization of heat shock proteins 90 (HSP83?) and 70 in tropical strains of Bombyx mori. Proteomics 10: 2734–2745. [DOI] [PubMed] [Google Scholar]
- Sumitani M., Yamamoto D. S., Oishi K., Lee J. M., Hatakeyama M., 2003. Germline transformation of the sawfly, Athalia rosae (Hymenoptera: Symphyta), mediated by a piggyBac-derived vector. Insect Biochem. Mol. Biol. 33: 449–458. [DOI] [PubMed] [Google Scholar]
- Takasu Y., Kobayashi I., Beumer K., Uchino K., Sezutsu H., et al. , 2010. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem. Mol. Biol. 40: 759–765. [DOI] [PubMed] [Google Scholar]
- Takasu Y., Sajwan S., Daimon T., Osanai-Futahashi M., Uchino K., et al. , 2013. Efficient TALEN construction for Bombyx mori gene targeting. PLoS ONE 8: e73458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Thibert C., Royer C., Kanda T., Abraham E., et al. , 2000. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat. Biotechnol. 18: 81–84. [DOI] [PubMed] [Google Scholar]
- Tamura T., Kuwabara K., Uchino K., Kobayashi I., Kanda T., 2007. An improved DNA injection method for silkworm eggs drastically increases the efficiency of producing transgenic silkworms. J. Insect Biotechnol. Sericol. 76: 155–159. [Google Scholar]
- Tanaka H., Fujita K., Sagisaka A., Tomimoto K., Imanishi S., et al. , 2009. shRNA expression plasmids by a novel method efficiently induce gene-specific knockdown in a silkworm cell line. Mol. Biotechnol. 41: 173–179. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Sagisaka A., Fujita K., Furukawa S., Ishibashi J., et al. , 2012. BmEts upregulates promoter activity of lebocin in Bombyx mori. Insect Biochem. Mol. Biol. 42: 474–481. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Kobayashi I., Uchino K., Sezutsu H., Iizuka T., et al. , 2010. Construction of a binary transgenic gene expression system for recombinant protein production in the middle silk gland of the silkworm Bombyx mori. Transgenic Res. 19: 473–487. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Sezutsu H., Tamura T., 2012. Utilization of transgenic silkworms for recombinant protein production. J. Biotechnol. Biomater. S9: 004. [Google Scholar]
- The International Silkworm Genome Consortium , 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38: 1036–1045. [DOI] [PubMed] [Google Scholar]
- Theodoraki M., Tatari M., Chrysanthis G., Zacharopoulou A., Mintzas A. C., 2008. Structural characterization of the medfly hsp83 gene and functional analysis of its proximal promoter region in vivo by germ-line transformation. Arch. Insect Biochem. Physiol. 67: 20–35. [DOI] [PubMed] [Google Scholar]
- Thomas J.-L., Rocha M. D., Besse A., Mauchamp B., Chavancy G., 2002. 3xP3-EGFP marker facilitates screening for transgenic silkworm Bombyx mori L. from the embryonic stage onwards. Insect Biochem. Mol. Biol. 32: 247–253. [DOI] [PubMed] [Google Scholar]
- Tsubota T., Shimomura M., Ogura T., Seino A., Nakakura T., et al. , 2010. Molecular characterization and functional analysis of novel carboxyl/cholinesterases with GQSAG motif in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 40: 100–112. [DOI] [PubMed] [Google Scholar]
- Tsubota T., Uchino K., Kamimura M., Ishikawa M., Hamamoto H., et al. , 2014. Establishment of transgenic silkworms expressing GAL4 specifically in the haemocyte oenocytoid cells. Insect Mol. Biol. 23: 165–174. [DOI] [PubMed] [Google Scholar]
- Tungjitwitayakul J., Tatun N., Singtripop T., Sakurai S., 2008. Characteristic expression of three heat shock-responsive genes during larval diapause in the bamboo borer Omphisa fuscidentalis. Zoolog. Sci. 25: 321–333. [DOI] [PubMed] [Google Scholar]
- Uchino K., Imamura M., Sezutsu H., Kobayashi I., Kojima K., et al. , 2006. Evaluating promoter sequences for trapping an enhancer activity in the silkworm Bombyx mori. J. Insect Biotechnol. Sericol. 75: 89–97. [Google Scholar]
- Uchino K., Sezutsu H., Imamura M., Kobayashi I., Tatematsu K. I., et al. , 2008. Construction of a piggyBac-based enhancer trap system for the analysis of gene function in silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38: 1165–1173. [DOI] [PubMed] [Google Scholar]
- Uhlířová M., Asahina M., Riddiford L. M., Jindra M., 2002. Heat-inducible transgenic expression in the silkmoth Bombyx mori. Dev. Genes Evol. 212: 145–151. [DOI] [PubMed] [Google Scholar]
- Vijaybhaskar V., Subbiah V., Kaur J., Vijayakumar P., Siddiqi I., 2008. Identification of a root-specific glycosyltransferase from Arabidopsis and characterization of its promoter. J. Biosci. 33: 185–193. [DOI] [PubMed] [Google Scholar]
- Wilson C., Pearson R. K., Bellen H. J., O’Kane C. J., Grossniklaus U., et al. , 1989. P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 3: 1301–1313. [DOI] [PubMed] [Google Scholar]
- Wu C., 1984. Two protein-binding sites in chromatin implicated in the activation of heat shock genes. Nature 309: 229–234. [DOI] [PubMed] [Google Scholar]
- Xiao H., Lis J. T., 1989. Heat shock and developmental regulation of the Drosophila melanogaster hsp83 gene. Mol. Cell. Biol. 9: 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Zou Q., Zheng H., Zhang F., Tang B., et al. , 2011. Three heat shock proteins from Spodoptera exigua: Gene cloning, characterization and comparative stress response during heat and cold shocks. Comp. Biochem. Physiol. Part B 159: 92–102. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Denlinger D. L., 2010. Molecular characterization of heat shock protein 90, 70 and 70 cognate cDNAs and their expression patterns during thermal stress and pupal diapause in the corn earworm. J. Insect Physiol. 56: 138–150. [DOI] [PubMed] [Google Scholar]
- Zhang S., Yu J., Cheng X., Ding L., Heng F., et al. , 1999. Regulation of human hsp90α gene expression. FEBS Lett. 444: 130–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.