Abstract
We focus on the application of constraint-based methodologies and, more specifically, flux balance analysis in the field of metabolic engineering, and enumerate recent developments and successes of the field. We also review computational frameworks that have been developed with the express purpose of automatically selecting optimal gene deletions for achieving improved production of a chemical of interest. The application of flux balance analysis methods in rational metabolic engineering requires a metabolic network reconstruction and a corresponding in silico metabolic model for the microorganism in question. For this reason, we additionally present a brief overview of automated reconstruction techniques. Finally, we emphasize the importance of integrating metabolic networks with regulatory information—an area which we expect will become increasingly important for metabolic engineering—and present recent developments in the field of metabolic and regulatory integration.
Keywords: Flux balance analysis, Metabolic engineering, Metabolic reconstructions, Automated network reconstruction, Gene regulatory networks
Introduction
Organisms natively use metabolic, mostly enzymatically catalyzed reactions to convert raw materials into the essential substances that are needed for the survival of their cells. As such, they represent a tremendous resource of existing biological machinery to carry out biochemical transformations. Metabolic engineering involves the process of modifying the metabolic potential and genetics of a microorganism to our advantage to increase the production of a specific substance of interest [91]. The objective of metabolic engineering is thus to reroute metabolism towards a pathway of interest to improve production of commercially valuable chemicals on an industrial scale. This has been achieved for several commodities, including fuels, pharmaceuticals, drinks such as wine and beer, fine chemicals and diesels. In short, many biotechnological products are being produced using microbial strains as cell factories [3, 9, 19, 37, 53, 79], with an increasing number on the horizon [35, 70, 104, 107, 109].
Traditionally, metabolism was altered using classical breeding and random mutagenesis, followed by selection and screening [65]. More recently, however, the introduction of recombinant DNA techniques has allowed the application of targeted genetic changes [47, 111] through gene knockouts, overexpression, and expression of heterologous genes [50]. In large part owing to the advent of genomics and systems biology, we nowadays have a number of new tools that generate a wealth of data for analysis, contributing to our understanding of metabolism and cellular behavior. Improved knowledge and new analytical tools [14, 67, 68] are increasingly available for use in the development of novel microbial strains with phenotypes that allow production of various bulk chemicals [74, 113]. Successful applications, for example using the model organisms Escherichia coli, Saccharomyces cerevisiae and Corynebacterium glutamicum (for amino acid production mainly) as production hosts, have been reported widely in the literature [52, 103].
Metabolic engineering focuses on altering the function of enzymes, transporters, or regulatory proteins informed by existing knowledge of the metabolic network, enzymes, their encoding genes, and overall regulation [59]. Strategies focus on either introducing new metabolic enzyme functions and pathways or altering existing metabolic pathways to optimize production of the chemical of interest [47]. For either strategy, detailed understanding of the network and a way to determine the distribution of flux [96] are necessary. Metabolic analysis methods are powerful analytical tools that can be utilized extensively in metabolic engineering, as they allow exploration and detailed consideration of the structure and design of a metabolic network [83]. Stoichiometric methods in particular, which are based on collecting all the available biochemical knowledge surrounding a particular metabolic network of an organism, have helped to construct a collection of metabolic models for an expanding number of microorganisms based on annotated genome sequences. Such models allow researchers to conduct simulations based on all known reactions occurring in the metabolic network of an organism using only the knowledge of the stoichiometry of the network as input and, thus, make computational predictions for achievable metabolic states of an organism under varying conditions. These predictions can encompass the outcomes of genetic manipulations, including but not limited to removal or addition of reactions to the network. The capability to perform such manipulations and simulate the results computationally forms the basis for rational metabolic engineering [61] and provides an aid for prospective study design [30, 44].
Here, we review applications and successes of genome-scale modeling for metabolic engineering, provide an overview of the metabolic reconstruction process (particularly the tools for automated reconstructions), and briefly offer our view on future developments of the field.
The flux balance analysis (FBA) formulation
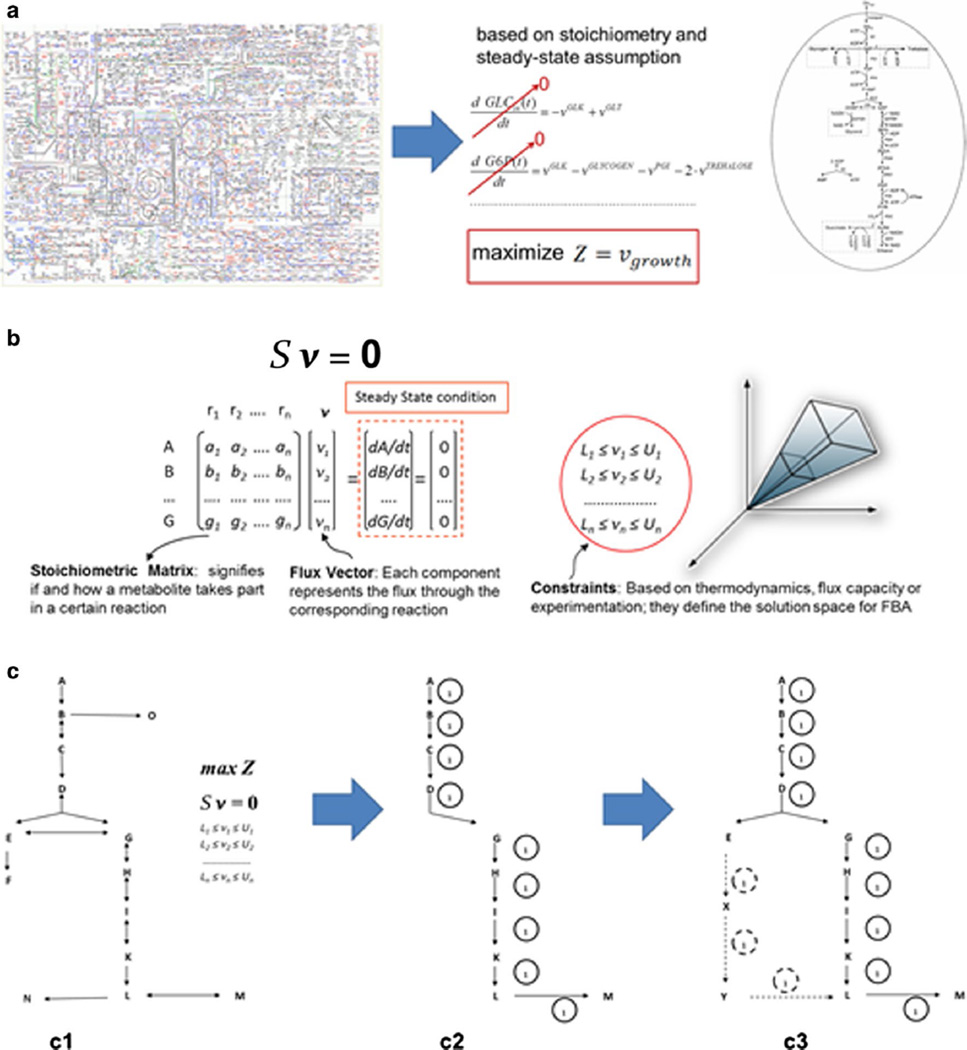
Flux balance analysis (FBA) (Fig. 1) can trace its foundations as far back as in the late 1960s [85, 102] and was popularized in the early 1990s [80, 81, 98–100]. FBA is a constraint-based optimization approach that can be used to simulate ranges of achievable reaction rates (referred to typically in this field as metabolic “fluxes”) in the metabolic network of an organism. The available stoichiometric information for a metabolic network is incorporated into a stoichiometric matrix S, in which rows represent metabolites and columns represent reactions. Typically, the network is assumed to exist in a quasi-steady state, represented by Sv = 0, where vector v represents the fluxes through each reaction. Lower and upper bounds can be applied wherever additional information is available for the fluxes of the reactions, or to impose directionality and capacity requirements for some or all reactions.
Fig. 1.
Conceptual illustration of flux balance analysis formulation and solution. a Reconstruction of a genome-scale metabolic network is performed by mathematically representing the flux through the reactions of the network. b The stoichiometric matrix for the system is constructed to represent the stoichiometry of all reactions, and the mathematical formulation for FBA is based on the steady-state condition. These stoichiometric constraints coupled with minimum and maximum bounds on reaction rates define the steady-state solution space. c FBA provides a method for calculating achievable fluxes through the system (c2), based only on the knowledge of the stoichiometry of a metabolic network (c1). Through simulations, alternative solutions can also be identified and/or the effects of alterations to the network, such as gene deletions or additions, can be predicted (c3). The “1” in the graph signifies that a reaction is “on”, i.e., there is flux through it
The system typically remains under-determined, with many alternative solutions for flux distribution that satisfy the imposed constraints. An optimal distribution is selected by optimizing an objective function, which usually describes the maximization of biomass production, based on the assumption that cells use the available food sources to optimize cellular growth. FBA formulations are often characterized by degeneracy, meaning that there exist multiple equivalent, non-unique optimal solutions [65, 73] to the problem. A typical FBA formulation maximizes the selected objective function (a subset of the fluxes in the system) subject to stoichiometric constraints and any necessary bounds on system fluxes:
Maximize → wv
- subject to:
- Sv = 0
- vmin ≤ v ≤ vmax
Vector w incorporates weights that represent the relative contribution of each reaction to the objective function. FBA formulations constitute linear programming (LP) problems, which makes the FBA approach suitable for application to very large metabolic networks. Typically, genome-scale metabolic networks consist of hundreds or a few thousand reactions. LP solvers are capable of solving problems with tens of thousands or more variables. The solution of an FBA problem is unique for the optimal value of the objective function, and also results in a non-unique (except in trivial cases) calculation of a flux distribution through every reaction in the system. Subsequently, patterns of consumption and production of each metabolite can be determined for systems with thousands or tens of thousands of components. Crucially, kinetic information or enzyme concentrations are not required for the analysis; although such information can be incorporated for increased accuracy. This lack of a high number of parameters greatly reduces the opportunities for overfitting models—although some overfitting certainly still exists, for example in the choices during model construction—and makes the resulting models amenable to very broad use across a wide range of organisms at the genome scale. Additional methods such as Flux Variability Analysis [55] or Monte Carlo sampling of solution spaces [4, 73] can address the variability possible in each of these reaction fluxes, providing insight to the full range of achievable metabolic states of a system given physico-chemical constraints and a finite set of biological measurements.
Genome-scale reconstructions
The reconstruction of genome-scale metabolic models requires the construction of an S matrix that closely represents the biochemistry of the organism. Models for an ever increasing number of bacteria have been published in recent years [58] (examples: [6, 12, 25, 31, 32, 34, 57, 60, 63, 82, 86, 110]) and more papers describing both new reconstructions and improvements upon previous iterations are published regularly. Most reconstructions are now available in a standard format such as the systems biology markup language (SBML) [36]. The SBML files can easily be imported into most software applications for FBA, such as the COBRA Toolbox [10]. Nevertheless, wherever a preexisting model is not readily available (including when the existing model is not of the necessary quality or does not cover the required elements of metabolism for the intended analysis), a new reconstruction is needed. This process is data intensive and involves gathering species-specific information from genome annotations, high-throughput experiments, the literature and/or publically available databases, such as KEGG [41], EcoCyc [42], BKM-react [46], or BRENDA [84]. Gap-filling methodologies are subsequently applied [13, 75] to improve connectivity to the point where the model can simulate phenotypes. As labor intensive as manual reconstruction is, the process has been well developed and described [95].
Automated reconstructions
As pointed out above, the construction of a genome-scale model is a complex task; but tools for improving and accelerating this process are becoming increasingly available. To reduce the painstaking process of manual annotation, draft metabolic models can be built by utilizing and integrating the resources available in various biological databases in an automated manner. Several such automated methods have been reported in the literature; for example, Model SEED [24, 33] is an online resource designed to simplify the construction of a genome-scale model by utilizing an automated framework. Model SEED can be used to create genome-scale metabolic models in a high-throughput manner, by automating the annotation of the genome, producing a preliminary reconstruction of the metabolic network, performing automatic gap filling of reactions necessary for cellular growth, and, when such data are available, incorporating array and gene essentiality data to improve the quality of the reconstruction. BioNetBuilder [8] is a Cytoscape plugin with a user-friendly interface to create biological networks integrated from several databases. ReMatch [71] is a web-based framework that reconstructs a metabolic network by integrating user-developed models into a database collected from several comprehensive metabolic data resources, including KEGG, MetaCyc and CheBI. The SuBliMinaL Toolbox [93] is a framework for reconstructing metabolic networks by providing independent modules that can be used individually or in a pipeline, and can perform tasks that are common in every reconstruction process, such as generating a draft, determining metabolite protonation states, mass-balancing reactions, compartmentalizing the cell, adding transport reactions, creating a biomass function and exporting the reconstruction in a format readable by software packages (typically SBML). Reyes et al. [77] presented an automatic method for the reconstruction of genome-scale metabolic models for any organism implemented in COPABI. Dale et al. [23] developed a method for predicting metabolic pathways that relies on machine learning approaches to reconstruct the network of an organism. In addition to automated tools, there have also been instances of semi-automated tools in the literature, for example reconstruction, analysis and visualization of metabolic networks (RAVEN) [2] is a toolbox for semi-automated reconstruction of genome-scale models, which accesses published models and the KEGG database to build a draft reconstruction, coupled with extensive gap filling and quality control. Microbes Flux [28] and a method presented by Zhou [112] both make extensive use of KEGG to achieve the construction of a draft metabolic model. Finally, Benedict et al. [13] presented a likelihood-based gap filling method that can automatically improve the quality of metabolic reconstructions by incorporating alternative potential gene annotations. This method assigns a score to gene annotations based on sequence homology, selects the most likely pathways for gap filling using an mixed integer linear programming (MILP) formulation and identifies orphaned reactions. The likelihood-based approach performs better both quantitatively and qualitatively when compared to pre-existing algorithms.
While automated methods significantly decrease the time and effort required for reconstructing a new metabolic model, there is still need for user feedback and manual curation to improve the quality and accuracy of the metabolic model. This is especially true during the final stages of the reconstruction, as the resulting model is being validated against experimental data. The curator is responsible for assessing the precision and accuracy of the model, and for evaluating if there is further need for gap filling, removing futile cycles and improvement of the biomass reaction. Semi-automated methods permit greater flexibility for user intervention during the reconstruction process and constitute a good compromise for refining an initial draft model to further elevate the quality of the reconstruction up to the required standards.
Once a working model has been constructed and improved to a satisfactory level, in silico experiments can predict flux distribution ranges and phenotypic behavior under conditions of the user’s choice. Targets for possible genetic manipulation to improve strain performance can be identified through comparative studies under both genetic and environmental perturbations. The model can then be used to calculate knockout lethality or growth rates, and results can be compared to experimental observations, which allows for the model to be iteratively tested and improved [40]. Several computational approaches for network manipulation and phenotypic simulation have been developed, such as the COBRA Toolbox for MATLAB [10], a popular FBA simulator.
Successes of genome-scale modelling
Flux balance analysis and related constraint-based methods can be used to predict the optimal set of gene knockout and overexpression targets to increase an organism’s ability to produce a chemical of interest. Here, we present various applications of genome-scale modeling to gage the impact this computational approach has had on metabolic engineering efforts. Table 1 summarizes examples of successes of genome-scale modeling in the context of metabolic engineering.
Table 1.
Examples of recent developments and successes of genome-scale modeling in metabolic engineering
| Publication | Year | Target | Organism |
|---|---|---|---|
| Lee et al. [49] | 2002 | Succinic acid production | E. coli |
| Alper et al. [5] | 2005 | Lycopene production | E. coli |
| Bro et al. [16] | 2006 | Decrease glycerol and increase ethanol yield | S. cerevisiae |
| Lee et al. [48] | 2007 | Threonine production | E. coli |
| Park et al. [66] | 2007 | l-valine production | E. coli |
| Song et al. [90] | 2008 | Optimize media and succinic acid production | M. succiniciproducens |
| Meijer et al. [56] | 2009 | Succinic acid production | A. niger |
| Ohno et al. [62] | 2013 | Butanol, propanol, propanediol production | E. coli |
| Sun et al. [93] | 2014 | Terpenoid biosynthesis | S. cerevisiae |
| Borodina et al. [15] | 2015 | 3-Hydroxypropionic acid biosynthesis | S. cerevisiae |
An exhaustive search of all feasible knockouts in an organism, especially with an experimental approach, to identify the exact genotype with the optimal production profile for a substance of interest, is a painstakingly tedious and often practically infeasible process. Genome-scale metabolic models can be a valuable tool for understanding the inner workings of metabolic networks, which cannot always be intuitively discerned. Such insight may be used to design strains with specific properties in a manner faster by many scales of magnitude, and therefore much more desirable. Genome-scale modeling has been applied in various metabolic engineering contexts and has been successfully used to predict genetic modifications for improved strains.
Lee et al. [49] constructed a metabolic model for E. coli, which was successfully used to develop and implement a strategy for increased succinic acid production. The authors proposed optimal metabolic pathways for the production of succinic acid based on the results of the metabolic flux analyses. For increasing succinic acid production, the pyruvate carboxylation pathway was selected as optimal for increasing the production in E. coli. Experimental validation of the proposed pathway was performed by comparing the yield of succinic acid with traditional succinic acid producing pathways. The experimental results suggested that the novel pathway selected through the computational analysis is more efficient than conventional pathways.
Alper et al. [5] used a genome-scale model for E. coli and identified and experimentally confirmed seven gene deletion strains that showed increased lycopene production. The E. coli iJE660 model [76] served as the basis for this approach. Targets for single gene knockouts were initially selected, and the ones that resulted in the highest production of lycopene were chosen as candidates. Then, a second knockout was computationally predicted and then performed on the best performing single gene mutants, and the double mutants with the highest yield were selected once more. This process produced knockout mutants with progressively increasing yields. The selected single, double, and triple knockout strains were constructed experimentally and were shown to significantly improve the yield of lycopene, with the top selected strain producing a yield almost 40 % higher than an engineered, high-producing parental strain.
Bro et al. [16] used an FBA model of Saccharomyces cerevisiae to identify a strategy for metabolic engineering of the redox metabolism that would lead to decreased glycerol and increased ethanol yields on glucose under anaerobic conditions. Several suggested mutants were suggested computationally that eliminated formation of glycerol and increased ethanol yield. One of the most promising results was selected and constructed experimentally. The resulting strain had a 40 % decrease and 3 % increase in glycerol and ethanol yields, respectively, without affecting the maximum specific growth rate.
Lee et al. [48] reported a strategy for increased threonine production in E. coli. A threonine producing strain was re-engineered based on transcriptome profiling and flux analysis simulations. The resulting strain produced threonine with a high yield of 0.393 g per gram of glucose and 82.4 g/l threonine by fed-batch culture. Similarly, Park et al. [66] constructed a genetically well-defined E. coli strain based on known metabolic information, transcriptome analysis, and in silico genome-scale knockout simulation. The authors identified the necessary gene knockouts for the construction of an E. coli strain with increased l-valine production. Genes ilvA, leuA, and panB were deleted to make more precursors available for l-valine biosynthesis, lrp and ygaZH were overexpressed and aceF, mdh, and pfkA were identified as knockout targets using gene knockout simulation. The resulting strain produced a high yield of 0.378 g per gram of glucose of l-valine, which is higher than industrial strains developed through random mutation and selection.
Another useful application of FBA is to identify optimal media composition for the growth of an organism and production of a desired metabolite [90]. Song et al. used a genome-scale metabolic network and flux balance analysis to identify two amino acids and four vitamins as essential compounds to be supplemented to a minimal medium that would improve the growth of Mannheimia succiniciproducens and the production of succinic acid. The optimized media increased the yield of succinic acid by 15 % compared to growth on a complex medium. The optimal, chemically defined medium also lowered by-products by 30 %.
Meijer et al. [56] presented a metabolic engineering approach for increased production of succinic acid with Aspergillus niger, a microorganism that is well established industrially, making it an interesting target for engineering of the production of specific chemicals. A deletion strategy based on simulations with a genome-scale stoichiometric model of the organism was devised. The gene producing citrate lyase (acl) was identified as a deletion target through in silico tests with a genome-scale metabolic model of the organism. The authors found that the mutant strain tripled the yield of succinic acid compared to the wild type, along with an overall increase in the production of organic acids in the mutant strain.
In 2013, Ohno et al. [62] demonstrated that the production of many valuable compounds, such as L-butanol, L-propanol, and 1,3-propanediol, can be improved using a triple gene knockout strategy. In silico screening was performed and the metabolic potential of all possible sets of triple knockouts were evaluated using a reduced metabolic model of Escherichia coli, based on the iAF1260 genome-scale model [27]. The use of a reduced model was preferred in this study, as it significantly lowered the computational costs. The results demonstrated the applicability of multiple deletion strategies, since in many cases the effects of the deletions were only observable when multiple genes were simultaneously disrupted. Traditional screening methods would have missed these opportunities. Such results are indicative of the possibility to develop industrially viable strains through metabolic engineering that utilizes genome-scale modeling.
Sun et al. [93] presented a study that identified knockout targets for improving terpenoid biosynthesis in S. cerevisiae. Terpenoids have important pharmacological activity, but the production of sufficient amounts is challenging. A constraint-based approach was used to identify knockout sites with the potential to improve terpenoid production (specifically, sesquiterpene amorphadiene). Based on the simulation results, a single mutant was constructed and engineered to produce amorphadiene. Production of amorphadiene was measured to assess the effects of gene deletions on the production of terpenoids. Ten novel gene knockout targets were described. The yield of amorphadiene produced by most single mutants increased 8- to 10-fold compared to the wild type.
Borodina et al. [15] engineered a synthetic pathway for de novo biosynthesis of 3-Hydroxypropionic acid, using a genome-scale model of S. cerevisiae to evaluate the metabolic capabilities of two promising routes. 3-Hydroxypropionic acid (3HP) is a potential chemical building block for sustainable production of superabsorbent polymers and acrylic plastics. Simulations suggested β-alanine biosynthesis as the most economically attractive route. A synthetic pathway for de novo biosynthesis of β-alanine and its subsequent conversion into 3-Hydroxypropionic acid was engineered, using a novel β-alanine-pyruvate aminotransferase discovered in Bacillus cereus. The expression of the critical enzymes in the pathway was optimized and aspartate biosynthesis was increased to obtain a high 3-Hydroxypropionic acid producing strain.
In addition to the growing number of studies that demonstrate the applicability of genome-scale modeling to rational metabolic engineering efforts by performing analyses and producing strains that improve the production of chemicals of interest, several computational approaches for automatic selection of gene knockout candidates have been developed. Such frameworks make FBA a tool that is now available to a much wider audience. In Zomorrodi et al. [114], the authors review computational tools that utilize mathematical optimization and were designed to assist in metabolic network analyses and redesign of metabolism. For example, OptKnock [18] is a framework that exploits duality theory to search for multiple gene knockout candidates, by solving a bi-level optimization problem: the inner problem optimizes biomass production, while the outer problem optimizes target chemical yield. The problem is formulated as a single MILP problem. Sets of gene knockouts for improved succinate, lactate, and propanediol production in E. coli were predicted by the authors.
The OptKnock framework suffers from certain limitations, for example the intractability of the problem when very large sets of knockouts are considered. To address such issues, researchers have developed extended and improved frameworks that identify deletion candidates, such as OptGene and RobustKnock. OptGene [67] utilizes a genetic algorithm to rapidly identify gene deletion strategies for optimization of a strain. The advantages of Opt- Gene are that it also allows the optimization of nonlinear objective functions, and can be much faster than an MILP approach, but unlike with MILP formulations, the identified solution is not guaranteed to be a global optimum. Opt-Gene has been used to predict sets of gene knockouts for improved production of vanillin, succinate, and glycerol in S. cerevisiae. RobustKnock [94] extends OptKnock by accounting for the presence of competing pathways in the network that may reroute metabolic flux away from the chemical of interest. The framework removes reactions from the network, so that the production of the chemical of interest becomes part of the model’s biomass production requirement. RobustKnock was used to predict sets of gene knockouts for improving the production of hydrogen, acetate, formate and fumarate in E. coli.
Although frameworks like OptKnock and OptGene are powerful in their ability to predict knockouts, the possible modifications are restricted by the selection of reactions included in the metabolic reconstruction. The possibility of adding new reactions that are not part of the original metabolic network is not considered with these methods. Opt- Strain [68] overcomes this problem with the use of a database of known biotransformations to maximize the yield of a pathway from substrate to target product, by including heterologous reactions. The number of non-native reactions is minimized, and the selected non-native reactions are incorporated into the host. In addition to the above tools, OptReg [69] and EMILiO(Enhancing Metabolism with Iterative Linear Optimization) [108] are frameworks that not only identify gene targets selected for deletion, but also identify genes that can be up or downregulated. Such computational tools have been used for several metabolic engineering applications, including the production of lactic acid in E. coli [29], vanillin production in yeast [17] and sesquiterpene production in S. cerevisiae [7]. For researchers and engineers that wish to apply genome-scale modeling methods and the automated gene knockout selection frameworks described here, several software options exist that are now freely available, including the COBRA toolbox [10], OptFlux [78], CellNetAnalyzer [45] other Systems Biology Research Tool [106], to name but a few.
Transcriptional regulation
Genome-scale modeling is not without its limitations; one of the major issues with the predictions made with this analysis method is that it does not consider the effects of gene regulation. In reality, however, the effect of regulation is very significant and one of the major reasons for failed predictions of the metabolic effect of gene modifications. For this reason, there is great motivation to look beyond just the metabolic network and attempt to integrate the effects of regulation on the metabolic reactions of an organism. Integrated models can significantly improve prediction accuracy, though again there is still much room for improvement. Machado and Herrgård have performed a systematic comparison of methods of transcriptomic data integration with genome-scale modeling [54].
In its simplest form, transcriptional regulation can be added to a stoichiometric model using a Boolean representation to map the effects of transcription factors (activating or repressing) on the expression of enzyme encoding genes. Such a representation forces the specific enzyme-catalyzed reaction to be either on or off, depending on the presence or absence of the controlling transcription factors. The implementation of this idea is known as regulatory Flux Balance Analysis (rFBA) [22]. rFBA offers the possibility of considering some basic regulatory effects on the metabolic network, but it is constrained by the fact that the genes that are controlled by transcriptional factors can only be either fully active or completely off. This prohibits good predictions in cases where a transcriptional factor knockout only has a partial effect on target genes. Another limitation of rFBA is that it arbitrarily chooses one metabolic steady state from a space of possible solutions, excluding a whole space of possible profiles. Instead, Steady-state Regulatory Flux Balance Analysis, or SR-FBA [88], enabled a comprehensive characterization of steady-state behaviors in an integrated model of metabolism and regulation. SR-FBA was used to characterize the flux distribution and gene expression levels of Escherichia coli across different growth media. Around 50 % of metabolic genes’ flux activity was found to be determined by metabolic constraints, whereas regulatory constraints determined the flux activity of 15–20 % of genes. The integrated model was then used to identify specific genes for which regulation is not optimally tuned for cellular flux demands.
Probabilistic regulation of metabolism (PROM) [20, 89] is another method that overcomes the limitations of rFBA by implementing a probabilistic approach for predicting the state of a gene, based on the level of expression of a transcription factor. The probability for the state of a gene is determined based on microarray data information, and the bounds on the flux of the relevant reaction are adjusted using this probability estimation. In addition, PROM requires little manual annotation compared to rFBA, because the process can be automated to a large degree. Still, the accuracy of all such methods needs to be improved, and there is substantial need to expand the repertoire of captured regulatory events related to metabolism beyond simple transcriptional effects.
Similarly, E-Flux [21] is an approach that incorporates transcript level measurements to the reaction flux constraints that define the maximum achievable flux through each reaction. The bounds on the fluxes of the system are determined based on the level of expression for the corresponding coding gene. The method was tested on Mycobacterium tuberculosis to predict the impact of drugs, drug combinations, and nutrient conditions. E-flux predicted seven of the eight known fatty acid inhibitors and made accurate predictions regarding the specificity of these compounds for fatty acid biosynthesis.
An important disadvantage of previous methods is that they often require a user-defined expression threshold over (or under) which a gene is considered “on” (or “off”). Metabolic adjustment by differential expression (MADE) [38] aims to overcome the problem of selecting arbitrary thresholds by comparing measurements across multiple conditions. MADE uses the statistical significant changes in gene expression measurements across sequential conditions to determine instances of high and low expression for various reactions. For this reason, MADE requires expression data from more than one experimental conditions. The solutions for all conditions are solved simultaneously to maximize agreement with the predicted patterns.
Other approaches for integrated simulation use mRNA expression data to construct a functional metabolic model for the organism. Gene Inactivity Moderated by Metabolism and Expression (GIMME) [11] utilizes user-supplied gene expression data, a genome-scale model and presupposed metabolic objectives to produce a context-specific reconstruction. GIMME performs an FBA run on the starting metabolic model to identify the maximum possible flux through the network. Then, experimental mRNA transcript levels are compared to a threshold and any reactions that fall below this threshold are removed from the network, unless their removal impacts the metabolic objectives, in which case an LP problem is solved that reintroduces inactive reactions in a way that minimizes deviation from the expression data. The algorithm also provides a quantitative inconsistency score indicating how consistent a set of gene expression data is with a particular metabolic objective.
The integrative Metabolic Analysis Tool (iMAT) [115] on the other hand is a web-based tool based on Shlomi et al. [87], which does not require prior knowledge of a defined metabolic functionality. iMAT enables the prediction of metabolic states in specific conditions using protein (or gene) expression data as input, integrating them with transcriptomic information and a genome-scale metabolic model. The web tool outputs a prediction for the flux state and a set of confidence values for all the reactions in the network. Additionally, iMAT can report predicted upregulated and downregulated genes post-transcriptionally. The main difference to GIMME is that instead of presupposed metabolic objectives, iMAT requires the existence of a minimum flux through reactions that correspond to the highly expressed genes in the dataset. This difference gives iMAT an advantage in cases where clear metabolic objectives cannot be established.
The first model that can be considered “whole-cell” was developed for Mycoplasma genitalium [43], a human pathogen, by combining all the biochemical components and all the interactions in the system. Modules with diverse characteristics were built, representing distinct cellular functions and combined into a dynamic framework. This integrative approach enabled the inclusion of physiologically and mathematically diverse processes and experimental measurements. The model was used to examine areas of cellular function that had not been studied in conjunction before, such as protein–DNA associations and the interactions between DNA replication and the initiation of replication. This whole-cell model represents an important advancement in the development of integrated genome-scale modeling.
The more biochemically accurate a model is, the more detailed the simulations of an organism’s phenotypic behavior we should be able to produce by varying genetic and environmental parameters. With the combination of Metabolism and gene Expression, an ME model was produced; an integrated model of Thermotoga maritima [51] that considerably improves the prediction accuracy of the genome-scale metabolic model of the organism, along with the added capability of gene expression prediction. The ME model represents the next generation of constraint-based models: stoichiometric models of metabolism that also explicitly consider gene transcription and translation. Thanks to the integration of additional levels of biological information, ME models can provide a basis for considering mRNA transcription, protein translation, protein complexing, reaction catalysis or molecule formation within the framework of genome-scale modeling. ME models represent a significant step in the effort to bridge the gap between molecular biology and cellular physiology.
Another important application of integration of transcriptome, proteome, and phenotypic data with metabolic reconstructions is to contextual generic metabolic reconstructions in higher organisms to contextualize those aspects of metabolism that are present in any particular tissue or cell type. A number of automatic reconstruction approaches have been built to achieve this. One such algorithm, the Model Building Algorithm (MBA) [39], was employed in the construction of a tissue-specific, hepatic model, from the generic human RECON1 model [26], integrating tissue-specific molecular data. The hepatic model was validated with flux measurements across various hormonal and dietary conditions. The advantage of MBA is that it eliminates the presence of superfluous metabolic reactions and streamlines the metabolic model to consist of metabolic reactions that are functional in the cell. Similarly, a method called metabolic Context-specificity Assessed by Deterministic Reaction Evaluation (mCADRE) [101] is able to infer a tissue-specific network based on gene expression data and metabolic network topology, along with evaluation of functional capabilities during model building. mCADRE produces models with similar functionality and achieves dramatic computational speed up over MBA using the network topology to set a deterministic ordering for reaction removal rather than computing a large ensemble of models based on random orderings. Using this method, a reconstruction of draft genome-scale metabolic models for 126 human tissue and cell types was performed. Finally, another approach is the INIT (integrative network inference for tissues) algorithm [1], which uses cell type specific information about protein abundances as its main source of evidence. INIT is formulated as an MILP problem and relies on evidence from the Human Protein Atlas [97] and tissue-specific gene expression data to decide on the presence or absence of metabolic enzymes in each cell type, while metabolomics data from the Human Metabolome Database [105] are used as constraints that force the ability to produce a specific metabolite by adding the necessary reactions, if said metabolite has been observed in a tissue. INIT was used to generate genome-scale models for 69 healthy human cell types and 16 cancer cell types.
Cells contain thousands of molecular components including transcripts, proteins and metabolites, and regulation plays a very important role in every cellular process (gene expression, protein transcription, enzymatic reactions). For these reasons, precise estimation of the metabolic states and comprehension of the way regulation works are crucial factors for accurate simulation of cellular processes. Approaches that integrate transcriptional regulation with more traditional constraint-based metabolic simulation make several assumptions, particularly since the transcription of genes and the way it correlates with flux are still not perfectly understood. As a result, predictions made with these approaches are not highly accurate, and while these methods have been successfully applied to specific example organisms, wide application is still problematic. Nevertheless, integrated approaches constitute an initial step in the effort to effectively correlate genotype with phenotype and often offer improved predictions compared to stand-alone FBA simulations.
Conclusions
In the current microbial metabolic engineering field, many tools and applications have been developed that facilitate genetic engineering of model organisms. Here, we summarized the genome-scale modeling approach, which, thanks to its simplicity and the fact that it offers large amounts of biochemical information for an organism’s reactions, is well suited for application in systematic metabolic engineering for bio-production using microorganisms. Metabolic design using genome-scale modeling is already widely used, as it enables prediction of the knockout or amplification target genes for enhancement of productivity. In this review, we offered an overview of genome-scale modeling and flux balance analysis, and focused particularly on the challenge of metabolic reconstructions, and on the developments that the various efforts for automatic reconstruction have achieved. We reviewed several successful studies in the area of genome-scale modeling for metabolic engineering. Techniques for metabolome analysis have made progress in recent years, and researchers can now have direct access to several tools that automate the selection of gene deletions, additions and modifications to produce mutants that would facilitate the production of specific chemicals. Finally, we summarized the importance of studying and understanding the regulatory mechanisms of the cell and presented studies that focused on integration of regulation and metabolism. In the future, we expect that integrated models of metabolism will become particularly important in the field of metabolic engineering.
Acknowledgments
The authors gratefully acknowledge funding from the Luxembourg Centre for Systems Biomedicine (ES), and the DOE ARPA-E program (DE-AR0000426), an NIH Center for Systems Biology (2P50 GM076547) and the Camille Dreyfus Teacher-Scholar Program (NDP). We also thank Julie Bletz and Ben Heavner for critical readings of the manuscript, and James Eddy for assistance with the illustrations.
Contributor Information
Evangelos Simeonidis, Email: evangelos.simeonidis@uni.lu, Institute for Systems Biology, 401 Terry Avenue, North Seattle, WA 98109, USA; Luxembourg Centre for Systems Biomedicine, University of Luxembourg, 7, avenue des Hauts-Fourneaux, L-4362 Esch-Sur-Alzette, Luxembourg.
Nathan D. Price, Email: nathan.price@systemsbiology.org, Institute for Systems Biology, 401 Terry Avenue, North Seattle, WA 98109, USA.
References
- 1.Agren R, Bordel S, Mardinoglu A, Pornputtapong N, Nookaew I, Nielsen J. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput Biol. 2012;8(5):17. doi: 10.1371/journal.pcbi.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agren R, Liu L, Shoaie S, Vongsangnak W, Nookaew I, Nielsen J. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput Biol. 2013;9(3):21. doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427(6977):839–843. doi: 10.1038/nature02289. doi:101038/nature02289. [DOI] [PubMed] [Google Scholar]
- 5.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng. 2005;7(3):155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Andersen MR, Nielsen ML, Nielsen J. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol Syst Biol. 2008;4(178):25. doi: 10.1038/msb.2008.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, Nielsen J. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng. 2009;11(6):328–334. doi: 10.1016/j.ymben.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Avila-Campillo I, Drew K, Lin J, Reiss DJ, Bonneau R. BioNetBuilder: automatic integration of biological networks. Bioinformatics. 2007;23(3):392–393. doi: 10.1093/bioinformatics/btl604. [DOI] [PubMed] [Google Scholar]
- 9.Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol. 2012;23(4):631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2(3):727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 11.Becker SA, Palsson BO. Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol. 2008;4(5):e1000082. doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benedict MN, Gonnerman MC, Metcalf WW, Price ND. Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A. J Bacteriol. 2012;194(4):855–865. doi: 10.1128/JB.06040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benedict MN, Mundy MB, Henry CS, Chia N, Price ND. Likelihood-based gene annotations for gap filling and quality assessment in genome-scale metabolic models. PLoS Comput Biol. 2014;10(10):e1003882. doi: 10.1371/journal.pcbi.1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blazeck J, Alper H. Systems metabolic engineering: genome-scale models and beyond. Biotechnol J. 2010;5(7):647–659. doi: 10.1002/biot.200900247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borodina I, Kildegaard KR, Jensen NB, Blicher TH, Maury J, Sherstyk S, Schneider K, Lamosa P, Herrgård MJ, Rosenstand I, Öberg F, Forster J, Nielsen J. Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine. Metabolic Engineering. 2015;27:57–64. doi: 10.1016/j.ymben.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Bro C, Regenberg B, Forster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng. 2006;8(2):102–111. doi: 10.1016/j.ymben.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Brochado AR, Matos C, Moller BL, Hansen J, Mortensen UH, Patil KR. Improved vanillin production in baker’s yeast through in silico design. Microb Cell Fact. 2010;9(84):1475–2859. doi: 10.1186/1475-2859-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84(6):647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 19.Burgess CM, Smid EJ, van Sinderen D. Bacterial vitamin B2, B11 and B12 overproduction: an overview. Int J Food Microbiol. 2009;133(1–2):1–7. doi: 10.1016/j.ijfoodmicro.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Chandrasekaran S, Price ND. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107(41):17845–17850. doi: 10.1073/pnas.1005139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colijn C, Brandes A, Zucker J, Lun DS, Weiner B, Farhat MR, Cheng TY, Moody DB, Murray M, Galagan JE. Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput Biol. 2009;5(8):28. doi: 10.1371/journal.pcbi.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Covert MW, Schilling CH, Palsson B. Regulation of gene expression in flux balance models of metabolism. J Theor Biol. 2001;213(1):73–88. doi: 10.1006/jtbi.2001.2405. [DOI] [PubMed] [Google Scholar]
- 23.Dale JM, Popescu L, Karp PD. Machine learning methods for metabolic pathway prediction. BMC Bioinform. 2010;11(15):1471–2105. doi: 10.1186/1471-2105-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devoid S, Overbeek R, DeJongh M, Vonstein V, Best AA, Henry C. Automated genome annotation and metabolic model reconstruction in the SEED and Model SEED. Methods Mol Biol. 2013;985:17–45. doi: 10.1007/978-1-62703-299-5_2. [DOI] [PubMed] [Google Scholar]
- 25.Dobson PD, Smallbone K, Jameson D, Simeonidis E, Lanthaler K, Pir P, Lu C, Swainston N, Dunn WB, Fisher P, Hull D, Brown M, Oshota O, Stanford NJ, Kell DB, King RD, Oliver SG, Stevens RD, Mendes P. Further developments towards a genome-scale metabolic model of yeast. BMC Syst Biol. 2010;4(145):0509–1752. doi: 10.1186/1752-0509-4-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104(6):1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3(121):26. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng X, Xu Y, Chen Y, Tang YJ. MicrobesFlux: a web platform for drafting metabolic models from the KEGG database. BMC Syst Biol. 2012;6:94. doi: 10.1186/1752-0509-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong SS, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BO. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng. 2005;91(5):643–648. doi: 10.1002/bit.20542. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh A, Zhao H, Price ND. Genome-scale consequences of cofactor balancing in engineered pentose utilization pathways in Saccharomyces cerevisiae. PLoS One. 2011;6(11):4. doi: 10.1371/journal.pone.0027316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonnerman MC, Benedict MN, Feist AM, Metcalf WW, Price ND. Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746. Biotechnol J. 2013;8(9):1070–1079. doi: 10.1002/biot.201200266. [DOI] [PubMed] [Google Scholar]
- 32.Heavner BD, Smallbone K, Price ND, Walker LP. Version 6 of the consensus yeast metabolic network refines biochemical coverage and improves model performance. Database. 2013;9(10) doi: 10.1093/database/bat059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28(9):977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 34.Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Bluthgen N, Borger S, Costenoble R, Heinemann M, Hucka M, Le Novere N, Li P, Liebermeister W, Mo ML, Oliveira AP, Petranovic D, Pettifer S, Simeonidis E, Smallbone K, Spasic I, Weichart D, Brent R, Broomhead DS, Westerhoff HV, Kirdar B, Penttila M, Klipp E, Palsson BO, Sauer U, Oliver SG, Mendes P, Nielsen J, Kell DB. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26(10):1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong KK, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci. 2012;69(16):2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 37.Jang YS, Park JM, Choi S, Choi YJ, Seung do Y, Cho JH, Lee SY. Engineering of microorganisms for the production of biofuels and perspectives based on systems metabolic engineering approaches. Biotechnol Adv. 2012;30(5):989–1000. doi: 10.1016/j.biotechadv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Jensen PA, Papin JA. Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics. 2011;27(4):541–547. doi: 10.1093/bioinformatics/btq702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol. 2010;6(401):56. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joyce AR, Palsson BO. Predicting gene essentiality using genome-scale in silico models. Methods Mol Biol. 2008;416:433–457. doi: 10.1007/978-1-59745-321-9_30. [DOI] [PubMed] [Google Scholar]
- 41.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:12. doi: 10.1093/nar/gkm882. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33(19):6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Jr, Assad-Garcia N, Glass JI, Covert MW. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150(2):389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King ZA, Feist AM. Optimal cofactor swapping can increase the theoretical yield for chemical production in Escherichia coli and Saccharomyces cerevisiae. Metab Eng. 2014;24:117–128. doi: 10.1016/j.ymben.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Klamt S, Saez-Rodriguez J, Gilles ED. Structural and functional analysis of cellular networks with Cell NetAnalyzer. BMC Syst Biol. 2007;1:2. doi: 10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang M, Stelzer M, Schomburg D. BKM-react, an integrated biochemical reaction database. BMC Biochem. 2011;12(42):1471–2091. doi: 10.1186/1471-2091-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8(6):536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 48.Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for l-threonine production. Mol Syst Biol. 2007;3(149):4. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SY, Hong SH, Moon SY. In silico metabolic pathway analysis and design: succinic acid production by metabolically engineered Escherichia coli as an example. Genome Inform. 2002;13:214–223. [PubMed] [Google Scholar]
- 50.Lee SY, Lee DY, Kim TY. Systems biotechnology for strain improvement. Trends Biotechnol. 2005;23(7):349–358. doi: 10.1016/j.tibtech.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Lerman JA, Hyduke DR, Latif H, Portnoy VA, Lewis NE, Orth JD, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Zengler K, Palsson BO. In silico method for modelling metabolism and gene product expression at genome scale. Nat Commun. 2012;3:929. doi: 10.1038/ncomms1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Redden H, Alper HS. Frontiers of yeast metabolic engineering: diversifying beyond ethanol and Saccharomyces. Curr Opin Biotechnol. 2013;24(6):1023–1030. doi: 10.1016/j.copbio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 53.Ma F, Hanna MA. Biodiesel production: a review. Bioresour Technol. 1999;70(1):1–15. [Google Scholar]
- 54.Machado D, Herrgard M. Systematic evaluation of methods for integration of transcriptomic data into constraint-based models of metabolism. PLoS Comput Biol. 2014;10(4):e1003580. doi: 10.1371/journal.pcbi.1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahadevan R, Schilling CH. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5(4):264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Meijer S, Nielsen ML, Olsson L, Nielsen J. Gene deletion of cytosolic ATP: citrate lyase leads to altered organic acid production in Aspergillus niger. J Ind Microbiol Biotechnol. 2009;36(10):1275–1280. doi: 10.1007/s10295-009-0607-y. [DOI] [PubMed] [Google Scholar]
- 57.Milne CB, Eddy JA, Raju R, Ardekani S, Kim PJ, Senger RS, Jin YS, Blaschek HP, Price ND. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst Biol. 2011;5:130. doi: 10.1186/1752-0509-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monk J, Nogales J, Palsson BO. Optimizing genome-scale network reconstructions. Nat Biotechnol. 2014;32(5):447–452. doi: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- 59.Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2008;72(3):379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci U S A. 2012;109(7):2678–2683. doi: 10.1073/pnas.1117907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5(320):3. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno S, Furusawa C, Shimizu H. In silico screening of triple reaction knockout Escherichia coli strains for overproduction of useful metabolites. J Biosci Bioeng. 2013;115(2):221–228. doi: 10.1016/j.jbiosc.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 63.Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BO. A comprehensive genome-scale reconstruction of Escherichia coli metabolism–2011. Mol Syst Biol. 2011;7(535):65. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papin JA, Price ND, Palsson BO. Extreme pathway lengths and reaction participation in genome-scale metabolic networks. Genome Res. 2002;12(12):1889–1900. doi: 10.1101/gr.327702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000;54(3):287–301. doi: 10.1007/s002530000403. [DOI] [PubMed] [Google Scholar]
- 66.Park JH, Lee KH, Kim TY, Lee SY. Metabolic engineering of Escherichia coli for the production of l-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci U S A. 2007;104(19):7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patil KR, Rocha I, Forster J, Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinform. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pharkya P, Burgard AP, Maranas CD. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14(11):2367–2376. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pharkya P, Maranas CD. An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Eng. 2006;8(1):1–13. doi: 10.1016/j.ymben.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 70.Philp JC, Ritchie RJ, Allan JE. Biobased chemicals: the convergence of green chemistry with industrial biotechnology. Trends Biotechnol. 2013;31(4):219–222. doi: 10.1016/j.tibtech.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 71.Pitkanen E, Akerlund A, Rantanen A, Jouhten P, Ukkonen E. ReMatch: a web-based tool to construct, store and share stoichiometric metabolic models with carbon maps for metabolic flux analysis. J Integr Bioinform. 2008;5(2):2008–2102. doi: 10.2390/biecoll-jib-2008-102. [DOI] [PubMed] [Google Scholar]
- 72.Price ND, Papin JA, Palsson BO. Determination of redundancy and systems properties of the metabolic network of Helicobacter pylori using genome-scale extreme pathway analysis. Genome Res. 2002;12(5):760–769. doi: 10.1101/gr.218002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price ND, Schellenberger J, Palsson BO. Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies. Biophys J. 2004;87(4):2172–2186. doi: 10.1529/biophysj.104.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ranganathan S, Suthers PF, Maranas CD. OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput Biol. 2010;6(4):1000744. doi: 10.1371/journal.pcbi.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A. 2006;103(46):17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/ GPR) Genome Biol. 2003;4(9):28. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reyes R, Gamermann D, Montagud A, Fuente D, Triana J, Urchueguia JF, de Cordoba PF. Automation on the generation of genome-scale metabolic models. J Comput Biol. 2012;19(12):1295–1306. doi: 10.1089/cmb.2012.0183. [DOI] [PubMed] [Google Scholar]
- 78.Rocha I, Maia P, Evangelista P, Vilaca P, Soares S, Pinto JP, Nielsen J, Patil KR, Ferreira EC, Rocha M. OptFlux: an open-source software platform for in silico metabolic engineering. BMC Syst Biol. 2010;4(45):0509–1752. doi: 10.1186/1752-0509-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329(5989):305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 80.Savinell JM, Palsson BO. Optimal selection of metabolic fluxes for in vivo measurement. I. Development of mathematical methods. J Theor Biol. 1992;155(2):201–214. doi: 10.1016/s0022-5193(05)80595-8. [DOI] [PubMed] [Google Scholar]
- 81.Savinell JM, Palsson BO. Optimal selection of metabolic fluxes for in vivo measurement. II. Application to Escherichia coli and hybridoma cell metabolism. J Theor Biol. 1992;155(2):215–242. doi: 10.1016/s0022-5193(05)80596-x. [DOI] [PubMed] [Google Scholar]
- 82.Schellenberger J, Park JO, Conrad TM, Palsson BO. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinform. 2010;11(213):1471–2105. doi: 10.1186/1471-2105-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schilling CH, Schuster S, Palsson BO, Heinrich R. Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era. Biotechnol Prog. 1999;15(3):296–303. doi: 10.1021/bp990048k. [DOI] [PubMed] [Google Scholar]
- 84.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shapiro HM. Input-output models of biological systems: formulation and applicability. Comput Biomed Res. 1969;2(5):430–445. doi: 10.1016/0010-4809(69)90008-1. [DOI] [PubMed] [Google Scholar]
- 86.Shinfuku Y, Sorpitiporn N, Sono M, Furusawa C, Hirasawa T, Shimizu H. Development and experimental verification of a genome-scale metabolic model for Corynebacterium glutamicum. Microb Cell Fact. 2009;8(43):1475–2859. doi: 10.1186/1475-2859-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26(9):1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 88.Shlomi T, Eisenberg Y, Sharan R. Ruppin E A genome-scale computational study of the interplay between transcriptional regulation and metabolism. Mol Syst Biol. 2007;3:101. doi: 10.1038/msb4100141. Epub 2007 Apr 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simeonidis E, Chandrasekaran S, Price ND. A guide to integrating transcriptional regulatory and metabolic networks using PROM (probabilistic regulation of metabolism) Methods Mol Biol. 2013;985:103–112. doi: 10.1007/978-1-62703-299-5_6. [DOI] [PubMed] [Google Scholar]
- 90.Song H, Kim TY, Choi BK, Choi SJ, Nielsen LK, Chang HN, Lee SY. Development of chemically defined medium for Mannheimia succiniciproducens based on its genome sequence. Appl Microbiol Biotechnol. 2008;79(2):263–272. doi: 10.1007/s00253-008-1425-2. [DOI] [PubMed] [Google Scholar]
- 91.Stephanopoulos G. Metabolic fluxes and metabolic engineering. Metab Eng. 1999;1(1):1–11. doi: 10.1006/mben.1998.0101. [DOI] [PubMed] [Google Scholar]
- 92.Sun Z, Meng H, Li J, Wang J, Li Q, Wang Y, Zhang Y. Identification of Novel Knockout Targets for Improving Terpenoids Biosynthesis in Saccharomyces cerevisiae. PLoS One. 2014;9(11):e112615. doi: 10.1371/journal.pone.0112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swainston N, Smallbone K, Mendes P, Kell D, Paton N. The SuBliMinaL Toolbox: automating steps in the reconstruction of metabolic networks. J Integr Bioinform. 2011;8(2):2011–2186. doi: 10.2390/biecoll-jib-2011-186. [DOI] [PubMed] [Google Scholar]
- 94.Tepper N, Shlomi T. Predicting metabolic engineering knockout strategies for chemical production: accounting for competing pathways. Bioinformatics. 2010;26(4):536–543. doi: 10.1093/bioinformatics/btp704. [DOI] [PubMed] [Google Scholar]
- 95.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trinh CT, Carlson R, Wlaschin A, Srienc F. Design, construction and performance of the most efficient biomass producing E coli bacterium. Metab Eng. 2006;8(6):628–638. doi: 10.1016/j.ymben.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 97.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L. Ponten F Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. doi:10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 98.Varma A, Palsson BO. Metabolic capabilities of Escherichia coli: I. synthesis of biosynthetic precursors and cofactors. J Theor Biol. 1993;165(4):477–502. doi: 10.1006/jtbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 99.Varma A, Palsson BO. Metabolic capabilities of Escherichia coli: II. optimal growth patterns. J Theor Biol. 1993;165(4):503–522. doi: 10.1006/jtbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 100.Varma A, Palsson BO. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol. 1994;60(10):3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, Eddy JA, Price ND. Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Syst Biol. 2012;6(153):0509–1752. doi: 10.1186/1752-0509-6-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Watson MR. Metabolic maps for the Apple II. Biochem Soc Trans. 1984;12:1093–1094. [Google Scholar]
- 103.Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006;9(3):268–274. doi: 10.1016/j.mib.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 104.Wijffels RH, Kruse O, Hellingwerf KJ. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr Opin Biotechnol. 2013;24(3):405–413. doi: 10.1016/j.copbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 105.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wright J, Wagner A. The Systems Biology Research Tool: evolvable open-source software. BMC Syst Biol. 2008;2(55):0509–1752. doi: 10.1186/1752-0509-2-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng. 2012;14(3):233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang L, Cluett WR, Mahadevan R. EMILiO: a fast algorithm for genome-scale strain design. Metab Eng. 2011;13(3):272–281. doi: 10.1016/j.ymben.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 109.Yin J, Chen J-C, Wu Q, Chen G-Q. Halophiles, coming stars for industrial biotechnology. Biotechnology Advances. 2014 doi: 10.1016/j.biotechadv.2014.10.008. (in press) [DOI] [PubMed] [Google Scholar]
- 110.Yoshikawa K, Kojima Y, Nakajima T, Furusawa C, Hirasawa T, Shimizu H. Reconstruction and verification of a genome-scale metabolic model for Synechocystis sp. PCC6803. Appl Microbiol Biotechnol. 2011;92(2):347–358. doi: 10.1007/s00253-011-3559-x. [DOI] [PubMed] [Google Scholar]
- 111.Zhou H, Cheng JS, Wang BL, Fink GR, Stephanopoulos G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng. 2012;14(6):611–622. doi: 10.1016/j.ymben.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 112.Zhou T. Computational reconstruction of metabolic networks from KEGG. Methods Mol Biol. 2013;930:235–249. doi: 10.1007/978-1-62703-059-5_10. [DOI] [PubMed] [Google Scholar]
- 113.Zhuang K, Bakshi BR, Herrgard MJ. Multi-scale modeling for sustainable chemical production. Biotechnol J. 2013;8(9):973–984. doi: 10.1002/biot.201200272. [DOI] [PubMed] [Google Scholar]
- 114.Zomorrodi AR, Suthers PF, Ranganathan S, Maranas CD. Mathematical optimization applications in metabolic networks. Metab Eng. 2012;14(6):672–686. doi: 10.1016/j.ymben.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 115.Zur H, Ruppin E, Shlomi T. iMAT: an integrative metabolic analysis tool. Bioinformatics. 2010;26(24):3140–3142. doi: 10.1093/bioinformatics/btq602. [DOI] [PubMed] [Google Scholar]