Abstract
Trinucleotide repeat (TNR) expansion is the root cause for many known congenital neurological and muscular disorders in human including Huntington's disease, Fragile X syndrome and Friedreich's ataxia. The stable secondary hairpin structures formed by TNR may trigger fork stalling during replication, causing DNA polymerase slippage and TNR expansion. Srs2 and Sgs1 are two helicases in yeast that resolve TNR hairpins during DNA replication and prevent genome expansion. Using single molecule fluorescence, we investigated the unwinding mechanism by which Srs2 and Sgs1 resolves TNR hairpin and compared it to unwinding of duplex DNA. While Sgs1 unwinds both structures indiscriminately, Srs2 displays repetitive unfolding of TNR hairpin without fully unwinding it. Such activity of Srs2 shows dependence on the folding strength and the total length of TNR hairpin. Our results reveal disparate molecular mechanism of Srs2 and Sgs1 that may contribute differently to efficient resolving of the TNR hairpin.
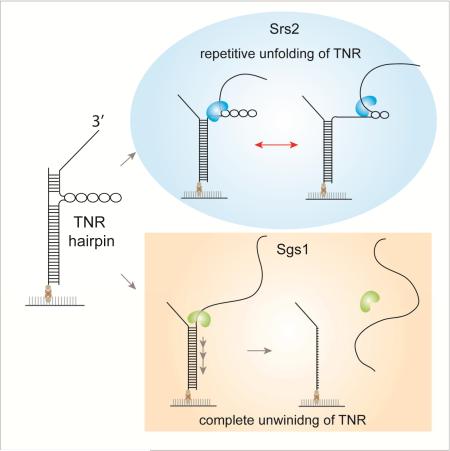
Graphical Abstract
INTRODUCTION
Trinucleotide repeats (TNR) are successive triplet DNA sequences made up of CTG, CAG, CGG or CCG that can develop into a secondary DNA structure known as hairpins (Mirkin, 2007). These hairpin structures can occasionally arise during aberrant DNA replication or error-prone DNA repair and act as toxic intermediates that can either stall the main replication machinery or trap proteins involved in the DNA repair pathways (Lahue and Slater, 2003; Liu et al., 2010; Mirkin, 2007; Pelletier et al., 2003; Samadashwily et al., 1997; Voineagu et al., 2009). If left unresolved, such TNR hairpins can lead to genome expansion and chromosomal instability (Cleary et al., 2002) that can give rise to numerous neurodegenerative diseases in human including myotonic dystrophy, Huntington's disease, fragile X syndrome and Friedreich's ataxia (Freudenreich et al., 1997; Gatchel and Zoghbi, 2005; Mirkin and Mirkin, 2014; Mirkin, 2006).
Due to the deleterious effects that can arise from the easily expanded TNR hairpins, many studies have focused on searching for proteins that can help destabilize the formation of hairpins. The genetic screening performed in Saccharomyces Cerevisiae revealed that DNA helicases Srs2 and Sgs1 are potential inhibitors of TNR expansions (Anand et al., 2012; Bhattacharyya and Lahue, 2004; Dhar and Lahue, 2008; Kerrest et al., 2009). Consistently, cells lacking the gene for Srs2 showed a significant increase (up to 40 folds) in TNR expansions and contractions, resulting in chromosomal fragility (Anand et al., 2012; Bhattacharyya and Lahue, 2004; Kerrest et al., 2009). Deletion of Sgs1 also caused contractions of CTG repeats and increased fragility (Kerrest et al., 2009). Furthermore, double mutant cells lacking both Srs2 and Sgs1 resulted in cell death (Gangloff et al., 2000), suggesting that the two proteins cooperated to help reduce the stalled replication forks due to TNR hairpins and also reduced the accumulation of toxic DNA intermediates (Fabre et al., 2002). Interestingly, this activity of Srs2 at TNR during replication was not dependent on Rad51 (Bhattacharyya and Lahue, 2004), suggesting a role of Srs2 unrelated to its anti-recombinase function.
We employed single molecule fluorescence assays to investigate the mechanisms used by Srs2 and Sgs1 in resolving/unfolding TNR hairpins and compared it to their activity in unwinding double strand (ds) DNA. The single molecule approach enabled us to clearly distinguish between the two distinct modes of unwinding mechanism adopted by the two proteins. First, we found that a monomer of Sgs1 is sufficient for unwinding duplex DNA while the monomer unit of Srs2 cannot achieve the same unwinding. Second, Sgs1 completely unwinds both duplex DNA and TNR hairpin non-discriminately whereas Srs2 exhibits a unique repetitive unfolding cycles of TNR hairpin. We also show that the TNR unfolding frequency of Srs2 is modulated by the folding strength and the total length of the TNR hairpin. These results suggest that Srs2's repetitive motions may lead to destabilization of TNR hairpins for an extended period whereas the robust unwinding activity of Sgs1 rapidly resolves the hairpin structures completely. In this way, the disparate TNR unfolding mechanism of Srs2 and Sgs1 can contribute to resolve TNR hairpin in a cooperative and complementary manner.
RESULTS
Unwinding of duplex DNA by Srs2 and Sgs1
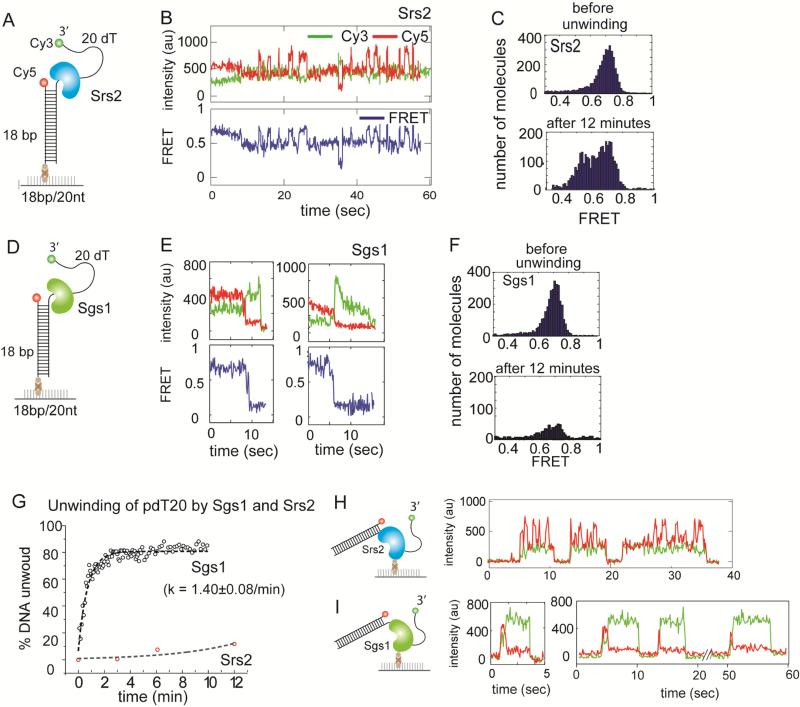
Prior to testing unwinding of DNA with TNR, we sought to compare the dsDNA unwinding activity between the Srs2 and Sgs1. We prepared a partially duplexed DNA substrate consisting of 18 basepairs (bps) and 20 nucleotide (nt) of polythymine DNA tail (pdT20). The Cy3 (green) and Cy5 (red) fluorescent dyes were located near the 3’ and 5’ end of ssDNA, respectively such that it produces a FRET value of 0.7 when excited with a green (532nm) laser (Roy et al., 2008) (Figure 1A). This DNA substrate enables us to detect both the unwinding of the duplex DNA and the possible motion of the protein on ssDNA (Qiu et al., 2013). Complete unwinding of duplex will result in the disappearance of FRET due to the dissociation of Cy3 strand whereas the motion of protein on ssDNA can be tracked by FRET change.
FIGURE 1. Unwinding of duplex DNA by Srs2 and Sgs1, also see Figure S1.
(A, D) Schematic of partial duplex DNA with donor (Cy3) and acceptor (Cy5) fluorophores used for unwinding. (B, E) Representative single molecule intensity (top) and FRET (bottom) traces obtained from applying 10nM Srs2 or Sgs1 to the unwinding substrate, respectively. (C) FRET histogram taken before and after unwinding reaction by Srs2 showing no unwinding. (F) FRET histogram taken before and after unwinding reaction by Sgs1 showing rapid unwinding. (G) Unwinding rate of Sgs1 (black circle) and Srs2 (red circle) and the first exponential fit. Data are represented as mean ± S.E.M.. (H, I) Histidine-tagged Srs2 or flag-tagged Sgs1 proteins (0.5-1nM) were each immobilized on a surface treated with biotinylated Ni–NTA (nitroloacetic acid) or biotinylated anti-flag antibody, respectively and the representative single molecule traces obtained in each case are shown.
We applied the same concentration of Srs2 (10nM) or Sgs1 (10nM) to the pdT20 substrate (Figure 1A, D). Here, we employed the full-length Srs2 protein instead of the C-terminal deletion mutant, Srs2CΔ276 used in our previous study, although both have been shown to have similar helicase activities (Qiu et al., 2013). The concentration of Srs2 used here is comparable to previous studies (Anand et al., 2012; Bhattacharyya and Lahue, 2004) where we do not anticipate significant unwinding of the dsDNA (Lytle et al., 2014). When Srs2 (10nM) and ATP (1mM) was added to the pdT20 substrate, we observed a rapid FRET fluctuation between two FRET states (Figure 1B), consistent with the previously reported repetitive movement of Srs2 on single strand (ss) DNA, fueled by ATP hydrolysis (Qiu et al., 2013). The FRET values collected from over one thousand single molecules were built into FRET histograms before (Figure 1C, top) and after 12 minutes of reaction (Figure 1C, bottom). The single high FRET peak (0.7) that arises from DNA molecules before the reaction (top) shifts into 2 peaks (bottom) upon addition of Srs2 and ATP. This is from the compilation of the two FRET states seen in the single molecule traces, such as in Figure 1B, and is due to the repetitive motion of Srs2 on ssDNA. To measure the unwinding activity, we counted the number of DNA molecules on the experimental surface over time. Over 12 minutes, the number of molecules exhibiting FRET (with both Cy3 and Cy5 signals) remained approximately the same (Figure S1A, C), indicating a negligible unwinding activity by Srs2 (10nM). Conversely, when the same concentration of Sgs1 (10nM) and ATP (1mM) were added to the same DNA construct (pdT20, Figure 1D), we observed a rapid FRET decrease in the majority of single molecule traces (Figure 1E), followed by the loss of Cy3 signal on the experimental surface. (Figure S1B, D). The FRET histograms taken before and after 12 minutes of unwinding (Figure 1F) indicates that the major population of high FRET molecules disappeared as a result of active unwinding by Sgs1 (10nM). To quantify the unwinding kinetics, we counted the number of Cy3 molecules every 5-10 seconds after the addition of the proteins and converted the decrease in Cy3 count as unwinding percentages for both Srs2 and Sgs1 (Figure 1G). The imaging area was switched every 5-10 seconds to minimize the loss of DNA molecule signals due to photobleaching. The unwinding rate of Sgs1 was estimated to be 1.40±0.08min−1 whereas the Srs2-induced unwinding was negligible. We have shown previously that 200nM of Srs2 unwound the same DNA at the rate of 0.3 min−1 (Qiu et al., 2013), which is still substantially slower than the rate observed for 10nM Sgs1.
We have also demonstrated in our earlier work that a monomer of Srs2CΔ276 is responsible for the repetitive motion on DNA which results in FRET fluctuation (Qiu et al., 2013). Here we adopted the same platform for testing the monomer unwinding activity of full length Srs2 and Sgs1 proteins. Histidine-tagged Srs2 or flag-tagged Sgs1 (0.5-1nM) were each immobilized on a surface treated with biotinylated Ni–NTA (nitroloacetic acid) or biotinylated anti-flag antibody, respectively (Figure 1H, I). This platform enables one to immobilize monomer proteins on surface and detect monomeric protein activity. To this platform, we applied non-biotinylated version of the same FRET DNA, pdT20 and ATP to initiate unwinding. In this reverse configuration, we do not capture any signal until the labeled substrate binds the protein. For Srs2, we obtained FRET fluctuations occurring in successive bursts (Figure 1H) representing a repetitive cycle of Srs2 motion per one DNA binding (Qiu et al., 2013). For Sgs1, we observed an initial high FRET (DNA binding) which immediately transitions to low FRET due to unwinding, followed by a disappearance of the Cy3 (green) signal indicating the release of the unwound strand due to complete unwinding of the dsDNA (Figure 1I). Taken together, we demonstrate that Sgs1, but not Srs2 can unwind duplex DNA as a monomer. This result combined with the requirement of 50-200nM of Srs2 for efficient dsDNA unwinding suggests that multimers of Srs2 is needed for DNA unwinding (Lytle et al., 2014; Qiu et al., 2013).
TNR hairpin targeting by Srs2 and Sgs1
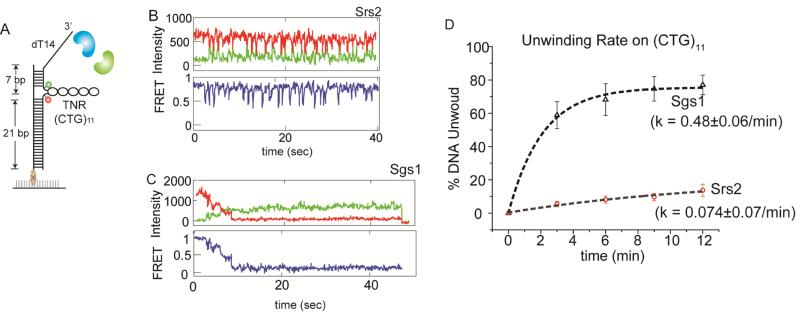
Next, we asked whether Srs2 and Sgs1 differ in processing a DNA substrate that contains a TNR hairpin within dsDNA. We prepared a DNA substrate (Figure 2A) consisting of 14 nt ssDNA (dT14) and 32 bp dsDNA with 11 repeats of CTG inserted in the middle of the duplex used previously (Anand et al., 2012). Two fluorophores, Cy3 and Cy5 were located at either end of the CTG sequence such that when a hairpin forms, the fluorophores will be brought to close proximity (Figure 2A, S2A) to yield a high FRET signal. When the DNA (annealed in 10mM MgCl2 buffer to promote the TNR hairpin formation) was applied to imaging surface, the resulting FRET histogram showed a narrow peak at 0.8 for DNA alone (Figure S2B C, upper histogram), suggesting a formation of the expected TNR hairpin by CTG repeats. When Srs2 (10nM) and ATP (1mM) was added to this DNA, we observed an unexpected high-to-mid FRET fluctuations (Figure 2B) indicating a repetitive opening of the CTG hairpin by Srs2. This FRET fluctuation pattern was not seen in the absence of ATP (Figure S2D, E). Although this FRET pattern may appear to be similar to what we observed previously in Figure 1B, based on the positioning of the dyes on the TNR substrate, the FRET fluctuation seen here represents a repetitive unfolding of the TNR hairpin rather than a repetitive motion of Srs2 on ssDNA (Qiu et al., 2013). In contrast, Sgs1 applied in the same condition (10nM) with 1mM ATP, induced a gradual FRET decrease followed by a disappearance of Cy3 signal (Figure 2C), suggesting a complete unwinding of the entire DNA construct. The gradual FRET decrease corresponds to the unwinding of TNR hairpin whereas the subsequent low FRET state that lasts until Cy3 disappears represents the unwinding of dsDNA. We note that we do not detect re-zipping of the unwound strand in Sgs1 mediated reaction although such may occur after the strand is released after the complete unwinding, which cannot be detected in our set up (TIRF microscope).
FIGURE 2. TNR hairpin unwinding by Srs2 and Sgs1, also see Figure S2.
(A) Schematic of FRET DNA construct with TNR hairpin. Representative single molecule traces of Srs2 (B) and Sgs1 (C). (D) Unwinding rate of TNR containing DNA by Sgs1 (black triangle) and Srs2 (red circle). Data are represented as mean ± S.E.M..
To quantitate and compare the unwinding activity of Srs2 and Sgs1, we counted single molecules that exhibit both Cy3 and Cy5 signals over time. The overall FRET histogram taken before and after the unwinding reaction clearly shows that Sgs1 unwinds actively while Srs2 does not induce substantial unwinding (Figure S2A-C). We calculated the rate of unwinding in the same manner as before (Figure 2D). We note that this rate, calculated from over one thousand molecules, includes both binding (Kon) and unwinding of TNR hairpin and dsDNA, comparable to a biochemical rate that can be measured in bulk solution. Therefore, this rate cannot be directly compared to the rate of FRET decrease observed in Figure 2D which only represents TNR hairpin unwinding of a single molecule. The resulting rate of Sgs1 unwinding of TNR-DNA (0.48 min−1) is approximately three times lower than the rate at which Sgs1 unwound a partial duplex DNA (Figure 1G). This difference is likely due to the combined effect of longer length of the dsDNA and a possible barrier effect imposed by TNR hairpin. In contrast, the Srs2 induced unwinding of the TNR substrate was negligible (Figure 2D). To check that the translocation activity of Srs2 was not perturbed by the dyes near the hairpin junction, we prepared an alternate DNA with modified dye positions. The same FRET fluctuations were obtained in this construct, suggesting that the dyes did not interfere with the hairpin opening (Figure S2F, G). Our previous study also confirms that the dye located on DNA did not disrupt the repetitive translocation of Srs2 (Figure S2H, I) (Qiu et al., 2013). Taken together, our results point to a clear difference between the unwinding pattern of Sgs1 and Srs2. Sgs1 unwinds duplex DNA regardless of the presence of the TNR hairpin whereas Srs2 displays a strong propensity to remain at the site of the TNR hairpin while repeatedly resolving its folded structure (indicated by repetitive FRET fluctuation in Figure 2B). It is interesting to note that Srs2 only partially unfolds the TNR hairpin but does not proceed to unwinding the full length of TNR and dsDNA. Such property of Srs2 may be suited to allow the replication or repair proteins to gain access to the unfolded hairpin region.
Both Srs2 and Sgs1 unwind TNR hairpin before duplex
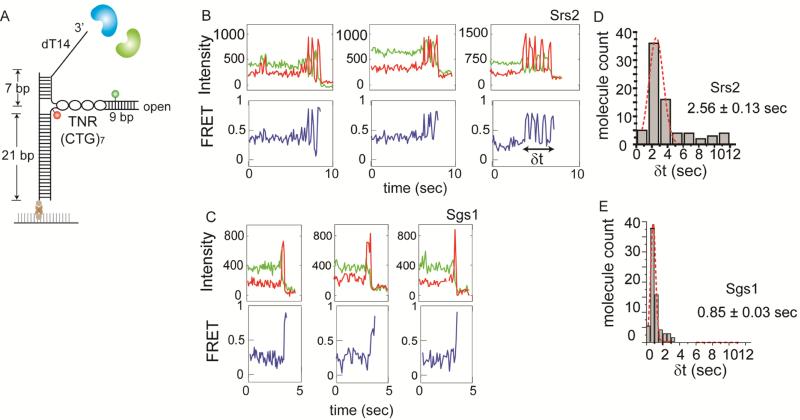
We asked if the unfolding of the hairpin occurs prior to unwinding of the DNA duplex, especially in the case of Sgs1. We prepared an open-ended TNR hairpin similar to a previous study (Dhar and Lahue, 2008) where the hairpin consists of 7 repeats of CTG, with a short, 9 base pair dsDNA to hold the end of the hairpin stem closed (Figure 3A). The 9 bp duplex was inserted at the end of TNR only to hold the two strands together, not requiring an active unwinding activity. The right upper strand is labeled with Cy3 and the right lower strand, with Cy5. In this configuration, if the protein proceeds through the hairpin completely, we will observe a loss of only the Cy3 signal as the right top strand is released from the DNA (Figure S3A). On the other hand, if the protein bypasses the hairpin region and unwinds the dsDNA, we will observe a disappearance of both Cy3 and Cy5 signals as both strands on the right are released. The addition of Srs2 or Sgs1 to this open-hairpin TNR construct in the absence of ATP induced neither FRET fluctuations nor loss of fluorescent signals due to DNA unwinding. When we applied Srs2 or Sgs1 to this substrate with 1mM ATP, we observed a loss of Cy3 signals when excited with the green (532nm) laser, but not the Cy5 signals when excited with the red (635nm) laser (Figure S3B, C), indicating that both proteins process through the TNR hairpin. The representative single molecule FRET traces obtained for Srs2 showed a short duration of lowto-high FRET fluctuations before the complete unwinding of the open hairpin (Figure 3B). In contrast, Sgs1 induced a rapid transition from high to low FRET, reflecting a fast unwinding of the TNR hairpin (Figure 3C). We measured the dwell time corresponding to the duration of hairpin unwinding (denoted as δt) by Srs2 and Sgs1 and found that Srs2 remained in the hairpin structure three times longer than Sgs1 (Figure 3D, E). In the case of Srs2, the complete unfolding of the TNR hairpin, which is not seen in the closed-loop hairpin substrate, is likely due to the short and open-ended hairpin which is held together only by a short DNA duplex that can be destabilized easily. To test if the dye position induced any difference in hairpin unfolding, we prepared an alternate DNA where the two dyes were positioned across the hairpin junction (Figure S3D). When the same experiments were performed, we obtained the same result with the comparable dwell time distribution for both Srs2 and Sgs1 (Figure S3E, F). This is a clear indication that both proteins proceed through the TNR hairpin and that the Sgs1 induced unwinding of dsDNA seen in Figure 2C occurs after unfolding the TNR. The short duration of FRET fluctuations observed only in Srs2 suggests that Srs2 has an inherent tendency to remain at the TNR hairpin while Sgs1 simply unwinds the hairpin in the same way it unwinds the dsDNA.
FIGURE 3. Open ended TNR targeted by Srs2 and Sgs1, also see Figure S3.
(A) Schematic of open-ended TNR DNA with FRET pair dyes. Single molecule traces obtained for Srs2 (B) and Sgs1 (C). Dwell time histogram of hairpin unwinding by Srs2 (D) and Sgs1 (E). Data are represented as mean ± S.E.M..
To test if ssDNA tail is required for unwinding by Sgs1 and repetitive unfolding by Srs2, we prepared a (CTG)11 containing DNA without the 3’ ssDNA (Figure S3G). At 10nM, Sgs1 induced about 60% unwinding with the rate of 0.3/min whereas Srs2 displayed the similar repetitive unfolding of TNR without unwinding duplex as seen before (Figure S3H-J). This data indicates that the unwinding by Sgs1 and the TNR unfolding by Srs2 may occur in the context of dsDNA without the ssDNA tail.
Repetitive unwinding by Srs2 is altered by the hairpin strength
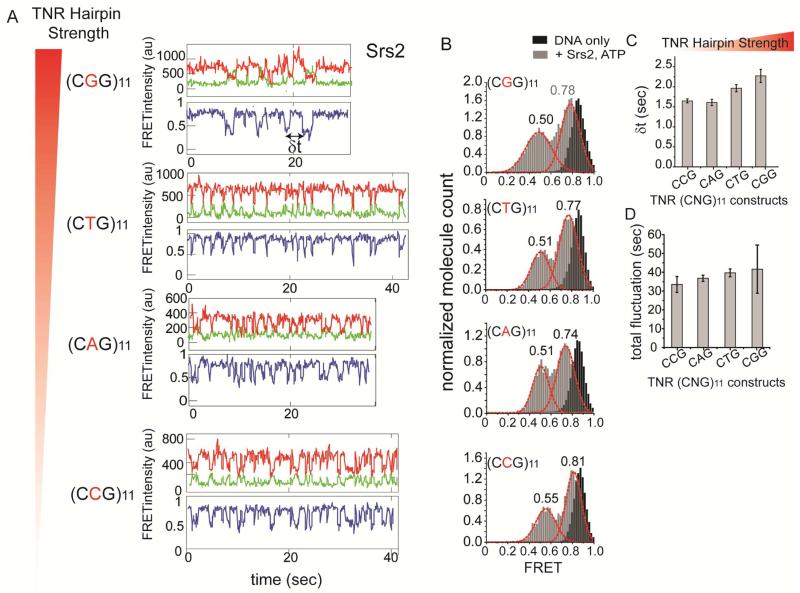
Next, we sought to investigate if the repetitive unfolding of TNR hairpin by Srs2 is affected by the strength of the hairpins. Previous studies indicate that the stability of TNR hairpin depends on the sequence of the triplet, with CGG being the strongest, followed by CTG, CAG and CCG (Mirkin, 2007). We prepared the four TNR DNA substrates mentioned above while keeping the repeat length at 11. When we applied Srs2 and Sgs1 to these substrate separately, we observed loss of both Cy3 and Cy5 signals for Sgs1 but not Srs2 (Figure S4A, B), suggesting that Sgs1 unwound the entire TNR DNA while Srs2 did not. The single molecule traces show that Srs2 exhibits the repetitive unfolding on all four TNR hairpins regardless of the hairpin strength (Figure 4A). To test whether or not the stability of hairpin affects the extent to which the hairpin is opened by Srs2, we collected traces showing FRET fluctuations and compiled the FRET values into FRET histograms for all four TNR sequences (Figure 4B). Overall FRET histograms can report on the different FRET states that the hairpins undergo during Srs2 mediated repetitive unfolding. If the stronger hairpin is unfolded less, the change in FRET will be less. Similarly, if the weakest hairpin is unfolded more, it will result in greater change in FRET due to a larger separation between the two FRET dyes. The FRET arising from DNA-only traces exhibit a single high FRET peak at 0.9 (Figure 4B, shown in black), whereas the FRET histograms taken after the addition of Srs2 and ATP showed two peaks (Figure 4B, shown in gray) arising from the FRET fluctuations between approximately 0.75 to 0.5. The same FRET distribution of 0.75-0.5 observed in all four TNR constructs, suggest that Srs2 repetitively unfolds approximately an equal length of hairpin regardless of the hairpin strength. The MD simulation of the (CAG)11 displays a double helical structure that resembles double strand (ds) DNA. Based on this structure, the unfolding of TNR hairpin will be similar to the unwinding of dsDNA where two strands of ssDNA will be splayed open by the protein situated in between (Figure S4C). In this regard, the lowest FRET value of 0.5 obtained at the most unfolded state indicates that the two dyes are approximately 6 nanometers away, which can arise from approximately 9-12 base pair separation.
FIGURE 4. Repetitive unfolding of Srs2 on varying strength of TNR hairpin, also see Figure S4.
(A) Single molecule traces obtained for varying sequence of TNR hairpin ranging from strongest to weakest folding strength, top to bottom. (B) FRET histogram of DNA only (in black) and after addition of Srs2 and ATP (in gray) and the FRET peak values for each FRET peak. (C) Average dwell time (δt marked in (A)) collected for all DNA constructs marked with standard error bars. (D) Total time of FRET fluctuation duration collected from all DNAs and standard error bars. Data are represented as mean ± S.E.M..
Although the degree of repetitive unfolding by Srs2 remained similar for different TNR hairpins, we observed apparent differences in the periodicity of FRET fluctuation, which represents the frequency at which Srs2 unfolds the hairpin. To make a quantitative comparison, we collected dwell times between successive FRET valleys for each TNR (δt in Figure 4A). The average dwell times collected from over 250 events are plotted from the strongest (CGG) to the weakest (CAG/CGG) hairpin (Figure 4C). It shows that the time interval of FRET fluctuations is slightly lengthened as the stability of the hairpin increases, reflecting a less frequent unfolding activity of Srs2 on a more stable hairpin. However, the total durations of these FRET fluctuations measured for all four TNR substrates remained similar regardless of the triplet sequence (Figure 4D). Taken together, the results indicate that the stability of the hairpin affected the frequency of the hairpin unfolding but did not affect the degree of hairpin unfolding or the total duration of repetitive activity by Srs2.
Repetitive unfolding by Srs2 is affected by the hairpin length
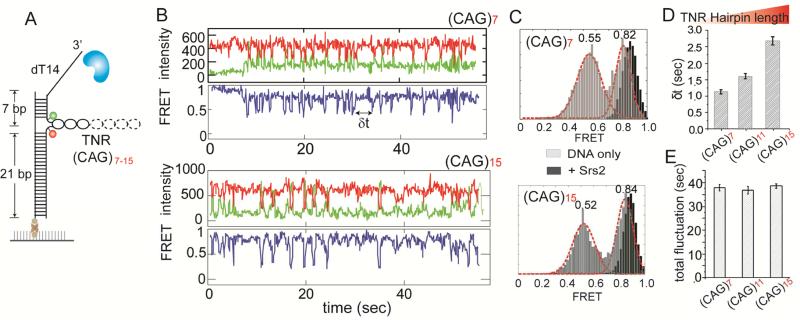
To investigate if the length of TNR influences the ability of Srs2 to resolve the hairpin structure, we varied the CAG triplet sequence length from 7 to 15 repeats (Figure 5A) and compared them to the 11 repeats CAG hairpin tested previously. The single molecule traces obtained for both 7 and 15 repeats showed repetitive hairpin unfolding (indicated by FRET fluctuations) by Srs2 (Figure 5B). In addition, the cumulative FRET histograms reveal the same range of FRET fluctuations as seen in 11 repeats, suggesting that Srs2 unfolds (CAG)7, (CAG)11 and (CAG)15 hairpins to a similar degree (Figure 5C). This indicates that Srs2 unfolds only a limited length of the TNR regardless of the total hairpin length. Such may represent a well-defined distance from the entry of the hairpin where the unfolding may have an important impact on the subsequent biological processes including the replication fork progression.
FIGURE 5. Repetitive unfolding of Srs2 on varying length of TNR hairpin.
(A) Schematic of TNR DNA in which TNR length was changed from seven to fifteen. (B) Single molecule traces obtained for 7 and 15 repeats of CAG. (C) FRET histogram of DNA only (black) and Srs2 with ATP added (gray) for 7 and 15 CAG repeats. (D) Average dwell time (δt) with standard error bars for 7, 11 and 15 CAG repeats. (E) Average dwell time of total FRET fluctuation durations for 7, 11 and 15 CAG repeats, with standard error bars. Data are represented as mean ± S.E.M..
Next, we looked at the frequency of FRET fluctuation amongst different repeat lengths. As before, we collected the dwell times between the two successive unfolding moments (denoted as δt) from over 250 events and plotted the average time for (CAG)7,(CAG)11 and (CAG)15 (Figure 5D). The dwell time for the longest TNR, (CAG)15 was more than two folds higher than that of the shortest TNR, (CAG)7, reflecting the difficulty of Srs2 in invading into the longer TNR hairpin, likely due to the higher thermal stability provided by the longer TNR hairpin. The total duration of the FRET fluctuations remained the same for all three hairpin lengths (Figure 5E). This showed that the length of the triplet repeats in TNR hairpins only affected the frequency of hairpin unfolding by Srs2. This is in agreement with the less frequent unfolding observed for the more stable hairpin (CGG > CTG > CAG = CCG) shown previously (Figure 4C).
DISCUSSION
Previous biochemical studies showed that the deletion of Srs2 or Sgs1 resulted in varying degrees of triplet repeats expansions and contractions that lead to increasing chromosomal fragility and replication fork stalling (Anand et al., 2012; Bhattacharyya and Lahue, 2004; Kerrest et al., 2009). Using 2-dimensional gel-electrophoresis, Srs2 was shown to facilitate the progression of replication fork by unwinding TNR hairpins that may act as a structural barrier (Anand et al., 2012). Interestingly, such activity was independent of Rad51, suggesting that the role of Srs2 in the context of TNR is not related to its role as an anti-recombinase (Bhattacharyya and Lahue, 2004; Kerrest et al., 2009; Qiu et al., 2013). In this study, we sought to probe the mechanisms by which Srs2 and Sgs1 unfold the TNR hairpin structures. We used DNA constructs that contain folded TNR hairpins in dsDNA, similar to those that can form in the process of replication. Such a stable hairpin structure is expected to stall replication machinery unless it is resolved by a helicase such as Srs2 and Sgs1 in yeast.
Our results revealed that Sgs1 and Srs2 are inherently different even for unwinding a duplex DNA without the TNR hairpin. Sgs1 is capable of unwinding dsDNA immediately after the addition of a low concentration of protein (10nM) whereas Srs2, when applied at the same condition, exhibits repetitive movement on single strand DNA without unwinding the duplex DNA (Qiu et al., 2013). We note that Srs2, when applied at a much higher concentration (50-200nM), is capable of unwinding the same DNA efficiently in a tail length-dependent manner, albeit at a lower unwinding rate than Sgs1 (Lytle et al., 2014; Qiu et al., 2013). Furthermore, the immobilized protein assay demonstrates that not Srs2, but a monomer of Sgs1 is sufficient for unwinding the DNA duplex. This is consistent with the previous finding that multimers of Srs2 are required for efficient unwinding (Qiu et al., 2013), with an unwinding concentration threshold (50nM), below which unwinding is limited (Lytle et al., 2014).
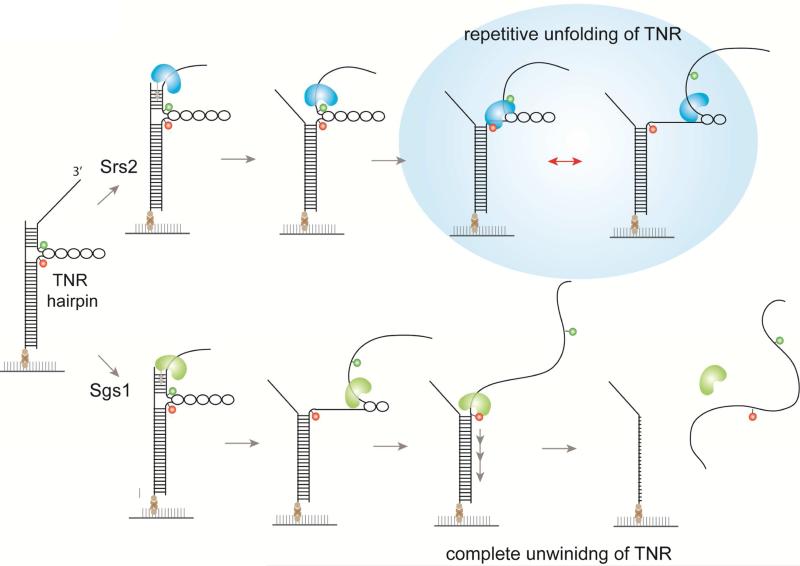
When encountering the TNR hairpins, Sgs1 and Srs2 display a disparate mechanism to resolve this secondary structure (Figure 6). Sgs1 unfolds the TNR hairpin in the same manner that it unwinds the duplex DNA. By adopting the open-ended TNR hairpin structure (Dhar and Lahue, 2008), we showed that Sgs1 does not bypass the TNR hairpin, but unwinds it, likely by tracking the single strand DNA from 3’ to 5’ direction (Bennett et al., 1999; Cejka and Kowalczykowski, 2010; Sun et al., 1999). In contrast, we observe that Srs2 remains at the site of the hairpin while repeatedly unfolding the TNR structure. Based on the range of the fluctuating FRET values, Srs2 is likely acting near the entry of the TNR hairpin. This repetitive unfolding activity can persist for 30-40 seconds without dissociation. We posit that the weak unwinding activity of Srs2 enables it to primarily focus on destabilizing the TNR hairpin rather than proceeding to dsDNA unwinding. This is in agreement with the previous studies reporting that Srs2 is more efficient at resolving TNR than Sgs1 (Anand et al., 2012; Bhattacharyya and Lahue, 2004; Dhar and Lahue, 2008). The repetitive TNR unfolding activity exhibited by Srs2 may provide an efficient mechanism for allowing replication fork to proceed in the presence of DNA secondary structures.
FIGURE 6. Summary of Srs2 and Sgs1 unwinding of TNR hairpin.
When encountering TNR hairpin, Sgs1 unwinds it by tracking single strand DNA in 3’ to 5’ direction and lead to duplex DNA unwinding. In contrast, Srs2 remains near the entry of the TNR hairpin and unfold it repetitively. Based on the different mode of TNR processing, Sgs1 and Srs2 can play a complementary role in resolving TNR structures.
The repetitive activity of SF1 and SF2 helicases have been previously reported (Myong et al., 2007; Myong et al., 2009; Myong et al., 2005; Park et al., 2010; Qi et al., 2013; Qiu et al., 2013). Although it is not clear if this activity is present in cells, based on the diverse array of biological pathways in which they participate, it is plausible to predict that the repetitive activity is conserved for functional purposes. In several cases, it was demonstrated that the protein's repetitive translocation activity serves to keep the single stranded DNA clear of other proteins from binding (Myong et al., 2005; Park et al., 2010; Qiu et al., 2013). In the context of TNR hairpin, the repetitive unfolding by Srs2 may serve to keep the hairpin open to allow the replication machinery to proceed. The repeated action, rather than a single round of unfolding activity, may be more efficient in maintaining the open structure of the hairpin for an extended period of time while waiting for the arrival of a replication complex, for example. In contrast, the complete unwinding of TNR hairpin displayed by Sgs1 may not serve in this capacity since the hairpin can reform easily after the protein has unwound the hairpin. To further test if Sgs1 and Srs2 can be loaded directly to the TNR hairpin without the 3’ ssDNA overhang tail, we tested a control DNA where the ssDNA tail was removed. Interestingly, Sgs1 induced complete unwinding of 60% of this DNA. Srs2, on the other hand, exhibited little-to-no unwinding as before, but showed strong repetitive FRET fluctuations similar to the TNR-DNA with a 3’ tail. Such activity of Srs2 may be more relevant to a genomic locus where most of the DNA are in double-stranded form. It is possible that both Sgs1 and Srs2 act in conjunction with each other, where Sgs1 acts as the forerunner of the initial opening of the hairpin, and Srs2 follows to maintain that opening.
The analysis of Srs2 on varying sequence and length of TNR hairpin reveals some similarities and interesting differences in its ability to resolve the hairpin. The outstanding similarity found in all DNAs we examined is that Srs2 exhibits a repetitive unfolding that unfold similar length of all hairpins. First, the fluctuating FRET signal exhibited in all cases indicates the universality of the repetitive nature of Srs2's hairpin destabilizing activity. Second, the similar level of FRET fluctuation range shown in all cases (FRET histogram peaks) points to the same degree to which Srs2 resolve the secondary structure formed by TNR. This is reminiscent of the repetitive movement of Srs2 seen on single strand (ss) DNA after its removal of Rad51 filament (Qiu et al., 2013). Regardless of the length of ssDNA, Srs2 scrunches a well-defined length of ssDNA. We reasoned that the position of such activity may be crucial in preventing the reformation and nucleation of Rad51 filament. Likewise, the repetitive unfolding of Srs2 at the entry of the hairpin may be advantageous in reducing the critical energetic barrier for the replication machinery to pass through the otherwise tightly structured TNR hairpin. In summary, our study revealed an intrinsic unwinding mechanism of Srs2 and Sgs1 that may lead to differential regulation of TNR hairpin.
EXPERIMENTAL PROCEDURES
Oligonucleotides
Custom oligonucleotides were purchased from Integrated DNA technologies (Coralville, IA). The oligonucleotides with end-labeled dyes are ordered pre-labeled. The oligonucleotides with closed hairpin structure and one or more internally-labeled dyes are ordered with internal amino modifiers at marked at (C6 dT) locations and subsequently labeled using NHS-ester Cy3 and/or Cy5 monofunctional NHS esters simultaneously as described (Joo and Ha, 2008). When two dyes are used for labeling, the ratio of Cy3 to Cy5 dyes used is 1:2.
DNA Sequences
pdT20
5’-GCCTCGCTGCCGTCGCCA-biotin-3’ + 5’-TGGCGACGGCAGCGAGGC-(T)20-3’; Internal amino modifier is represented as (C6 dT), this can be used to label DNA with an internal Cy3 or Cy5 dye.
(CGG)11
5’-GTGTAGCACCGAGGTTTAGGCTGGCACGGTCG-biotin-3’ + 5’-CGACCGTGCCAGCCTAAACC(C6 dT) (CGG)11 (C6 dT)GCTACACTTGCCCGTTTTAT T -3’
(CTG)11
5’-GTGTAGCACCGAGGTTTAGGCTGGCACGGTCG-biotin-3’ + 5’-CGACCGTGCCAGCCTAAACC(C6 dT) (CTG)11 (C6 dT)GCTACACTTGCCCGTTTTAT T -3’
(CAG)11
5’-GTGTAGCACCGAGGTTTAGGCTGGCACGGTCG-biotin-3’ + 5’-CGACCGTGCCAGCCTAAACC(C6 dT) (CAG)11 (C6 dT)GCTACACTTGCCCGTTTTAT T -3’
(CCG)11
5’-GTGTAGCACCGAGGTTTAGGCTGGCACGGTCG-biotin-3’ + 5’-CGACCGTGCCAGCCTAAACC(C6 dT) (CCG)11 (C6 dT)GCTACACTTGCCCGTTTTAT T -3’
(CAG)7
5’-GTGTAGCACCGAGGTTTAGGCTGGCACGGTCG-biotin-3’ + 5’-CGACCGTGCCAGCCTAAACC(C6 dT) (CAG)7 (C6 dT)GCTACACTTGCCCGTTTTAT T -3’
(CAG)15
5’-GTGTAGCACCGAGGTTTAGGCTGGCACGGTCG-biotin-3’ + 5’-CGACCGTGCCAGCCTAAACC(C6 dT) (CAG)15 (C6 dT)GCTACACTTGCCCGTTTTAT T -3’
3-stranded DNA
5-CGACCGTGCCAGCCTAAACCACTGCTGCTGCTGCCGAGAGCC-3’ + 5’-TGGCTC (C6 dT) CGGCTGCTGCTGCTGTGCTACACTTGCCCGTTTTATT-3’ + 5’-Cy5-TGTGTAGCATGCTGGTTTAGGCTGGCACGGTCG-biotin-3’
DNA Substrate Preparation
Partial duplex DNA substrates were prepared by mixing the appropriate biotinylated and nonbiotinylated oligonucleotides in a 1:2 molar ratio at 10 μM concentraion in DNA annealing buffer (10mM MgCl2, 10mM Tris-HCl (pH 8.0)). Partial duplex DNA substrates for tethered-protein experiments were prepared using non-biotinylated strands of oligonucleotides with the same sequences as the biotinylated oligos. The annealing reaction was performed by incubating the two strands at 95°C for 2 minutes followed by slow cooling to room temperature. Three-stranded oligonucleotide mixtures were annealed using method described in (Dhar and Lahue, 2008).
Proteins
The full-length Srs2 protein was overexpressed and purified as described (Antony et al., 2009). The yeast wildtype Sgs1 protein was provided by Prof. Patrick Sung's lab (New Haven, Ct).
Reaction Conditions for Srs2 and Sgs1
Standard reaction buffer was 40 mM Tris-HCl (pH 7.5), 50 mM KCl, 2 mM MgCl2, with an oxygen scavenging system containing 0.8% v/v dextrose, 1 mg/ml glucose oxidase, 0.03 mg/ml catalase(Joo and Ha, 2008), and 2-mercaptoethanol (1% v/v), all items were purchased from Sigma Aldrich (St. Louis, MO). The measurements were performed at room temperature (21°C ± 1°C). 1 mM ATP was used in all experiments unless otherwise specified.
Single-Molecule Fluorescence Assay
smFRET and smPIFE measurements were done using a wide-field total internal reflection fluorescence microscope(Hwang et al., 2011; Roy et al., 2008). Cy3 (donor) on DNA was excited by an Nd:YAG laser (532nM, 75mW, Coherent CUBE) via total internal reflection. The fluorescence signals from Cy3 and Cy5 were collected through an objective (Olympus Uplan SApo; X100 numerical aperture; 1.4 oil immersion) and detected at 100ms time resolution using an EMCCD (electron multiplying charge-coupled device) camera (iXon DU-897ECS0-#BV; Andor Technology). The camera was controlled using home-made C++ program. Single-molecule traces were extracted from the recorded video file by IDL software.
Srs2 and Sgs1 Unwinding partial-duplex DNA
Yeast Srs2 or Sgs1 was each mixed in reaction buffer and ATP to 10 nanomolar conentration and added to a flow imaging chamber that had 100pM partial duplex DNA specifically immobilized on a PEG-coated quartz surface through biotin-NeutrAvidin linkage (Hanahan and Weinberg, 2011). For counting unwound DNA molecules (loss of Cy3 signals), short movies (5-10 seconds) were taken for over 12 minutes.
Srs2 and Sgs1 Unwinding TNR DNA
Yeast Srs2 or Sgs1 was mixed at 10nM with reaction buffer and ATP and added to immobilized TNR DNA as described previously. For counting unwound DNA molecules (loss of Cy3 signals), long movies (3 minutes) were taken for over 15 minutes.
Tethered Srs2 Protein
The full-length Srs2 protein has 9x histadine tags which are tethered to the PEG-coated quartz surface through neutravidin-biotin-tris-NTA linkage. Biotin-tris-NTA was a generous gift from Prof. Paul J. Hergenrother, Department of Chemistry at University of Illinois Urbana-Champaign (Murphy et al., 2005).
The full-length Sgs1 protein has 6x histidine tags which are tethered to the PEG-coated quartz surface via biotinylated anti-his antibody. Anti-6X His tag antibody (Biotin) is obtained through Abcam (Cambridge, MA).
For Srs2 translocation experiments, biotin-tris-NTA (20nM) was mixed NiCl2 (50nM) in T50 buffer (50mM Tris pH 7.5, 50mM NaCl) and incubated on ice for 15 minutes. The mixture is then added to a flow chamber that already had NeutrAvidin immobilized to the PEG-coated surface, then allowed to incubate for 10 minutes at room temperature. 0.5-1nM of Srs2 in T50 buffer were then added to the flow chamber and incubated for 5 minutes at room temperature. Finally, non-biotinylated partial duplex DNA substrate in reaction buffer and ATP was added to the flow chamber to initiate the reaction.
For Sgs1 translocation experiments, biotinylated anti-his antibody (10nM) was added to a flow chamber that already had neutravidin immobilized to the PEG-coated surface, then allowed to incubate for 10 minutes at room temperature. 0.5-1nM of Sgs1 in T50 buffer were then added to the flow chamber and incubated for 5 minutes at room temperature. Finally, non-biotinylated partial duplex DNA substrate in reaction buffer and ATP was added to the flow chamber to initiate the reaction.
Data Analysis
Single molecule traces were analyzed using codes written in Matlab. FRET efficiency values were calculated as a ratio between acceptor intensity and total donor and acceptor intensity (Roy et al., 2008).
For various dwell time analyses: FRET valley-to-valley dwell time analysis to obtain δt was measured manually from individual FRET traces within Matlab, and the resulting histograms and fittings were generated using Origin (OriginLab Corporation, Northampton, MA). Binning sizes vary based on the type and range of data collected. Fluctuation duration is measured as the time that DNA is occupied by the protein before leaving. DNA unwinding time by Srs2 or Sgs1 is measured as the time it took from high FRET fluctuations (protein occupying DNA) to go to low FRET before signal disappearance due to DNA unwinding.
Supplementary Material
Highlights (less than 85 characters with space).
Srs2 unfolds trinucleotide repeat (TNR) hairpin repetitively.
Sgs1 unwinds TNR hairpin completely by translocation mediated unwinding.
Srs2 activity depends on the folding strength and the total length of TNR hairpin.
Disparate mechanism of Srs2 and Sgs1 may cooperatively resolve TNR hairpin.
ACKNOWLEDGEMENTS
We thank the Myong lab members for helpful discussions and Jordan Billingsley for editing the manuscript.
FUNDING
Support for this work was provided by NIH Director's New Innovator Award (343 NIH 1 DP2 GM105453 A) and American Cancer Society (Research Scholar Grant; RSG-12-066-01-DMC) for S. M. and Y. Q.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Y.Q purified Srs2 protein, conducted all the experiments and analysis and participated in writing and editing the manuscript. S.M supervised the study and wrote the manuscript. H.N purified the Sgs1 protein under the guidance of P.S. L.V performed the MD simulation.
REFERENCES
- Anand RP, Shah KA, Niu H, Sung P, Mirkin SM, Freudenreich CH. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Keck JL, Wang JC. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J Mol Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Lahue RS. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol Cell Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J Biol Chem. 2010;285:8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication- mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- Dhar A, Lahue RS. Rapid unwinding of triplet repeat hairpins by Srs2 helicase of Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:3366–3373. doi: 10.1093/nar/gkn225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci U S A. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol Cell Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature genetics. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Ha T. Single-molecule FRET with total internal reflection microscopy. In: Selvin PR, Ha T, editors. Single- molecule techniques : a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2008. pp. 3–35. [Google Scholar]
- Kerrest A, Anand RP, Sundararajan R, Bermejo R, Liberi G, Dujon B, Freudenreich CH, Richard GF. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat Struct Mol Biol. 2009;16:159–167. doi: 10.1038/nsmb.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue RS, Slater DL. DNA repair and trinucleotide repeat instability. Front Biosci. 2003;8:s653–665. doi: 10.2741/1107. [DOI] [PubMed] [Google Scholar]
- Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nature chemical biology. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle AK, Origanti SS, Qiu Y, VonGermeten J, Myong S, Antony E. Context-dependent remodeling of Rad51-DNA complexes by Srs2 is mediated by a specific protein-protein interaction. J Mol Biol. 2014;426:1883–1897. doi: 10.1016/j.jmb.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. To switch or not to switch: at the origin of repeat expansion disease. Mol Cell. 2014;53:1–3. doi: 10.1016/j.molcel.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Murphy RT, Thaman R, Blanes JG, Ward D, Sevdalis E, Papra E, Kiotsekoglou A, Tome MT, Pellerin D, McKenna WJ, Elliott PM. Natural history and familial characteristics of isolated left ventricular non-compaction. European heart journal. 2005;26:187–192. doi: 10.1093/eurheartj/ehi025. [DOI] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM. Replication and expansion of trinucleotide repeats in yeast. Molecular and cellular biology. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Pugh RA, Spies M, Chemla YR. Sequence-dependent base pair stepping dynamics in XPD helicase unwinding. eLife. 2013;2:e00334. doi: 10.7554/eLife.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nature communications. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nature genetics. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol. 2009;16:226–228. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.