Abstract
Merkel cell polyomavirus (MCV) is the etiological agent of Merkel cell carcinoma (MCC), a rare and highly lethal human skin cancer. A natural component of skin flora, MCV becomes tumorigenic only after integration into the host DNA together with specific mutations to the viral genome. Research on MCV large T (LT) and small T (sT) antigens, the only viral products expressed in MCC, shows that these major oncoproteins not only possess biochemical functions found in common with other polyomavirus T antigens, but also demonstrate new cellular targets not described in previous polyomavirus models. This review provides a map of the relevant functional motifs and domains in MCV T antigens that have been identified, highlighting their roles in tumorigenesis.
Introduction
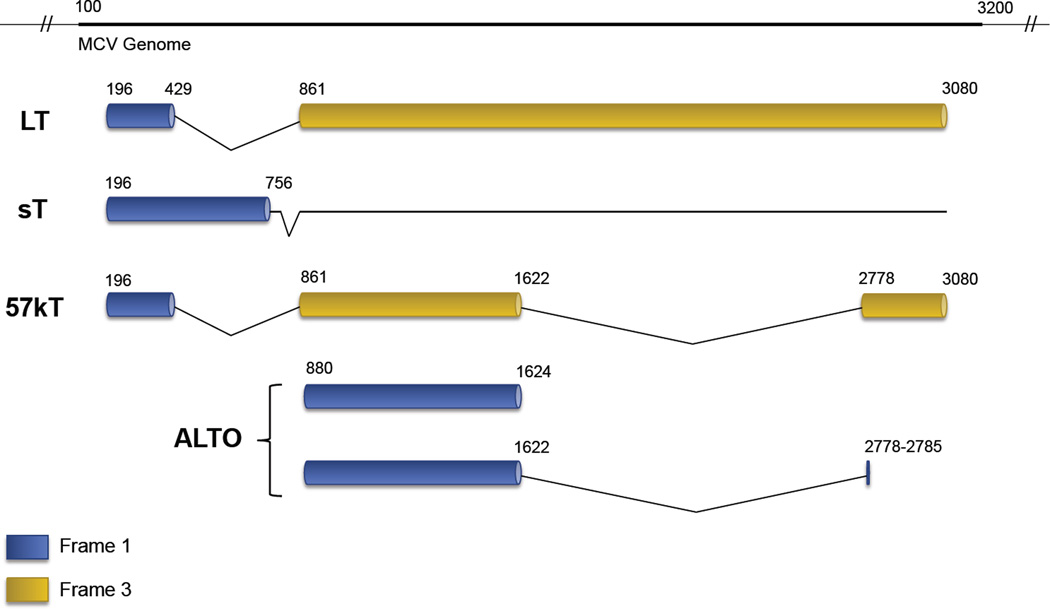
Merkel cell polyomavirus (MCV) is the newest member of the short list of human cancer viruses [1,2], and is the only known human polyomavirus confirmed to be oncogenic [3–17]. While MCV was only discovered in 2008, polyomavirus research dates back over a half-century, beginning with the isolation of murine polyomavirus (MuPyV) [18] and later simian vacuolating virus 40 (SV40) [19,20]. These polyomaviruses have provided invaluable insights into our mechanistic understanding of tumor and cell biology. Polyomaviruses have small genomes (~5kb) comprised of early and late coding regions, separated by a noncoding regulatory region (NCRR). The early region contains the T (“Tumor”) antigen gene locus [21], from which multiple, alternatively-spliced RNA transcripts are generated. MCV expresses four unique gene products from this early coding region: the large T (LT), small (sT), and 57kT antigens along with a product from an alternate frame of the LT open reading frame (ALTO) [22] (Figure 1). In natural polyomavirus lytic infection, a sequential expression of early antigens followed by late capsid proteins is seen. By contrast, MCV-associated tumorigenesis is characterized and mediated by the sole expression of LT and sT antigens [21,23]. This review will present a biochemical map of the functionally relevant motifs and domains within LT and sT, the two major oncoproteins of MCV.
Large T Antigen
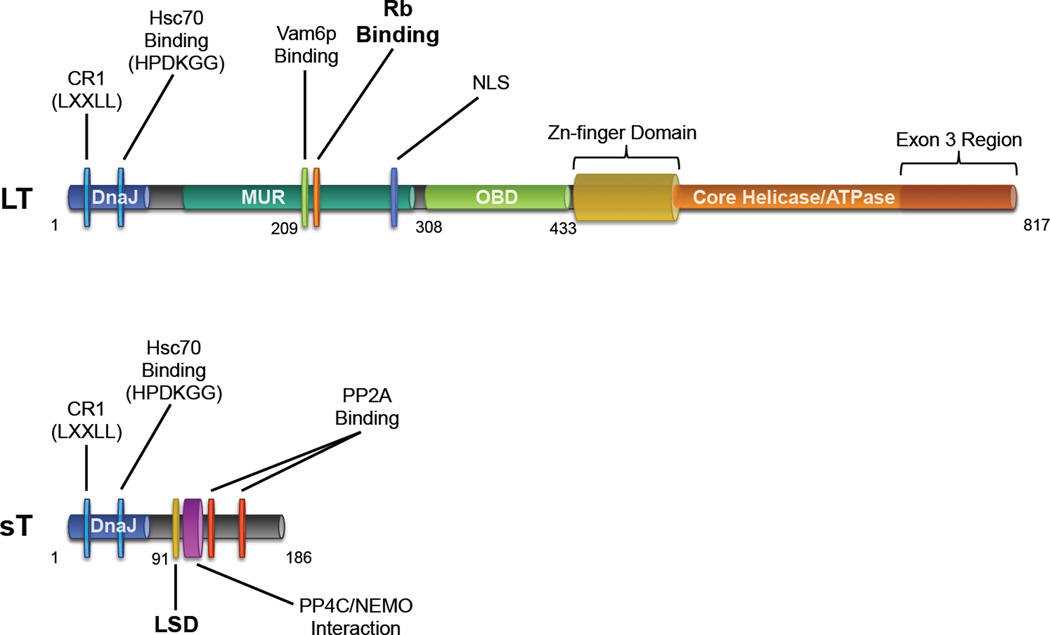
The LT antigens of polyomaviruses contain a number of common motifs and domains important for facilitating the viral life cycle [24]. In the context of oncogenesis, some of these elements also have the effect of disabling tumor suppressor pathways, for example by targeting Rb and p53 [21]. The LT antigen of MCV encodes many of these conserved features as well as a few unique ones (Figure 2).
The N-terminal end of MCV LT (1–70 aa) contains the DnaJ domain [24] comprised of the CR1 (13–17 aa) motif followed by the HPDKGG hexapeptide sequence responsible for Hsc70 binding (42–47 aa) [24,27]. Kwun et al. confirmed that MCV LT interacts with Hsc70, and by disrupting this interaction with a point mutation, they showed the necessity of the DnaJ domain for MCV replication in vitro [28].
Between the first exon and the OBD (~100–300 aa) lies a stretch of sequences that contains a conserved LXCXE motif and nuclear localization signal [29], but otherwise bears little homology to other polyomaviruses. This region, designated the MCV T antigen unique region (MUR) contains a binding motif for the vacuolar sorting protein Vam6p. The LT-Vam6p interaction, which can be ablated by mutation of a single tryptophan residue at position 209, results in the nuclear sequestration of this cytosolic protein and disrupts lysosomal clustering [30]. Although Vam6p interaction appears to be unique to MCV, the site of this interaction parallels the site for Bub1 interaction in SV40 LT, which also depends on the presence of tryptophan residues and modulates SV40 LT-mediated transformation by overriding the mitotic spindle checkpoint [21,31]. In an in vitro replication assay using an infectious molecular clone of MCV mutated at position 209, Feng et al. demonstrated that loss of LT-Vam6p binding leads to enhanced viral replication compared to a wild-type control [32]. It is possible that in the natural life cycle of MCV, LT-Vam6p interaction inhibits or minimizes viral reactivation, a potential form of ‘viral latency’. SV40 miRNA has been proposed to serve a similar autoregulatory function by inhibiting SV40 LT expression [33]. MCV encodes an miRNA that may have a similar function and may augment Vam6p-related replication silencing [34,35]. Whether or not Vam6p targeting is also important in tumorigenesis is not presently known.
The Rb-binding (LXCXE) motif of LT (212–216 aa) embedded in the MUR lies directly adjacent to the Vamp6-binding site (209 aa). This highly conserved sequence across polyomaviruses allows for dysregulation of E2F-mediated transcription and drives cells into S-phase [21,25]. Rb-binding and subsequent upregulation of E2F target genes such as cyclin E has been confirmed for MCV LT [27,36]. Houben et al. showed the importance of the LXCXE motif in the proliferation of MCC tumors cells, since complementation with an Rb-binding mutant of LT after RNAi-mediated T antigen depletion in MCC cell lines was incapable of rescuing cell growth [37]. A novel biochemical function was mapped to this region when Arora et al. observed that upregulation of the anti-apoptotic protein survivin upon LT expression is dependent on the presence of and intact LXCXE motif [36]. Targeting LT-mediated survivin upregulation represents a viable therapeutic option, supported by in vivo studies where pharmacologic inhibition of survivin lengthened the survival of mice bearing MCC xenografts [36,38].
The C-terminal portion of MCV LT contains several critical elements required for viral replication. The OBD of LT (308–433 aa) is responsible for recognition and binding of a minimum 71-bp origin of replication in the MCV NCRR [28,39]. Diaz et al. observed that phosphorylation of LT upstream of the OBD at residues T297 and T299 decreases the origin-binding affinity of LT and negatively affects replication initiation [40]. Following the OBD is a zinc-finger motif (437–528 aa) that is contained in the core polyomavirus helicase/ATPase domain (441–817 aa), both of which are important for LT oligomerization and initiation of replication [25]. In addition to replicase activity, the helicase domain in SV40 contains a bi-partite sequence that can directly bind p53 and prevent transcription of its target genes, allowing for the evasion of senescence or apoptosis [21,24,25,41–43]. Currently, there is no evidence that either full length or truncated forms of MCV LT directly bind p53 [44,45].
One of the most striking features of the LT antigen of MCV is that the C-terminal domains are consistently truncated by tumor associated polymorphisms, but and the N terminus up to the Rb-binding motif are always intact [27,46]. The short NLS of MCV LT that lies between these two regions (277–280 aa) was initially believed to also be conserved in tumors [29], but recent evidence indicates that the NLS is not preferentially preserved in MCC [44,47]. In the cases where the NLS is eliminated by tumorspecific mutations to LT (as in cell lines MCC350, MCCL-11, and MCCL-12) nuclear localization is not lost, but rather both nuclear and cytoplasmic distribution are observed [44].
This signature pattern of C-terminal truncation indicates a selective pressure for the elimination of viral replicative elements in tumor development since a full-length LT could initiate unlicensed replication at the site of integration in the host genome leading to a DNA damage response (DDR) and cell death [27]. This is not unlike other polyomaviruses (or other tumor viruses) in which replication competency is lost or suppressed in the setting of tumorigenesis [48–50]. This notion is supported by Li et al. who observed a DDR induced by MCV infection which was mapped to the core helicase region of LT. The expression of a LT with intact and functional helicase induces ATR and subsequent p53 activation in multiple cell lines [51] leading to cell cycle arrest and decreased proliferation. There exists additional pressure for Cterminal truncation of LT that has been mapped to the very C-terminal end of the protein, corresponding to the third exon of 57kT, which is shared by LT (717–817 aa). Upon retroviral transduction of an exon 3 construct, Cheng et al. noticed retardation of cell growth in an established MCC cell line as well as in human fibroblasts immortalized by SV40. The mechanism underlying this phenotype is presently unknown [45]. Altogether, this evidence suggests that the C-terminal portion of LT generally has an antiproliferative effect, necessitating its deletion in tumorigenesis.
Small T Antigen
Although MCV LT and sT share exon 1 of the T antigen locus, and the DnaJ domain of LT is required for replication, the functional significance of this domain in sT is unknown. It has been shown that mutating Hsc70 binding on sT does not interfere with its effect on MCV viral replication [28] or the in vitro transformative activity of sT [52]. This is similar to SV40 in which the DnaJ domain of LT sT is known to bind Hsc70 [53], but this function also does not impact tumorigenesis [21,54].
The primary functions of sT antigens in the prototypic polyomaviruses SV40 and MuPyV has been long attributed to modifying Akt-mTOR signaling via their interactions with protein phosphatase 2A (PP2A) [55–57], a heterotrimeric complex comprised of ABC subunits. sT antigens bind to the A and C subunits of PP2A to displace a variety of modular B subunits [21,57], but the consequences of these interactions and their requirement in tumorigenesis varies between different polyomavirus species [58]. Both SV40 and MuPyV sT antigens have been shown to have broad effects on cell cycle progression, survival, and differentiation that are dependent on PP2A [59,60]. The primacy of PP2A targeting in other polyomavirus sT antigens contrasts directly with MCV sT, whose PP2A interacting domains (119–124 and 147–152 aa) are dispensable for its in vitro and in vivo transformative activity [52,61]. MCV sT instead possesses other functionalities in addition to PP2A binding that make it a key oncoprotein for MCC. MCV sT expression alone, independent of LT, is responsible for in vitro transformation of rodent fibroblasts in soft agar and focus formation assays [52], and sT is required for continued cell proliferation in MCC [62]. Recent work by Verhaegen et al. has shown that MCV sT has in vivo proliferative activity when expressed in mice [61].
A major feature of MCV sT is a domain spanning 74–98 aa in intron 1 that is predicted to encode for an exposed, unstructured loop remotely located from PP2A binding sites [63]. Scanning alanine mutagenesis in this loop has identified a region from 91–95 aa termed the LT-Stabilization Domain (LSD). This domain is responsible for inhibiting the SCFFbw7 E3 ubiquitin ligase [63]. Since LT is a target of Fbw7, co-expression of sT prevents LT degradation, increases steady-state LT levels and has been shown to enhance viral replication [63]. sT targeting of Fbw7 not only regulates LT levels during natural infections, sT also stabilizes truncated tumor T antigens and other SCFFbw7 substrates, including c-Myc and cyclin E, contributing to carcinogenesis. The importance of the LSD is evidenced by the fact that when mutated, sT loses its in vitro transformation activity [63].
In addition to Fbw7, an intact sT LSD domain is required for targeting 4E-BP1, the major regulator of eukaryotic cap-dependent translation. In its active, hypophosphorylated form, 4E-BP1 directly binds and inhibits eIF4E from recruiting 40S ribosomal assembly at the 5’ cap of mRNA and inhibits translation. Mammalian target of rapamycin complex 1 (mTOR1)-mediated phosphorylation of 4E-BP1 releases 4E BP1 from eIF4E allowing translation initiation to proceed. sT expression has been shown to result in increased hyperphosphorylated 4E-BP1 that independent of mTORC1 signaling [52]. The precise mechanism for sT-mediated 4E-BP1 hyperphosphorylation is not defined at present, but is independent of PP2A and Fbw7 interaction [63].
Griffiths et al. through a mass spectrometric approach have revealed that MCV sT also interacts with protein phosphatase 4C (PP4C) at a region (95–111 aa) directly adjacent to the LSD at 91–95 aa [64]. MCV sT binding of PP4C or PP2A Aβ targets the NF-κB essential modulator (NEMO) protein and prevents nuclear translocation of NF-κB, inhibiting NF-κB-mediated transcription[64]. PP4C binding is also important for the induction of a highly motile cell phenotype that may correspond to the highly metastatic nature of MCC [65].
Conclusions
In six short years, the map of MCV has transitioned from a simple sketch to a more intricate schematic with detailed characterizations of the viral gene products and how they impact tumorigenesis. Unearthing the functional aspects of the T antigens has aided in identifying novel targets for MCC treatment, spurring the development of MCV-directed vaccines and therapeutics [66–69]. Still, there exist uncharted activities associated with MCV infection that warrant additional characterization for a more comprehensive understanding of this virus and its role in MCC. As previous research on polyomaviruses has contributed fundamental concepts in cell and cancer biology, continued efforts in elucidating the mechanisms of MCV pathogenesis may uncover similar discoveries that extend beyond the context of MCC.
Acknowledgements
We would like to thank the members of the Chang-Moore lab for helpful discussions during the construction of this review.This work was supported by the National Institutes of Health CA136363, CA136806, CA170354 grants (P.S.M. and Y.C), the American Cancer Society (P.S.M. and Y.C.), and by the Pittsburgh and UPMC Foundations. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References and recommended reading
- 1.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore PS, Chang Y. The conundrum of causality in tumor virology: the cases of KSHV and MCV. Semin Cancer Biol. 2014;26:4–12. doi: 10.1016/j.semcancer.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 4.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 5.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. Identification of a third human polyomavirus. J Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6:e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus. J Virol. 2011;85:4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O, Manuguerra JC, Richardson J, Lecuit M, Burguiere A, et al. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg Infect Dis. 2011;17:1364–1370. doi: 10.3201/eid1708.110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol. 2012;86:10321–10326. doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu G, Greninger AL, Isa P, Phan TG, Martinez MA, de la Luz Sanchez M, Contreras JF, Santos-Preciado JI, Parsonnet J, Miller S, et al. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS One. 2012;7:e49449. doi: 10.1371/journal.pone.0049449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA. Complete genome sequence of a tenth human polyomavirus. J Virol. 2012;86:10887. doi: 10.1128/JVI.01690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology. 2013;436:295–303. doi: 10.1016/j.virol.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One. 2013;8:e58021. doi: 10.1371/journal.pone.0058021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, Kapoor A, Briese T, Faust PL, Lipkin WI. Identification of a Novel Polyomavirus in a Pancreatic Transplant Recipient With Retinal Blindness and Vasculitic Myopathy. J Infect Dis. 2014 doi: 10.1093/infdis/jiu250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross L. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc Soc Exp Biol Med. 1953;83:414–421. doi: 10.3181/00379727-83-20376. [DOI] [PubMed] [Google Scholar]
- 19.Sweet BH, Hilleman MR. The vacuolating virus, S.V. 40. Proc Soc Exp Biol Med. 1960;105:420–427. doi: 10.3181/00379727-105-26128. [DOI] [PubMed] [Google Scholar]
- 20.Eddy BE. Tumors Produced in Hamsters by Sv40. Federation Proceedings. 1962;21 930-&. [Google Scholar]
- 21.Gjoerup O, Chang Y. Update on human polyomaviruses and cancer. Adv Cancer Res. 2010;106:1–51. doi: 10.1016/S0065-230X(10)06001-X. [DOI] [PubMed] [Google Scholar]
- 22.Carter JJ, Daugherty MD, Qi X, Bheda-Malge A, Wipf GC, Robinson K, Roman A, Malik HS, Galloway DA. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc Natl Acad Sci U S A. 2013;110:12744–12749. doi: 10.1073/pnas.1303526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pipas JM. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. An P, Saenz Robles MT, Pipas JM. Large T antigens of polyomaviruses: amazing molecular machines. Annu Rev Microbiol. 2012;66:213–236. doi: 10.1146/annurev-micro-092611-150154. This article reviews the shared features of the LT antigens of various polyomaviruses
- 26.DeCaprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nat Rev Microbiol. 2013;11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol. 2009;83:12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T, Sato Y, Watanabe D, Ito H, Shimonohara N, Tsuji T, Nakajima N, Suzuki Y, Matsuo K, Nakagawa H, et al. Nuclear localization of Merkel cell polyomavirus large T antigen in Merkel cell carcinoma. Virology. 2010;398:273–279. doi: 10.1016/j.virol.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Hein J, Richardson SC, Basse PH, Toptan T, Moore PS, Gjoerup OV, Chang Y. Merkel cell polyomavirus large T antigen disrupts lysosome clustering by translocating human Vam6p from the cytoplasm to the nucleus. J Biol Chem. 2011;286:17079–17090. doi: 10.1074/jbc.M110.192856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cotsiki M, Lock RL, Cheng Y, Williams GL, Zhao J, Perera D, Freire R, Entwistle A, Golemis EA, Roberts TM, et al. Simian virus 40 large T antigen targets the spindle assembly checkpoint protein Bub1. Proc Natl Acad Sci U S A. 2004;101:947–952. doi: 10.1073/pnas.0308006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J Clin Virol. 2011;52:272–275. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 36. Arora R, Shuda M, Guastafierro A, Feng H, Toptan T, Tolstov Y, Normolle D, Vollmer LL, Vogt A, Domling A, et al. Survivin is a therapeutic target in Merkel cell carcinoma. Sci Transl Med. 2012;4:133ra156. doi: 10.1126/scitranslmed.3003713. This article identifies the Rb-dependent upregulation of survivin by LT and how this could be a useful therapeutic target in MCC.
- 37. Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer. 2012;130:847–856. doi: 10.1002/ijc.26076. This article provides evidence that the Rb-binding motif of LT is critical in MCC tumor maintenance..
- 38. Dresang LR, Guastafierro A, Arora R, Normolle D, Chang Y, Moore PS. Response of Merkel cell polyomavirus-positive merkel cell carcinoma xenografts to a survivin inhibitor. PLoS One. 2013;8:e80543. doi: 10.1371/journal.pone.0080543. This article shows the responsiveness of MCC xenografts to survivin inhibition using the compound YM155
- 39.Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J Mol Biol. 2011;409:529–542. doi: 10.1016/j.jmb.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz J, Wang X, Tsang SH, Jiao J, You J. Phosphorylation of large T antigen regulates merkel cell polyomavirus replication. Cancers (Basel) 2014;6:1464–1486. doi: 10.3390/cancers6031464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bargonetti J, Reynisdottir I, Friedman PN, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 42.Segawa K, Minowa A, Sugasawa K, Takano T, Hanaoka F. Abrogation of p53-mediated transactivation by SV40 large T antigen. Oncogene. 1993;8:543–548. [PubMed] [Google Scholar]
- 43.Pipas JM, Levine AJ. Role of T antigen interactions with p53 in tumorigenesis. Semin Cancer Biol. 2001;11:23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- 44.Borchert S, Czech-Sioli M, Neumann F, Schmidt C, Wimmer P, Dobner T, Grundhoff A, Fischer N. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J Virol. 2014;88:3144–3160. doi: 10.1128/JVI.02916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J Virol. 2013;87:6118–6126. doi: 10.1128/JVI.00385-13. This article describes the anti-proliferative effect mediated by the C-terminal end of LT.
- 46.Fischer N, Brandner J, Fuchs F, Moll I, Grundhoff A. Detection of Merkel cell polyomavirus (MCPyV) in Merkel cell carcinoma cell lines: cell morphology and growth phenotype do not reflect presence of the virus. Int J Cancer. 2010;126:2133–2142. doi: 10.1002/ijc.24877. [DOI] [PubMed] [Google Scholar]
- 47.Houben R, Angermeyer S, Haferkamp S, Aue A, Goebeler M, Schrama D, Hesbacher S. Characterization of functional domains in the Merkel cell polyoma virus Large T antigen. Int J Cancer. 2014 doi: 10.1002/ijc.29200. [DOI] [PubMed] [Google Scholar]
- 48.zur Hausen H. A specific signature of Merkel cell polyomavirus persistence in human cancer cells. Proc Natl Acad Sci U S A. 2008;105:16063–16064. doi: 10.1073/pnas.0808973105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manos MM, Gluzman Y. Simian virus 40 large T-antigen point mutants that are defective in viral DNA replication but competent in oncogenic transformation. Mol Cell Biol. 1984;4:1125–1133. doi: 10.1128/mcb.4.6.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadaja M, Sumerina A, Verst T, Ojarand M, Ustav E, Ustav M. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 2007;26:2180–2191. doi: 10.1038/sj.emboj.7601665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li J, Wang X, Diaz J, Tsang SH, Buck CB, You J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J Virol. 2013;87:9173–9188. doi: 10.1128/JVI.01216-13. This article shows that the C-terminal end of LT induces a DDR that results in cell cycle arrest and reduced cell proliferation.
- 52.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyapati A, Wilson M, Yu J, Rundell K. SV40 17KT antigen complements dnaj mutations in large T antigen to restore transformation of primary human fibroblasts. Virology. 2003;315:148–158. doi: 10.1016/s0042-6822(03)00524-5. [DOI] [PubMed] [Google Scholar]
- 55.Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez-Viciana P, Collins C, Fried M. Polyoma and SV40 proteins differentially regulate PP2A to activate distinct cellular signaling pathways involved in growth control. Proc Natl Acad Sci U S A. 2006;103:19290–19295. doi: 10.1073/pnas.0609343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang JH, Jiang T, Kulkarni S, Faure N, Schaffhausen BS. Protein phosphatase 2A isoforms utilizing Abeta scaffolds regulate differentiation through control of Akt protein. J Biol Chem. 2013;288:32064–32073. doi: 10.1074/jbc.M113.497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrabi S, Hwang JH, Choe JK, Roberts TM, Schaffhausen BS. Comparisons between murine polyomavirus and Simian virus 40 show significant differences in small T antigen function. J Virol. 2011;85:10649–10658. doi: 10.1128/JVI.05034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pores Fernando AT, Andrabi S, Cizmecioglu O, Zhu C, Livingston DM, Higgins JM, Schaffhausen BS, Roberts TM. Polyoma small T antigen triggers cell death via mitotic catastrophe. Oncogene. 2014 doi: 10.1038/onc.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hwang JH, Pores Fernando AT, Faure N, Andrabi S, Hahn WC, Schaffhausen BS, Roberts TM. Polyomavirus small T antigen interacts with yes-associated protein to regulate cell survival and differentiation. J Virol. 2014;88:12055–12064. doi: 10.1128/JVI.01399-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Verhaegen ME, Mangelberger D, Harms PW, Vozheiko TD, Weick JW, Wilbert DM, Saunders TL, Ermilov AN, Bichakjian CK, Johnson TM, et al. Merkel Cell Polyomavirus Small T Antigen is Oncogenic in Transgenic Mice. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.446. This article provides evidence for in vivo transformative activity of sT with respect to its functional domains.
- 62.Shuda M, Chang Y, Moore PS. Merkel cell polyomavirus-positive Merkel cell carcinoma requires viral small T-antigen for cell proliferation. J Invest Dermatol. 2014;134:1479–1481. doi: 10.1038/jid.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe. 2013;14:125–135. doi: 10.1016/j.chom.2013.06.008. This article identified the LSD of sT as playing a key role in MCV replication and tumorigenesis.
- 64. Griffiths DA, Abdul-Sada H, Knight LM, Jackson BR, Richards K, Prescott EL, Peach AH, Blair GE, Macdonald A, Whitehouse A. Merkel cell polyomavirus small T antigen targets the NEMO adaptor protein to disrupt inflammatory signaling. J Virol. 2013;87:13853–13867. doi: 10.1128/JVI.02159-13. This article identifies a PP4C interaction for sT that results in an immune subversive mechanism for MCV
- 65.Knight LM, Stakaityte G, Wood JJ, Abdul-Sada H, Griffiths DA, Howell GJ, Wheat R, Blair GE, Steven NM, Macdonald A, et al. Merkel cell polyomavirus small T antigen mediates microtubule destabilisation to promote cell motility and migration. J Virol. 2014 doi: 10.1128/JVI.02317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomez BP, Wang C, Viscidi RP, Peng S, He L, Wu TC, Hung CF. Strategy for eliciting antigenspecific CD8+ T cell-mediated immune response against a cryptic CTL epitope of merkel cell polyomavirus large T antigen. Cell Biosci. 2012;2:36. doi: 10.1186/2045-3701-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomez B, He L, Tsai YC, Wu TC, Viscidi RP, Hung CF. Creation of a Merkel cell polyomavirus small T antigen-expressing murine tumor model and a DNA vaccine targeting small T antigen. Cell Biosci. 2013;3:29. doi: 10.1186/2045-3701-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyngaa R, Pedersen NW, Schrama D, Thrue CA, Ibrani D, Met O, Thor Straten P, Nghiem P, Becker JC, Hadrup SR. T-cell responses to oncogenic merkel cell polyomavirus proteins distinguish patients with merkel cell carcinoma from healthy donors. Clin Cancer Res. 2014;20:1768–1778. doi: 10.1158/1078-0432.CCR-13-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willmes C, Adam C, Alb M, Volkert L, Houben R, Becker JC, Schrama D. Type I and II IFNs inhibit Merkel cell carcinoma via modulation of the Merkel cell polyomavirus T antigens. Cancer Res. 2012;72:2120–2128. doi: 10.1158/0008-5472.CAN-11-2651. [DOI] [PubMed] [Google Scholar]