Abstract
We have recently recapitulated metastasis of human PTEN/TP53-mutant PC in mouse using the RapidCaP system. Surprisingly, we found that this metastasis is driven by Myc-, and not Akt-activation.
Here, we show that cell-cell communication by Il6 drives the Akt-Myc switch through activation of the Akt-suppressing phosphatase Phlpp2, when Pten and p53 are lost together, but not separately. Il6 then communicates a downstream program of Stat3-mediated Myc-activation, which drives cell proliferation. Similarly in tissues, peak proliferation in Pten/Trp53 mutant primary and metastatic PC does not correlate with activated Akt, but with Stat3/Myc activation instead. Mechanistically, Myc strongly activates the Akt phosphatase Phlpp2 in primary cells and PC metastasis. We show genetically that Phlpp2 is essential for dictating proliferation of Myc-mediated Akt-suppression.
Collectively, our data reveal competition between two proto-oncogenes: Myc and Akt, which ensnarls the Phlpp2 gene to facilitate Myc-driven PC metastasis after loss of Pten and Trp53.
INTRODUCTION
Prostate cancer (PC) is one of the most prevalent cancers among men and it is estimated that in 2014 there will be 233,000 new PC cases, making up 14% of all new cancer cases, causing an estimated 29,480 deaths in the United States alone (1). Although mortality rates have been decreasing, PC is still among the most common causes of cancers-related death and malignancy in men in developed countries (2). The prognosis for men with metastatic PC is worst, especially due to resistances that arise during treatment (3, 4). Although a large-scale recent study suggested that early (PSA-based) detection reduces mortality, the screening-associated interventions have negative side effects that challenge the low efficiency of the screening effort and can result in over-treatment of the disease (5). Thus, there is an unmet need for causative biomarkers of potentially lethal PC and for more effective treatment options.
Recent advances in the genomic analysis of PC have revealed a wealth of data about gene alterations in the disease. Commonly observed events include: TMPRSS2-ERG gene fusions (6), deletions of 8p and gains in 8q chromosomal arms (7) and point mutations in the SPOP gene (8). Regarding progression to lethal PC, a most notable and potentially actionable principle that has emerged is the observed increase and predictive value of copy number alterations in disease progression (9). Metastatic PC commonly harbors MYC and AR amplifications, and deletions of PTEN, RB1 and TP53 tumor suppressor genes (9, 10).
To perform functional analyses of lethal metastatic PC, we recently generated the RapidCaP mouse model, which allowed us to demonstrate that loss of Pten and Trp53 genes suffices to cause PC metastasis at near complete penetrance (11). These data strongly suggested that in Pten-deficient mouse PC the progression from indolent to metastatic disease requires disruption of the p53-senescence response (12). Although there is much progress in understanding the role that genetic alterations play within the metastatic PC cell it has remained less well understood how these aberrations affect cell-cell communication. Emerging evidence supports the idea that inflammation contributes to initiation and progression of PC (13), and that inflammatory chemokines and interleukins affect cell motility and proliferation in the transition from normal to Benign Prostatic Hyperplasia (BPH) to PC (reviewed here (14)).
Thus, we set out to define hallmarks of cell secretion that are associated with the metastatic gene signature of Pten and Trp53 loss. Our findings reveal that Il6 secretion is integral to metastasis of Pten-deficient PC because it promotes the Myc proto-oncogene to drive proliferation and disease progression.
RESULTS
Co-deletion of Pten and Trp53 triggers Il6 secretion
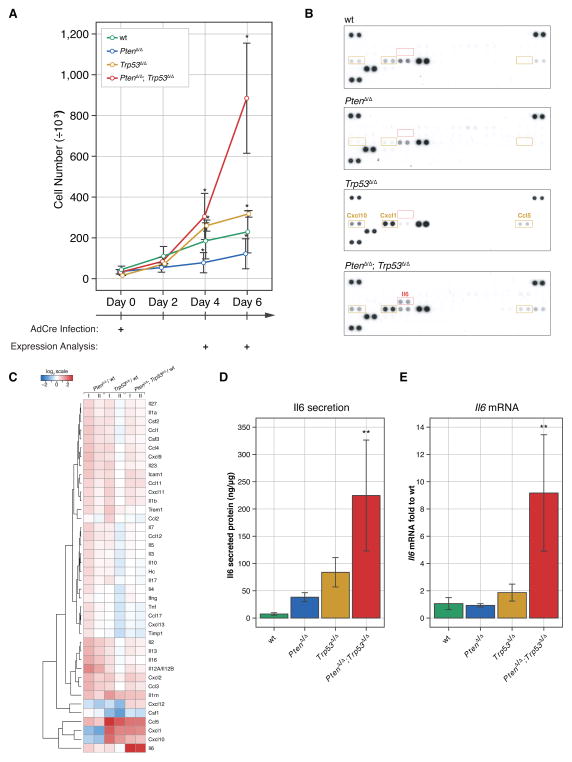
Functional analysis in genetically engineered mouse (GEM) models of PC has confirmed the need for alterations in Trp53 (the murine TP53 ortholog) in disease progression after Pten-loss (12, 15). Moreover, we recently showed that Pten/Trp53 co-deletion in only a few prostate cells can trigger highly penetrant endogenous metastasis in the RapidCaP mouse model (11). Collectively, these observations emphasized the need for a transition to ‘post-senescence’ in PTEN-mutant PC metastasis. Our analysis of human PTEN and TP53 deletions using three recently published whole genome studies on metastatic PC (Supplementary Fig. S1A–C) confirmed that PTEN/TP53 co-deletion is significantly associated with metastasis (9, 16, 17), thus underscoring the value of studying the biology of this specific genetic setting. To investigate the effect of Pten/Trp53 loss on secretion, we used primary mouse embryonic fibroblasts (MEFs), PtenloxP/loxP; Trp53loxP/loxP; loxP-stop-loxP-tdTom (lsl-tdTom) (Supplementary Fig. S1D), and infected them with Cre recombinase-expressing adenovirus (AdCre). This approach yielded over 95% infection efficiency, as measured by Cre-dependent tdTomato fluorescent protein activation and simultaneous deletion of Pten/Trp53 (PtenΔ/Δ; Trp53Δ/Δ; tdTom+/+) (Supplementary Fig. S1E–F). Efficient recombination of target genes was evident by day 4 post infection (p.i., see Supplementary Fig. S2A). Cells were analyzed up to day 6 p.i. without antibiotic treatment to minimize selection for spontaneous gene alterations. As shown in Fig. 1A, PtenΔ/Δ MEFs grew significantly slower than wild type (wt) MEFs due to induction of senescence (Supplementary Fig. S2B) consistent with previous results (12, 18, 19). Deletion of Trp53 alone caused only slightly increased growth compared to wt MEFs. In contrast, combined loss of Pten and Trp53 led to significant growth acceleration (Fig. 1A). To study the differences in secreted proteins between post-senescent cells (PtenΔ/Δ; Trp53Δ/Δ - double mutant) and the other three genotypes (wt, PtenΔ/Δ, Trp53Δ/Δ), we collected conditioned medium and analyzed secreted cytokines and chemokines using mouse specific cytokine profiling arrays (Fig. 1B). Amongst 40 profiled cytokines and chemokines, Il6 stood out by being secreted specifically in the PtenΔ/Δ; Trp53Δ/Δ cells. Other proteins detected in the conditioned medium included Ccl5 (RANTES), Cxcl1 (KC) and Cxcl10 (IP-10), however, these did not respond to Pten status and were also up-regulated by loss of Trp53Δ/Δ alone. The heat map for hierarchical clustering analysis of quantified data from multiple cytokine arrays confirmed induction of Il6 specifically after co-deletion of Pten and Trp53 (Fig. 1C). We next validated these results using Il6 specific ELISA assays on supernatant from the cells (Fig. 1D). Using RT-qPCR analysis we found that Il6 up-regulation occurred at the RNA level (Fig. 1E). Taken together our results revealed that Il6 secretion is a specific response to combined Pten and Trp53-loss.
Figure 1. Il6 secretion is a hallmark of post-senescence.
(A) Combined loss of the Pten and Trp53 genes causes primary MEFs to proliferate more aggressively when compared to the single genotypes. Error bars are SD, n≥3, * p<0.05 with ANOVA, Dunnett’s post-hoc test on all genotypes vs wt at days 4 and 6.
(B) Il6 is secreted specifically after simultaneous loss of Pten and Trp53 genes. A threshold of 3-fold was used to filter up-regulated proteins specific for Trp53Δ/Δ, PtenΔ/Δ and PtenΔ/Δ; Trp53Δ/Δ cells. Ccl5 (RANTES), Cxcl1 (KC), and Cxcl10 (IP-10) proteins were up-regulated in both Trp53Δ/Δ and PtenΔ/Δ; Trp53Δ/Δ cells.
(C) Protein secretion represented as log2-transformed fold expression relative to wt and normalized to total cellular protein content. Note that there are no proteins specifically secreted in PtenΔ/Δ MEFs that exceed the 3-fold cut-off. n=3. Note that we cannot exclude stimulation of protein secretion in senescence (i.e. in the Pten-null cells) below the threshold criteria.
(D) ELISA results confirm increased expression of Il6 in conditioned medium from PtenΔ/Δ; Trp53Δ/Δ MEFs. ANOVA, Dunnett’s post-hoc test, **p<0.01, vs wt, error bars are SD, n=3.
(E) Increased transcription of Il6 is observed in PtenΔ/Δ; Trp53Δ/Δ MEFs compared to wt. ANOVA, Dunnett’s post-hoc test, **p<0.01, vs wt, error bars are SD, n=4.
Il6 secretion drives cell proliferation through Stat3/Myc after loss of Pten/Trp53
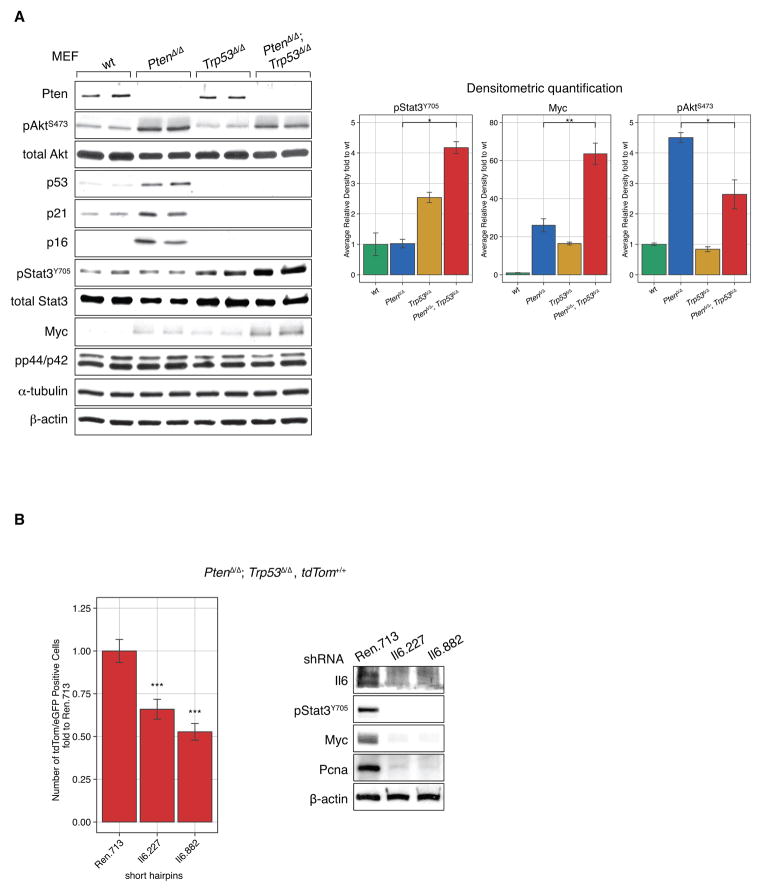
We next studied Pten and p53 signaling pathway components. As shown (Fig. 2A, left panel), deletion of Pten alone activated Akt and p53/p21/p16 signaling pathways, which cause the senescence phenotype (Supplementary Fig. S2B) (12, 15, 18, 19). Accordingly, upon co-deletion of Pten and Trp53, the p21 and p16 tumor suppressors were no longer activated. Furthermore, we observed increased Stat3 phosphorylation and significantly increased expression of its transcriptional target, Myc when compared to deletion of Pten or Trp53 alone (Fig. 2A, Densitometric quantification). Note that we did not detect p44/42 activation, which was shown previously to increase Myc protein stability through phosphorylation (20).
Figure 2. Il6 activation contributes to activation of Stat3/Myc signaling in PtenΔ/Δ; Trp53Δ/Δ cells.
(A) Analysis of Pten/Trp53-dependent signaling pathways by western blot of cell lysates from tested genotypes of primary MEFs. Loss of Pten and Tp53 results in Il6-dependent Stat3/Myc activation as confirmed by western blot quantification (right). ANOVA, Bonferrroni’s post-hoc test, *p<0.05, **p<0.01, error bars are SD, n=2.
(B) short hairpins against Il6 decreased proliferation of Pten/Trp53 negative cells and western blot analysis confirm efficient knock-down of Il6 and decrease in activation of Stat3/Myc signaling, Pcna western blot confirms decreased proliferation. ANOVA, Dunnett’s post-hoc test, ***p<0.001, vs Ren.713, error bars are SD, n=4.
Intracellular signaling downstream of PI 3-Kinase has previously been implicated in Stat3 activation through the TEC kinase family member BMX, which contains a PIP3 sensitive PH domain (reviewed in (21)). Therefore, we used shRNA against Il6 to test if its secretion contributed in auto-/paracrine fashion to proliferation of Pten/p53-lacking cells as scored in MEFs with conditional tdTomato alleles (PtenΔ/Δ; Trp53Δ/Δ; tdTom+/+ MEFs). As shown in Fig. 2B (left), addition of sensor-screened mirE based shRNA (22, 23) against Il6 decreased proliferation of the double mutant MEFs. Molecular pathway analysis furthermore confirmed that sh-Il6 positive cells significantly impaired Stat3 activation and Myc expression (Fig. 2B, right). In a concentration dependent manner, Il6-neutralizing antibodies also decreased proliferation of the double mutant MEFs (Supplementary Fig. S3A) and suppressed Stat3 and Myc activation (Supplementary Fig. S3B). Activation of Stat3 in the PtenΔ/Δ; Trp53Δ/Δ cells depended at least in part on PI 3-kinase activity as the pan-PI 3-Kinase inhibitor LY294002, but not rapamycin, suppressed Stat3 activation (Supplementary Fig. S3C). Taken together, these data suggested that in addition to intracellular PI 3-Kinase signaling through Akt, auto-/paracrine Il6 signaling significantly contributed to Stat3 activation and growth of Pten/Trp53-deficient cells.
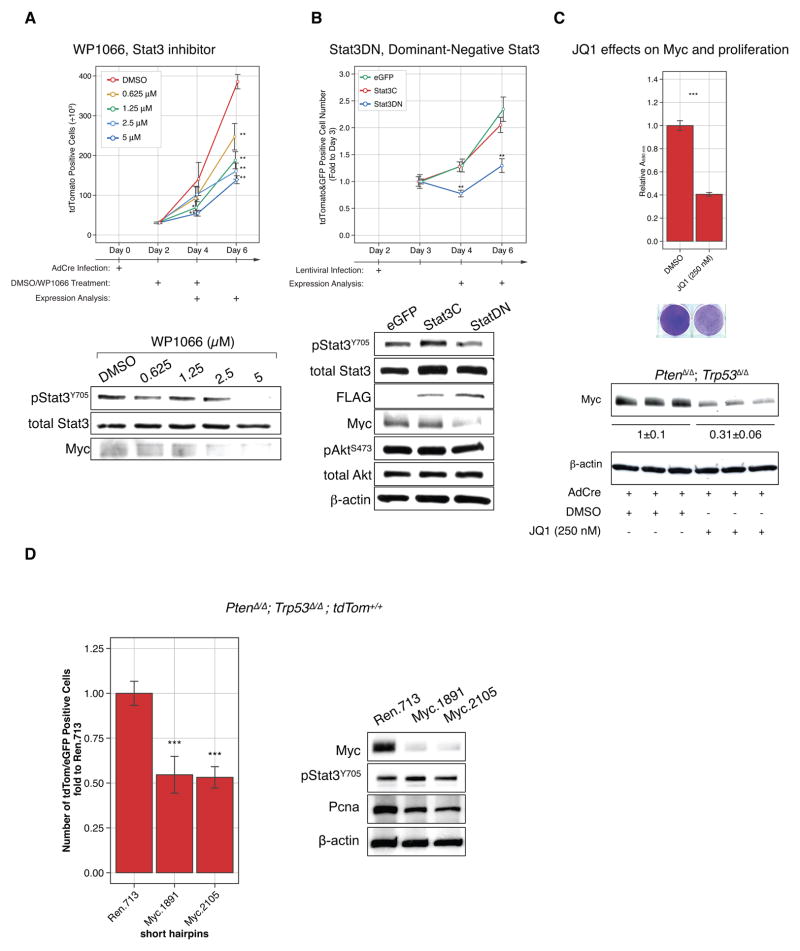
Pharmacological targeting of Stat3 activation also suppressed proliferation: the Stat3 inhibitor WP1066 (24) reduced PtenΔ/Δ; Trp53Δ/Δ; tdTom+/+ cell proliferation by over 62.5% compared to DMSO treatment (Fig. 3A, top). Western blot analysis confirmed WP1066-dependent reduction of phospho-Stat3 levels and decreased phosphorylation of Stat3 correlated with marked reduction of its transcriptional target Myc (Fig. 3A, bottom). Stat3 was previously shown to bind the Myc promoter (25) and to affect the activity of Myc (reviewed in (26)). Next, we targeted Stat3 genetically by over-expressing a dominant negative isoform, Stat3DN, which harbors a point mutation in a tyrosine phosphorylation site (Y705F) that is critical for its activation and nuclear translocation (27, 28). The PtenloxP/loxP; Trp53loxP/loxP; lsl-tdTom MEFs were first infected with AdCre, and then with a lentiviral plasmid containing green fluorescent protein and Stat3DN or with a control lentivirus containing green fluorescent protein alone (29). Double infected cells thus expressed both red and green fluorescent proteins and were counted by flow cytometry (Supplementary Fig. S3D). Our results (Fig. 3B, top) showed that over-expression of the dominant negative Stat3DN significantly reduced proliferation of the Pten/p53-deleted cells compared to the control plasmid. Western blotting analysis confirmed the reduction of both Stat3-phosphorylation and Myc levels. Note that the constitutively active Stat3C mutant showed only minor activation of Stat3/Myc beyond the observed activation in the Pten/p53-deficient background (Fig. 3B, bottom), and accordingly, it had no effect on cell proliferation (Fig. 3B, top). Collectively, our data revealed that Stat3 activation after loss of Pten and Trp53 significantly contributes to cell proliferation, and suggested that in this setting, paracrine signaling through Il6 can mediate activation of Myc in cell culture.
Figure 3. Stat3/Myc signaling is downstream of Il6 signaling in PtenΔ/Δ; Trp53Δ/Δ cells and is responsible for proliferation.
(A) Treatment with the Stat3 inhibitor WP1066 decreases proliferation of PtenΔ/Δ; Trp53Δ/Δ primary MEFs compared to DMSO treatment and decreases phosphorylation of Stat3. ANOVA, Dunnett’s post-hoc test, **p<0.01, *p<0.05 vs DMSO, error bars are SD, n=3. (B) Overexpression of dominant negative Stat3DN blocks proliferation of PtenΔ/Δ; Trp53Δ/Δ primary MEFs., dominant negative Stat3DN inhibits the Stat3 transcriptional target Myc. ANOVA, Dunnett’s post-hoc test, **p<0.01, vs GFP at prospective day, error bars are SD, n=3.
(C) JQ1, a Brd4 inhibitor, blocks proliferation of PtenΔ/Δ; Trp53Δ/Δ primary MEFs as measured using crystal violet (upper panel) and Myc expression (bottom panel), Student’s t-Test, ***p < 0.001, error bars are SD, n=3.
(D) short hairpins against Myc decreased proliferation of Pten/Trp53 negative cells and western blot analysis confirm efficient knock-down of Myc and decrease in activation of Stat3/Myc signaling, Pcna western blot confirms decreased proliferation but to less extend than Il6. ANOVA, Dunnett’s post-hoc test, ***p<0.001, vs Ren.713 at prospective day, error bars are SD, n=4.
Next we asked if Myc activation contributes to proliferation of the Pten/Trp53-deficient cells. First we used the Brd4-inhibitor JQ1, which among other targets suppresses Myc transcription (30, 31). As shown (Fig. 3C), the drug indeed suppressed Myc activation and suppressed proliferation of the primary Pten/Trp53-null MEFs. Similarly, two separate shRNAs targeting Myc also strongly reduced proliferation of the double-mutant Pten/Trp53-deficient MEFs (Fig. 3D). Importantly, RNA interference selectively blocked Myc, but not Stat3 activation, consistent with Myc being downstream of Stat3 action.
Taken together, our results using primary MEFs with defined genetic alterations suggested that: activation of Myc via Il6/Stat3 is (A) a hardwired response to co-deletion of Pten and Trp53 genes, and (B) critical for driving cell proliferation in this genotype (see also cartoon, Supplementary Fig. S7) and (C) presents a targeting opportunity for drugs.
Ptenpc−/−; Trp53pc−/− prostate epithelia secrete Il6 and activate stromal proliferation through Stat3/Myc
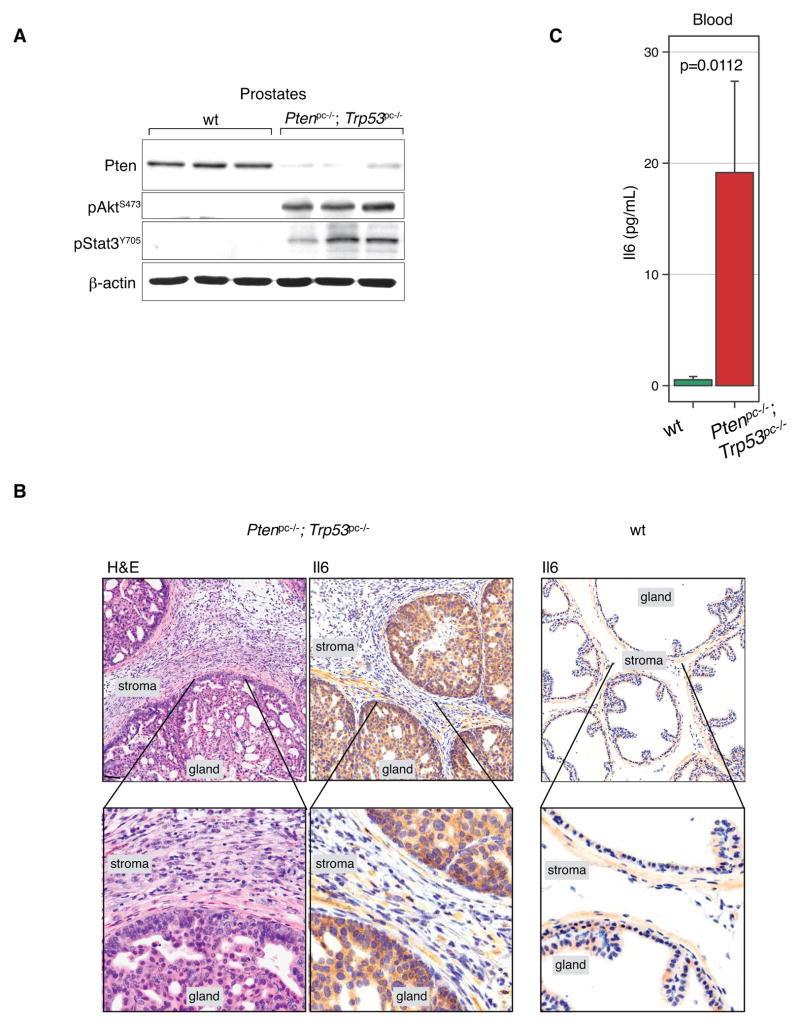
Next we tested the relevance of our findings to prostate tumorigenesis in vivo. As previously (12, 32, 33), we used the probasin promoter to generate prostate conditional knockout mice, by prompting Cre recombination in the prostate epithelium, Ptenpc−/−; Trp53pc−/−; Pb-Cre4. First, we harvested prostates from these mice at 11 weeks (approximately 4 weeks after Pten/Trp53-recombination) to test Stat3 status in vivo. Western Blotting analysis of Ptenpc−/−; Trp53pc−/− prostates (see Fig. 4A, each lane corresponds to different animal) confirmed strong Akt activation as well as increased Stat3 phosphorylation compared to normal prostate, fully consistent with our observations in MEF cells (Fig. 2A). Using prostate immunohistochemistry (IHC) analysis we observed markedly elevated Il6 levels in Ptenpc−/−; Trp53pc−/− prostate epithelial cells (Fig. 4B, gland), consistent with their role as sender cells, analogous to the PtenΔ/Δ; Trp53Δ/Δ MEFs identified earlier. Notably, we were able to detect significantly higher Il6 levels in the blood of the Ptenpc−/−; Trp53pc−/−; Pb-Cre4 mice than in wt mice (see Fig. 4C), suggesting that a systemic readout of the Pten/p53-mutant state might be possible.
Figure 4. Secretion of Il6 and Stat3/Myc signaling is specific to the in vivo Ptenpc−/−; Trp53pc−/− genotype.
(A) Western blot analysis of anterior prostate tissue from Ptenpc−/−; Trp53pc−/− mice at 11 weeks shows lack of Pten, activation of Akt, and Stat3.
(B) H&E analysis of 11-week-old prostates shows expansion of stroma, and high cytoplasmic Il6 levels in Pten/Trp53 deleted prostate but not in wt prostate. Scale Bar, 100 μm.
(C) Il6 levels are increased in blood serum taken from Ptenpc−/−; Trp53pc−/− animals (n=9) reaching almost 20 pg/mL levels, in contrast to serum from wt animals (n=4) that was below the detection level (< 7.8 pg/mL). Mann-Whitney Test, p=0.0112 vs wt, error bars are SEM.
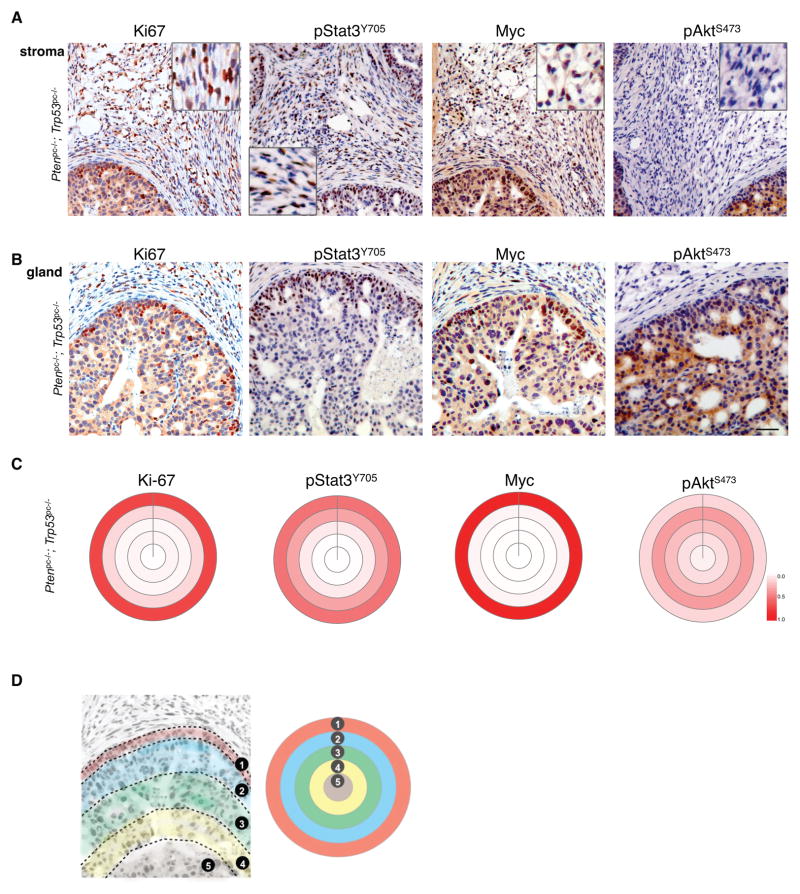
As seen in Fig. 4B (H&E), Ptenpc−/−; Trp53pc−/− prostates showed a massive expansion of stromal fibroblasts compared to the few stromal cells between prostate glands of wt animals (Fig. 4B, Il6, far right). This striking response was absent from Pten- or Trp53-deficient prostate glands (Supplementary Fig. S4A). Furthermore, IHC analysis of the proliferation marker Ki67 revealed strong staining of stromal fibroblasts specifically surrounding Ptenpc−/−; Trp53pc−/− mutant epithelia (Fig. 5A, Ki67). Next we tested whether stromal cells also had activated Stat3. As shown (Fig. 5A, pStat3Y705), stromal fibroblasts were pStat3Y705 positive and also showed high levels of Myc. In contrast, stromal fibroblasts were negative for pAktS473, consistent with loss of Pten/Trp53 being restricted to the prostate epithelium (see Pten staining in Supplementary Fig. S4B). These data revealed that the activation of Stat3 and Myc is conserved in Ptenpc−/−; Trp53pc−/− mutant prostate epithelium and strongly suggested that in vivo Il6 secretion acts in a paracrine fashion on the surrounding stromal cells to trigger proliferation in the tumor environment in the absence of Akt activation and Pten-loss. Please note that this stromal activation phenotype is unusual in human, yet is a characteristic of the Ptenpc−/−; Trp53pc−/− mutant prostate model (34) and lacking when disease initiation is more focal, as in the RapidCaP system (11). Examination of cell proliferation in the Ptenpc−/−; Trp53pc−/− mutant prostate epithelium revealed that it was strongest in the periphery of glands (Fig. 5B, Ki67). This high proliferation zone also had most of the phosphorylated Stat3 and Myc staining (Fig. 5B, pStat3Y705, Myc). To our surprise, however, IHC staining for phosphorylated Akt revealed an inverse correlation with Ki67 staining: inside the gland, where Ki67 staining was relatively infrequent, there was a strong pAktS473 signal. Conversely, the gland’s outermost cell layer showed weakest pAktS473 staining, where proliferation and pStat3Y705/Myc were strongest. Quantification of the radial distribution of IHC signal in the glands confirmed these results (Fig. 5C–D, see also Methods). Myc and pStat3Y705 correlated with the distribution of the proliferation marker Ki67, whereas pAktS473 was inversely correlated with Ki67 (Fig. 5C). In Pten-deficient prostate glands, however, Ki67 and pAktS473 staining did correlate: both were found throughout the gland, consistent with a lack of Il6/Stat3/Myc signaling activation when Pten is deleted alone and not together with Trp53 (Supplementary Fig. S4C). Western blotting analysis also consistently revealed these results: pAktS473 staining was weaker when Myc was activated in PtenΔ/Δ; Trp53Δ/Δ MEFs than in the PtenΔ/Δ cells (Fig. 2A, see pAktS473 and Myc, see Western and densitometry).
Figure 5. Prostate stroma and gland analysis show specific interactions between Myc and pAktS473 activation.
(A) IHC analysis of stroma reveals paracrine activation of Myc/Stat3 signaling in Ptenpc−/−; Trp53pc−/− prostate. Note that Akt is not activated in the stroma.
(B) Epithelial Ptenpc−/−; Trp53pc−/− glands show correlation of the proliferation marker Ki67, Myc, and pStat3Y705 in the periphery of the glands, where phosphorylated Akt is weak. Scale bar (A & B), 50 μm.
(C) Target plot analyses visualize the average distance of IHC positive epithelial cells to stroma and confirm the correlation between Ki-67, pStat3Y705 and Myc, and their inverse correlation with pAktS473.
(D) Schematic of target plot generation from IHC data (note that only entire glands were recorded and analyzed). Track 1 represents epithelial cells along the epithelial border defined as 0–25 μm from the stroma/fibromuscular layer, Track 2: cells within 25% of the maximum possible distance of a cell in a particular gland from the border, Track 3: cells that are within 25–50% of this maximum possible distance, Track 4 represents cells that are within 50–75% of this maximum possible distance, and Track 5 represents cells within 75–100% of the maximum possible distance. Color intensity represents the relative number of stained cells.
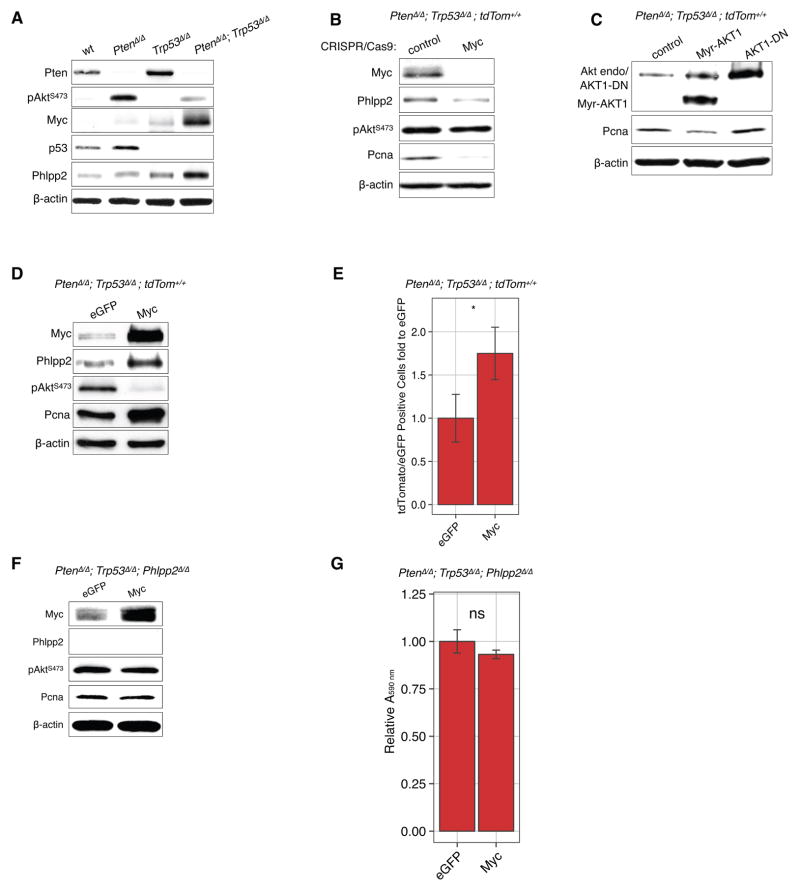
Next we probed the mechanism responsible for this suppression of Akt phosphorylation. We showed previously that the Akt phosphatase Phlpp2 (35, 36), but not the Phlpp1 homolog, is specifically induced by Akt pathway activation to limit the signaling output ((15), and reviewed in (37)). As shown in Fig. 6A, we confirmed these results in Pten-deficient MEFs but found that combined loss of Pten/Trp53 resulted in even higher Phlpp2 protein levels, which correlated inversely with AktS473 phosphorylation and thus could mechanistically explain Akt suppression in this genotype. Since the increase in Phlpp2 protein correlated with the increase in Myc in PtenΔ/Δ; Trp53Δ/Δ MEFs, we next tested whether Myc causes these elevated Phlpp2 levels. As shown in Fig. 6B, the CRISPR/Cas9 based knock-out of Myc led to decreased expression of Phlpp2 in the PtenΔ/Δ; Trp53Δ/Δ MEFs. The overexpression of constitutively active AKT1 (Myr-AKT1) in the Pten/p53-negative cells led to reduced, not increased expression of the proliferating cell nuclear antigen (Pcna) and AKT1-DN did not affect levels of Pcna (Figure 6C.). This experiment shows that proliferation is not increased even by high levels of active Akt in the context of Pten/Trp53-loss and spontaneous activation of Myc. Conversely, the over-expression of Myc led to increased expression of Phlpp2 in the PtenΔ/Δ; Trp53Δ/Δ MEFs (Fig. 6D) and a corresponding suppression of pAktS473. Importantly, cell proliferation, measured by cell count (Fig. 6E) or Pcna levels, in both of these cases correlated with Myc levels, and not those of phospho-Akt. Therefore, these data confirmed that Myc dictates proliferation and is necessary and sufficient for suppressing Akt-activation. Subsequently we tested whether Phlpp2 is essential for suppression of Akt activation downstream of Myc. To this end, we transduced Cre into PtenloxP/loxP; Trp53loxP/loxP; Phlpp2loxP/loxP triple-mutant MEFs (generated from Phlpp2-mutant mice, see Methods). As shown in Fig. 6F, overexpression of Myc in these cells no longer suppressed Akt phosphorylation, consistent with the loss of its phosphatase, Phlpp2. To our surprise however, Myc overexpression did not trigger increased cell proliferation in these triple-mutant MEFs, as shown by Pcna staining and cell number counts in proliferation assays (Fig. 6F–G).
Figure 6. Myc decreases a phosphorylation of pAktS473 that is mediated by Phlpp2 phosphatase.
(A) Activation of the Akt-phosphatase Phlpp2 is found after Myc activation and pAkt-reduction after Pten/Trp53 loss.
(B) Knock-out of Myc using the CRISPR/Cas-9 system leads to decrease in Phlpp2 expression and reduced levels of the proliferation marker Pcna.
(C) The overexpression of constitutively active AKT1 (Myr-AKT1) in the Pten/Trp53-recombined MEF cells while AKT1-DN did not noticeably affect Pcna levels.
(D) Overexpression of Myc triggers an increase in Phlpp2 and in proliferation (Pcna), but reduces Akt phosphorylation.
(E) Overexpression of Myc in Pten/Trp53 negative cells causes a sharp increase in proliferation compared to the eGFP control (* p<0.05, Student’s t-test, n=4).
(F) Deletion of Phlpp2 uncouples Myc from causing suppression of pAkt.
(G) Deletion of Phlpp2 uncouples Myc from causing increased proliferation (not significant - ns, p>0.05, Student’s t-test, n=4).
Collectively these data strongly suggested that activation of Myc and the resulting Phlpp2-mediated suppression of phospho-Akt are critical for the proliferation of Pten/Trp53-deficient cells.
Myc suppresses Akt in prostate metastasis via the Akt phosphatase Phlpp2
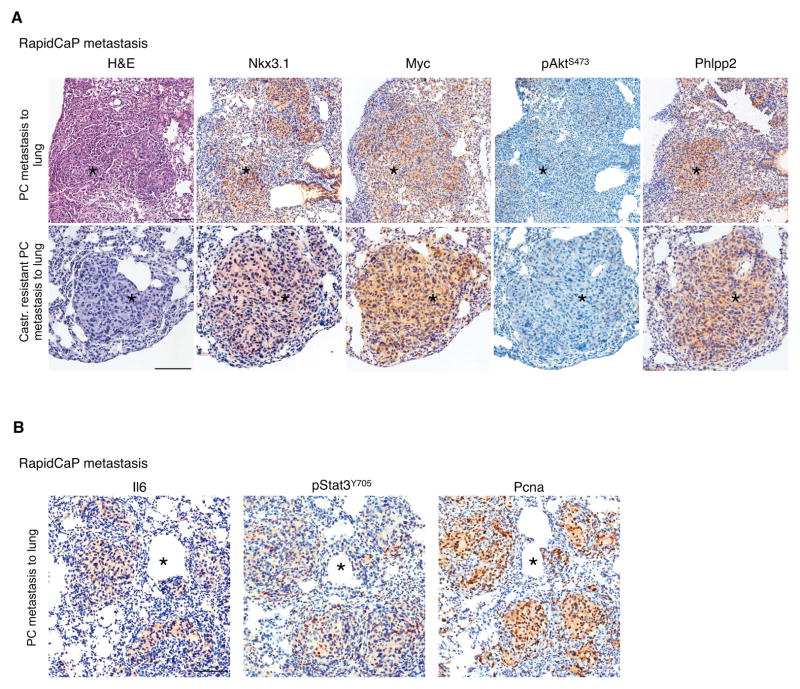
We next tested the relevance of these findings to PC metastasis by using our recently developed RapidCaP GEM model, which features endogenous Pten/Trp53-deficient PC metastasis at 50% penetrance within 4 months. Myc can drive initiation and maintenance of metastasis as revealed by treatment with the Brd4 antagonizing bromodomain inhibitor JQ1 (31, 38) and by using Myc as a transgenic driver of metastasis in RapidCap. Castration of RapidCaP mice suppresses primary and metastatic disease. However, this invariably results in lethal relapse, which presents with even stronger Myc expression, Myc gene amplification, and with growth at much faster rates than the pre-castration lesions (11). As shown in Fig. 7A, metastatic prostate cancer nodules, which can be found in the lungs of untreated or castrated RapidCaP mice, are positive for the Nkx3.1 prostate marker and show high levels of Myc, yet no or very faint staining for pAktS473. Importantly, we validated that the metastatic nodules consistently presented with elevated levels of the Phlpp2 phosphatase, which inversely correlated with the low/absent pAktS473 staining. These patterns were not observed in wild type lungs (Supplementary Fig. S5A). We also confirmed increased expression of Stat3/Il6 in these Pten/Trp53-deficient metastatic nodules and their proliferating potential using Pcna (Fig. 7B). Il6 staining was present in all metastatic nodules with the extent typically varying between 20% and 70% positive cells per nodule (Supplementary Fig. S5B). We found no Il6 activation in the p53-deficient prostate and the Pten-null glands, which were free of hyperplasia/neoplasia, while the proliferating Pten-deficient prostate glands showed Il6 staining, consistent with spontaneous breaking of the p53 response in parts of the tissue, as published previously (12) (Supplementary Fig. S5C). These results were in agreement with our in vitro (see Fig. 1), and in vivo ELISA analysis, where Pten/p53 double mutant probasin-driven animals had high Il6 levels in blood and tissue (Fig. 4A, B). Note that we found no Il6 changes in blood from Trp53-null animals and either undetectable or strong Il6 increase in the Pten-null animals (not shown), consistent with their variable spontaneous evolution mentioned above.
Figure 7. Analysis of lung metastasis in RapidCaP model points a role of Phlpp2 in metastasis.
(A) IHC analysis of RapidCaP metastasis to lung (in castrated and non-castrated animals) validates the positive correlation between Myc and Phlpp2 and their inverse correlation with pAkt staining. Scale Bars, 100 μm.
(B) IHC analysis of RapidCaP lung metastasis (non-castrated animals) reveals correlation of increased Il6/Stat3 with proliferation (Pcna). Scale Bars, 50 (upper panels) and 100 μm (lower panels).
IHC analysis of the Ar status in RapidCaP lesions showed no detectable (nuclear) Ar staining (Supplementary Figure 6A) but positive staining for Nkx3.1 as published (11), (see Figure 7A). In contrast, the primary prostates from both RapidCaP and Probasin-Cre animals showed strong Ar staining (Supplementary Figure 6B). Note that we previously showed that spontaneous loss of Ar can also be found in some prostate epithelial cells of the RapidCaP mice (11) and we showed that some metastases strongly respond to castration suggesting that both IHC-positive and IHC-negative Ar cells can metastasize. It is important to note that the histopathology based IHC analysis requires lesions that are large enough to be unambiguously identified by pathology. These advanced IHC-negative Ar cells in metastatic lesions are very likely resistant to castration, similar to the IHC-negative Ar cells of the castration resistant prostate lesions (11).
Collectively (see Supplementary Fig. S6) these data are consistent with a model where Pten/Trp53-deficient prostate metastasis progresses through an Akt-independent pathway, which requires the activation of Myc by Il6, and Phlpp2-mediated suppression of Akt to drive proliferation and castration-resistance.
DISCUSSION
Our findings identify a critical role for Il6/Stat3 signaling in promoting Myc as a driver of Pten/Trp53 mutant metastasis. The MYC gene is frequently amplified in PC (as curated at the cBio portal (39)) and functional validation established it as a driver of PC (40) and its metastasis, as seen using the RapidCaP system (11). Human genome analysis revealed that Myc alterations usually stem from broad amplifications on chromosome 8q and correlate with metastatic PC (57% in metastasis vs. 15% in primary PC, (9)). Co-deletion of PTEN and TP53 is also a hallmark of PC metastasis as previously published and shown in Supplementary Fig. S1A, by using additional recent PC metastasis genome studies.
Our data support two novel concepts for metastatic PC (Supplementary Fig. S7). Firstly, they present a molecular mechanism that links MYC activation to loss of both PTEN and TP53, but not to loss of either separately. The combined loss marks a genetic condition that is tightly linked to metastatic PC, as discussed above. This regulatory signaling to Myc occurs within 4 days of Pten/Trp53-loss in our in vitro experiments, thus likely preceding Myc gene amplification. If validated in human, this could indicate a therapeutic window of opportunity before MYC gene amplification is detected as a driver of late stage disease at a point when it may respond less well to anti-MYC therapy.
These data suggest that cell-cell communication is intimately linked to genetic changes of PTEN and TP53. It is known that chemokines and interleukins change their expression patterns during PC progression (14). Now, our in vitro and in vivo data specifically link the gene loss of PTEN and TP53 to IL6 secretion. The Probasin-Cre driven PtenΔ/Δ; Trp53Δ/Δ model illustrates the effect that this communication can have on normal stroma, by triggering Myc expression and proliferation (and possibly a sarcoma phenotype of this classic model (34)). Our data suggest that the Il6/Stat3/Myc module shown in Figure S6 becomes independent of p-Akt in metastasis. It remains to be seen if the Myc module still depends on PI 3-Kinase activity in metastasis, as it has been shown that the PI 3-K family member TEC-kinases such as BMX can activate Stat3 downstream of PIP3 (reviewed in (21)). Alternatively, Il6/Stat3 may be increased through a PI 3-Kinase pathway autonomous mechanism such as gene amplification (IL6 amplification is observed in 38% of metastatic samples of the published MSKCC cohort). It remains to be seen if Il6 signaling of prostate cells at the metastatic site is strictly needed to enhance fitness and proliferation of the cancer cells or also helps disease progression by landscaping the microenvironment and immune response. ELISA based detection of Il6 in the blood of PtenΔ/Δ; Trp53Δ/Δ mice points to a potential avenue for identifying patients with PTEN/TP53 mutant metastasis. Elevated levels of IL6 have been associated with advanced tumor stages of various cancers including prostate, multiple myeloma, non-small cell lung carcinoma, colorectal, renal, breast and ovarian cancer. Anti-IL6 therapies using a neutralizing antibody (e.g. siltuximab) or anti-JAK/STAT3 signaling (e.g. ruxolitinib). Siltuximab has shown mixed results in metastatic castration-resistant prostate cancer (41–44). We hope that future trials on IL6/STAT3 pathway inhibition may benefit from our modeling results by pointing to MYC for patient selection and monitoring of drug efficacy.
Secondly, our results surprisingly suggest that both Myc activation and Akt suppression are required to drive proliferation in Pten/p53-deficient cells. It is particularly intriguing that a bona fide tumor suppressing Akt phosphatase, Phlpp2, is co-opted by Myc to drive proliferation by suppressing Akt activity. Several factors could explain why metastatic and castration-resistant disease favors Myc over Akt activity. Apart from its role in promoting ribosome biogenesis, Myc has been implicated in de-differentiation of cells (reviewed in (45)), which may explain the observed loss of epithelial cell characteristics in the Myc positive Pten/p53-deficient RapidCaP lung metastases and in the castration-resistant PC (11). Furthermore, several studies have previously identified isoform-specific anti-migratory and cancer suppressing roles for the Akt suggesting several mechanisms by which Akt may impede prostate metastasis (46–51) reviewed in (52)). Indeed, this switch may be critical for resistance to hormone therapy: using the RapidCaP system, we consistently observed the highest levels of Myc protein and highest percentage of Myc gene amplification (32%) in relapsed castration-resistant prostate tumors, and activation of Myc also correlated with loss of androgen receptor (Ar) staining (11). Thus, enhanced reliance of lethal PC lesions on MYC could create a novel tumor sensitivity to drugs that target MYC.
METHODS
Mouse Embryonic Fibroblasts (MEFs)
Animals with different combination of loxP alleles (PtenloxP/loxP, Trp53loxP/loxP Phlpp2loxP/loxP) or PtenloxP/loxP; Trp53loxP/loxP; lsl-tdTomato were used. At 13.5 day embryos were harvested by sacrificing a female using CO2. Embryos were minced and trypsinized for 15 minutes in a 37°C in water bath. Cells were plated on 10 cm dish and cultured in 10% DMEM medium and 1xPS, this was called passage 0. 3 days after plating, cells were split to 10 cm dish at 900,000 cells/dish per 15 mL volume. Cells were cultured for the next 3 days, collected, frozen in 20% Dimethyl Sulfoxide, 80% FBS and and stored in batches of 1,000,000 cells/cryovial in liquid nitrogen. All protocols for mouse experiments were in accordance with the institutional guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC).
Mutant mice
Mutant mice were generated using animals containing homozygous alleles for: PtenloxP/loxP (33) and Trp53loxP/loxP (53). Female PtenloxP/loxP; Trp53loxP/loxP were crossed with a male carrying a heterozygous allele for Pb-Cre4. Embryonic stem (ES) cells were purchased from the Mouse Phenotype Consortium (Phlpp2 - IKMC Project: 47191). The Phlpp2 gene gives rise to three wild type transcripts, of which two are protein-coding. Initially, mice were crossed to a flippase mouse to remove the lacZ/loxP/neo cassette, which gave rise to loxP conditional mice. Following removal of the floxed region, two transcripts were predicted to produce truncated protein products, which may be subject to nonsense-mediated decay (NMD). Our expression analysis confirmed knock-outs of Phlpp2 at the protein level. For genotyping, DNA from the tail was extracted and PCR analyses were performed as previously described (53). PtenΔ/Δ; Trp53Δ/Δ RapidCaP animals were generated by intraprostatic delivery of virus and the PC metastases to lung analyzed as previously described (11).
Cell proliferation assays
Total cell counts were done using the Guava System (Millipore). Briefly, cells were trypsinized and mixed with medium in a 1 to 5 ratio, then 150 μL were loaded into 96-well plates and analyzed using the Guava easyCyte 8HT Benchtop Flow Cytometer (Millipore). Cell proliferation was also measured using the crystal violet method. Briefly, cells were fixed in 10% formalin for 15 minutes, washed with sterile water and stained with 0.1% crystal violet solution. Solutions were aspirated, washed with water and then plates were air dried over night and 10% acetic acid was added and mixed with water in a 1:3 ratio. Absorbance was measured at 590 nM using a plater reader (Synergy H4 Hybrid Multi-Mode Microplate Reader, Biotek).
Cell Treatments
For Il6 neutralizing experiments Il6 neutralizing antibody (R&D, AB-406-NA) and control IgG antibody (R&D, AB-108) were used to assess effects of secreted Il6 on the proliferation of MEFs. The Stat3 inhibitor WP1066 (Selleckchem, S2796), PI3K inhibitor LY294002 (Cell Signaling, #9901), Rapamycin (Cell Signaling, #9904) and JAK inhibitor I (Calbiochem, 420099) were resuspended in DMSO. JQ1 was a kind gift from Dr. J. Bradner (Harvard Medical School) and was used as described previously (31). Note that the IC50 value for inhibition of proliferation was 1.4 μM for Pten/Trp53-deleted cells, and 3.3 μM for wt cells. These results are in similar range to earlier reported, WP1066 effects on cell survival, proliferation and Stat3 phosphorylation (54–56).
cDNA plasmids
Adenoviral plasmid, Ad5CMVCre was obtained from the Gene Transfer Core, University of Iowa. FUGW backbone lentiviral plasmids were purchased from Addgene (plasmid numbers in brackets): GFP (14883), Sta3C (24983), Stat3DN (24984) (29), Akt1-DN (53597), Myr-Akt1 (53583), Myr-Akt3 (53596) (57). Retroviral Plasmids were kind gifts from Dr. C. Vakoc (CSHL): control (GFP) and Myc that was cloned into pMSCV-PGK-Puro-IRES-GFP plasmid as published (31).
shRNA plasmids
Retroviral short hairpins against Il6 and Myc are using the optimized “miR-E” backbone and were identified using a high-throughput RNAi Sensor assay and expressed from the optimized “miR-E” backbone that improves pri-miRNA processing efficiency (23). The following 97-mer sequences were used:
shRen.713(control):TGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGA
sh_Il6.277:TGCTGTTGACAGTGAGCGACACTTGCAGAAAACAATCTGATAGTGAAGCCACAGATGTATCAGATTGTTTTCTGCAAGTGCTGCCTACTGCCTCGGA
sh_Il6.882:TGCTGTTGACAGTGAGCGCCACTTGAAATGTTATATGTTATAGTGAAGCCACAGATGTATAACATATAACATTTCAAGTGATGCCTACTGCCTCGGA
sh_Myc.1891:TGCTGTTGACAGTGAGCGCACGACGAGAACAGTTGAAACATAGTGAAGCCACAGATGTATGTTTCAACTGTTCTCGTCGTTTGCCTACTGCCTCGGA
sh_Myc.2105:TGCTGTTGACAGTGAGCGCCTGCCTCAAACTTAAATAGTATAGTGAAGCCACAGATGTATACTATTTAAGTTTGAGGCAGTTGCCTACTGCCTCGGA
CRISPR/Cas-9 plasmids
The guide RNAs (gRNAs) were designed using the R package CRISPRseek (58). The three highest-ranking gRNAs were chosen. Among these was guide Myc.87 (5′-GCTGTACGGAGTCGTAGTCG-3′), a gRNA that targets Myc in the first exon. The gRNA oligonucleotides were purchased from Sigma-Aldrich, and phosphorylated and annealed according to previously published protocols (59). The duplex oligonucleotides were subsequently cloned into the lentiCRISPR_V1 plasmid (Addgene #49535), replacing the 2 kb filler.
Retrovirus Production
Phoenix cells were plated at a density of 2 × 10E6 cells per 10 cm dish the day before calcium chloride transfection. 15.5 μg of plasmid DNA were combined with 5 μg helper constructs and 2M calcium chloride. The mixture was added drop-wise to plates after treatment with 2.5 μL of 100 mM chloroquine. Medium was changed after 14 hours and viral supernatants were collected at 48 and 72 hours post-transfection. Supernatants were then centrifuged and filtered with a 0.45 μm filter before infection of cells. Ecotropic Phoenix cells were a kind gift from Dr. Scott W. Lowe. They were not authenticated.
Lentivirus Production
293FT cells were plated at a density of 10 × 10E6 per 10 cm dish the day before calcium chloride transfection. 27 μg of target DNA were combined with packaging constructs, 8 μg of pMD2.G and 20 μg of psPAX2 and 2 M calcium chloride. The mixture was then added drop-wise to cells which were treated with 6.7 μL 100 mM chloroquine. Medium was changed after 14 hours and viral supernatants were collected at 48 and 72 hours post-transfection. Supernatants were then centrifuged and filtered with a 0.45 μm filter before infection of cells. 293FT cells were a kind gift from Dr. Scott W. Lowe. They were not authenticated.
Antibody Arrays
Medium was collected and snap-frozen in liquid nitrogen and stored at −20°C until analyzed. Expression of cytokines and chemokines was measured using the Mouse Cytokine Array Panel A (R&D, ARY006) which enables simultaneous profiling of multiple secreted proteins. For data analysis, the signal intensity was determined using the ImageJ 1.38x software, backgrounds were subtracted using the intensity from negative control spots on the strip. All genotypes were normalized to wild-type and these ratios were log2-transformed in order to determine relative secretion levels.
Western blotting
To determine protein expression in our system we used SDS-PAGE (6%, 8% and 12% reducing gels, 5% 2-β mercaptoethanol). Gels were loaded with 20 μg protein per well (Bradford assay). Antibodies used: beta-actin (1:3000, Sigma), pAkt(S473) (193H12, 1:2000, Cell Signaling), total Akt (40D4, 1:300, Cell Signaling), p44/p42 (1:1000, Cell Signaling), Myc (14721-1, 1:1000, Epitomics), pStat3(Tyr705) (9145, 1:2000, Cell Signaling), Pcna (Santa Cruz, 1:6000), Pten (6H2.1, Cascade Bioscience, 1:1000), total Stat3 (9139, 1:2000, Cell Signaling), and p16 (M-156, Santa Cruz, 1:1000), p21 (sc-397, 1:200, Santa Cruz), p53 (IMX25, 1:500, Leica). ECL chemoluminescence was used with Amersham Hyperfilm ECL (Amersham Bioscience). For data analysis, the signal intensity was determined using the ImageJ 1.38x software.
Histology and Immunohistochemistry analysis
Tissues were fixed in 10% buffered formalin for 24 hrs before transfer to PBS. Paraffin-embedded tissues were sectioned 6 μm thick, placed on charged glass slides and stained with hematoxylin & eosin, or the appropriate immunohistochemical stains. Antigen retrieval was performed by incubating the slides in 0.01 M citric acid buffer (pH 6.0) at 95°C for 15 min. Slides were then cooled to room temperature for 20 min in citric acid buffer. The slides were transferred to TBS (pH 7.4) for 5 min after a wash with deionized water. The following detection and visualization procedures were performed according to manufacturer’s protocol. Slides were counterstained in Mayer’s hematoxylin, dehydrated, cleared, and cover slipped. Negative control slides were stained with secondary antibody only. Antibodies: Ar (5153, 1:200), pAktS473 (4060, 1:800, Cell Signaling), total Akt (40D4, 1:300, Cell Signaling), Myc (14721-1, 1:50, Epitomics), pStat3Tyr705 (9145, 1:800, Cell Signaling), total Stat3 (9139, 1:400, Cell Signaling), Ki-67 (1:2000, Novus), Il6 (1:50, Cell Signaling), Pcna (3110, 1:8000, Cell Signaling), Phlpp2 (1:200, Bethyl). For human, archival formalin-fixed paraffin-embedded tissue from resection of a prostate cancer brain metastasis was retrieved under an Institutional Review Board approved protocol from the files of the Department of Pathology & Laboratory Medicine at Weill Cornell Medical College. Five micron thick sections were cut and stained as above, except for c-myc staining, which used a rabbit monoclonal antibody (clone Y69; Epitomics, Burlingame, California, USA) and the BOND III Autostainer and supplied reagents (Leica Microsystems, Buffalo Grove, IL, USA). Pretreated sections were blocked with 5% normal horse serum and 1% BSA (in TBS) for 1 hour at room temperature. Primary antibodies were diluted as suggested by the manufacturer and incubated overnight at 4°C. Following three 10 min. washes with TBS, sections were incubated with biotinylated secondary antibody for 30 min. at room temperature and rinsed three times with TBS for 10 min. Sections were then treated with diaminobenzidine for 3 min and rinsed with distilled water, mounted on gelatin-coated slides, air-dried, dehydrated with 70%–100% alcohol, cleared with xylene and then cover-slipped for microscopic observation. Stained slides were digitally scanned using Aperio ScanScope software (Vista, California).
Target plots for IHC staining patterns
‘Target plots’ were used to quantify and summarize the distance of positively stained cells in a gland from the stroma, using ImageJ. The first circle layer represents epithelial cells along the epithelial border, defined as being within 0–25 μm from the stroma/fibromuscular layer. The second layer represents cells that are within 25% of the maximum possible distance of a cell in a particular gland from the border. The third layer represents cells that are within 25–50% of this maximum possible distance, the fourth layer represents cells that are within 50–75% of this maximum possible distance, and the fifth layer represents cells within 75–100% of the maximum possible distance.
In vitro Enzyme-Linked Immunosorbent Assay (ELISA)
Medium was collected and snap-frozen in liquid nitrogen, and then stored at −20°C until analysis. Il6 concentration was measured using the mouse Quantikine IL-6 ELISA (R&D Systems, ARY006) according to the manufacturer’s protocol. IL-6 levels were normalized to total protein concentration from the same well.
In vivo Enzyme-Linked Immunosorbent Assay (ELISA)
Around 0.8 mL of blood was collected from mouse and blood samples were left to clot for 2 hours at room temperature. After 2 hours incubation, samples were centrifuged for 20 minutes at 2,000 × rcf at 4°C and serum was delicately removed and aliquoted 200 μL in 1 mL cryovials and were snap frozen in liquid nitrogen. Samples were stored as serum at −20°C until used. The concentration of Il6 was measured using mouse Quantikine IL-6 ELISA (R&D Systems, ARY006) according to the manufacturer’s protocol.
RTqPCR analysis
Total RNA from MEFs: wt, PtenloxP/loxP, Trp53loxP/loxP and PtenloxP/loxP; Trp53loxP/loxP was isolated using the RNeasy Plus mini kit (Qiagen). cDNA was reverse transcribed using the SuperScript® VILO™ cDNA Synthesis Kit (Life Technologies), and RT-PCR for the Il6 mRNA was performed using a LigthCycler 480 SYBR Green I Master (Roche). The forward primer used for mouse Il6 was 5′-TAGTCCTTCCTACCCCAATTTCC-3′ and the reverse primer was 5′-TTGGTCCTTAGCCACTCCTTC-3′. Hprt mRNA was measured as an internal control; the oligos used were: forward primer: 5′-TCAGTCAACGGGGGACATAA-3′ and reverse primer: 5′-GGGGCTGTACTGCTTAACCAG-3′.
Data Analysis
For multiple comparisons, statistical analyses were carried out on raw data using the one-way ANOVA test (Dunnett’s post) and p<0.05 was considered statistically significant. To compare two groups, Student’s t-test or Mann-Whitney test were used, p<0.05 was considered statistically significant. Values are expressed as the means (SD) unless otherwise stated. Graphs and heatmap analyses were performed using the R software and ggplot2 and heatmap.2 packages (60) on Apple Macintosh computers.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Our data identify Il6 detection as a potential causal biomarker for Myc driven metastasis after loss of Pten and p53.
Second, our finding that Myc then must supersede Akt to drive cell proliferation, points to Myc inhibition as a critical part of PI 3-Kinase pathway therapy in lethal PC.
Acknowledgments
We would like to thank Mikala Egeblad, Tim Kees, Daniel Ferrante, Chris Vakoc, Junwei Shi and Jay Bradner and members of the Trotman lab for valuable discussions and help with experimental protocols and reagents, the CSHL Animal Resources team, Lisa Bianco, Lotus Altholtz, Jodi Coblentz, Michael Cahn for help with animal work, Aigoul Nourjanova, Raisa Puzis, Denise Hoppe, Afsar Barlas and Katia Manova with histopathology procedures and Pamela Moody and Jordan Ratcliff for cell sorting and FACS advice and Dorothy Tsang for help with the manuscript.
GRANT SUPPORT
L.C.T. is a Young Investigator of the Pershing Square Sohn Cancer Research Alliance and a Research Scholar of the American Cancer Society. We are grateful for the generous funding of this work through grants to LCT from the NIH (CA137050), the Pershing Square Sohn Cancer Research Alliance, the Department of Defense (W81XWH-14-1-0247), the STARR Foundation (I8-A8-112), the Robertson Research Fund of Cold Spring Harbor Laboratory, and from the NIH to the CSHL Cancer Center through Support Grant 5P30CA045508 for funding of animal shared resources, histology, flow cytometry and gene sequencing. We are also very thankful for the generous donations from the local Foundations, Long Island Cruisin’ for the Cure and Glen Cove Cares.
ABBREVIATIONS
- AR
Androgen receptor
- BPH
Benign prostatic hyperplasia
- Brd4
Bromodomain containing 4
- Cas9
CRISPR-associated protein 9
- Ccl5
Chemokine (C-C motif) ligand 5
- CRISPR
Clustered regularly interspaced short palindromic repeats
- Cxcl1
Chemokine (C-X-C motif) ligand 1; KC
- Cxcl10
Chemokine (C-X-C motif) ligand 10; IP-10
- ELISA
Enzyme-linked immunosorbent assay
- ERG
ETS-related gene
- FP
Fluorescent protein
- GEM
Genetically engineered mouse
- IHC
Immunohistochemistry
- Il6
Interleukin 6
- IP-10
Interferon gamma-induced protein 10; Cxcl10
- JAK
Janus kinase
- KC
Keratinocyte chemoattractant; Cxcl1
- loxP
locus of X-over P1
- lsl-tdTomato
loxP-STOP-loxP tdTomato
- MEF
Mouse Embryonic Fibroblast
- Myc
Myelocytomatosis oncogene
- Nkx3.1
NK3 homeobox 1
- p53
Tumor protein p53
- PC
Prostate Cancer
- Pten
Phosphatase and tensin homolog
- Δ/Δ
Homozygous knock-out
- Pcna
Proliferating cell nuclear antigen
- Phlpp2
PH domain and leucine rich repeat protein phosphatase 2
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- RANTES
Regulated on activation, normal T cell expressed and secreted
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- shRNA
Short hairpin RNA
- Stat3
Signal transducer and activator of transcription 3
- Stat3C
Stat3 Constitutive Active isoform
- Stat3DN
Stat3 Dominant Negative isoform
- tdTom
tandem dimer Tomato
- TMPRSS2
Transmembrane protease, serine 2
- TMPRSS2-ERG
TMPRSS2-ERG fusion gene
- TP53
Tumor protein p53
Footnotes
The authors disclose no potential conflicts of interest
Bibliography
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al. SEER Cancer Statistics Review, 1975–2011. 2013 [Google Scholar]
- 2.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–11. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroder FH, Hugosson J, Roobol MJ, Tammela TLJ, Ciatto S, Nelen V, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981–90. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJC, Kollermann J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- 8.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labbe DP, Nowak DG, Deblois G, Lessard L, Giguere V, Trotman LC, et al. Prostate cancer genetic-susceptibility locus on chromosome 20q13 is amplified and coupled to androgen receptor-regulation in metastatic tumors. Mol Cancer Res. 2014;12:184–9. doi: 10.1158/1541-7786.MCR-13-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H, Herzka T, Zheng W, Qi J, Wilkinson JE, Bradner JE, et al. RapidCaP, a novel GEM model for metastatic prostate cancer analysis and therapy, reveals myc as a driver of Pten-mutant metastasis. Cancer Discov. 2014;4:318–33. doi: 10.1158/2159-8290.CD-13-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Trotman LC, Shaffer D, Lin H-K, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigene-sis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantovani A. Molecular pathways linking inflammation and cancer. Current molecular medicine. 2010;10:369–73. doi: 10.2174/156652410791316968. [DOI] [PubMed] [Google Scholar]
- 14.Vindrieux D, Escobar P, Lazennec G. Emerging roles of chemokines in prostate cancer. Endocr Relat Cancer. 2009;16:663–73. doi: 10.1677/ERC-09-0109. [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O’Neill A, et al. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell. 2011;20:173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J-S, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol Cell Biol. 2007;27:662–77. doi: 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–93. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogt PK, Hart JR. PI3K and STAT3: a new alliance. Cancer Discov. 2011;1:481–6. doi: 10.1158/2159-8290.CD-11-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–46. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–13. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 24.Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–44. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 25.Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, et al. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J Exp Med. 1999;189:63–73. doi: 10.1084/jem.189.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–56. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 27.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE. Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–8. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amin HM, McDonnell TJ, Ma Y, Lin Q, Fujio Y, Kunisada K, et al. Selective inhibition of STAT3 induces apoptosis and G(1) cell cycle arrest in ALK-positive ana-plastic large cell lymphoma. Oncogene. 2004;23:5426–34. doi: 10.1038/sj.onc.1207703. [DOI] [PubMed] [Google Scholar]
- 29.Hillion J, Dhara S, Sumter TF, Mukherjee M, Di Cello F, Belton A, et al. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an achilles heel for hematopoietic malignancies? Cancer Res. 2008;68:10121–7. doi: 10.1158/0008-5472.CAN-08-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–9. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 33.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1 doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73:2718–36. doi: 10.1158/0008-5472.CAN-12-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–31. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol. 2014;54:537–58. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M, Nowak DG, Trotman LC. Molecular pathways: PI3K pathway phospha-tases as biomarkers for cancer prognosis and therapy. Clin Cancer Res. 2014;20:3057–63. doi: 10.1158/1078-0432.CCR-12-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bro-modomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 41.Dorff TB, Goldman B, Pinski JK, Mack PC, Lara PN, Van Veldhuizen PJ, et al. Clinical and correlative results of SWOG S0354: a phase II trial of CNTO328 (sil-tuximab), a monoclonal antibody against interleukin-6, in chemotherapy-pretreated patients with castration-resistant prostate cancer. Clin Cancer Res. 2010;16:3028–34. doi: 10.1158/1078-0432.CCR-09-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karkera J, Steiner H, Li W, Skradski V, Moser PL, Riethdorf S, et al. The anti-interleukin-6 antibody siltuximab down-regulates genes implicated in tumorigene-sis in prostate cancer patients from a phase I study. Prostate. 2011;71:1455–65. doi: 10.1002/pros.21362. [DOI] [PubMed] [Google Scholar]
- 43.Fizazi K, De Bono JS, Flechon A, Heidenreich A, Voog E, Davis NB, et al. Ran-domised phase II study of siltuximab (CNTO 328), an anti-IL-6 monoclonal antibody, in combination with mitoxantrone/prednisone versus mitoxan-trone/prednisone alone in metastatic castration-resistant prostate cancer. Eur J Cancer. 2012;48:85–93. doi: 10.1016/j.ejca.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 44.Hudes G, Tagawa ST, Whang YE, Qi M, Qin X, Puchalski TA, et al. A phase 1 study of a chimeric monoclonal antibody against interleukin-6, siltuximab, combined with docetaxel in patients with metastatic castration-resistant prostate cancer. Invest New Drugs. 2013;31:669–76. doi: 10.1007/s10637-012-9857-z. [DOI] [PubMed] [Google Scholar]
- 45.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hutchinson JN, Jin J, Cardiff RD, Woodgett JR, Muller WJ. Activation of Akt-1 (PKB-alpha) can accelerate ErbB-2-mediated mammary tumorigenesis but suppresses tumor invasion. Cancer Res. 2004;64:3171–8. doi: 10.1158/0008-5472.can-03-3465. [DOI] [PubMed] [Google Scholar]
- 47.Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, et al. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–34. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539–50. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, Radisky DC, Nelson CM, Zhang H, Fata JE, Roth RA, et al. Mechanism of Akt1 inhibition of breast cancer cell invasion reveals a protumorigenic role for TSC2. Proc Natl Acad Sci U S A. 2006;103:4134–9. doi: 10.1073/pnas.0511342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maroulakou IG, Oemler W, Naber SP, Tsichlis PN. Akt1 ablation inhibits, whereas Akt2 ablation accelerates, the development of mammary adenocarcinomas in mouse mammary tumor virus (MMTV)-ErbB2/neu and MMTV-polyoma middle T transgenic mice. Cancer Res. 2007;67:167–77. doi: 10.1158/0008-5472.CAN-06-3782. [DOI] [PubMed] [Google Scholar]
- 51.Chin YR, Yuan X, Balk SP, Toker A. PTEN-deficient tumors depend on AKT2 for maintenance and survival. Cancer Discov. 2014;4:942–55. doi: 10.1158/2159-8290.CD-13-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chin YR, Toker A. Akt isoform-specific signaling in breast cancer: uncovering an anti-migratory role for palladin. Cell Adh Migr. 2011;5:211–4. doi: 10.4161/cam.5.3.15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 54.Verstovsek S, Manshouri T, Quintas-Cardama A, Harris D, Cortes J, Giles FJ, et al. WP1066, a novel JAK2 inhibitor, suppresses proliferation and induces apop-tosis in erythroid human cells carrying the JAK2 V617F mutation. Clin Cancer Res. 2008;14:788–96. doi: 10.1158/1078-0432.CCR-07-0524. [DOI] [PubMed] [Google Scholar]
- 55.Horiguchi A, Asano T, Kuroda K, Sato A, Asakuma J, Ito K, et al. STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell carcinoma. British journal of cancer. 2010;102:1592–9. doi: 10.1038/sj.bjc.6605691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatiboglu MA, Kong L-Y, Wei J, Wang Y, McEnery KA, Fuller GN, et al. The tumor microenvironment expression of p-STAT3 influences the efficacy of cyclophos-phamide with WP1066 in murine melanoma models. Int J Cancer. 2012;131:8–17. doi: 10.1002/ijc.26307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie R, Cheng M, Li M, Xiong X, Daadi M, Sapolsky RM, et al. Akt isoforms differentially protect against stroke-induced neuronal injury by regulating mTOR activities. J Cereb Blood Flow Metab. 2013;33:1875–85. doi: 10.1038/jcbfm.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu LJ, Holmes BR, Aronin N, Brodsky MH. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R Core Team. R: A Language and Environment for Statistical Computing. 2014 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.