Abstract
Circulating tumor cells (CTCs) have become an important biomarker for early cancer diagnosis, prognosis, and treatment monitoring. Recently, a replication-competent recombinant adenovirus driven by a human telomerase gene (hTERT) promoter was shown to detect live CTCs in blood samples of cancer patients. Here, we report a new class of adenoviruses containing regulatory elements that repress the hTERT gene in normal cells. Compared to the virus with only the hTERT core promoter, the new viruses showed better selectivity for replication in cancer cells than in normal cells. In particular, Ad5GTSe, containing three extra copies of a repressor element, displayed a superior tropism for cancer cells among leukocytes and was thus selected for CTC detection in blood samples. To further improve the efficiency and specificity of CTC identification, we tested a combinatory strategy of microfiltration enrichment using flexible micro spring arrays (FMSAs) and Ad5GTSe imaging. Our experiments showed that this method efficiently detected both cancer cells spiked into healthy blood and potential CTCs in blood samples of breast and pancreatic cancer patients, demonstrating its potential as a highly sensitive and reliable system for detection and capture of CTCs of different tumor types.
Keywords: Circulating tumor cells, Telomerase expression, Recombinant adenoviruses
Introduction
Cancers shed their tumor cells into the peripheral blood, generating metastases that confer lethality. Circulating tumor cells (CTCs) have been detected in patients with many types of cancers, even those with no signs of clinically overt metastases [1]. Because of their easy accessibility, CTCs are clinically relevant to cancer detection and prognosis. Repeated sampling of CTCs may be used for real-time assessment of cancer progression and treatment response [2].
CTC detection and capture are technically demanding. One of the challenges is their extremely low abundance in the peripheral blood: the clinically relevant numbers of CTCs are as low as one CTC per 105–107 nucleated blood cells. CTCs are also highly variable and their genetic and epigenetic characteristics differ not only among tumors of different tissue origins but also within the same cancer type, or even within a patient. Most current detection strategies are based on the epithelial markers like epithelial cell adhesion molecule (EpCAM) and cytokeratins. However, evidence is accumulating that, in certain tumor types, these epithelial markers may be down-regulated during tumor cell dissemination, potentially hampering the detection of CTCs [3]. To date, the CellSearch assay for breast, prostate, and colon cancer patients, based on EpCAM expression, is the only CTC detection method cleared by the FDA. We and others have shown that, in at least some cases, CellSearch detection of CTC is less efficient than detection after enrichment by microfiltration [4, 5]. Thus, simple and failsafe techniques for efficient CTC enumeration and capture are still needed for clinical applications.
Most adult cancers are of epithelial origin and CTCs in these patients are larger than blood leukocytes. CTC enrichment by microfiltration, based on intrinsic physical parameters such as size and deformability, is cost-effective [6–8]. To further improve the efficiency of microfiltration and the viability of enriched cells, we previously described a flexible micro spring array (FMSA) device, a new design of microfilters that can enrich CTCs from 7.5 ml of blood samples with 104-fold enrichment, 90% capture efficiency, and over 80% viability within 10 minutes [9]. Thus, filtration by FMSA is less disruptive to cell integrity than other microfiltration devices, providing a platform capable of enrichment and analysis of viable CTCs from clinically relevant volumes of blood samples [4].
Telomerase expression is a hallmark of cancer [10] and is required for limitless proliferation of tumor cells. The catalytic subunit of human telomerase, hTERT, is silenced in normal human somatic cells but activated in the majority of cancers. This common trait of cancer cells was used in the development of the GFP-containing recombinant adenovirus OBP-401, in which an hTERT promoter drives the viral E1 “master regulator” gene. This virus infects CTCs ex vivo without prior enrichment of blood samples, providing a new logic for CTC detection [11]. However, evidence regarding the specificity of this virus for cancer cells is limited [12, 13]. Although the strategy of CTC detection and enumeration by adenovirus infection remains promising, questions regarding its cancer cell specificity persist and the recombinant virus is not readily available for most academic scientists. Here, we report the development of a class of recombinant adenoviruses with DNA elements that repress hTERT promoter in normal cells [14]. Our results show that multiple copies of the repressive element rendered the virus more selective for replication in cancer cells than in normal cells, especially leukocytes, as determined by GFP imaging. Furthermore, we developed a combinatory strategy for CTC detection and enumeration, by taking advantage of both \ efficient FMSA enrichment technique and the specificity and sensitivity conferred by imaging cells infected with the improved adenovirus. This combinatory approach generated a simple and fail-safe, yet highly efficient, system for CTC analysis in the clinical setting without labor-intensive cytological staining procedures.
Material and methods
Recombinant adenoviruses
Adenoviral constructs were derived from pAdZ5, a bacterial artificial chromosome (BAC) containing a wild type adenovirus type 5 genome [15]. The E3 gene in pAdZ5 was first replaced by a GFP cassette from pEGFP-N1 (Clontech, Mountain View, CA), resulting in pAd5G. The hTERT promoter-containing adenoviral constructs were created by replacing the 179-bp E1 promoter with three versions of the hTERT promoter, using the hTERT ATG codon as the E1a initiation codon (Figure 1). BAC modifications were performed using a two-step recombineering strategy [16]. Viruses were generated by transfecting linearized BACs into 293 packaging cells and purified by CsCl2 gradient ultracentrifugation. The structures of viral DNAs, prepared by Hirt’s method, were verified by restriction enzyme digestion and direct sequencing of the junctions. Viral titers were determined by plaque-forming assays using Hela cells.
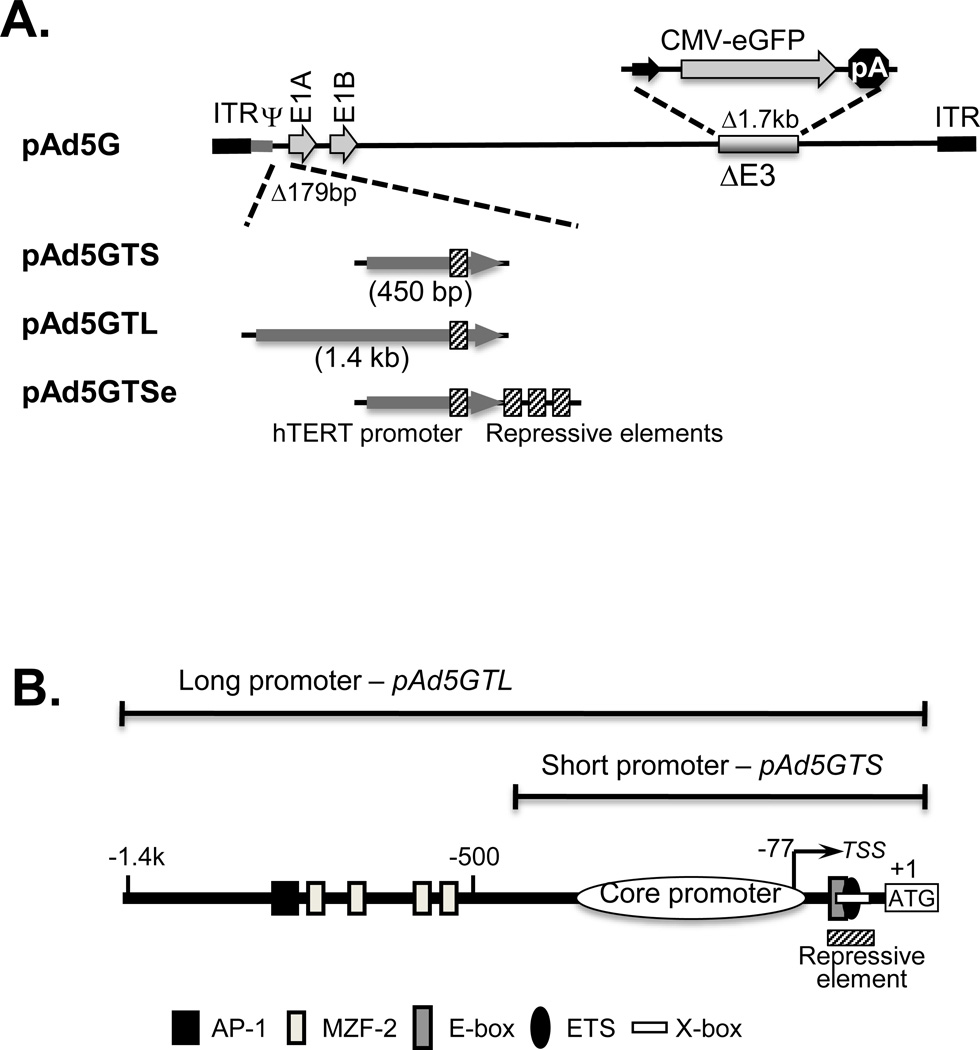
Figure 1.
Recombinant adenoviruses. A. A schematic illustration of recombinant adenoviruses. The upper diagram shows the genome of adenovirus type 5 (~35kb) and the positions of the E1 and E3 genes. In Ad5G, the E3 gene is replaced by a CMV-eGFP expression cassette. In Ad5GTS and Ad5GTL, the 179-bp E1 promoter is replaced by a 450-bp and a 1.4-kb hTERT promoter fragment, respectively, with the hTERT ATG codon serving as the E1A initiation codon. Ad5GTSe contains the short hTERT promoter fragment and three extra copies of a repressor element. ITR, inverted terminal repeat; Ψ, packaging signal; pA, polyA signal. B. The hTERT promoter. The hTERT core promoter, depicted as an oval, contains five GC-boxes and an initiator element. The long and short promoter fragments are within the brackets and the position of a repressor element is indicated by a hatched bar below. ATG, initiation codon; TSS, transcriptional start site.
Cell culture and telomerase expression
Normal human foreskin fibroblasts (NHF) were obtained from Dr. Thea Tlsty’s laboratory at UCSF in 1995 and the fifth passage of this stock was used. Normal human breast epithelial cells (HBECs) were obtained in 2011 from Dr. Andrea Manni’s laboratory at Hershey Medical Center and cultured in MEGM™ mammary epithelial cell growth medium (Lonza, Walkersville, MD). All cancer cell lines were obtained in 2011 from Dr. El-Deiry’s laboratory where they were regularly authenticated by growth, morphological observation, and protein expression that was monitored by Western blotting. The cells were used within two passages in the recommended culture conditions without further authentication after they were transferred. Human colon cancer cell lines HCT-116 and HT-29 were cultured in McCoy’s 5A medium with 10% heat-inactivated fetal bovine serum (FBS). Breast cancer cell lines MDA-MB-231 and T-47D were cultured in DMEM/F12 medium with 5% FBS and RPMI-1640 medium with 10% FBS, respectively. MCF-7 and 293 cells were cultured in DMEM with 10% FBS. MCF-10A cells were cultured in DMEM/F12 containing 10ul/ml insulin, 30ng/ml EGF, 100ng/ml cholera toxin, and 0.5ug/ml hydrocortisone. Human fibroblast lines 167b and NHF were cultured in MEM with 10% FBS. HX-98 cells were immortalized from HBECs using retrovirus pBABE-hTERT [17]. mCherry-expressing cells were obtained by infecting cells with pQCXIP-mCherry or pBABEneo-mCherry (for HX-98 cells) retroviruses, followed by selection with puromycin or neomycin and further enriched by flow-cytometry (BD FACSAria cell sorter). Telomerase activities and hTERT mRNA expression in human cell lines were determined by TRAP assay and real-time RT-PCR analyses, as described previously [18, 19].
White blood cell (WBC) preparation and adenoviral infection
Whole blood was centrifuged at 2000xrpm for 5min and the plasma phase was removed. Erythrocytes were lysed by incubating the blood cells twice with red blood cell lysis buffer containing ammonium chloride at 37°C for 5–10 min, followed by centrifugation and suspension in PBS. mCherry-marked cancer cells, mixed with or without WBCs, were infected with adenoviruses and plated at 37°C in non-tissue culture 96-well plates.
Fluorescence signal detection and immunostaining
Cells in 96-well flat clear bottom black plates were infected with adenoviruses for 72h. GFP and mCherry fluorescence was quantitated every 12h using a microplate reader (Spectra Max M5, Molecular Devices, Sunnyvale, CA) with excitation/emission at 485nm/528nm and 585nm/610nm, respectively. Fluorescent images were captured with an EVOS FL microscope. To identify leukocytes, mixtures of WBCs and cancer cells were fixed in BD fixation/permeabilization solution (BD™ Cytofx/Cytoperm™ Kit), transferred onto glass slides by cytospin, and stained sequentially with a CD45 antibody (Cell Signaling Technology, Danvers, MA) and Alexa Fluor 594-conjugated antibody (Invitrogen, Grand Island, NY).
FMSA filtration
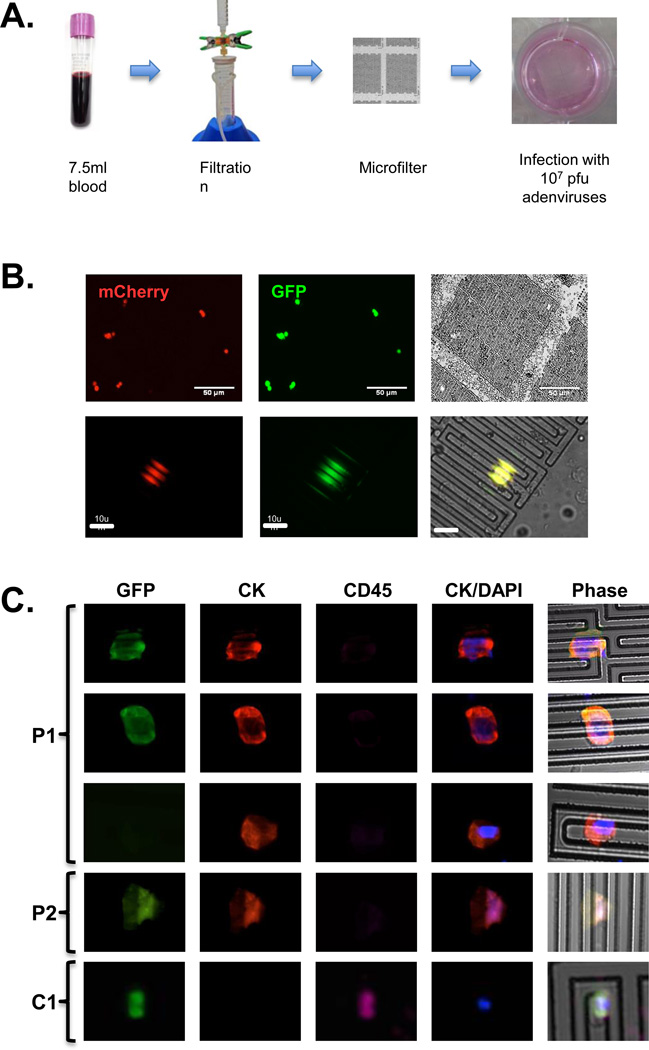
7.5 ml of blood samples were passed through a FMSA filtration device under gravity flow, followed by washing with 7.5 ml of PBS. During filtration, the FMSA device was retained between two sealing O-rings (polydimethylsiloxane, DOW Corning) and clamped within a housing consisting of two plastic jigs [4]. A syringe barrel served as a sample-loading chamber at the top of the filtration device and the bottom was connected to a waste trap. Immediately after filtration, the microfilters with the attached O-rings were transferred to a well of a six-well plate containing 1.5 ml of medium. Ad5GTSe virus was added to the top chamber formed between the O-ring and FMSA, followed by incubation at 37°C for 24h. For immuno-staining, cells on the FMSA devices were fixed with 4% paraformaldehyde and permeabilized by addition of 0.3% Triton X-100. After blocking in 5% goat serum, cells were incubated sequentially with anti-cytokeratin 8/18/19 (clone 2A4, Abcam) and goat anti-mouse IgG conjugated to DyLight 550 (Thermo Scientific). Cells on the devices were blocked again with 5% goat serum, incubated with an anti-CD45 antibody conjugated to Alexa Fluor 647 (clone 35-Z6, Santa Cruz Biotech.), and nuclei were stained with 4’, 6-diamidino-2-phenylindole (DAPI). Each fluorophore was imaged separately using a monochromatic camera (QImaging, Retiga EXi Blue) and composite images were created with pseudocolor added using QCapture Pro 7 software (QImaging). Each mCherry+ and/or GFP+ cell was confirmed to be DAPI+ (data not shown).
For comparison, 7.5 ml of blood samples was also subjected to the CellSearch test at the Penn State Cancer Center. The CellSearch Analyzer II software pre-selects events that are potential CTCs based on EpCAM and CK positivity in close proximity to a DAPI signal. The standard CellSearch procedure was performed, and candidate CTCs were scored by a trained operator.
Statistics
Statistical analyses were performed using GraphPad Prism 6, and results are reported as mean ± SD.
Results
Construction of recombinant adenoviruses
Recombinant adenoviruses were generated from the BAC construct pAdZ5 [15]. First, the E3 gene, dispensable for virus replication in cell culture, was replaced by a GFP cassette to generate pAd5G. Three telomerase-specific adenoviral vectors were engineered from pAd5G (Figure 1). pAd5GTS contains a 450-bp hTERT promoter, which is active in many cell types [20, 21]. This DNA fragment, which contains the hTERT initiation codon, replaced a 179-bp sequence 5’ of and including the E1a initiation codon. This configuration retains the viral packaging signal. In pAd5GTL, a 1.4-kb hTERT promoter substituted for the E1a promoter. The additional sequence in this long promoter contained four MZF-2 sites and an AP-1 site, which are important for repression of hTERT expression in normal cells [22, 23]. pAd5GTSe is similar to pAd5GTS except that it consists of three extra copies of a 20-bp negative control element (CGCACGTGGGAAGCCCTGG), containing overlapping E-box, Ets, and X-box sites [14, 24, 25]. The replication of these recombinant viruses was examined in MCF7 cells (Figure S1). At a multiplicity of infection (MOI) of 10, GFP-positive cells were detected as early as 12h after infection and the numbers of infected cells continued to increase for at least 2 days after infection.
Adenoviral replication and telomerase expression
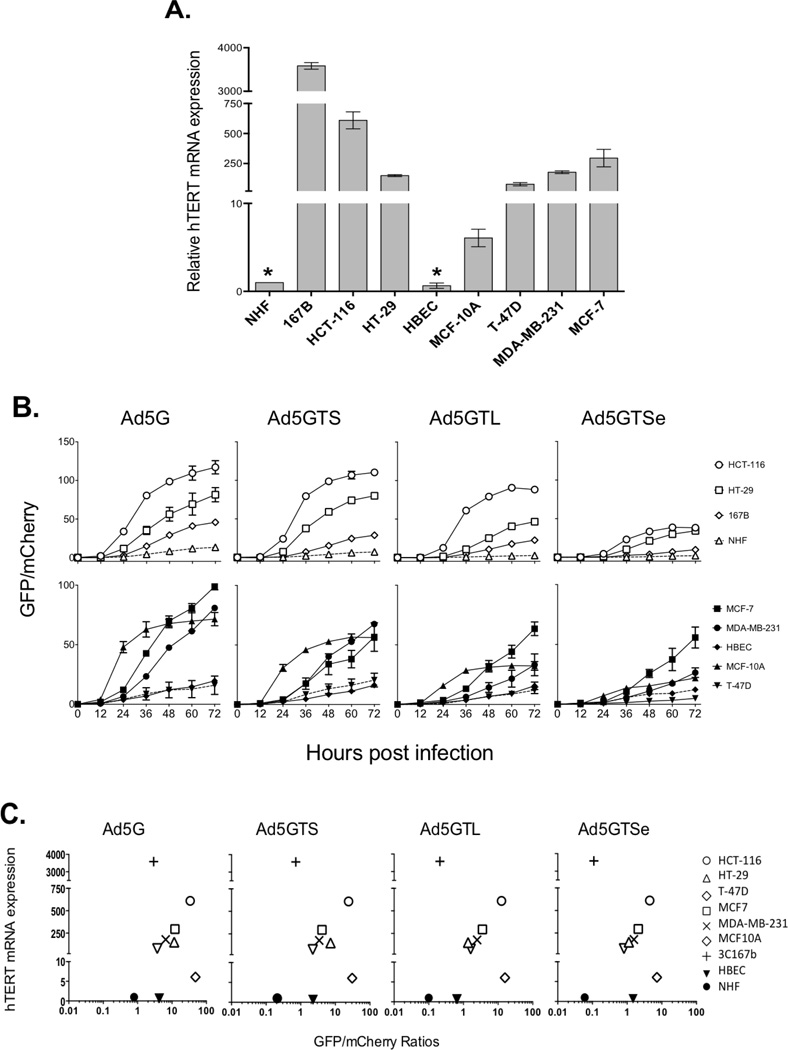
To determine the relationship between viral replication and hTERT expression, we used a collection of normal and cancer-derived cells of lung, breast, and colon origins. As shown in Figure 2A & S2, all the cancer cell lines tested expressed high levels of hTERT mRNA and telomerase activity. Fibroblast cell line 3C167b also contained abundant hTERT mRNA and telomerase activity. Further, immortalized but untransformed breast epithelial MCF10A cells expressed detectable hTERT mRNA and telomerase activity. In contrast, normal human fibroblasts (NHFs) and HBECs had little hTERT mRNA and undetectable telomerase.
Figure 2.
hTERT expression and replication of recombinant adenoviruses. A. hTERT expression in cancer cell lines and normal cells. hTERT mRNA levels were determined by quantitative RT-PCR. Values were normalized to the levels of 18S ribosomal RNA. *, normal cells with limited life spans. B. Replication of recombinant adenoviruses. 12h prior to infection, 1.5×104 cells were plated into individual wells of 96-well plates and infected with recombinant adenoviruses at MOI 10. GFP and mCherry fluorescent signals were recorded every 12h post infection using a microplate reader. Viral replication was reported as the ratio of GFP signals to those of mCherry. C. Relationship of hTERT mRNA levels and adenoviral replication. The data in panel A were plotted against the levels of fluorescent signals at 24h in panel B. All data were collected as averages of triplicates.
An important objective of this work was to evaluate whether cancer cells mixed with normal cells would be preferentially labeled by virus infection of the mixed population (see below). Accordingly, we needed to establish a means of tracking the cancer cells in the mixtures. This strategy would also permit us to control for any differences among the cells lines in their ability to express GFP. To do so, cells were transduced with retroviruses containing a mCherry protein and sorted by FACS, yielding over 98% mCherry-positive (mCherry+) cells. These cells were infected with adenoviruses and GFP/mCherry signal ratios were determined (Figure 2B). All of the recombinant adenoviruses replicated more efficiently in telomerase-positive cancer cells than in normal cells, although cellular hTERT mRNA levels were not strictly correlated to viral GFP signals (Figure 2C). The parental Ad5G virus replicated better than hTERT promoter-driven viruses in all of the cell lines. Ad5GTL and Ad5GTSe replicated even slower than Ad5GTS, especially in NHFs and HBECs, probably because the repressive elements reduced the rate of accumulation of E1a gene products and consequently delayed the viral lytic cycle.
Labeling cancer cells and white blood cells by adenoviruses
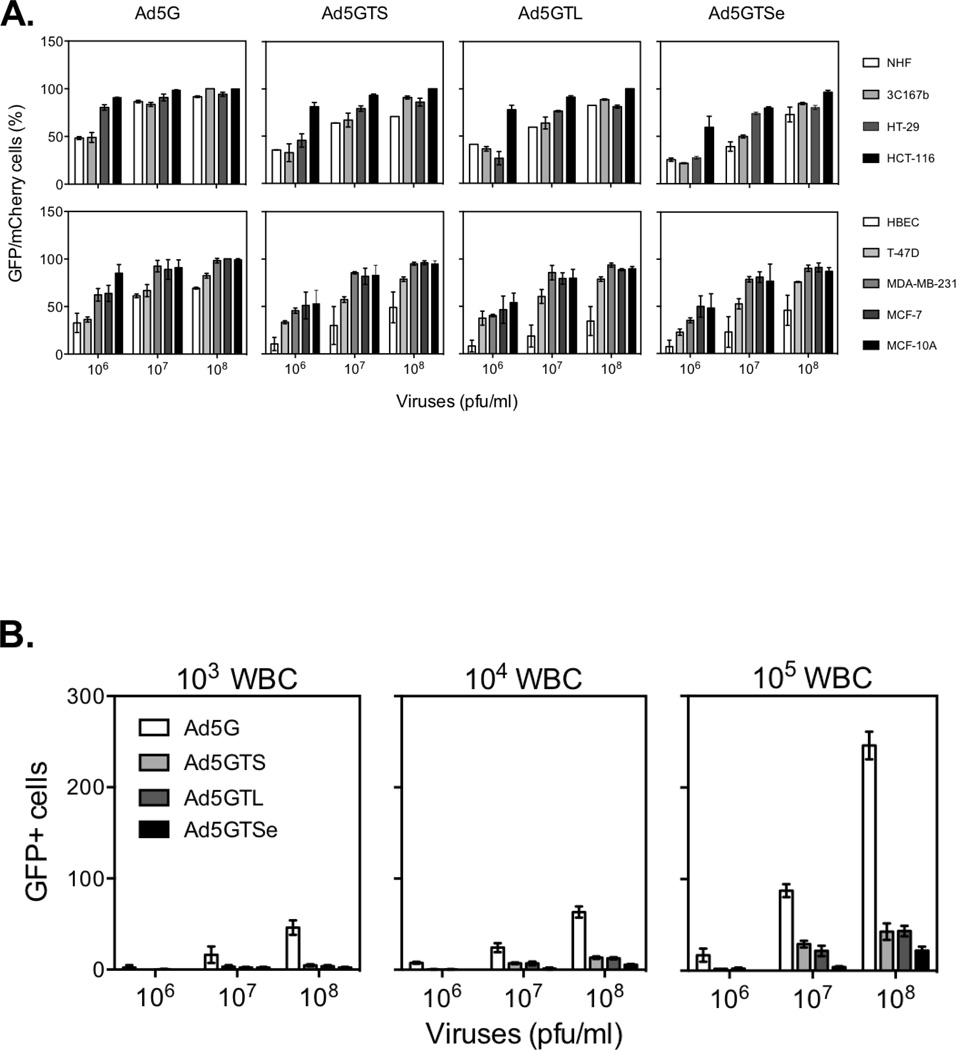
To determine the abilities of the recombinant adenoviruses to label cancer cells, about 200 cells in a single well of 96-well plates were infected with an individual adenovirus at different doses and GFP+ cells were quantified 24h later. As shown in Figure 3A, over 80% of all immortal and cancer cells displayed GFP signals when infected with 108 pfu/ml of any of the viruses. With 107 pfu/ml of virus, the labeling efficiencies were still above 80% for most cell lines, except for 3C167b and T-47D lines. For normal and telomerase-non-expressing NHFs and HBECs, the labeling efficiencies were lower than for cancer cell lines in most cases. Among different viruses, Ad5G infected both cancer cell lines and normal cells most efficiently, whereas the infection efficiencies by Ad5GTSe were lower than those by other viruses, requiring at least 107 pfu/ml to label 70–80% of the cancer cells. Nevertheless, our data showed that 107 pfu/ml Ad5GTSe was best suited for further investigations of differentially labeling cancer cells, although none of the viruses was strictly specific for cancer cells.
Figure 3.
Adenoviral infection of different cell types. A. Efficiency of fluorescent labeling of normal and cancer cells by infection with recombinant adenoviruses. Approximately 200 cells were mixed with 100µl of adenovirus stock and added to individual wells of a 96-well plate with no tissue culture coating. GFP-expressing cells were counted 24h post infection. Total numbers of cells were determined by counting mCherry+ cells. B. Infection of white blood cells (WBCs) by adenoviruses. WBCs (103, 104, or 105) were infected with 100µl of adenovirus stock for 24h.
CTC preparations contain large amounts of blood cells even after enrichment, so we examined the ability of the adenoviruses to infect white blood cells (WBCs). WBCs were infected with adenoviruses and GFP-labeled cells were examined 24h post infection. Less than 5% of the WBCs were labeled at the highest Ad5G dose tested (108 pfu/ml) (Figure 3B), much less than Ad5G-infected epithelial or fibroblast cells. The hTERT promoter-driven viruses infected WBCs at efficiencies much lower than Ad5G did. Ad5GTSe was particularly ineffective in labeling WBCs at 107 pfu/ml dose. The data indicate that Ad5GTSe is suitable for distinguishing cancer cells from WBCs.
Preferential infection of cancer cells in mixtures with WBCs
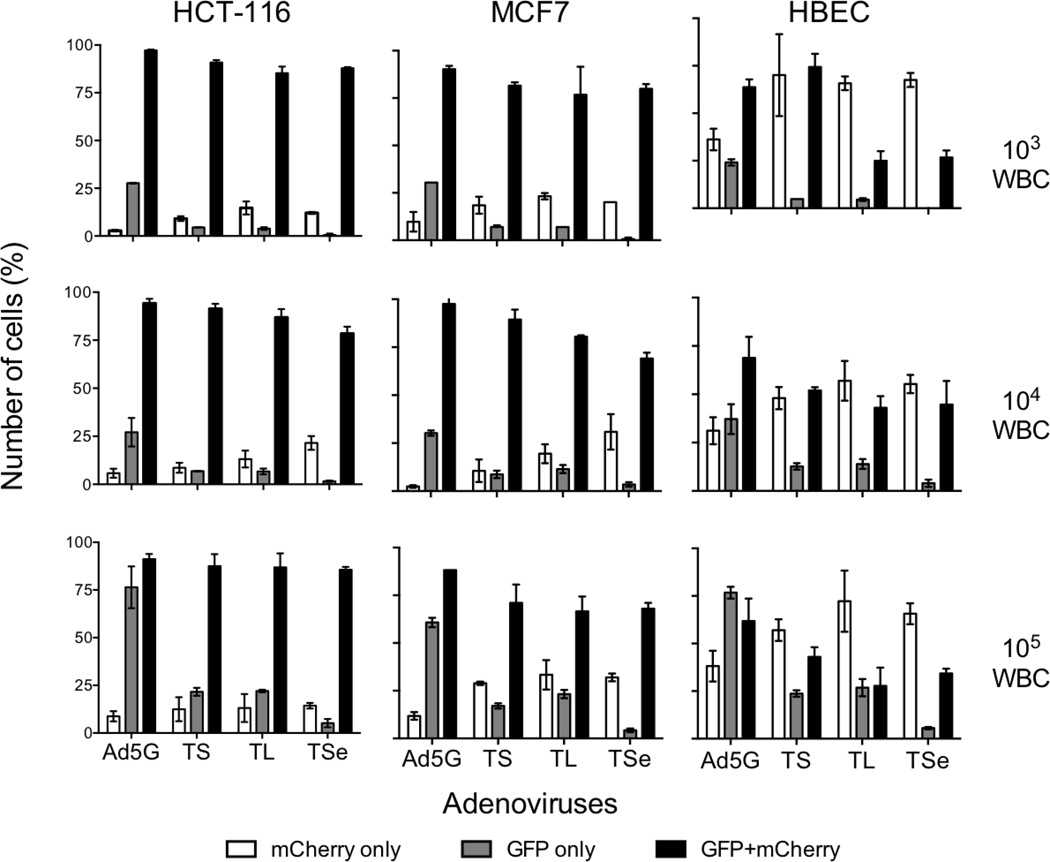
To establish the feasibility of CTC detection using adenovirus-mediated imaging, mCherry+ HCT-116 colon cancer cells were mixed with WBCs and infected with 107 pfu/ml of an adenovirus for 24h (Figure S3). When 100 or 1000 HCT-116 cells were mixed with 103 to 105 WBCs, similar numbers of mCherry+ and GFP+ cells were observed. When 10 cancer cells were added to the WBCs, there were more GFP+ cells than mCherry+ cells especially after infection with Ad5G, indicating that Ad5G must have infected significant numbers of WBCs. Ad5GTSe-infected cells were a notable exception: similar numbers of GFP+ and mCherry+ cells were always observed, suggesting that very few WBCs if any were labeled by this virus. To illustrate the specificities of infection of HCT-116 cells by adenoviruses, GFP+ only, mCherry+ only, and double positive cells in the mixtures containing 100 HCT-116 cells were also counted (Figure 4, left column). Most of mCherry+ cells were labeled by GFP, indicating that HCT-116 cells were efficiently infected by all four viruses. Conversely, upon infection by hTERT promoter-driven viruses, fewer GFP+mCherry− cells were detected than those in Ad5G-infected mixtures. In the case of Ad5GTSe, about 5 GFP+mCherry− cells were found even when 105 WBCs were used, indicating a strong tropism of this virus for HCT-116 cells. To check the infection of WBCs in the mixtures directly, one of the Ad5GTSe-infected cell mixtures (1000 HCT-116 cells and 105 WBCs) was stained with an antibody against CD45, a common leukocyte antigen. In this experiment, unlabeled HCT-116 cells were used so that the mCherry fluorescence would not interfere with the visualization of CD45 staining in the red channel. Figure S4 shows that no GFP+/CD45+ cells were observed. We conclude that Ad5GTSe is highly selective for labeling of HCT-116 cells in mixtures with WBCs.
Figure 4.
Adenovirus-mediated labeling of mixtures of cancer and normal cells with WBCs. 100 mCherry+ HCT-116 (left charts), MCF7 (left charts), or HBECs (right charts) were mixed with 103, 104, or 105 WBCs, and infected with 107 pfu/ml adenoviruses in 100µl. GFP, mCherry, and doubly-labeled cells were scored after 24h incubation.
To further evaluate cancer cell imaging using the recombinant adenoviruses, similar experiments were performed to compare the imaging efficiencies of cancer and normal breast epithelial cells, MCF7 and HBEC, respectively, in mixtures with WBCs. 100 cells were mixed with varying numbers of WBCs, infected with 107 pfu/ml adenoviruses, and imaged 24h post infection. GFP-/mCherry+ cells were uninfected epithelial cells and GFP+/mCherry− cells were most likely infected WBCs. As shown in the middle column of Figure 4, there were significant numbers of GFP+/mCherry− MCF7 cells in the Ad5G-infected mixtures in all conditions tested (grey bars), indicating that this virus produced significant false positive signals due to its infection of WBCs. Ad5GTS and Ad5GTL labeled some mCherry− cells, especially when 105 WBCs were present. Notably, Ad5GTSe labeled over 70% MCF7 cells in all conditions, but infected no more than 5 mCherry− cells even when 105 WBCs were used. As above, we did not detect any Ad5GTSe-labeled cells that expressed CD45 in a mixture of MCF7 cells and 105 WBCs (Figure S5).
The data in Figures 2 and 3 showed that the recombinant adenoviruses also infect normal human cells. Indeed, these viruses infected and labeled many HBECs in the mixtures (Figure 4, right column). Fewer cells were labeled by infection with Ad5GTL or Ad5GTSe, likely due to the negative regulatory elements in the hTERT promoter in these viruses. The results support the conclusion that, because it displayed the best selectivity in labeling cancer cells over both HMECs and WBCs, Ad5GTSe is a viable and attractive candidate for improved CTC detection.
Enrichment and imaging of cancer cells using FMSA microfilters
Because none of the viruses was absolutely specific for cancer cells, enrichment of cancer cells prior to adenoviral imaging may reduce false-positive signals. To explore the possibility of combining microfiltration enrichment and adenoviral imaging in CTC detection, approximately 300 mCherry-labeled colon cancer HCT-116 or breast cancer MCF7 cells were added to 7.5 ml of blood from healthy donors and the mixtures were passed through a FMSA filtration device (Figures 5A&B). The microfilters were then transferred into single wells of a 6-well plate and infected with 107 pfu/ml Ad5GTSe for 24h. As shown in Table 1, the labeling efficiencies of cancer cells ranged from 75% to 93% in four experiments. However, there were 3–5 GFP+/mCherry− cells, either rare mCherry− cancer cells or infected WBCs, in each experiment. This result demonstrated that Ad5GTSe could detect live cancer cells following their enrichment on FMSA.
Figure 5.
A combinatory strategy to detect cancer cells in blood samples. A. Schematic of the procedure. 7.5 ml of blood was passed through a FMSA filtration device and the microfilter was transferred to a single well of a 6-well plate. Cells captured on the microfilters were infected with Ad5GTSe. B. Infection of HCT116 and MCF7 cells by Ad5GTSe on a FSMA. 7.5 ml of whole blood from healthy donors mixed with approximately 300 mCherry+ cancer cells was enriched on FMSA and infected with 107 pfu Ad6GTSe virus for 24h at 37°C. Cancer cells were detected in fluorescent images, but WBCs could only be visualized by phase microscopy. Images of HCT 116 cells on FSMA are shown. C. Expression of surface markers by Ad5TSe-infected CTCs in cancer patient samples. 7.5 ml of patient blood was passed through a FSMA device and the captured cells were infected with 107 pfu of Ad5GTSe. Cells on the microfilters were stained with antibodies against cytokeratin8/18/19 (CK, DyLight 550) and CD45 (Alexa Fluor 647), and visualized in their respective wavelengths. Nuclei were stained with DAPI. P1, P2, & C1 refer to Patients #1, #2, and control #1 (see Table 2).
Table 1.
Labeling efficiencies of HCT116 and MCF7 cells retained on microfilters.
| Cell lines | Total mCherry+ cells |
GFP+mCherry+ cells |
Infection efficiency (GFP+/mCherry+) |
GFP+mCherry− cells |
|---|---|---|---|---|
| HCT-116 | 239 | 221 | 93% | 5 |
| 277 | 252 | 91% | 3 | |
| MCF-7 | 236 | 178 | 75% | 3 |
| 267 | 220 | 82% | 5 |
Experiments were performed as described in Figure 5B.
Identification of CTCs in patient blood samples
Next, we applied this combinatory strategy in the analyses of blood samples from patients with breast and pancreatic cancers. Two 7.5 ml blood samples were collected from each patient. One sample was passed through a FMSA device and cells retained on the microfilter were infected with 107 pfu/ml Ad5GTSe. To distinguish cells of epithelial origin from those of hematopoietic origin, the cells on FMSA were then stained with antibodies against cytokeratin (CK), an epithelial marker, and CD45 (Figure 5C). As shown in Table 2, GFP+ cells were identified in all patient samples, ranging from 19 in patient #4 to 32 in patient #3. Most of these GFP+ cells were CK+ and CD45−, consistent with an epithelial origin characteristic of most breast and pancreatic cancer cells. Interestingly, most CK+/CD45− cells were also infected by Ad5GTSe, as indicated by GFP expression in these cells, indicating that most of the CTCs in the clinical samples were actually viable.
Table 2.
Detection of potential CTCs in blood samples of cancer patients.
| Patients | Patient1 | Patient2 | Patient3 | Patient4 | Control1 | Control2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GFP+ | GFP− | GFP+ | GFP− | GFP+ | GFP− | GFP+ | GFP− | GFP+ | GFP− | GFP+ | GFP− | |
| CK+/CD45− | 18 | 4 | 17 | 2 | 30 | 3 | 13 | 4 | 0 | 0 | 0 | 0 |
| CK+/CD45+ | 0 | ND | 0 | ND | 2 | ND | 4 | ND | 0 | ND | 0 | ND |
| CK−/CD45+ | 0 | tmtc | 1 | tmtc | 0 | tmtc | 2 | tmtc | 1 | tmtc | 6 | tmtc |
| CK−/CD45− | 3 | ND | 5 | ND | 0 | ND | 0 | ND | 5 | ND | 1 | ND |
| Total GFP+ cells | 21 | 23 | 32 | 19 | 6 | 7 | ||||||
| Veridex counting | 8 | 0 | 1 | 0 | ND | ND | ||||||
| Cancer types | Breast (Triple−) | Breast (ER+) | Pancreatic (Stage IV) | Pancreatic (Stage IIB) | No | No | ||||||
| Metastasis | Bone | Bone | Liver | No | No | No | ||||||
Numbers indicate the quantities of cells detected on each microfilter. ND, nnot determined; tmtc, too many to count.
Although a few GFP+ cells were detected in blood samples from two healthy donors (Controls #1 and #2), none of them were CK+/CD45−, suggesting that the GFP+/CK+/CD45− cells in the samples from cancer patients were tumor cells. The other 7.5 ml of blood sample from each patient was subjected in parallel to the CellSearch® CTC test. Eight and one positive cells were detected in 7.5 ml of blood from patients #1 and #3, respectively, but none was found in other two samples. These data suggest that Ad5GTSe selectively detected CTCs in breast and pancreatic cancer patients with potentially higher sensitivity than Veridex CellSearch® system. The results demonstrate proof of principle that the combinatory strategy of microfiltration and adenoviral imaging using Ad5GTSe is likely an extremely sensitive and specific method for CTC detection and enumeration for cancer patients.
Discussion
Major challenges in CTC detection are to process large amounts of blood cells in a short period of time and to employ sensitive and specific methods to identify rare and often-variable tumor cells. Most reported CTC methods are based on antibodies against cell surface antigens. Recently, the recombinant adenovirus OBP-401 was used to image viable CTCs in patient blood samples [11]. Although it was reported that this virus was specific for telomerase-expressing cancer cells, data supporting this conclusion have thus far been limited [12, 13]. Based on extensive studies of hTERT gene regulation, the promoter fragment used in OBP-401 should provide only limited cancer cell-specificity [20, 21]. Here, we generated a new class of replication-competent adenoviruses, Ad5GTS, Ad5GTL, and Ad5GTSe, with variants of the hTERT promoter. Our data demonstrated that Ad5GTL and Ad5GTSe, with extra repressive elements, were more selective for replication in cancer cells versus normal cells than Ad5GTS, which contains an hTERT promoter fragment comparable to OBP-401. Among the new viruses, Ad5GTSe was best suited for CTC detection because this virus labeled the fewest number of WBCs.
Expression of the E1 genes of Ad5GTS and OBP-401 is driven by 450 or 455-bp of the hTERT promoter upstream of the ATG codon, respectively. Based on the similarity of their genetic organization and sequences, these two viruses are expected to have comparable replication efficiencies and cancer cell specificities. The region controlling E1 expression in the Ad5GTL genome has an additional 1kb of sequence upstream of the core hTERT promoter, including several DNA elements involved in hTERT repression in normal cells (Figure 1B) [22, 23]. Instead of this region, the Ad5GTSe genome has three extra copies of a composite element known to repress the hTERT promoter in normal but not cancer cells [14]. The E-box site in this element mediates hTERT repression in differentiated cells by the Myc antagonist Mad1 [25]. A putative X-box-binding protein NFX1-91 represses hTERT expression in epithelial cells. As part of its oncogenic activity, the HPV E6 protein activates hTERT expression by targeting NFX1-91 for ubiquitin-mediated degradation [24]. As predicted, replication of the two viruses containing additional repressive elements, especially Ad5GTSe, was less efficient in normal cells than in most cancer cell lines. Two notable exceptions were 3C167b fibroblasts and T-47D breast cancer cells. The hTERT gene is translocated and amplified in some immortal cell lines and cancer cells [26] and is rearranged in 3C167b cells [27]. Such alterations of the hTERT locus might contribute to the atypical response of these cell lines to viral infection and also help explain why we observed poor correlation between viral replication, measured by GFP expression, and endogenous hTERT mRNA levels (Figure 2). In addition, many cancer cells contain mutations at the hTERT locus, such as promoter mutations in familial and sporadic melanoma patients [28, 29]. These mutations of at the endogenous hTERT gene could also alter hTERT transcription and contribute to the miscorrelation between cellular hTERT mRNA levels and viral GFP signals in our study.
Another new observation in this study is that the hTERT promoter-driven adenoviruses replicate much less efficiently in WBCs than the wild type Ad5G (Figure 3B). The data suggest that the hTERT promoter has a low activity in normal blood cells. This low activity results in the poor replication of the hTERT promoter-containing viruses in WBCs and is likely the molecular basis for selective CTC detection in patient blood samples by these viruses [11].
We found that Ad5GTSe was best suited for discriminating between CTCs and WBCs in blood samples; 107 infectious particles of this virus labeled about 5 WBCs per 105 cells in all experiments. Thus, the lower limit of CTC detection by Ad5GTSe is about one in 104 WBCs. While this was the best among all the recombinant adenoviruses, such an efficiency is unlikely to achieve accurate CTC detection in cancer patients due to the overwhelmingly large numbers of WBCs in blood samples. Therefore, enrichment of cancer cells was required to reduce false positive signals. To this end, we designed a novel strategy combining microfiltration enrichment using FMSA devices and adenoviral infection [4]. In this combinatory approach, FMSA filtration was employed to enrich the CTCs against the WBCs by a factor of ~104. In a sample of 7.5 mL undiluted blood, the total number of WBCs on the filter was thus reduced to less than 104–5, falling within the range in which an optimal detection sensitivity and specificity of CTCs could be achieved by using Ad5GTSe.
Unlike microfilter devices used previously, the FMSA device has flexible micro springs that deform in response to pressure, allowing a higher flow-through rate under low differential pressure, minimizing cellular mechanical stress, and thus preserving viability [9]. With this combined strategy, we identified more potential CTCs than the CellSearch assay in our experiments using a limited number of cancer patient samples (Table 1). Previously, Lin et al. also reported that immunostaining of patient samples enriched by a different type of microfilter identified potential CTCs in more cancer patients as well as more CTCs in most patients [5]. Thus, the superior recovery rate of CTCs by microfiltration in combination with adenoviral infection of live cells may lead to a faster and more efficient strategy of viable CTC detection and enumeration.
In summary, our current combinatory strategy using imaging of adenovirus-infected cells has the potential to be an excellent method for detecting CTCs in clinical settings. First, adenoviral infection and imaging are simple procedures, requiring no sophisticated equipment or highly technical manipulations. Second, the method labels only live cells, because infection and replication in living cancer cells is required to obtain GFP expression. Third, the hTERT promoter-dependent viruses should infect many types of cancer cells, regardless of their origins and development stages. This broad host range is an important feature, since cells in most tumors are highly heterogeneous. Susceptibility to adenovirus infection is unlikely to be affected by changes in the molecular features of tumor cells, such as EMT, a transition from epithelial tumor cells to an invasive and mesenchymal cell-like phenotype, and so virus infection should provide a reliable CTC evaluation. Finally, though Kojima et al. reported the detection of CTCs by OBP-401 without enrichment [11], our study revealed that even the improved virus Ad5GTSe labeled normal leukocytes in patient samples. However, while the combinatory strategy promises to result in a more sensitive detection with fewer false positives than straight adenoviral infection, additional studies will be needed to test its efficacy in various clinical settings.
Supplementary Material
Acknowledgement
We thank Dr. Richard Stanton of Cardiff University for adenoviral BAC constructs. The work was supported by the NIH Grants R03CA165182 and R01GM071725 to J.Z and DP2CA174508 to S.Z. In addition, J.Z. was also supported by W.W. Smith Charitable Trust. D.J.S. thanks Richard Courtney and members of the Department of Microbiology and Immunology for their support.
Footnotes
None of the authors have professional or financial affiliations that could be perceived to bias the presentation of this manuscript.
References
- 1.Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell. 2013;23(5):573–581. doi: 10.1016/j.ccr.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin H, Balic M, Zheng S, Datar R, Cote RJ. Disseminated and circulating tumor cells: Role in effective cancer management. Crit Rev Oncol Hematol. 2010;77(1):1–11. doi: 10.1016/j.critrevonc.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Balic M, Williams A, Lin H, Datar R, Cote RJ. Circulating tumor cells: from bench to bedside. Annu Rev Med. 2013;64:31–44. doi: 10.1146/annurev-med-050311-163404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harouaka RA, Zhou MD, Yeh YT, Khan WJ, Das A, Liu X, et al. Flexible Micro Spring Array Device for High-Throughput Enrichment of Viable Circulating Tumor Cells. Clin Chem. 2014;60(2):323–333. doi: 10.1373/clinchem.2013.206805. [DOI] [PubMed] [Google Scholar]
- 5.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16(20):5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, et al. Isolation by size of epithelial tumor cells : a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156(1):57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162(2):154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 8.Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13(1):203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harouaka RA, Zhou MD, Yeh TY, Khan WJ, Allerton J, Zheng SY. Viable circulating tumor cell enrichment by flexible micro spring array. Conf Proc IEEE Eng Med Biol Soc. 2012;2012:6269–6272. doi: 10.1109/EMBC.2012.6347427. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Kojima T, Hashimoto Y, Watanabe Y, Kagawa S, Uno F, Kuroda S, et al. A simple biological imaging system for detecting viable human circulating tumor cells. J Clin Invest. 2009;119(10):3172–3181. doi: 10.1172/JCI38609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kishimoto H, Kojima T, Watanabe Y, Kagawa S, Fujiwara T, Uno F, et al. In vivo imaging of lymph node metastasis with telomerase-specific replication-selective adenovirus. Nat Med. 2006;12(10):1213–1219. doi: 10.1038/nm1404. [DOI] [PubMed] [Google Scholar]
- 13.Maida Y, Kyo S, Sakaguchi J, Mizumoto Y, Hashimoto M, Mori N, et al. Diagnostic potential and limitation of imaging cancer cells in cytological samples using telomerase-specific replicative adenovirus. Int J Oncol. 2009;34(6):1549–1556. doi: 10.3892/ijo_00000284. [DOI] [PubMed] [Google Scholar]
- 14.Horikawa I, Cable PL, Mazur SJ, Appella E, Afshari CA, Barrett JC. Downstream E-Box-mediated Regulation of the Human Telomerase Reverse Transcriptase (hTERT) Gene Transcription: Evidence for an Endogenous Mechanism of Transcriptional Repression. Mol Biol Cell. 2002;13(8):2585–2597. doi: 10.1091/mbc.E01-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanton RJ, McSharry BP, Armstrong M, Tomasec P, Wilkinson GW. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques. 2008;45(6):659–662. 664–668. doi: 10.2144/000112993. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol Biotechnol. 2009;42(1):110–116. doi: 10.1007/s12033-009-9142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Wang H, Bishop JM, Blackburn EH. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc Natl Acad Sci USA. 1999;96(7):3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia W, Wang S, Horner JW, Wang N, Wang H, Gunther EJ, et al. A BAC transgenic reporter recapitulates in vivo regulation of human telomerase reverse transcriptase in development and tumorigenesis. Faseb J. 2011;25(3):979–989. doi: 10.1096/fj.10-173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathew R, Jia W, Sharma A, Zhao Y, Clarke LE, Cheng X, et al. Robust activation of the human but not mouse telomerase gene during the induction of pluripotency. Faseb J. 2010;24(8):2702–2715. doi: 10.1096/fj.09-148973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, Newbold RF, et al. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61(20):7594–7602. [PubMed] [Google Scholar]
- 21.Wang S, Zhu J. Evidence for a relief of repression mechanism for activation of the human telomerase reverse transcriptase promoter. Journal of Biological Chemistry. 2003;278(21):18842–18850. doi: 10.1074/jbc.M209544200. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto K, Kyo S, Takakura M, Kanaya T, Kitagawa Y, Itoh H, et al. Identification and characterization of negative regulatory elements of the human telomerase catalytic subunit (hTERT) gene promoter: possible role of MZF-2 in transcriptional repression of hTERT. Nuc Acid Res. 2000;28(13):2557–2562. doi: 10.1093/nar/28.13.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takakura M, Kyo S, Inoue M, Wright WE, Shay JW. Function of AP-1 in transcription of the telomerase reverse transcriptase gene (TERT) in human and mouse cells. Mol Cell Biol. 2005;25(18):8037–8043. doi: 10.1128/MCB.25.18.8037-8043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gewin L, Myers H, Kiyono T, Galloway DA. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004;18(18):2269–2282. doi: 10.1101/gad.1214704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D, Popov N, Hou M, Wang Q, Bjorkholm M, Gruber A, et al. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci USA. 2001;98(7):3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Zhao Y, Wang S. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein & Cell. 2010;1(1):22–32. doi: 10.1007/s13238-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Wang S, Popova EY, Grigoryev SA, Zhu J. Rearrangement of upstream sequences of the hTERT gene during cellular immortalization. Genes Chromosomes Cancer. 2009;48(11):963–974. doi: 10.1002/gcc.20698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 29.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.