Abstract
Abstract
In the present study, artificial neural network (ANN) modelling coupled with particle swarm optimization (PSO) algorithm was used to optimize the process variables for enhanced low density polyethylene (LDPE) degradation by Curvularia lunata SG1. In the non-linear ANN model, temperature, pH, contact time and agitation were used as input variables and polyethylene bio-degradation as the output variable. Further, on application of PSO to the ANN model, the optimum values of the process parameters were as follows: pH = 7.6, temperature = 37.97 °C, agitation rate = 190.48 rpm and incubation time = 261.95 days. A comparison between the model results and experimental data gave a high correlation coefficient (). Significant enhancement of LDPE bio-degradation using C.lunata SG1by about 48 % was achieved under optimum conditions. Thus, the novelty of the work lies in the application of combination of ANN–PSO as optimization strategy to enhance the bio-degradation of LDPE.
Graphical Abstract
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0522-z) contains supplementary material, which is available to authorized users.
Keywords: Low density polyethylene, Modelling, Optimization, Waste utilization
Introduction
Synthetic polymers like low-density polyethylene (LDPE) are mostly inert towards microorganisms. In the initially produced form, they undergo very slow biodegradation in both stabilized and un-stabilized forms [1]. The long-term properties of the synthetic and natural polymers have attracted more interest during the last decade as environmental concerns regarding the disposal of these commodity plastics have increased. It is generally proposed that oxo-biodegradation of LDPE films is initiated by UV irradiation [1] or thermal exposure [2]. These treatments generate a macro radical in the amorphous region of the polymer substrate. The free radical is then rapidly oxidized through a series of chain reactions to produce a carbonyl group [3, 4]. Microbial attack is started where the carboxylic acid group is generated through Norrish Type I and II mechanism during oxidation [5]. The microbiological process, which leads to the degradation of natural polymers such as starch, cellulose and proteins during exposure to soil, is well understood. However for the commercialized synthetic polymers there are conflicting claims concerning their susceptibility to microbial attacks [3, 5, 6]. The manner and rate are dependent on the mechanism of degradation and on the acceleration of the process, respectively. The mechanism consists of generation of terminal carboxylic acid group, which then undergoes beta oxidation. In most studies, fungi were considered for the degradation of LDPE due to their ability to form hydrophobic proteins that can attach to the polymer surface [7, 8], their generation of degrading enzymes that are well-matched to the insoluble LDPE [9], the faster growth of fungal biomass in soil compared to bacteria [10], and the growth extension and penetration into other locations through the distribution of hyphae. Also, fungi survive environments with low nutrient, pH and moisture availability. However, there are still many unexplored areas of research in the degradation of LDPE by fungi. Characterization of LDPE degradation by solid waste-source fungi is among these attractive topics because of their compatibility with a waste-rich environment (such as landfill and composting) that contains a variety of discarded polymers. Among various statistical optimization techniques for media components along with environmental parameters, response surface methodology (RSM) has been extensively employed in the optimization of various bio-processes. However, in some cases, complex non-linear biological interactions cannot be completely described by using second-order polynomial model based on RSM [11, 12]. Hence, a more advanced modelling and optimization technique such as artificial neural network modelling coupled with genetic algorithm has been successfully implemented to optimize multivariate non-linear bio-processes [13, 14]. The merits of ANN based models were discussed in earlier reports [13, 15]. Since genetic algorithm suffers from one major shortcoming as it destroys previous information between successive generations, a more robust algorithm that can deal with relatively small population size and can help converge at the optimal solutions very quickly while memorizing the previously known good solutions between generations is in great demand. Particle swarm optimization (PSO) is one such population-based evolutionary algorithm that can be more effectively used for solving a non-linear problem involving multiple variables. The particles in PSO follow a similar trend to share information among them and thereby, develop an evolutionary advantage [16]. Constructive cooperation, information sharing, inexpensive computation (requires low memory and CPU speed requirements), and easy implementation are few other attractive properties of PSO [12, 17]. Owing to its robustness, PSO is now being applied to solve various non-linear problems with multiple variables in majority of engineering disciplines and has gained the status of a potential competitor of frequently used genetic algorithm [15]. Since there are no literatures on the use of ANN–PSO optimization technique to enhance LDPE bio-degradation, the present work is thus aimed at optimizing the process parameters by ANN modelling coupled with PSO algorithm to maximize LDPE bio-degradation.
Methods
Substrate and Pre-treatment
Standard commercial polyethylene (grade-LD103) films were procured from commercial market, Gunupur, India. The average molecular weight (Mw) was about 100,000. Raw material composition was 30 micron thickness, 132 °C melting point, density 0.923 g/cm3, melt flow index 2 g/10 min. They were cut in pieces of approximately 8 cm × 2 cm size. All the chemicals were procured from (HIMEDIA Laboratories, India). Photo and thermal oxidation were used as pre-treatment steps of the pre-weighed LDPE samples where one set of LDPE strips were thermally pre-treated (TP) at 80 °C in an hot air oven (Sigma, USA) for 120 h. Another set of LDPE films were UV irradiated (UV) under 20 W ultraviolet lamp (ZSZ-D, Changsha Guangming Co. Ltd). The samples were placed 15 cm away from the lamp, where the light intensity was measured using a UV intensity meter (UV-I, Beijing Shida Ltd) at wavelength (354 nm) for 50 h. The third set was the combined pre-treatment of thermal and UV-irradiation (TP-UV).
Microorganism, Medium and Growth Conditions
The fungus Curvularia lunata SG1 isolated from soil samples of garbage dumping site of Gunupur, India, was cultured and maintained in Sabouraud’s broth (SB) [Dextrose-20, Peptone-10, pH −5.6 ± 0.2 (in g L−1)] at 37 °C for 7 days for spore formation. Spores were harvested and suspended in 1 % (v/v) Tween 80 (T-80). Spore suspension was then centrifuged at 4000 rpm for 20 min. The spores were suspended in distilled water to prepare a spore inoculum of 1 × 107 spores/mL. About 10 % (v/v) inoculum was developed in 250 mL of SB in 1 L Erlenmeyer flask containing pre-treated LDPE and T-80 as surfactant and incubated in an incubator shaker (Remi, RIS 24 BL) at 100 rpm and 37 °C for 90 days. Initial screening of different media showed that SB supplemented with T-80 exhibited good LDPE degradation and hence SB was used as medium for further studies. Flasks with the pure culture but without the polymer films served as the biotic control. Control was treated the same way as the test (samples and fungus) samples. Samples were withdrawn every 15 days under aseptic conditions, washed in sterile water and air-dried before further analysis.
Study of Fungal Viability
The viability of the microorganism (live and dead cells) present on the LDPE films were determined using Live/Dead® FungaLight™ yeast viability Kit (Invitrogen, Germany). The kit consists of SYTO9 and propidium iodide (PI) dyes. These two dyes differ in their ability to stain the fungal cells. SYTO9 (green colour) stains the live as well as the dead cells and, PI (red colour) stains only the dead cells. When the later is added it reduces the fluorescence of SYTO9 by penetrating into the dead cells. Hence live cells fluorescence green and dead cells fluorescence red [18]. LDPE films removed at regular time intervals were stained with FungaLight, incubated for 10–15 min in the dark and then the images were captured under a fluorescence microscope (Leica DM5000, Germany) with a blue filter at an excitation of 475 nm.
Measurement of Biological Properties
The culture broth was harvested after fermentation and the cells were separated by centrifuging at 10,000 rpm for 15 min. The supernatant was used to analyze the total extracellular protein concentration using Bradford assay [19] with Bovine serum albumin as the standard. Oxidase activity was determined as reported method by Seong et al. [20]. The total carbohydrates were estimated as suggested by Dubois et al. [21] with glucose as the standard. Fungal biomass is a direct measure of its growth in the medium; and its amount was estimated as per a reported method by Trishul and Doble [22].
Polymer Characterization
In order to shed further light on the nature of bio-degraded product, the product associated with fungal cells was characterized by gravimetric weight loss, tensile strength (TS), percentage elongation at break (EAB), contact angle (CA), scanning electron microscopy (SEM) FT-IR and XRD. The bio-degradation of LDPE films were evaluated by their weight loss. The tensile properties of the samples were measured as per the test method: ASTMD 882, speed: 50 mm/min, gauge length: 12 cm, type of test specimen: rectangle (8 cm length–2 cm width) on a universal testing machine (Instron 1195). The strips were subjected to TS tests; where the TS was calculated by dividing the maximum load by the original cross sectional area and EAB was calculated by dividing the elongation at the moment of rupture of the specimen by the initial gauge length of the specimen and multiplying by 100 according to Yabannavar and Bartha [23].
The CA of the samples was analyzed using a Camtel Goniometer (Royston, FT200). The surface morphologies of bio-degraded samples were examined by SEM (Hitachi S3400-N) operated at 200 kV accelerating voltage. FT-IR (Nicolet Magna 460) spectrophotometer was used to study the spectrum character of bio-degraded samples associated with or without pre-treatment and fungal exposure. Measurement range was 4000–500 cm−1, with a 4 cm−1 resolution, 0.475 cm−1/s scan speed and 32 scans. The technique used was attenuated total reflectance (ATR) with an Avantar multibounce HATR accessory with ZnSe crystal at 45°. Also XRD of the bio-degraded samples were performed to understand the progress of bio-degradation using X-ray diffractometer (Phillips, PW-1710) and X-ray generator (Philips, PW-1729) using Cu Kα (λ = 1.542 Å). The incident angle (2θ) was taken between 10° and 60°.
Optimization of Bio-degradation in Batch Ferrmentor
Statistical Experimental Design
LDPE degradation was carried out in a 3.7 L fermentor (Model: KLF-2000; Make: Bio Engineering, Wald, Switzerland) with a working volume of 2 L. Each degradation experiment contained a mixture of pre-treated LDPE, inoculum, SB and T-80. The range and the levels of the four critical process variables, namely, pH, temperature, incubation time and agitation are given in Table 1. A central composite design (CCD) was employed for four factors and the experimental design (Table S1) was obtained by using Design Expert version 7.0. The process parameters were varied on the basis of the experimental design and controlled automatically. Sampling was done every 30 days for bio-degradation analysis.
Table 1.
Experimental range and levels of independent variables
| Variables | Component | Level | ||||
|---|---|---|---|---|---|---|
| −2 | −1 | 0 | +1 | +2 | ||
| A | Temperature (°C) | 37 | 45 | 50 | 55 | 60 |
| B | pH | 4 | 5 | 6 | 7 | 8 |
| C | Incubation time (days) | 0 | 30 | 60 | 90 | 120 |
| D | Agitation (rpm) | 0 | 50 | 100 | 150 | 200 |
Artificial Neural Network (ANN) Modelling
Artificial neural network is a good inspiration of human brain and nerve systems that are known for their extreme ability to learn and classify data [24]. For this study, a feed forward error back propagation network has been chosen as it helps in training data in more accurate way. A feed forward network with back propagation contains three layers those are input layers, hidden layers and output layers which altogether makes the model of network. Input layers and output layers depends on the input data and reason of the network, like if the network is used as a regressor it will have one output or if it is used for classification it will have all classes as its output. Hidden layers were chosen according to rules of thumb. According to one of its rule, hidden layers will be less than twice of its input layers. Feed forward network is trained using average square error as performance index. During training, ANN calculates the input weights, layer weights and biases to be used for data set that is to be calculated by mean square error (MSE).
| 1 |
Feed forward error back propagation network will have to train for some n number of times to reach the approximate precision level. Functioning of neural network proceed in two stages, viz., learning or training and testing or inferences. Operation performed by the network during training is to sum up the weights and inputs for a neuron with its bias by some activation function. Input layer receives information from the external sources and passes this information to the network for processing. Hidden layer accept information from the input layer, does all the information processing, output layer receives processed information from the network, and sends the results out to an external receptor. The input signals are modified by inter connection weight known as weight factor (Wij), which represents the interconnection of ith node of the first layer to jth node of the second layer. The sum of modified signals (total activation) is then modified by a sigmoid transfer function. Similarly, outputs signal of hidden layer are modified by interconnection weight (Wij) of kth node of output layer to jth node of hidden layer. All data are normalized in the 0.1–0.9 range to avoid the scaling effect of parameter values. Therefore, all of the data (Xi) are converted to normalized values. Where Xi is ith input or output variable X. More details regarding construction of ANN can be found in the quoted Refs [25–28].
| 2 |
Wij weights from input layer i to hidden layer j, Xi input, bj bias to hidden layer, Zj activation function.
The choice of optimal neural network architecture and topology is vital for successful application of ANN [11]. Hence, various network topologies were investigated for their predictability. The ANN model developed was used as fitness function in PSO algorithm to identify the optimal set of input conditions that can yield maximum LDPE degradation.
Particle Swarm Optimization
This technique uses population of particles that evolves during its search for best fit condition. PSO process randomly initialises the system and the particles in the system move through multi-dimensional search space with a certain velocity associated with each particle. Every particle moves through the entire search space and keeps track of its best value. This best value associated with each particle is known as their personal best. Apart from its own personal best value it also has information about the overall best value or the global best value of the objective function or fitness function. Each particle updates its velocity and position according to its most successful neighbouring particles to follow their successful positions or the particle will move back to its previous best value position if it is found to be the global best value position. Following Eqs. (3, 4) are equated for updating velocity and position respectively.
| 3 |
| 4 |
k current iteration, C1, C2 learning factors, R1, R2 uniformly distributed random variables range from [0–1]. Lk−1i local best solution of particle, Gk−1i global best solution of system, Wk−1 inertia weights, Vk−1i velocity of particle i at k−1 iteration, Pk−1i position of particle i at k−1 iteration. At last each particle will be accumulated at an optimum value for the system.
Results and Discussion
Biological Properties
The initial abiotic step involves the oxidation of the polymer chain which leads to the formation of carbonyl groups. Carbonyl group decreases during microbial integration. Microorganisms secrete catalytic agents (i.e. enzymes and free radicals) able to cleave polymeric molecules reducing progressively their molecular weight. This process generates oligomers, dimers and monomers. Microorganisms act by mechanical, chemical and/or enzymatic means [29]. Growth of C. lunata SG1 secretes oxidase enzyme which leads to removal of two carbon fragments, acetyl CoA. The highest production (478 U/mL) of oxidase was seen with TP-UV treated followed by UV-treated (345 U/mL) and TP LDPE (197 U/mL) with C. lunata SG1 for 90 days. These extracellular enzymes convert polymer into oligomers, dimmer and monomer, which can enter the cell and then be utilized as the energy source.
The amount of total carbohydrates in terms of reducing sugars (such as glucose) increased throughout the study period. The highest carbohydrate was observed in TP-UV treated LDPE with C. lunata SG1. It is higher in thermally treated samples when compared to untreated samples. The carbohydrate in the medium in the presence of the polymer was higher than in its absence (−ve control) indicating that the polymer was used by the organisms for their growth. Starch provides higher oxygen permeability as it is consumed by microorganism. Highest permeability helps in the release of degradation products from the samples, thus making the matrix hollow, increasing the surface to volume ratio [30].
Fungal biomass is a direct measure of the growth of the strains in the medium and it will contain live and dead cells. There is an increase in the biomass as a function of time (direct measurement of fungal biomass is hampered because fungi penetrate into and bind themselves tightly to the solid-substrate particles. Biomass measured based on the content of certain cell components like chitin). The formation of a biofilm on the polymer surface is the first crucial step prior to the onset of bio-degradation. Since the bio-degradation proceeds at a slow rate, the biofilm on the surface should remain active having live microorganisms for a long period of time. After 90 days of treatment with C. lunata SG1, both live and dead organisms were observed on the LDPE surface. More live organisms were observed on TP-UV treated LDPE surface when compared to that on the TP or untreated LDPE surface. Similar results showing highest biomass was observed in supernatant of TP-UV treated LDPE with C. lunata SG1 after 90 days (60 mg/L). Biomass produced by the fungal strain was significantly influenced by the pre-treatment strategy. Pre-treatments of the polymer lead to its oxidation and subsequent breakdown assisting in the easy assimilation by the fungus. Hence UV or thermal treatment can be effectively used as a strategy before subjecting the polymer to bio-degradation [22, 31]. Hence the biomass content is high in the pre-treated samples than in the un-pretreated samples.
Bio-degraded Sample Characterization
Bio-deterioration Analyses
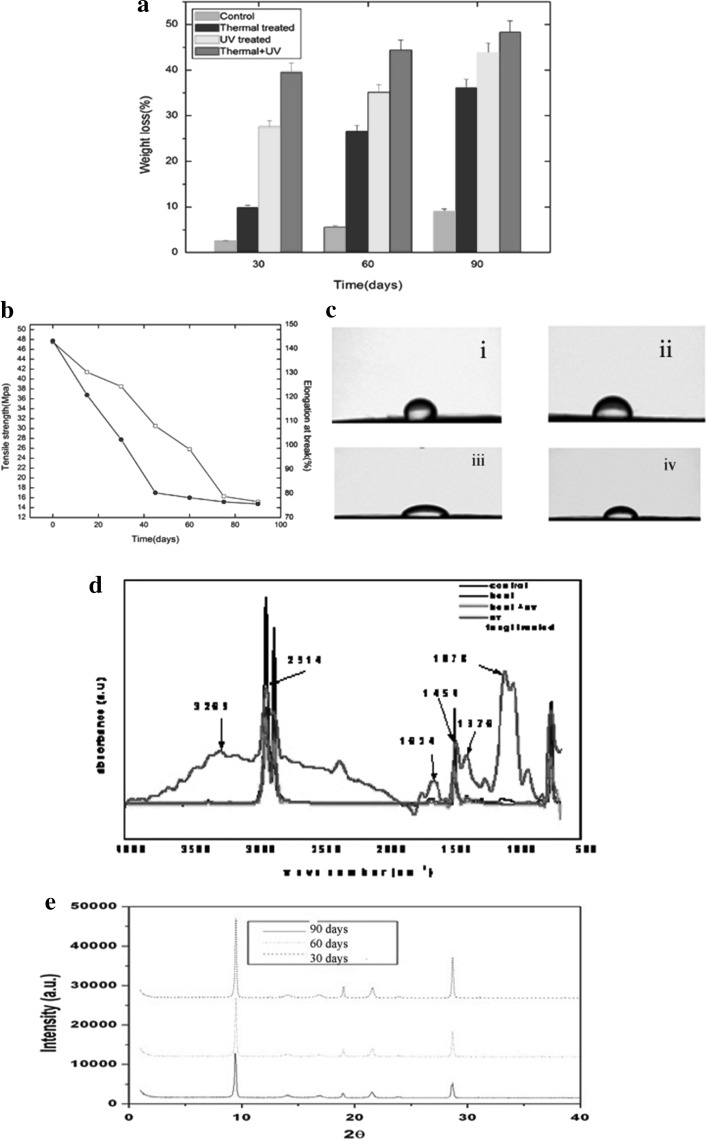
To quantify LDPE degradation by C. lunata SG1, weight loss of the pre-treated LDPE films were measured at different time intervals. The results (Fig. 1a) showed a time dependent and effect of pre-treatment on weight loss of LDPE films. A decrease in weight loss was observed in the pre-treated LDPE films after they were exposed to C. lunata SG1 in SB, T-80 medium at 100 rpm and 37 °C and this decrease over a 90 days period was highest (48.40 %) with combined TP-UV pre-treated samples, while less (43.76 %) and least (36.20 %) with individual photo and thermal pre-treatment respectively (Fig. 1a). However, no weight loss was evident in the control experiment where in incubation was carried out at the same temperature for a similar length of time but without C. lunata SG1 (data not shown). 6.2 % weight loss was reported for UV irradiation of 60 h. as compared to untreated LDPE with Bravebacillus brostelensis for 30 days [32]. UV irradiation for 500 h. increased the biodegradation of polyethylene by 25 % using Penicillium simlicissimum YK [33]. A 17 % weight loss was observed with TP LDPE exposed to Bacillus sphericus, GC subgroup IV (Alt) in 6 months, while the weight loss was only 10 % with untreated LDPE [34]. The weight loss of the LDPE films can be attributed to the breakdown of carbon backbone due to enzymatic degradation by the fungus. Taken together, the above results indicate that C. lunata SG1 is capable of degrading LDPE. The TS of the pre-treated LDPE films decreased from 47.5 ± 0.0003 to 15.2 ± 0.00011 Mpa in 90 days (Fig. 1b). This may be due to a combination of bio-degradation and bio-deterioration. Bio-degradation brings about chemical changes in the substrate due to action of micro and macro fouling whereas bio-deterioration is more of a physical change in terms of changes in the integrity of the substrate surface. Similar reports were also seen that after 12 months exposure of polypropylene to mixed soil culture, the TS of the films decreased from 23 to 17.3 Mpa in UV treated and from 32 to 18.8 Mpa in thermal treated during the study period [35]. Thermal and photo-treatment was found to be working in synergy with biotic treatment resulting in higher % weight loss and higher decrease in TS during the study period. Figure 2b shows the changes in % crystallinity of the pre-treated LDPE samples as a function of time. During the 90 days period, EAB decreased from 143 ± 3 to 75 ± 3 %. CA is an indication of the hydrophobicity or wettability of the surface, higher the CA higher is the hydrophobicity. The increasing oxidation of pre-treated LDPE was also shown by the change in the wettability of the surface of the LDPE samples. The initial CA of the untreated LDPE film was 85.4 ± 3.5° (Fig. 1ci), which decreased to 34.8 ± 3.5° as a result of combined thermal and UV-pre-treatment and incubation with C. lunata SG1 for a period of 90 days. Analysis of the thermal and UV-pre-treated LDPE films exposed to C. lunata SG1 for 30, 60 and 90 days showed that the wetability and the associated hydrophilicity of the polymer surface increased further, with the CA decreasing to 65.7 ± 2.5°, 45.3 ± 1.5° and 34.8 ± 3.5° respectively (Fig. 1cii–iv). Previous studies account the ability of the fungal strains (Fusarium sp. AF4, Aspergillusterreus AF5 and Penicillum sp. AF6) to form biofilm on polyethylene was attributed to the gradual decline in hydrophobicity of its surface [36]. There was, however, no such decrease in the CA for the control set of sample. In the earlier studies, biodegradation of starch and metal ions blended pre-treated polypropylene by P. chrysosporium NCIM 1170 and E. album MTP09 for 1 year showed a decrease in CA of the polymer by 10° [37].
Fig. 1.
a Percentage weight loss of the pre-treated and untreated LDPE incubated with C. lunata SG1. b Changes in tensile strength and EAB of LDPE c contact angle: (i) 85.4° (ii) 65.7° (iii) 45.3° (iv) 34.8° d FTIR of pre-treated LDPE films exposed to Curvularia lunata SGI for 90 days e XRD of pre-treated LDPE films exposed to Curvularia lunata SGI for (i) 30 day, (ii) 60 day (iii) 90 day
Fig. 2.
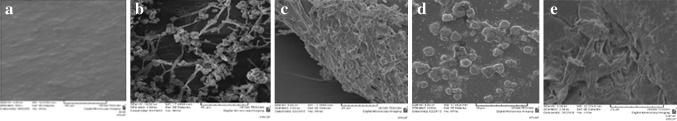
SEM micrographs of LDPE films before and after incubation with Curvularia lunata SG1 for 90 days. a Control (no pre-treatment, no incubation), b after pre-treatment and incubation with C. lunata (expansion of hyphae on the LDPE matrix), c after pre-treatment and incubation with C. lunata (biofilm formation on LDPE matrix), d erosion on the LDPE surface, e disintegratiom of LDPE film
Spectral Analyses
A comparison of FT-IR spectra of pre-treated LDPE incubated with C. lunata SG1 may provide further information about the nature of possible interaction of functional groups of LDPE and biomass. The main band of 2914 cm−1 was indicative of the C–H stretch and 1454 cm−1 of CH2 asymmetric bending and CH2 rocking at 718 cm−1 (Fig. 1d). From the IR spectroscopy it can be stated that the fungal contamination leads to a substantial decrease in the C–H stretch band of the polyethylene at 2914 cm−1. The increase in carbonyl absorption band at 1624 cm−1 region was primarily due to the formation of carbonyl bond through oxidation of the polyethylene moieties during the thermal and photo treatment. Typical degradation of PE and formation of bands at 1620–1640 and 840–880 cm−1 was also reported by Onodera et al. [38], attributed to oxidation of polyethylene. Generally, polyethylene degradation is a combined photo- and bio-degradation process. First, either by abiotic oxidation (UV light exposure) or heat treatment, necessary abiotic precursors are obtained. Secondly, selected microorganisms degrade the low molar mass oxidation products to complete the biodegradation [2]. Several reports state that the fungal action may cause a decrease in the carbonyl absorption band [24]. Appearance of bands in between 1662 and 1550 cm−1 is attributed to NH bending of primary and secondary amide from the peptide bond coupled with COO anion [36]. As observed, the broad absorption bands appeared at ~3500 and 3269 cm−1 are respectively attributed to OH and NH stretching modes of polysaccharides and proteins. Similar stretching has also been reported by Dhal et al. [39]. A strong peak at 1078 cm−1 corresponds to stretching vibration of C=O bond of polysaccharides. The absorbance at 1376 cm−1 is due to the methyl group and in the present study its intensity decreases as a function of time, which indicates that the oxidation takes place at the primary position of the polymer chain which can further decompose to produce ketones and esters [40]. More insight into the nature of bio-degraded product was obtained from XRD analysis. XRD patterns of the untreated and pre-treated LDPE films incubated with C. Lunata SG1 in SB T-80 medium after 90 days (Fig. 1e) showed distinguished peaks at 9.8 and 29.2 of the angular position 2θ. A decrease in percentage crystallinity is observed in LDPE films after they are exposed to C. lunata SG1 and this decrease over a 90 days period is higher with thermally and UV pre-treated LDPE films (Fig. 1e). There were no significant differences in degree of crystalinity between corresponding films incubated in the absence of C. Lunata SG1. The obtained data coincide with the results indicating that crystalinity and the crystal sizes for UV-irradiated films decreased during the process with the selected microorganisms [41].
Microscopic Analyses
The surfaces of the untreated LDPE films are smooth without cracks and free from any defects (Fig. 2a). Expansion of hyphae (Fig. 2b) and formation of bio-films of C. lunata SG1 (Fig. 2c) was evident on the surface and was considered to be a result of the surface moistness. Water could spread smoothly on the surface of thermal and photo-oxidised LDPE films because the surface had been modified to be hydrophilic. Therefore, microorganisms could also expand their colonies over the surface and form a fungal bio-film. These findings are consistent with the results of Shah et al. [9]. Such colonization and adhesion by microorganisms are a fundamental pre-requisite for bio-degradation of the polymer. Pits were observed on the surface, suggesting that the fungi penetrate into the LDPE matrix during degradation. Figure 2d indicates penetration of fungal hyphae into the LDPE matrix and presence of surface deformation after bio-film removal. The samples possessed pitted and eroded surfaces (Fig. 2e). The surface of the polymer after biological attack was physically weak and readily disintegrated under mild pressure. Also the extent of colonization on TP-UV treated are more than on UV treated LDPE films. The colonization is more on UV treated than on TP LDPE. These results indicate that pre-treatment induces oxidation and, hence, the polymer becomes brittle, which eventually leads to cracks due to the action of fungi. Microorganisms that colonize the polymer surface can probably adhere by means of extracellular polymeric substances, which mainly constitutes of polysaccharides. This forms a sheath that is bonded to the polymer. This plays an important role in transporting the depolymerising enzymes to its surface [42]. These changes are probably due to surface degradation. Earlier studies have suggested that the fungal strain, especially Fusarium sp. AF4, was able to adhere to the surface of LDPE and can cause surface damage [43]. In a study by Bonhomme et al. [5], SEM evidence confirmed that microorganisms (fungi) build up on the surface of the polymer (polyethylene) and after removal of the microorganisms; the surface became physically pitted and eroded.
Prediction of LDPE Bio-degradation Using ANN–PSO Optimization
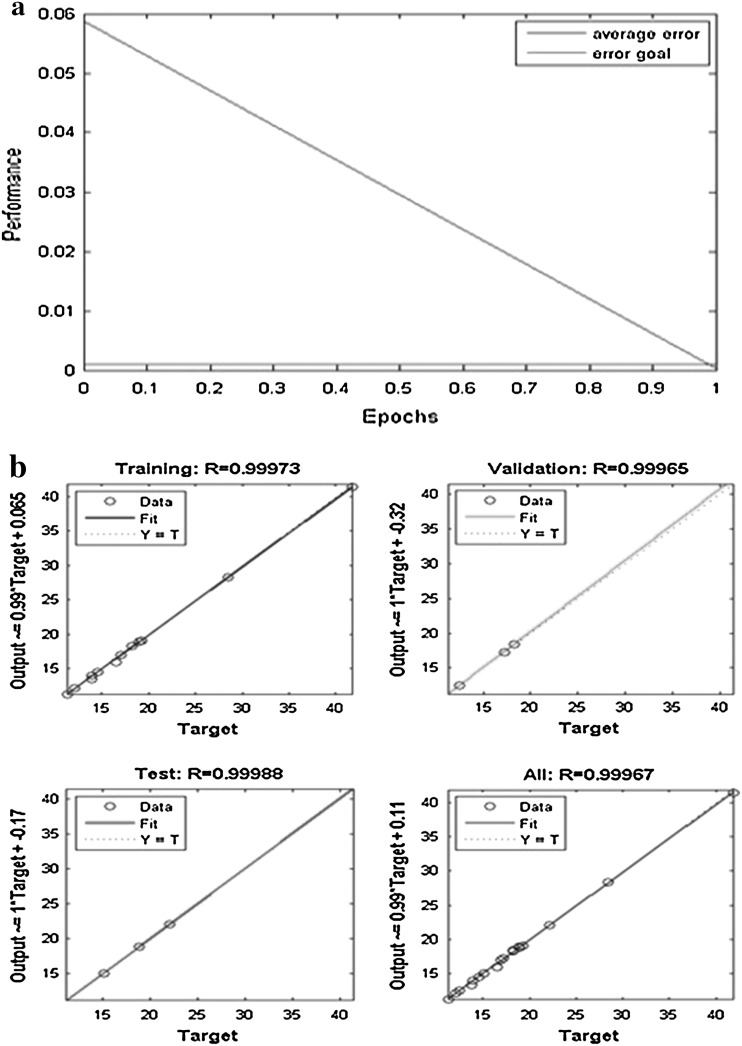
For ANN–PSO optimization our experimental data has to be first passed onto ANN training algorithm to create a model and then this model will be passed as a fitness function to PSO to calculate the optimum values for each parameter that has been trained. Thus, the experimental data that were obtained by performing CCD (Table S1) were used to develop an ANN model by splitting the 19 data into three sets (70 % for training, 15 % for testing and 15 % for validating). Purpose of training is to achieve a model with low value for mean square error and higher value for determinant coefficient (R2). While evaluating the best model it is possible that our model may show higher value for determinant coefficient and also has high MSE value, this case should not be considered. Evaluation should be done carefully by either increasing the number of hidden layers or by increasing the iterations for training. We have used 4-7-1 model (Fig.S1.a supplementary) following the rule of thumb as discussed. By training this model several numbers of times, at a point the value of MSE was observed to be equal to the error goal with (R2) values for the model to be 0.99967 (Fig.S1.b supplementary) and hence this model was considered for our study. Figure 3 a shows that at epoch value zero the MSE value was less than 0.06 which has come down linearly to achieve the error goal at epoch value 1 and remained constant thereafter. Figure 3 b depicts the (R2) values for each part of training and overall training with their corresponding values for (R2) with no over fitting of data.
Fig. 3.
a Performance profile of epochs and b regression of experimental data with ANN predicted data
As it is obvious that there is no part with value <0.999 for (R2), it can be concluded that there is no chance of over fitting of data. Just by looking into any one value at the regression plot will not give us good result. Training will update the initial weights, layer weights and biases value at each iteration which is the important factor for PSO. This model is fitness function for PSO optimization. We have used a PSO toolbox provided by MATLAB. For PSO there are two important factors: the representation of solution and the fitness function. Unlike GA it takes real numbers as input. Optimization is considered for population of 40 with maximum number of iteration of 500, no changes has been made to the toolbox parameters besides the two mentioned. Fitness function to be included is the model that has been tested to its best precision value. Equation 5 is the fitness function for PSO:
| 5 |
Y optimization output, LWj layer weights from hidden to output, IWij initial weights from input layer i to hidden layer j, Xj input to ANN model, Btj bias value for hidden layer j for tangent-sigmoid function (t), Bp bias used for purelin function.
This fitness function when passed into optimizer it displays a wait bar with the global best value on the top of it. By observing wait bar it can be seen that it takes less than half iterations to find a stable global best value. Global best value achieved by PSO toolbox was −521.1571.
Conclusions
This study has demonstrated that the cells of C. lunata SG1, successfully applied for bio-degradation of LDPE films. From these results it could be indicated that the oxidase produced from C. lunata SG1 may act as a promising tool in the treatment process of these LDPE films due to its bioactivity and thermo-stability. Moreover, this enzyme may be used for further investigations as immobilized form to ensure the bio-degradability of these LDPE films in environment. Temperature, pH, agitation and contact time closely affected the bio-degradation capacity of C. lunata SG1. Bio-degradation kinetic data were successfully described with ANN because of the lowest MSE and the highest determination of coefficient values between network prediction and corresponding experimental data. PSO gives a unique model to calculate bio-degradation percentage by C. lunata SG1 under studied conditions to describe both kinetic and equilibrium data. Results of this model showed that contact time was the most efficient parameter, followed by pH for the bio-degradation process. Bio-degradation of LDPE indicated a great potential to remove waste plastic as an eco-friendly process, which was well described by PSO and ANN. It has been previously reported that a glucoamylase from C. lunata is able to hydrolyze the terminal 1,2-linked rhamnosyl residues of carbon chains at C-3 position of steroidal saponins. Cytochrome P450 enzymes (CYPs) have a function with the conversion of hydrophobic intermediates of metabolisms and the detoxification of natural and environmental pollutants [44] With a broader knowledge of Curvularia genetics and enzymatic activities, sophisticated molecular breeding can produce strains and biotechnological processes, which could eliminate many types of contaminants in a cheap and environmentally friendly manner.
Electronic supplementary material
Acknowledgments
We extend our sincere gratitude to Prof. Basudam Adhikari, Head, Materials Science Center, IIT, Kharagpur, West Bengal and management of GIET, Gunupur for allowing us to use the available facilities to carry out this research work.
References
- 1.Ragnarsson L, Albertsson AC. Total luminescence intensity as a tool to classify degradable polyethylene films by early degradation detection and changes in activation energy. Biomacromolecules. 2003;4:900–907. doi: 10.1021/bm025752v. [DOI] [PubMed] [Google Scholar]
- 2.Scott G. Degradable polymers: principles and Application. Netherlands: Kluwer Academic Press; 2002. pp. 27–70. [Google Scholar]
- 3.Albertsson AC, Karlsson S. Degradable polymers for future. Acta Polym. 1995;46:114–123. doi: 10.1002/actp.1995.010460203. [DOI] [Google Scholar]
- 4.Albertsson AC, Andersson SO, Karlsson S. The mechanism of biodegradation of Polyethylene. Polym Degrad Stab. 1987;18:73. doi: 10.1016/0141-3910(87)90084-X. [DOI] [Google Scholar]
- 5.Bonhomme S, Cuer A, Delort AM, Lemaire J, Sancelme M, Scott G. Environmental biodegradation of polyethylene. Polym Degrad Stab. 2003;81:441–452. doi: 10.1016/S0141-3910(03)00129-0. [DOI] [Google Scholar]
- 6.Arnaud R, Davin P, Lemaire J, Malaika S, Al Chohan M, Coker M, Scott G, Fauve A, Maaroufi A. Photo oxidation and biodegradation of commercial photodegradable polyethylene. Polym Degrad Stab. 1994;46:211–221. doi: 10.1016/0141-3910(94)90053-1. [DOI] [Google Scholar]
- 7.Kim DY, Rhee YH. Biodegradation of microbial and synthetic polyesters by Fungi. Appl Microbiol Biotechnol. 2003;61:300–308. doi: 10.1007/s00253-002-1205-3. [DOI] [PubMed] [Google Scholar]
- 8.Seneviratne G, Tennakoon NS, Weerasekara MLMAW, Nanadasena KA. LDPE biodegradation by a developed Penicillium-Bacillus biofilm. Curr Sci. 2006;90:20–21. [Google Scholar]
- 9.Kershaw MJ, Talbot NJ. Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet Biol. 1998;23:18–33. doi: 10.1006/fgbi.1997.1022. [DOI] [PubMed] [Google Scholar]
- 10.Shah AA, Hasan F, Hameed A, Ahmed S. Biological degradation of plastics: a comprehensive review. Biotechnol Adv. 2008;26:246–265. doi: 10.1016/j.biotechadv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Ebrahimpour A, Rahman RNZRA, Ch’ng DHE, Basri M, Salleh AB. A modeling study by response surface methodology and artificial neural network on culture parameters optimization for thermostable lipase production from a newly isolated thermophilic Geobacillus sp. strain ARM. BMC Biotechnol. 2008;8:96. doi: 10.1186/1472-6750-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Sun J, Xu WB, Du GC, Chen J. Modelling and optimization of microbial hyaluronic acid production by Streptococcus zooepidemicus using radial basis function neural network coupling quantum-behaved particle swarm optimization algorithm. Biotechnol Prog. 2009;25:1819–1825. doi: 10.1002/btpr.278. [DOI] [PubMed] [Google Scholar]
- 13.Sivapathasekaran C, Mukherjee S, Ray A, Gupta A, Sen R. Artificial neural network modeling and genetic algorithm based medium optimization for the improved production of marine biosurfactant. Bioresour Technol. 2010;101:2884–2887. doi: 10.1016/j.biortech.2009.09.093. [DOI] [PubMed] [Google Scholar]
- 14.Sivapathasekaran C, Sen R. Performance evaluation of an ANN-GA aided experimental modeling and optimization procedure for enhanced synthesis of marine biosurfactant in a stirred tank reactor. J Chem Technol Biotechnol. 2013;88:794–799. doi: 10.1002/jctb.3900. [DOI] [Google Scholar]
- 15.Huang J, Mei LH, Xia J. Application of artificial neural network coupling particle swarm optimization algorithm to biocatalytic production of GABA. Biotechnol Bioeng. 2007;96:924–931. doi: 10.1002/bit.21162. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy J, Eberhart R (1995) Particle swarm optimization. In: Proceedings of IEEE International Conference on Neural Networks, 1942–1948
- 17.Cockshott AR, Hartman BE. Improving the fermentation medium for Echinocandin B production part II: particle swarm optimization. Process Biochem. 2001;36:661–669. doi: 10.1016/S0032-9592(00)00261-2. [DOI] [Google Scholar]
- 18.Boulos L, Pre´ vost M, Barbeau B, Coallier J, Desjardins R. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods. 1999;37:77–86. doi: 10.1016/S0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for quantification method of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Seong BL, Son HJ, Mheen TI, Han MH. Microbial transformation of rifamycin B: a new synthetic approach to rifamycin derivatives. J Antibiot. 1983;36:1402–1404. doi: 10.7164/antibiotics.36.1402. [DOI] [PubMed] [Google Scholar]
- 21.Dubois et al. (1956) Colorimetric method for determination of sugars and related substances. Division of Biochemistry, University of Minnesota, St. Paul, Minn. 28:350–356
- 22.Trishul A, Doble M. Biodegradation of physicochemically treated polycarbonate by fungi. Biomacromolecules. 2010;11:20–28. doi: 10.1021/bm9008099. [DOI] [PubMed] [Google Scholar]
- 23.Yabannavar AV, Bartha R. Methods for assessment of biodegradability of plastic films in soil. Appl Environ Microbiol. 1994;60:3608–3614. doi: 10.1128/aem.60.10.3608-3614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy PK, Titus S, Surekha P, Tulsi E, Deshmukh C, Rajagopal C. Degradation of abiotically aged LDPE films containing pro-oxidant by bacterial consortium. Polym Degrad Stab. 2008;93:1917–1922. doi: 10.1016/j.polymdegradstab.2008.07.016. [DOI] [Google Scholar]
- 25.lshihri MM, Azmy AM, El-Bisy MS. Neural networks for predicting compressive strength of structural light weight concrete. Constr Build Mater. 2009;23:2214–2219. doi: 10.1016/j.conbuildmat.2008.12.003. [DOI] [Google Scholar]
- 26.Trtnik G, Kavcˇicˇ F, Turk G. Prediction of concrete strength using ultrasonic pulse velocity and artificial neural networks. Ultrasonics. 2009;49:53–60. doi: 10.1016/j.ultras.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Topcu IB, Sarıdemir M. Prediction of mechanical properties of recycled aggregate concretes containing silica fume using artificial neural networks and fuzzy logic. Comput Mater Sci. 2008;42:74–82. doi: 10.1016/j.commatsci.2007.06.011. [DOI] [Google Scholar]
- 28.Parichatprecha R, Nimityongskul P. Analysis of durability of high performance concrete using artificial neural networks. Constr Build Mater. 2009;23:910–917. doi: 10.1016/j.conbuildmat.2008.04.015. [DOI] [Google Scholar]
- 29.Gu JD. Microbiological deterioration and degradation of synthetic polymeric materials: recent research advances. Int Biodeter Biodegr. 2003;52:69–91. doi: 10.1016/S0964-8305(02)00177-4. [DOI] [Google Scholar]
- 30.Rutkowska M, Heimowska A, Krasowska K, Janik H. Biodegradability of polyethylene starch blends in sea water. Pol J Environ Sci. 2002;11:267–274. [Google Scholar]
- 31.Sudhakar M, Artham T, Doble M, Sureshkumar K, Syed JS, Inbakandan D, Viduthalai RR, Umadevi VR, Sriyutha M, Venkatesan R. Biofouling and biodegradation of polyolefins in ocean waters. Polym Degrad Stab. 2007;92:1743–1752. doi: 10.1016/j.polymdegradstab.2007.03.029. [DOI] [Google Scholar]
- 32.Hadad D, Geresh S, Sivan A. Biodegradation of polyethylene by the thermophillic bacterium Brevibacillus borstelensis. J Appl Microbiol. 2005;98:1093–1100. doi: 10.1111/j.1365-2672.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 33.Onodera KY, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y. Degradation of polyethylene by a fungus Penicillium simplicissimum YK. Polym Degrad Stab. 2001;72:323–327. doi: 10.1016/S0141-3910(01)00027-1. [DOI] [Google Scholar]
- 34.Sudhakar M, Doble M, Murthy PS, Venkatesan R. Marine microbe-mediated biodegradation of low- and high-density polyethylenes. Int Biodeterior Biodegrad. 2008;61:203–213. doi: 10.1016/j.ibiod.2007.07.011. [DOI] [Google Scholar]
- 35.Arkatkar A, Arutchelvi J, Bhaduri S, Uppara PV, Doble M. Degradation of unpretreated and thermally pretreated polypropylene by soil consortia. Int Biodeterior Biodegradation. 2009;63:106–111. doi: 10.1016/j.ibiod.2008.06.005. [DOI] [Google Scholar]
- 36.Gilan I, Hadar Y, Sivan A. Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl Microbiol Biotechnol. 2004;65:97–104. doi: 10.1007/s00253-004-1584-8. [DOI] [PubMed] [Google Scholar]
- 37.Jeyakumar D, Chirsteen J, Doble M. Synergistic effects of pretreatment and blending on fungi mediated biodegradation of polypropylenes. Bioresour Technol. 2013;148:78–85. doi: 10.1016/j.biortech.2013.08.074. [DOI] [PubMed] [Google Scholar]
- 38.Yamada-Onodera K, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y. Degradation of polyethylene by a fungus Penicillium simplicissimum YK. Poly Degrad Stab. 2001;72:323–327. doi: 10.1016/S0141-3910(01)00027-1. [DOI] [Google Scholar]
- 39.Dhal B, Thatoi HN, Das NN, Pandey BD. Reduction of hexavalent chromium by Bacillus sp. isolated from chromite mine soils and characterization of reduced product. J Chem Technol Biotechnol. 2010;85:1471–1479. [Google Scholar]
- 40.Gijsman P, Heinekens J. The mechanism of the low-temperature oxidation of polypropylene. Polym Degrad Stab. 1993;42:95–105. doi: 10.1016/0141-3910(93)90031-D. [DOI] [Google Scholar]
- 41.El-Rehim HAA, Hegazy ESA, Ali AM, Rabie AM. Synergistic effect of combining UV-sunlight-soil burial treatment on the biodegradation rate of LDPE/starch blends. J Photochem Photobiol. 2004;163:547–556. doi: 10.1016/j.jphotochem.2004.02.003. [DOI] [Google Scholar]
- 42.Sepulveda TV, Casta ~ neda GS, Rojas MG, Manzur A, Torres EF. Thermally treated low density polyethylene biodegradation by Penicillium pinophilum and Aspergillus niger. J Appl Polym Sci. 2002;83:305–314. doi: 10.1002/app.2245. [DOI] [Google Scholar]
- 43.Shah A A (2007) Role of microorganisms in biodegradation of plastics, Ph. D. thesis. Quaid-i- Azam University, Islamabad, Pakistan
- 44.Crešnar B, Petrič S. Cytochrome P450 enzymes in the fungal kingdom. Biochimica et Biophysica Acta (BBA) Proteins and Proteomics. 2011;1814:29–35. doi: 10.1016/j.bbapap.2010.06.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.