Abstract
Muscle stem cells (MuSCs, satellite cells) are the major contributor to muscle regeneration. Like most adult stem cells, long-term expansion of MuSCs in vitro is difficult. The in vivo muscle regeneration abilities of MuSCs are quickly lost after culturing in vitro, which prevents the potential applications of MuSCs in cell-based therapies. Here, we establish a system to serially expand MuSCs in vitro for over 20 passages by mimicking the endogenous microenvironment. We identified that the combination of four pro-inflammatory cytokines, IL-1α, IL-13, TNF-α, and IFN-γ, secreted by T cells was able to stimulate MuSC proliferation in vivo upon injury and promote serial expansion of MuSCs in vitro. The expanded MuSCs can replenish the endogenous stem cell pool and are capable of repairing multiple rounds of muscle injuries in vivo after a single transplantation. The establishment of the in vitro system provides us a powerful method to expand functional MuSCs to repair muscle injuries.
Keywords: muscle stem cells, T cells, muscle regeneration, inflammation, long-term expansion, stemness maintenance
Introduction
Muscle atrophy caused by disuse, malnutrition and diseases such as muscular dystrophies, diabetes1, cancer2 and HIV infection3, affects millions of patients. There is no effective cure for muscle atrophy caused by diseases thus far. MuSCs are the major force that drives postnatal muscle injury repair4. Upon injury, quiescent MuSCs are activated and undergo proliferation and differentiation to form new myofibers and regenerate functional skeletal muscles5,6. Due to their outstanding abilities to regenerate muscles7, MuSCs have great potentials in cell-based therapies to treat muscle atrophy. To retrieve MuSCs from small muscle biopsies followed by in vitro expansion has been considered to be a promising strategy to treat muscle atrophy. However, the development of the real therapy has been long hampered by inability to expand functional MuSCs in vitro. Similar to most of the adult stem cells, MuSCs can barely be efficiently expanded in vitro. Ex vivo cultured MuSCs differentiate to myoblast progenitor cells in a few days and quickly lost their abilities to regenerate muscles in vivo8. The conventional in vitro culture condition for MuSCs does not amplify their injury reparation abilities, and was considered to be “empty amplification”8. Although the cell number is increased by conventional culturing condition, these cells cannot be utilized to treat muscle atrophies due to the loss of muscle injury reparation abilities in vivo. Consistent with the results in mice, transplantation of myoblast progenitors failed to rescue muscle atrophy in clinical trials9,10. Thus, establishment of an in vitro system to efficiently expand functional MuSCs in vitro will break this bottleneck and facilitate the stem cell-based therapies.
The lack of critical niche components in culturing system is the major reason why most types of adult stem cells are difficult to be maintained and serially expanded in vitro. By recapitulating the in vivo microenvironment, the in vitro adult stem cell culture system could be improved. For example, by mimicking the rigidity of endogenous niche in dish, the proliferation ability of isolated MuSCs is increased11. Other than biophysical properties, soluble factors present in the microenvironment can also regulate the activation, proliferation and differentiation of MuSCs. It has been previously shown that Wnt7 stimulates the symmetric divisions of MuSCs12,13 and Notch maintains the quiescent stage of MuSCs and promotes myoblast proliferation at a later stage of muscle regeneration14,15,16. Treating MuSCs with forskolin has been reported to promote MuSC proliferation17. However, the conditions for long-term MuSC expansion in vitro have not been characterized. Identification of the critical microenvironment components at various stages of muscle regeneration would shed light on optimizing the in vitro MuSC culturing and expansion system.
Here we describe an in vitro culture system to maintain and serially expand functional MuSCs for numerous passages to obtain a large amount of MuSCs capable of efficient muscle injury reparation. The establishment of this cell propagation system sheds new light on development of MuSC-based therapies from small muscle biopsies to treat muscle atrophy.
Results
T cells facilitate muscle regeneration
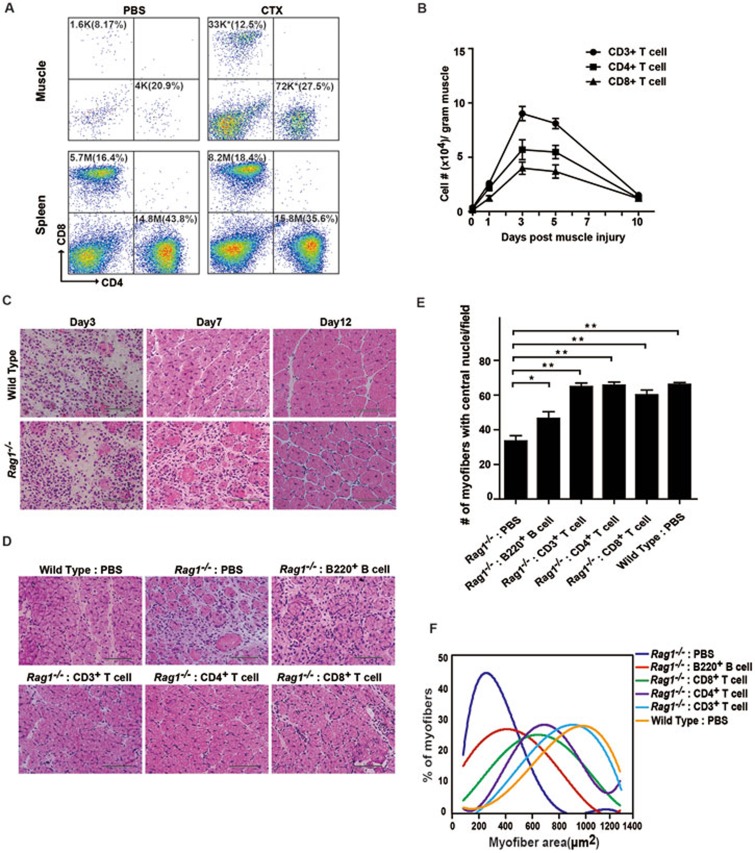
To identify the environment promoting MuSC proliferation, we characterize the events after muscle injury. Shortly after muscle injury, large scale lymphocyte infiltration was observed at the injury site. Flow cytometry (FACS) analysis was performed to analyze the components of the infiltrated lymphocytes. Muscle injury was induced by cardiotoxin (CTX) injection. A large amount of CD3+ T cells infiltrated the local injury site, and reached the peak at 3-5 days post injury (Figure 1A and 1B). Both CD4+ and CD8+ subtypes of T cells infiltrated the local injury site after the occurrence of muscle injury (Figure 1A and 1B). The change of T cell number was limited to the injury site as the T cell distribution in other lymphatic organs such as spleen remained unchanged (Figure 1A).
Figure 1.
T cells are required for muscle regeneration. (A) FACS analysis of CD4+ and CD8+ T lymphocytes in the TA muscle or the spleen on day 3 after CTX-induced muscle injury. The total number of CD4+ or CD8+ cells is indicated on top of each panel. The percentage of CD4+ or CD8+ cells is indicated in parenthesis. The change of CD4+ and CD8+ T lymphocyte cell number was statistically significant after CTX injection in muscle. * indicates statistically significant, P < 0.01. The percentage changes were not statistically significant. Statistical analysis is based on 3 independent experiments. K indicates 103 cells. M indicates 106 cells. (B) The total number of CD3+, CD4+ and CD8+ T cells infiltrating into the TA muscle on various days post injury. Error bars are based on 3 independent experiments. (C) Representative HE staining of muscle sections on days 3, 7 and 12 after CTX injection in wild-type or Rag1−/− mice. Scale bar, 200 μm. (D) Activated T or B lymphocytes were injected into the TA muscle of Rag1−/− mice right after CTX injection. Muscle regeneration was monitored by HE staining 7 days post injury. Scale bar, 200 μm. (E) Statistical analysis of number of myofibers containing centrally located nuclei per visual field. Error bars are based on 3 independent experiments. **P < 0.01P < 0.05. (F) Statistical analysis of the myofiber size 12 days post injury. The graph is based on the average myofiber area distribution from 3 independent experiments. See also Supplementary information, Figures S1 and S2.
We next further investigated the functions of infiltrated T cells in muscle regeneration using Rag1−/− mice that lack both T and B cells but have intact macrophages18. The uninjured Rag1−/− mice displayed the similar myofiber size to wild type (Supplementary information, Figure S1A). MuSCs are primarily responsible for the reparation of muscle injury. We then examined the status of MuSCs in uninjured Rag1−/− mice. CD34+ integrin-α7+ CD31− CD45− CD11b− Sca1− MuSCs were analyzed by FACS as previously described7. Similar amounts of MuSCs were present in both Rag1−/− and wild-type muscle (Supplementary information, Figure S1B and S1C). The niche occupation of MuSCs in both Rag1−/− and wild-type mice was also similar (Supplementary information, Figure S1D). The Rag1−/− MuSCs could differentiate normally in vitro as well. Together, these observations suggested that there were no obvious intrinsic defects in Rag1−/− MuSCs.
After muscle injury was induced by CTX injection in Rag1−/− mice, the muscle regeneration progress was monitored by hematoxylin and eosin (HE) staining at 3, 7 and 12 days post injury. Remarkably, Rag1−/− mice showed significantly delayed muscle regeneration compared to their wild-type littermates (Figure 1C). Normal macrophage infiltration post injury was observed in Rag1−/− mice at the local injury site (Supplementary information, Figure S1E). Together, these observations suggest that T and B lymphocytes are required for proper muscle regeneration in a macrophage-independent manner. To identify the major contributor to muscle regeneration, lymphocyte transplantations were performed. After muscle injury, either activated T or B cells were transplanted into Rag1−/− mice. The muscle regeneration was monitored by HE staining of muscle sections 7 days post injury. The transplantation of activated CD3+ T cells rescued the muscle regeneration defects of Rag1−/− mice to the level of wild-type mice as indicated by the muscle tissue organization, the number of myofibers with centrally located nuclei, and the myofiber size (Figure 1D-1F). Both CD4+ and CD8+ subtypes of T cells could rescue the muscle regeneration defects in Rag1−/− mice (Figure 1D-1F). In contrast, B cell transplantation only marginally rescued the muscle regeneration defects in Rag1−/− mice (Figure 1D-1F). These results suggest that T cells can facilitate muscle regeneration and are required for timely regeneration of muscles.
To further confirm the facilitation functions of T cells in muscle regeneration, muscle injury and reparation experiments were performed in FoxN1−/− mice that lacked T cells, but possessed intact B cells (Supplementary information, Figure S2A and S2B). B cells could infiltrate the injury site normally, whereas T cells were absent (Supplementary information, Figure S2C and S2D). Consistent with the observations in Rag1−/− mice, FoxN1−/− mice also displayed delayed muscle regeneration (Supplementary information, Figure S2E-S2G). Overall, these observations suggest that T cell infiltration to the local injury site is required for proper muscle regeneration independent of macrophages. Since no obvious intrinsic defects were observed in MuSCs isolated from Rag1−/− mice, T cells may be required to provide a suitable microenvironment for MuSCs.
Long-term expansion of MuSCs in T cell conditional medium
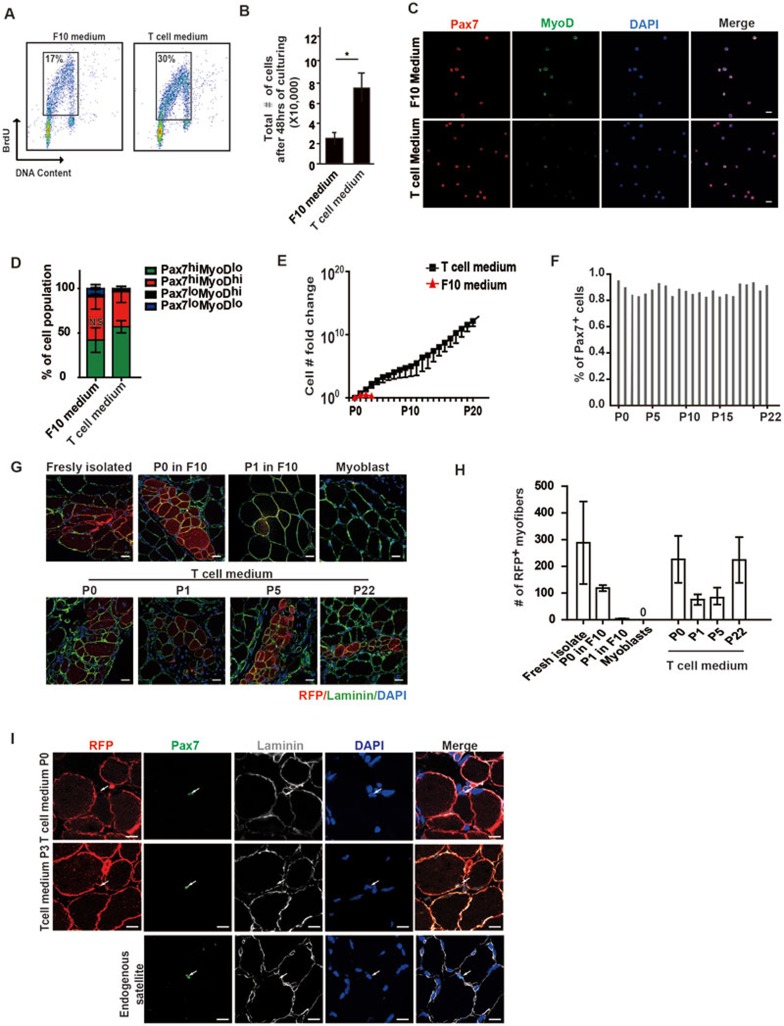
MuSCs were difficult to be expanded in vitro. We next explored whether improvement of MuSC expansion could be achieved by the presence of T cells, which mimics the in vivo microenvironment components. MuSCs were isolated and co-cultured with activated T cells. The proliferation abilities of MuSCs improved dramatically by either co-culturing with activated T cells or in T cell conditional medium. Indeed, T cell conditional medium displayed superior ability to promote MuSC growth to co-culturing with activated T cells, suggesting that direct cell-cell interactions between T cells and MuSCs were not required. In T cell conditional medium, MuSCs displayed improved proliferative abilities as indicted by higher percentage of BrdU+ cells compared to cells grown in conventional F10 medium (Figure 2A and Supplementary information, Figure S3A). Consistent with this, more MuSCs were obtained after 48 h of in vitro culturing in conditional medium when the same amounts of cells were seeded (Figure 2B and Supplementary information, Figure S3B). MuSCs cultured in T cell conditional medium expressed Pax7, the MuSC marker, at a comparable level to that in MuSCs cultured in F10 medium, while MyoD expression level was lower than that in MuSCs cultured in F10 medium (Figure 2C and Supplementary information, Figure S3C). Higher percentage of Pax7highMyoDlow MuSCs was detected in cells cultured in T cell conditional medium (Figure 2D). Altogether, these observations indicate that cells cultured in T cell conditional medium are likely to be the Pax7highMyoDlow activated MuSCs.
Figure 2.
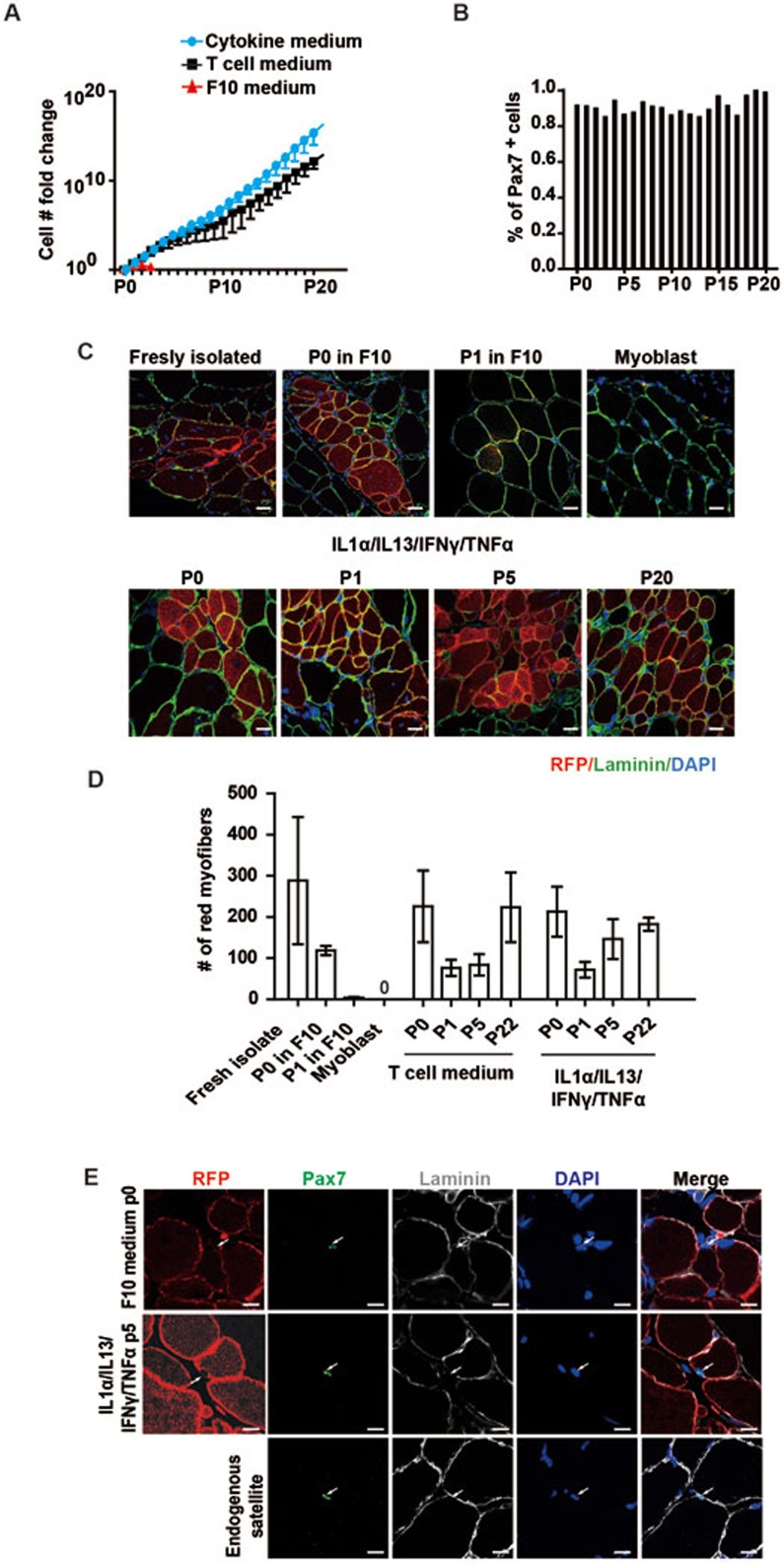
T cell conditional medium promotes the expansion of MuSCs. (A) BrdU pulse labeling of MuSCs grown in F10 medium or T cell medium. Logarithmically proliferating MuSCs in F10 or T cell medium were pulse labeled by BrdU for 2 h. The labeled MuSCs were analyzed by FACS. (B) More MuSCs were obtained after culturing in T cell medium for 48 h compared to culturing in F10 medium. * indicates statistically significant, P < 0.01. Error bars are based on 3 independent experiments. (C) Immunofluorescent staining of Pax7 and MyoD in MuSCs cultured in either F10 or T cell conditional medium. Red indicates Pax7 staining. Green indicates MyoD staining. Blue indicates DAPI staining of nuclei. “Merge” indicates merged images of Pax7, MyoD and DAPI staining. Scale bar, 20 μm. (D) Statistical analysis of the percentages of Pax7highMyoDlow (more quiescent MuSCs), Pax7highMyoDhigh (active MuSCs), Pax7lowMyoDhigh (myoblasts) and Pax7lowMyoDlow (more differentiated muscle cells) subpopulations in MuSCs cultured in F10 or T cell conditional medium. Error bars are based on 4 independent experiments. (E) MuSC number increases after 20 serial expansions in T cell conditional medium. F10 medium could not support long-term serial expansion of MuSCs as indicated by the red line. (F) Statistical analysis of the percentage of Pax7+ cells in MuSCs serially expanded in T cell medium. The statistics are based on the quantification of 3 independent Pax7 immunofluorescent staining at each passage. The average percentage at each passage is shown. (G) Representative images of engraftment of MuSCs expanded in T cell medium. MuSCs isolated from RFP transgenic mice were expanded in T cell conditional medium. They were transplanted into injured wild-type non-fluorescent recipient TA muscle. Engraftments of RFP+ MuSCs were analyzed by confocal microscopy. Red indicates engrafted RFP+ cells. Green indicates immunofluorescent staining of Laminin. Blue indicates DAPI staining of nuclei. Only merged images of RFP, laminin and DAPI staining are shown. The source of MuSCs used for transplantation is indicated on top of each panel. Scale bar, 20 μm. (H) Statistical analysis of engraftment efficiency indicated by the number of RFP-expressing myofibers generated from freshly isolated MuSCs and MuSCs expanded in F10 medium or T cell conditional medium. Error bars are based on 4 independent experiments. (I) Representative images of Pax7 immunofluorescent staining 30 days after CTX injection when the injury was completely repaired. Red indicates RFP. The RFP is membrane localized. Green indicates Pax7 immunofluorescent staining. Grey indicates laminin immunofluorescent staining. Blue indicates DAPI staining of nuclei. Merge indicates the merged images of RFP, Pax7, laminin and DAPI stainings. Arrow indicates the position of Pax7+ MuSCs located underneath laminin. Scale bar, 20 μm. See also Supplementary information, Figure S3.
Like most adult stem cells, the long-term serial expansion of MuSCs in vitro was challenging. We next examined whether T cell conditional medium could support long-term MuSC expansion ex vivo. Strikingly, T cell conditional medium was able to support serial expansion of MuSCs for at least 22 passages (Supplementary information, Figure S3D). By serial expansion, the initial isolated MuSCs were amplified 1 × 1015-fold in vitro (Figure 2E). At each passage, these expanded MuSCs showed normal cell morphologies (Supplementary information, Figure S3D). After long-term in vitro expansion, cells from each passage maintained > 80% of Pax7+ populations (Figure 2F and Supplementary information, Figure S3E), suggesting that the stemness of the cells was maintained after serial expansions. Cells of all passages were able to differentiate in vitro with close to 100% differentiation efficiency (Supplementary information, Figure S3F and S3G), suggesting that the differentiation potentials of MuSCs remained intact after serial expansions.
Previous reports have raised the concern that the regenerative potentials of MuSCs declined dramatically after being cultured in vitro7,8. Once the MuSCs differentiated to myoblast progenitors, the engraftment efficiency was extremely low7,8. To evaluate the in vivo regenerative potentials of MuSCs cultured in T cell conditional medium, cell transplantation experiments were performed. MuSCs were isolated from mice expressing RFP and then cultured and expanded in T cell conditional medium. The same amount of freshly isolated or in vitro expanded RFP-expressing MuSCs was transplanted into irradiated anterior tibialis (TA) muscles of non-RFP-expressing recipients. Four weeks after transplantation, the engraftment of RFP cells was examined in muscle sections. Engraftment could be observed in all transplantations with distinct passages of MuSCs cultured and expanded in T cell conditional medium (Figure 2G and 2H), suggesting that these MuSCs maintained their muscle regeneration abilities. Furthermore, single-nucleus RFP+ cells could be detected at the niche of endogenous MuSCs after completion of muscle regeneration (Figure 2I), suggesting that the transplanted MuSCs are capable of proper homing. All the observations suggest that functional MuSCs could be greatly propagated while culturing in T cell conditional medium.
Four cytokines are the minimum effective components in T cell conditional medium
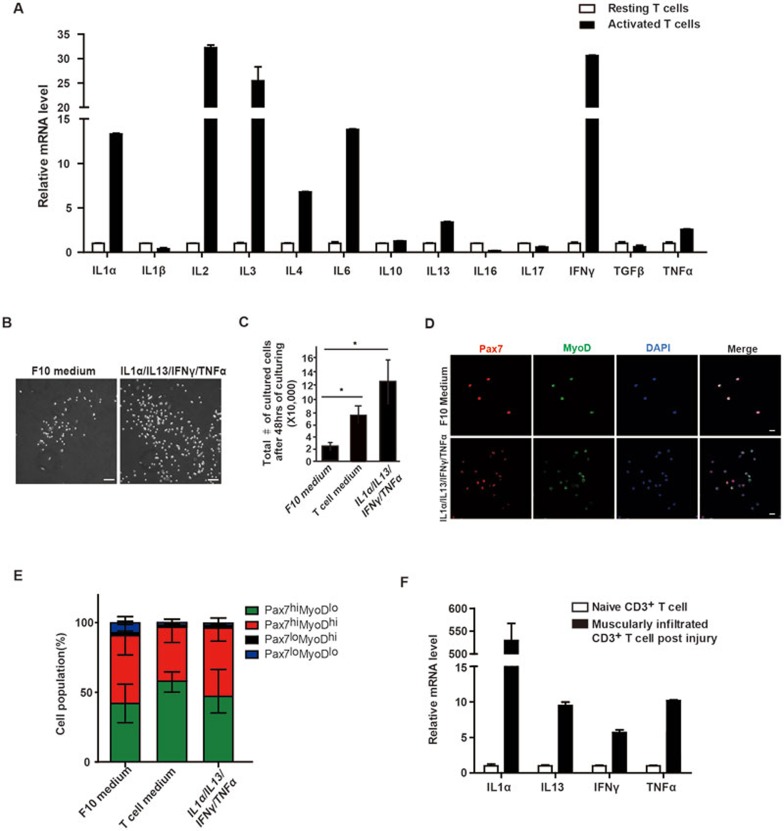
To identify the effective components present in T cell conditional medium, we first examined mRNA levels of the cytokines suggested by previous reports to be secreted by T cells, including IL-1α19, IL-1β19, IL-219,20,21,22, IL-319,23, IL-419,20, IL-619,24, IL-1019,25,26, IL-1327, IL-1628,29,30, IL-1731,32, IFN-γ19,20,21,22, TGF-β19,33 and TNF-α19,21,22. Among them, mRNA levels of IL-1α, IL-2, IL-3, IL-4, IL-6, IL-13, IFN-γ and TNF-α were upregulated in activated T cells compared to resting T cells (Figure 3A). To further detect the cytokine proteins, cytokine array was performed with T cell conditional medium. IL-1α, IL-2, IL-3, IL-10, IL-13, IL-16, IL-17, IFN-γ and TNF-α proteins were present in T cell conditional medium, while IL-4 and IL-6 proteins could not be detected in T cell medium even though their mRNAs were detectable in activated T cells (Supplementary information, Figure S4A). We next compared the mRNA and protein profiles of cytokines in T cell medium, and selected those showing upregulation at mRNA level in activated T cells and detectable protein level in T cell medium, namely IL-1α, IL-2, IL-3, IL-10, IL-13, IFN-γ and TNF-α as the initial pool to start screening (Supplementary information, Figure S4B).
Figure 3.
Four cytokines are the minimum effective components in T cell conditional medium. (A) mRNA levels of cytokines in resting and activated T cells. (B) Phase-contrast pictures of MuSCs cultured in F10 medium or medium supplemented with cytokine cocktail. Scale bar, 200 μm. (C) Total number of MuSCs in medium with or without cytokine cocktail supplementation after 48 h of culturing. Error bars are based on 3 independent experiments. * indicates statistically significant, P < 0.01. (D) Immunofluorescent staining of MuSCs cultured in F10 or cytokine medium. Red indicates Pax7 immunofluorescent staining. Green indicates MyoD immunofluorescent staining. Blue indicates DAPI staining of nuclei. Merge indicates merged images of Pax7, MyoD and DAPI staining. Scale bar, 20 μm. (E) Statistical analysis of the percentages of Pax7highMyoDlow (more quiescent MuSCs), Pax7highMyoDhigh (active MuSCs), Pax7lowMyoDhigh (myoblasts) and Pax7lowMyoDlow (more differentiated muscle cells) subpopulations in MuSCs cultured in F10, T cell conditional medium or cytokine medium. Error bars are based on 4 independent experiments. (F) The mRNA levels of the four cytokines increased in T cells infiltrating into injured TA muscles. See also Supplementary information, Figure S4 and Table S1.
We pooled the above 7 enriched cytokines together to treat MuSCs and assessed their abilities to promote proliferation. The dispensable cytokines were removed from the recipe until the minimum cytokine combination was achieved. After multiple rounds of screening, a cocktail of four cytokines (IL-1α, IL-13, IFN-γ and TNF-α) was shown to be necessary and sufficient to promote MuSC proliferation and maintain their undifferentiated status (Figure 3B-3E). The cocktail of the above four cytokines promoted the proliferation of MuSCs to a slightly higher extent compared to the T cell conditional medium. More MuSCs were obtained after 48 h of in vitro culturing compared to those cultured in T cell medium (Figure 3C). Consistent with this, higher percentage of MuSCs were labeled by BrdU in cytokine medium (Supplementary information, Figure S4C and S4D). When any one of the four cytokines was removed from the culture system, the cells were either less proliferative, unable to maintain their undifferentiated status or incapable of expansion (Supplementary information, Table S1). These data suggest that IL-1α, IL-13, IFN-γ and TNF-α were all required to promote MuSC proliferation and expansion, and maintain their undifferentiated status. Consistent with the above results, IL-1α, IL-13, IFN-γ and TNF-α mRNA levels were dramatically higher in infiltrated T cells isolated from the injured muscle compared to those in resting T cells isolated from uninjured muscles (Figure 3F), suggesting that infiltrated T cells produced the four cytokines locally at the injury site. Consistent with the above notion, the protein levels of the four cytokines during muscle regeneration were lower in Rag1−/− mice compared to those in their wild-type littermates (Supplementary information, Figure S4G), further indicating that a critical portion of the four cytokines were produced by T cells.
Among the four identified factors, IFN-γ promotes muscle regeneration34. TNF-α receptor knockout mice (p55−/−p75−/−) exhibited muscle regeneration deficiency35. In addition, TNF-α has been shown to be highly expressed in injured myofibers, inflammatory cells, endothelial cells and fibroblasts during muscle regeneration36. It has been suggested that TNF-α facilitated the quiescent satellite cells to enter cell cycle and accelerated their G1-to-S transition in vivo37. IL-1 receptor knockout mice had smaller body size and less muscle mass in adult, suggesting that there is a postnatal muscle growth defect38. IL-13 and IL-4 share the common receptor IL-4 receptor. IL-4 receptor knockout mice displayed muscle regeneration defects39. All the above genetic evidence supports the notion that the cocktail of four cytokines could facilitate muscle regeneration in vivo. However, none of the four factors have been pinpointed to facilitate the expansion of MuSCs. Furthermore, addition of each of the four cytokines alone only showed marginal effects on the proliferation of MuSCs (Supplementary information, Figure S4E and S4F); and the MuSCs treated with each individual cytokine could not maintain their undifferentiated status well, represented by the presence of significantly higher number of morphologically more differentiated cells in cultures (Supplementary information, Figure S4E, as indicated by arrows). Removal of any one of the four cytokines affected MuSC expansion (Supplementary information, Table S1). Together, these results suggest that the four cytokines constitute the minimum combination required to promote MuSC proliferation.
The cytokine cocktail could facilitate MuSC expansion in vivo
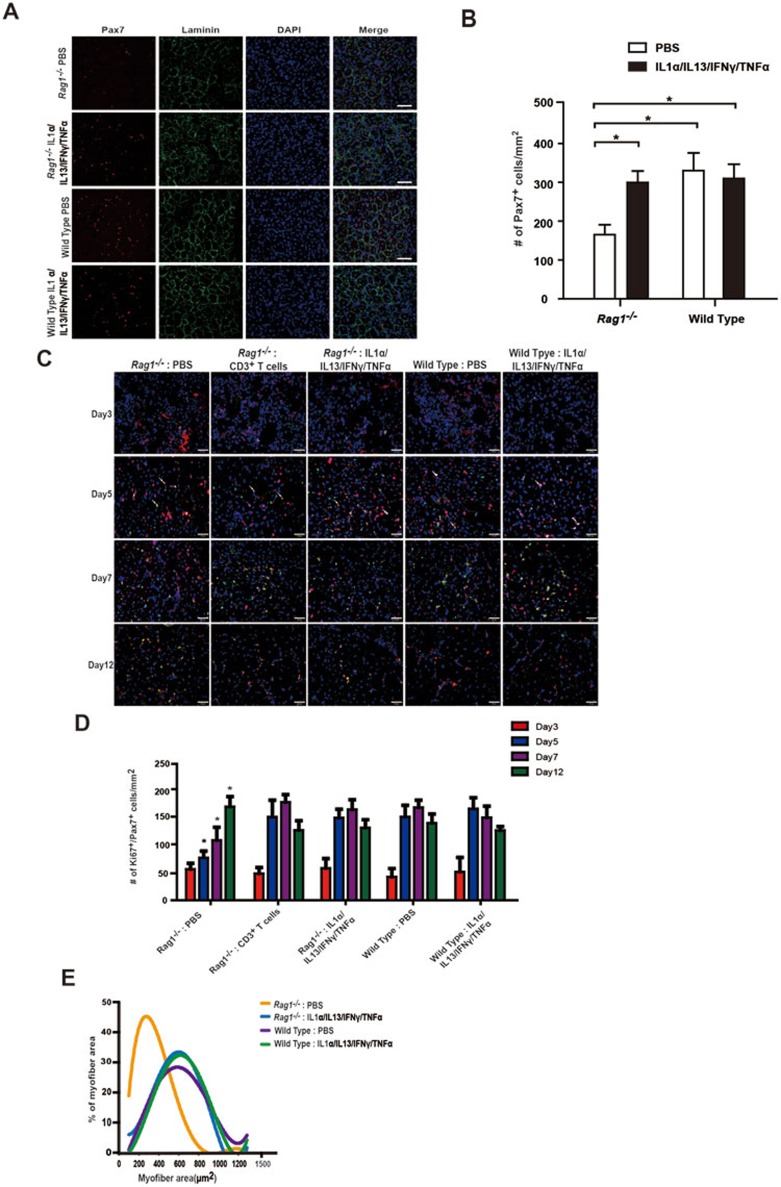
To further explore the functions of the four cytokines produced by T cells in vivo, cytokine injection experiments were performed. We first examined whether injection of the four cytokines could rescue the muscle regeneration defects caused by the absence of T cells in Rag1−/− mice. IL-1α, IL-13, IFN-γ and TNF-α were injected intramuscularly into the TA muscle of Rag1−/− mice after muscle injury. Rag1−/− mice showed significantly lower number of MuSCs after muscle injury compared to the wild-type mice (Figure 4A and 4B), while the number of MuSCs in uninjured muscles remained similar in Rag1−/− and wild-type mice (Supplementary information, Figure S1C). These observations suggest a defect in MuSC proliferation in Rag1−/− mice. When the four cytokines were supplemented to Rag1−/− muscle, the proliferation defect of Rag1−/− mice was rescued to the level of wild type (Figure 4A and 4B). Taken together, these results suggest that supplementation of the four cytokines is sufficient to rescue the MuSC proliferation defect in Rag1−/− mice. We next investigated the dynamic changes of the number of proliferating MuSCs (Ki67+ Pax7+) in Rag1−/− mice during muscle regeneration. The kinetics of MuSC proliferation in vivo was rescued to normal by muscular injection of the four cytokines or T cell transplantation (Figure 4C and 4D), suggesting that supplementation of four cytokines could replace T cell transplantation to promote endogenous MuSC proliferation. Both muscular injection of the four cytokines and T cell transplantation could rescue the myofiber size to the level of wild type (Figure 4E), further supporting that the injection of the four cytokines could replace T cell transplantation to rescue the muscle regeneration defects of Rag1−/− mice. Taken together, these results indicate that supplementation of the four cytokines could circumvent the requirement of T cells for proper muscle regeneration, and infiltrated T cells at the injury site facilitate muscle regeneration by producing the four cytokines.
Figure 4.
The cytokine cocktail could facilitate MuSC expansion in vivo. (A) Immunofluorescent staining of Pax7 to detect the presence of MuSCs in TA muscle sections 5 days after muscle injury. TA muscle samples were harvested from Rag1−/− mice, Rag1−/− mice with cytokine cocktail muscular injection, wild-type mice and wild-type mice with cytokine cocktail muscular injection. Red indicates Pax7 immunofluorescent staining. Green indicates laminin immunofluorescent staining. Blue indicates DAPI staining of nuclei. Merge indicates merged images of Pax7, laminin and DAPI staining. Scale bar, 20 μm. (B) Statistical analysis of the number of Pax7+ cells 5 days after muscle injury in TA muscle from Rag1−/− mice, Rag1−/− mice with cytokine cocktail muscular injection, wild-type mice and wild-type mice with cytokine cocktail muscular injection. Rag1−/− PBS injection sample displayed significantly lower number of MuSCs when compared to samples of Rag1−/− with cytokine cocktail injection, wild type and wild type with cytokine cocktail injection. There were no statistically significant changes between samples of Rag1−/− with cytokine injection, wild type and wild type with cytokine cocktail injection. Error bars are based on 3 independent experiments. * indicates statistically significant, P < 0.01. (C) Representative images of the time course of MuSC proliferation post injury. TA muscles were harvested on days 3, 5, 7 and 12 post muscle injury from Rag1−/− mice, Rag1−/− mice with cytokine cocktail muscular injection, wild-type mice and wild-type mice with cytokine cocktail muscular injection. Red indicates Pax7 immunofluorescent staining. Green indicates Ki67 immunofluorescent staining. Blue indicates DAPI staining of nuclei. The merged images of Pax7, Ki67 and DAPI staining are shown. Scale bar, 20 μm. (D) Statistical analysis of the number of proliferating MuSCs in vivo. Ki67+Pax7+ cells indicates the proliferating MuSCs. Error bars are based on 5 independent experiments. * indicates statistically significant, P < 0.01. (E) Statistical analysis of the myofiber size 12 days post injury.
Four cytokines promote the long-term expansion of MuSCs in vitro
If the four cytokines represent the effective components of T cell medium, MuSCs cultured in medium supplemented with the cytokine cocktail should display similar features of MuSCs cultured in T cell medium. To test this hypothesis, MuSCs treated with the cytokine cocktail were further characterized. Similar to MuSCs in T cell medium, MuSCs grown in cytokine medium were also able to be serially expanded in vitro for over 20 passages and showed normal cell morphologies (Figure 5A and Supplementary information, Figure S5A). Via serial expansion, the initial isolated MuSCs were amplified approximately 1 × 1018-fold (Figure 5A), indicating a potential to generate sufficient amount of MuSCs for regenerative medicine under a more defined culture condition. Similar to cells grown in T cell medium, over 80% of MuSCs expanded in cytokine medium maintained Pax7 expression up to passage 20 (Figure 5B and Supplementary information, Figure S5B). Every passage of the long term cultured MuSCs efficiently differentiated into myotubes in vitro (Supplementary information, Figure S5C and S5D). These observations demonstrate that MuSCs cultured in medium supplemented with the cytokine cocktail displayed similar features of stem cell marker expression, proliferation and expansion abilities, and differentiation potentials as those cultured in T cell conditional medium, further confirming that the four cytokines represent the effective components in T cell medium.
Figure 5.
Four cytokines promote the long-term expansion of MuSCs. (A) Statistical analysis of the MuSC number fold change after serial expansion in F10, T cell and cytokine cocktail medium. Error bars are based on 3 independent experiments. (B) Statistical analysis of the percentage of Pax7+ MuSCs based on quantification of Pax7 immunofluorescent staining. (C) Representative images of the engraftment of MuSCs expanded in medium containing the four cytokines. MuSCs isolated from RFP transgenic mice were expanded in medium containing cytokine cocktail. In vitro expanded RFP+ MuSCs were transplanted into injured wild-type non-fluorescent recipient TA muscle. Engraftments of RFP+ MuSCs were analyzed by confocal microscopy. Red indicates engrafted RFP+ cells. Green indicates immunofluorescent staining of Laminin. Blue indicates DAPI staining of nuclei. Only merged images of RFP, laminin and DAPI staining are shown. The source of MuSCs used for transplantation is indicated on top of each panel. “Freshly isolated”, “P0 in F10”, “P1 in F10” and “myoblast” controls were the same images as those in Figure 2G to allow direct comparison of the engraftment of MuSCs expanded in cytokine medium and those in T cell medium. All transplantation experiments were performed at the same time to allow direct comparison with each other. Scale bar, 20 μm. (D) Statistical analysis of the engraftment efficiency indicated by the number of RFP-expressing myofibers generated from freshly isolated MuSCs and MuSCs expanded in F10 medium, T cell conditional medium or cytokine medium. Error bars are based on 4 independent experiments. (E) Representative images of Pax7 immunofluorescent staining 30 days after CTX injection when the injury was completely repaired. Red indicates RFP. The RFP is membrane localized. Green indicates Pax7 immunofluorescent staining. Grey indicates laminin immunofluorescent staining. Blue indicates DAPI staining of nuclei. Merge indicates the merged images of RFP, Pax7, laminin and DAPI stainings. Arrow indicates the position of Pax7+ MuSCs located underneath laminin. Scale bar, 20 μm. See also Supplementary information, Figure S5.
Consistent with the above notion, the expression of the receptors for the four cytokines in MuSCs was easily detected by RT-PCR (Supplementary information, Figure S5E). Blocking any of the receptors of the four cytokines inhibited MuSC proliferation (Supplementary information, Figure S5F). Together, these results suggest that the four-cytokine cocktail is necessary to promote MuSC proliferation and expansion.
To evaluate the ability of MuSCs expanded in cytokine medium to repair muscle injury in vivo, cell transplantations were performed similar to cells expanded in T cell medium. MuSCs from passages 1 to 20 were able to repair the muscle injury in vivo as indicated by the presence of engrafted red myofibers (Figure 5C). Indeed, MuSCs expanded in cytokine medium displayed higher engraftment efficiency than those in T cell conditional medium. The engraftment efficiency of MuSCs expanded in cytokine medium was close to that of freshly isolated MuSCs (Figure 5D), suggesting that these cells maintained close to intact stemness even after long-term in vitro culture. Single-nucleus RFP+ cells could also be detected at the niche of endogenous MuSCs after completion of muscle regeneration (Figure 5E), suggesting that the MuSCs expanded in cytokine medium are capable of proper homing after transplantation. All these results suggest that the four-cytokine cocktail could replace T cell medium to promote long-term expansion of MuSCs in vitro.
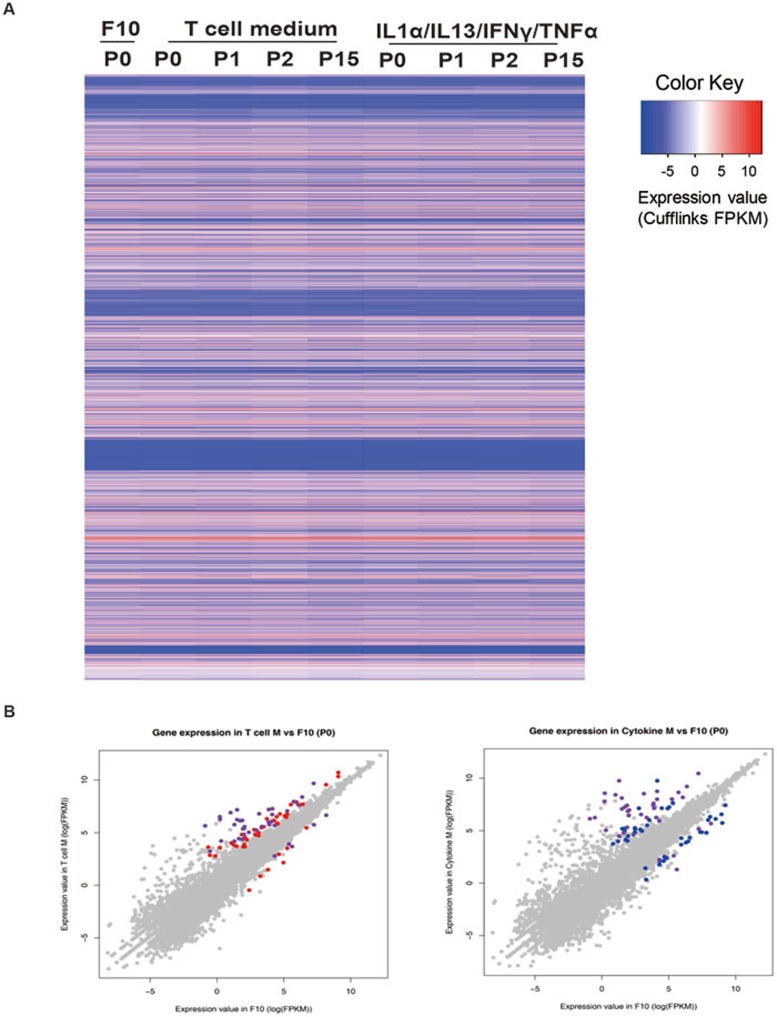
We next analyzed the genome-wide gene expression profiles of MuSCs expanded in T cell medium or cytokine medium by mRNA-seq. The gene expression profiles of MuSCs expanded in T cell medium and cytokine medium were quite similar to each other. They all displayed over 99% similarity when compared to MuSCs cultured in conventional F10 medium for 12 h (Figure 6A and 6B), suggesting that expansion in T cell medium or cytokine medium does not change the cell identity. We further examined the MuSC-specific genes in the mRNA-seq profiles. Most of the MuSC markers, including Pax7, integrin-α7, Itgb1, Vcam1, Cdh15 (M-cadherin), Desmin and c-Met, remain largely unchanged (Supplementary information, Figure S6). A few of them were slightly upregulated or downregulated (Supplementary information, Figure S6). The genes predominantly expressed in quiescent MuSCs were either unchanged or downregulated (Supplementary information, Figure S6), consistent with the notion that cells expanded in T cell or cytokine medium are activated MuSCs. Myogenin was the only differentiation marker gene detected in the datasets. It was downregulated in MuSCs cultured in cytokine medium (Supplementary information, Figure S6), suggesting that the cytokine medium prevents premature MuSC differentiation. Consistent with the RT-PCR results (Supplementary information, Figure S5E), receptors of the four cytokines showed abundant expression in the mRNA-seq datasets (Supplementary information, Figure S6). Dystrophy-, apoptosis- and oncogenesis-related genes all remained largely unchanged in MuSCs expanded in T cell or cytokine medium (Supplementary information, Figure S6). All together, the genome-wide gene expression profile analysis further confirmed that cells expanded in T cell or cytokine medium maintain their MuSC identity well.
Figure 6.
MuSCs expanded in T cell or cytokine medium displayed similar gene expression profiles to MuSCs cultured in conventional medium. (A) An overview of the MuSC transcriptome in F10, T cell and cytokine medium by heatmap. Each line represents the expression value of a RefSeq gene (a total of 22 498) as measured by log2(FPKM). MuSCs were cultured in F10 medium for 12 h. (B) A scatter plot showing the comparison of MuSC transcriptome in T cell (left) or cytokine (right) medium versus the conventional F10 medium. The majority of the genes did not display differential expression (grey dots); The red (left) and blue (right) dots indicate the genes that were differentially expressed in T cell and cytokine medium, versus conventional medium, respectively; purple dots are the genes that were differentially expressed in both T cell and cytokine medium compared to F10 medium. See also Supplementary information, Figures S6 and S7.
MuSCs expanded in cytokine medium support the secondary injury repair
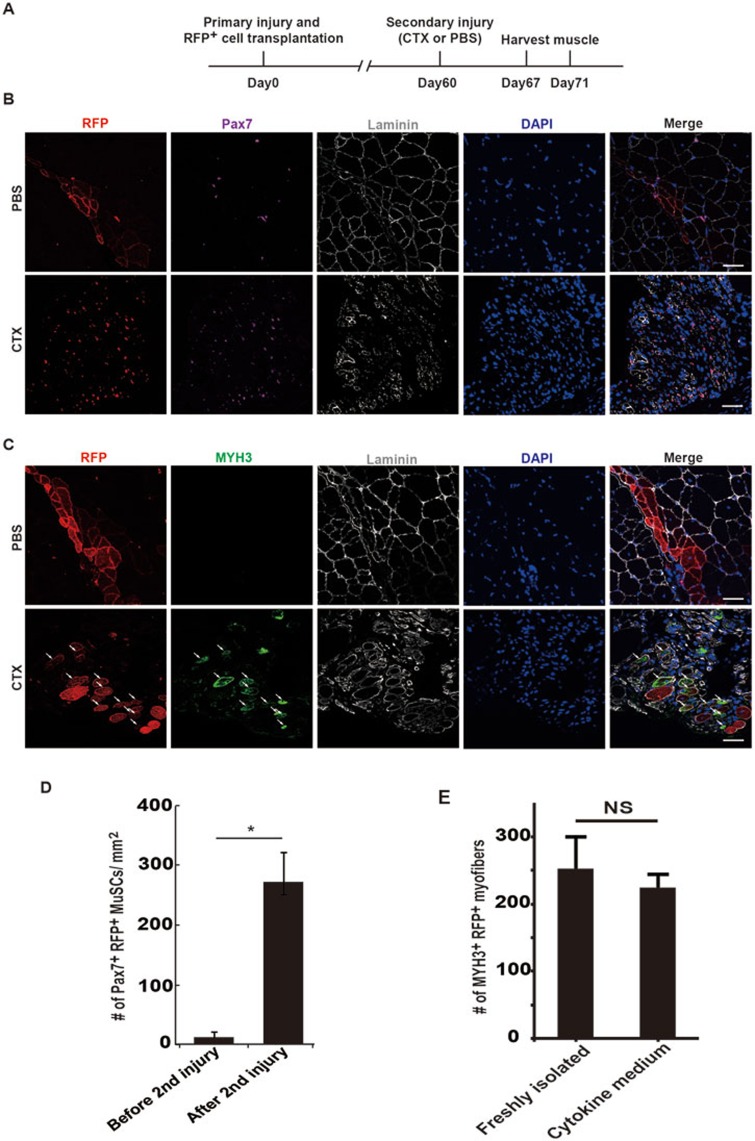
Stem cells with intact stemness could integrate into the endogenous stem cell pool and function in reparation of the secondary injury after a single transplantation. It is considered to be the “golden standard” of stem cells. We next investigated whether MuSCs expanded in cytokine medium could repair the secondary injury after initial transplantation. As shown in Figure 5E, the transplanted MuSCs were capable of proper homing, indicating that they could be re-activated and contribute to the muscle regeneration upon the secondary injury after a single transplantation. To test this hypothesis, MuSCs expanded in cytokine medium were transplanted to repair the first injury. Muscle injury was re-introduced 60 days after the first injury when it has been completely repaired. No more MuSC transplantation was performed and muscle regeneration was monitored at different time points after the secondary injury (Figure 7). The transplanted MuSCs contribute to the first muscle injury reparation as indicated by the presence of RFP+ myofibers after the first injury (Figure 7B, PBS panel). After the secondary injury, the RFP+ myofibers were destructed and the number of single-nucleus Pax7+ RFP+ MuSCs increased dramatically (Figure 7B and 7D). In contrast, in mice without the secondary injury, RFP+ myofibers remained intact and only few Pax7+ RFP+ single-nucleus cells could be detected (Figure 7B and 7D). Together, these observations suggest that similar to the endogenous MuSCs, the transplanted RFP+ MuSCs were able to quickly respond to the secondary injury to be activated and proliferate.
Figure 7.
MuSCs treated with cytokine cocktail support the secondary injury repair after a single transplantation. (A) Strategy to examine the abilities of MuSCs to repair the secondary injury with a single transplantation. Primary injury and MuSC transplantation were performed at TA muscles in hindlimb using wild-type recipient mice. The secondary injury was induced 60 days later. Muscle tissues were harvested 7 and 11 days after the secondary injury to analyze the proliferative capabilities of RFP+ cells and their abilities to repair injury. (B) Pax7 staining of sections from TA muscle 7 days after the secondary injury. RFP+ MuSCs expanded in cytokine medium were transplanted into injured wild-type non-fluorescent recipient TA muscle as described in Figure 5C. The secondary injury was induced 60 days later. The proliferation ability of RFP+ MuSCs upon the secondary injury stimulation was analyzed by immunofluorescent staining of Pax7 and observed by confocal microscopy. Red indicates RFP; purple indicates Pax7 immunofluorescent staining; grey indicates laminin immunofluorescent staining; blue indicates DAPI staining of nuclei; merge indicates merged images of Pax7, Laminin, DAPI staining and RFP. Scale bar, 20 μm. (C) MYH3 staining of sections from TA muscle 11 days after the secondary injury. The newly formed myofibers were monitored by MYH3 immunofluorescent staining. Newly formed myofibers derived from transplanted MuSCs should be positive for both MYH3 and RFP. Red indicates RFP; green indicates MYH3 immunofluorescent staining; grey indicates laminin staining; blue indicates DAPI staining of nuclei; merge indicates merged images of MYH3, laminin, DAPI staining and RFP. Scale bar, 20 μm. (D) Statistical analysis of the number of RFP+ Pax7+ MuSCs 7 days after the secondary injury. Error bars are based on 4 independent experiments. * indicates statistically significant, P < 0.01. (E) Statistical analysis of the reparation efficiency after the secondary injury. Error bars are based on 4 independent experiments. NS indicates no statistically significant difference.
To further confirm that the transplanted MuSCs expanded in cytokine cocktail medium were able to form new myofibers after the secondary injury, immunofluorescent staining of myosin heavy chain 3 (embryonic myosin heavy chain, MYH3), which labels the newly formed myofibers, was performed. Many RFP+ cells simultaneously displayed MYH3 expression in the TA muscle subjected to the secondary injury, whereas no MYH3-positive newly formed myofibers was detected in PBS mock injury mice (Figure 7C), suggesting that these newly formed myofibers were originated from the MuSCs transplanted two months earlier. The reparation efficiency of MuSCs expanded in cytokine medium was comparable to that of freshly isolated MuSCs (Figure 7E). Overall, our data revealed that MuSCs expanded in medium containing the four-cytokine cocktail could replenish the endogenous stem cell pool and repair the secondary muscle injuries after a single transplantation. They meet the “golden standard” of stem cells and are authentic stem cells. These observations suggest that IL-1α, IL-13, IFN-γ and TNF-α were sufficient to promote long-term expansion of MuSCs and maintain their stemness; therefore, the four cytokines represent components of a functional microenvironment to facilitate MuSC propagation.
Discussion
Adult stem cell represents an important source for regenerative medicine. For example, hematopoietic stem cells have been utilized to treat leukemia and other diseases for decades. However, the application of adult stem cells other than hematopoietic stem cells has been hampered by the lack of a system to obtain sufficient amount of cells without causing major damages to the donors. The strategy of isolating adult stem cells from small biopsies followed by in vitro amplification is the most ideal one. However, most adult stem cells, including hematopoietic stem cells and muscle stem cells, could not be efficiently expanded in vitro. This has been a major obstacle to hamper the research and application of adult stem cells. Here we identified IL-1α, IL-13, INF-γ and TNF-α produced by infiltrated T cells at the injury site as the minimum effective components to promote MuSC proliferation and serial expansion both in vitro and in vivo upon muscle injury. By supplementing these four factors to the culturing system, we established an in vitro system to serially expand functional MuSCs. The long-term cultured MuSCs were capable of homing to the right niche in vivo and repairing multiple rounds of injuries after a single transplantation. Using this method, sufficient amount of functional MuSCs could be yielded from limited number of cells isolated from small muscle biopsies. This sheds new lights on the application of MuSCs in regenerative medicine to treat muscle atrophy.
It has been well known that inflammation could worsen the injury and clear the tissue debris generated from wounds. However, emerging evidence starts to support the previously unappreciated positive role of acute inflammation in tissue regeneration. M2 macrophages and eosinophils have been shown to facilitate muscle injury reparation39,40,41,42. Recently, regulatory T cells (Treg) have also been suggested to play a role in muscle regeneration43. Here we showed the direct link between T cells and MuSC proliferation at the early stage of muscle regeneration. The effects of T cells we observed and the effects of Treg described by Burzyn et al.44 may represent highly context-dependent functions of T cells at distinct stages of muscle regeneration. Distinct phases of inflammatory cascades are activated orderly during muscle regeneration. T cells facilitate MuSC expansion by producing four proinflammatory cytokines, suggesting that inflammation can directly contribute to stem cell proliferation. Though the four cytokines are not exclusively secreted by T cells, the portion made by T cells is indispensible. The four proinflammatory cytokines can directly promote MuSC proliferation and expansion. They could also function to further activate other lymphocytes such as macrophages to further facilitate MuSC proliferation in vivo besides the direct proliferation effects of the four cytokines.
Stem cell transplantation has led to many therapies to treat various diseases ranging from degenerative diseases to cancer45,46. Despite many advantages of stem cell transplantation, the cost is high and the stem cell culturing technology is challenging. Development of therapies to improve the functions of the endogenous stem cells will bring the stem cell therapies to a new level. As indicated by our results, the combination of four cytokines could serve as the endogenous MuSC proliferation stimulator and improve muscle regeneration. It has great potentials in developing cell therapies without cell transplantation.
Proinflammatory signaling has been shown to play important roles in cell fate determination in both embryo and adult stages39,47. Some of the signaling pathways such as Notch and NFκB have been shown to be important for stem cell generation47. NFAT and JAK-STAT signaling pathways downstream of cytokine activation have been reported to play critical roles in muscle hypertrophy and myoblast recruitment during development48,49. To further investigate the detailed mechanism of how acute inflammation-induced signaling regulates MuSC proliferation will provide new insights into defining the link between inflammation and muscle regeneration. IL-13 and IFN-γ are known to activate the JAK-STAT signaling pathway; TNF-α and IL-1α can activate NF-κB and MAPK signaling pathways. In our GO analysis of the RNA-seq data, none of the above pathways were predominantly enriched. A few components of the JAK-STAT signaling pathway were upregulated such as JAK2 and STAT4 (Supplementary information, Figure S7), but almost all the known target genes of the JAK-STAT signaling pathway remained unchanged. That could be due to the crosstalk between cytokine signaling and their downstream signaling pathways. Further detailed mechanism study will shed lights on the stemness maintenance of MuSCs.
Muscle is considered to be an important cytokine secretary organ50,51,52. MuSCs probably serve not only as passive receivers of the signals originated from the immune system, but also as active players to regulate immune responses. Inflammatory stimulus at the injury site could stimulate injured muscle cells to release cytokines, chemokines and soluble ICAM-1, which further act on T lymphocytes, macrophages or neutrophils to regulate innate and adaptive immune responses51,53,54. MuSCs and immune cells are thus tightly coupled and mutually regulate each other to orchestrate the timely muscle regeneration. If the mutual regulation was disrupted, muscle regeneration disorder will occur. Indeed, persisted inflammation causes muscle atrophy and reduced wound healing55,56,57,58. Acute inflammation has also been reported to help initiate the neuronal regenerative process after damage in zebrafish59. Since injury is always accompanied by acute inflammation, the direct link between immune responses and adult stem cells may widely exist in many tissues besides muscle. Further research in this direction may facilitate identification of the pro-proliferation microenvironment for other adult stem cells.
Materials and Methods
Animals and muscle injury
IL2r−/− and FoxN1−/− mice were purchased from Jackson Laboratories. Rag1−/− mice were purchased from Animal Model Research Center of Nanjing University. Animal care and use were in accordance with the guidelines of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. Muscle injury was induced by the injection of CTX (Sigma) as previously described39,60 to TA muscle. Briefly, 20 μl of 10 μM CTX was injected at each injection site using 28 gauge needles. For TA muscle, 3 injections were performed for one piece of muscle. The needle was inserted parallel to the muscle fiber and injected CTX. Two injections were located close to each end of the TA muscle. The third injection was performed at the middle of the TA muscle.
Isolation and activation of T and B cells from muscles
Mononuclear cells were harvested from muscle tissues by collagenase digestion. Monocytes were collected by Histopaque Ficoll 1.077 (Sigma) gradient centrifugation. Cells were stained with anti-CD3-PE (eBioscience) and CD3+ T cells were collected by FACS sorting (Influx, BD Biosciences). T cells were incubated at 4 × 106 cells/ml in RPMI 1640 medium (RPMI 1640 (Invitrogen) supplemented with 10% FBS, 1% ℒ-glutamine, 1% penicillin and streptomycin, and 50 μM β-mercaptoethanol) and activated with 2.5 μg/ml concanavalin A (ConA; Sigma) for 2 days. Isolated T cells were further stained with anti-CD4-PE (eBioscience) and anti-CD8-FITC (eBioscience) to distinguish the CD4+ and the CD8+ T cell subpopulations. The monocytes collected from the Ficoll gradient centrifugation were stained with anti-B220-FITC (eBioscience) to harvest B220+ B cells by FACS sorting. 1 mg/ml of lipopolysaccharides (LPS; Sigma) was used to activate isolated B cells. Anti-F4/80 (eBioscience) and anti-CD11b (eBioscience) staining were performed to detect macrophages. The antibody-stained cells were analyzed by FACS (FACS Calibur, BD Biosciences).
MuSC isolation, expansion and differentiation
MuSCs were isolated as previously described7,11. Briefly, TA muscles from 1-month-old mice were dissected and dissociated with collagenase (Roche). The cells were negatively selected by biotinylated CD45, CD11, CD31 and Sca1 antibodies. The muscle cells in the flow-through were subjected to CD34-FITC (BD Biosciences) and integrin-α7-allophycocyanin (R&D systems) staining. The viable PI−CD34+ integrin-α7+ MuSCs were collected by FACS sorting. MuSCs were cultured on collagen-coated dishes in F10 basal medium (F10 medium containing 15% FBS and 2.5 ng/ml FGF (Invitrogen)), T cell conditional medium (F10 medium with 10% FBS:T cell medium = 50:50) or cytokine cocktail medium (F10 medium containing 10% FBS, 5 ng/ml IL-1α, 5 ng/ml IL-13, 10 ng/ml IFN-γ and 10 ng/ml TNF-α (R&D Systems), and 2.5 ng/ml FGF). For serial expansion, 10 000 cells were seeded in a 3.5-cm dish, and the cells were expanded every 2 days. MuSCs were differentiated in differentiation medium (DMEM (Invitrogen) with 2% horse serum (Sigma)).
Isolation of splenocytes
Spleen was dissected from mice and grinded in PBS. The cell suspension was filtered through 40 μm strainer. Red blood cells were lysed by red blood cell lysis buffer. The rest of the cells were analyzed by FACS for T cell percentage.
Conditional medium
The supernatant was collected from activated T cell culture and used as T cell conditional medium.
Immune cell transplantation
1 × 106 activated T or B cells were injected into the TA muscle of Rag1−/− mice intramuscularly.
Immunofluorescent staining
The protocol for Pax7 staining on cryosections was kindly provided by Dr Margaret Buckingham (Pasteur Institute, Paris, France)61. Briefly, the muscle sections were fixed with 4% paraformaldehyde and permeabilized with cold methanol. The sections were demasked by 0.01 M citric acid treatment at 80 °C for 5 min. Anti-Pax7 (Developmental Studies Hybridoma Bank) was applied after blocking with M. O. M. reagent for 1 h. Alexa 595 anti-mouse antibody (Invitrogen) was next applied as the secondary antibody. For other immunofluorescent stainings, anti-MYH3 (Developmental Studies Hybridoma Bank), anti-MYH1 (Upstate) and anti-laminin (Abcam) antibodies were used as primary antibody, respectively. The cells and sections were next stained with Alexa 488-, 561- or 647-labeled anti-mouse or -rabbit antibodies (Invitrogen). All images were acquired by Leica SP8 confocal microscope.
Fluorescent intensity analysis and histological image acquisition
Fluorescent intensity was measured by Leica analysis software Leica LAS AF Lite. For each sample, signals from at least 100 cells from 3 individual visual fields were collected for analysis. The histological images were acquired by Olympus DP73 microscope.
BrdU labeling
10 μM BrdU (Sigma) was added to logarithmically proliferating MuSCs for 2 h at 37 °C. MuSCs were then harvested and fixed with ice-cold ethanol at 4 °C overnight. MuSCs were then permealized in 2 N HCl/0.5% Triton for 30 min at room temperature, followed by washing in 0.1 M Na2B4O7. The cells were next stained with BrdU-FITC antibody and PI (BD Biosciences), and analyzed by FACS.
Myofiber diameter measurement
Myofiber area was measured by Adobe Acrobat 9 pro software. For each sample, at least 100 fibers from three individual visual fields were measured for analysis.
Cytokine array
Proteome profiler-mouse cytokine array panel A (R&D Systems) was subjected to analysis. All protocols were performed according to the manufacturer's instructions. Briefly, 1 ml of freshly collected T cell conditional medium was applied to pre-blocked cytokine array membrane. After 1 h incubation, detection antibody cocktail (R&D systems) was added and incubated with the membrane overnight at 4 °C. Streptavidin-HRP antibody (R&D systems) was used as the secondary antibody. The signals were detected by Las4000 X-ray device (GE Healthcare) and the pixel density was analyzed by Las4000 software (GE Healthcare).
Double antibody sandwich ELISA assays
96-well microplates were coated by 0.2 μg antibodies against IL-1α (R&D systems), IL-13 (R&D systems), IFN-γ (eBioscience) and TNF-α (eBioscience), respectively. Total proteins extracted from TA muscles were applied to the microplates. Biotinylated goat antibodies against IL-1α (R&D systems), IL-13 (R&D systems), IFN-γ (eBioscience) and TNF-α (eBioscience) were applied as detection antibodies, respectively. Streptavidin-HRP antibody (R&D systems) was used as the secondary antibody. The signals were collected by microplate reader (Thermo MK3, Thermo Fisher) and quantified by comparison with standard solution containing recombinant mouse IL-1α (R&D systems), IL-13 (R&D systems), IFN-γ (eBioscience) and TNF-α (eBioscience), respectively. The standard curves were made according to the manufacturer's instructions.
Cytokine injection
50 μl cytokine cocktail (50 ng/ml IL-1α, 50 ng/ml IL-13, 100 ng/ml IFN-γ and 100 ng/ml TNF-α) was injected intramuscularly to TA muscle together with CTX. The second dose of 50 μl cytokine cocktail was administered 2 days later. TA muscle samples were harvested 7 days after CTX injection.
mRNA-seq and analysis
MuSCs were cultured in F10 medium for 12 h. Or MuSCs were expanded in T cell medium and cytokine medium. RNA was extracted from MuSCs by TRIZOL (Sigma) and processed for mRNA-seq by TruSeq Stranded mRNA sample prep kit (Illumina) according to the manufacturer's instructions. The sequencing data were collected by HiSeq 2000 (Illumina). After obtaining the raw RNA-seq reads, we first trimmed the sequencing adaptors as well as low quality base pairs, then removed the PCR duplicated reads using in-house programs. The preprocessed reads were then aligned to the mouse reference genome (NCBI 37/UCSC mm9) using Tophat2 (version 2.0.10) guided by the RefSeq genes and allowing a maximum of two mismatches. Transcriptome profiling was thereafter performed using Cufflinks (version 2.1.1) against RefSeq genes for each sample. Differentially expressed genes were detected using Cuffdiff (version 2.1.1) against RefSeq genes. Gene Ontology (GO) Biological Process (BP) together with KEGG pathway databases62,63 were used for identification of functional enrichment among gene groups.
RT-qPCR
Frozen TA muscles were homogenized with a mortar and pestle in liquid nitrogen. 1 μl of TRIZOL (Invitrogen) per 100 mg tissue was directly added to the homogenized tissues. Total RNA was extracted by phenol:chloride (24:1) for 3 times. 1 μg of total RNA from each sample was reverse transcribed to cDNA using MMLV transcriptase (Promega) according to the manufacturer's instruction. Briefly, RNA was first denatured at 70 °C for 3 min. MMLV transcriptase was then added and incubated at 42 °C for 1 h. qPCR was performed in triplicate using a SYBR Green PCR master mix in an iCycler thermocycler system (BioRad) and analyzed by iQ5 optical system software (BioRad).
The PCR primers used are listed as below: IL-1α-F, 5′-TTCTGCCATTGACCATCTCTCTC-3′, IL-1α-R, 3′-TGCTTGACGTTGCTGATACTGT-5′ IL-1β-F, 5′-CTGGTACATCAGCACCTCAC-3′, IL-1β-R, 5′-AGAAACAGTCCAGCCCATAC-3′ IL-2-F, 5′-GGAGCAGCTGTTGATGGACCTAC-3′, IL-2-R, 5′-AATCCAGAACATGCCGCAGAG-3′ IL-4-F, 5′-GGTCTCAACCCCCAGCTAGT-3′, IL-4-R, 5′-GCCGATGATCTCTCTCAAGTGAT-3′ IL-6-F, 5′-TGTATGAACAACGATGATGCACTT-3′, IL-6-R, 5′-ACTCTGGCTTTGTCTTTCTTGTTATCT-3′ IL-10-F, 5′-CAGGGATCTTAGCTAACGGAAA-3′, IL-10-R, 5′-GCTCAGTGAATAAATAGAATGGGAAC-3′ IL-12-F, 5′-TGGTTTGCCATCGTTTTGCTG-3′, IL-12-R, 5′-ACAGGTGAGGTTCACTGTTTCT-3′ IL-17-F, 5′-GTTCTGCTGCCTGTCACATCATC-3′, IL-17-R, 5′-CGGGCAATTACTATCAGTTCCTGTC-3′ IL-23-F, 5′-CAGCGCCCCCTTCTCCGTTC-3′, IL-23-R, 5′-CTCAGGCCACGCAGCTGCTT-3′ TNF-α-F, 5′-AGTGACAAGCCTGTAGCCC-3′, TNF-α-R, 5′-GAGGTTGACTTTCTCCTGGTAT-3′ IFN-γ-F, 5′-ATGAACGCTACACACTGCATC-3′, IFN-γ-R, 5′-CCATCCTTTTGCCAGTTCCTC-3′ TGF-β-F, 5′-CACTGATACGCCTGAGTG-3′, TGF-β-R, 5′-CTCCCGTGGCTTCTAGTGC-3′ TNFRSF1α-F, 5′-ACCAAGTGCCACAAAGGAAC-3′, TNFRSF1α-R, 5′-TCAGCTTGGCAAGGAGAGAT-3′ IFNgr2-F, 5′-CTCCCCTCCCTTTGATGTGT-3′, IFNgr2-R, 5′-TGCTGTTGTTTCGTGACAGG-3′ IL1rα-F, 5′-GAATGACCCTGGCTTGTGTT-3′, IL1rα-R, 5′-TGTGCTCTTCAGCCACATTC-3′ IL13rα1-F, 5′-AGCCTGGAGAAAAGTCGTCA-3′, IL13rα1-R, 5′-GGCACCATTTTTGAGGAGAA-3′.
Accession number
The complete mRNA-seq data used to produce intensity maps have been uploaded to the GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo) website under accession number GSE58465.
Acknowledgments
We thank M Buckingham for providing Pax7 immunofluorescent staining protocol, W Bian from SIBCB cell biology core facility for helping with FACS sorting, Drs G Wang, D Li, C Xu, G Xu and S Wu for critical reading of the manuscript. This work was sponsored by the Ministry of Science and Technology of China (2014CB964700 to PH and 2012CB910800 to HW), the “Strategic Priority Research Program” of Chinese Academy of Sciences (XDA01010204 to PH), the National Natural Science Foundation of China (31171420 to PH and 31422018 to HW), One Hundred Talents Program of the Chinese Academy of Sciences (2012OHTP02 to PH), the Science and Technology Commission of Shanghai Municipality (13JC1406500 to PH), the Cancer Center of Xuhui Central Hospital (CCR2012005 to HW), Instrument Developing Project of the Chinese Academy of Sciences (YZ201339 to HW), and General Research Funds (GRF) from the Research Grants Council (RGC) of the Hong Kong SAR, China (476310 to HW and 476113 to HS).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
MuSCs displayed the similar properties in uninjured muscles from Rag1−/− and wild type mice, Related to Figure 1
FoxN1−/− mice displayed muscle regeneration defects, Related to Figure 1
Serial expaned MuSCs in T cell conditional medium displayed normal cell morphologies and differentiation abilities, Related to Figure 2
The combination of four cytokines was the minimus effective components in T cell medium, Related to Figure 3
MuSCs ser ially expanded in cytokine medium displayed normal cell morphologies and differentiation potentials, Related to Figure 4
Heat map of expression levels of the selected genes in MuSCs expanded in various mediums, related to Figure 6
Analysis of upregulated or downregulated pathways from mRNA-seq, Related with Figure 6
Removal of any one of the components from the four cytokine cocktail led to defects in MuSC expansion, related to Figure 3.
References
- Workeneh B, Bajaj M. The regulation of muscle protein turnover in diabetes. Int J Biochem Cell Biol. 2013;45:2239–2244. doi: 10.1016/j.biocel.2013.06.028. [DOI] [PubMed] [Google Scholar]
- Johns N, Stephens NA, Fearon KC. Muscle wasting in cancer. Int J Biochem Cell Biol. 2013;45:2215–2229. doi: 10.1016/j.biocel.2013.05.032. [DOI] [PubMed] [Google Scholar]
- Thibault R, Cano N, Pichard C. Quantification of lean tissue losses during cancer and HIV infection/AIDS. Curr Opin Clin Nutr Metab Care. 2011;14:261–267. doi: 10.1097/MCO.0b013e3283455d60. [DOI] [PubMed] [Google Scholar]
- Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Rando TA. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell. 2012;10:504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Rudnicki MA. Satellite cells, the engines of muscle repair. Nat Rev Mol Cell Biol. 2012;13:127–133. doi: 10.1038/nrm3265. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Vilquin JT. Myoblast transplantation: clinical trials and perspectives. Mini-review. Acta Myol. 2005;24:119–127. [PubMed] [Google Scholar]
- Mendell JR, Kissel JT, Amato AA, et al. Myoblast transfer in the treatment of Duchenne's muscular dystrophy. N Engl J Med. 1995;333:832–838. doi: 10.1056/NEJM199509283331303. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, von Maltzahn J, Soleimani VD, Yin H, Rudnicki MA. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell. 2013;12:75–87. doi: 10.1016/j.stem.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F, Jones AE, Seale V, Scime A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, Rando TA. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Bi P, Liu W, Asakura A, Keller C, Kuang S. Constitutive Notch activation upregulates Pax7 and promotes the self-renewal of skeletal muscle satellite cells. Mol Cell Biol. 2012;32:2300–2311. doi: 10.1128/MCB.06753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Tabebordbar M, Iovino S, et al. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lang PA, Christianson SW, et al. NOD/LtSz-Rag1null mice: an immunodeficient and radioresistant model for engraftment of human hematolymphoid cells, HIV infection, and adoptive transfer of NOD mouse diabetogenic T cells. J Immunol. 2000;164:2496–2507. doi: 10.4049/jimmunol.164.5.2496. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Aso H, Miyazawa K, et al. Differential cytokine gene expression in CD4+ and CD8+ T cell subsets of calves. Vet Immunol Immunopathol. 2007;118:84–91. doi: 10.1016/j.vetimm.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Williams ME, Chang TL, Burke SK, Lichtman AH, Abbas AK. Activation of functionally distinct subsets of CD4+ T lymphocytes. Res Immunol. 1991;142:23–28. doi: 10.1016/0923-2494(91)90006-5. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Joshi NS, Kaech SM. Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J Immunol. 2008;180:1309–1315. doi: 10.4049/jimmunol.180.3.1309. [DOI] [PubMed] [Google Scholar]
- Niemeyer CM, Sieff CA, Mathey-Prevot B, et al. Expression of human interleukin-3 (multi-CSF) is restricted to human lymphocytes and T-cell tumor lines. Blood. 1989;73:945–951. [PubMed] [Google Scholar]
- Van Snick J. Interleukin-6: an overview. Annu Rev Immunol. 1990;8:253–278. doi: 10.1146/annurev.iy.08.040190.001345. [DOI] [PubMed] [Google Scholar]
- Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O'Garra A. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haben I, Hartmann W, Specht S, et al. T-cell-derived, but not B-cell-derived, IL-10 suppresses antigen-specific T-cell responses in Litomosoides sigmodontis-infected mice. Eur J Immunol. 2013;43:1799–1805. doi: 10.1002/eji.201242929. [DOI] [PubMed] [Google Scholar]
- de Waal Malefyt R, Abrams JS, Zurawski SM, et al. Differential regulation of IL-13 and IL-4 production by human CD8+ and CD4+ Th0, Th1 and Th2 T cell clones and EBV-transformed B cells. Int Immunol. 1995;7:1405–1416. doi: 10.1093/intimm/7.9.1405. [DOI] [PubMed] [Google Scholar]
- Baier M, Bannert N, Werner A, Lang K, Kurth R. Molecular cloning, sequence, expression, and processing of the interleukin 16 precursor. Proc Natl Acad Sci USA. 1997;94:5273–5277. doi: 10.1073/pnas.94.10.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DM, Zhang Y, Parada NA, et al. Processing and release of IL-16 from CD4+ but not CD8+ T cells is activation dependent. J Immunol. 1999;162:1287–1293. [PubMed] [Google Scholar]
- Keane J, Nicoll J, Kim S, et al. Conservation of structure and function between human and murine IL-16. J Immunol. 1998;160:5945–5954. [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- Hall BM, Verma ND, Tran GT, Hodgkinson SJ. Distinct regulatory CD4+T cell subsets; differences between naive and antigen specific T regulatory cells. Curr Opin Immunol. 2011;23:641–647. doi: 10.1016/j.coi.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Cheng M, Nguyen MH, Fantuzzi G, Koh TJ. Endogenous interferon-gamma is required for efficient skeletal muscle regeneration. Am J Physiol Cell Physiol. 2008;294:C1183–C1191. doi: 10.1152/ajpcell.00568.2007. [DOI] [PubMed] [Google Scholar]
- Ceafalan LC, Popescu BO, Hinescu ME. Cellular players in skeletal muscle regeneration. Biomed Res Int. 2014;2014:957014. doi: 10.1155/2014/957014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz E. Satellite cell proliferative compartments in growing skeletal muscles. Dev Biol. 1996;175:84–94. doi: 10.1006/dbio.1996.0097. [DOI] [PubMed] [Google Scholar]
- Allbrook DB, Han MF, Hellmuth AE. Population of muscle satellite cells in relation to age and mitotic activity. Pathology. 1971;3:223–243. doi: 10.3109/00313027109073739. [DOI] [PubMed] [Google Scholar]
- Relaix F, Marcelle C. Muscle stem cells. Curr Opin Cell Biol. 2009;21:748–753. doi: 10.1016/j.ceb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Heredia JE, Mukundan L, Chen FM, et al. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold L, Henry A, Poron F, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell D, Mourkioti F, Gambardella A, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynes MW, Riches DW. Induction of macrophage insulin-like growth factor-I expression by the Th2 cytokines IL-4 and IL-13. J Immunol. 2003;171:3550–3559. doi: 10.4049/jimmunol.171.7.3550. [DOI] [PubMed] [Google Scholar]
- Burzyn D, Kuswanto W, Kolodin D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D, Benoist C, Mathis D.Regulatory T cells in nonlymphoid tissues Nat Immunol 2013141007–1013.24048122 [Google Scholar]
- Dooley D, Vidal P, Hendrix S. Immunopharmacological intervention for successful neural stem cell therapy: new perspectives in CNS neurogenesis and repair. Pharmacol Ther. 2014;141:21–31. doi: 10.1016/j.pharmthera.2013.08.001. [DOI] [PubMed] [Google Scholar]
- van Laar JM, Sullivan K. Stem cell transplantation in systemic sclerosis. Curr Opin Rheumatol. 2013;25:719–725. doi: 10.1097/01.bor.0000434669.32150.ac. [DOI] [PubMed] [Google Scholar]
- Espin-Palazon R, Stachura DL, Campbell CA, et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell. 2014;159:1070–1085. doi: 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- Serrano AL, Baeza-Raja B, Perdiguero E, Jardi M, Munoz-Canoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008;7:33–44. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Kawaguchi Y, Harigai M, et al. Increased CD40 expression on muscle cells of polymyositis and dermatomyositis: role of CD40-CD40 ligand interaction in IL-6, IL-8, IL-15, and monocyte chemoattractant protein-1 production. J Immunol. 2000;164:6593–6600. doi: 10.4049/jimmunol.164.12.6593. [DOI] [PubMed] [Google Scholar]
- Marino M, Scuderi F, Provenzano C, Bartoccioni E. Skeletal muscle cells: from local inflammatory response to active immunity. Gene Ther. 2011;18:109–116. doi: 10.1038/gt.2010.124. [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol. 2012;8:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- Gallucci S, Provenzano C, Mazzarelli P, Scuderi F, Bartoccioni E. Myoblasts produce IL-6 in response to inflammatory stimuli. Int Immunol. 1998;10:267–273. doi: 10.1093/intimm/10.3.267. [DOI] [PubMed] [Google Scholar]
- Pedersen BK. Muscle as a secretory organ. Compr Physiol. 2013;3:1337–1362. doi: 10.1002/cphy.c120033. [DOI] [PubMed] [Google Scholar]
- Acharyya S, Villalta SA, Bakkar N, et al. Interplay of IKK/NF-κB signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. J Clin Invest. 2007;117:889–901. doi: 10.1172/JCI30556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, Ferrucci L, Ragno E, et al. Chronic inflammation and the effect of IGF-I on muscle strength and power in older persons. Am J Physiol Endocrinol Metab. 2003;284:E481–E487. doi: 10.1152/ajpendo.00319.2002. [DOI] [PubMed] [Google Scholar]
- Porter JD. Extraocular muscle: cellular adaptations for a diverse functional repertoire. Ann NY Acad Sci. 2002;956:7–16. doi: 10.1111/j.1749-6632.2002.tb02804.x. [DOI] [PubMed] [Google Scholar]
- Taaffe DR, Marcus R. Musculoskeletal health and the older adult. J Rehabil Res Dev. 2000;37:245–254. [PubMed] [Google Scholar]
- Kyritsis N, Kizil C, Zocher S, et al. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012;338:1353–1356. doi: 10.1126/science.1228773. [DOI] [PubMed] [Google Scholar]
- Hu P, Geles KG, Paik JH, DePinho RA, Tjian R. Codependent activators direct myoblast-specific MyoD transcription. Dev Cell. 2008;15:534–546. doi: 10.1016/j.devcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reference Genome Group of the Gene Ontology C. The gene ontology's reference genome project: a unified framework for functional annotation across species. PLoS Comput Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MuSCs displayed the similar properties in uninjured muscles from Rag1−/− and wild type mice, Related to Figure 1
FoxN1−/− mice displayed muscle regeneration defects, Related to Figure 1
Serial expaned MuSCs in T cell conditional medium displayed normal cell morphologies and differentiation abilities, Related to Figure 2
The combination of four cytokines was the minimus effective components in T cell medium, Related to Figure 3
MuSCs ser ially expanded in cytokine medium displayed normal cell morphologies and differentiation potentials, Related to Figure 4
Heat map of expression levels of the selected genes in MuSCs expanded in various mediums, related to Figure 6
Analysis of upregulated or downregulated pathways from mRNA-seq, Related with Figure 6
Removal of any one of the components from the four cytokine cocktail led to defects in MuSC expansion, related to Figure 3.