Abstract
Phycobilisomes (PBSs) are light-harvesting antennae that transfer energy to photosynthetic reaction centers in cyanobacteria and red algae. PBSs are supermolecular complexes composed of phycobiliproteins (PBPs) that bear chromophores for energy absorption and linker proteins. Although the structures of some individual components have been determined using crystallography, the three-dimensional structure of an entire PBS complex, which is critical for understanding the energy transfer mechanism, remains unknown. Here, we report the structures of an intact PBS and a PBS in complex with photosystem II (PSII) from Anabaena sp. strain PCC 7120 using single-particle electron microscopy in combination with biochemical and molecular analyses. In the PBS structure, all PBP trimers and the conserved linker protein domains were unambiguously located, and the global distribution of all chromophores was determined. We provide evidence that ApcE and ApcF are critical for the formation of a protrusion at the bottom of PBS, which plays an important role in mediating PBS interaction with PSII. Our results provide insights into the molecular architecture of an intact PBS at different assembly levels and provide the basis for understanding how the light energy absorbed by PBS is transferred to PSII.
Keywords: phycobilisome, photosynthesis, phycobiliprotein, electron microscopy
Introduction
Phycobilisomes (PBSs) are attached to the cytoplasmic surface of thylakoid membranes and are responsible for the majority of light capture for photosynthesis in cyanobacteria and red algae1,2,3. A hemidiscoidal PBS, which is the most common PBS type, has two parts: the core and the peripheral rods. The core of this type of PBS consists of two, three, or five cylindrical substructures, and the peripheral rods radiate from the core. PBSs from Anabaena sp. strain PCC 7120 (Anabaena 7120, also known as Nostoc sp. PCC 7120) have a five cylinder core and eight peripheral rods with a molecular mass of ∼6 000 kDa4,5,6.
PBSs are composed of phycobiliproteins (PBPs) and linker proteins. PBPs are brilliantly colored, water-soluble proteins bearing different numbers of chromophores, which are open-chain tetrapyrroles covalently bound to cysteine residues via thioether bonds22. Based on the bilin energy level, PBPs are mainly categorized into three types: phycoerythrins (PEs) or phycoerythrocyanins at the core-distal ends of rods (which absorb high-energy light), phycocyanins (PCs) at the core-adjacent portions of rods (which absorb intermediate-energy light), and allophycocyanins (APCs), the major components of the core (which absorb low-energy light)1. Although PBPs have different absorption spectra due to different bilin energy levels, they have similar crystal structures7,8 and assemble into PBSs through a common hierarchical organization. Two different subunits of PBPs, α and β, initially form a heterodimer (αβ), conventionally called the (β) monomer, which subsequently assembles into the (αβ)3 trimer. The trimers are the fundamental assembly unit of PBSs, and stack face to face to form a hexamer with or without linker proteins in its central cavity2,9. APCs in the core have a few variants, such as αLCM (the α domain in LCM; LCM is coded by apcE), αAPC-like variant (denoted αAP-B, coded by apcD), and βAPC-like variant (denoted β18.5, coded by apcF) (Supplementary information, Figure S1A)10.
Linker proteins are mostly colorless polypeptides that are involved in the assembly of PBSs at different levels2. These proteins include LR/CpcC (PecC; involved in rod extension), LRT/CpcD (for distal termination of rods), LRC/CpcG (mediating the association between rods and the core), LC (participating in the assembly of the core), and LCM/ApcE (involved in the assembly of the core and interactions between the core and the thylakoid membrane)2,11. There are two types of conserved domains in linkers: Pfam00427 and Pfam01383. The Pfam00427 domain makes up the N-termini of LR and LRC. LCM contains two to four (depending on the species) repetitive (REP) domains, which are homologs of the Pfam00427 domain. In Anabaena 7120, four copies of REP domains are present in LCM6. The Pfam01383 domain constitutes the majority of LRT and LC, and the C-terminus of LR (Supplementary information, Figure S1). The crystal structures of some isolated components of PBSs from different species have been determined. These include PEs12,13, PCs8,14,15,16,17, APCs10,17,18,19, and linker proteins10,20 (Supplementary information, Figure S1). The structural layouts of various PBSs have also been suggested in electron microscopic images21,22. However, the precise position of PBPs and the linker proteins in the PBS remains largely unclear.
How light energy is transferred from PBS to the reaction center is another important unknown question. PBS can transfer energy to both photosystem II (PSII) and photosystem I (PSI)2, and energy distribution between PSII and PSI is regulated by a process called state transition23,24. Early reports showed that state transitions occurred in a time range of subseconds to seconds25 and that this process is controlled by the redox state of electron carriers between PSI and PSII26. Liu et al.27 recently reported the isolation of a PBS-PSII-PSI mega-complex, and suggested that PBS does not need to change its spatial position during state transitions. In this study, we investigated the three-dimensional (3D) structures of the intact PBS and PBS-PSII complexes from Anabaena 7120 using single-particle electron microscopy (EM) analysis.
Results
Overall structures of the PBS and PBS-PSII complexes
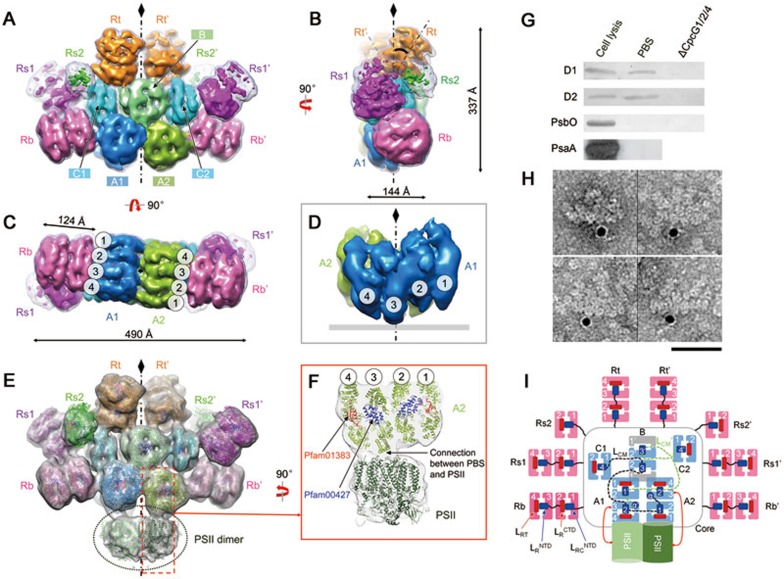
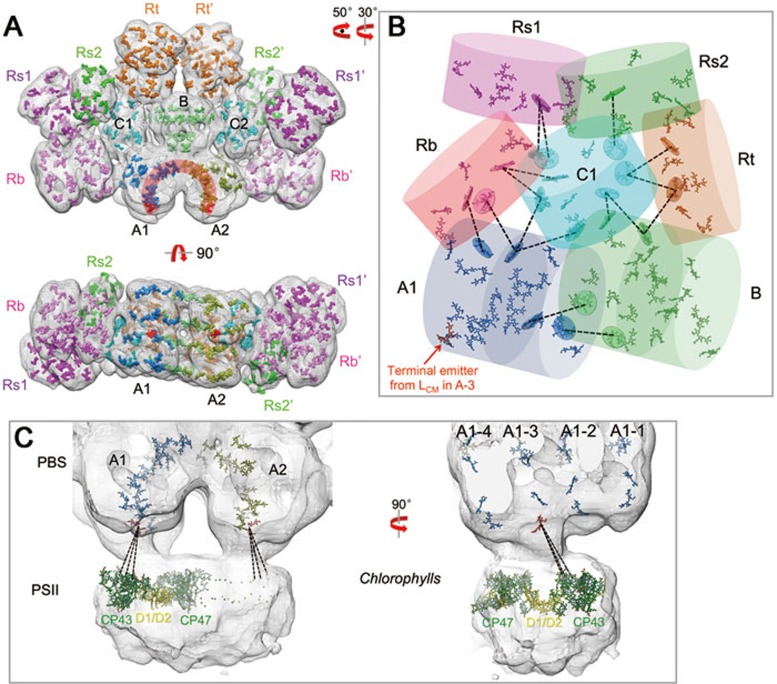
PBSs from Anabaena 7120 were purified (Supplementary information, Figure S2A-S2E) and negative-stain EM imaging of the samples showed clear side, top, and oblique views of the isolated PBS (Supplementary information, Figure S2F). The end-on-view averages indicate a two-fold symmetry, which was verified using a rotational cross-correlation coefficient analysis (Supplementary information, Figure S3A-S3B). We reconstructed the 3D structure of the intact PBS (Figure 1A-1C) using 32 966 particles, achieving a 21 Å resolution according to the FSC0.5 criterion (Supplementary information, Figure S3C-S3F). The hemidiscoidal structure has a length of 49.0 nm, height of 33.7 nm, and thickness of ∼20.0 nm (Figure 1A-1C). All PBP trimers of the core and the rods could be unambiguously distinguished and located in the map (Figure 1A-1C and 1I). The core consists of three complete cylinders A1, A2, and B (each formed by four layers of APC trimers) and two half cylinders C1 and C2 (each formed by two APC trimers). Two basal core cylinders A1 and A2 are arranged in an antiparallel fashion (Figure 1C) and core cylinder B is located on top of the two A cylinders (Figure 1A). The two half core cylinders C1 and C2 are unique. In contrast to half core cylinders arranged in a side-by-side manner with other core cylinders in early models5, C1 and C2 are arranged perpendicularly to the other core cylinders (Figure 1A). In addition, these half core cylinders have connections to all other PBS components (core cylinders A1, A2, and B, and all peripheral rods), suggesting that they are important in the overall organization of this PBS.
Figure 1.
Overall structures of the intact PBS and the PBS-PSII complex. (A-C) EM density map of the PBS from front (A), side (B), and bottom (C) views. Cylinders of the core (A1, A2, B, C1 and C2) and rods (Rb, Rs1, Rs2 and Rt) are shown in different colors. The low-pass filtered (to 20 Å) map is shown in mesh at the lower threshold to show the side rods, Rs1 and Rs2. The tilted angle of Rt with the symmetry axis is indicated at 35° (B). (D) Extracted density map of core A1 and A2 showing the protrusion of disc 3 to the membrane. (E) EM density map of the PBS-PSII complex. Densities corresponding to PSII are circled. Available crystal structures of APC or PC trimers, linker domains (Pfam00427 and Pfam01383) and the PSII dimer are docked into the density map. (F) Zoomed view of the connection between PBS and PSII through disc 3 of the A2 cylinder. (G) Immunoblotting demonstrating that the reaction center subunits D1 and D2 of PSII can be detected in the PBS sample. Lane 1, cell lysis as a positive control; lane 2, PBS sample used for EM analysis in this study; lane 3, PBS sample taken from the ΔcpcG1/2/4 mutant (in which all core-rod association-related genes are deleted) as a negative control. (H) Immunogold labelling EM of PBS preparation using antibody against PSII subunit D1. Scale bar, 500 Å. (I) Schematic model of the PBS-PSII architecture. Dashed lines indicate connections between the REP domains of LCM, which have not been experimentally verified. Blue box: the Pfam00427 domain; red box: the Pfam01383 domain.
Eight peripheral rods emanate out of the hemidiscoidal plane from the core at departure angles between 5° and 30° (Figure 1A-1C and Supplementary information, Figure S4), which could increase the cross-section areas of PBS in light harvesting. Each rod in the bottom pair (Rb and Rb′), top pair (Rt and Rt′), and one of the side pairs (Rs1 and Rs1′) has two hexamers, whereas the densities of the other pair of side rods (Rs2 and Rs2′) were weak and small and each rod likely contains only one hexamer (Figure 1A). The length of the core cylinder (A1, A2, or B) formed by four APC trimers is 14.4 nm, which is longer than that of the peripheral rod formed by four PC trimers (12.4 nm; Figure 1B and 1C), reflecting differences in their organization. In both the bottom and side views, the trimeric discs of the two bottom core cylinders (A1 and A2) are stacked in a staggered manner, with disk 3 protruding towards the thylakoid membrane (Figure 1C and 1D). This structural feature is important for energy transfer from PBSs to PSII complexes that are embedded in the membranes (see Discussion further below).
We noted that a portion of the raw particles and two-dimensional (2D) class averages showed additional density attached to the bottom of the PBS in a position where PSII should bind28,29,30,31,32 (Supplementary information, Figure S5A). We screened these particles and obtained a 3D reconstruction, in which we can observe a clear density with a volume in agreement with the size of a PSII dimer (Figure 1E and Supplementary information, Figure S5B). To test whether the density is contributed by PSII, we first performed an immunoblotting experiment, which showed that the PBS sample contained the reaction center subunits (D1 and D2) of PSII33,34 but no PsaA (a PSI protein) or PsbO (a PSII peripheral protein required for normal oxygen evolution) (Figure 1G). Second, we performed immunogold labelling experiments using antibody against D1 and observed gold particle labelling at the bottom of the PBS, which is consistent with the assignment of the density as PSII (Figure 1H). Assigning this density as PSII was further supported by a satisfactory docking of the crystal structure of PSII34 into the 3D map (Figure 1E-1F). The above evidence suggests that these particles represent PBS-PSII complexes. Notably, the PSII density directly connects to the protrusions of disc 3 of the core substructures A1 and A2 (Figure 1E and 1F). Coincidentally, the protrusion of disc 3 fits well with the cavity at the PSII surface (Supplementary information, Figure S5C and S5D).
Core and rod assembly
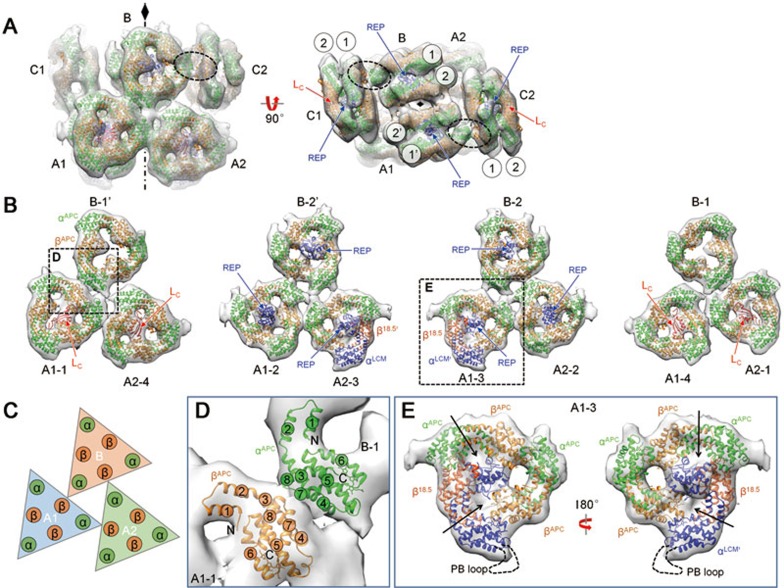
The crystal structure of APC in complex with the linker protein Lc (the Pfam01383 domain) from Mastigocladus laminosus (PDB code: 1B33)10 and a recently reported crystal structure of the N-terminal domain of linker LR (the Pfam00427 domain) from Synechocystis sp. PCC 6803 (PDB code: 3NPH)20 were used for docking analysis of the core. The crystal structures docked well into the density maps of the core, with a cross-correlation coefficient of ∼0.8 (Figure 2A, Supplementary information, Table S1 and Movie S1). Layers 1-2 of the four trimeric discs of cylinder A1 formed a face-to-face hexamer, and layers 3-4 did the same. The two hexamers were further assembled in a back-to-back orientation to form the cylinder. All hexamers in the core are similarly packed, as suggested via crystallography (Supplementary information, Figure S6A and S6B). Within each cylinder A, all of the trimers have additional densities occupying their central holes. The central density of the trimer of disc 3 is significantly larger than that of the other three trimers (Figure 2B and 2E), suggesting different components in this trimer. Together with the fact that disc 3 directly connects to PSII, which suggests that αLCM is located in disc 3, we hypothesize that the extra density in disc 3 could be partially contributed by the loop between αLCM and REP1 and/or the inserted PB loop in αLCM2,35 (Figure 2E and Supplementary information, Figure S1A). Six Pfam01383 domains from LC linker proteins can be well fitted into the central densities of the outer APC trimers of cylinders A (four copies) and C (two copies). In contrast, no such central densities corresponding to the Pfam01383 domain were present in cylinder B (Figure 2A and 2B). Eight Pfam00427 domains, likely derived from two LCM proteins as each LCM contains four REP domains (Supplementary information, Figure S1A), can be fitted into the densities of the inner APC trimers of cylinders A (four copies), B (two copies), and C (two copies) (Figure 2A and 2B). The connections of these REP domains mediated by loops36 cannot be clearly observed at the current resolution. The Pfam01383 domains are located inside the trimer plane, as in the crystal structure10, whereas the Pfam00427 domains protrude from the trimer plane (Figure 2B and Supplementary information, Figure S6A). APC trimers in the core (cylinders A1, A2, and B) are associated via a specific pattern, in which a corner of the triangle-shaped trimer directly contacts the edge of another trimer (Figure 2B and 2C). This corner-edge association pattern, which is the basic connection mode found in the core, is likely mediated by the interaction between and β subunits (Figure 2D).
Figure 2.
Assembly of the core. (A) Extracted density map of the core with fitted crystal structures shown in side and top views. Connections between cylinders C and B via α-β subunit interactions are circled. (B) The inter-cylinder interaction in cylinders A1, A2 and B is shown layer by layer. (C) Schematic model showing the interaction of the inter-cylinder trimers. (D) Zoomed view of α-β interaction mediating the interaction of inter-cylinder trimers. Helices of each subunit are numbered from the N- to C-terminus. (E) Details of disc 3 docking, where the terminal emitter from LCM locates. The position of PB loop is indicated by a dashed line. The filled holes of the trimer are indicated with arrows.
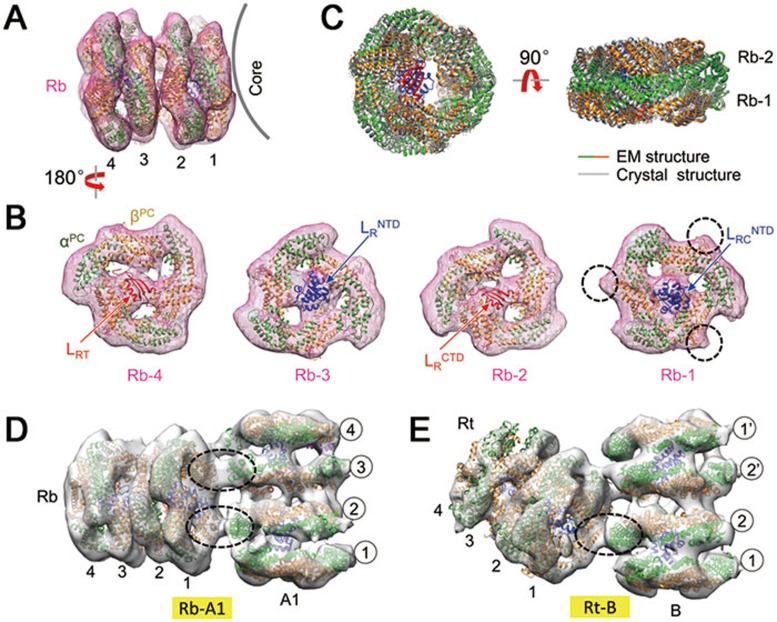
For the docking analysis of the peripheral rods, the crystal structures of PCs (PDB code: 1JBO)14, the Pfam01383 domain (isolated from the APC-trimer-Lc complex, PDB code: 1B33)10, and the Pfam00427 domain (PDB code: 3NPH)20 were used (Figure 3A and 3B). The shape of the density maps of the PC trimer was similar to that of the APC trimer with a small difference due to an additional loop in the PC17 that can be distinguished in the density map (dashed circles for Rb-1 in Figure 3B). A typical rod cylinder in our reconstruction contains two PC hexamers associated with two Pfam00427 domains and two Pfam01383 domains. The PC hexamer is formed by a face-to-face interaction of PC trimers in the same manner as in the crystal structure of the PC hexamer (Figure 3C and Supplementary information, Figure S6C and S6D).
Figure 3.
Assembly of the rods. (A, B) Docking of the bottom rod (Rb). Additional loops in the crystal structure of PC trimers (compared with the APC trimer) are distinguishable in the EM density map and are indicated by dashed circles for Rb-1. (C) Comparison of EM and crystal structures of the PC hexamer. (D, E) Connections between the bottom/top rod (Rb/Rt) and the core cylinder (A1/B). The α-β inter-trimer interactions, which mediate rod-core connections, are circled.
In the core, the Pfam01383 domains were found in the outer trimers and the Pfam00427 domains were found in the inner trimers. In contrast, the Pfam01383 and Pfam00427 domains in the rods appear alternatingly from the terminal trimer (Figure 3B). LRC binds to the innermost trimer (Rb-1) through its N-terminal Pfam00427 domain (Figure 3B), whereas its C-terminus (LRCCTD) is likely anchored to the core. However, the density corresponding to LRCCTD cannot be clearly distinguished in the map, indicating that the C-terminus of LRC does not fold into a globular domain as does its N-terminus. LR connects the two PC hexamers by binding to the external hexamer (Rb-3) through its N-terminal Pfam00427 domain and to the internal hexamer (Rb-2) through its C-terminal Pfam01383 domain (Figure 3B). LRT (the Pfam01383 domain) terminates the rod extension by binding to the outermost PC trimer (Rb-4) (Figure 3B).
A careful analysis of the interactions between rods Rt and Rb and the core components suggests that they also use the corner-to-edge interaction mode for rod-core assembly (Figure 3D and 3E). The corner-to-edge interaction pattern, which is likely mediated by the inter-trimer α-β subunit interaction19,37, may also provide a major force holding rods and core together. Although Rb and Rt are similarly geometrically oriented relative to their interacting core cylinders A1 and B, respectively (Figure 3D and 3E), this geometric orientation is not found between Rs1/Rs2 (and Rs1′/Rs2) and their associated half core cylinders C1 and C2, suggesting a variable association geometry between rods and the core38.
Interactions between the rods and the core
We next investigated the roles of the linker protein LRC in the rod-core association. LRCs are encoded by cpcG genes (Supplementary information, Figure S1A). Four cpcG genes (cpcG1-4) exist in Anabaena 7120. CpcG3 (also named cpcL39) is equivalent to CpcG2 from Synechocystis sp. PCC 7002 and plays a role in the formation of a special PBS complex40,41. A recent study demonstrated that CpcG3 is important for the association of a special PBS with PSI in Anabaena 712039. CpcG1, CpcG2, and CpcG4 of Anabaena 7120 are functionally similar to CpcG1 of Synechocystis 7002 and are responsible for the rod-core association4,5.
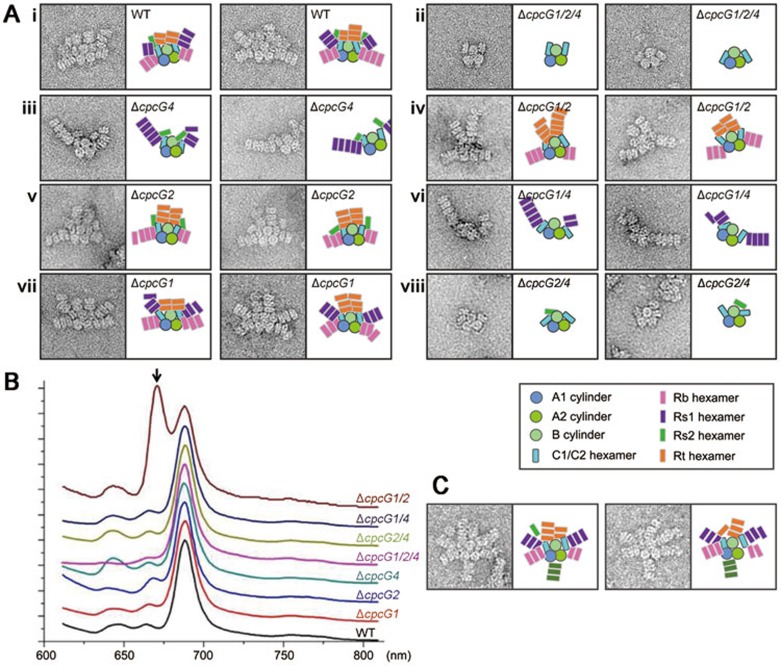
To study the specific roles of these LRC proteins, we generated a series of cpcG mutant strains, which covered all combinations of deletions of three linker genes involved in the rod-core association, i.e., cpcG1, cpcG2, and cpcG4 (Figure 4A and Supplementary information, Figure S7A-S7C). We isolated PBS complexes from these mutant strains and examined their assembly (Figure 4A) and energy transfer ability (Figure 4B). The role of each LRC was assigned based on the EM images of PBSs isolated from the strains mentioned above (Figure 4A and Supplementary information, Figure S7E). CpcG4 is responsible for the connections of both the top and bottom pairs (Rt and Rb) of rods to the core. CpcG2 is necessary for connection of the larger side rod Rs1 to the core, whereas CpcG1 is required for the association of the smaller side rod Rs2 to the core (Figure 4A). Thus, these LRC proteins are responsible for recognizing the specific sites on the core, and each individual rod may interact with the core independently.
Figure 4.
Interaction of rods with the core. (A) Typical negative-stain EM images of a wild-type and mutant PBSs. The schematic models for each sample are shown to the right of the images. (B) 77K fluorescence spectra of wild-type and mutant PBSs under 590 nm excitation. The unique peak at approximately 670 nm in the ΔcpcG1/2 mutant (indicated with an arrow) may be caused by a leak of energy from the half core cylinders that cannot efficiently transfer the energy within the core. (C) Negative-stain EM images of PBSs from the ΔapcF mutant. Note that a rod is attached at the bottom of the PBS.
The EM images of PBSs from the ΔcpcG1/2/4 mutant strain, which had all three cpcG genes deleted, showed that no rods were attached and that the densities of the half core cylinders C1 and C2 became faint and flexible (Supplementary information, Figure S7D), suggesting that rod binding may help stabilize the half core cylinders. Figure 4B shows fluorescence emission spectra of the wild-type and mutant PBSs. The spectrum of the ΔcpcG1/2 mutant, in which no peripheral rods were attached to the half core cylinders C1 and C2, had an additional peak at 670 nm, indicating that some energy presumably escaped from C1 and C2. In wild-type and mutant PBSs, no peripheral rods were found at the bottom side of the PBS core. However, when we studied the PBS from a mutant lacking the key core gene apcF (encoding β18.5), we frequently observed one or two peripheral rods attached to the bottom of the cores (Figure 4C). This result suggests that an extra site for peripheral rod attachment might be provided in the ΔapcF mutant, where ApcF (β18.5) is possibly replaced by ApcB (βAPC).
Pigment arrangement and energy transfer
The global distribution of chromophores in the PBS was investigated using results of docking analyses (Figure 5A and Supplementary information, Movie S1). The Rb and Rt rods have structural connections with the core cylinders A and B, respectively. They also have connections with the half core cylinders C1 and C2. The side rods Rs1 and Rs2, however, are only spatially attached to the half core cylinders C1 and C2. Based on the analysis of distances between bilins (Supplementary information, Figure S8A-S8D), energy harvested by the bottom rods Rb could be transferred to the core cylinders A directly or through the half core cylinders C1 and C2. The top rods Rt could transfer their energy to the core cylinders A through either core cylinder B or the half core cylinders C1 and C2. However, side rods (Rs1 and Rs2) could only transfer their energy via half core cylinders C1 and C2 (Figure 5B). This structural feature suggests that the half core cylinders C1 and C2 play an important role in connecting the rods with the core during energy transfer.
Figure 5.
Pigment arrangement and energy transfer pathways. (A) Pigment arrangement in the intact PBS. The pigments are presented as surfaces and colored according to their cylinders shown in Figure 1A. Pigments from A1 and A2 (front view) form an inverted 'U' shape (colored dark orange), in which the terminal emitters from disc 3 are colored red. (B) Energy transfer from the rods to the core. All pigments in core cylinders A1, B, and C1 and the pigments from the core-connected PC trimers are shown. The distances of one or two nearest pigment (from rods to the core) are indicated by dashed lines. The disc covering a pigment is the plane defined by all the atoms of that pigment. (C) Energy transfer from PBS to PSII. The EM density map of the PBS-PSII complex is shown in mesh. Pigments from core cylinders A1 and A2 are presented, with the terminal emitter from disc 3 in red. Chlorophylls from one PSII monomer (35 chlorophylls) are presented as a stick model (chlorophylls from CP43/CP47 and D1/D2 are colored dark green and yellow, respectively). Close connections between A1-3 and CP43 are indicated using dashed lines.
Pigments in the basal core cylinders (A1 and A2) form an arch when viewed from the face side. The two copies of chromophores bound to ApcE (terminal emitters) are located at the bottom of the arch (Figure 5A). This arrangement could efficiently transfer and converge absorbed light energy to the terminal emitters that in turn transfer energy to PSII. The other subunit containing a far-red-emitting chromophore, ApcD, which is required for energy transfer to PSI, could not be unambiguously identified in the PBS core structure. Analyses of the distance of the bilins of the PBSs closest to the chlorophylls in PSII suggest that disc 3 is spatially close to chlorophylls in the antenna protein CP43 of PSII34 (Figure 5C). The nearest chlorophyll a to disc 3 (with an estimated distance of 48 Å, Supplementary information, Figure S8E) is located at the periphery of PSII and is the only chlorophyll a with a central magnesium atom that is coordinated by an asparagine residue, CP43-Asn39 (others are coordinated via either water or a histidine residue34).
Discussion
The primary functions of PBS are the absorption of photons and the transfer of the energy to photosynthetic reaction centers. Highly ordered chromophores in PBS are very important for energy absorption and transfer. These functions are achieved via a hierarchical spatial organization of PBPs, in which linker proteins play critical roles. Extensive efforts have been made to determine the 3D structures of intact PBSs, but limited progress has been made. Adir and colleagues recently crystallized two intact PBSs; however, the overall structure of the PBS remained unsolved due to random positioning of the core components in the crystal38. The density maps in previous EM studies of intact PBSs were of insufficient quality to determine the accurate docking of crystal structures of PBPs and linker proteins21,22. To improve PBS particle quality for EM studies, we isolated PBSs from different types of cyanobacteria. We found that PBSs from Anabaena 7120 yielded the best results with negative-stain EM. We also performed a cryo-EM study, but obtained poorer results due to the low contrast (possibly because of the high salt concentration in the buffer) and the preferred orientation (only the end-on view shown in vitreous ice).
In our 3D reconstruction, the structures of the PBP trimers and the two linker protein domains (Pfam00427 and Pfam01383) can be unambiguously identified in both the core cylinders and the peripheral rods (Figure 1I), including the LRC (CpcG) linker proteins that are required for the attachment of the peripheral rods to the core42. In hemidiscoidal PBSs, the core structures are organized by LCM proteins, which have two, three, and four linker domain (REP domain) repeats in so-called two, three, and five cylinder cores, respectively. The presence of C cylinders in the core of PBS from Anabaena 7120 provides two additional attachment sites for peripheral rods compared with the PBSs, which have cores of three cylinders. Furthermore, two additional CpcG linkers are utilized for rod attachment. Our results suggest that CpcG4 is the primary rod-core linker that connects the majority of rods to the core, and CpcG1 and CpcG2 are likely involved in the attachment of additional rods. The fact that different linker proteins are required for different rod attachment indicates that the linker proteins play a direct role in rod attachment to the core.
Three important PBPs are located in the A cylinders of the PBS core, namely LCM, β18.5 and αAP-B, which are encoded by apcE, apcF, and apcD, respectively. The density map (Figure 2) and physical position of the chromophores suggest that LCM/ApcE and β18.5/ApcF in disk 3 of cylinder A are responsible for the protrusion of this trimer at the bottom of the PBS core. This protrusion fits well with the hole on the cytoplasmic side of PSII and forms a tight interaction between PBS and PSII, which is likely required for an efficient energy transfer from PBS to PSII. The tight association between PBS and PSII agrees with a recent report that demonstrated the formation of the PBS-PSII-PSI megacomplex in vivo via protein cross-linking and mass spectrometry analyses27. It should also be noted that EM imaging showed peripheral rods attached to the bottom of the cores of PBSs isolated from the ΔapcF mutant (Figure 4C). ApcF (18.5) is critical for energy transfer from PBS to PSII41,43 and has a C-terminal extension compared with ApcB (βAPC). This intriguing observation suggests that ApcB may not fully substitute for ApcF function in the PBS core.
The EM structures and the pseudo-atomic model were used to map the global distribution of chromophores and thereby provide the basis for understanding energy transfer from the rods to the core and from PBS to PSII. Many questions remain to be answered. For instance, the position of ApcD, the terminal emitter for energy transfer to PSI43, could not be unambiguously identified in our PBS model (Supplementary information, Figure S9), and further study is needed to determine whether ApcD is located on disc 1 or 444.
Materials and Methods
PBSs preparation
Anabaena sp. PCC 7120 was cultured in BG11 liquid media, bubbled with air containing 1% CO2. Cells (3-5 g) were collected and every 1 g pellet of cells was resuspended in 8 ml buffer C (0.8 M K/Na-PO4, pH 8.0). Samples were disrupted through a French Pressure at 20 000 psi at 4 °C for three times. Undisrupted cells and debris were removed by centrifugation at 3 300 g for 10 min. Triton X-100 (20% (w/v)) was added to the supernatant to a final concentration of 2%. The mixture was gently shaked at room temperature for 30 min, and then centrifuged. The supernatant containing PBSs was loaded to the top of sucrose density gradients. The sucrose gradients were made from buffer C by adding sucrose to these concentrations: 0.25, 0.4, 0.55, 0.7, 0.85, and 1.0 M. Sucrose solution at each concentration was then manually added to a centrifuge tube layer by layer with volume of 3, 3, 3, 4, 4, and 5 ml, respectively. The samples were centrifuged at 35 000 rpm. for 6 h using a Ti-70 rotor on Beckman Optima L-80XP centrifuge and resulted in a visible bands of PBSs, with band 1 as the main layer of intact PBSs (Supplementary information, Figure S2A).
Negative-stain EM
PBSs of Anabaena sp. PCC 7120 (4 μl, 500 μg/ml) separated from the sucrose gradient centrifugation were applied to glow-discharged EM grids with continuous carbon film. After 30-s incubation, excess sample was blotted, and the grid was washed five to six times with buffer C to remove sucrose in the sample. After washing, the remaining liquid was removed with the filter paper. Then, 2% (w/v) uranyl acetate was added onto the grid from one side continuously, and was removed with filter paper from the other side simultaneously. This process removed the deposition formed by phosphates and uranyl acetate immediately after its formation. Deposition was thoroughly removed from the grid, and the stain was left on the grid for 30 s. Excess stains were removed and the grid was dried for EM imaging.
Samples were examined on a Tecnai F20 electron microscope (FEI) at 200 kV. Images were recorded on a Gatan 4k × 4k CCD camera at a nominal magnification of 50 000, yielding a pixel size of 2.23 Å at the sample level. Defocuses were set at about −1 200 nm.
Data analysis and 3D reconstruction
For data analysis, particles were manually selected using the EMAN45 program boxer. In total, 32 966 particles were picked from 2 108 micrographs. The 2D classification was done using the EMAN program refine2d.py. Rotational cross-correlation coefficient values (Supplementary information, Figure S3B) were calculated by 'CC C' program in SPIDER46. The initial PBS model was determined using common lines method (startAny program in EMAN).
For reconstruction of the structure of the PBS-PSII complex, two initial models were used for multirefinement in EMAN using the multirefine program. The first initial model was PBS reconstructed with all the particles, while the second one was a combination of the first initial model with a PSII density map transformed from its crystal structure and added up to the bottom of PBS randomly. Both initial models were low-pass filtered to 60 Å before multirefinement. Finally, 2 363 particles were classified to the second group of PBS-PSII complex, reaching a resolution of ∼34 Å by FSC = 0.5 criterion.
Docking analysis and measurements of distances, axes, and angles
For docking analyses, the crystal structure of the APC trimer (PDB code: 1B33) was fitted onto the density map using 'fit in map' program in UCSF Chimera47. Map segmentation and visualization were performed using Chimera.
Centroids of each trimer and axes of each cylinder (Supplementary information, Figure S4) were determined by 'centroid' and 'define axis' commands in Chimera, respectively, based on the docked crystal structures. Distances of pigment with pigment were measures by 'distance' command in Chimera. Angles (Figure 1B and Supplementary information, Figure S4C and S4D) were measured by 'angle' command in Chimera.
Construction of PBS mutants
In Anabaena sp. PCC 7120, four LRC genes are located at the 3′ portion of the cpc operon, tandemly in the sequence cpcG1-4. To construct ΔcpcG1/2/4 mutant, cpcG flank sequences were amplified by PCRs, using the total genomic DNA as template. The 3.2-kb upper stream sequence was amplified by primer pair PN-up and PC-up (Supplementary information, Table S2), digested with XhoI and EcoRI and ligated into pBS at the same restriction sites, and resulted in pBS-Gup. The 3.3-kb downstream sequence was amplified by PN-down and PC-down, digested with NotI and SacI and ligated to pBS-Gup at the same restriction sites, and resulted in PBS-Gup-Gdown. A cartridge of Emr was amplified by PN-EmS and PC-Em, digested with EcoRI and NotI, and ligated into pBS-Gup-Gdown at the same restriction sites. The result plasmid was pBS-Gup-Em-Gdown. The 7.6-kb XhoI-SacI fragment from pBS-Gup-Em-Gdown was cloned into the pRL277 plasmid for transformation of Anabaena sp. PCC 7120 by conjugation (Supplementary information, Figure S7A).
In construction of other LRC mutant strains, combinations of cpcG fragments were amplified by corresponding primers and ligated with each other and later with cartridge of Emr (Supplementary information, Table S2). The combined sequences were ligated into pBS-Gup-Gdown as former single cartridge of Emr, and later to pRL277 plasmid in the same manner as above to generate corresponding plasmids for transformation (Supplementary information, Figure S7A).
To construct apcF mutant strain, a 7.5-kb sequence including apcF gene and its flanking sequence were amplified by primer pair PN-Fup and PC-Fdown (Supplementary information, Table S2) and cloned into Promega pGEM-T easy vector to generate pTapcF-1. Primer pair PN-Fdown and PC-Fup (Supplementary information, Table S2) was used to amplify the 9.9-kb fragment by reverse PCR from pTapcF-1. A cartridge of Emr was ligated with the 9.9-kb fragment to generate pTapcF-2. The 8.4-kb XhoI-BglII fragment of pTapcF-2 was cloned into the pRL277 plasmid for transformation.
Segregation of mutants was confirmed by PCR analysis (Supplementary information, Figure S7B). The PBSs of mutant strains were prepared in the same way as wild-type ones, with differences in layer patterns and main layer positions for their PBSs.
Absorption and fluorescence spectra
Before spectra analysis, OD730 nm of all PBS samples were adjusted to 0.8 with fresh BG11 medium. The absorption spectra between 400-800 nm were acquired by Shimadzu UV-2501PC. 77K fluorescence emission and excitation spectra were acquired with PTI fluorescence spectrofluorometer. PBS samples for tests were in thermal equilibrium with liquid nitrogen in a Dewar vacuum flask.
Immunoblotting
PBSs from sucrose density gradient centrifugation were not suitable for direct use in SDS-PAGE because of high concentration of phosphate. Desalting was performed with ultrafiltration using Milipore Amicon Ultra centrifugal filters. The desalted samples were measured by Bradford method and then diluted to 1 mg/ml. Approximately 4 μg of sample was loaded to each lane and resolved by SDS-PAGE. The samples were probed with antibodies against D1, D2, PsbO, and PsaA from Arabidopsis thaliana. All antibodies are rabbit polyclonal antibodies against recombinant proteins expressed from E.coli.
Immunogold labeling EM
Collodion/carbon coated grids (400 meshes) were glow discharged and floated on drops of PBS in buffer C for 1-2 min. PBS was fixed by floating the grids on drops of 4% paraformaldehyde for 10-15 min. After washing with 200 μl buffer C, PBS bounded to the grids was incubated with the primary antibody (antibodies against D1 subunit of PS II in Arabidopsis thaliana, which also recognize cyanobacteria D148,49,50) in buffer C for 25-30 min at the room temperature. The grids were then washed by six drops of 0.01 M PBS (pH 7.4) and incubated with 10 nm immunegold labeled second antibody (anti-rabbit IgG) for 25-30 min at the room temperature. The grids were washed with 0.01 M PBS (pH7.4) and negative stained with 2% uranyl acetate.
Data deposition
Electron density maps have been submitted to the Electron Microscopy Data Bank (http://www.emdatabank.org/) under accession numbers 2821 and 2822.
Acknowledgments
We thank Jianlin Lei for setting up a semi-automated program for data collection, Lixin Zhang for providing antibodies against D1 and D2, Ying-Chun Hu for technical assistance in immunogold labeling experiments, and Hong-Wei Wang and Da-Neng Wang for critical reading of the manuscript. We acknowledge the Tsinghua University Branch of China National Center for Protein Sciences Beijing for providing EM resources. This work was supported by the National Basic Research Program of China (2011CB910500 and 2010CB833706 to SFS, and 2009CB118500 and 2015CB150100 to JZ) and the National Natural Science Foundation of China (31230016 and 31370717 to SFS).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Structural elements and available crystal structures of subunits in the PBS from Anabaena sp. strain PCC 7120.
Sample preparation of intact PBSs from Anabaena sp. strain PCC 7120.
Negative staining of intact PBS from Anabaena sp. strain PCC 7120.
Coordinate system building and the orientation of each cylinder.
Association of PSII to the PBS.
Comparison of hexamers.
Molecular reconstitution and purification of LRC mutated PBSs.
Pigment distribution and energy transfer pathway.
Two models of PSI in the complex PBS-PSII-PSI.
Cross-correlation coefficient (CCC) of crystal structures and the EM density map.
Primers used in this study.
Structures of the intact PBS and the PBS-PSII complex.
References
- MacColl R. Cyanobacterial phycobilisomes. J Struct Biol. 1998;124:311–334. doi: 10.1006/jsbi.1998.4062. [DOI] [PubMed] [Google Scholar]
- Sidler WA.Phycobilisome and phycobiliprotein structures. In: Bryant DA, ed. The Molecular Biology of Cyanobacteria. The Netherlands: Kluwer Academic Publishers, 1994:139–216.
- Adir N. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res. 2005;85:15–32. doi: 10.1007/s11120-004-2143-y. [DOI] [PubMed] [Google Scholar]
- Glauser M, Bryant DA, Frank G, et al. Phycobilisome structure in the cyanobacteria Mastigocladus laminosus and Anabaena sp. PCC 7120. Eur J Biochem. 1992;205:907–915. doi: 10.1111/j.1432-1033.1992.tb16857.x. [DOI] [PubMed] [Google Scholar]
- Ducret A, Sidler W, Wehrli E, Frank G, Zuber H. Isolation, characterization and electron microscopy analysis of a hemidiscoidal phycobilisome type from the cyanobacterium Anabaena sp. PCC 7120. Eur J Biochem. 1996;236:1010–1024. doi: 10.1111/j.1432-1033.1996.01010.x. [DOI] [PubMed] [Google Scholar]
- Ducret A, Muller SA, Goldie KN, et al. Reconstitution, characterisation and mass analysis of the pentacylindrical allophycocyanin core complex from the cyanobacterium Anabaena sp. PCC 7120. J Mol Biol. 1998;278:369–388. doi: 10.1006/jmbi.1998.1678. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Toole CM. A model for early events in the assembly pathway of cyanobacterial phycobilisomes. Mol Microbiol. 1998;30:467–474. doi: 10.1046/j.1365-2958.1998.01081.x. [DOI] [PubMed] [Google Scholar]
- Schirmer T, Bode W, Huber R. Refined three-dimensional structures of two cyanobacterial C-phycocyanins at 2.1 and 2.5 Å resolution. A common principle of phycobilin-protein interaction. J Mol Biol. 1987;196:677–695. doi: 10.1016/0022-2836(87)90040-4. [DOI] [PubMed] [Google Scholar]
- Schirmer T, Huber R, Schneider M, Bode W, Miller M, Hackert ML. Crystal structure analysis and refinement at 2.5 Å of hexameric C-phycocyanin from the cyanobacterium Agmenellum quadruplicatum. The molecular model and its implications for light-harvesting. J Mol B. 1986;188:651–676. doi: 10.1016/s0022-2836(86)80013-4. [DOI] [PubMed] [Google Scholar]
- Reuter W, Wiegand G, Huber R, Than ME. Structural analysis at 2.2 Å of orthorhombic crystals presents the asymmetry of the allophycocyanin-linker complex, AP.LC7.8, from phycobilisomes of Mastigocladus laminosus. Proc Natl Acad Sci USA. 1999;96:1363–1368. doi: 10.1073/pnas.96.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Qin S, Zhao F, Zhang X, Tang X. Phycobilisomes linker family in cyanobacterial genomes: divergence and evolution. Int J Biol Sci. 2007;3:434–445. doi: 10.7150/ijbs.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk KE, Harrop SJ, Jankova L, et al. Evolution of a light-harvesting protein by addition of new subunits and rearrangement of conserved elements: crystal structure of a cryptophyte phycoerythrin at 1.63-Å resolution. Proc Natl Acad Sci USA. 1999;96:8901–8906. doi: 10.1073/pnas.96.16.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter S, Hiller RG, Wrench PM, Welte W, Diederichs K. Crystal structure of a phycourobilin-containing phycoerythrin at 1.90-Å resolution. J Struct Biol. 1999;126:86–97. doi: 10.1006/jsbi.1999.4106. [DOI] [PubMed] [Google Scholar]
- Nield J, Rizkallah PJ, Barber J, Chayen NE. The 1.45 Å three-dimensional structure of C-phycocyanin from the thermophilic cyanobacterium Synechococcus elongatus. J Struct Biol. 2003;141:149–155. doi: 10.1016/s1047-8477(02)00609-3. [DOI] [PubMed] [Google Scholar]
- David L, Marx A, Adir N. High-resolution crystal structures of trimeric and rod phycocyanin. J Mol Biol. 2011;405:201–213. doi: 10.1016/j.jmb.2010.10.036. [DOI] [PubMed] [Google Scholar]
- Stec B, Troxler RF, Teeter MM. Crystal structure of C-phycocyanin from Cyanidium caldarium provides a new perspective on phycobilisome assembly. Biophys J. 1999;76:2912–2921. doi: 10.1016/S0006-3495(99)77446-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A, Adir N. Allophycocyanin and phycocyanin crystal structures reveal facets of phycobilisome assembly. Biochim Biophys Acta. 2013;1827:311–318. doi: 10.1016/j.bbabio.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Liu JY, Jiang T, Zhang JP, Liang DC. Crystal structure of allophycocyanin from red algae Porphyra yezoensis at 2.2-Å resolution. J Biol Chem. 1999;274:16945–16952. doi: 10.1074/jbc.274.24.16945. [DOI] [PubMed] [Google Scholar]
- Murray JW, Maghlaoui K, Barber J. The structure of allophycocyanin from Thermosynechococcus elongatus at 3.5 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:998–1002. doi: 10.1107/S1744309107050920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang N, Wei TD, et al. Crystal structure of the N-terminal domain of linker L(R) and the assembly of cyanobacterial phycobilisome rods. Mol Microbiol. 2011;82:698–705. doi: 10.1111/j.1365-2958.2011.07844.x. [DOI] [PubMed] [Google Scholar]
- Arteni AA, Ajlani G, Boekema EJ. Structural organization of phycobilisomes from Synechocystis sp. strain PCC6803 and their interaction with the membrane. Biochim Biophys Acta. 2009;1787:272–279. doi: 10.1016/j.bbabio.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Yi ZW, Huang H, Kuang TY, Sui SF. Three-dimensional architecture of phycobilisomes from Nostoc flagelliforme revealed by single particle electron microscopy. FEBS Lett. 2005;579:3569–3573. doi: 10.1016/j.febslet.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Murata N. Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim Biophys Acta. 1969;172:242–251. doi: 10.1016/0005-2728(69)90067-x. [DOI] [PubMed] [Google Scholar]
- Fork DC, Satoh K. State I-state II transitions in the thermophilic blue-green alga (cyanobacterium) Synechococcus lividus. Photochem Photobiol. 1983;37:421–427. [Google Scholar]
- Mohanty P, Govindjee Light-induced changes in the fluorescence yield of chlorophyll a in Anacystis nidulans II. The fast changes and the effect of photosynthetic inhibitors on both the fast and slow fluorescence induction. Plant Cell Physiol. 1973;14:611–629. [Google Scholar]
- Mullineaux CW, Allen JF. State 1-State 2 transitions in the cyanobacterium Synechococcus 6301 are controlled by the redox state of electron carriers between Photosystems I and II. Photosynth Res. 1990;23:297–311. doi: 10.1007/BF00034860. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang H, Niedzwiedzki DM, et al. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science. 2013;342:1104–1107. doi: 10.1126/science.1242321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Wasmann C, Staehelin LA. Structure of the thylakoids and envelope membranes of the cyanelles of Cyanophora paradoxa. Plant Physiol. 1983;71:409–419. doi: 10.1104/pp.71.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux CW. Phycobilisome-reaction center interaction in cyanobacteria. Photosynth Res. 2008;95:175–182. doi: 10.1007/s11120-007-9249-y. [DOI] [PubMed] [Google Scholar]
- Barber J, Morris EP, da Fonseca PC. Interaction of the allophycocyanin core complex with photosystem II. Photochem Photobiol Sci. 2003;2:536–541. doi: 10.1039/b300063j. [DOI] [PubMed] [Google Scholar]
- Chereskin BM, Clement-Metral JD, Gantt E. Characterization of a purified photosystem II-phycobilisome particle preparation from Porphyridium cruentum. Plant Physiol. 1985;77:626–629. doi: 10.1104/pp.77.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt E, Clement-Metral JD, Chereskin BM.Methods in Enzymology. San Diego: Academic Press, 1988.
- Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- Ajlani G, Vernotte C. Deletion of the PB-loop in the L(CM) subunit does not affect phycobilisome assembly or energy transfer functions in the cyanobacterium Synechocystis sp. PCC6714. Eur J Biochem. 1998;257:154–159. doi: 10.1046/j.1432-1327.1998.2570154.x. [DOI] [PubMed] [Google Scholar]
- Capuano V, Braux AS, Tandeau de Marsac N, Houmard J. The “anchor polypeptide” of cyanobacterial phycobilisomes. Molecular characterization of the Synechococcus sp. PCC 6301 apce gene. J Biol Chem. 1991;266:7239–7247. [PubMed] [Google Scholar]
- Satyanarayana L, Suresh CG, Patel A, Mishra S, Ghosh PK. X-ray crystallographic studies on C-phycocyanins from cyanobacteria from different habitats: marine and freshwater. Acta Crystallogr Sec F Struct Biol Cryst Commun. 2005;61:844–847. doi: 10.1107/S1744309105025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Prado M, Arteni AA, Elmlund DA, Blankenship RE, Adir N. Structural studies show energy transfer within stabilized phycobilisomes independent of the mode of rod-core assembly. Biochim Biophys Acta. 2014;1837:385–395. doi: 10.1016/j.bbabio.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Semchonok DA, Webber-Birungi MT, et al. Attachment of phycobilisomes in an antenna-photosystem I supercomplex of cyanobacteria. Proc Natl Acad Sci USA. 2014;111:2512–2517. doi: 10.1073/pnas.1320599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbi M, Rosinski J, Siegelman HW, Sutherland JC. Cyanobacterial phycobilisomes: Selective dissociation monitored by fluorescence and circular dichroism. Proc Natl Acad Sci USA. 1980;77:1961–1965. doi: 10.1073/pnas.77.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindt YM, Zhou J, Bryant DA, Sauer K. Core mutations of Synechococcus sp. PCC 7002 phycobilisomes: a spectroscopic study. J Photochem Photobiol B. 1992;15:75–89. doi: 10.1016/1011-1344(92)87007-v. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Stirewalt VL, Glauser M, Frank G, Sidler W, Zuber H. A small multigene family encodes the rod-core linker polypeptides of Anabaena sp. PCC7120 phycobilisomes. Gene. 1991;107:91–99. doi: 10.1016/0378-1119(91)90301-q. [DOI] [PubMed] [Google Scholar]
- Dong C, Tang A, Zhao J, Mullineaux CW, Shen G, Bryant DA. ApcD is necessary for efficient energy transfer from phycobilisomes to photosystem I and helps to prevent photoinhibition in the cyanobacterium Synechococcus sp. PCC 7002. Biochim Biophys Acta. 2009;1787:1122–1128. doi: 10.1016/j.bbabio.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Bryant DA.Genetic analysis of phycobilisome biosynthesis, assembly, structure, and function in the Cyanobacterium Synechococcus sp. PCC 7002In: Stevens SE, Bryant DA. ed. Light-Energy Transduction in Photosynthesis: HigherPlant and Bacterial Models. Rockville, Maryland, USA: American Society of Plant Physiologists 1988:62–90.
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Tichy M, Lupinkova L, Sicora C, et al. Synechocystis 6803 mutants expressing distinct forms of the Photosystem II D1 protein from Synechococcus 7942: relationship between the psbA coding region and sensitivity to visible and UV-B radiation. Biochim Biophys Acta. 2003;1605:55–66. doi: 10.1016/s0005-2728(03)00064-1. [DOI] [PubMed] [Google Scholar]
- Bajkan S, Varadi G, Balogh M, et al. Conserved structure of the chloroplast-DNA encoded D1 protein is essential for effective photoprotection via non-photochemical thermal dissipation in higher plants. Mol Genet Genomics. 2010;284:55–63. doi: 10.1007/s00438-010-0549-0. [DOI] [PubMed] [Google Scholar]
- Toth T, Zsiros O, Kis M, Garab G, Kovacs L. Cadmium exerts its toxic effects on photosynthesis via a cascade mechanism in the cyanobacterium, Synechocystis PCC 6803. Plant Cell Environ. 2012;35:2075–2086. doi: 10.1111/j.1365-3040.2012.02537.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural elements and available crystal structures of subunits in the PBS from Anabaena sp. strain PCC 7120.
Sample preparation of intact PBSs from Anabaena sp. strain PCC 7120.
Negative staining of intact PBS from Anabaena sp. strain PCC 7120.
Coordinate system building and the orientation of each cylinder.
Association of PSII to the PBS.
Comparison of hexamers.
Molecular reconstitution and purification of LRC mutated PBSs.
Pigment distribution and energy transfer pathway.
Two models of PSI in the complex PBS-PSII-PSI.
Cross-correlation coefficient (CCC) of crystal structures and the EM density map.
Primers used in this study.
Structures of the intact PBS and the PBS-PSII complex.