Characterization of maize autophagy through detailed transcriptome studies and phenotypic and 15N-partitioning analyses of atg12 mutants reveal a central role for autophagy in nitrogen remobilization.
Abstract
Autophagy is a primary route for nutrient recycling in plants by which superfluous or damaged cytoplasmic material and organelles are encapsulated and delivered to the vacuole for breakdown. Central to autophagy is a conjugation pathway that attaches AUTOPHAGY-RELATED8 (ATG8) to phosphatidylethanolamine, which then coats emerging autophagic membranes and helps with cargo recruitment, vesicle enclosure, and subsequent vesicle docking with the tonoplast. A key component in ATG8 function is ATG12, which promotes lipidation upon its attachment to ATG5. Here, we fully defined the maize (Zea mays) ATG system transcriptionally and characterized it genetically through atg12 mutants that block ATG8 modification. atg12 plants have compromised autophagic transport as determined by localization of a YFP-ATG8 reporter and its vacuolar cleavage during nitrogen or fixed-carbon starvation. Phenotypic analyses showed that atg12 plants are phenotypically normal and fertile when grown under nutrient-rich conditions. However, when nitrogen-starved, seedling growth is severely arrested, and as the plants mature, they show enhanced leaf senescence and stunted ear development. Nitrogen partitioning studies revealed that remobilization is impaired in atg12 plants, which significantly decreases seed yield and nitrogen-harvest index. Together, our studies demonstrate that autophagy, while nonessential, becomes critical during nitrogen stress and severely impacts maize productivity under suboptimal field conditions.
INTRODUCTION
Available nitrogen (N) is a major external factor influencing plant growth, and its supply as fertilizer is the largest single cost in crop management with 114 million metric tons expected to be used globally to augment production in 2015 (http://faostat3.fao.org/). As might be expected, N utilization involves sophisticated mechanisms for uptake, translocation, assimilation, and remobilization, which are tightly associated with the plant’s life cycle (reviewed in Chardon et al., 2012; Xu et al., 2012). During early vegetative stages, developing roots and leaves function as sinks for the uptake of inorganic N and its subsequent incorporation into amino acids via the nitrate/nitrite reductases and the ammonia assimilation pathways. The accumulating amino acids then provide building blocks for synthesizing the myriad of enzymes and structural and regulatory proteins required for photosynthesis, growth, and other metabolic activities. During the latter phases of plant development, leaf proteins are catabolized and the released amino acids and other nitrogenous compounds are exported to reproductive and storage organs (Masclaux-Daubresse et al., 2008). In cereals, 50 to 90% of seed N is derived from these remobilized stores, a process that is strongly dependent on genotype (Kichey et al., 2007). As examples, 20 to 50 g of N is required by maize (Zea mays), wheat (Triticum aestivum), and rice (Oryza sativa) to produce 1 kg of grain (Robertson and Vitousek, 2009).
As a major source of remobilized N, leaf proteins, especially those in the chloroplasts, are rapidly degraded before the onset of reproduction, which culminates in the export of most available N during the final senescence program of the leaf (Patrick and Offler, 2001). Although the routes for protein turnover in aging leaves are not fully understood, plastid resident proteases, senescence-associated vacuoles (SAVs), and macroautophagy (hereafter referred to as autophagy) are considered to be three important routes (Otegui et al., 2005; Ishida et al., 2008; Wada et al., 2009; Roberts et al., 2012). As examples, the transcript levels of multiple chloroplast proteases and various components of the autophagic machinery are known to rise early during leaf senescence in Arabidopsis thaliana and various cereals (Parrott et al., 2007; Liu et al., 2008; Phillips et al., 2008; Ruuska et al., 2008; Breeze et al., 2011; Avila-Ospina et al., 2014; Penfold and Buchanan-Wollaston, 2014) and is concomitant with the export of major protein stores like Rubisco and glutamine synthetase into SAVs with intense proteolytic activity (Otegui et al., 2005; Martínez et al., 2008). In addition, direct connections between autophagy and the turnover of chloroplast and mitochondrial constituents was provided by the study of autophagy mutants during leaf senescence (Ishida et al., 2008; Wada et al., 2009; Izumi et al., 2010; Li et al., 2014). For chloroplast turnover in particular, a special type of autophagy has been proposed whereby Rubisco-containing bodies bud from stromule projections and are then delivered to vacuoles for breakdown (Ishida et al., 2008; Spitzer et al., 2015).
Autophagic recycling is accomplished by the sequestration of cytoplasmic material into a double membrane-bound compartment called the autophagosome. Its outer membrane fuses with the tonoplast to release the inner vesicle as an autophagic body into the vacuolar lumen for breakdown. Studies, first with yeast and subsequently in plants and animals, defined a suite of autophagy-related (ATG) proteins that direct the process in response to internal and external cues (Li and Vierstra, 2012; Liu and Bassham, 2012; Feng et al., 2014). Included are factors that help recruit membranes to the emerging cup-shaped phagophore, choose appropriate cargo for capture, close phagophores to form autophagosomes, deliver the vesicles to the vacuole (lysosome in animals), and degrade the resulting autophagic bodies. Central components include the ATG1 kinase complex that responds to upstream nutritional cues provided by TOR and other sensor kinases, the ATG2/9/18 transmembrane complex that likely supplies membranes to the phagophore, the ATG6/Vacuolar Protein Sorting-Associated Protein 34 (VPS34)/ATG14/VPS15 phosphatidylinositol-3 (PI3) kinase complex that assists with vesicle nucleation, and the ATG8/ATG12 conjugation pathway that promotes cargo capture, vesicle expansion and closure, and fusion of autophagosomes with the vacuole. Cargo includes unwanted or damaged mitochondria (mitophagy; Li et al., 2014), chloroplasts (chlorophagy; Ishida et al., 2008; Wada et al., 2009), peroxisomes (pexophagy; Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013), large protein complexes such as ribosomes (ribophagy; Hillwig et al., 2011) and proteasomes (proteaphagy; Marshall et al., 2015), insoluble protein aggregates that become toxic if not eliminated (aggrephagy; Zhou et al., 2013), and even invading pathogens (xenophagy; Gutierrez et al., 2004; Nakagawa et al., 2004). Bulk cytoplasm can also be engulfed indiscriminately should nutrient supply become substantially limiting.
The ubiquitin-fold proteins ATG8 and ATG12 are signature components of the ATG autophagic system (Li and Vierstra, 2012; Ohsumi, 2014). Through conjugation cascades analogous to those used for ubiquitylation, ATG8 and ATG12 are activated by a common ATP-dependent E1 activating enzyme, ATG7, and subsequently transferred to their respective E2 conjugating enzymes, ATG3 and ATG10. Activated ATG12 is donated to the ATG5 protein and becomes covalently attached via an isopeptide bond between its C-terminal glycine and a conserved lysine in ATG5. After associating with ATG16, the ATG12-ATG5 adduct provides the ligase activity that couples the C-terminal glycine from activated ATG8 to phosphatidylethanolamine (PE). This ATG8-PE adduct then coats expanding phagophores and serves as a docking platform for factors that promote vesicle closure and tonoplast fusion, as well as for numerous autophagy receptors that recruit appropriate cargo (Li and Vierstra, 2012; Klionsky and Schulman, 2014; Rogov et al., 2014). These receptors have separate recognition sites for specific cargo and ATG8-PE, using a short ATG8-interacting motif (AIM) for ATG8 binding. Included are plant and animal Next to BRCA1 Gene 1 Protein (NBR1), mammalian p62/SEQUESTOSOME1, and yeast (Saccharomyces cerevisiae) Cue5/Tollip proteins that bind ubiquitylated aggregates, yeast ATG30 and ATG36 for pexophagy, mammalian BCL2/Adenovirus E1B 19-kD-Interacting Protein 3 for mitophagy, and Arabidopsis Tryptophan-rich Sensory Protein, ATG8-Interacting Protein 1, and Regulatory Particle non-ATPase 10 (RPN10) that identify free porphyrins, chloroplast proteins, and inactivated proteasomes, respectively (Svenning et al., 2011; Vanhee et al., 2011; Zientara-Rytter et al., 2011; Liu et al., 2012; Michaeli et al., 2014; Rogov et al., 2014; Marshall et al., 2015).
Genetic analyses with yeast, mammalian cells, and Arabidopsis have demonstrated that autophagy is the major contributor to cellular housekeeping and recycling and is thus essential for alleviating nutrient stress (Li and Vierstra, 2012; Liu and Bassham, 2012; Ohsumi, 2014). For Arabidopsis in particular, most autophagy-associated loci are transcriptionally upregulated by nutrient starvation and during the senescence of various tissues, and mutants abrogating the system are hypersensitive to N- and fixed-carbon (C)-limiting conditions and display accelerated senescence even under nutrient-rich conditions (Doelling et al., 2002; Chung et al., 2010; Thomas, 2013). As a consequence, Arabidopsis autophagy mutants have reduced fecundity that results in lower seed yield and reduced seed N (Doelling et al., 2002; Guiboileau et al., 2012). In addition, autophagy is activated in response to other extra- or intracellular stress signals, such as endoplasmic reticulum stress, heat shock, and pathogen infection, presumably to clear abnormal/dysfunctional organelles and proteins that accumulate (Hayward and Dinesh-Kumar, 2011; Liu et al., 2012; Zhou et al., 2013). For rice, the ATG system has also been implicated in pollen development through control(s) on tapetal lipid metabolism (Kurusu et al., 2014).
Given that autophagy likely affects many important agronomic processes, including nutrient remobilization from senescing leaves into seeds and other storage organs, pathogen defense, and responses to various abiotic challenges (Li and Vierstra, 2012; Liu and Bassham, 2012; Avila-Ospina et al., 2014), it should have a major impact on crop productivity, especially under suboptimal field conditions. While our understanding of how autophagy contributes to plant physiology has been largely gained from studies with Arabidopsis, defining its importance to crops is now needed. Previously, we described the maize ATG8/12 conjugation pathways and showed that the transcript levels of maize Atg genes and the lipidation status of ATG8 are tightly regulated during development and in response to nutrient availability (Chung et al., 2009). Here, we more fully characterized the maize ATG system through detailed transcriptome analysis, the exploitation of a fluorescent ATG8 reporter to track autophagic vesicles, and phenotypic and 15N partitioning studies with atg12 mutants to show that autophagy plays a central role in maize N utilization. Given this impact, controlling nutrient recycling by altering the timing and strength of autophagy might offer novel strategies to improve N utilization by maize and other crops.
RESULTS
Genome-Wide Identification of Atg Genes in Maize
Prior studies by Chung et al. (2009) defined the maize genomic loci that encode components of the ATG8 and ATG12 conjugation cascades, which culminate in the formation of the ATG8-PE adduct. Here, we extended this description to predict a number of upstream complexes/factors using BLAST searches against the maize B73 inbred genome (http://www.maizegdb.org/) combined with relevant Arabidopsis queries (Li and Vierstra, 2012). By aligning cDNA, EST, and/or RNA-seq data (see below) with the complete B73 genomic sequence, the exact gene models based on the most dominant transcripts were built (Figure 1). Included in the list were 13 genes encoding components of the ATG1 kinase complex (Atg1a, b, c, and t, Atg11a and b, Atg13a-f, and Atg101), 12 for the ATG9/2/18 complex (Atg2, Atg9, and Atg18a-j), five components of the PI3 kinase complex (Atg6a and b, Vps34, and Vps15a and b), and two for the ubiquitin cargo receptor NBR1 (Nbr1a and b), which can now be added to the 12 loci described previously as encoding factors involved in ATG8/ATG12 conjugation (Atg3, Atg4a and b, Atg5, Atg7, Atg8a-e, Atg10, and Atg12). The only uncertainties were Atg18i, Atg18j, and Nbr1b, whose gene models could not yet be supported by mRNA sequence information. In the absence of transcripts, the intron/exon junctions and coding regions for these three loci were predicted from nucleotide alignments with available paralogs (Figure 1). We also discovered a likely ortholog of ATG16, which constitutes part of the ATG8 ligation complex (Figure 1). Plant versions of Atg16 had escaped our prior searches due to low sequence homology with their yeast counterpart; the Arabidopsis (At5g50230) and maize (GRMZM2G078252) loci were eventually discovered by alignments with human and mouse ATG16L1.
Figure 1.
Annotation of Genes Encoding Upstream Components of the Maize ATG8-Mediated Autophagic System.
Included are genes encoding components from the ATG1 kinase complex, the ATG9/2/18 complex, the PI3 kinase complex, the ubiquitin receptor NBR1 that helps recognize ubiquitylated substrates during selective autophagy, and ATG16 that associates with the ATG12-ATG5 conjugate and promotes formation of the ATG8-PE adduct. The amino acid sequence length and maize genome GRMZM accession number of each protein/gene are on the right. Colored and gray boxes denote coding regions and untranslated regions, respectively. Lines indicate introns. The conserved lysine (K) within the ATP hydrolysis site in the ATG1 kinase family is indicated by arrowheads. The kinase domain (KD) and regulatory domain (RD) in ATG1, the ATG11 domain in ATG11, and the Phox and Bem1p (PB1), ZZ-type zinc finger (ZZ), four tryptophan (FW), and ubiquitin-associated (UBA) domains in NBR1 are shown. See Supplemental Table 1 for more information on each gene.
As shown in Supplemental Table 1 online, the identified maize Atg genes are 30 to 50% identical and 40 to 70% similar to their Arabidopsis orthologs except for the highly conserved ATG8 and ATG12 proteins, with 85%/95% identity/similarity. Whereas Arabidopsis expresses two of the nine ATG8 isoforms (ATG8h and i) with the C-terminal glycine required for conjugation exposed, and thus can act without cleavage by the ATG4 protease, all five isoforms of maize ATG8 are capped with additional residues beyond this glycine and thus require ATG4 processing. Like Arabidopsis, several maize ATG components are encoded by single genes (Atg2, Atg3, Atg5, Atg7, Atg9, Atg10, Atg12, Atg101, and Vps34), which make them practical targets for reverse genetics, whereas others are encoded by small gene families that imply redundancy/subfunctionalization (Atg1, Atg8, Atg13, and Atg18) (Figure 1; Supplemental Table 1). For the Atg1 gene family in particular, the Atg1t paralog in maize and other plants encodes a truncated protein containing the complete kinase domain but missing the C-terminal regulatory domain, which is relevant to ATG1 function in other organisms by fostering interactions with ATG13 and ATG11/FIP200 (Cheong et al., 2008; Hara and Mizushima, 2009). Strikingly, the maize ATG1t protein terminates close to or at the same amino acid position as those found in Arabidopsis and other plant species (Suttangkakul et al., 2011), implying that this conserved truncation predates the seed plant lineage and is likely relevant to a novel activity for this isoform.
The Atg18 gene family in maize and likely in other plants was also unusual. In line with Atg18j being a possible nontranscribed pseudogene, it is also unique by being devoid of introns, suggesting that it arose by retrotransposition of an Atg18 mRNA. The Atg18 family could also be grouped by the size of the encoded protein. Whereas most paralogs (Atg18a-e and Atg18i-j) encode relatively short proteins of ∼370 to 450 amino acids that align reasonably well with their Arabidopsis, yeast, and animal orthologs, Atg18f-h encode proteins more than double this length with the extra amino acids introduced by novel regions interspersed among the conserved regions and by a long C-terminal extension (Figure 1). Arabidopsis and other plants, but not fungi or animals, express similarly large ATG18 isoforms (Xiong et al., 2005), suggesting that they have unique plant-specific function(s).
RNA-seq Analysis of Maize Atg Gene Expression
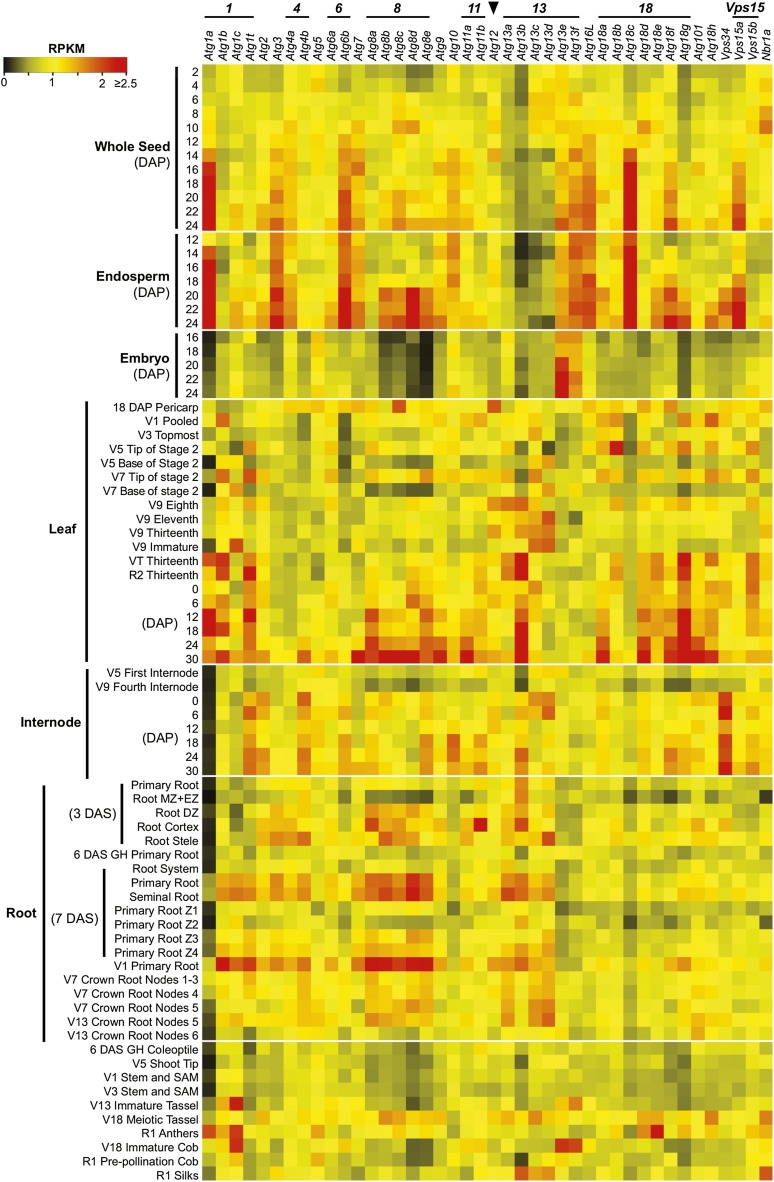
To investigate the spatial and temporal expression patterns of Atg genes in maize, we quantified transcript abundance from an extensive RNA-seq data set generated from a broad range of tissues (80 total), including whole seeds, endosperm, embryo, leaf, root, stem, internode, anther, tassel, silk, and ear (Figure 2). Previous studies with Arabidopsis showed that many ATG genes are expressed ubiquitously and that their transcript levels are upregulated during senescence and by nutrient stress (Contento et al., 2004; Sláviková et al., 2005; Thompson et al., 2005; Rose et al., 2006; Breeze et al., 2011). A majority of maize Atg loci displayed a similarly widespread pattern of expression (Figure 2). Notably, many Atg transcripts were upregulated in older leaves and in the leaf tip, which represents the most mature zone of a maize leaf (30 and 27 loci, respectively). This age-related increase was subsequently confirmed by quantitative RT-PCR on select transcripts over the full set of leaves and the base, middle, and tip regions of the seventh leaf for plants beginning pollination (Supplemental Figure 1). Together, the transcriptomics imply a central role for autophagy in directing breakdown/recycling as maize leaves age and during their final senescence program. Similarly, we found that the mRNA levels for 28 Atg genes were significantly higher in the more mature zone of roots (primary root region Z4 harvested 7 d after sowing [DAS]; Figure 2) than in the younger regions that encompass the root cap (Z1), division zone (Z2), and elongation zone (Z3). Interestingly, we also detected increased transcript abundance during seed development (29 loci), which appeared to be confined to the endosperm and not embryo (e.g., four of the five Atg8 loci, Atg1a, Atg3, Atg18c, and Vps15a), strongly suggesting that autophagy participates in the maturation and death of the endosperm during seed development (Figure 2). Despite widespread coverage over numerous tissues and developmental states, no expression evidence for Atg18i, Atg18j, and Nbr1b was gathered, suggesting that they are transcriptionally inactive, although restricted expression patterns in tissues not yet examined cannot be discounted (Supplemental Table 2).
Figure 2.
Developmental and Tissue-Specific Expression Profiles of Maize Atg Genes.
RNA-seq experiments representing 80 developmentally or anatomically distinct maize samples were analyzed for autophagy-related genes based on reads per kilobase per one million reads (RPKM). Gene families encoding individual ATG factors are highlighted on the left. The arrowhead locates transcript values from the single maize Atg12 locus. The color indicates the degree of fold change: red, high; black, low. Vegetative (V1 to 18) and reproductive (R1 and 2) growth stages were defined based on the Maize Field Guide published by Iowa State University Extension (Abendroth et al., 2011). Dissected primary root: Z1, zone 1 (first centimeter of root tip); Z2, zone 2 (from end of Z1 to the point of root hair/lateral root initiation); Z3, zone 3 (lower half of differentiation zone); Z4, zone 4 (upper half of differentiation zone). DZ, differentiation zone; EZ, elongation zone; MZ, meristematic zone; SAM, shoot apical meristem. See Supplemental Table 2 for full descriptions of the tissues analyzed.
With respect to the maize Atg gene families, our RNA-seq analyses also revealed expression differences and potential tissue-specific functions for the corresponding protein isoforms. For example, Atg1a is dominantly expressed during endosperm development and in leaves but is generally less active in other tissues as compared with its paralogs (Figure 2). The Atg18 family, which helps transport lipids to the expanding autophagic membranes (Li and Vierstra, 2012; Liu and Bassham, 2012), could be subdivided into four groups based on mRNA patterns (Figure 2; Supplemental Figure 2). The first group, encompassing Atg18a, Atg18c, and Atg18g, had ubiquitous expression patterns, whereas the second group, containing Atg18e, Atg18f, and Atg18h, displayed elevated expression in the whole seed and endosperm, suggesting a role in seed development. All six genes in these two groups were also highly expressed in the leaf tip and older leaves. The third group included only Atg18b; its transcripts were most abundant in the endosperm and increased in number as this nutritive tissue matured. Atg18d by itself formed the last group: It was expressed preferentially in anthers above all other tissues, implying a highly specific role for this isoform during anther and/or pollen development. Grouping the expression patterns via the R statistical package identified several coexpression clusters that might be relevant, including Atg1a/Atg18b, Atg1b/Atg13a, Vsp15b/Atg11b, and Atg101/Atg18c, suggesting that specific isoforms of the autophagic machinery work in concert through coexpression (Supplemental Figure 3 and Supplemental Data Set 1).
The RNA-seq studies also extended our prior observations (Chung et al., 2009) that many maize Atg transcripts frequently undergo alternative splicing (26 of 41 loci; Supplemental Table 1). Most genes generated a dominant splice variant (20 loci) that accounted for >70% of total transcripts, whereas a few genes (six including three Atg1 loci) produced multiple splice variants at more equal levels. Interestingly, we also discovered that several Atg genes are alternatively spliced in tissue- and/or growth-specific patterns that could have developmental consequences (Supplemental Figure 4). For example, of the three annotated splice isoforms of Atg1b, the T01 isoform increased in abundance during endosperm maturation and as leaves aged, whereas the T02 isoform dropped in abundance during endosperm maturation. Likewise, the two splice forms of Atg1t showed reciprocal changes in abundance during endosperm development and leaf aging.
Identification of Maize Mutants Inactivating Atg12
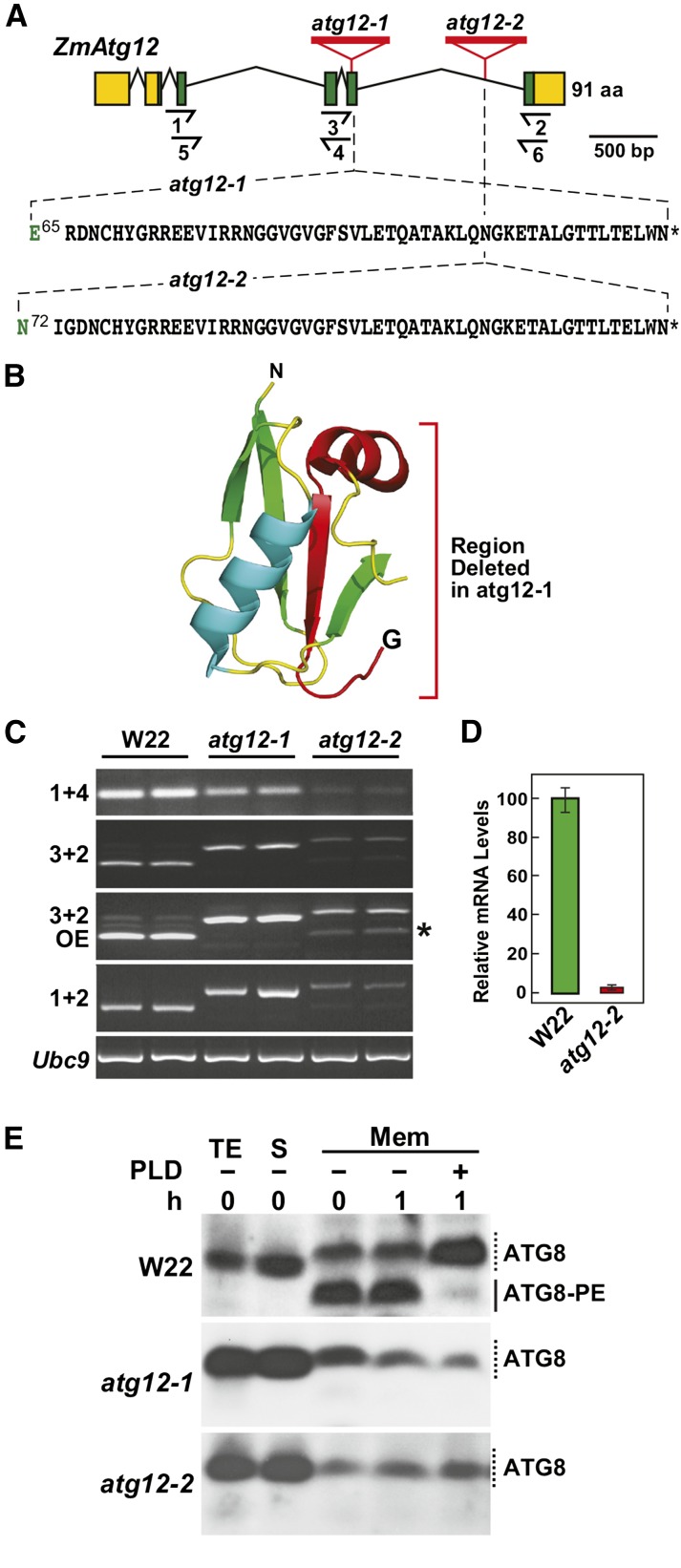
To help appreciate autophagy in crop species and its potential impact on productivity, we used the list of annotated maize autophagy genes to dissect the system via reverse genetics. From searches of the various transposon insertion populations, including the UniformMu (McCarty et al., 2005) and Ac/Ds collections (Vollbrecht et al., 2010), we identified potentially useful alleles affecting a number of Atg loci (e.g., Atg3, Atg5, Atg10, Atg12, and Nbr1a). Of relevance here are two UniformMu insertions (atg12-1 and atg12-2) impacting the single gene encoding ATG12. Sequencing genomic PCR and RT-PCR products spanning the Atg12 locus revealed that the two lines harbored a Mu element within the fifth exon and intron, respectively, which drove the accumulation of longer Mu-containing mRNAs compared with their wild-type W22 parent (Figures 3A and 3C). The atg12-1 transcript contained a 156-bp insertion after Glu-65 that encoded a random 51-residue sequence followed by a stop codon, whereas the atg12-2 transcript contained a 159-bp insertion after Asn-72 that encoded a similar 52-amino acid sequence followed by a stop codon (Figure 3A; Supplemental Figure 5). Besides introducing a long stretch of additional amino acids, the insertions also eliminated the C-terminal end if the mutant atg12 polypeptides were translated. As the absent regions bear the terminal glycine essential for conjugation of ATG12 to ATG5, the resulting atg12-1 and atg12-2 proteins should be nonfunctional when derived from these dominant transcripts (Figure 3B).
Figure 3.
Genetic Description of Maize atg12 Mutants
(A) Gene diagram of the maize Atg12 locus showing its exon/intron organization and the positions of atg12-1 and atg12-2 UniformMu insertion mutations. Lines represent introns and the green and yellow boxes indicate coding and untranslated regions, respectively. Half arrows underneath locate positions of the primers used for RT-PCR in (C) and quantitative RT-PCR in (D). Amino acids in black identify the codons appended after Glu-65 in atg12-1 and Asn-72 in atg12-2, respectively. Asterisks indicate stop codons.
(B) Three-dimensional ribbon diagram of Arabidopsis ATG12 (PDB 1WZ3). The polypeptide region deleted by the atg12-1 insertion is colored in red.
(C) RT-PCR analysis demonstrating the accumulation of longer, Mu element-containing transcripts in homozygous atg12-1 and atg12-2 seedlings. Wild-type W22 was included as a control. Positions of the primers used are shown in (A). OE, overexposure of the ethidium bromide-stained gel for the PCR products revealed that a wild type-like transcript (asterisk) accumulates in atg12-2 plants. RT-PCR of the UBC9 locus was used to verify the analysis of equal amounts of RNA. Two independent experiments gave similar results.
(D) Quantitative real-time RT-PCR analysis of wild type-like atg12 transcripts in homozygous atg12-2 plants. The locations of primers 5 and 6 used for the PCR are shown in (A). Each bar represents mean (±sd) of three biological replicates.
(E) ATG8 lipidation is blocked in the atg12 mutants. Seedlings were grown hydroponically on high-N liquid medium for 10 d and then exposed to low-N medium for 2 d before sampling. Total protein extracts (TE) prepared from root tissues were separated into soluble (S) and membrane fractions (Mem) by centrifugation, and the membrane fractions were solubilized with Triton X-100 and incubated with or without PLD for 1 h. Samples were subjected to SDS-PAGE in the presence of 6 M urea and immunoblotted with antibodies against Arabidopsis ATG8a. Solid and dashed lines locate ATG8-PE and free ATG8, respectively.
Besides the longer Mu element-containing transcript, a shorter wild type-like transcript also accumulated in homozygous atg12-2 plants presumably through excision of the modified intron (Figure 3C). Although a normal ATG12 protein could be synthesized from this minor RNA, its abundance as measured by quantitative real time RT-PCR was ∼50-fold lower than the wild-type mRNA expressed in the W22 parent (Figure 3D). Taken together, we concluded that the atg12-1 allele represents a functionally null mutation, whereas the atg12-2 allele is a strong knockdown mutation.
Loss of ATG12 Blocks ATG8 Lipidation
ATG12 in combination with ATG5 and ATG16 provide the essential ligase activity that attaches ATG8 to PE, with the resulting adduct then decorating the expanding phagophore. To evaluate the impact of the atg12-1 and atg12-2 mutations on autophagy, we assessed the lipidation state of ATG8 in three times backcrossed, selfed homozygous lines using the phospholipase D (PLD) assay developed previously (Chung et al., 2009, 2010). We first collected membranes from N-starved roots by centrifugation of total protein extracts, solubilized the membrane-anchored ATG8-PE adducts with Triton X-100, and then incubated the adducts with or without PLD. As shown in Figure 3E, ATG8-PE conjugates were readily detected immunologically in the membrane fraction from wild-type W22 roots as faster migrating species during SDS-PAGE in the presence of 6 M urea, whose abundance was sensitive to PLD cleavage. These faster migrating species were absent in the atg12-1 and atg12-2 mutants, demonstrating that the lipidation of ATG8 is severely attenuated, if not abolished, by either UniformMu insertion.
Autophagic Vesicles Are Not Generated in the atg12 Mutants
Prior studies showed that ATG8 bearing N-terminal fluorescent tags are effectively lipidated in vivo and provide excellent fluorescent markers for visualizing autophagy (Yoshimoto et al., 2004; Thompson et al., 2005; Klionsky et al., 2012). For example, autophagic bodies deposited in the vacuole and possible autophagosomes accumulating in the cytoplasm can be readily detected in wild-type Arabidopsis lines expressing GFP-ATG8a and pretreated with the V-ATPase inhibitor concanamycin A (ConA) as bright ∼1-μm puncta, which are absent in the strong atg mutant backgrounds (Phillips et al., 2008; Chung et al., 2010). ConA does not block autophagic transport but aids autophagic body detection by raising the vacuolar pH above the optima for resident hydrolases, which in turn extends the lifetime of these vesicles (Yoshimoto et al., 2004; Klionsky et al., 2012).
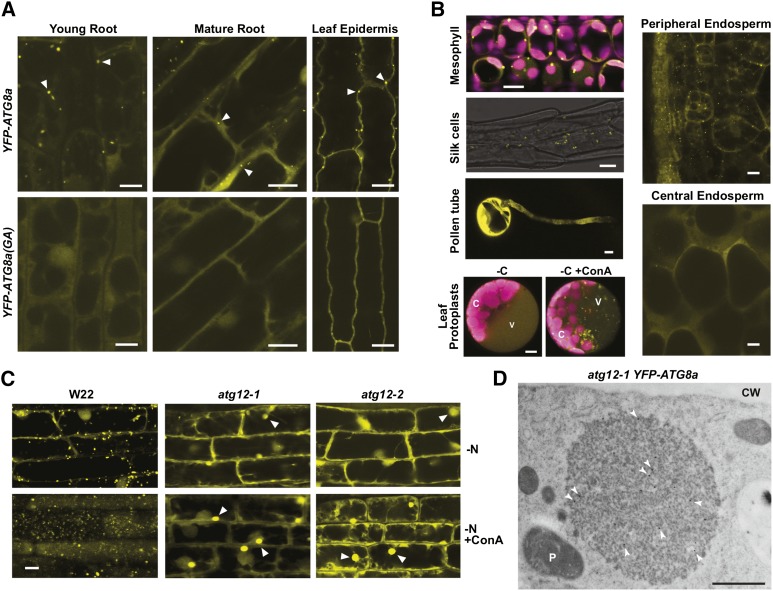
To detect comparable autophagic structures in maize and assess the strength of the atg12 mutants, we introduced transgenes that express YFP fused to ATG8a or an ATG8a variant missing the C-terminal glycine (Gly-117) and thus cannot be lipidated [ATG8a(GA)]. For this study, the transgenes were under the control of the maize Ubq1 promoter and following transformation were introgressed by repeated backcrosses from the transformable HiII parent into both the B73 and W22 inbred backgrounds. From confocal microscopy analysis of the YFP-ATG8a lines grown under well-fed conditions without ConA, fluorescent signal was mostly confined to the cytoplasm, with phagophore- and/or autophagosome-like structures often seen as bright dots (Figures 4A, 4B, and 5C). These puncta were common in the leaf epidermal cells and more abundant in young versus mature root cells and were readily detected in the leaf mesophyll, silk, and peripheral endosperm cells. In all cases, these YFP-labeled puncta were absent in YFP-ATG8a(GA) plants, implying that they are autophagic membranes containing the ATG8-PE adduct (for examples, see Figure 4A). While pollen grains and central endosperm cells strongly expressed the YFP-ATG8a reporter, few or no puncta were evident, suggesting that autophagy is relatively inactive in these tissues (Figure 4B).
Figure 4.
Autophagic Vesicles Detected Using the YFP-ATG8a Reporter Are Absent in atg12 Mutants.
(A) and (B) Confocal fluorescence microscopy of maize B73 tissues expressing YFP-ATG8a or the YFP-ATG8a(GA) mutant. In (A), seedlings were grown hydroponically for 12 d in high-N liquid medium for imaging root cells or on soil for 14 d for imaging leaf epidermal cells. Possible autophagosomes observed in YFP-Atg8a plants are located by the arrowheads. In (B), images of mesophyll cells were captured from leaves of 2-week-old soil-grown seedlings. Chlorophyll fluorescence is shown in magenta. Silk, peripheral, and central endosperm tissues were collected 18 DAP from greenhouse-grown plants. For pollen tubes, fresh pollen grains were dusted on solid pollen germination medium and allowed to germinate for 4 h at room temperature before microscopy. For maize leaf protoplasts, the protoplasts from 2-week-old, soil-grown seedlings expressing YFP-ATG8a were incubated in Suc-free medium supplemented with 1 μM ConA for 16 h before microscopy. C, chloroplast; V, vacuole. Bars = 10 μm.
(C) Maize atg12 mutants fail to accumulate autophagic bodies. Wild-type W22, atg12-1, and atg12-2 seedlings expressing YFP-ATG8a were grown hydroponically in high-N liquid medium for 10 d, transferred to low-N liquid medium for 32 h, and then treated for 16 h with 1 μM ConA or an equal volume of DMSO. Root cells were imaged by confocal fluorescence microscopy. The large amorphous cytoplasmic structures containing YFP-ATG8a that accumulate in atg12 plants are indicated by arrowheads. Bar = 10 μm.
(D) Electron microscopy image of an atg12-1 root cell pretreated with ConA showing a YFP-ATG8a aggregate. Root tips were fixed, sliced into thin sections, and immunogold-labeled with anti-GFP antibodies (arrowheads). CW, cell wall; P, plastid. See Supplemental Figure 6A for additional images. Bar = 1 μm.
Figure 5.
Autophagy Is Accelerated by Nitrogen or Fixed-Carbon Starvation in Maize.
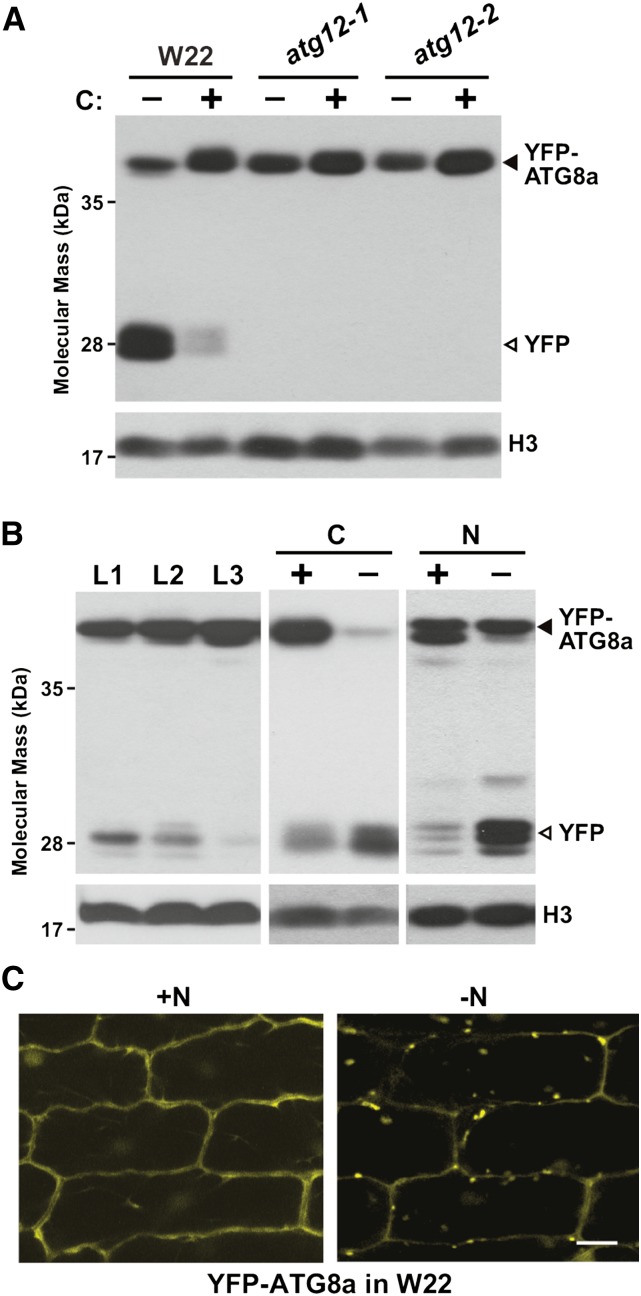
(A) atg12 mutants block the release of free YFP from YFP-ATG8a. Wild-type W22, atg12-1, and atg12-2 seedlings expressing YFP-ATG8a were grown on soil for 2 weeks and the second leaf blade (L2) was either kept in the light (+) or subjected to fixed-carbon (C) starvation by covering a section with aluminum foil for 2 d (−). Total protein extracted from a mix of three to six leaves was subjected to SDS-PAGE followed by immunoblot analysis with anti-GFP antibodies. YFP-ATG8a and free YFP are indicated by closed and open arrowheads, respectively. Immunodetection of histone H3 was used to confirm near equal protein loading. Two independent experiments gave similar results.
(B) Vacuolar release of free YFP from YFP-ATG8a is increased as leaves age or upon nitrogen (N) or fixed-C starvation. Tissue analyzed included the L1, L2, and L3 leaves (numbers correspond to the order of emergence) of plants grown in N-rich soil (left panel), the L2 leaf blade subjected to fixed-C starvation as in (A), and roots from plants grown hydroponically in high-N liquid medium for 10 d and then either kept on high-N (+) or transferred to low-N (−) liquid medium for 2 d (right panel). Total protein extracts were subjected to SDS-PAGE followed by immunoblot analysis as in (A). YFP-ATG8a and free YFP are indicated by closed and open arrowheads, respectively.
(C) N starvation induces the accumulation of autophagic vesicles. YFP-ATG8a plants were grown hydroponically in high-N liquid medium for 10 d and then either kept on high-N (+N) or transferred to low-N (−N) liquid medium for 2 d. Leaf epidermal cells were examined for autophagosomes and autophagic bodies by confocal fluorescence microscopy. Bar = 10 μm.
Consistent with an autophagic function, the frequency of the vacuolar YFP-ATG8a puncta increased dramatically in root cells and leaf protoplasts when starved for N or fixed-C and pretreated with ConA (Figures 4B and 4C). However, when atg12-1 and atg12-2 plants expressing YFP-ATG8a were examined similarly, we found that the block in ATG8 modification also negated the accumulation of these autophagic vesicles. No bright puncta were evident in root cells even after exposing the plants to N starvation and pretreating the roots with ConA (Figure 4C). By all criteria, the YFP-ATG8a-labeled puncta behaved as bona fide autophagosomes and autophagic bodies with the response of the mutants demonstrating that their assembly and delivery to the vacuole requires the ATG12-ATG5 conjugate and its downstream ATG8-PE adduct.
In addition to a diffuse YFP-ATG8a signal, we detected large cytoplasmic foci of YFP-ATG8a in the atg12 mutant cells, which became more obvious upon ConA addition (Figure 4C). These amorphous ∼10-μm structures could also be seen by immuno-electron microscopy using labeling with anti-GFP antibodies for confirmation, but did not appear to be confined by a membrane (Figure 4D). These structures were also enriched in ubiquitin, as detected by immuno-electron microscopy with antiubiquitin antibodies, implying that they also contained ubiquitylated proteins (Supplemental Figure 6A). One likely possibility is that they represent aggregates of YFP-ATG8a that have coalesced into aggresome-like structures awaiting autophagic turnover (Kuma et al., 2007) but could not be enveloped into autophagosomes without ATG12.
In addition to microscopy, the YFP-ATG8a reporter can also be used to monitor autophagy more quantitatively through its cleavage in planta. In this case, it has been well documented that the GFP/YFP moieties fused to ATG8 and various autophagic substrates often become detached after autophagic transport and accumulate as free forms inside the vacuole because of their resistance to further digestion (Chung et al., 2010; Klionsky et al., 2012; Li et al., 2014; Marshall et al., 2015). The ratio of free GFP/YFP versus the tagged protein can then be used as a measure of autophagic flux. As shown in Figure 5A, leaves from W22 plants expressing YFP-ATG8a accumulated free YFP, with its level rising dramatically as autophagy was induced by fixed-C starvation. However, this appearance of free YFP was completely abolished in the atg12-1 and atg12-2 mutants, consistent with a block in autophagic transport of the reporter into the vacuole. That this block in the atg12-2 plants was indistinguishable from that of the atg12-1 plants also implied that the atg12-2 allele approaches the strength of a null mutant despite accumulating the wild type-like Atg12 transcript.
Subsequent assays based on this free-YFP/fused YFP ratio showed that autophagy is significantly impacted in maize by developmental stage and nutrient availability. For example, the ratio rose as leaves from soil-grown 2-week-old maize seedlings matured (L1 versus L3) and in leaves following exposure to fixed-C and N stress (Figure 5B). Free YFP appearance in leaves during N starvation was coincident with the appearance of YFP-ATG8a-decorated cytoplasmic and vacuolar puncta even without ConA as detected by confocal fluorescence microcopy (Figure 5C).
Physiology of ATG12-Deficient Plants
Previous studies with Arabidopsis revealed that autophagy-defective plants develop normally and are fertile but senesce early and are generally hypersensitive to nutrient limitations (Hanaoka et al., 2002; Doelling et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Chung et al., 2010; Suttangkakul et al., 2011; Guiboileau et al., 2012, 2013; Masclaux-Daubresse et al., 2014), while more recent studies with rice have connected autophagy to fixed-C recycling and male gametogenesis (Kurusu et al., 2014). To reveal how autophagy impacts maize, we studied the growth and development of atg12-1 and atg12-2 plants in the W22 background under both nutrient rich and poor conditions. When grown on soil supplemented with N-rich fertilizer (16 mM NO3–), both mutants appeared relatively normal phenotypically and exhibited a similar growth rate as compared with wild-type W22 (Figures 6A, 6C, and 6D). No significant differences compared with the wild type were seen in terms of total dry matter production (see Figure 8B). The mutants produced healthy pollen grains, as judged by pollination trials, morphology, and vital staining with Alexander stain (Alexander, 1969; Supplemental Figure 7A), and were fertile when grown in a greenhouse as judged by seed number per plant (Table 1). However, a significant decrease in the seed yield (grams of seeds per plant) was observed under N-rich conditions, which when compared with the wild type was 28 and 25% lower for atg12-1 and atg12-2 plants, respectively (Table 1). This drop was not paralleled by lower seed N concentration or lower seed number per plant (Table 1).
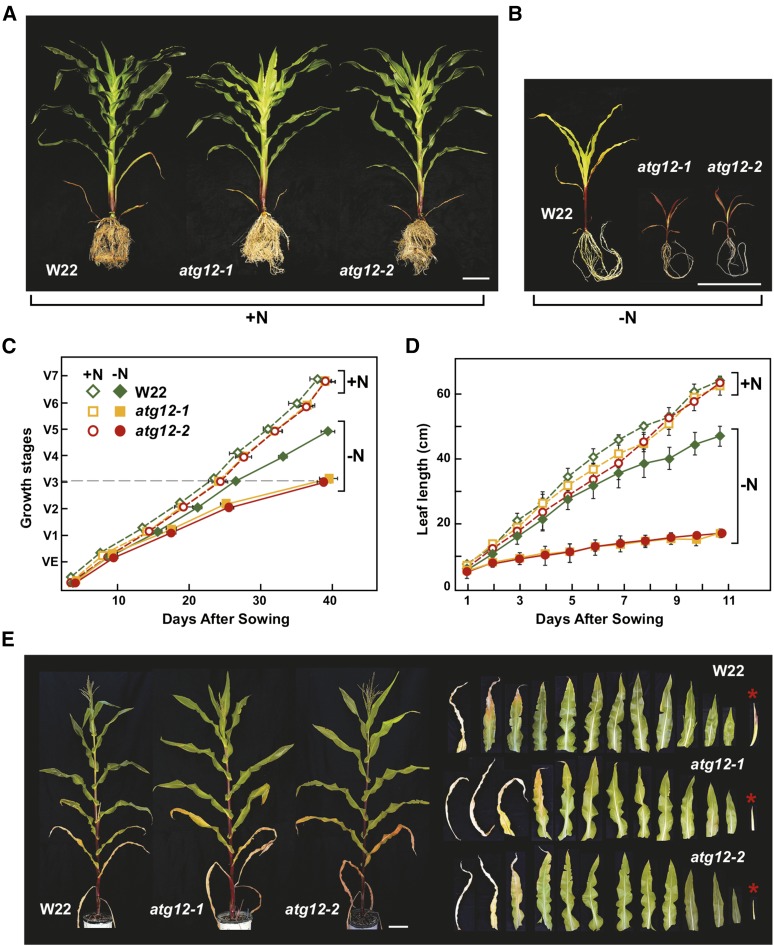
Figure 6.
Growth of Maize atg12 Mutants Is Hypersensitive to Nitrogen Deprivation.
(A) and (B) Representative plants grown continuously on high nitrogen (+N) or low-N (−N) soil for 7 weeks. Quantitative measurements of growth are shown in (C) and (D). Bar = 20 cm.
(C) Growth rate is slower for atg12 mutants grown on low-N soil. Shown is the maturation index for maize as determined by leaf appearance, e.g., VE = emergence of seedlings and V7 = final maturation of the seventh leaf.
(D) Leaf elongation is slower in atg12 mutants grown on low-N soil. Length of the fifth leaf was measured over an 11-d time course beginning at emergence. Each point in (C) and (D) represents the adjusted mean (±sd) from two biological repeats analyzing six plants each (n = 12).
(E) N deprivation accelerates leaf senescence and retards yield. Plants were grown for 5 weeks on high-N soil and then deprived of N thereafter. Representative wild-type W22, atg12-1, and atg12-2 plants at maturity are shown on the left. On the right shows the full complement of leaves and ears (asterisks) in developmental sequence from these plants. Note the smaller ears in atg12 plants. Bar = 20 cm.
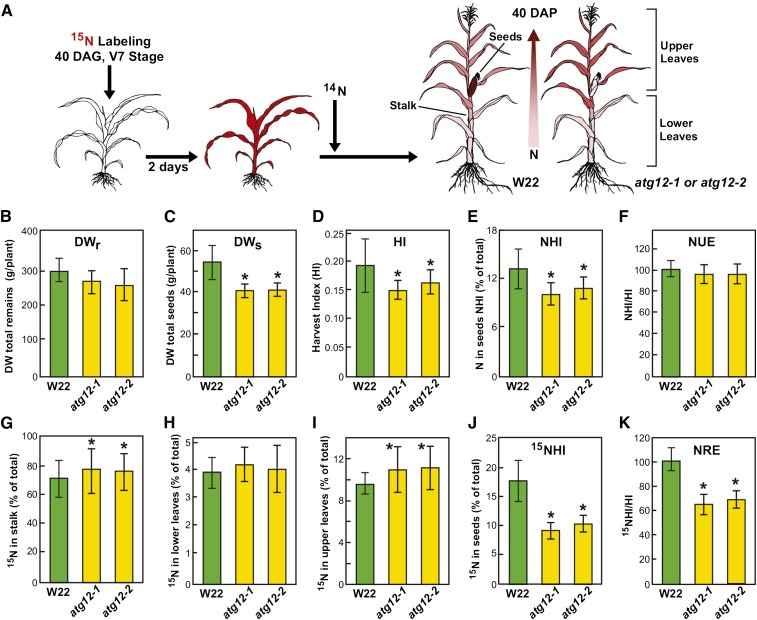
Figure 8.
Nitrogen Remobilization Efficiency Is Reduced in Maize atg12 Mutants.
(A) Overview of 15N labeling and subsequent N partitioning. Wild-type W22, atg12-1, and atg12-2 plants were grown on soil under high-14N conditions, pulse labeled at 40 DAG with 15NO3− for 2 d, and then grown on high-14N thereafter. At maturity (110 DAG), total N and the 15N/14N ratio of various tissues were analyzed by mass spectrometry. Two biological replicates each containing six plants were used for data analysis (n = 12). Values are adjusted means (±sd). Asterisks highlight values for atg12-1 and atg12-2 plants that are significantly different from the wild type as determined by Student’s t test (P < 0.05). See Supplemental Figure 9 for 15N labeling efficiency at 47 DAG.
(B) Biomass accumulation as measured by DW of remains (DWr = DW of stalk + upper leaves + lower leaves).
(C) DWs of seeds.
(D) HI as measured by the DW ratio of seeds to the aboveground portions of the plants.
(E) NHI as measured by the partitioning of total plant nitrogen in seeds.
(F) NHI:HI ratio as an estimate of NUE.
(G) to (J) Partitioning of total 15N in stalk, upper leaves, lower leaves, and seeds, respectively.
(K) 15NHI:HI ratio as an indicator of NRE.
Table 1. Seed Yield and N Content Are Decreased in Maize atg12 Mutants.
| Seed Yield (g/plant) | Seed Number/Plant | Thousand Seed Weight (g) | Seed N Concentration (%DWseed) | |
|---|---|---|---|---|
| Wild type (W22) | 54.2 ± 8.2 (100) | 248 ± 37 (100) | 218 ± 15 (100) | 1.90 ± 0.10 (100) |
| atg12-1 | 39.2 ± 3.6 (72)a | 240 ± 22 (97) | 163 ± 9 (75)a | 1.85 ± 0.09 (97) |
| atg12-2 | 40.9 ± 3.9 (75)a | 236 ± 25 (95) | 173 ± 15 (79)a | 1.83 ± 0.11 (96) |
Plants were grown in high-N (16 mM NO3–) fertilized soil under a 16-h-light/8-h-dark photoperiod. Values represent the mean (±sd) from two independent experiments analyzing six plants for each genotype. Numbers in parentheses are percent values expressed compared to the wild type.
Statistically significant difference as determined by Student’s t test (P < 0.05).
A novel autophagic transport route has been tentatively connected to the vacuolar transport of storage proteins in maize aleurone cells (Reyes et al., 2011). Electron microscopy analysis of developing atg12-1 seeds revealed no ultrastructural defects in the aleurone or starchy endosperm and showed a normal appearance and number of protein storage vesicles and protein bodies (Supplemental Figure 6B), further supporting the conclusion that the autophagy-like transport of seed storage proteins into maize aleurone storage compartments does not involve the ATG8/ATG12 conjugation system. Collectively, the results indicated that ATG12, and by inference the entire ATG system, is not essential to the growth and development of maize under N-rich conditions, but does compromise seed yield, as first seen in Arabidopsis (Doelling et al., 2002).
Strikingly, when fed low N fertilizer (0.1 mM NO3–) starting from planting, the growth and development of homozygous atg12 plants were severely challenged as compared with W22 plants. Dramatic reductions in root and shoot growth and enhanced leaf senescence were clearly evident, indicative of severe N stress (Figure 6B). For example, both wild-type and atg12 plants exposed to N-rich conditions had similar growth rates and simultaneously reached the V7 growth stage at 40 DAS. However, under N deprivation, wild-type plants reach the V5 stage within the same time period, whereas the atg12-1 and atg12-2 plants only reached the V3 stage (Figures 6B and 6C). The mutant leaves elongated much more slowly, with the length of the 5th leaf expanding to only a third of that produced by the 5th leaf from wild-type plants at 11 d after leaf initiation (Figure 6D). We also attempted to study the response of the atg12 plants to phosphate and sulfur stress. However, soil-grown atg12-1 and atg12-2 plants supplemented with phosphate- or sulfur-deficient fertilizer grew similarly to wild-type plants (Supplemental Figure 7B), leaving a role for autophagy in recycling these nutrients unresolved, as small but sufficient amounts could have originated from the soil.
This hypersensitivity to N deprivation was also apparent when the atg12-1 and atg12-2 plants were well fertilized for the first 5 weeks after germination and then fed N-poor fertilizer thereafter. Soon after anthesis, the older leaves rapidly senesced and ear development stalled markedly in both mutants (Figure 6E). To examine the phenotypes of field-grown atg12 plants, we cultivated them at the West Madison Agricultural Research Station (Verona, WI) where the soil was pretreated with 200 kg per acre of urea (46-0-0) before sowing. Under such relatively high N conditions, the mutants exhibited similar growth rates to that of wild-type W22, flowered slightly later, and were only marginally smaller at maturity. However, during flowering and seed fill, the lower leaves from the atg12 mutants senesced prematurely (Supplemental Figure 8), similar to well-described autophagy mutants in Arabidopsis, including atg9-1 (Hanaoka et al., 2002), atg7-1 (Doelling et al., 2002), atg4a/b (Yoshimoto et al., 2004), atg5-1 (Thompson et al., 2005), and atg10-1 (Phillips et al., 2008). Together, these data demonstrate that the ATG8/ATG12 autophagic system provides much needed recycled N during maize development and seed fill that strongly augments yield under both N-limiting and field conditions.
Lack of Autophagy Affects Leaf Protein Profiles
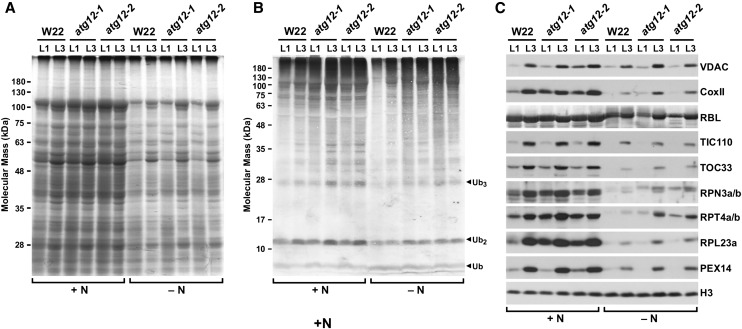
To assess how a block in autophagic recycling might impact maize leaf protein content, we examined the protein profiles of young wild-type W22 and atg12 leaves exposed to high- and low-N conditions. Here, the seedlings were grown for only 2 weeks, long before the lack of N noticeably slowed development. Even after this short time, we observed the effects of N deprivation. Whereas the L1 and L3 leaves from the wild type and the atg12-1 and atg12-2 lines grown on high-N medium retained their overall protein profile as judged by SDS-PAGE of total seedling extracts, identical leaves from N-starved plants had much less protein, with the L1 leaves containing considerably less (Figure 7A), indicating that both N availability and age have dramatic consequences on maize leaf protein content. The effects of autophagy could then be seen when specific cytosolic and organellar proteins were examined individually. For example, the ATG system has been implicated in the removal of ubiquitylated protein aggregates through receptors such as NBR1 (Svenning et al., 2011; Zhou et al., 2013). Consistent with this route, we found that older L1 leaves from atg12-1 and atg12-2 seedlings grown under N-limiting conditions accumulated more ubiquitin conjugates than those from wild-type seedlings, suggesting that autophagy represents a major route for clearing these aggregates and other targets of the ubiquitin/proteasome system as maize leaves age under N stress (Figure 7B). This effect is not restricted to N starvation, as young L3 leaves from N-fed atg12-1 and atg12-2 plants also accumulated slightly more ubiquitin conjugates (Figure 7B).
Figure 7.
Effects of the atg12 Mutations on Maize Protein and Ubiquitin Conjugate Profiles.
The older L1 and younger L3 leaves were collected from wild-type (W22), atg12-1, and atg12-2 seedlings after 14 d of hydroponic growth on high-nitrogen (+N) or low-N (−N) liquid medium.
(A) Profile of total seedling extracts separated by SDS-PAGE and stained for protein with Coomassie blue.
(B) and (C) Immunoblot analysis of the extracts in (A) with antibodies against ubiquitin (B), VDAC, CoxII, the large subunit of Rubisco (RBL), TIC110, TOC33, RPN3a/b, RPT4a/b, RPL23a, and PEX14 (C). Total protein extracts were obtained from a mix of at least five leaves. Immunodetection of histone H3 was used to confirm near-equal protein loading. Two independent experiments gave similar results.
Interestingly, examination of markers for mitochondria, chloroplasts, peroxisomes, and the ribosome and proteasome complexes in the cytoplasm revealed a complex pattern of autophagic elimination. In all cases, the levels of each protein were higher in younger leaves and were dramatically reduced by N stress, consistent with the negative impacts of both age and nutrient availability (Figure 7C). Levels of the chloroplast marker Rubisco large subunit and the inner and other envelope proteins TIC110 and TOC33 in atg12-1 and atg12-2 leaves, respectively, were indistinguishable to W22 for both N-fed and N-starved plants, suggesting that ATG8-mediated autophagy has little impact on the clearance of chloroplast constituents either before or after N stress (Figure 7C). Conversely, the mitochondrial markers, voltage-dependent anion channel (VDAC; outer membrane) and cytochrome c oxidase subunit II (inner membrane), and the peroxisomal marker PEX14 were present at higher levels in the atg12 backgrounds compared with wild type, suggesting that autophagy helps in the turnover of these organelles (Figure 7C). Likewise, levels of the proteasome subunits RPN3 and RPT4 as well as ribosome subunit RPL23a were elevated by the atg mutants, thus connecting their degradation to the ATG system (Figure 7C). This effect was especially evident for RPL23a in N-fed L1 leaves, suggesting that autophagy is strongly responsible for reducing ribosome levels as maize leaves age and their translational needs diminish.
Nitrogen Remobilization to the Seeds Is Impaired in atg12 Plants
To further study the impact of autophagy on N recycling, we employed pulse-chase with 15NO3− to monitor nitrogen movements. Using the 15N tracing strategy developed previously for Arabidopsis (Masclaux-Daubresse and Chardon, 2011; Guiboileau et al., 2012), we first grew the plants to 40 d after germination (DAG; V7 stage) on soil with 16 mM 14NO3−, labeled them by a 2-d pulse of 15NO3− fed to the roots, exhaustively washed out the remaining soil 15NO3−, and then refed the plants with 16 mM 14NO3− thereafter (Figure 8A). Half of the plants were harvested 7 d after labeling to monitor 15N uptake. The other half was harvested 40 d after pollination (DAP). Both sets of samples were dissected into various organs, dried, weighed, and analyzed for 15N and the 14N/15N ratio by isotopic ratio mass spectrometry. Several features related to yield and N utilization were calculated, including dry weight (DW) of seeds, stalks, and leaves, total N content, and 15N abundance, which were then used to assess N use efficiency (NUE) and N remobilization efficiency (NRE). When the 15N tracer was applied in this way, only the roots, leaves, and stalks were labeled initially, whereas the seeds could acquire 15N only if it moved from N stores assimilated by these other organs.
When wild-type, atg12-1, and atg12-2 plants were analyzed only 7 d after 15NO3− labeling, no significant differences were seen in the total DW and 15N content in each vegetative tissue (Supplemental Figure 9), consistent with the normal growth of atg12 plants under well-fed conditions (Figure 6). For all genotypes, the stalk was the major N sink, as it accounted for ∼59% of the assimilated 15N, with the remaining 15N partitioned to the upper leaves (17 to 18%), lower leaves (8%), and roots (15%). When the plants were analyzed at 40 DAP, the role of autophagy in DW accumulation and N movement became obvious. A marked decrease in seed DW was measured in both atg12 mutants as described above (Table 1), which is similar to Arabidopsis mutants lacking core components of the ATG system (Doelling et al., 2002; Guiboileau et al., 2012). This was coupled with a slight decrease in the DW of the roots and a significant decrease in the DW of the shoots (Figures 8B and 8C). The harvest index (HI = seed DW/whole plant DW), which is an important gauge of productivity, was also lower in the atg12 mutants compared with wild-type W22 plants (Figure 8D).
To determine NUE, we first used DW and N content data to calculate the N harvest index (NHI = seed N/whole plant N), which is a key indicator of N allocation efficiency into seeds (Masclaux-Daubresse and Chardon, 2011; Guiboileau et al., 2012). As shown in Figure 8E, the NHI observed in both atg12 mutants was significantly lower than that in the wild type. We then used the NHI/HI ratio as a measure of NUE performances between the atg12 and wild-type plants, as NHI and HI may vary depending on genetic background (Chardon et al., 2012). Neither of the atg12 mutants showed significant differences for NHI/HI compared with the wild type, indicating that autophagy might not be essential for seed NUE under N-rich conditions, although its absence reduces seed yield and NHI.
Using the abundance of 15N and the 14N/15N ratio as determined by isotopic ratio mass spectrometry, we then tracked the partitioning of N within the wild-type and atg12 plants when combined with the DW data (Figures 8G to 8K). Stalks still contained most of the total N even after seed fill, based on the fact that wild-type and atg12 stalks had the highest 15N values, with 70% of measured 15N in wild-type and ∼76% in atg12 stalks, respectively. Conversely, lower leaves were minor sinks for 15N; they only stored ∼4% of measured 15N and there was no significant difference between the wild type and the atg12 mutants. The partitioning of 15N into upper leaves was generally ∼2- to 3-fold higher than that in lower leaves, representing 9.4 and 11% in the wild type and atg12 mutants, respectively. Many of the upper leaves were apparent but not yet fully elongated during the labeling period at 40 DAG. We presume that these immature upper leaves acquired most of their 15N during this period and not from remobilized lower leaf stores during subsequent development until 40 DAP. Interestingly, the partitioning of 15N into all three of these vegetative tissues was generally higher in atg12-1 and atg12-2 plants than in wild-type W22 plants, with the difference being significant in stalks and upper leaves (Figures 8G to 8I), suggesting that a block in autophagy restricts stored N mobilization. In contrast, the partitioning of 15N into seeds (15NHI) and the ratio of 15NHI to HI, which was used to calculate NRE, were substantially lower in both atg12 mutants than in the wild type (Figures 8J and 8K). Taken together, our 15N partitioning studies showed that inactivation of ATG8-mediated autophagy through loss of ATG12 significantly reduces NRE during seed fill, which in turn detrimentally impacts seed yield by weight.
DISCUSSION
Given the importance of autophagy to nutrient recycling, cellular housekeeping, survival in adverse environments, and defense against pathogens in plants (Li and Vierstra, 2012; Liu and Bassham, 2012), its core machinery and modes of regulation have drawn increasing research attention. Whereas most of our current understanding comes from Arabidopsis, recent studies have also begun to focus on crop species to understand its consequences to nutrient management and agronomic yield. This study and that by Chung et al. (2009) with maize along with parallel work in rice (Xia et al., 2011; Kurusu et al., 2014), wheat (Kuzuoglu-Ozturk et al., 2012), and tomato (Solanum lycopersicum; Zhou et al., 2014) revealed that a mechanistically similar autophagic system exists in crops and that most genes within the pathway are tightly regulated transcriptionally during development, are activated by nutrient limitation and other stresses, and are upregulated during leaf senescence. Unfortunately, the male-sterile phenotype of the first rice atg mutant described that eliminated ATG7 (Kurusu et al., 2014) limited an appreciation of autophagy with respect to N economy and yield.
In this study, we expanded our molecular description of the maize ATG system and identified 31 new core autophagy genes and two Nbr1 cargo receptor-encoding genes that can be added to the 12 previously described genes encoding components of the ATG8/ATG12 conjugation pathways (Chung et al., 2009). We also discovered a likely ortholog of yeast Atg16, thus completing the list of factors necessary for ATG8 lipidation based on the yeast paradigm. In-depth transcriptome analysis of all 42 expressed loci demonstrated that most of the maize ATG system components are upregulated in senescing leaves, maturing endosperms, and differentiating root cells, implying a central role for autophagy in the development, housekeeping, nutrient management, and/or possible programmed cell death of these tissues.
Importantly, we also provided a phenotypic description of a maize mutant deficient in the ATG8/ATG12-mediated pathway with the study of two functionally null mutant alleles affecting the single gene encoding ATG12. These mutants abrogated synthesis of the ATG8-PE adduct and blocked the assembly of ATG8-decorated autophagosomes and their subsequent deposition into the vacuole as autophagic bodies. Phenotypic analyses of both atg12 alleles showed that loss of this autophagic pathway is not detrimental to maize grown under N-rich conditions, and unlike the similarly compromised rice atg7 mutant (Kurusu et al., 2014), the plants generated viable pollen and were fully fertile. However, like comparable mutants in Arabidopsis (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Chung et al., 2010; Suttangkakul et al., 2011; Guiboileau et al., 2012; Masclaux-Daubresse et al., 2014), homozygous maize atg12 plants are strongly hypersensitive to N stress and display accelerated leaf senescence even under nutrient-rich field conditions. The atg12 plants also had lower total seed weights but had a normal number of seeds when well fertilized with N. N remobilization studies using 15N implied that this reduced seed yield was at least partially caused by a compromised ability to remobilize N assimilated by vegetative tissue to the developing seed, presumably because of impaired N recycling by autophagy.
One striking feature of maize Atg genes is their highly coordinated expression, which is often controlled by developmental state and growth conditions. Previous studies with Arabidopsis demonstrated that the transcript abundance of numerous ATG genes increased in senescing tissues and under nutrient-limiting growth conditions (Contento et al., 2004; Sláviková et al., 2005; Thompson et al., 2005; Phillips et al., 2008). As with the ATG8/12 conjugation cascade (Chung et al., 2009), our in-depth RNA-seq analysis of the maize genes that synthesize (1) the ATG1 kinase complex that regulates autophagy in response to nutritional cues, (2) the ATG2/9/18 complex that delivers membranes to the expanding phagophores, (3) the PI3 kinase complex that decorates autophagic membranes with PI3 phosphate, and (4) the NBR1 receptor for ubiquitylated cargo often showed similar transcription patterns, implying that an autophagy regulon exists that coordinates the expression patterns of the system.
In addition to increased mRNA abundance in senescing leaves and maturing endosperms, we also detected upregulation during the maturation of primary roots. Intriguingly, root maturation in Arabidopsis was also coincident with elevated expression of several ATG8 genes and increased autophagosome accumulation (Sláviková et al., 2005; Yano et al., 2007). Because the maturation zone is where xylem and aerenchyma, used for water transport and gas exchange, respectively, differentiate, one possibility is that autophagy contributes to the programmed cell death pathways that ultimately eliminate the cytoplasm from their progenitor cells. It is also notable that within the maize Atg gene families, there is often at least one locus that shows strong developmental upregulation, whereas others appear to have more constitutive expression patterns, implying that subfunctionalization within the families exists (e.g., the Atg1, Atg13, and Atg18 families; Figure 2).
The transcription factor networks that activate ATG gene expression in plants are not yet known. In mammalian cells, recent studies have revealed a complex autophagy network encompassing tens of transcription factors, along with epigenetic events involving microRNAs and histone modifications (Füllgrabe et al., 2014), with the conserved FORKHEAD BOX-O3 transcription factor being a central player (Webb and Brunet, 2014). Clearly, a systems analysis of autophagy gene expression in plants is now needed to fully appreciate the contributions of such a regulon to their biology.
In agreement with previous studies with yeast, mammalian cells, and Arabidopsis, we confirmed that YFP-ATG8 fusions are excellent markers for monitoring autophagic activity in maize, with demonstrations that the frequencies of YFP-ATG8a decorated vesicles increase dramatically upon nutrient starvation and/or ConA treatment. Whereas the detection of cytosolic phagophores/autophagosomes has been challenging in Arabidopsis expressing comparable fluorescent ATG8 fusions (e.g., GFP-ATG8 fusions; Thompson et al., 2005; Phillips et al., 2008), cytoplasmic puncta that likely represent these structures could be easily detected in maize. The formation of these structures requires ATG8 lipidation based on their absence in atg12 plants expressing YFP-ATG8a and in wild-type plants expressing the YFP-ATG8a(G-A) mutant that a priori cannot be modified. The maize YFP-ATG8a puncta detected here look remarkably similar in size and number to GFP-ATG5 labeled structures concluded by Le Bars et al. (2014) to represent phagophores and their resulting autophagosomes emerging from endoplasmic reticulum nucleation sites in Arabidopsis.
In addition, we readily detected autophagic bodies inside the vacuole from YFP-ATG8a-expressing lines, whose appearance requires ATG12. This delivery was also supported by the release of free YFP from the fusion, which accumulates via an ATG8/ATG12-dependent route as a somewhat stable form in the vacuole (Meiling-Wesse et al., 2002; Chung et al., 2010). Both the appearance of YFP-ATG8a decorated puncta inside the vacuole and the release of free YFP were increased by N and fixed-C starvation and in leaves as they aged, thus providing a facile measure of autophagic flux in maize.
Unexpectedly, we also detected large fluorescent foci in the cytoplasm of atg12 roots expressing the YFP-ATG8a reporter. These foci were more abundant in N-starved plants and appeared brighter upon prior treatment of the roots with ConA. Using electron microscopy, they appeared as amorphous aggregates without a delineating membrane and tested positive for both YFP-ATG8a and ubiquitin. In mice, it has been reported that the ATG8 ortholog LC3 will concentrate into cytoplasmic protein aggregates or ubiquitin-positive inclusion bodies when transiently overexpressed in autophagy-deficient cells (Kuma et al., 2007). Our finding of comparable YFP-ATG8a-containing aggregates in maize suggests that this reporter could be aggregation prone, which then coalesces in ubiquitin-containing, aggresome-like structures awaiting turnover. Aggresome substrates are often ubiquitylated, which then promotes their recognition by AIM-containing ubiquitin receptors such as NBR1 for eventual deposition into and clearance by autophagic vesicles (Rogov et al., 2014). Consequently, examining these maize structures in nbr1 mutant backgrounds could be informative. Notwithstanding, it should be emphasized that we did not detect similarly large fluorescent foci in wild-type maize expressing YFP-ATG8a. Therefore, while YFP/GFP-ATG8 fusions remain useful markers for autophagy, their cytoplasmic fluorescence patterns should be carefully interpreted in autophagy-defective plants.
Plants employ multiple strategies to cope with environmental stress, such as nutrient starvation, and to maintain fecundity. One general approach involves the rapid degradation of N and C-rich source tissues and remobilization of these stores to support the growth of new vegetative tissues and reproductive organs and seeds (Chardon et al., 2012; Xu et al., 2012). Previous studies with Arabidopsis revealed that autophagy plays a central role in nutrient recycling under limiting conditions and during leaf senescence, with many organelles and protein complexes becoming targets, including chloroplasts, peroxisomes, mitochondria, ribosomes, and even proteasomes (Ishida et al., 2008; Wada et al., 2009; Hillwig et al., 2011; Farmer et al., 2013; Guiboileau et al., 2013; Kim et al., 2013; Shibata et al., 2013; Li et al., 2014; Marshall et al., 2015). Our preliminary studies with the maize atg12 lines starved for N indicated that the removal of ubiquitylated proteins, possibly via aggresomes, requires in part ATG8-mediated autophagy, as does the loss of ribosomes, mitochondria, and proteasomes. In contrast, the abundance of chloroplast proteins appeared to be unaffected in the maize atg12 backgrounds with or without N. Given that multiple routes might be evoked to eliminate chloroplasts and their contents, including chloroplast-resident proteases, SAVs, and chloroplast vesiculation in addition to autophagy via Rubisco-containing bodies (Otegui et al., 2005; Ishida et al., 2008; Wada et al., 2009; Roberts et al., 2012; Michaeli et al., 2014; Wang and Blumwald, 2014), one scenario is that these other pathways assume responsibility in the absence of the ATG8-mediated autophagic route. Another is the possibility that ATG8/ATG12-independent autophagic routes become relevant (Nishida et al., 2009; Reyes et al., 2011). Alternatively, given the fact that autophagic turnover of chloroplast constituents by RBC is stimulated by fixed-C and not by N stress (Izumi et al., 2010; Izumi and Ishida, 2011), measuring chloroplast turnover in limiting fixed-C might be required to detect the effects of the atg12 mutations.
Seed yield is strongly influenced by N availability, which is for the most part acquired from existing organic stores via remobilization and not from inorganic soil sources (Chardon et al., 2012; Xu et al., 2012). Here, we confirmed a crucial role for autophagy in this process that was first observed with Arabidopsis (Doelling et al., 2002; Guiboileau et al., 2012; Masclaux-Daubresse et al., 2014) by showing that maize plants defective in ATG8-mediated autophagy have lower yield even under well fertilized conditions and are attenuated in their remobilization of stored N from vegetative tissues to developing seeds. Consequently, the manipulation of autophagy in efforts to improve N and fixed-C recycling might be a beneficial strategy to enhance the yield of seed-bearing food and biofuel crops, especially under suboptimal growth conditions or in situations where soil fertilization is cost prohibitive or ecologically unsound.
METHODS
An Expanded Catalog of Maize Atg Genes
Maize (Zea mays) autophagy genes/proteins were identified by reciprocal TBLASTN searches (e-value threshold of <10−4) of the MaizeGDB genome sequence database (http://www.maizegdb.org/) using the Arabidopsis thaliana (Columbia-0 ecotype) orthologs as queries. Maize Atg genes previously described by Chung et al. (2009) as involved in the ATG8/ATG12 conjugation pathway were included to confirm previous findings. For further support of the various protein classes, we used the functional annotations provided by MaizeGDB and the domain architectures predicted by Pfam (http://pfam.sanger.ac.uk) and SMART (http://smart.embl-heidelberg.de/) to identify additional signatures. The current list of maize autophagy proteins is summarized in Supplemental Table 1.
Expression Patterns and Splicing Analyses
Expression patterns for maize Atg genes were extracted from the RNA-seq transcriptome profiles generated by Sekhon et al. (2013) and S. Stelpflug and H.F. Kaeppler (unpublished data) with the B73 inbred background. To quantify expression, reads per kilobase per one million reads were used to normalize transcript levels from each data set. Transcript levels for each Atg gene at the specific tissue or developmental stages were normalized to the average transcript level from all samples. Expression heat maps were generated using R 3.1.1 statistics software (Comprehensive R Archive Network; http://www.R-project.org). RNA splicing events were identified by mapping the RNA-seq-derived transcripts to the maize B73 V2 pseudomolecules (AGPv2; http://ftp.maizesequence.org) using Bowtie version 0.12.7 and TopHat version 1.4.1 as previously described (Salvo et al., 2014)
For RT-PCR analyses, total RNA was extracted from 100 mg of maize tissues using the RNeasy Plant Mini Kit (Qiagen). One microgram of RNA was subsequently used to synthesize the RNA/DNA hybrid with SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. RT-PCR analyses were performed using EconoTaq Plus Green 2X Master Mix (Lucigen) and the indicated primer pairs, with the analysis of Ubc9 used as an internal control. All critical primers employed in the study are listed in Supplemental Table 3. Quantitative real-time RT-PCR analyses were performed as previously described (Li et al., 2014), using the levels of maize Ubc9 transcript for normalization. The relative abundance of each mRNA was determined by the comparative cycle threshold method (Chung et al., 2009).
Plant Materials and Growth Conditions
The maize atg12-1 (UFMu-02975) and atg12-2 (UFMu-02196) mutants were derived from the UniformMu transposon population constructed in the W22 inbred background (McCarty et al., 2005) and were obtained from the Maize Genetics Cooperation Stock Center. The positions of the UniformMu insertions were verified by genomic PCR using a gene-specific primer and the UniformMu border-specific primer TIR6. The PCR products were sequenced to locate the exact insertion sites. Each mutant was backcrossed three times to the W22 inbred and then self-pollinated to obtain homozygous lines before the various phenotypic analyses.
For the genetic manipulations and various nutrient starvation studies, plants were grown in a greenhouse under a 16-h-light/8-h-dark photoperiod provided by natural lighting supplemented with halogen lamps. For N-limitation studies, the W22 and atg12 mutant plants were germinated and grown in pots (15 liter) containing Metro-Mix 360 (Sun Gro Horticulture). The pots were irrigated with 500 mL high-N or low-N solutions twice a week, starting either at 1 DAS or the V7 leaf stage, as indicated. A modified Hoagland solution was used for high-N treatments (16 mM N) and contained 5 mM KNO3, 1 mM KH2PO4, 2 mM MgSO4, 5 mM Ca(NO3)2, 0.05 mM Fe-EDTA, and 1× micronutrients (Caisson Laboratories). The low-N solution (0.1 mM N) contained 0.1 mM KNO3, 1 mM KH2PO4, 2 mM MgSO4, 0.05 mM Fe-EDTA, 1× micronutrients, 4.9 mM KCl, and 5 mM CaCl2. Plants were watered with distilled water between the fertilizations depending on water status in the pot. To assess the atg12 mutant phenotype for field-grown maize, the plants were grown at the West Madison Agricultural Research Station during summer 2014 under well-fertilized soil conditions as previously described (Sekhon et al., 2011).
Construction of Transgenic Maize Expressing YFP-ATG8a
The plasmid pMCG1005-YFP-Atg8a expressing YFP fused to the N terminus of wild-type maize ATG8a and expressed under the control of the Ubq1 promoter was as described previously (Reyes et al., 2011). The Gly117-Ala mutation was introduced into the Atg8a cDNA (Chung et al., 2009) by PCR amplification using the primers ZmAtg8a_F1 and ZmAtg8a_R2; the resulting Atg8a(GA) cDNA was cloned into pENTR/D-TOPO (Invitrogen) and then recombined by an LR Clonase (Invitrogen) reaction into the pMCG1005-YFP-Gateway vector to generate pMCG1005-YFP-Atg8a(GA). Maize transformation using microprojectile biolistic DNA delivery was performed as described previously (McGinnis et al., 2005). To minimize the number of insertion events, immature embryos derived from self-pollinated, greenhouse-grown HiIIA × HiIIB plants were transformed with a low concentration of plasmid DNA (0.4 μg plasmid DNA/1 mg microcarriers). YFP-Atg8a and YFP-Atg8a(GA) F1 plants expressing strong YFP signals were backcrossed at least four times into the B73 and W22 inbred backgrounds. The YFP-ATG8a reporters were then introgressed into the atg12 mutants by crossing followed by a self-cross to obtain stable homozygous lines.
Confocal Laser Scanning Microscopy and Image Analysis
Transgenic lines expressing YFP-ATG8a or YFP-ATG8a(GA) were imaged with a Zeiss 510-Meta laser scanning confocal microscope, using 488-nm light combined with the band-pass 500-550 IR filter. Captured cell images were processed using the LSM510 image browser (Zeiss) and converted to TIFF format. For leaf cells, leaf sections harvested from 2-week-old soil-grown plants were used. To image cells of silk, and the peripheral and central endosperm tissues (18 DAP), they were dissected from plants grown in a greenhouse to maturity. To image pollen tubes, pollen grains were germinated for 4 h at room temperature on solid pollen germination medium [18% Suc, 0.01% boric acid, 1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, and 0.5% phytoagar (Sigma-Aldrich)]. To detect YFP-ATG8a signals in the atg12 mutants, the plants were grown hydroponically under a 16-h-light/8-h-dark photoperiod in 1× Murashige and Skoog (MS) + N liquid medium for 10 d, transferred to MS-N liquid medium for 32 h, and then treated with 1 μM ConA or an equivalent volume of DMSO for an additional 16 h (Chung et al., 2009). Lateral roots were imaged for YFP by confocal fluorescence microscopy. To detect fixed-C starvation-induced autophagy, protoplasts were isolated from leaves of 2-week-old, soil-grown seedlings as described (Lee et al., 2002). Protoplasts were resuspended in Suc-free medium (1.5 mM MES-KOH, pH 5.6, 150 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 5 mM mannitol) supplemented with 1 μM ConA for 24 h before observation.
Electron Microscopy
Root tips from 12-d-old atg12-1 mutant seedlings expressing YFP-ATG8a and wild-type W22 and atg12-1 endosperm at 20 DAP were high-pressure frozen in 0.1 M Suc using a Baltec HPM 010 unit (Technotrade). Endosperm samples were substituted in 2% OsO4 in anhydrous acetone at –80°C for 72 h and embedded in Epon resin (Ted Pella) for structural analysis. Root tips were substituted in 0.2% uranyl acetate (Electron Microscopy Sciences) and 0.2% glutaraldehyde in acetone at −80°C for 72 h and embedded in Lowicryl HM20 (Electron Microscopy Sciences) for immunolocalization. Root sections were mounted on Formvar-coated nickel grids and blocked for 20 min with a 10% (w/v) solution of nonfat milk in TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween 20). The sections were incubated with the primary anti-GFP (Torrey Pines; 1:10) or antiubiquitin antibodies (van Nocker et al., 1996; 1:25) for 1 h, rinsed in TBS containing 0.5% Tween 20, and then transferred to the secondary antibody (anti-rabbit IgG 1:10) conjugated to 15-nm gold particles for 1 h. Controls omitted the primary antibodies.
Immunoblot Analyses
In most cases, the immunoblot analyses were performed according to Chung et al. (2009). For analysis of the ATG8-PE adduct, roots from 2-week-old hydroponically grown plants were homogenized in TNPI buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM phenylmethanesulfonyl fluoride, and 10 mM iodoacetamide). After filtering through four layers of cheesecloth and centrifugation at 2000g for 5 min, the clarified crude extract was centrifuged at 100,000g for 1 h to collect the membrane fraction. The membranes were solubilized in TNPI buffer containing 0.5% (v/v) Triton X-100 and incubated with or without phospholipase D (Enzo Life Sciences) followed by SDS-PAGE in the presence of 6 M urea (Chung et al., 2009). YFP-ATG8a and free YFP were assayed in total protein extracts prepared by homogenizing tissues with 2 mL/g fresh weight of SDS-PAGE sample buffer. The extracts were clarified by centrifugation at 16,000g, and the proteins in the supernatants were solubilized by heating for 5 min before electrophoresis. Antibodies against Arabidopsis ATG8a (Phillips et al., 2008), ubiquitin (van Nocker et al., 1996), RPN3 and RPT4 (Marshall et al., 2015), TIC110 (Lübeck et al., 1996), VDAC (Subbaiah et al., 2006), and Rubisco large subunit (Phillips et al., 2008) were as described previously. Antibodies against histone H3 (ab1791), GFP (ab290), and RPL23a (ab157110) were obtained from Abcam. Antibodies against CoxII (AS04 053A), PEX14 (AS08 372), and TOC33 (AS07 236) were obtained from Agrisera.
To measure autophagic flux via YFP-ATG8a cleavage, seeds of W22 and atg12 mutants expressing YFP-ATG8a were germinated and grown on Metro-Mix 360 for 2 week at 25°C with a 16-h-light/8-h-dark photoperiod. YFP-ATG8a and free YFP were detected by immunoblot analyses with anti-GFP antibodies following SDS-PAGE of total protein extracts. To detect fixed-C-induced flux, the second leaf blades from 14-DAG seedlings were wrapped with aluminum foil for 2 d. To detect N starvation-induced flux, YFP-seedlings were grown hydroponically with 1× MS liquid medium for 10 d and then transferred to MS-N medium for 2 d.
15N-Labeling and Determination of 15N Abundance
Wild-type W22 and atg12 plants were grown in a greenhouse as above in 15-liter pots of Metro-Mix 360 with a 16-h-light/8-h-dark photoperiod. Plants were watered with 500 mL high-N Hoagland solution (16 mM N) twice a week and distilled water between fertilizations depending on water status. At the V7 leaf stage (∼40 DAG), plants were labeled with 15N by soaking the surrounding soil for 48 h with a nutrient solution containing 10 atom% excess K[15N]O3. Afterwards, the pots were rinsed thoroughly with least 20 volumes of distilled water to remove the remaining 15N, and high-N Hoagland solution was then applied twice a week for the rest of the experiment. For the 15N uptake measurements, six plants of each genotype were harvested 7 d after 15N labeling, and separated into root, stalk, lower leaves (fully expanded leaves before labeling), and upper leaves (young leaves when labeled). To assess nitrogen remobilization, plants were grown to maturity; at 40 DAP, the plants were separated into seed, stalk, lower leaves, and upper leaves. Unlabeled plants were also collected to determine the natural 15N abundance.
Collected samples (labeled and unlabeled) were dried, weighed, and pulverized into a fine powder. A DW subsample of each powder (4 to 20 mg) was assayed for total N content and 15N abundance using a PDZ Europa ANCA-GSL elemental analyzer interfaced with a PDZ Europa 20-20 isotope ratio mass spectrometer (Sercon) at the University of Wisconsin-Madison Soil Science Stable Isotope Facility. The 15N abundance of each sample was expressed as a percent of total N and calculated as atom% or A%sample = 100×(15N)/(15N+14N). The natural abundance of 15N obtained from unlabeled samples was used as the Acontrol% (∼0.367) to calculate 15N enrichment of labeled samples (E%= A%sample − Acontrol%).
NUE and NRE Calculations
Factors used to estimate NUE and NRE were calculated as described by Guiboileau et al. (2012). Briefly, HI was used to evaluate yield and was defined as the DWseed/(DWseed+DWstalk+DWlower+DWupper). NHI used to assess grain filling with nitrogen was calculated as: N%seed × DWseed/(N%seed × DWseed + N%stalk × DWstalk + N%lower × DWlower + N%upper × DWupper). The NUE was then expressed as the ratio of NHI/HI to compare NUE performance among genotypes. To determine the efficiency of nitrogen remobilization to the seeds, 15NHI was used and defined as (E%seed × N%seeds × DWseeds)/[(E%seed × N%seedsxDWseeds)+(E%stalkxN%stalk × DWstalk) + (E%upper × N%upper × DWupper) + (E%lower × N%lower × DWlower)]. The 15NHI:HI ratio was used to compare NRE performances among genotypes. 15N-labeling data were compiled from two biological replicates involving six plants for each genotype.
Accession Numbers
atg12-1 and atg12-2 mRNA sequences have been submitted to NCBI GenBank under accession numbers KP780167 and KP780168, respectively.
Supplemental Data
Supplemental Table 1. Collection of Arabidopsis and Maize Atg Genes.
Supplemental Table 2. List of Tissues Analyzed by RNA-seq in This Study.
Supplemental Table 3. Oligonucleotide Primers Used in This Study.
Supplemental Figure 1. Transcript Abundance from Selected Maize Atg Genes Increases as Leaves Mature or Senesce.
Supplemental Figure 2. Developmental and Tissue-Specific Expression Profiles of Maize Atg18 Genes.
Supplemental Figure 3. Developmental and Tissue-Specific RNA-seq Expression Profiles of Maize Autophagy Genes Clustered by Coexpression.
Supplemental Figure 4. Developmental and Tissue-Specific Alternative Splicing of Maize Atg Gene Isoforms in Endosperm and Leaf Tissues.
Supplemental Figure 5. Nucleotide and Amino Acid Sequence Alignment of Wild-Type ATG12 and the atg12-1 and atg12-2 Mutants.
Supplemental Figure 6. Electron Microscopy Images of Cells from atg12 Mutants.
Supplemental Figure 7. atg12 Mutants Produce Viable Pollen and Are Not Hypersensitive to Phosphorus or Sulfur Limitation.
Supplemental Figure 8. Field-Grown atg12 Mutants Display Early Leaf Senescence.
Supplemental Figure 9. 15N Labeling Efficiency of Wild-Type, atg12-1, and atg12-2 Maize Plants.
Supplemental Data Set 1. Expression Levels of All Analyzed Maize Atg Genes.
Supplementary Material
Acknowledgments
We thank Wisconsin colleagues Joseph M. Walker, Christopher Lee, and Richard S. Marshall for technical assistance and reagents, Xiao Huang for the schematic representations of maize, Harry W. Read and Jonathan Pauli for help with the 15N measurements, and Sarah J. Swanson for advice on confocal microscopy imaging. Donald McCarty and Karen Koch via the Maize Genetics Cooperation Stock Center provided the UniformMu stocks harboring the ATG12 mutations. We thank Karen McGinnis for kindly providing the pMCG1005-YFP-Gateway vector. This work was supported by grants from the USDA-National Institute of Food and Agriculture (Grant 2008-02545) to R.D.V. and by the National Science Foundation Plant Genome Programs (Grant IOS-1339325) to R.D.V. and M.S.O.
AUTHOR CONTRIBUTIONS
F.L. designed and performed the bulk of the research, analyzed the data, and wrote the article. T.C. contributed genetic materials and cell biological reporters. M.L.F. and H.F.K. contributed to new cell biological reporters. J.G.P. and M.S.O. performed the electron microscopy analyses. S.M.K. contributed RNA-seq data. R.D.V. contributed to experimental design and data analysis and wrote the article.
Glossary
- SAV
senescence-associated vacuole
- DAS
days after sowing
- PLD
phospholipase D
- ConA
concanamycin A
- VDAC
voltage-dependent anion channel
- DAG
days after germination
- DAP
days after pollination
- DW
dry weight
- NUE
N use efficiency
- NRE
N remobilization efficiency
- HI
harvest index
- NHI
N harvest index
- MS
Murashige and Skoog
References
- Abendroth L.J., Elmore R.W., Boyer M.J., Marlay S.K. (2011). Corn Growth and Development. (Ames, IA: Iowa State University Extension; ). [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Avila-Ospina L., Moison M., Yoshimoto K., Masclaux-Daubresse C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65: 3799–3811. [DOI] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F., Noël V., Masclaux-Daubresse C. (2012). Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J. Exp. Bot. 63: 3401–3412. [DOI] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D.J. (2008). The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 19: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149: 220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493. [DOI] [PubMed] [Google Scholar]
- Contento A.L., Kim S.J., Bassham D.C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135: 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Farmer L.M., Rinaldi M.A., Young P.G., Danan C.H., Burkhart S.E., Bartel B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., Klionsky D.J. (2014). The machinery of macroautophagy. Cell Res. 24: 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J., Klionsky D.J., Joseph B. (2014). The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 15: 65–74. [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Avila-Ospina L., Yoshimoto K., Soulay F., Azzopardi M., Marmagne A., Lothier J., Masclaux-Daubresse C. (2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199: 683–694. [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Yoshimoto K., Soulay F., Bataillé M.P., Avice J.C., Masclaux-Daubresse C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194: 732–740. [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753–766. [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Mizushima N. (2009). Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy 5: 85–87. [DOI] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. (2011). What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49: 557–576. [DOI] [PubMed] [Google Scholar]
- Hillwig M.S., Contento A.L., Meyer A., Ebany D., Bassham D.C., Macintosh G.C. (2011). RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. USA 108: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Ishida H. (2011). The changes of leaf carbohydrate contents as a regulator of autophagic degradation of chloroplasts via Rubisco-containing bodies during leaf senescence. Plant Signal. Behav. 6: 685–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Wada S., Makino A., Ishida H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 154: 1196–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichey T., Hirel B., Heumez E., Dubois F., Le Gouis J. (2007). In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crops Res. 102: 22–32. [Google Scholar]
- Kim J., Lee H., Lee H.N., Kim S.H., Shin K.D., Chung T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Schulman B.A. (2014). Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 21: 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., et al. (2012). Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Matsui M., Mizushima N. (2007). LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3: 323–328. [DOI] [PubMed] [Google Scholar]
- Kurusu T., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10: 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuoglu-Ozturk D., Cebeci Yalcinkaya O., Akpinar B.A., Mitou G., Korkmaz G., Gozuacik D., Budak H. (2012). Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta 236: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Le Bars R., Marion J., Le Borgne R., Satiat-Jeunemaitre B., Bianchi M.W. (2014). ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 5: 4121. [DOI] [PubMed] [Google Scholar]
- Lee K.H., Kim D.H., Lee S.W., Kim Z.H., Hwang I. (2002). In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol. Cells 14: 388–397. [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Li F., Chung T., Vierstra R.D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhou Y., Zhou G., Ye R., Zhao L., Li X., Lin Y. (2008). Identification of early senescence-associated genes in rice flag leaves. Plant Mol. Biol. 67: 37–55. [DOI] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübeck J., Soll J., Akita M., Nielsen E., Keegstra K. (1996). Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 15: 4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D. (2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell , doi/10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D.E., Costa M.L., Guiamet J.J. (2008). Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol. (Stuttg.) 10 (suppl. 1): 15–22. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Chardon F. (2011). Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J. Exp. Bot. 62: 2131–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Orsel M. (2008). Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. (Stuttg.) 10 (suppl. 1): 23–36. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Clément G., Anne P., Routaboul J.M., Guiboileau A., Soulay F., Shirasu K., Yoshimoto K. (2014). Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell 26: 1857–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. [DOI] [PubMed] [Google Scholar]
- McGinnis K., Chandler V., Cone K., Kaeppler H., Kaeppler S., Kerschen A., Pikaard C., Richards E., Sidorenko L., Smith T., Springer N., Wulan T. (2005). Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 392: 1–24. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K., Barth H., Thumm M. (2002). Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett. 526: 71–76. [DOI] [PubMed] [Google Scholar]
- Michaeli S., Honig A., Levanony H., Peled-Zehavi H., Galili G. (2014). Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26: 4084–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., Hamada S., Yoshimori T. (2004). Autophagy defends cells against invading group A Streptococcus. Science 306: 1037–1040. [DOI] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. (2009). Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461: 654–658. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2014). Historical landmarks of autophagy research. Cell Res. 24: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Noh Y.S., Martínez D.E., Vila Petroff M.G., Staehelin L.A., Amasino R.M., Guiamet J.J. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41: 831–844. [DOI] [PubMed] [Google Scholar]
- Parrott D.L., McInnerney K., Feller U., Fischer A.M. (2007). Steam-girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence-associated genes. New Phytol. 176: 56–69. [DOI] [PubMed] [Google Scholar]
- Patrick J.W., Offler C.E. (2001). Compartmentation of transport and transfer events in developing seeds. J. Exp. Bot. 52: 551–564. [PubMed] [Google Scholar]
- Penfold C.A., Buchanan-Wollaston V. (2014). Modelling transcriptional networks in leaf senescence. J. Exp. Bot. 65: 3859–3873. [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F.C., Chung T., Holding D., Jung R., Vierstra R., Otegui M.S. (2011). Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell 23: 769–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I.N., Caputo C., Criado M.V., Funk C. (2012). Senescence-associated proteases in plants. Physiol. Plant. 145: 130–139. [DOI] [PubMed] [Google Scholar]
- Robertson G.P., Vitousek P.M. (2009). Nitrogen in agriculture: balancing the cost of an essential resource. Annu. Rev. Environ. Resour. 34: 97–125. [Google Scholar]
- Rogov V., Dötsch V., Johansen T., Kirkin V. (2014). Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53: 167–178. [DOI] [PubMed] [Google Scholar]
- Rose T.L., Bonneau L., Der C., Marty-Mazars D., Marty F. (2006). Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell 98: 53–67. [DOI] [PubMed] [Google Scholar]
- Ruuska S.A., Lewis D.C., Kennedy G., Furbank R.T., Jenkins C.L., Tabe L.M. (2008). Large scale transcriptome analysis of the effects of nitrogen nutrition on accumulation of stem carbohydrate reserves in reproductive stage wheat. Plant Mol. Biol. 66: 15–32. [DOI] [PubMed] [Google Scholar]
- Salvo S.A., Hirsch C.N., Buell C.R., Kaeppler S.M., Kaeppler H.F. (2014). Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS ONE 9: e111407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563. [DOI] [PubMed] [Google Scholar]
- Sekhon R.S., Briskine R., Hirsch C.N., Myers C.L., Springer N.M., Buell C.R., de Leon N., Kaeppler S.M. (2013). Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Oikawa K., Yoshimoto K., Kondo M., Mano S., Yamada K., Hayashi M., Sakamoto W., Ohsumi Y., Nishimura M. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25: 4967–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sláviková S., Shy G., Yao Y., Glozman R., Levanony H., Pietrokovski S., Elazar Z., Galili G. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56: 2839–2849. [DOI] [PubMed] [Google Scholar]
- Spitzer C., Li F., Buono R., Roschzttardtz H., Chung T., Zhang M., Osteryoung K.W., Vierstra R.D., Otegui M.S. (2015). The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Palaniappan A., Duncan K., Rhoads D.M., Huber S.C., Sachs M.M. (2006). Mitochondrial localization and putative signaling function of sucrose synthase in maize. J. Biol. Chem. 281: 15625–15635. [DOI] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. (2013). Senescence, ageing and death of the whole plant. New Phytol. 197: 696–711. [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23: 785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Deveraux Q., Rechsteiner M., Vierstra R.D. (1996). Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc. Natl. Acad. Sci. USA 93: 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., et al. (2010). Genome-wide distribution of transposed dissociation elements in maize. Plant Cell 22: 1667–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Blumwald E. (2014). Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26: 4875–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A.E., Brunet A. (2014). FOXO transcription factors: key regulators of cellular quality control. Trends Biochem. Sci. 39: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K., Liu T., Ouyang J., Wang R., Fan T., Zhang M. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546. [DOI] [PubMed] [Google Scholar]
- Xu G., Fan X., Miller A.J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63: 153–182. [DOI] [PubMed] [Google Scholar]
- Yano K., Suzuki T., Moriyasu Y. (2007). Constitutive autophagy in plant root cells. Autophagy 3: 360–362. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J., Yu J.Q., Chen Z. (2014). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J., Cheng Y., Chi Y.J., Fan B., Yu J.Q., Chen Z. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawrzyńska A., Sirko A. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.