Functional analysis identifies amino acid substitutions causing PEBP functional evolution and shows the importance of coding sequence divergence in driving functional divergence of duplicated genes.
Abstract
Gene duplication provides resources for novel gene functions. Identification of the amino acids responsible for functional conservation and divergence of duplicated genes will strengthen our understanding of their evolutionary course. Here, we conducted a systemic functional investigation of phosphatidylethanolamine binding proteins (PEBPs) in soybean (Glycine max) and Arabidopsis thaliana. Our results demonstrated that after the ancestral duplication, the lineage of the common ancestor of the FLOWERING LOCUS T (FT) and TERMINAL FLOWER1 (TFL1) subfamilies functionally diverged from the MOTHER OF FT AND TFL1 (MFT) subfamily to activate flowering and repress flowering, respectively. They also underwent further specialization after subsequent duplications. Although the functional divergence increased with duplication age, we observed rapid functional divergence for a few pairs of young duplicates in soybean. Association analysis between amino acids and functional variations identified critical amino acid residues that led to functional differences in PEBP members. Using transgenic analysis, we validated a subset of these differences. We report clear experimental evidence for the functional evolution of the PEBPs in the MFT, FT, and TFL1 subfamilies, which predate the origin of angiosperms. Our results highlight the role of amino acid divergence in driving evolutionary novelty after duplication.
INTRODUCTION
Gene duplication is a common phenomenon in all life forms and is particularly prevalent in plants (Wendel, 2000; Kondrashov et al., 2002; Conant and Wolfe, 2008). It can occur on various scales via independent mechanisms, including tandem and segmental duplications arising from recombination or DNA replication and whole-genome duplications (WGD) (Ramsey and Schemske, 1998). Some of the gene pairs that formed by duplication have a brief life span; only one copy may be maintained as a single copy, while the other copy becomes lost or pseudogenized. However, some gene pairs survive after duplication. These surviving duplicated genes and their subsequent divergence provide the raw genetic resources required for adaptive evolution and play a central role in the evolution of novel gene functions (Flagel and Wendel, 2009).
Many theoretical models have been proposed to account for the mechanism of evolution of duplicated genes (Innan and Kondrashov, 2010). Neofunctionalization presumes that after duplication, one copy is left to maintain its original function, while most of the extra copies will be pseudogenized via negative selection. Meanwhile, duplicated genes may occasionally acquire new functions via the accumulation of substitutions (Ohno, 1970). The subfunctionalization model (Hughes, 1994), together with the duplication-degeneration-complementation model (Force et al., 1999), postulates that the two copies are subfunctionalized during evolution. One copy is insufficient to fulfill the original function; thus, selection maintains both copies. The dosage-balance model suggests that duplicated copies have an optimum dosage dependency on each other among dosage-sensitive genes. Thus, such genes will not be duplicated unless their interaction partners are also duplicated (Papp et al., 2003; Sémon and Wolfe, 2007; Veitia et al., 2008). Several other models have also been proposed, such as the adaptive radiation and permanent heterozygote models (Innan and Kondrashov, 2010). Each model is supported by appropriate research. For instance, the evolution of eosinophil cationic protein and eosinophil-derived neurotoxin in primates supported the neofunctionalization model (Zhang et al., 1998), while the evolution of engrailed in zebra fish and ZAG1 and ZMM2 in maize (Zea mays) supported the subfunctionalization model (Force et al., 1999).
Mutations are generally believed to play major roles in coding-sequence evolution (Hughes, 1994; James and Tawfik, 2003). Theoretically, functional comparison of young and old duplicated genes with their ancestral single-copy genes would allow us to identify the substitutions critical for functional conservation and divergence and in turn provide insights into the evolutionary course of duplicated genes. Nevertheless, in practice, the original ancestral copy is difficult to determine. Alternatively, the systematic examination of functional divergence among duplicated genes in two closely related species may partially elucidate this question (Innan and Kondrashov, 2010; Jiao et al., 2011). Due to the complexity and time-consuming nature of functional analyses, it is almost impossible to functionally characterize duplicated genes at a genome-wide level. Instead, comprehensive comparisons of a conserved duplicated gene family between species could strengthen our understanding of the evolution of duplicated genes.
The phosphatidylethanolamine binding protein (PEBP) family is an ancient protein family found across the biosphere and plays important roles in regulating flowering (Banfield et al., 1998; Hengst et al., 2001). In plants, the PEBP family can be divided into three subfamilies, TERMINAL FLOWER1 (TFL1)-like, FLOWERING LOCUS T (FT)-like, and MOTHER OF FT AND TFL1 (MFT)-like (Danilevskaya et al., 2008). Interestingly, although the PEBP members share high degrees of amino acid sequence similarity, their functions diverged from each other after gene duplication. For instance, TFL1 and FT control flowering time and plant architecture in Arabidopsis thaliana, but they have opposite activities: TFL1 functions as a repressor, while FT functions as an activator (Bradley et al., 1997; Kardailsky et al., 1999; Kobayashi et al., 1999). MFT has weak FT-like activity (Yoo et al., 2004), but it mainly plays a critical role in regulating seed germination via the abscisic acid (ABA) and gibberellic acid signaling pathways (Xi et al., 2010). More interestingly, one amino acid substitution (H88Y) results in the functional conversion of TFL1 and FT (Hanzawa et al., 2005).
It has been suggested that most eudicot plants descended from an ancient hexaploid ancestor, followed by one or more rounds of lineage-specific ploidizations in some taxa (Jaillon et al., 2007; Jiao et al., 2011). Arabidopsis is believed to have undergone at least two rounds of tetraploidizations (Blanc et al., 2003; Bowers et al., 2003), and soybean (Glycine max) underwent multiple WGD events (Schlueter et al., 2007; Gill et al., 2009; Schmutz et al., 2010). As multiple homologs can be found in soybean for each single-copy gene in Arabidopsis (Jung et al., 2012), the PEBPs should have become a larger family in soybean compared with Arabidopsis. However, the functional conservation and divergence of this family and its evolutionary course in soybean has remained unclear, as did the relationship between soybean and Arabidopsis. The complex history of genome duplications in soybean and Arabidopsis provides a good opportunity to address these questions. A comprehensive functional investigation of this family in soybean and Arabidopsis may lead us to understand the evolution of PEBP functions after multiple duplication events and identify the critical molecular mechanisms underlying such processes.
In this study, we systemically analyzed the functional conservation and divergence of dozens of PEBP family members in soybean and Arabidopsis through a series of complementary tests, protein interaction assays, subcellular localization detection, and expressional profiling. In combination with molecular evolution analysis, we show that amino acid substitutions play an important role in driving functional divergence of PEPB members. The candidate key domain/amino acid residues responsible for functional divergence of PEBP members were identified, and one subset was experimentally verified.
RESULTS
Evolutionary History of the PEBP Family in Arabidopsis and Soybean
In Arabidopsis, the PEBP family consists of six members: MFT, FT, TFL1, TWIN SISTER OF FT (TSF), Arabidopsis thaliana CENTRORADIALIS homolog (ATC), and BROTHER OF FT AND TFL1 (BFT) (Carmona et al., 2007; Hedman et al., 2009). Through a homology search against the soybean genome (Schmutz et al., 2010), a total of 23 PEBP gene models were identified and grouped into the FT-like, TFL1-like, and MFT-like subfamilies (Supplemental Figure 1A). A classical PEBP gene is composed of four exons (Danilevskaya et al., 2008). We observed that although the gene lengths for individual members were divergent, most members in Arabidopsis and soybean, with the exception of three gene models, Glyma02g07650, Glyma18g53670, and Glyma08g05650, have the conserved gene structure (Supplemental Figure 2).
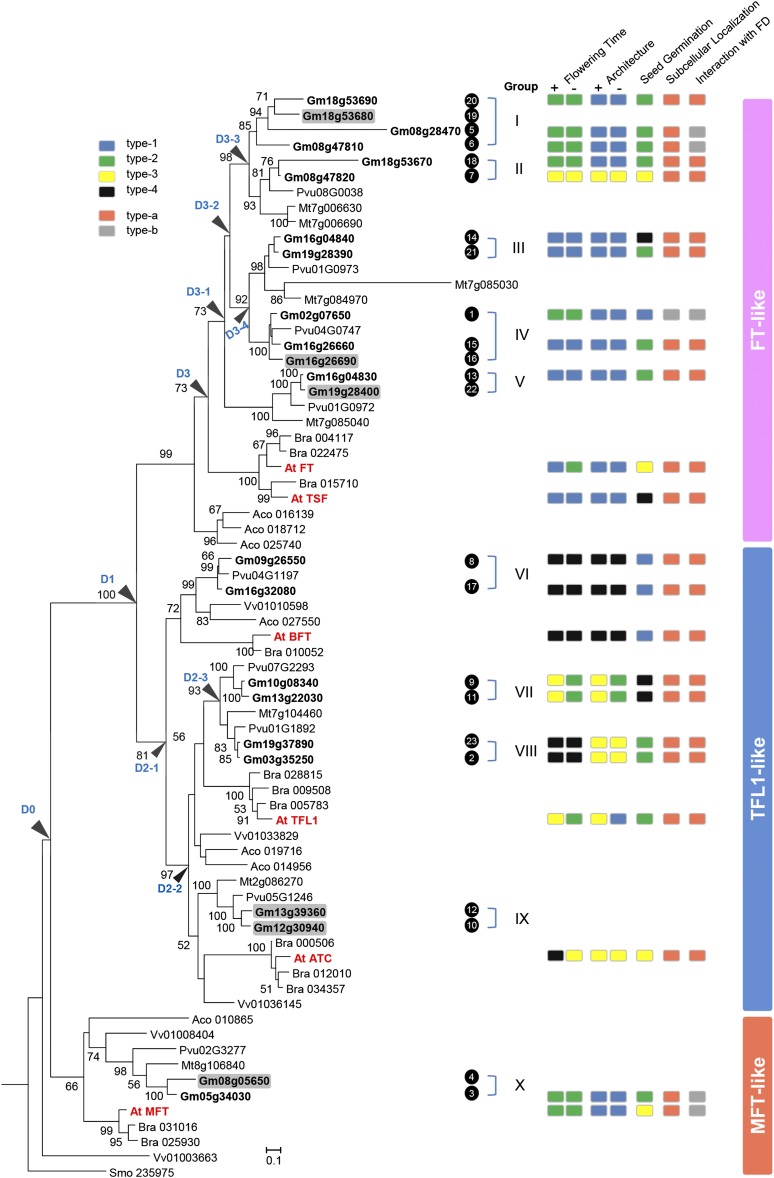
Using previously reported WGD information (Schmutz et al., 2010; Du et al., 2012), we found that all the PEBP homologs except for Gmly08g28470 have duplicated counterparts generated from the last WGD in soybean (Supplemental Figure 1B). To elucidate the evolution history of PEBPs, we reconstructed the maximum likelihood phylogenetic relationships of PEBP genes in six embryophyte species using Selaginella moellendorfii as an outgroup (Figure 1). According to the phylogeny, the PEBP members in soybean were classified into 10 groups that mainly contained copies derived from the recent WGD.
Figure 1.
Evolution of PEBP Members and Summary of the Functional Analyses of PEBP Members in Soybean and Arabidopsis.
The left panel shows the phylogenetic tree of PEBP members in seven species. The PEBP members from soybean are classified into 10 groups based on their phylogeny, and the groups are labeled after each gene. The PEBP members from Arabidopsis are labeled in red. The number after each gene represents the series number of the gene and is consistent with the number shown in other figures. The right panel indicates the functional variation of different members from soybean and Arabidopsis. Gm, Glycine max; At, Arabidopsis thaliana; Pvu, Phaseolus vulgaris; Mt, Medicago truncatula; Bra, Brasscia rapa; Aco, Aquilegia coerulea; Vv, Vitis vinifera; Smo, Selaginella moellendorfii. The flowering phenotypes are designated as follows: type-1, earlier than tfl1-1; type-2, same as tfl1-1; type-3, same as Col-0; and type-4, later than Col-0. The architecture phenotypes are designated as follows: type-1, determinate; type-2, determinate but higher; type-3, indeterminate; and type-4, indeterminate with bush. (−) Represents the TFL1pro:CDS->tfl1-1 construct, and (+) represents the TFL1pro:CDS:3′UTR->tfl1-1 construct. The variations in seed germination are characterized in MFTpro:CDS->mft-2 transgenic lines. The order of seed germination rate from low to high is 1 < 2 < 3 < 4. For subcellular localization, type-a represents localized in the nucleus and cytoplasm, and type-b represents localized in the nucleus. Regarding the interaction with Arabidopsis FD, type-a indicates an interaction, and type-b indicates no interaction. The gene models shaded by a gray box indicate genes that could not be amplified in this study.
The gene tree revealed two ancient PEBP duplication events in the lineage leading to the common ancestor of angiosperms after its split with gymnosperms. The first duplication (D0) gave rise to the MFT-like subfamily and the ancient lineage of the TFL1-like and FT-like subfamilies, which experienced a second duplication (D1) to create the two subfamilies (Figure 1). The TFL1 ancestor underwent two separate duplication events (D2-1 and D2-2) in the common ancestor of angiosperms, which created three daughter lineages corresponding to BFT, TFL1, and ATC in Arabidopsis and Group VI, Groups VII-VIII, and Group XI in soybean (Figure 1). Groups VII and VIII were further derived from a TFL1 duplication (D2-3) in the lineage leading to the common ancestor of the Papilionoideae.
The FT ancestor experienced different duplication events in the lineages of Eurosids I and II after they diverged (Figure 1). There was a lineage-specific duplication (D3) in Brassicaceae that gave rise to FT and TSF in Arabidopsis (Figure 1). In soybean, the FT-like copies constituted five groups that experienced more complicated duplication events because they contain an ancient Eurosids I-specific tandem duplication, as supported by the adjacent pairs in all three Eurosids I genomes (Gm08g47810 and Gm08g47820, Gm18g53670 and Gm18g53680, Glyma16g04830 and Glyma16g04840, Gm19g28390 and Gm19g28400, Pvu01G0973 and Pvu01G0972, and Mt7g085040 and Mt7g085030). A recent tandem duplication also occurred between Glyma18g53680 and Glyma18g53690. Group V was separated from the other four groups by the duplication event D3-1. Subsequently, the duplication event D3-2 gave rise to two well-supported sister clades, Groups I and II, which were further separated by duplication D3-3, and Groups III and IV, which were further separated by duplication D3-4.
The members within each group in soybean were generated from the recent WGD at 13 million years ago, and the duplication events D3-3, D3-4, and D2-3 may correspond to the WGD that occurred at 59 million years ago. The members within group (termed as within-group), between groups in each subfamily (termed as within-subfamily), and between subfamilies (termed as between-subfamilies) could be considered to be the young, middle-aged, and old duplicates, respectively. Taken together, these data suggest that the soybean PEBP family underwent ancient and recent WGDs as well as ancient and recent tandem duplications. With so many duplication events, the functional consequences of PEBP proteins are intriguing.
Detection of the Functional Evolution of PEBP Proteins Using Rescue Experiments
To detect functional evolution of PEBPs at the protein level, we performed a series of rescue experiments. The coding sequences (CDSs) of individual Arabidopsis and soybean PEBP genes were transformed into the Arabidopsis tfl1-1, mft-2, and ft-10 mutants under the control of the endogenous promoters of Arabidopsis TFL1, MFT, and FT, respectively (Supplemental Figure 4). Because the 3′ untranslated region (UTR) is known to be critical for the regulation of Arabidopsis TFL1 function (Kaufmann et al., 2010), two sets of constructs containing the CDS with or without the 3′-UTR were used for transforming tfl1-1 (Supplemental Figures 4A and 4B). Among the 23 PEBPs in soybean, six members, Glyma16g26690, Glyma19g28400, Glyma18g53680, Glyma13g39360, Glyma12g30940, and Glyma08g05650, of which CDSs could not be amplified from samples encompassing all soybean developmental stages, were excluded from further functional analyses in this study.
Compared with wild-type Arabidopsis (Columbia-0 [Col-0]), mft-2 showed greater reduction in seed germination rate in the presence of exogenous ABA (Xi et al., 2010), tfl1-1 exhibited phenotypes of earlier flowering and determination (Bradley et al., 1997), and ft-10 showed greatly delayed flowering time (Kardailsky et al., 1999; Kobayashi et al., 1999). Accordingly, we examined the germination rate under ABA treatment, flowering time and architecture, and flowering time for the mft-2, tfl1-1, and ft-10 transformants, respectively.
Flowering time of the tfl1-1 transformants could be classified into four main categories: earlier than tfl1-1, the same as tfl1-1, similar to Col-0, and later than Col-0. The architecture was also classified into four main categories: determinate architecture the same as tfl1-1, determinate architecture but taller than tfl1-1, indeterminate architecture the same as Col-0, and indeterminate architecture with a bush phenotype. Detailed phenotypic characterizations of each PEBP member revealed that neither gene from the MFT-like subfamily rescued the tfl1-1 defects (Figure 1; Supplemental Figures 5 and 6 and Supplemental Table 1), suggesting that the MFT-like members have no or a weak function in regulating flowering time or plant architecture. All the transgenic lines of the TFL1-like members with the 3′-UTR exhibited the same or later flowering time than Col-0, which was accompanied by the indeterminate or indeterminate with bush architecture (Figure 1; Supplemental Figures 5Q to 5V and Supplemental Table 1). However, almost all the transgenic lines of the FT-like members flowered earlier or same as tfl1-1 and displayed the same determinate architecture as the wild type (Figure1; Supplemental Figures 5E to 5P and Supplemental Table 1). The only exceptional FT-like member was Glyma08g47820, which exhibited the same flowering time as Col-0 and indeterminate architecture in the tfl1-1 background (Figure 1; Supplemental Figure 5H and Supplemental Table 1).
When only the CDS were transformed into tfl1-1 (Supplemental Figure 6 and Supplemental Table 1), transgenic lines of four TFL1-like members (At-ATC, At-TFL1, Glyma13g22030, and Glyma10g08340) flowered earlier than their counterparts possessing the corresponding CDS with 3′-UTRs and showed either determinate or determinate but taller architectures (Figure 1; Supplemental Figure 7). In contrast, the transgenic line expressing At-FT flowered at the same as tfl1-1 but later than its counterpart with the CDS and 3′-UTR (Figure 1; Supplemental Figures 7G and 7N and Supplemental Table 1). As the CDS with the 3′-UTR is more likely to represent the authentic function of a protein-coding gene, the observed phenotypic difference between PEBP transgenic lines with and without 3′-UTRs suggested that the TFL1-like genes mainly repress flowering and cause changes in the indeterminate architecture, while the FT-like genes mainly activate flowering; such functions may have been lost or partially lost for some CDSs due to the accumulation of degenerative mutations.
The germination rates of the mft-2 transformants could be classified into four categories: lower than mft-2, the same as mft-2, higher than mft-2 but lower than Col-0, and similar to or higher than Col-0 (Figure 1; Supplemental Figure 8). Of the PEBP genes from the MFT-like subfamily, At-MFT partially rescued the mft-2 phenotype with a germination rate higher than that of mft-2 but lower than that of Col-0, whereas Glyma05g34030 did not rescue the germination rate. The TFL1-like and FT-like genes affected seed germination rates to variable extents in the mft-2 background. At-ATC and the two soybean Group VII genes (Glyma13g22030 and Glyma10g08340) from the TFL1-like subfamily and At-TSF, At-FT, Glyma0847820 (Group II), and Glyma16g04840 (Group III) from the FT-like family partially or fully rescued the germination rate of mft-2. In contrast, transgenic lines of At-BFT, Glyma09g26550, and Glyma16g32080 from soybean Group VI and Glyma02g07650 from Group IV displayed even lower germination rates than mft-2.
Due to the difficulty in obtaining transgenic ft-10 lines, only the coding sequences of several representative gene models were successfully transformed into the ft-10 mutant. The gene models included At-FT, At-TFL1, Glyma16g26660, Glyma03g35250, and Glyma09g26550. Of the five ft-10 transformants, At-FT and Glyma16g26660 from the FT-like subfamily rescued the late flowering phenotype, whereas the remainder of the genes from the TFL1-like subfamily did not (Supplemental Figure 9 and Supplemental Table 2).
Protein Interaction and Subcellular Localization Assay of PEBP Members
Previous studies have suggested that in Arabidopsis, both FT and TFL1 are involved in an FD-dependent transcriptional complex. FD is required for FT activity to activate the expression of floral marker genes (Abe et al., 2005; Wigge et al., 2005), and TFL1 interacts with FD to repress FD-dependent events (Hanano and Goto, 2011). In the rescue experiments, some PEBP genes could not rescue the early flowering phenotype of tfl1-1. To determine the involvement of an FD-dependent transcriptional complex, we investigated the interactions of PEBP proteins with At-FD using a yeast two-hybrid assay (Supplemental Figure 10). Among the 23 PEBP proteins surveyed, only At-MFT and Glyma05g34030 from the MFT-like subfamily and Glyma02g07650, Glyma08g47810, and Glyma08g28470 from the FT-like subfamily could not interact with At-FD (Figure 1; Supplemental Figure 10). These genes consistently failed to rescue the early flowering phenotype of tfl1-1, confirming that the PEBP-FD interaction is necessary for regulating flowering time. The lack of an interaction between At-FD and the proteins encoded by the two MFT-like genes confirmed that the MFT-like subfamily members, unlike the TFL1-like and FT-like genes, do not regulate flowering time. However, several members that could interact with At-FD also failed to rescue the phenotype of tfl1-1, such as Glyma18g53670 and Glyma18g53690, suggesting that FD itself may not be sufficient for the regulation.
Arabidopsis FT localizes in the nucleus and cytoplasm (Abe et al., 2005; Wigge et al., 2005). We further identified the subcellular distribution of each PEBP member to determine whether the lack of rescue was due to changes in protein subcellular localization. Our analyses demonstrated that all the PEBP proteins except for Gm02g07650 localized in the nucleus and cytoplasm (Figure 1; Supplemental Figure 11). Gm02g07650 could only be detected in the nucleus (Figure 1; Supplemental Figure 11), coincident with its failure to rescue the early flowering of tfl1-1. These results suggest that the localization of PEBP proteins in the cytoplasm may be also necessary for the flowering-associated function, although subcellular localization is not associated with the functional variations of most PEBP genes.
Inference of Amino Acids Critical for the Functional Divergence between Subfamilies and Groups
Taken together, the functional analyses showed that PEBPs are multifunctional proteins whose subfamilies have diverged in function. In addition, despite the PEBP members within the same subfamily having similar functions, their individual functions differ to variable extents (Figure 1). The functional divergence measured as euclidean distance was related to duplicate age, which was supported by the increase of divergence as clade got older (between-subfamilies > within-subfamily > within-group; Supplemental Figure 3A), and the positive relationship between pairwise functional divergence and synonymous substitution rate (dS) (Supplemental Figure 3B).
To identify the amino acids that drove the functional divergence of the PEBPs, we checked the amino acid residue frequency in each subfamily and group based on the alignment, in which Arabidopsis TFL1 was used as the reference. Highly truncated members with sequence lengths <80% of the total length of the alignment were excluded for the analysis (Supplemental Figure 12). First, we identified 46 residues that exhibited high conservation with >95% identity in the 61 PEBP members on survey (Table 1; Supplemental Figure 12). About 20% of these residues (9/46) have been reported to result in functional loss when being mutated (Bradley et al., 1997; Kobayashi et al., 1999; Tian et al., 2010; Ho and Weigel, 2014), suggesting that these conserved amino acids might be critical to maintain the basic function of the PEBP family.
Table 1. Candidate Conserved Amino Acids Needed to Maintain PEBP Function.
| Site | Amino Acid | Ratio | Related Mutants |
|---|---|---|---|
| 11 | P/T/S | 59/1/1 | |
| 12 | L | 61 | |
| 15 | G/K/S | 59/1/1 | |
| 17 | V/L/I | 59/1/1 | |
| 19 | G/E | 60/1 | |
| 21 | V/I | 60/1 | |
| 42 | N | 60/1 | |
| 46 | P/S | 60/1 | |
| 55 | P | 61 | |
| 61 | G/D/S | 58/2/1 | |
| 69 | T | 61 | tfl1-14 (Bradley et al., 1997) |
| 70 | L/M | 58/3 | |
| 74 | D/N | 59/2 | |
| 76 | D | 61 | (Ho and Weigel, 2014) |
| 78 | P | 61 | |
| 80 | P/R | 60/1 | |
| 81 | S/G/D | 58/2/1 | |
| 83 | P/A/R | 59/1/1 | |
| 87 | E | 61 | tfl1-13 (Bradley et al., 1997); ft-4 (Kobayashi et al., 1999) |
| 90 | H/L/Y | 59/1/1 | |
| 91 | W | 61 | |
| 93 | V/I/L | 59/1/1 | |
| 96 | I | 61 | |
| 97 | P/Q | 60/1 | ft-6 (Kobayashi et al., 1999) |
| 105 | G | 61 | tfl1-1 (Bradley et al., 1997) |
| 107 | E/V | 58/3 | |
| 114 | P | 61 | |
| 116 | P | 61 | Gmtfl1-ab (Tian et al., 2010) |
| 119 | G/W | 60/1 | |
| 120 | I | 61 | |
| 121 | H | 61 | |
| 122 | R | 61 | ft-3 (Kobayashi et al., 1999) |
| 127 | L/V | 60/1 | |
| 128 | F/L/Y | 59/1/1 | |
| 130 | Q/M | 60/1 | (Ho and Weigel, 2014) |
| 141 | P | 61 | |
| 143 | R | 61 | |
| 146 | F | 61 | |
| 148 | T/S | 58/3 | |
| 151 | F | 61 | |
| 157 | L | 61 | |
| 160 | P/D/- | 59/1/1 | |
| 161 | V | 61 | |
| 162 | A/G/S | 59/1/1 | |
| 167 | N | 61 | |
| 177 | R/H/- | 59/1/1 | (Ho and Weigel, 2014) |
A >95% frequency at a single amino acid site corresponds to conserved. The amino acid sites were calculated based on the sequence of Arabidopsis MFT.
Subsequently, we inferred candidate amino acids that may drive the functional divergence by checking the differentiation of nonsynonymous substitution frequencies between subfamilies. The MFT-like and TFL1/FT-like clades were prominently diverged in their interactions with FD. Seven amino acid substitutions (Figure 2A) were found to exhibit high frequency differences between the MFT-like and TFL1/FT-like clades. Residues 64 and 169 have been verified in the soybean mutants Gmtfl1-ta and Gmtfl1-bb previously (Tian et al., 2010). Additionally, substitutions at these residues were detected in Glyma08g47810 and Glyma08g28470 (Supplemental Figure 12), which failed to interact with At-FD, further supporting the importance of these residues. Following this methodology, we identified eight candidate amino acids that might be responsible for the functional divergence between the TFL1-like and FT-like subfamilies (Figure 2B), among which residues 88, 142, 144, and 156 were reported to play important roles in regulating flowering time (Hanzawa et al., 2005; Ho and Weigel, 2014).
Figure 2.
Frequency of Candidate Amino Acid Residuals Responsible for the Functional Divergence between Subfamilies and Subgroups.
Frequency of candidate amino acid residuals responsible for the functional divergence between MFT-like and FT/TFL1-like subfamily (A), FT-like and TFL1-like subfamily (B), Group VI and other TFL1 subfamily groups (C), and Groups VII and VIII (D). The gray color in each pie indicates the frequency of nonlabeled amino acid.
Likewise, we inferred the amino acids responsible for the functional divergence between groups within the TFL1-like and FT-like clades, respectively, based on the association between nonsynonymous substitution frequency differentiation and phenotypic variation. Changes at residues 117, 131, and 156 may be responsible for the functional divergence between Group VI and the other TFL1-like groups (Figure 2C) in regulating branch, and changes at the residue 107 may be responsible for the functional divergence between Groups VII and VIII (Figure 2D) in regulating architecture and seed germination. In addition, we found that the weaker activation of flowering by Groups I and II compared with Groups III, IV, and V might be due to substitutions at the conserved amino acid residues 43, 49, 80, 81, and 119 (Supplemental Figure 12).
Amino Acid Residues Responsible for Rapid Functional Divergence in Several Young Duplicated PEBP Family Members
Although most of the PEBP members within groups shared conserved functions, there were some exceptions consisting of rapid functional divergence (Figure 1). For example, Gm08g47820 exhibited later flowering and indeterminate architecture compared with other members in Groups I and II. The genetic basis of its functional divergence cannot be explained by the above substitutions.
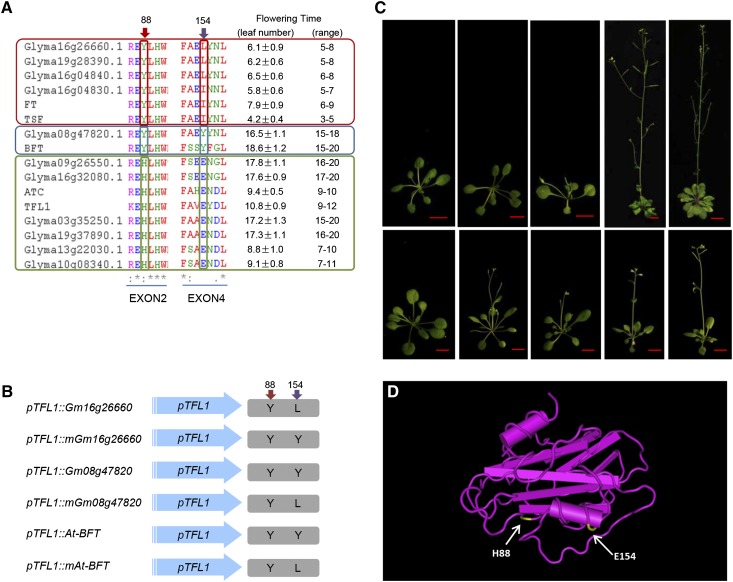
To identify amino acid residues explaining its functional divergence, we carefully investigated the association between the amino acid residue variations and functional divergence. Amino acid residue H88Y determined the functional conversion of Arabidopsis TFL1 and FT. The Tyr amino acid residue activates flowering, while His represses flowering (Hanzawa et al., 2005). Our transgenic results showed that most of the transgenic lines containing Tyr at this location were consistent with this prediction (Figure 1; Supplemental Figure 12), confirming the importance of this amino acid residue. However, Gm08g47820 and At-BFT resulted in later flowering in the transgenic lines even though they contain Tyr at this location (Figure 3A). These inconsistent results indicated that other amino acid residues in addition to H88Y may play important roles in regulating flowering. To identify these amino acid residues, we searched for amino acids that are shared by Gm08g47820 and At-BFT but are not shared by other FT members. We identified amino acid residue 154 as a candidate. Both Gm08g47820 and At-BFT contain Tyr at this location, while the other FT subfamily members contain Leu or Ile. Therefore, the Tyr at 154 amino acid residue might repress flowering.
Figure 3.
L154Y Is Responsible for Flowering Regulation.
(A) Analysis of flowering time and critical amino acid residues in the FT subfamily members (red panel on the top), BFT and Gm08g47820 (blue panel in the middle), and TFL1 subfamily members (green panel at the bottom). The red and purple arrows indicate the critical sites 88H/Y and 154L/Y, respectively.
(B) Schematic diagram of the transgenic constructs in which wild-type and mutant forms of Gm16g26660, BFT, and Gm08g47820 are driven by the Arabidopsis TFL1 promoter. The red and purple arrows indicate the critical sites 88H/Y and 154L/Y, respectively.
(C) Phenotypic analysis of transgenic plants. The upper panels, from left to right, are 2-week-old Col-0, 2-week-old tfl1-1, 2-week-old TFL1pro:Gm16g26660, 5-week-oldTFL1pro:Gm08g47820, and 6-week-old TFL1pro:At-BFT; the bottom panels, from left to right, are 3-week-old Col-0, 3-week-old tfl1-1, 3-week-old TFL1pro:mGm16g26660, 3-week-old TFL1pro:mGm08g47820, and 3-week-old TFL1pro:mAt-BFT. The plants were grown under long-day conditions at 22°C. Bar = 1 cm.
(D) Predicted crystal structure of the TFL1 protein. The arrows indicate the critical amino acid positions His-88 and Leu-154.
To validate this hypothesis, we constructed point mutants of several proteins, including Gm16g26660, Gm08g47820, and At-BFT (Figure 3B). The Leu residue was mutated to Tyr in TFL1pro:mGm16g26660, and the Tyr residues were mutated to Leu inTFL1pro:mGm08g47820 and TFL1pro:mAt-BFT (Figure 3B). The transgenic lines from these point mutation constructs were compared with the non-point-mutation transgenic lines (Figure 3C). As expected, the point mutation of Leu→Tyr in TFL1pro:mGm16g26660 delayed flowering compared with TFL1pro:Gm16g26660, and the point mutation of Tyr→Leu in TFL1pro:mGm08g47820 and TFL1pro:mAt-BFT promoted flowering compared with TFL1pro:Gm08g47820 and TFL1pro:At-BFT, respectively (Figure 3C; Supplemental Table 3). These results confirmed that amino acid residue 154 affects flowering. However, this conserved residue cannot completely explained flowering variations, as the TFL1pro:mGm08g47820 and TFL1pro:mAt-BFT transgenic lines did not flower as early as TFL1pro:Gm16g26660 even though these lines shared the same amino acid residues at these two residues. Similarly, TFL1pro:mGm16g26660 did not flower as late as TFL1pro:Gm08g47820 and TFL1pro:At-BFT. In addition, although the TFL1-like subfamily members contained the same amino acid residues at these two positions, the corresponding transgenic lines exhibited divergent flowering times (Figure 3A). These results indicated that other amino acid residues may also affect flowering.
Using a similar strategy, we found that amino acid residues 10 and 113 may be also responsible for the flowering variation. In addition, Gm02g07650, Glyma08g47810, and Glyma08g28470 lost the ability to interact with At-FD (Figure 1). We found they contained many substitutions at the conserved amino acid residues of the TFL1/FT clade (Supplemental Figure 12), suggesting that they may have become pseudogenes or acquired new functions.
Structural Distribution of the Residues Putatively Responsible for the Functional Divergence
It was presumed that the fourth exon plays a critical role, and variation in the sequence of this exon could affect the crystal structure (Ahn et al., 2006). Our alignment results showed that in addition to the fourth exon, many conserved and deduced functional amino acid residues were located in the second and third exons (Supplemental Figure 12). It was reported that the potential ligand binding pocket was also important for the functional conversion of Arabidopsis FT (Ho and Weigel, 2014). Amino acid residue 154, which we determined to regulate flowering time in the above analysis, was located near the α-helix (Figure 3D). We then investigated the location of each functional amino acid residue based on the predicted crystal structure of TFL1 (Supplemental Figure 13). We found that most of the candidate critical amino acid residues were located on the loops near the core structure of the β-sheet or α-helix (Supplemental Figure 13), suggesting that the loops were critical for the function of PEBP members.
Expression Pattern Divergence of the PEBP Members
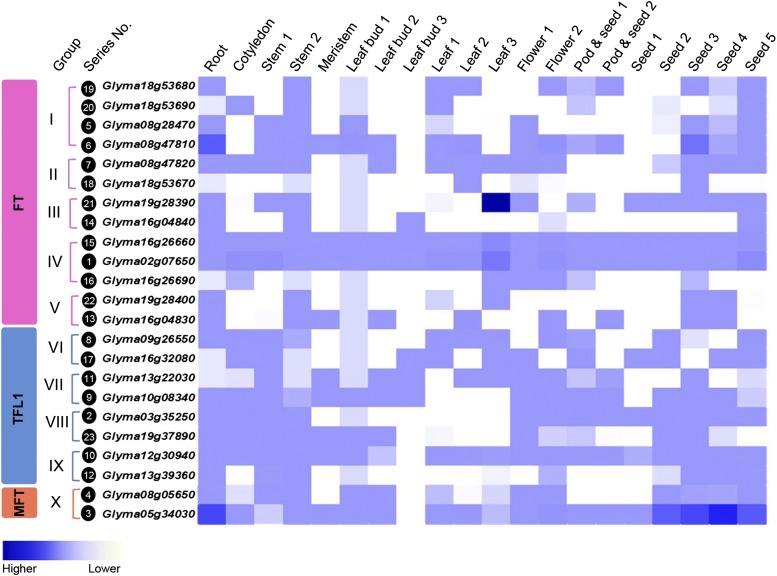
In addition to the functional divergence at the protein level, expression pattern divergent is another important force in driving the functional division of duplicated genes (Force et al., 1999; Oakley et al., 2006). It has been reported that the expression patterns of the three main PEBP members in Arabidopsis have diverged: TFL1 is mainly expressed in young inflorescence tissues (Bradley et al., 1997), FT expression is limited to the vasculature (Adrian et al., 2010), and MFT expression gradually increases in germinated seeds (Xi et al., 2010). Our previous study suggested that the divergent expression patterns of two recently duplicated PEBP genes, Glyma19g37890 and Glyma03g35250, led to different dominance in controlling stem determinacy in soybean (Tian et al., 2010).
To detect the expression divergence of soybean PEBP members, transcriptional profiling was performed by real-time PCR (Figure 4). We found that most of the duplicated genes, including the recently duplicated gene pairs, exhibited divergence in expression pattern. For instance, no expression of Glyma16g04840 was detected during the early stages of seed development, whereas relatively high expression of its paired gene, Glyma19g28390, was detected (Figure 4). We assessed the differences between gene expression profiles using euclidean distances. No significant differences were detected for the three classes, within-group, within-subfamily, or between-subfamilies (Supplemental Figure 3C). Consistent with this, the pairwise euclidean distance did not correlate with the dS (Supplemental Figure 3D).
Figure 4.
Expression Patterns of PEBP Members in Soybean.
Expression analyses of the 23 soybean PEBP members in 20 different tissues as detected via quantitative PCR. The values are the means of four independent replicates.
DISCUSSION
Polyploidy is common in flowering plants and is a major force in promoting genome evolution (Adams and Wendel, 2005). The subsequent divergence of surviving duplicated genes provides raw genetic resources for adaptive evolution and the gain of novel gene function (Flagel and Wendel, 2009). Clarifying how the duplicated genes diverge is important to understanding the evolution of duplicated genes. A particular challenge is to identify the substitutions responsible for functional conservation and divergence (reviewed in Conant and Wolfe, 2008; Freeling, 2009; Innan and Kondrashov, 2010; Wang et al., 2012). In this study, we clarified the evolutionary processes of the PEBPs in soybean and Arabidopsis and highlight the important role of amino acid substitution in the functional divergence of duplicated genes. Our results also suggested that a systematic evolutionary functional assay of a duplicated gene family would be a powerful approach to identify functional critical amino acids.
Functional Evolution of PEBPs in Soybean
The PEBP family is one of the most ancient gene families, with a highly conserved gene structure and high protein sequence similarities across species (Banfield et al., 1998; Hengst et al., 2001; Karlgren et al., 2011). This gene family is also well known for its two members, TFL1 and FT, whose amino acids share a high degree of similarity but act in opposite manners in controlling flowering time in Arabidopsis (Kardailsky et al., 1999; Kobayashi et al., 1999). Studies in Arabidopsis have indicated that the functions of members of different subfamilies differ significantly, whereas within each subfamily, they showed functional redundancy. For instance, TFL1 functions as a floral repressor, while FT functions as a floral activator (Kardailsky et al., 1999; Kobayashi et al., 1999). MFT mainly plays a critical role in regulating seed germination via the ABA and gibberellic acid signaling pathways (Xi et al., 2010). TSF, a homolog of FT, was reported to regulate flowering via a mechanism similar to that of FT (Yamaguchi et al., 2005). ATC and BFT, two members of the TFL1-like subfamily, possess similar activity as TFL1 in regulating inflorescence meristem development (Mimida et al., 2001; Chung et al., 2010; Yoo et al., 2010). However, the evolutionary process of PEBP members remains unclear, and much less of the amino acids critical for their functional divergence.
Through a systematic investigation of the functional variation of PEBPs in two species, Arabidopsis and soybean, our results showed that the degree of functional divergence between individual PEBP members at the protein level appeared to associate with duplication age. PEBP members within the same subfamily have similar functions, whereas those in different subfamilies exhibit prominent functional divergence. Subsequently, through association analysis, we identified the substitutions corresponding to the functional divergence of each duplication event. Many of these candidate amino acids have been verified by previous functional studies. In addition, we validated a rare allele responsible for the functional divergence in flowering time in this study. Together, the findings in this study illuminated a clear functional evolutionary history of the PEBP family (Figure 5).
Figure 5.
A Deduced Evolutionary Process of the PEBP Family.
Each duplication event is deduced based on the phylogenetic analysis of PEBP members in seven species. The differently selected amino acids in duplications are labeled above each corresponding duplication event. The functional variants of different subclades after duplications are described below the corresponding clade.
The MFT-like subfamily mainly affects seed germination. After the duplication event of D0, seven amino acids were differentially selected in two subclades, which led to the common ancestor of the TFL1-like and FT-like families gaining an interaction with FD in addition to affecting seed germination. Subsequently, the TFL1-like and FT-like families were separated by the D1 duplication event, during which eight amino acids were further differentially selected. The selection of these amino acids, or some of these amino acids, led to the divergence in regulating flowering time: FT-like functions as an activator, while TFL1-like functions as a repressor.
Subsequently, in the TFL1-like subfamily, rapid divergence occurred following several duplication events. The duplication of D2-1 separated Group VI and At-BFT from the ancestor of the other members. Unlike transgenic lines corresponding to other TFL1-like members, Group VI transgenic lines exhibit a branching phenotype. This phenotypic difference might be caused by Group VI members gaining a new function, or in contrast, other members showing a weak architecture phenotype after the duplication D2-1. During duplication D2-1, three amino acids were differentially selected, which could be responsible for the phenotypic divergence following this event. Duplication D2-3 separated Group VII from Group VIII in soybean. The functions of repressing flowering time and architecture were weaker in Group VII than Group VIII, whereas the function in seed germination was stronger. The function variation might be due to the differences of amino acid 103. In the FT-like subfamily, Groups I and II members showed decreased function in active flowering after the D3-1 and D3-2 duplications, which may be caused by the substitutions at four amino acid residues. In addition, we found several recently duplicated members, Gm08g28470, Gm08g47810, and Gm02g07650, that obtained substitutions in critical amino acids, which resulted in losing the interaction with FD and in turn losing the ability to activate flowering. In contrast, Gm08g47820 gained the ability to repress flowering, which might be caused by the substitution of the amino acid Tyr-150.
Therefore, our study demonstrates a clear evolutionary process of PEBP members. Although functional analyses of the PEBP family have been thorough in Arabidopsis, most of them were performed separately, which failed to give a comprehensive survey of how the functions of different subfamilies or members diverged or even converted. Our results answered the questions of the subfamily and group formation after different duplication events, the subsequent functional divergence, and critical amino acids responsible for functional divergence, which enhance our understanding of the mechanisms for duplicated gene evolution and highlights the role of natural selection in driving evolutionary novelty after duplication.
A Comprehensive Functional Assay Leads to the Identification of Critical Amino Acid Residues Responsible for Duplicated Gene Evolution
The identification of critical amino acid residues for functional divergence of duplicated genes or families helps to elucidate their function. Amino acid residues are typically identified via mutant screening, point mutation assays, and sequence alignments. Using a point mutation assay, Hanzawa et al. (2005) identified a single amino acid that was critical for the functional conservation of Arabidopsis TFL1 and FT. By comparing the crystal structures of TFL1 and FT, Ahn et al. (2006) determined that the fourth exon was important for the functional divergence of TFL1 and FT. However, our understanding of the amino acid residues that are essential for functional conservation and divergence among different PEBP members remains limited.
Here, our results revealed that a comprehensive rescue assay using an endogenous promoter is another powerful method by which to identify critical amino acids responsible for the functional divergence of duplicated genes. The sequence alignment in this study indicated that some amino acid residues are conserved in almost all members, and the substitutions in these conserved amino acids could result in a loss of function, suggesting their critical role in the maintenance of the basic function of this family. Furthermore, in the rescue experiments using an endogenous promoter, only the coding sequences differed, while the constructs and transformation were identical, indicating that the phenotypic variations were associated with the differences in the amino acid sequences. This method led us to identify specific amino acid residues that are critical for the functional divergence of the PEBP family members, such as amino acid residue 154, which is important for regulating flowering time. In addition to previously reported amino acid residues, we identified many new residues that may play important roles in the functional divergence or even conversion of different subfamilies or groups, indicating that the function of the PEBPs may be more complicated than anticipated. The regions/amino acid residues or their combinations in each PEBP protein may play different roles. A detailed functional assay may help clarify the functional regulatory network of each PEBP member. The results from this study will be highly valuable for future detailed functional analyses of PEBPs.
In summary, through a systemic investigation of the functional conservation and divergence of PEBP family members in soybean and Arabidopsis, we clarified the evolutionary process of PEBPs and determined domains/amino acid residues critical for the functional divergence of duplicated genes. This study provides insights into the identification of critical amino acids for duplicated gene evolution and strengthens our understanding of the evolutionary course of duplicated genes.
METHODS
Plant Material, Growth Conditions, and Transgenic Plant Analysis
The soybean (Glycine max) cultivar Williams 82 was grown at the Experimental Station of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, in Beijing from May to September. The tissues were collected during different developmental stages and quickly frozen in liquid nitrogen. Arabidopsis thaliana plants were grown at 22°C under a 16-h-light/8-h-dark photoperiod at 100 μmol m–2 s–1 in soil or on Murashige and Skoog medium (Sigma-Aldrich) containing 1.5% sucrose and 0.8% agar. The wild-type Arabidopsis Col-0 accession was used in this study. The tfl1-1, mft-2, and ft-10 mutants are in the Col-0 background.
Transgenic plants were generated using the floral dip method for Agrobacterium tumefaciens transformation (Clough and Bent, 1998). All T0 transgenic seeds were sterilized with bleach (10%) for 10 min, washed five times with sterilized water, selected on hygromycin-supplemented Murashige and Skoog plates, and transferred to soil after 10 d.
For the flowering time analysis, at least 20 plants were selected to monitor the flowering time by counting the rosette leaves on the main inflorescence from each transgenic and nontransgenic Col-0, tfl1-1, and ft-10 plant. The seed germination assay was performed as previously described (Xi et al., 2010).
Identification of the PEBP Genes and Phylogenetic Analysis
PEBP homologs were identified in soybean by similarity searches using BLAST analysis (Altschul et al., 1990). The protein sequences of the individual PEBP family members from Arabidopsis were used as queries against the soybean genome (Schmutz et al., 2010). Duplicated gene pairs of PEBP members were identified based on previous whole-genome analyses of duplicated regions and genes (Schmutz et al., 2010; Du et al., 2012).
Phylogenetic analysis was performed following a previously reported method (Jiao et al., 2011). Briefly, multiple sequence alignments were conducted using MUSCLE (Edgar, 2004) with the default parameters, and then the sequences were trimmed using TRIMAL 1.2 (Capella-Gutiérrez et al., 2009) with the option of “automated1.” The corresponding coding sequences were aligned. The phylogenetic tree was constructed using the maximum-likelihood method using RAxML (Stamatakis, 2014). The numbers of synonymous substitution sites (dS) and nonsynonymous substitution sites (dN) between duplicates were estimated using PAML (Yang, 1997). The alignment corresponding to Figure 1 is shown in Supplemental Data Set 1, and the alignment corresponding to Supplemental Figure 1 is shown in Supplemental Data Set 2.
Gene Expression Assay
Total RNA of the 20 samples was isolated using the RNeasy Plant Mini Kit (Qiagen 74904). After treatment with DNase I (New England M0303S) at 37°C for 30 min, 1 μg of RNA was used for first-strand cDNA synthesis using SuperScript II reverse transcriptase following the manufacturer’s instructions (Invitrogen 18064-014). Real-time PCR was performed on a Roche LightCycler 480 system with LightCycler 480 SYBR Green I Master Mix (Roche 04887352001). Soybean ACTIN-11 was used as a control, and the relative expression level was calculated using the 2−ΔΔCT method.
Plasmid Construction
The primers used in this study are listed in Supplemental Table 4.
For theTFL1pro:PEBP constructs, a 2.2-kb 5′-UTR region of Arabidopsis TFL1 was amplified and ligated into the pCAMBIA1391 vector at the BamHI and SpeI sites to construct backbone I. Then, the full coding regions of the different homologous genes were amplified using specific primers (PEBP-CDS-SpeI and PEBP-CDS-PmlI; Supplemental Table 4) and ligated into the pZERO-blunt vector; the correct clone was then cleaved with SpeI and PmlI and ligated into backbone I.
For the TFL1pro:PEBP:3′-UTR constructs, a 4.7-kb 3′-UTR region of Arabidopsis TFL1 was amplified (Kaufmann et al., 2010) and ligated into backbone I at the BstEII site to construct backbone II. Then, the full coding regions of the different homologous genes were amplified using specific primers (PEBP-CDS-SpeI and PEBP-CDS-KpnI; Supplemental Table 4), digested with SpeI and KpnI, and ligated into backbone II.
For the MFTpro:PEBP constructs, a 1.8-kb 5′-UTR fragment of Arabidopsis MFT (Xi et al., 2010) was amplified using specific primers and ligated into pCAMBIA1391. The final constructs were constructed in a manner similar to that used for the At-TFL1pro:PEBP constructs.
For the FTpro:PEBP constructs, the 8.1-kb FTpro-pDONR207 and GW-MCS-NOS-pGREEN (Corbesier et al., 2007; Adrian et al., 2010) plasmids were used. The coding regions of the PEBPs were amplified using specific primers and ligated into the multiple cloning site of GW-MCS-NOS-pGREEN. The 8.1-kb Arabidopsis FT promoter was introduced into the correct clone through an LR reaction to produce the final constructs.
For the yeast two-hybrid assay constructs, the full-length coding regions of the different PEBP homologs were amplified using specific primers and ligated into pGADT7 at the SmaI and KpnI sites. The Arabidopsis FD full-length coding region was amplified using specific primers and ligated into pGBKT7.
For the subcellular location assay, the full-length coding regions of the different PEBP homologous genes were amplified using specific primers, fused with green fluorescent protein (GFP) at the C terminus, and cloned into the above pUC19 vector at the SpeI and KpnI sites to produce the 35Spro:PEBP:GFP constructs.
Yeast Two-Hybrid Assay
The yeast two-hybrid analysis was performed using the Matchmaker GAL4 Two-Hybrid System 3 according to the supplier’s instructions (Clontech). The PEBP family genes were fused with the activation domain of GAL4 in the yeast vector pGADT7 and cotransformed into the yeast strain AH109 with the pGBKT7 vector-fused FD genes. The resulting yeast cotransformants were screened on synthetic complete (SC) medium with galactose lacking leucine and tryptophan (SC/-Leu/-Trp) and on SC medium with galactose lacking leucine, tryptophan, adenine, and histidine (SC/-Leu/-Trp/-Ade/-His) at 28°C.
Subcellular Localization of PEBP Family Proteins
For the subcellular localization, the PEBPs and 35Spro:PEBP:GFP constructs were transformed into Arabidopsis protoplast cells as described by Yoo et al. (2007). After overnight incubation at 25°C, GFP fluorescence was observed using a confocal laser scanning microscope (LSM510; Carl Zeiss).
Critical Amino Acid Residues Analysis
Three-dimensional structures of At-TFL1 (PDB ID, 1WKO; MMDB ID, 33641) were obtained from the MMDB (http://www.ncbi.nlm.nih.gov/Structure/mmdb), and all the critical amino acid residues were visualized and marked by Cn3D macromolecular structure viewer (version 4.3).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At-TFL1 (AT5G03840), At-FT (AT1G65480), At-MFT (AT1G18100), At-TSF (AT4G20370), At-BFT (AT5G62040), At-ATC (AT2G27550), At-FD (AT4G35900), Gm-DT1 (Glyma19g37890), and Gm-Actin11 (Glyma18g52780).
Supplemental Data
Supplemental Figure 1. PEBP Members in Arabidopsis and Soybean, and Duplications of PEBP Members in Soybean.
Supplemental Figure 2. Gene Structures of 29 PEBP Members in Soybean and Arabidopsis.
Supplemental Figure 3. Divergence of Different PEPB Members from Within-Group, Within-Subfamily, and Between-Subfamilies.
Supplemental Figure 4. Schematic Diagram of the Constructs Used in the Rescue Experiment.
Supplemental Figure 5. Phenotypes of the TFL1pro:PEBPs:3UTR Transgenic Plants.
Supplemental Figure 6. Phenotypes of the TFL1pro:PEBPs Transgenic Plants.
Supplemental Figure 7. Phenotype Comparison of Representative TFL1pro:PEBPs and TFL1pro:PEBPs:3UTR Transgenic Plants.
Supplemental Figure 8. Germination Rates of the MFTpro:PEBPs Transgenic Plants.
Supplemental Figure 9. Phenotypes of the FTpro:PEBPs Transgenic Plants.
Supplemental Figure 10. Protein-Protein Interaction Assay of PEBPs and Arabidopsis FD.
Supplemental Figure 11. Subcellular Localization of the PEBP Members in Soybean and Arabidopsis.
Supplemental Figure 12. Alignments of the Amino Acid Sequences of the PEBP Proteins in Seven Species.
Supplemental Figure 13. Critical Amino Acid Residues Marked in the At-TFL1 Predicted Crystal Structures.
Supplemental Table 1. Phenotyping of the Transgenic Lines in the tfl1-1 Background.
Supplemental Table 2. Phenotyping of the Transgenic Lines in the ft-10 Background.
Supplemental Table 3. Flowering Times of Point Mutation Transgenic Lines.
Supplemental Table 4. Primers Used in This Study.
Supplemental Data Set 1. Text File of Alignment Corresponding to the Phylogenetic Analysis in Figure 1.
Supplemental Data Set 2. Text File of Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 1A.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for their constructive comments. We thank Chung-I Wu (Beijing Institute of Genomics, Chinese Academy of Sciences), Wenfeng Qian (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), and Wei Qian (Institute of Microbiology, Chinese Academy of Sciences) for their valuable suggestions in the evolutionary analysis. We also thank Yu Hao (National University of Singapore) for the kind donation of the Arabidopsis mft-2 seeds, Ji Hoon Ahn (Korea University) for the kind donation of the Arabidopsis ft-10 seeds, and Franziska Turck (Max Planck Institute for Plant Breeding Research) for the kind donation of the Arabidopsis FT promoter 8.1kbFTpro-pDONR207 and GW-MCS-NOS-pGREEN plasmids. This work was supported by the National Natural Science Foundation of China (Grants 91131005 and 31222042) and by a special program from the State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences.
AUTHOR CONTRIBUTIONS
Z.W., J.M., T.T., and Z.T. conceived this project and designed all of the experiments. Z.W., Y.L., Y.S., M. Wang, T.L., Y.J., C.L., Q.L., M. Wu, and Z.T. performed the experiments. Z.Z., T.T., Z.W., T.L., C.F., and Z.T. analyzed the data. J.M., T.T., Z.W., and Z.T. wrote the article.
Glossary
- WGD
whole-genome duplication
- ABA
abscisic acid
- CDS
coding sequence
- UTR
untranslated region
- Col-0
Columbia-0
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- Adams K.L., Wendel J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J.H., Miller D., Winter V.J., Banfield M.J., Lee J.H., Yoo S.Y., Henz S.R., Brady R.L., Weigel D. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25: 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Banfield M.J., Barker J.J., Perry A.C., Brady R.L. (1998). Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 6: 1245–1254. [DOI] [PubMed] [Google Scholar]
- Blanc G., Hokamp K., Wolfe K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438. [DOI] [PubMed] [Google Scholar]
- Bradley D., Ratcliffe O., Vincent C., Carpenter R., Coen E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona M.J., Calonje M., Martínez-Zapater J.M. (2007). The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 63: 637–650. [DOI] [PubMed] [Google Scholar]
- Chung K.S., Yoo S.Y., Yoo S.J., Lee J.S., Ahn J.H. (2010). BROTHER OF FT AND TFL1 (BFT), a member of the FT/TFL1 family, shows distinct pattern of expression during the vegetative growth of Arabidopsis. Plant Signal. Behav. 5: 1102–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Hou Z., Ananiev E.V., Simmons C.R. (2008). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146: 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Tian Z., Sui Y., Zhao M., Song Q., Cannon S.B., Cregan P., Ma J. (2012). Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183: 557–564. [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453. [DOI] [PubMed] [Google Scholar]
- Gill N., Findley S., Walling J.G., Hans C., Ma J., Doyle J., Stacey G., Jackson S.A. (2009). Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 151: 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Goto K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y., Money T., Bradley D. (2005). A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 102: 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman H., Källman T., Lagercrantz U. (2009). Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 70: 359–369. [DOI] [PubMed] [Google Scholar]
- Hengst U., Albrecht H., Hess D., Monard D. (2001). The phosphatidylethanolamine-binding protein is the prototype of a novel family of serine protease inhibitors. J. Biol. Chem. 276: 535–540. [DOI] [PubMed] [Google Scholar]
- Ho W.W., Weigel D. (2014). Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26: 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L. (1994). The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256: 119–124. [DOI] [PubMed] [Google Scholar]
- Innan H., Kondrashov F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108. [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al. ; French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467. [DOI] [PubMed] [Google Scholar]
- James L.C., Tawfik D.S. (2003). Conformational diversity and protein evolution—a 60-year-old hypothesis revisited. Trends Biochem. Sci. 28: 361–368. [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Jung C.H., Wong C.E., Singh M.B., Bhalla P.L. (2012). Comparative genomic analysis of soybean flowering genes. PLoS ONE 7: e38250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Karlgren A., Gyllenstrand N., Källman T., Sundström J.F., Moore D., Lascoux M., Lagercrantz U. (2011). Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol. 156: 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kondrashov F.A., Rogozin I.B., Wolf Y.I., Koonin E.V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3: RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimida N., Goto K., Kobayashi Y., Araki T., Ahn J.H., Weigel D., Murata M., Motoyoshi F., Sakamoto W. (2001). Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6: 327–336. [DOI] [PubMed] [Google Scholar]
- Oakley T.H., Ostman B., Wilson A.C. (2006). Repression and loss of gene expression outpaces activation and gain in recently duplicated fly genes. Proc. Natl. Acad. Sci. USA 103: 11637–11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. (New York: Springer; ). [Google Scholar]
- Papp B., Pál C., Hurst L.D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197. [DOI] [PubMed] [Google Scholar]
- Ramsey J., Schemske D. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501. [Google Scholar]
- Schlueter J.A., Lin J.Y., Schlueter S.D., Vasylenko-Sanders I.F., Deshpande S., Yi J., O’Bleness M., Roe B.A., Nelson R.T., Scheffler B.E., Jackson S.A., Shoemaker R.C. (2007). Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Sémon M., Wolfe K.H. (2007). Consequences of genome duplication. Curr. Opin. Genet. Dev. 17: 505–512. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Wang X., Lee R., Li Y., Specht J.E., Nelson R.L., McClean P.E., Qiu L., Ma J. (2010). Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. USA 107: 8563–8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia R.A., Bottani S., Birchler J.A. (2008). Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24: 390–397. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang X., Paterson A.H. (2012). Genome and gene duplications and gene expression divergence: a view from plants. Ann. N. Y. Acad. Sci. 1256: 1–14. [DOI] [PubMed] [Google Scholar]
- Wendel J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Xi W., Liu C., Hou X., Yu H. (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22: 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46: 1175–1189. [DOI] [PubMed] [Google Scholar]
- Yang Z. (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoo S.J., Chung K.S., Jung S.H., Yoo S.Y., Lee J.S., Ahn J.H. (2010). BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 63: 241–253. [DOI] [PubMed] [Google Scholar]
- Yoo S.Y., Kardailsky I., Lee J.S., Weigel D., Ahn J.H. (2004). Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol. Cells 17: 95–101. [PubMed] [Google Scholar]
- Zhang J., Rosenberg H.F., Nei M. (1998). Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.