Abstract
Advancing the quest for new drug targets demands the development of innovative plasma membrane proteome research strategies applicable to small, functionally defined tissue samples. Biotinylation of acute tissue slices and streptavidin pull-down followed by shotgun proteomics allowed the selective extraction and identification of >1,600 proteins of which >60% are associated with the plasma membrane, including (G-protein coupled) receptors, ion channels and transporters, and this from mm3-scale tissue.
The plasma membrane (PM) physically separates a cell from its external environment and is composed out of a lipid bilayer and associated proteins1,2,3. The PM proteome is very dynamic because of extensive trafficking between the PM and the endomembrane compartment of eukaryotic cells via exocytosis, endocytosis and recycling processes4,5. PM proteins (PMPs) like (G-protein coupled) receptors, ion channels and transporters are crucial for a wide variety of fundamental physiological processes6. Targeted profiling of this PM proteome, and specifically the proteome exposed at the cell surface, is key to e.g. the identification of cell surface biomarkers or the isolation of tissue-specific cell types2,7,8,9. Their role in cell-cell interactions, molecular transport and signalling explains their potential as important therapeutic targets1,10,11.
PMPs exist in two main forms, the integral cell surface proteins spanning the lipid bilayer and the peripheral proteins, anchored to the PM1. This heterogeneity, the low overall abundance and hydrophobic nature, which results in poor solubility, few trypsin cleavage sites and difficult accessibility for proteases, make proteomic analysis of PMPs challenging1,12. Traditional isolation of PMPs from biological tissue samples by subcellular fractionation based on ultracentrifugation suffers from weak enrichment and contamination from other cellular compartments1,7. It also requires high sample loads, being a major disadvantage particularly in e.g. the field of neuroscience research where, usually, sample quantities are limited6,13.
It has been demonstrated that biotinylation of cell surface-exposed proteins followed by affinity purification from cell lines or cell cultures offers a usable alternative to the classical ultracentrifugation for the specific extraction and enrichment of PMPs3,7,14,15. In 2003, Thomas-Crusells and colleagues developed and optimized a comparable method for the biotinylation of such cell surface proteins in acute brain slices16. This, in combination with standard immunoblotting for predefined PMPs17,18,19, created the opportunity to study PMP trafficking in a more natural and physiologically relevant experimental setting16,20. Simultaneous ex vivo slice experiments such as electrophysiological recordings can be performed16. To our knowledge, biotinylation of acute tissue slices in conjunction with the proteomic profiling of the PM proteome has not yet been reported. Nevertheless, it holds the potential to solve both the problem of poor extraction efficiency and of high sample consumption characteristic to the more common tissue extraction protocols based on ultracentrifugation used in plasma membrane proteomics today.
Results and Discussion
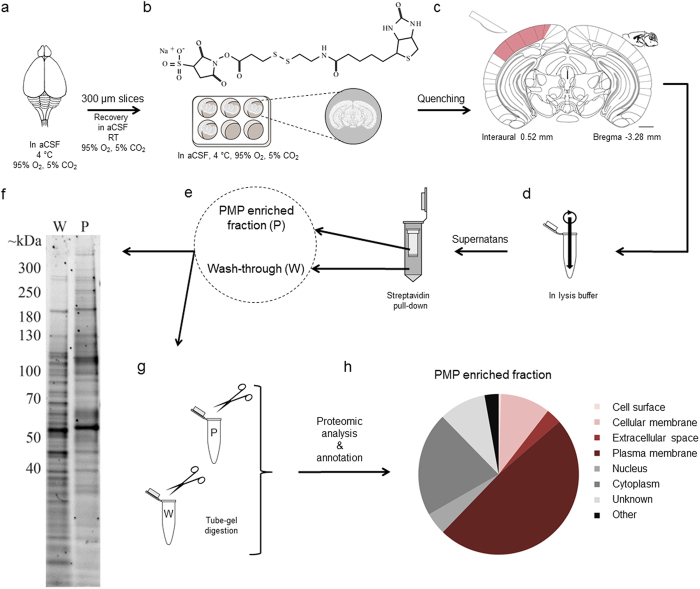
In this study, we performed an ‘acute slice biotinylation assay’ (ASBA) on mouse coronal brain slices (Fig. 1a–d) followed by streptavidin pull-down to separate cell surface-associated proteins in a subfraction termed the ‘PMP enriched fraction’, from the rest of the proteome termed the ‘wash-through fraction’ (Fig. 1e). Traditionally, biotinylation of acute slices and affinity purification is used in combination with immunoblotting to investigate trafficking of receptors and transporters in and out the PM in anatomically or functionally delineated regions of interest in a tissue16 such as mouse visual cortex in the forebrain17. With the intention to verify the applicability of ASBA in combination with proteomic analysis independent of a priori assumptions about the identity of PMPs of potential biological interest, and to a lot smaller tissue samples, we also isolated mouse visual cortex tissue (Fig. 1c; red), but on mm3-scale as study sample.
Figure 1. Workflow for plasma membrane proteomic analysis of small tissue samples.
(a) Dissect organ of interest, like mouse brain, in artificial cerebrospinal fluid (aCSF). (b) Make slices, allow recovery, label with EZ-Link Sulfo-NHS-SS-Biotin. After quenching, dissect region of interest like the visual cortex (c, red) and mechanically homogenize (d). (e) Separate the plasma membrane protein (PMP) enriched fraction (P) from the rest of the proteome (wash-through, W) by streptavidin pull-down. Panel (f) illustrates SDS-PAGE for P and W. After digestion (g) analyse the protein samples and annotate (h). c adapted from30. Scale bar: 1 mm.
To judge the reproducibility of ASBA and streptavidin pull-down, a total protein stain was performed on 1 μg of proteins separated on SDS-PAGE belonging to the PMP enriched fractions and the wash-through fractions (Fig. 1f) derived from 5 different brain samples. The resulting pattern of protein bands, with a predominant location in the higher Mw regions, appeared identical for each of the 5 PMP enriched fractions and differed markedly from the pattern of protein bands, identical between all 5 wash-through fractions. For each of these protein samples we calculated the relative proportion of protein quantity in its PMP enriched fraction to the initial total protein content, that is the sum of the wash-through and PMP enriched fraction. The percentage of proteins in the PMP enriched fraction from each of the extracts ranged between 6.0 and 7.2%.
The clear dissimilarity in band pattern between a PMP enriched fraction and wash-through of one and the same ASBA extract (Fig. 1f), is indicative of a clear difference between the proteins retained on the Streptavidin agarose resin versus those in the eluent. This prompted us to identify the proteins present in the two fractions of each of the 5 brain samples using shotgun proteomics (Table 1 and Supplementary Data 1 and 2). An intermediate step of tube-gel digestion on 25 μg proteins per fraction improved solubilisation and digestion efficiency of membrane proteins21, and facilitated the removal of detergents prior to mass spectrometric analysis of 1 μg samples (Fig. 1g,h). Next, we used IPA to categorize each identified protein present in the 5 PMP enriched fractions into their subcellular compartment, as an extra validation of the capability of the ASBA method to truly enrich PMPs from a proteomic sample. The percentage of proteins categorised as PMPs by IPA in the 5 separate PMP enriched fractions ranged between 26.8 and 28.8%, illustrating enrichment in and reproducibility of our workflow (Table 1).
Table 1. Percentage of PMPs in the 5 PMP enriched fractions.
| sample | # of ≠ IDs | # of ≠ IDs annotated by IPA | # of PMPs (1° IPA) | % of PMPs | |
|---|---|---|---|---|---|
| PMP ENRICHED FRACTION | |||||
| 1 | 968 | 934 | 250 | 26.8 | |
| 2 | 927 | 896 | 242 | 27.0 | |
| 3 | 872 | 846 | 244 | 28.8 | |
| 4 | 922 | 890 | 254 | 28.5 | |
| 5 | 993 | 968 | 270 | 27.9 | |
| merge 1,2,3,4,5 | 1,698 | 1,625 | 417 | 25.7 | |
| WASH-THROUGH FRACTION | |||||
| 1 | 1,594 | ||||
| 2 | 1,587 | ||||
| 3 | 1,640 | ||||
| 4 | 1,494 | ||||
| 5 | 1,584 | ||||
| merge 1,2,3,4,5 | 2,872 | ||||
For a more detailed analysis of the plasma membrane proteome, we then merged all identifications of the 5 PMP enriched fractions together into 1 protein list. We also merged the identification lists of the 5 wash-through fractions. This resulted in respectively 1,698 and 2,872 discrete proteins that were identified in that PMP enriched fraction and wash-through fraction (Table 1). Of these 1,698 proteins identified in this PMP enriched fraction, IPA successfully annotated 1,625 proteins. Out of these 1,625, 417 proteins or 25.7% were classified as PMP (Table 1 and 2). Because IPA only provides one subcellular localization per protein and does not consider the additional cellular compartments in which a protein can occur15, secondary annotations were also checked in IPA and DAVID, in combination with an intensive literature search6. As such, a large number of proteins (372) could be additionally assigned to potentially reside in association with (peripheral proteins) or even be fully embedded within the plasma membrane (integral proteins), leading to a total of 789 or 48.6% of PMPs in the PMP enriched fraction (Table 2, Supplementary Table 1, Fig. 1h). Of note, this additional analysis classified an even larger subset of PMPs as ion channel, transmembrane receptor, transporter or G-protein coupled receptor (Table 2 and Supplementary Table 1). Together with these 789 annotated PMPs, another 8 proteins located at the cell surface (0.5%), 163 at the cellular membrane (10.0%) and 51 proteins in the extracellular space (3.1%) (Supplementary Table 2), this accounts for 1,011 or 62.2% proteins in the PMP enriched fraction that have been reported to reside in or near the cell surface (Fig. 1h). The remaining 37.8% proteins in the PMP enriched fraction might be the result of co-purification of large intracellular complexes, with the biotinylated proteins still associated to the plasma membrane or with the readily releasable vesicle pools. These proteins deserve attention in future research to either confirm or exclude their capacity to potentially reside at the PM. In sum, our yields are in agreement with or even higher than recent PMP enrichment studies based on aqueous two-phase affinity partitioning of a much larger tissue sample, a complete rat or mouse cerebellum13,22, or on biotinylation and affinity purification of cell surface proteins of cultured mouse cortical neurons14.
Table 2. 417 PMPs in the PMP enriched fraction (1° IPA annotation).
| Accession no. | Protein name | identified in # of samples |
|---|---|---|
| ION CHANNEL | ||
| IPI00113149 | syntaxin 1B | 5 |
| IPI00113244 | tweety family member 1 | 4 |
| IPI00113772 | gamma-aminobutyric acid (GABA) A receptor, alpha 1 | 5 |
| IPI00122300 | calcium channel, voltage-dependent, gamma subunit 3 | 5 |
| IPI00122974 | glycoprotein M6A | 5 |
| IPI00129491 | potassium voltage-gated channel, Shal-related subfamily, member 2 | 4 |
| IPI00129774 | potassium voltage-gated channel, shaker-related subfamily, member 2 | 5 |
| IPI00130253 | calcium channel, voltage-dependent, alpha 2/delta subunit 3 | 5 |
| IPI00130546 | gamma-aminobutyric acid (GABA) A receptor, beta 3 | 5 |
| IPI00136965 | glutamate receptor, ionotropic, AMPA 1 | 5 |
| IPI00315359 | potassium voltage-gated channel, shaker-related subfamily, beta member 2 | 4 |
| IPI00322698 | transient receptor potential cation channel, subfamily V, member 2 | 5 |
| IPI00323554 | gamma-aminobutyric acid (GABA) A receptor, beta 2 | 5 |
| IPI00338309 | ryanodine receptor 2 (cardiac) | 5 |
| IPI00410982 | calcium channel, voltage-dependent, alpha 2/delta subunit 1 | 5 |
| IPI00461322 | annexin A7 | 5 |
| IPI00608056 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | 5 |
| IPI00625961 | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | 1 |
| IPI00652101 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 5 |
| IPI00673613 | sodium channel, voltage-gated, type I, alpha subunit | 1 |
| IPI00751228 | glutamate receptor, ionotropic, AMPA 2 | 5 |
| IPI00761641 | sodium channel, voltage-gated, type II, alpha subunit | 5 |
| IPI00877256 | tweety family member 3 | 5 |
| IPI00895035 | glutamate receptor, ionotropic, AMPA 3 | 5 |
| IPI00930821 | potassium channel tetramerization domain containing 12 | 2 |
| IPI01019157 | potassium channel tetramerization domain containing 12 | 2 |
| IPI00110601 | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | 3 |
| IPI00119283 | gamma-aminobutyric acid (GABA) A receptor, beta 1 | 1 |
| IPI00119615 | potassium inwardly-rectifying channel, subfamily J, member 3 | 1 |
| IPI00120318 | glutamate receptor, ionotropic, kainate 3 | 1 |
| IPI00128826 | calcium channel, voltage-dependent, gamma subunit 8 | 4 |
| IPI00130455 | FXYD domain containing ion transport regulator 1 | 4 |
| IPI00131471 | glutamate receptor, ionotropic, AMPA 4 | 1 |
| IPI00132786 | calcium channel, voltage-dependent, gamma subunit 2 | 1 |
| IPI00133980 | hyperpolarization activated cyclic nucleotide-gated potassium channel 2 | 1 |
| IPI00228358 | gamma-aminobutyric acid (GABA) A receptor, gamma 2 | 3 |
| IPI00331064 | calcium channel, voltage-dependent, R type, alpha 1E subunit | 1 |
| IPI00421206 | potassium channel tetramerization domain containing 12 | 3 |
| IPI00473235 | calcium channel, voltage-dependent, beta 4 subunit | 3 |
| IPI00554917 | chloride channel, voltage-sensitive 6 | 1 |
| IPI00625414 | calcium channel, voltage-dependent, N type, alpha 1B subunit | 1 |
| IPI00751689 | calcium channel, voltage-dependent, alpha 2/delta subunit 2 | 1 |
| IPI00752080 | integrin, alpha V | 4 |
| IPI00775995 | calcium channel, voltage-dependent, L type, alpha 1F subunit | 1 |
| IPI00844657 | potassium voltage-gated channel, Shaw-related subfamily, member 3 | 1 |
| IPI00874658 | gamma-aminobutyric acid (GABA) A receptor, gamma 2 | 1 |
| IPI00875552 | tweety family member 1 | 1 |
| IPI00928524 | potassium voltage-gated channel, shaker-related subfamily, beta member 2 | 1 |
| IPI00930809 | calcium channel, voltage-dependent, gamma subunit 3 | 1 |
| TRANSMEMBRANE RECEPTOR | ||
| IPI00114939 | neuronal pentraxin receptor | 5 |
| IPI00119063 | low density lipoprotein receptor-related protein 1 | 5 |
| IPI00137311 | plexin A1 | 5 |
| IPI00229992 | plexin B1 | 5 |
| IPI00403079 | CD47 molecule | 5 |
| IPI00462790 | coagulation factor III (thromboplastin, tissue factor) | 4 |
| IPI00463489 | opioid binding protein/cell adhesion molecule-like | 5 |
| IPI00473582 | ciliary neurotrophic factor receptor | 5 |
| IPI00756275 | plexin B2 | 2 |
| IPI00876097 | plexin A4 | 5 |
| IPI00130995 | interleukin 18 receptor accessory protein | 1 |
| IPI00313025 | scavenger receptor class A, member 3 | 1 |
| IPI00315280 | semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) | 1 |
| IPI00351062 | cholinergic receptor, nicotinic, alpha 9 (neuronal) | 1 |
| IPI00463026 | interleukin 1 receptor accessory protein-like 1 | 1 |
| IPI00471022 | plexin D1 | 4 |
| IPI00480518 | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 1 |
| IPI00674255 | plexin C1 | 2 |
| IPI00754710 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | 1 |
| IPI00874523 | roundabout, axon guidance receptor, homolog 1 (Drosophila) | 2 |
| IPI00986716 | plexin B2 | 2 |
| IPI00989396 | roundabout, axon guidance receptor, homolog 1 (Drosophila) | 1 |
| TRANSPORTER | ||
| IPI00109153 | solute carrier family 17 (vesicular glutamate transporter), member 7 | 5 |
| IPI00111151 | rabphilin 3A | 5 |
| IPI00113869 | basigin (Ok blood group) | 5 |
| IPI00114279 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 5 |
| IPI00114641 | solute carrier family 3 (amino acid transporter heavy chain), member 2 | 5 |
| IPI00121550 | ATPase, Na+/K+transporting, beta 1 polypeptide | 5 |
| IPI00122048 | ATPase, Na+/K+transporting, alpha 3 polypeptide | 5 |
| IPI00123704 | ATPase, Na+/K+transporting, beta 2 polypeptide | 5 |
| IPI00124221 | ATPase, Na+/K+transporting, beta 3 polypeptide | 2 |
| IPI00125397 | solute carrier family 30 (zinc transporter), member 3 | 1 |
| IPI00125635 | synaptosomal-associated protein, 25 kDa | 5 |
| IPI00126796 | solute carrier family 27 (fatty acid transporter), member 4 | 3 |
| IPI00127713 | ATPase, Ca++transporting, plasma membrane 2 | 5 |
| IPI00134191 | solute carrier family 2 (facilitated glucose transporter), member 3 | 5 |
| IPI00135130 | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 5 |
| IPI00136372 | synapsin I | 5 |
| IPI00137194 | solute carrier family 16 (monocarboxylate transporter), member 1 | 1 |
| IPI00221456 | synaptic vesicle glycoprotein 2B | 5 |
| IPI00227928 | solute carrier family 6 (neurotransmitter transporter), member 1 | 4 |
| IPI00230289 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 1 |
| IPI00268433 | solute carrier family 8 (sodium/calcium exchanger), member 1 | 3 |
| IPI00308691 | solute carrier family 2 (facilitated glucose transporter), member 1 | 5 |
| IPI00311682 | ATPase, Na+/K+transporting, alpha 1 polypeptide | 5 |
| IPI00314289 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | 4 |
| IPI00322156 | solute carrier family 38, member 3 | 5 |
| IPI00331577 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 | 4 |
| IPI00403860 | neurexin 1 | 5 |
| IPI00407692 | ATPase, H+transporting, lysosomal 70 kDa, V1 subunit A | 5 |
| IPI00420244 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | 4 |
| IPI00420569 | ATPase, Na+/K+transporting, alpha 2 polypeptide | 5 |
| IPI00465769 | solute carrier family 12 (potassium/chloride transporter), member 5 | 5 |
| IPI00555118 | solute carrier family 4, sodium bicarbonate transporter, member 10 | 5 |
| IPI00556827 | ATPase, Ca++transporting, plasma membrane 1 | 5 |
| IPI00621162 | ATPase, Ca++transporting, plasma membrane 3 | 3 |
| IPI00648537 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | 3 |
| IPI00648633 | solute carrier family 44 (choline transporter), member 1 | 1 |
| IPI00750917 | ATPase, Ca++transporting, plasma membrane 4 | 2 |
| IPI00754989 | solute carrier family 39 (zinc transporter), member 12 | 5 |
| IPI00857092 | solute carrier family 4 (sodium bicarbonate cotransporter), member 4 | 5 |
| IPI00884508 | solute carrier family 2 (facilitated glucose transporter), member 1 | 5 |
| IPI00890144 | solute carrier family 4, sodium bicarbonate cotransporter, member 8 | 5 |
| IPI00970455 | neurexin 1 | 5 |
| IPI00125830 | Ly6/neurotoxin 1 | 2 |
| IPI00128152 | ATP-binding cassette, sub-family B (MDR/TAP), member 1B | 3 |
| IPI00128391 | megalencephalic leukoencephalopathy with subcortical cysts 1 | 1 |
| IPI00129395 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 | 1 |
| IPI00135632 | solute carrier family 7 (amino acid transporter light chain, L system), member 8 | 1 |
| IPI00135678 | X-linked Kx blood group | 1 |
| IPI00136867 | solute carrier family 6 (neurotransmitter transporter), member 11 | 3 |
| IPI00153278 | solute carrier family 29 (equilibrative nucleoside transporter), member 4 | 1 |
| IPI00165688 | solute carrier family 23 (ascorbic acid transporter), member 2 | 2 |
| IPI00170146 | ATP-binding cassette, sub-family A (ABC1), member 6 | 1 |
| IPI00172274 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 | 1 |
| IPI00221831 | solute carrier family 32 (GABA vesicular transporter), member 1 | 4 |
| IPI00221932 | mal, T-cell differentiation protein 2 (gene/pseudogene) | 3 |
| IPI00230290 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 4 |
| IPI00310247 | ANKH inorganic pyrophosphate transport regulator | 2 |
| IPI00338618 | ATPase, class V, type 10A | 1 |
| IPI00380273 | gap junction protein, alpha 1, 43 kDa | 3 |
| IPI00463589 | ATPase, Ca++transporting, plasma membrane 4 | 3 |
| IPI00623542 | solute carrier family 8 (sodium/calcium exchanger), member 1 | 2 |
| IPI00648270 | solute carrier family 44 (choline transporter), member 1 | 4 |
| IPI00652257 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | 1 |
| IPI00776182 | solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 | 1 |
| IPI00785299 | ATPase, Ca++transporting, plasma membrane 3 | 2 |
| IPI00788403 | ATP-binding cassette, sub-family A (ABC1), member 8 | 1 |
| IPI00927968 | copine VI (neuronal) | 2 |
| G-PROTEIN COUPLED RECEPTOR | ||
| IPI00132061 | purinergic receptor P2Y, G-protein coupled, 12 | 3 |
| IPI00135659 | oligodendrocyte myelin glycoprotein | 5 |
| IPI00136716 | glutamate receptor, metabotropic 3 | 5 |
| IPI00229528 | brain-specific angiogenesis inhibitor 1 | 4 |
| IPI00281619 | glutamate receptor, metabotropic 1 | 4 |
| IPI00407689 | gamma-aminobutyric acid (GABA) B receptor, 1 | 3 |
| IPI00465871 | G protein-coupled receptor 158 | 5 |
| IPI00762862 | glutamate receptor, metabotropic 2 | 5 |
| IPI00816879 | latrophilin 1 | 5 |
| IPI00881441 | latrophilin 3 | 3 |
| IPI01018412 | gamma-aminobutyric acid (GABA) B receptor, 1 | 3 |
| IPI00117887 | neuromedin B receptor | 1 |
| IPI00120115 | sphingosine-1-phosphate receptor 1 | 1 |
| IPI00126064 | olfactory receptor 1018 | 1 |
| IPI00127181 | olfactory receptor 1 | 1 |
| IPI00136713 | olfactory receptor 157 | 1 |
| IPI00153507 | vomeronasal 1 receptor 217 | 1 |
| IPI00229361 | glutamate receptor, metabotropic 6 | 1 |
| IPI00269278 | G protein-coupled receptor 119 | 1 |
| IPI00402890 | adenylate cyclase activating polypeptide 1 (pituitary) receptor type I | 1 |
| IPI00470960 | glutamate receptor, metabotropic 4 | 2 |
| IPI00474802 | glutamate receptor, metabotropic 7 | 2 |
| IPI00553387 | glutamate receptor, metabotropic 5 | 2 |
| IPI00605298 | G protein-coupled receptor 123 | 1 |
| IPI00675087 | vomeronasal 2, receptor 32 | 1 |
| IPI00755301 | gamma-aminobutyric acid (GABA) B receptor, 1 | 2 |
| IPI00867815 | glutamate receptor, metabotropic 5 | 3 |
| IPI00880691 | latrophilin 3 | 2 |
| IPI00944116 | adenosine A3 receptor | 1 |
| KINASE | ||
| IPI00125147 | membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) | 5 |
| IPI00129198 | EPH receptor A4 | 5 |
| IPI00314316 | membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) | 4 |
| IPI00337992 | EPH receptor A4 | 5 |
| IPI00351246 | membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) | 5 |
| IPI00626797 | discs, large homolog 4 (Drosophila) | 5 |
| IPI00672505 | discs, large homolog 1 (Drosophila) | 5 |
| IPI00762272 | discs, large homolog 2 (Drosophila) | 5 |
| IPI00776413 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | 2 |
| IPI00830221 | EPH receptor B4 | 2 |
| IPI00830635 | EPH receptor A5 | 3 |
| IPI00128360 | neurotrophic tyrosine kinase, receptor, type 2 | 4 |
| IPI00229334 | neurotrophic tyrosine kinase, receptor, type 2 | 1 |
| IPI00338094 | bone morphogenetic protein receptor, type II (serine/threonine kinase) | 1 |
| IPI00474411 | TYRO3 protein tyrosine kinase | 5 |
| IPI00474965 | epidermal growth factor receptor | 3 |
| IPI00655218 | phosphatidylinositol-4-phosphate 5-kinase, type I, gamma | 1 |
| IPI00808241 | EPH receptor B1 | 2 |
| IPI00875987 | G protein-coupled receptor kinase 6 | 1 |
| IPI00886325 | membrane protein, palmitoylated 1, 55 kDa | 1 |
| IPI00918777 | phosphatidylinositol-4-phosphate 5-kinase, type I, gamma | 1 |
| PEPTIDASE | ||
| IPI00627016 | ADAM metallopeptidase domain 22 | 5 |
| IPI00650001 | ADAM metallopeptidase domain 23 | 4 |
| IPI00798468 | ubiquitin specific peptidase 9, X-linked | 4 |
| IPI00881709 | dipeptidyl-peptidase 6 | 5 |
| IPI00118674 | nicastrin | 3 |
| IPI00169524 | thyrotropin-releasing hormone degrading enzyme | 1 |
| IPI00408232 | ADAM metallopeptidase domain 11 | 2 |
| IPI00621146 | transmembrane protease, serine 11c | 1 |
| IPI00648033 | ADAM metallopeptidase domain 23 | 1 |
| IPI00752133 | signal peptide peptidase like 2A | 1 |
| IPI00928374 | nicastrin | 2 |
| IPI01027504 | ubiquitin specific peptidase 9, X-linked | 1 |
| IPI01027684 | ubiquitin specific peptidase 9, X-linked | 1 |
| PHOSPHATASE | ||
| IPI00115626 | phosphatidic acid phosphatase type 2B | 2 |
| IPI00405703 | protein tyrosine phosphatase, receptor type, A | 1 |
| IPI00420590 | lipid phosphate phosphatase-related protein type 4 | 5 |
| IPI00465836 | protein tyrosine phosphatase, receptor type, D | 5 |
| IPI00627008 | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | 5 |
| IPI00875821 | SET binding factor 1 | 5 |
| IPI00876489 | signal-regulatory protein alpha | 5 |
| IPI00915502 | protein tyrosine phosphatase, receptor type, S | 5 |
| IPI01027153 | protein tyrosine phosphatase, receptor type, D | 5 |
| IPI00110264 | protein tyrosine phosphatase, receptor type, F | 1 |
| IPI00336550 | protein tyrosine phosphatase, receptor type, A | 2 |
| IPI00399905 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | 1 |
| IPI00475109 | protein phosphatase 3, catalytic subunit, beta isozyme | 4 |
| IPI00857748 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | 3 |
| IPI00881167 | protein tyrosine phosphatase, receptor type, G | 1 |
| TRANSCRIPTION REGULATOR | ||
| IPI00222057 | neogenin 1 | 3 |
| ENZYME | ||
| IPI00115429 | gamma-glutamyltransferase 7 | 5 |
| IPI00117176 | fatty acid amide hydrolase | 1 |
| IPI00120716 | guanine nucleotide binding protein (G protein), beta polypeptide 1 | 5 |
| IPI00121387 | guanine nucleotide binding protein (G protein), alpha 11 (Gq class) | 4 |
| IPI00123058 | contactin 1 | 5 |
| IPI00126551 | DIRAS family, GTP-binding RAS-like 2 | 5 |
| IPI00130949 | adenylate cyclase 1 (brain) | 5 |
| IPI00133218 | ADP-ribosylation factor-like 8B | 3 |
| IPI00138716 | RAP2B, member of RAS oncogene family | 5 |
| IPI00162780 | guanine nucleotide binding protein (G protein), beta polypeptide 2 | 1 |
| IPI00222125 | catechol-O-methyltransferase domain containing 1 | 2 |
| IPI00228618 | guanine nucleotide binding protein (G protein), q polypeptide | 5 |
| IPI00230192 | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O | 4 |
| IPI00230193 | guanine nucleotide binding protein (G protein), alpha z polypeptide | 5 |
| IPI00230194 | guanine nucleotide binding protein (G protein), gamma 2 | 2 |
| IPI00309113 | neuroligin 1 | 4 |
| IPI00378017 | guanine nucleotide binding protein (G protein), beta 5 | 5 |
| IPI00396701 | RAP2A, member of RAS oncogene family | 5 |
| IPI00467152 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 5 |
| IPI00468605 | neuroligin 2 | 5 |
| IPI00649388 | guanine nucleotide binding protein (G protein), alpha 13 | 2 |
| IPI00816946 | gephyrin | 4 |
| IPI00858047 | monoglyceride lipase | 5 |
| IPI00881278 | adenylate cyclase 9 | 5 |
| IPI00928550 | gamma-glutamyltransferase 7 | 5 |
| IPI00929787 | trans-2,3-enoyl-CoA reductase | 5 |
| IPI00115546 | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O | 1 |
| IPI00123623 | hyaluronan synthase 1 | 1 |
| IPI00126501 | carbonic anhydrase XIV | 5 |
| IPI00128097 | adenylate cyclase 4 | 1 |
| IPI00225670 | gephyrin | 1 |
| IPI00228295 | contactin 4 | 1 |
| IPI00272230 | RAB39B, member RAS oncogene family | 1 |
| IPI00315334 | neuroblastoma RAS viral (v-ras) oncogene homolog | 1 |
| IPI00331267 | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) | 1 |
| IPI00377311 | diacylglycerol lipase, alpha | 1 |
| IPI00403586 | neutral cholesterol ester hydrolase 1 | 1 |
| IPI00649078 | SH3-domain GRB2-like 2 | 2 |
| IPI00652606 | RAB2B, member RAS oncogene family | 2 |
| IPI00749677 | dynamin 2 | 1 |
| IPI00750570 | GNAS complex locus | 5 |
| IPI00758356 | guanine nucleotide binding protein (G protein), beta polypeptide 2 | 4 |
| IPI00856692 | diacylglycerol lipase, alpha | 1 |
| IPI00876486 | ectonucleotide pyrophosphatase/phosphodiesterase 7 | 1 |
| IPI00885337 | neuroligin 3 | 1 |
| IPI00886041 | neuroligin 3 | 1 |
| IPI00918346 | contactin 6 | 1 |
| IPI01027614 | dynamin 2 | 1 |
| OTHER | ||
| IPI00109727 | Thy-1 cell surface antigen | 5 |
| IPI00110451 | SLIT and NTRK-like family, member 1 | 5 |
| IPI00115827 | glioblastoma amplified sequence | 5 |
| IPI00117181 | flotillin 1 | 3 |
| IPI00118020 | cell adhesion molecule 3 | 4 |
| IPI00118075 | microtubule-associated protein 2 | 5 |
| IPI00119033 | intercellular adhesion molecule 5, telencephalin | 5 |
| IPI00119130 | BTB (POZ) domain containing 17 | 5 |
| IPI00119689 | adaptor-related protein complex 2, beta 1 subunit | 5 |
| IPI00119870 | catenin (cadherin-associated protein), alpha 2 | 4 |
| IPI00119970 | contactin 2 (axonal) | 5 |
| IPI00120302 | leucine-rich, glioma inactivated 1 | 5 |
| IPI00120793 | prion protein | 4 |
| IPI00120943 | cyclin and CBS domain divalent metal cation transport mediator 1 | 2 |
| IPI00121378 | activated leukocyte cell adhesion molecule | 5 |
| IPI00122971 | neural cell adhesion molecule 1 | 5 |
| IPI00128022 | protocadherin 7 | 2 |
| IPI00131376 | spectrin, beta, erythrocytic | 2 |
| IPI00134200 | leucine rich repeat and Ig domain containing 1 | 5 |
| IPI00134492 | synapsin II | 5 |
| IPI00136135 | catenin (cadherin-associated protein), delta 2 | 4 |
| IPI00137331 | CAP, adenylate cyclase-associated protein 1 (yeast) | 1 |
| IPI00153840 | cell adhesion molecule 4 | 5 |
| IPI00221540 | ER lipid raft associated 2 | 1 |
| IPI00227126 | tenascin R | 5 |
| IPI00227235 | ankyrin 2, brain | 4 |
| IPI00228617 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | 5 |
| IPI00228680 | neurexin III | 1 |
| IPI00229299 | erythrocyte membrane protein band 4.1-like 3 | 4 |
| IPI00229475 | junction plakoglobin | 2 |
| IPI00229703 | vesicle-associated membrane protein 2 (synaptobrevin 2) | 5 |
| IPI00230151 | myelin associated glycoprotein | 5 |
| IPI00230408 | microtubule-associated protein tau | 2 |
| IPI00263013 | proteolipid protein 1 | 2 |
| IPI00274767 | glycoprotein M6B | 3 |
| IPI00310916 | CD81 molecule | 3 |
| IPI00311405 | poliovirus receptor-related 1 (herpesvirus entry mediator C) | 5 |
| IPI00319830 | spectrin, beta, non-erythrocytic 1 | 5 |
| IPI00322617 | neural cell adhesion molecule 2 | 5 |
| IPI00323800 | neurofilament, medium polypeptide | 3 |
| IPI00329927 | neurofascin | 4 |
| IPI00331579 | synaptogyrin 3 | 5 |
| IPI00338983 | contactin associated protein 1 | 5 |
| IPI00400180 | amphiphysin | 4 |
| IPI00405736 | CD81 molecule | 1 |
| IPI00410985 | cell adhesion molecule 1 | 3 |
| IPI00420467 | poliovirus receptor-related 1 (herpesvirus entry mediator C) | 5 |
| IPI00420554 | contactin associated protein-like 2 | 5 |
| IPI00458574 | cadherin 13 | 5 |
| IPI00461199 | bassoon presynaptic cytomatrix protein | 5 |
| IPI00467747 | neuronal growth regulator 1 | 5 |
| IPI00471176 | hepatic and glial cell adhesion molecule | 5 |
| IPI00474209 | synaptosomal-associated protein, 91 kDa | 5 |
| IPI00620207 | dematin actin binding protein | 1 |
| IPI00648658 | clathrin, light chain A | 3 |
| IPI00649966 | synaptosomal-associated protein, 47 kDa | 2 |
| IPI00652675 | limbic system-associated membrane protein | 5 |
| IPI00652902 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | 5 |
| IPI00656204 | NCK-associated protein 1 | 5 |
| IPI00663736 | synaptic Ras GTPase activating protein 1 | 3 |
| IPI00670856 | cadherin 10, type 2 (T2-cadherin) | 1 |
| IPI00675985 | potassium channel tetramerization domain containing 16 | 5 |
| IPI00719927 | protocadherin 1 | 4 |
| IPI00751569 | DnaJ (Hsp40) homolog, subfamily C, member 5 | 5 |
| IPI00753793 | spectrin, alpha, non-erythrocytic 1 | 5 |
| IPI00756921 | tetraspanin 7 | 5 |
| IPI00757097 | SH3 and multiple ankyrin repeat domains 2 | 4 |
| IPI00757771 | neuroplastin | 5 |
| IPI00830223 | tropomyosin 1, alpha | 5 |
| IPI00831568 | L1 cell adhesion molecule | 5 |
| IPI00831624 | connector enhancer of kinase suppressor of Ras 2 | 1 |
| IPI00848690 | lymphocyte antigen 6 complex, locus H | 4 |
| IPI00850833 | cell adhesion molecule 2 | 4 |
| IPI00854028 | contactin associated protein 1 | 5 |
| IPI00855176 | protocadherin 9 | 4 |
| IPI00857329 | neurotrimin | 5 |
| IPI00858209 | LanC lantibiotic synthetase component C-like 2 (bacterial) | 5 |
| IPI00869430 | CAP, adenylate cyclase-associated protein 1 (yeast) | 1 |
| IPI00882293 | membrane bound O-acyltransferase domain containing 7 | 3 |
| IPI00882316 | CAP, adenylate cyclase-associated protein, 2 (yeast) | 5 |
| IPI00894724 | microtubule-associated protein 2 | 5 |
| IPI00918899 | syntaxin binding protein 5 (tomosyn) | 1 |
| IPI00929916 | tenascin R | 5 |
| IPI00990801 | regulating synaptic membrane exocytosis 1 | 2 |
| IPI01027487 | immunoglobulin superfamily, member 8 | 5 |
| IPI00112226 | angiopoietin-like 1 | 1 |
| IPI00118420 | stimulated by retinoic acid 6 | 1 |
| IPI00121091 | proteolipid protein 1 | 1 |
| IPI00121627 | cleft lip and palate associated transmembrane protein 1 | 2 |
| IPI00122032 | receptor accessory protein 2 | 1 |
| IPI00131762 | cyclin and CBS domain divalent metal cation transport mediator 2 | 1 |
| IPI00131896 | mitochondrial pyruvate carrier 2 | 1 |
| IPI00136021 | regulating synaptic membrane exocytosis 1 | 3 |
| IPI00154057 | protocadherin 1 | 1 |
| IPI00173032 | integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) | 1 |
| IPI00173248 | ankyrin 3, node of Ranvier (ankyrin G) | 1 |
| IPI00222908 | fibronectin leucine rich transmembrane protein 2 | 1 |
| IPI00228632 | catenin (cadherin-associated protein), delta 2 | 1 |
| IPI00230610 | proteolipid protein 1 | 2 |
| IPI00230751 | catenin (cadherin-associated protein), alpha 2 | 1 |
| IPI00273822 | lysosomal-associated membrane protein 3 | 1 |
| IPI00309419 | leucine rich repeat containing 4C | 3 |
| IPI00313492 | leucine rich repeat transmembrane neuronal 2 | 2 |
| IPI00321348 | immunoglobulin superfamily, member 8 | 2 |
| IPI00330250 | regulating synaptic membrane exocytosis 3 | 2 |
| IPI00336313 | protein phosphatase 1, regulatory subunit 9A | 4 |
| IPI00338880 | neuronal cell adhesion molecule | 2 |
| IPI00346482 | cadherin 10, type 2 (T2-cadherin) | 1 |
| IPI00351827 | SH3 and multiple ankyrin repeat domains 3 | 3 |
| IPI00380242 | desmoglein 4 | 1 |
| IPI00381088 | unc-13 homolog A (C. elegans) | 5 |
| IPI00405986 | erythrocyte membrane protein band 4.1-like 1 | 1 |
| IPI00420570 | neurofascin | 1 |
| IPI00460715 | neurexin III | 4 |
| IPI00461212 | oxysterol binding protein-like 8 | 1 |
| IPI00466076 | sidekick cell adhesion molecule 1 | 1 |
| IPI00468202 | trophoblast glycoprotein | 3 |
| IPI00473188 | annexin A8-like 1 | 1 |
| IPI00473968 | cadherin 10, type 2 (T2-cadherin) | 3 |
| IPI00648543 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 | 1 |
| IPI00648759 | stomatin (EPB72)-like 2 | 5 |
| IPI00649994 | CAP, adenylate cyclase-associated protein 1 (yeast) | 2 |
| IPI00653438 | trophoblast glycoprotein | 2 |
| IPI00653674 | KRIT1, ankyrin repeat containing | 1 |
| IPI00670114 | Ca++-dependent secretion activator | 1 |
| IPI00751974 | syntrophin, alpha 1 | 1 |
| IPI00756961 | netrin G2 | 1 |
| IPI00762484 | Down syndrome cell adhesion molecule like 1 | 1 |
| IPI00830145 | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A | 2 |
| IPI00831210 | neurofilament, medium polypeptide | 2 |
| IPI00831427 | synovial sarcoma, X breakpoint 2 interacting protein | 1 |
| IPI00831714 | leucine rich repeat containing 7 | 4 |
| IPI00849429 | cell adhesion molecule 1 | 2 |
| IPI00853863 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | 2 |
| IPI00856771 | podocalyxin-like 2 | 1 |
| IPI00867858 | protocadherin-related 15 | 3 |
| IPI00876257 | clathrin, light chain A | 1 |
| IPI00880812 | LanC lantibiotic synthetase component C-like 1 (bacterial) | 4 |
| IPI00889283 | SH3 and multiple ankyrin repeat domains 2 | 1 |
| IPI00889292 | FAT atypical cadherin 3 | 1 |
| IPI00921638 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | 1 |
| IPI00921642 | ankyrin 2, brain | 1 |
| IPI00928058 | FCH domain only 1 | 1 |
| IPI00928139 | DAB2 interacting protein | 1 |
| IPI00955069 | contactin 5 | 1 |
| IPI00987809 | spectrin, beta, erythrocytic | 1 |
| IPI00988904 | fibronectin leucine rich transmembrane protein 2 | 2 |
| IPI00989004 | cyclin and CBS domain divalent metal cation transport mediator 1 | 2 |
| IPI00990422 | synaptic Ras GTPase activating protein 1 | 4 |
| IPI01008331 | desmocollin 3 | 1 |
| IPI01008664 | receptor accessory protein 2 | 2 |
| IPI01027584 | flotillin 1 | 1 |
The efficiency of the presented workflow for the enrichment of PMPs was further validated using the DAVID web tool. This tool allows visualization of the specific protein enrichment in the PMP enriched and wash-through fractions by a gene ontology enrichment analysis on all protein identifications of each fraction relative to a background. The background was built by merging all proteins identified in the PMP enriched with those from the wash-through fraction into one background data set. We used the functional annotation charts of the DAVID web tool based on cellular component ontology and visualized the results in ReViGO treemaps (Supplementary Fig. 1 and 2). The treemap adapted from ReViGO for the PMP enriched fraction (Supplementary Fig. 1) is summarized in Table 3, illustrating an enrichment of proteins associated to the cell surface specific for neuronal cells in the clusters ‘plasma membrane’, ‘plasma membrane part’, ‘ion channel complex’, ‘intrinsic’ and ‘integral component of PM’, and the clusters ‘synapse’, ‘synapse part’, ‘neuron projection’, ‘dendritic spine’, and ‘cell junction’. The treemap adapted from ReViGO resulting from our wash-through data set (Supplementary Fig. 2) summarized in Table 4 shows clusters of proteins associated with different intracellular organelles, especially with mitochondrial function and the ‘respiratory chain’. This reflects the high energy demand and oxygen consumption of neurons, and thus the high metabolic rate of the tissue under study23,24,25,26,27. Other clusters contain proteins with a role at the envelope, within the endomembrane system, and within the membrane-enclosed lumen. Importantly, no clear cluster was suggested for cell surface-associated proteins for the wash-through fraction.
Table 3. Summary of ReViGO treemap showing the specific enrichment of proteins within the PMP enriched fraction.
| Cluster representative | % | |
|---|---|---|
| GO Term | ||
| GO:0031224 | intrinsic component of membrane | 16 |
| GO:0016021 | integral component of membrane | 12 |
| GO:0005886 | plasma membrane | 6 |
| GO:0044459 | plasma membrane part | 4 |
| GO:0034702 | ion channel complex | 1 |
| GO:0031225 | anchored component of membrane | 1 |
| GO:0031226 | intrinsic component of plasma membrane | 1 |
| GO:0005887 | integral component of plasma membrane | 1 |
| GO:0005832 | chaperonin-containing T-complex | |
| GO:0070469 | respiratory chain | |
| GO:0033178 | proton transporting two-sector ATPase complex, catalytic domain | |
| GO:0031090 | organelle membrane | 4 |
| GO:0005740 | mitochondrial envelope | 4 |
| GO:0044429 | mitochondrial part | 3 |
| GO:0042470 | melanosome | 2 |
| GO:0048770 | pigment granule | 2 |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | 3 |
| GO:0031982 | vesicle | 1 |
| GO:0031410 | cytoplasmic vesicle | 1 |
| GO:0045202 | synapse | 9 |
| GO:0044456 | synapse part | 7 |
| GO:0045111 | intermediate filament cytoskeleton | 3 |
| GO:0005882 | intermediate filament | 3 |
| GO:0044430 | cytoskeletal part | |
| GO:0043005 | neuron projection | 4 |
| GO:0043197 | dendritic spine | 1 |
| GO:0030054 | cell junction | 4 |
| GO:0031975 | envelope | 3 |
Table 4. Summary of ReViGO treemap showing the specific enrichment of proteins within the wash-through fraction.
| Cluster representative | % | |
|---|---|---|
| GO Term | ||
| GO:0005739 | mitochondrion | 21 |
| GO:0044429 | mitochondrial part | 12 |
| GO:0031967 | organelle envelope | 13 |
| GO:0031090 | organelle membrane | 11 |
| GO:0005829 | cytosol | 7 |
| GO:0031982 | vesicle | 2 |
| GO:0031410 | cytoplasmic vesicle | 2 |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | 2 |
| GO:0043228 | non-membrane-bounded organelle | 2 |
| GO:0043232 | intracellular non-membrane-bounded organelle | 2 |
| GO:0005759 | mitochondrial matrix | 1 |
| GO:0015630 | microtubule cytoskeleton | 1 |
| GO:0070013 | intracellular organelle lumen | 1 |
| GO:0005875 | microtubule associated complex | 1 |
| GO:0005768 | endosome | 1 |
| GO:0048770 | pigment granule | 1 |
| GO:0042470 | melanosome | 1 |
| GO:0005874 | microtubule | |
| GO:0005694 | chromosome | |
| GO:0044445 | cytosolic part | |
| GO:0005840 | ribosome | |
| GO:0031975 | envelope | 13 |
| GO:0070469 | respiratory chain | 2 |
| GO:0019898 | extrinsic component of membrane | 1 |
| GO:0016469 | proton-transporting two-sector ATPase complex | 1 |
| GO:0009898 | cytoplasmic side of plasma membrane | 1 |
| GO:0030529 | ribonucleoprotein complex | |
| GO:0000502 | proteasome complex | |
| GO:0044448 | cell cortex part | |
| GO:0031974 | membrane-enclosed lumen | 1 |
| GO:0012505 | endomembrane system | 1 |
| GO:0045177 | apical part of cell | |
| GO:0000267 | cell fraction | |
In conclusion, in this report we present the new combination of a procedure for the specific extraction of cell surface-associated proteins including PMPs originating from mm3-scale tissue derived from acute tissue slice preparations, with proteomic analysis. This reproducible and efficient enrichment methodology is undoubtedly applicable to many different tissue types and can significantly contribute to future differential plasma membrane proteomics research in many fields of application ranging from neuroscience, cancer, and immunology, to stem cell research.
Methods
Animals
Adult (n = 5) C57Bl/6J mice of either sex were housed under standard laboratory conditions with a daily photoperiod of 13 hours light and 11 hours darkness with water and food available ad libitum.
All experiments were approved by the ethical research committee of KU Leuven and were in strict accordance with the European Communities Council Directive of 22 September 2010 (2010/63/EU) and with the Belgian legislation (KB of 29 May 2013). Every possible effort was made to minimize animal suffering and to reduce the numbers of animals.
Acute slice biotinylation assay (ASBA)
Mice were killed by cervical dislocation. Brains were rapidly dissected in ice-cold (4 °C) artificial cerebrospinal fluid (aCSF; 124 mM NaCl, 4.9 mM KCl, 2 mM MgSO4, 2 mM CaCl2, 1.2 mM KH2PO4, 25.6 mM NaHCO3, and 10 mM D-glucose; pH 7.4) saturated with 95% O2 and 5% CO2. Subsequently, each brain was separated in half along the longitudinal fissure, and the left hemisphere was cut into 300 μm-thick coronal slices using a Vibratome (HM 650 V, Prosan). A thickness of tissue slices between 300 and 400 μm is essential to biotinylate an amount of functionally healthy and intact cells that will outnumber the sliced ones28,29. Four slices between Bregma level –2.70 and –4.16 of each hemisphere were placed in an incubation chamber (65-0076/BSC-PC, Harvard Apparatus by Warner Instruments) filled with aCSF and provided with a continuous flow of 95% O2 and 5% CO2 for 90 min in order to recover.
Next, the sections were put on ice and supplemented with CO2 and O2 throughout the whole biotinylation procedure. They were washed twice in ice-cold aCSF, incubated for 45 min with EZ-Link Sulfo-NHS-SS-Biotin (0.5 mg ml−1 in aCSF; Thermo Scientific) and washed twice with ice-cold aCSF complemented by 100 μM lysine (Sigma) to block the remaining reactive Sulfo-NHS-SS-biotin. It has already been demonstrated that these tissue slices stay viable with the cells intact during the complete process. Incubation on ice during biotin labelling will limit protein internalization to a minimum in order to create a snapshot of the cell surface proteome. The Sulfo-NHS-SS-biotin can reach all cell layers of a slice up to 350 μm-thick after incubation with minimum 100 μM for at least 45 min. Because of the intact cell membranes and the membrane impermeability of the biotin label, cell surface-exposed proteins will be biotinylated and labelling of intracellular proteins will be minimal16, as substantiated with our approach and data set.
The sections were washed and kept in ice-cold, saturated aCSF until dissection of the region of interest under a binocular microscope (ASZ30E; Bausch & Lomb). The borders of the visual cortex were determined based on the stereotaxic mouse brain atlas30. For each brain, the dissected visual cortex tissue from the four slices was collected in 100 μL lysis buffer (1% Triton X-100, 0.1% sodium dodecyl sulphate (SDS), 1 mM ethylenediaminetetraacetic acid (EDTA), 50 mM NaCl, 20 mM Tris; pH 7.5) and 4 μl of a mix of protease inhibitors (Roche). After mechanical homogenization and centrifugation (10,000g, 5 min, 4 °C) the supernatant was collected and stored at –80 °C. The total protein concentration of the 5 biotin-labelled samples was determined according to the QubitTM Quantification Platform (Invitrogen) using a QubitTM fluorometer (Invitrogen, Merelbeke, Belgium).
Isolation of plasma membrane proteins
Biotin-labelled proteins were separated from the rest of the proteome by a protocol based on biotin’s affinity for streptavidin. For this purpose, 150 μl of at room temperature (RT) calibrated Streptavidin agarose resin (Thermo Scientific) was loaded onto a Pierce® Spin Cups - Cellulose Acetate Filter (Thermo Scientific). After centrifugation, the column was centrifuged (500 g, 1 min) and washed four times by adding 700 μl of phosphate-buffered saline (PBS; 0.1 M H3PO4, 0.15 M NaCl; pH 7.2) and centrifugation (500 g, 1 min). The 5 biotinylated samples, each containing ±500 μg of proteins extracted from ±1 mm3 tissue, were loaded onto 5 prepared columns and all were shaken for 15 min at RT. After centrifugation (500 g, 1 min), the columns were washed three times with 700 μl PBS. All of the non-biotinylated proteins were eluted during the wash steps with the vast majority of proteins eluted in the first wash-through. Next, 5 μl 2% SDS, 45 μl 200 mM Triethylammonium bicarbonate (TEAB; Fluka Analytical), 45 μl MilliQ (Merck Millipore) and 5 μl 200 mM Tris (2-carboxyethyl) phosphine (TCEP; Thermo Scientific) were added to each column and they were incubated for 1 h at 55 °C for denaturation and reduction. Samples were alkylated for 30 min in the dark in 5 μl 375 mM iodoacetamide (IAA; Thermo Scientific), centrifuged (500 g, 1 min) and the 5 obtained eluents were collected. Subsequently, 25 μl 200 mM TEAB and 25 μl MilliQ were added to each column, they were centrifuged (500 g, 1 min), each eluent added to the corresponding previous one and these 5 combined samples, being the plasma membrane protein (PMP) enriched fractions, were stored at –20 °C, together with the 5 first wash-through fractions.
Protein quantity determination
The total protein concentration of the first wash-throughs and the PMP enriched fractions, were determined according to the QubitTM Quantification Platform (Invitrogen) as described above. For each sample, the relative proportion of protein quantity in the PMP enriched fraction was calculated to the total protein content of the initial sample, the wash-through and PMP enriched fraction combined. For the calculation of these percentages, original Qubit protein quantitation measurements were used and renumbered according to the total volume of each fraction.
Gel-electrophoresis and total protein stain
To 1 μg protein of each of the wash-throughs and 1 μg protein of each of the PMP enriched fractions, 5 μl of XT Sample Buffer solution (4x; Bio-Rad) and 1 μl of XT Reducing Agent (20x; Bio-Rad) was added. The protein samples were denatured and reduced for 10 min on 70 °C. Proteins were separated on a CriterionTM Precast GelXT 4–12% Bis-Tris (Bio-Rad) in the CriterionTM Cell (Bio-Rad) system and 4 μL of the SpectraTM Multicolor High range protein ladder (Thermo Scientific) was used as molecular weight standard. Next, we performed a total protein stain with Serva Purple (SERVA) according to manufacturer’s instructions. After the staining, the gel was scanned with the Bio-Rad ChemiDocTM MP Imaging System.
Proteomic analysis
For the 5 wash-throughs and the 5 PMP enriched fractions, 25 μg was diluted in MilliQ to a total volume of 100 μL. The samples were subsequently transformed into tube-gels by adding 25 μL of Acrylamide/Bis solution (40% 29:1; Bio-Rad), 1 μl SDS (10%), 0.5 μl APS (ammonium persulfate, 10%; Sigma) and 0.1 μl Temed (N, N, N’, N’ - Tetramethylethylenediamine; Fluka BioChemika) and incubation of 30 min at RT. Peptides were extracted from these tube-gels by in-gel trypsin digestion. The tube gels were cut in pieces of approximately 1 mm3. The pieces were washed twice with 50 μl MilliQ, followed by 3 × 50 μl acetonitrile. After three cycles of hydration with acetonitrile and rehydration with 100 mM ammonium bicarbonate, the gel pieces were vacuum dried in a vacuum concentrator. To start the enzymatic digestion, 25 μl of a solution containing 5 ng μl−1 trypsin (Promega), 50 mM ammonium bicarbonate and 5 mM calciumchloride was added to each gel piece and placed on 37 °C overnight. The next day, the tryptic peptides were extracted using 50 mM ammonium bicarbonate followed by an extraction with 50% acetonitrile and 5% formic acid. This step was repeated twice. Afterwards, the pooled extracts were vacuum dried and the peptides were stored at –20 °C. The equivalent of 1 μg of total protein was loaded and analysed by nanoLC-mass spectrometry. Liquid chromatography mass spectrometric analysis was performed on a Waters nanoAquity LC system connected to a Thermo Scientific LTQ Velos Orbitrap mass spectrometer. The equivalent of 2 μg of total protein of the digested sample was dissolved in 20 μl of 2% acetonitrile in HPLC-grade water. 10 μl of the sample was loaded onto the trapping column (Pepmap C18 300 μm x 20 mm, Dionex) with an isocratic flow of 2% acetonitrile in water with 0.1% formic acid at a flow rate of 5 μl min−1. After 2 min, the column-switching valve was switched, placing the pre-column online with the analytical capillary column, a Pepmap C18, 3 μm 75 μm x 150 mm nano column (Dionex). Separation was conducted using a linear gradient from 2% acetonitril in water, 0.1% formic acid to 40% acetonitril in water, 0.1% formic acid in 160 min. The flow rate was set at 400 nl min−1. The LTQ Orbitrap Velos (Thermo Scientific) was set up in a data dependent MS/MS mode where a full scan spectrum (350–5,000 m/z, resolution 60,000) was followed by a maximum of ten CID tandem mass spectrum (100 to 2,000 m/z). Peptide ions were selected as the twenty most intense peaks of the MS1 scan. Collision induced dissociation (CID) scans were acquired in the LTQ iontrap part of the mass spectrometer. The normalized collision energy used was 35% in CID. We applied a dynamic exclusion list of 45 s.
Data analysis
Proteome Discoverer 1.3.0.339 (Thermo Scientific) was used as a workflow manager to handle the data. The tandem MS data were searched using both SEQUEST and Mascot (Matrix science) against the Swissprot database (v 09/2012, 538010 sequences) for Mus musculus taxonomy. All tandem mass spectra in the range of 300 Da to 8,000 Da were interpreted.
Monoisotopic peak assignment, charge state determination, co-isolation interference, and delta mass calculation between the measured and theoretical monoisotopic masses were determined by Proteome Discoverer. The precursor mass tolerance was set at 5 ppm, while fragment mass tolerance was set to 0.5 Da. A maximum of two missed cleavages by trypsine was allowed for. A static modification of 57.021 Da on cysteine was defined to allow for carbamidomethylation. Further, a dynamic modification of 15.9955 Da was introduced to account for possible oxidation of methionine. The use of average precursor masses and average fragment masses was prohibited. Only first ranked PSMs were considered for further analysis. The false discovery rate is controlled by a target-decoy approach on a reversed database. Peptide spectrum matches were found significant at an FDR of 5%.
Protein discoverer 1.3.0.339 was used to combine the significant peptide annotation from Mascot and SEQUEST in a parsimonious protein list.
Protein grouping follows the rule of parsimony. Essentially only the minimal list of proteins that can explain all the peptides in the data set is reported.
Ingenuity Pathway Analyis (IPA® , QIAGEN Redwood City, www.qiagen.com/ingenuity) and Database of Annotation, Visualization and Integrated Discovery (DAVID, version 6.7) were used for cellular component assignment.
The enrichment of proteins in the PMP enriched fraction was investigated by analysing all identifications within this fraction relative to a background composed of all proteins identified in the wash-through and PMP enriched fraction together with DAVID31,32. DAVID summarized the cellular component ontology identified via a functional annotation chart and calculated a Fisher exact test for each ontology as a measure for enrichment within the fraction. Similarly, information about the enrichment of proteins in the wash-through fraction was retrieved. The GO-terms and corresponding p-values with Benjamini correction were subsequently submitted to ReViGO, a web server that Reduces and Visualizes long lists of Gene Ontology terms33, and visualized in treemaps that cluster ontologies with high semantic similarity. The size of these cluster representatives, which are joined in different superclusters each indicated by 1 colour, is proportional to the p-values derived by ReViGO. The treemap figures were then adapted by changing the colours of the superclusters.
Additional Information
How to cite this article: Smolders, K. et al. An effective plasma membrane proteomics approach for small tissue samples. Sci. Rep. 5, 10917; doi: 10.1038/srep10917 (2015).
Supplementary Material
Acknowledgments
We thank L. Geenen, K. Schildermans, E. Maes and B. Van de Plas for excellent technical assistance. We are grateful to M. Christiaens for helping with making illustrations. This work was supported by grants of the Research Council KU Leuven (GOA/12/008). Katrien Smolders is supported by a Ph.D. fellowship of the Agency for Innovation through Sience and Technology Flanders (IWT Vlaanderen, 101421).
Footnotes
Author Contributions L.A. and G.B. designed study. L.A., G.B. and K.S. designed experiments. K.S., N.L. and G.B. performed experiments. All authors analyzed and interpreted data. D.V. assisted in all bioinformatics and statistics analysis. K.S., N.L., G.B. and L.A. wrote the paper and revised the text. All authors discussed results and commented on the manuscript. L.A. and G.B. supervised the project.
References
- Tan S., Tan H. T. & Chung M. C. Membrane proteins and membrane proteomics. Proteomics. 8, 3924–3932 (2008). [DOI] [PubMed] [Google Scholar]
- Wu C. C. & Yates J. R. III. The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 21, 262–267 (2003). [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhang W., Kho Y. & Zhao Y. Proteomic analysis of integral plasma membrane proteins. Anal Chem. 76, 1817–1823 (2004). [DOI] [PubMed] [Google Scholar]
- Buckley K. M., Melikian H. E., Provoda C. J. & Waring M. T. Regulation of neuronal function by protein trafficking: a role for the endosomal pathway. J Physiol. 525 (Pt 1), 11–19 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyder S., De Craene J. O., Bar S., Bertazzi D. L. & Friant S. Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int J Mol Sci. 16, 1509–1525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler J., Lewandrowski U., Sickmann A., Friauf E. & Nothwang H. G. Proteomic analysis of brain plasma membranes isolated by affinity two-phase partitioning. Mol Cell Proteomics. 5, 390–400 (2006). [DOI] [PubMed] [Google Scholar]
- Elschenbroich S., Kim Y., Medin J. A. & Kislinger T. Isolation of cell surface proteins for mass spectrometry-based proteomics. Expert Rev Proteomics. 7, 141–154 (2010). [DOI] [PubMed] [Google Scholar]
- Leth-Larsen R., Lund R. R. & Ditzel H. J. Plasma membrane proteomics and its application in clinical cancer biomarker discovery. Mol Cell Proteomics. 9, 1369–1382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varady G., Cserepes J., Nemeth A., Szabo E. & Sarkadi B. Cell surface membrane proteins as personalized biomarkers: where we stand and where we are headed. Biomark Med. 7, 803–819 (2013). [DOI] [PubMed] [Google Scholar]
- Hopkins A. L. & Groom C. R. The druggable genome. Nat Rev Drug Discov. 1, 727–730 (2002). [DOI] [PubMed] [Google Scholar]
- Santoni V., Molloy M. & Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 21, 1054–1070 (2000). [DOI] [PubMed] [Google Scholar]
- Zheng Y. Z. & Foster L. J. Biochemical and proteomic approaches for the study of membrane microdomains. J Proteomics. 72, 12–22 (2009). [DOI] [PubMed] [Google Scholar]
- Schindler J., Lewandrowski U., Sickmann A. & Friauf E. Aqueous polymer two-phase systems for the proteomic analysis of plasma membranes from minute brain samples. J Proteome Res. 7, 432–442 (2008). [DOI] [PubMed] [Google Scholar]
- Pischedda F. et al. A cell surface biotinylation assay to reveal membrane-associated neuronal cues: Negr1 regulates dendritic arborization. Mol Cell Proteomics. 13, 733–748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes M. P. et al. Comparative analysis of techniques to purify plasma membrane proteins. J Biomol Tech. 21, 108–115 (2010). [PMC free article] [PubMed] [Google Scholar]
- Thomas-Crusells J., Vieira A., Saarma M. & Rivera C. A novel method for monitoring surface membrane trafficking on hippocampal acute slice preparation. J Neurosci Methods. 125, 159–166 (2003). [DOI] [PubMed] [Google Scholar]
- Gao M. et al. Rebound potentiation of inhibition in juvenile visual cortex requires vision-induced BDNF expression. J Neurosci. 34, 10770–10779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heynen A. J. et al. Molecular mechanism for loss of visual cortical responsiveness following brief monocular deprivation. Nat Neurosci. 6, 854–862 (2003). [DOI] [PubMed] [Google Scholar]
- Holman D. & Henley J. M. A novel method for monitoring the cell surface expression of heteromeric protein complexes in dispersed neurons and acute hippocampal slices. J Neurosci Methods. 160, 302–308 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel L. R., Wu S. & Melikian H. E. Brain slice biotinylation: an ex vivo approach to measure region-specific plasma membrane protein trafficking in adult neurons. J Vis Exp. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. & Zhu H. Tube-gel digestion: a novel proteomic approach for high throughput analysis of membrane proteins. Mol Cell Proteomics. 4, 1948–1958 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler J., Ye J., Jensen O. N. & Nothwang H. G. Monitoring the native phosphorylation state of plasma membrane proteins from a single mouse cerebellum. J Neurosci Methods. 213, 153–164 (2013). [DOI] [PubMed] [Google Scholar]
- Aoun M. & Tiranti V. Mitochondria: A crossroads for lipid metabolism defect in neurodegeneration with brain iron accumulation diseases. Int J Biochem Cell Biol. (2015). [DOI] [PubMed] [Google Scholar]
- Attwell D. & Laughlin S. B. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 21, 1133–1145 (2001). [DOI] [PubMed] [Google Scholar]
- Currais A. Ageing and inflammation - A central role for mitochondria in brain health and disease. Ageing Res Rev. 21C, 30–42 (2015). [DOI] [PubMed] [Google Scholar]
- Harris J. J., Jolivet R. & Attwell D. Synaptic energy use and supply. Neuron. 75, 762–777 (2012). [DOI] [PubMed] [Google Scholar]
- Wong-Riley M. T. Cytochrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 12, 94–101 (1989). [DOI] [PubMed] [Google Scholar]
- Collingridge G. L. The brain slice preparation: a tribute to the pioneer Henry McIlwain. J Neurosci Methods. 59, 5–9 (1995). [DOI] [PubMed] [Google Scholar]
- Suter K. J., Smith B. N. & Dudek F. E. Electrophysiological recording from brain slices. Methods. 18, 86–90 (1999). [DOI] [PubMed] [Google Scholar]
- Franklin G. & Paxinos K. The mouse brain in stereotaxic coordinates. (Elsevier, Amsterdam, The Netherlands, 2008). [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4, 44–57 (2008). [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N. & Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.