SUMMARY
Upon exposure to stress, tRNAs are enzymatically cleaved, yielding distinct classes of tRNA-derived fragments (tRFs). We identify a novel class of tRFs derived from tRNAGlu, tRNAAsp, tRNAGly, and tRNATyr that, upon induction, suppress the stability of multiple oncogenic transcripts in breast cancer cells by displacing their 3′UTRs from the RNA-binding protein YBX1. This mode of post-transcriptional silencing is sequence-specific, as these fragments all share a common motif that matches the YBX1 recognition sequence. Loss-of-function and gain-of-function studies, using antisense locked-nucleic acids (LNAs) and synthetic RNA mimetics respectively, revealed that these fragments suppress growth under serum-starvation, cancer cell invasion, and metastasis by breast cancer cells. Highly metastatic cells evade this tumor-suppressive pathway by attenuating the induction of these tRFs. Our findings reveal a tumor suppressive role for specific tRNA-derived fragments and describe a molecular mechanism for their action. This transcript displacement-based mechanism may generalize to other tRNA, ribosomal-RNA, and sno-RNA fragments.
INTRODUCTION
Transfer RNA-derived RNA fragments (tRFs) belong to a family of short non-coding RNAs (ncRNAs) present in most organisms. These RNAs can be both constitutively generated and produced in the context of stress. Constitutive tRFs are thought to arise from ribonucleolytic processing of tRNAs by Dicer (Cole et al., 2009) and RNase Z (Lee et al., 2009). The cleavage of stress-induced tRFs, also known as stress-induced fragments (tiRNAs), have been shown to occur via the action of specific ribonucleases such as Angiogenin (Fu et al., 2009). While tRNAs are one of the most abundant ncRNA molecules in the cell (~10% of total cellular RNA), only a small fraction of tRNAs are cleaved to produce tRFs (Thompson and Parker, 2009). Multiple classes of tRFs have been identified in various cell-types, organisms, and induced by various conditions. These classes are defined by the position of the tRNA cleavage site that gives rise to tRFs, and these classes include 5′- and 3′-tRNA halves (cleaved in the anticodon loop), 5′- and 3′-tRFs (also known as 3′CCA tRF), and 3′U tRFs, among others (Gebetsberger and Polacek, 2013).
Stress-induced tRFs have been reported to mediate a stress response, which results in stress granule assembly and inhibition of protein synthesis (Emara et al., 2010). Moreover, these tRFs can impact a number of cellular functions, such as cell proliferation and mediating RNA inactivation through Argonaute engagement (Gebetsberger and Polacek, 2013). In this study, we sought to investigate if tRFs could play a role in metastatic progression. We reasoned that tRFs could have roles in cancer progression analogous to that of specific microRNAs (Krol et al., 2010). We also reasoned that since hypoxia is a major stress encountered by cells during cancer progression, tRFs induced under hypoxic conditions may act to curb metastatic progression. By employing next-generation small-RNA sequencing, we identified a group of tRFs that were up-regulated under hypoxia in breast cancer cells as well as in non-transformed mammary epithelial cells. Interestingly, highly metastatic breast cancer cells did not display induction of these tRFs under hypoxia, suggesting a potential role for these molecules in cancer progression. We identified a common sequence motif present in these hypoxia-induced fragments, suggesting they may interact with a common trans factor. By using one of these tRFs (tRFGlu) as bait, we immunoprecipitated and identified the RNA-binding protein YBX1 as a trans factor whose mRNA-stabilizing activity is repressed by these fragments.
YBX1 is a versatile RNA-binding protein with a variety of interacting partners. It is involved in many key cellular pathways and its genetic inactivation leads to embryonic lethality (Uchiumi et al., 2006). Importantly, it is highly over-expressed in multiple cancer types (Jurchott et al., 2010; Matsumoto and Bay, 2005; Wu et al., 2012). By combining molecular, biochemical, and computational approaches, we find that tRFs bind YBX1 and displace a number of known oncogenic transcripts from YBX1, thereby antagonizing YBX1 activity. YBX1 stabilizes these oncogenic transcripts and mediates their enhanced expression. The displacement of these oncogenic transcripts by tRFs represses their stability and expression—thereby suppressing metastatic progression.
RESULTS
Systematic identification of tRNA-derived RNA fragments in breast cancer cells
Tumor cells encounter various cellular stresses during the course of cancer progression. A critical stress is reduced access to oxygen, a condition known as hypoxia (Moyer, 2012; Wilson and Hay, 2011). Multiple regulatory programs are co-opted by tumor cells to counteract the negative impacts of hypoxic stress (Bristow and Hill, 2008). For example, the stabilization and activation of the transcription factor HIF1α under hypoxia results in the activation of vascular endothelial growth factor (VEGF, angiogenesis; Shen and Kaelin, 2013), GLUT1 (glucose transport) and carbonic anyhydrase IX (CA9, pH regulation; Semenza, 1999). Recently, it was reported that tRFs are produced under hypoxia and during other stress conditions (Fu et al., 2009). Given the ability of hypoxia to significantly modulate the regulatory landscape of the cell at both transcriptional and post-transcriptional levels, we searched for tRNA fragments with potential regulatory roles that are modulated under hypoxic conditions in cancer cells. To do so, we performed small-RNA sequencing of breast cancer cells (MDA-MB-231, hereafter termed MDA-parental). We observed that a sizeable fraction (~4%) of the small-RNA population originated from tRNAs, and therefore could be categorized as tRNA-derived RNA fragments (tRFs). We observed >10 small-RNA reads mapping to each of more than 300 tRNA loci across these samples.
Transfer RNA-derived RNA fragments belong to a class of small RNAs that are generated through endonucleolytic cleavage of tRNAs (Gebetsberger and Polacek, 2013; Thompson and Parker, 2009). These fragments have been detected in bacteria, yeast and mammalian cells under normal and stress conditions (Gebetsberger and Polacek, 2013; Lee et al., 2009). While tRNA fragments were first detected in the urine of cancer patients more than three decades ago, and at the time proposed to be oncogenic molecules (Borek et al., 1977; Speer et al., 1979), their roles and mechanisms of action during cancer progression remain uncharacterized.
Consistent with their potential roles in stress response, tRF levels in breast cancer cells significantly increased under hypoxia (p<1e-6, Figure S1A). Interestingly, the induction of these fragments by hypoxia was significantly blunted in MDA-LM2 cells—a highly metastatic sub-population derived through in vivo selection from the MDA-parental population (Figure S1A; Minn et al., 2005). These findings suggested that highly metastatic cells evade the up-regulation of these fragments under hypoxic conditions. Sequence analysis of the tRNAs with hypoxia-induced fragmentation revealed the significant enrichment of a common linear sequence motif (SCUBYC; Figure 1A). This motif was not significantly enriched in the tRNA loci whose fragments were up-regulated in highly metastatic MDA-LM2 cells, further highlighting the absence of a concerted up-regulation of these tRFs in highly metastatic cells (Figures 1A and S1A). The identification of this sequence motif among hypoxia-induced tRFs in MDA-parental cells raised the possibility that this element serves as a binding site for a common trans factor that potentially interacts with these small ncRNAs in vivo. For example, among tRNAGlu-derived fragments, which were significantly up-regulated under hypoxia in MDA-parental but not MDA-LM2 cells, several tRF species carried instances of the SCUBYC sequence motif described above (Figure 1B). In order to identify the unknown trans factor that may recognize this sequence motif, we used synthetic oligonucleotides from tRNAGlu as bait in an in vitro co-precipitation experiment. A 21-nt 3′-biotinylated oligonucleotide carrying an instance of the identified motif was immobilized on streptavidin beads and was subsequently used to co-precipitate the interacting protein complexes. A scrambled RNA was processed in parallel as the control to measure the co-precipitation of proteins above background. In-solution digestion followed by mass-spectrometry was employed to determine the identity of the co-precipitated proteins. Gene-set enrichment analysis revealed that proteins annotated as components of “ribonucleoprotein complex” and “stress granule complex” were significantly over-represented among co-precipitated proteins (Figure 1C). YBX1, which showed five-fold enrichment above background (Figure S1B), was the only protein that shares both of these annotations (Figures 1C); As such, we chose to further study this RNA-binding protein as a candidate tRF interacting protein. To validate this interaction, we performed reciprocal co-immunoprecipitations and detected binding of YBX1 to an exogenously transfected 3′-biotinylated tRFGlu mimetic, but not to the scrambled RNA or tRFAla controls (Figure 1D). Consistently, we also detected the binding of endogenous YBX1 to the tRFGlu mimetic, but not the scrambled RNA control (Figure 1E).
Figure 1. Genome-wide profiling of tRNA-derived fragments in breast cancer MDA-231 cells under normal and hypoxic conditions.
(A) The linear motif SCUBYC was enriched in RNA fragments mapping to tRNA loci that were up-regulated in MDA-parental cells, but not MDA-LM2 cells, under hypoxic conditions. Shown are the mutual information values and their associated z-scores for the discovered motif in both cell-lines (Elemento et al., 2007). The enrichment score (positive for enrichment and negative for depletion), presented as logP (hypergeometric p-value), is also shown as a heatmap with blue showing depletion and yellow showing enrichment of the SCUBYC motif among the sequences in each cluster. The red border marks statistical significance of the enrichment score. (B) The levels of tRFs derived from tRNAGlu were significantly enhanced under hypoxic conditions in MDA-parental cells but not in MDA-LM2 cells. The log fold-change was calculated from the small-RNA sequencing data. The p-value was calculated using Wilcoxon rank sum test. Two exemplary tRFs that contain the SCUBYC motif are also indicated. (C) Streptavidin beads were used to co-precipitate proteins interacting with a 3′-biotinylated synthetic tRFGlu mimetic and scrambled oligonucleotide in vivo. YBX1 was identified as a potential interacting partner based on the identity of the annotated RNA-protein complexes enriched among the tRFGlu co-precipitated RNA binding proteins. (D) 3′-biotinylated synthetic oligonucleotides were used to co-precipitate YBX1. In addition to the scrambled RNA, a tRNAAla-derived fragment, which does not carry the identified motif, was also included as control. Western blotting was performed to detect YBX1 in the eluate from each sample. (E) MDA-LM2 cells were transfected with a 21-nt synthetic tRFGlu mimetic (unlabeled), also shown are scrambled transfected mimetic and untransfected cells as controls. After crosslinking immunoprecipitation of endogenous YBX1, and radiolabeling of the RNA population, a strong interaction between the transfected tRFGlu mimetic and endogenous YBX1 was observed. Error bars in all panels indicate s.e.m. unless otherwise specified.
Specific tRNA-derived fragments interact with YBX1
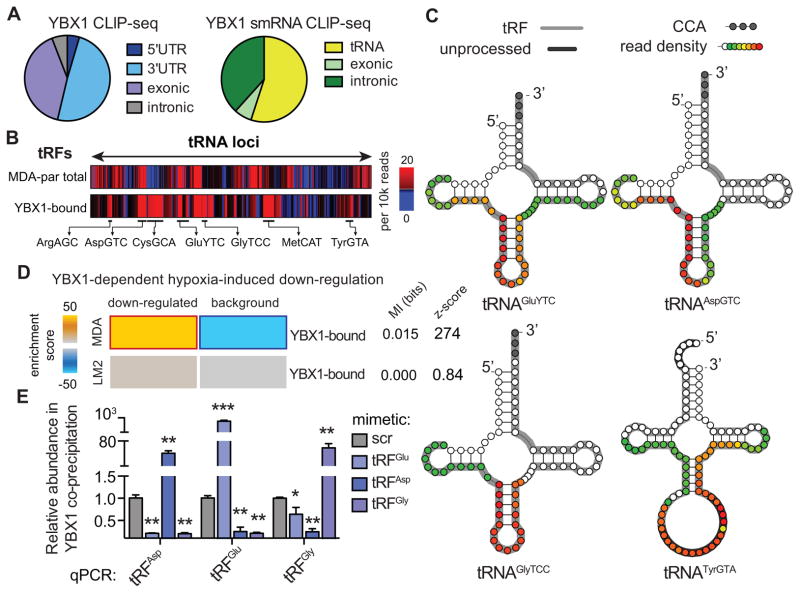
YBX1, a multifunctional RNA binding protein and a member of the Y-box binding protein family, has been implicated in various aspects of RNA biology. Importantly, it is a known modulator of RNA translation and stability and has been implicated in tiRNAAla-mediated inhibition of ribosomal activity in vivo (Ivanov et al., 2011). Consistently, while resolving YBX1-crosslinked RNAs on a polyacrylamide gel, we observed a prominent small-RNA band in addition to longer RNA species interacting with YBX1 (Figures 1E and S1C). We hypothesized that in addition to tRFGlu, which was used as bait to identify YBX1, YBX1 may also interact with a broader population of endogenous small-RNAs in the cell. To generate a detailed and precise snapshot of genome-wide YBX1-RNA interactions across long and short RNA species, we performed crosslinking immunoprecipitation of endogenous YBX1 followed by high-throughput sequencing (CLIP-seq) in human MDA-parental breast cancer cells. Analysis of the CLIP-seq data provided us with the first in vivo YBX1 transcript interactome revealing more than 4,000 endogenous transcripts bound by YBX1. The majority of YBX1 binding sites were localized to 3′ untranslated regions (3′UTRs) and exons, while minimal binding was detected in 5′-UTRs and in intronic sequences (Figure 2A). A large number of cellular processes and pathways were over-represented among the YBX1-bound transcripts, including RNA processing, translation, cell cycle, glucose catabolism, spindle organization, and additional key signaling and stress-response pathways (Figure S1D). The breadth and diversity of the YBX1 regulon (Figure S1E–F) highlight its role in RNA homeostasis and growth, as well as its importance for cellular response to internal and external stimuli.
Figure 2. Endogenous YBX1 interacts with a large regulon of transcripts and small-RNAs in vivo.
(A) Pie-charts depicting the annotation of YBX1 binding sites obtained from immunoprecipitation of endogenous YBX1 from RNase-treated lysate of UV-irradiated MDA-parental cells followed by high-throughput sequencing of both long- and small-RNAs. (B) Relative frequency of reads mapped to each tRNA-locus in MDA-parental small-RNA sequencing and YBX1 small-RNA CLIP-seq. The tRF species most abundantly bound by YBX1 are marked. (C) Based on the YBX1 small-RNA CLIP-seq results, four species of tRNA-derived RNA fragments (tRFs) bound by YBX1 in vivo were identified. Shown are examples of tRNA structures for each species depicting the boundaries of the identified tRFs along with the YBX1 binding region based on smRNA YBX1 CLIP-seq read density at each position (also see Figure S2A). The grey nucleotides at the 3′ end mark the presence of terminal CCA sequences. The dark grey highlights for tRNATyrGTA mark the leader and intronic sequences in the unprocessed tRNA. The longest identified form of each tRF based on our high-throughput sequencing results are also indicated (grey highlight) along with the YBX1 smRNA CLIP-seq read density at each position (overlaid as a heatmap) indicating the YBX1 binding site. (D) Gene-expression profiling of control and YBX1-knockdown cells was performed under normal and hypoxic conditions in both MDA-parental and MDA-LM2 backgrounds. The set of transcripts that was down-regulated under hypoxia in a YBX1-dependent manner was identified for each cell-line. While YBX1-bound transcripts were significantly enriched among YBX1-dependent hypoxia-induced downregulated transcripts in MDA-parental cells, this enrichment was absent in the highly metastatic MDA-LM2 cells. (E) qPCR-based validation of interactions between YBX1 and tRFAspGTC, tRFGluYTC, and tRFGlyTCC. Cells transfected with exogenous tRF mimetics were subjected to UV-crosslinking and YBX1 immunoprecipitation. The abundance of each tRF in the co-immunoprecipitated RNA population was then measured using a small-RNA qPCR-based quantitation assay (n=3–4). Statistical significance is measured using one-tailed Student’s t-test: *, p<0.05, **, p<0.01, and ***, p<0.001. Error bars in all panels indicate s.e.m. unless otherwise specified.
Consistent with our observation regarding the interaction between tRFGlu and YBX1 in vivo, high-throughput sequencing of the YBX1-crosslinked small RNAs (smRNA CLIP-seq) revealed that the majority of these CLIP-seq tags mapped to tRNA loci and represented tRNA-derived RNA fragments (Figures 2A and S2A–B). We observed that YBX1 interacts with a specific subset of tRFs present in these cells. For example, tRFGlyTCC and tRFAspGTC, both with relatively low cellular expression levels displayed substantial binding to YBX1, whereas highly expressed fragments, tRFLysTTT and tRFSerAGA, were absent among the YBX1 smRNA CLIP-seq tags (Figure S2C). A more global comparison is provided in Figure 2B in which the relative abundance of tRNA fragments mapping to each tRNA locus in MDA-parental cells is shown relative to those from the YBX1 smRNA CLIP-seq. These findings reveal that the interactions between tRFs and YBX1 were not simply a function of tRF abundance in the cell and that YBX1 binding to tRFs is specific and dependent on factors other than tRF levels (e.g. sequence specificity). Based on our YBX1 smRNA CLIP-seq experiment, we identified a number of specific tRFs as most abundantly bound by YBX1; chief among them, the tRNAGlu, tRNAAsp, and tRNAGly fragments which mapped to the anti-codon loops of these tRNAs, and a tRNATyr-derived fragment matching the intron-containing precursor of this tRNA (Figure 2C).
Transcriptomic profiling under normoxic and hypoxic conditions in both control and YBX1-knockdown cells revealed that in the MDA-parental breast cancer cells, YBX1-bound transcripts were significantly enriched among those down-regulated under hypoxia in a YBX1-dependent manner (Figure 2D). This observation suggested the presence of a hypoxia-induced and YBX1-mediated post-transcriptional regulatory program. Gene expression analysis of non-tumorigenic mammary epithelial MCF10a cells under hypoxia also showed an enrichment of YBX1-bound transcripts among the hypoxia-induced down-regulated genes, further strengthening this hypothesis (z-score=5.7; data not shown). More importantly, highly metastatic MDA-LM2 cells did not exhibit YBX1-dependent down-regulated expression of the target transcripts under hypoxia, consistent with their lack of induction of specific hypoxia-induced tRFs (Figure 2D). Given that YBX1 did not show a significant change in expression in MDA-LM2 cells relative to the MDA-parental line (Figure S2D), we hypothesized that the observed enrichment may be mediated by tRFs, which are induced in poorly metastatic but not highly metastatic cells (Figure S1A).
Our findings described above reveal a direct physical interaction between tRNAGlu fragments and YBX1 (Figures 1D–E). In order to validate the in vivo interaction between the other tRFs and YBX1, we developed a cell-based competition experiment, based on quantitative PCR (qPCR) assays of small-RNAs, in which chemically synthesized tRF mimetics were used to compete with endogenous fragments for YBX1 binding in vivo. We designed specific primers for reliable qPCR-mediated detection of tRFGlu, tRFAsp, and tRFGly in this competition assay. Under the assumption that exogenous tRFs effectively bind YBX1 in vivo (as was shown for tRFGlu), we predicted that the increase in cellular levels of a specific tRF would result in the subsequent displacement of other tRNA fragments from the endogenous YBX1-bound RNA population. To test this hypothesis, we UV-irradiated cells transfected with synthetic tRFs or scrambled controls, immunoprecipitated YBX1 under stringent CLIP-seq conditions (Ule et al., 2005) and used qPCR to detect the abundance of each specific tRF in the YBX1-bound fraction. Consistent with active binding of YBX1 to synthetic tRFs, we observed that exogenous transfection of a given tRF led to depletion of the other assayed endogenous tRF species in every case (Figure 2E). This quantitative assay demonstrates not only that these tRF species bind YBX1, but that they also compete for YBX1 binding in vivo. Moreover, as was the case for tRFGlu, we used 3′-biotinylated short oligonucleotides mimicking tRFAsp and tRFGly to co-immunoprecipitate interacting proteins from total cell lysates. We observed significant enrichment in endogenous YBX1 protein levels upon co-immunoprecipitation of the tRFs relative to a 3′-biotinylated scrambled oligonucleotide (Figure S2E). Importantly, we also observed a significant up-regulation in tRFGlu, tRFAsp, and tRFGly under hypoxic conditions, quantified using tRF-specific qPCR in both MDA–parental breast cancer cells and MCF10a non-transformed mammary epithelial cells (Figure S2F–G). Consistent with our prior findings, this induction was absent in metastatic MDA-LM2 cells (Figure S2H). While these YBX1-binding tRFs are constitutively expressed, their levels are enhanced in the context of hypoxic stress. This observation indicates that the tRFs identified here can be categorized as tRNA-derived stress-induced RNAs (tiRNAs) and likely play roles in stress-responses. More importantly, the absence of their induction in highly metastatic MDA-LM2 cells highlights the potential suppressive roles they may play during breast cancer metastasis, in which tRFs must be antagonized for metastasis to progress.
tRF-mediated post-transcriptional modulation through YBX1
Previous studies have established a role for another class of tRNA fragments—tRNA 5′-halves (e.g. 5′-tiRNAAla and 5′-tiRNACys) in translation inhibition. These fragments were found to cause translation initiation factors to disengage from mRNAs (Ivanov et al., 2011). This translational inhibition effect was shown to be YBX1 dependent. The molecular mechanism through which this previously identified class of tRFs modulates YBX1 interaction with translation initiation factors remains to be elucidated. YBX1 has also been implicated in other post-transcriptional regulatory programs, most notably transcript stability. We set out to test whether functional interactions between tRFs and YBX1 affect expression levels of endogenous transcripts. We envisioned two plausible molecular mechanisms through which tRFs may affect YBX1 binding to mRNAs in vivo (Figure S3A). First, in a tRF-mediated transcript engagement model, a given tRF may act as a guide RNA whereby the YBX1-tRF complex binds specific target transcripts based on sequence complementarity to the bound tRF, in a manner similar to miRNA-mediated binding of transcripts by Argonaute. In this scenario, reducing tRF levels would lead to reduced YBX1 binding of transcripts. Second, in a tRF-mediated transcript displacement model, YBX1 would interact with tRFs and mRNAs alike, in which case tRFs would be actively competing with endogenous transcripts for YBX1 binding. In this model, reducing tRF levels would lead to greater YBX1 binding of its target mRNAs. Importantly, in the transcript engagement model, mRNAs carrying the reverse-complement of the tRF sequence would be affected in terms of YBX1-binding abundance or expression, while in the transcript displacement model, only the transcripts that contain YBX1 binding motifs would be affected by tRF modulation. In order to distinguish between these two molecular mechanisms, we utilized synthetic antisense locked-nucleic acids (LNAs) targeting the YBX1 binding site (based on the smRNA CLIP-seq peaks; Figure S2A–B) on the most abundantly bound tRFs—namely, tRFAsp, tRFGly, tRFGlu, and tRFTyr (Figure 2B). We used specific antisense LNAs to bind and inhibit the endogenous forms of these tRFs individually in order to observe their effects on the transcriptome relative to a scrambled LNA control. Importantly, to specifically focus on transcripts impacted by tRF inhibition through their direct interaction with YBX1, we conducted whole-transcriptome profiling of both control and YBX1-knockdown cells transfected with antisense LNAs (Figures S3B).
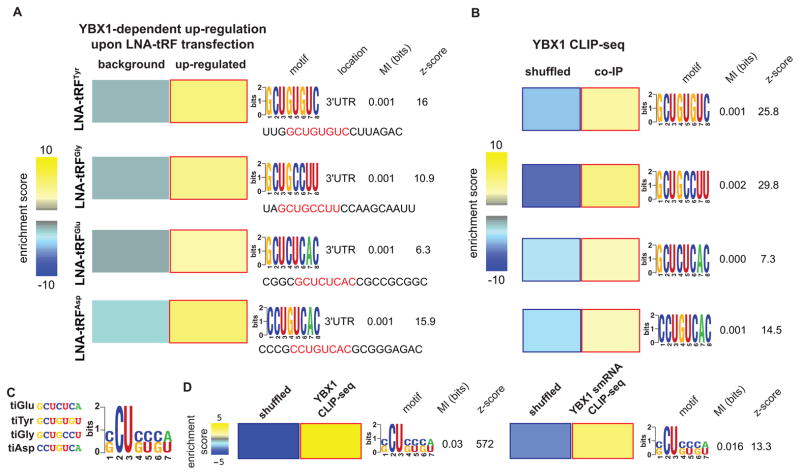
We first sought to identify the YBX1 binding sequences on these four tRFs in order to observe the behavior of mRNAs carrying these sequences (transcript displacement model) or their complementary sequence (transcript engagement model). We made no assumptions about the YBX1 binding site on RNAs, instead opting to test all possible 8-mers (and their reverse complements) along the identified binding sites on each of the four tRFs (sequences shown in Figure 3A). Each 8-mer was assessed for (i) its enrichment (or that of its reverse complement sequence) in 3′UTRs of transcripts that were deregulated in a YBX1-dependent manner in each experiment (see Methods) and (ii) its enrichment among YBX1 binding sites on endogenous transcripts (YBX1 CLIP-seq). For each tRF, we successfully identified an 8-mer that was both enriched in (i) the 3′ UTRs of transcripts up-regulated (not down-regulated) in a YBX1-dependent manner and (ii) in the YBX1 CLIP-sites on endogenous mRNAs (Figure 3A–B). Given that in each case, it was the specific 8-mer, rather than its reverse-complement, that was functionally bound by YBX1 in vivo, the YBX1 modulation of transcript levels observed here is consistent with the model wherein tRFs displace transcripts from YBX1. In this competition-based model, the binding of specific tRFs to YBX1 is inhibited by LNA transfection, allowing free YBX1 to interact with YBX1 binding sites on endogenous transcripts. Increased YBX1 binding would result in higher transcript abundances, most likely through enhanced mRNA stabilization by YBX1 in vivo. Subsequently, we provide additional genomic, molecular, and biochemical evidence that support this YBX1-dependent post-transcriptional mode of regulation.
Figure 3. YBX1 interacts with long- and small-RNAs via a specific linear sequence motif.
(A) The transcripts up-regulated upon antisense LNA transfections targeting each of the four identified tRFs were compared to the remainder of the transcriptome (background) to identify over-representation of specific sequence elements in their 3′ UTRs. Here, we have shown the enrichment of specific 8-mers along each YBX1-bound tRF in the 3′ UTRs of these transcripts as a heatmap, with yellow and blue showing the extent of enrichment and depletion respectively (red and blue borders mark statistical significane). Also shown are the associated mutual information values and z-scores (Elemento et al., 2007). We have provided the sequence of each tRF and highlighted the identified 8-mers. (B) These 8-mers were also required to be enriched among the YBX1 binding sites identified using CLIP-seq. We used a shuffled version of each YBX1-binding site to create a background set and tested the enrichment of each 8-mer in the YBX1 binding sites relative to shuffled controls. (C) In order to infer a consensus element for YBX1 on these tRFs, the four significant 8-mers were aligned and the possible nucleotides at each position were combined to build the CU-box element represented as a regular expression. (D) The CU-box motif showed a significant enrichment in both long- and small-RNA YBX1 CLIP-seq datasets relative to randomly shuffled sequences, indicating YBX1 binds a common linear sequence motif on both short and long endogenous RNAs. Error bars in all panels indicate s.e.m. unless otherwise specified.
In order to determine a consensus binding site for YBX1 on tRFs and mRNAs, we performed multiple alignments for the 8-mers that were independently identified for each tRF. The resulting sequence motif, named CU-box based on the prominence of a C and U at the second and third positions along the identified regular expression representation of the element (Figure 3C), was significantly enriched among CLIP-seq tags in both YBX1 small-RNA (p<0.002) and long RNA (mRNA) CLIP-seq datasets (p<10−80; Figure 3D). Importantly, this element resembles an in vitro YBX1 binding motif described previously (Figure S3C). Moreover, tracking crosslinking-induced mutation sites (CIMS; Zhang and Darnell, 2011), which mark protein-RNA interactions at single-nucleotide resolution, at and around CU-box elements (10 flanking nucleotides) across the YBX1 CLIP sites, revealed that the crosslinked nucleotides were most frequent at the site of the motif (Figure S3D). This observation further supports a direct physical interaction between YBX1 and CU-box elements. Taken together, our results indicate that YBX1 interacts with both endogenous target transcripts and tRFs via the CU-box element. It should also be noted that the SCUBYC motif identified in Figure 1A constitutes a specific subset of the CU-box element described here (CompareACE score of 0.85). The commonality of the binding site would enable tRFs to competitively modulate the levels of YBX1 available for transcript binding, which would in turn affect the expression of a large set of target transcripts.
Competitive tRF binding to YBX1 results in transcript destabilization
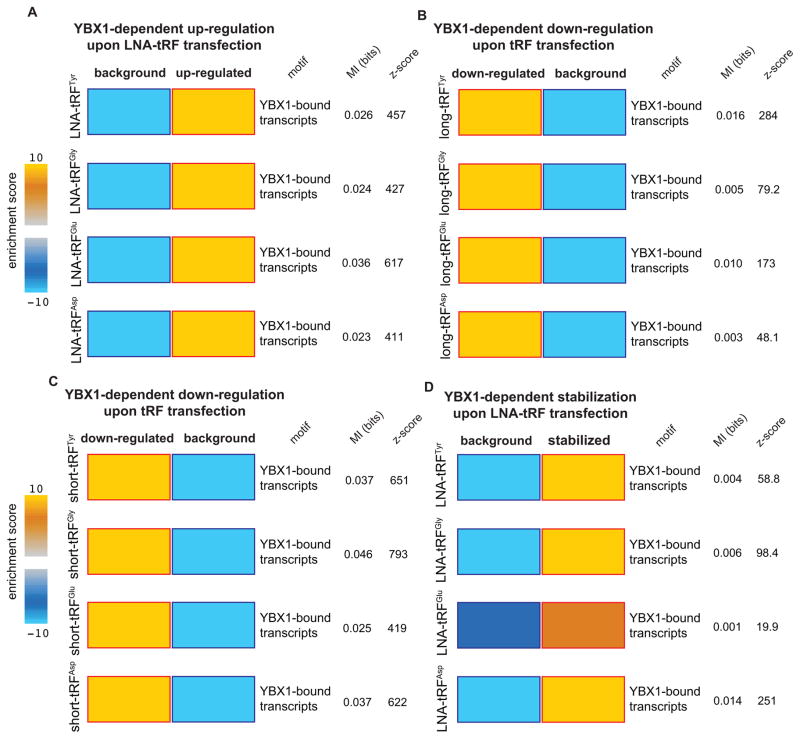
In addition to the antisense LNA-mediated tRF loss-of-function experiments followed by transcriptomic profiling, we also transfected control and YBX1-knockdown cells with synthetic tRF mimetics in gain-of-function experiments delineated in Figure S3E. For this, we performed two separate experiments; in one, we transfected control and YBX1-knockdown cells with the mimetics representing the long form of each identified tRF (Figures 2C and S2B), and in another, we used ~20 nucleotide short synthetic tRF mimetics containing the identified YBX1 binding sites. While YBX1-bound transcripts (and transcripts with CU-boxes in their 3′ UTRs) were significantly up-regulated upon LNA transfections (Figures 4A and S4A–B), these transcripts were significantly down-regulated in the context of tRF mimetic transfection in a YBX1 dependent manner (Figures 4B–C and S4C–D). These observations reveal that (i) exogenously transfected tRF mimetics act as modulators of the YBX1 regulon, and (ii) short tRF mimetics carrying the YBX1 binding site are sufficient for exerting this regulatory effect.
Figure 4. Endogenous transcripts bound by YBX1 are modulated by YBX1-bound tRFs.
In order to measure the post-transcriptional regulatory consequences of tRFs, gain-of-function and loss-of-fuction experiments were performed by transfecting synthetic tRF mimetics or inhibitory antisense LNAs, for each of the four YBX1-binding tRFs, in normal and YBX1 knockdown cells. Transcripts that were up- or down-regulated in a YBX1 dependent manner were identified by comparing the gene expression changes in normal cells relative to those in YBX1-knockdown cells (Figure S3). Transcripts that interact with YBX1 in vivo (determined from YBX1 CLIP-seq data) were significantly deregulated upon modulations of tRF levels: (A) they were up-regulated upon LNA-mediated inhibition of YBX1-binding tRFs; (B–C) they were down-regulated in the presence of exogenously added short and long tRF mimetics (~60 and ~20 nucleotides respectively; see Figure S2B), and (D) the observed up-regulation in the LNA-transfected cells coincided with a significant increase in their stability. Whole-genome transcript stability measurements were performed in LNA-transfected cells using α-amanitin-mediated inhibition of RNA polymerase II followed by RNA extraction and profiling at 0- and 8-hr time-points. In all datasets, the calculated mutual information values (in bits) and their associated p-values are provided. Also shown are the enrichment scores, presented as logP (positivie for enrichments and negative for depletions) where P is calculated from hypergeometric distribution (shown as a heatmap with blue and gold showing depletion and enrichment respectively). The red and blue borders mark statistical significance of the enrichment/depletions. Error bars in all panels indicate s.e.m. unless otherwise specified.
To determine if the observed YBX1-dependent tRF-mediated modulations in YBX1 target transcripts′ levels were occurring post-transcriptionally, we performed whole-genome transcript stability measurements. Through α-amanitin-based inhibition of RNA polymerase II in antisense LNA-transfected control and YBX1 cells, we found that the observed increase in YBX1-targeted transcript abundance resulted from a significant enhancement of their stabilities in a YBX1-dependent manner (Figures 4D, S4E–F). This observation further establishes the role of these YBX1-tRF interactions as a coherent and functional post-transcriptional regulatory program (Figure S4G). Importantly, the changes observed in the expression of YBX1-dependent transcripts upon transfection of antisense LNAs or tRF mimemtics were significantly anti-correlated (Figure S4H). Further supporting this model, we also observed a reduction in total RNA bound to YBX1 upon transient transfection of an exogenous tRFGlu mimetic (Figure S5A).
Target-specific validation of a role for tRF-YBX1 in transcript destabilization
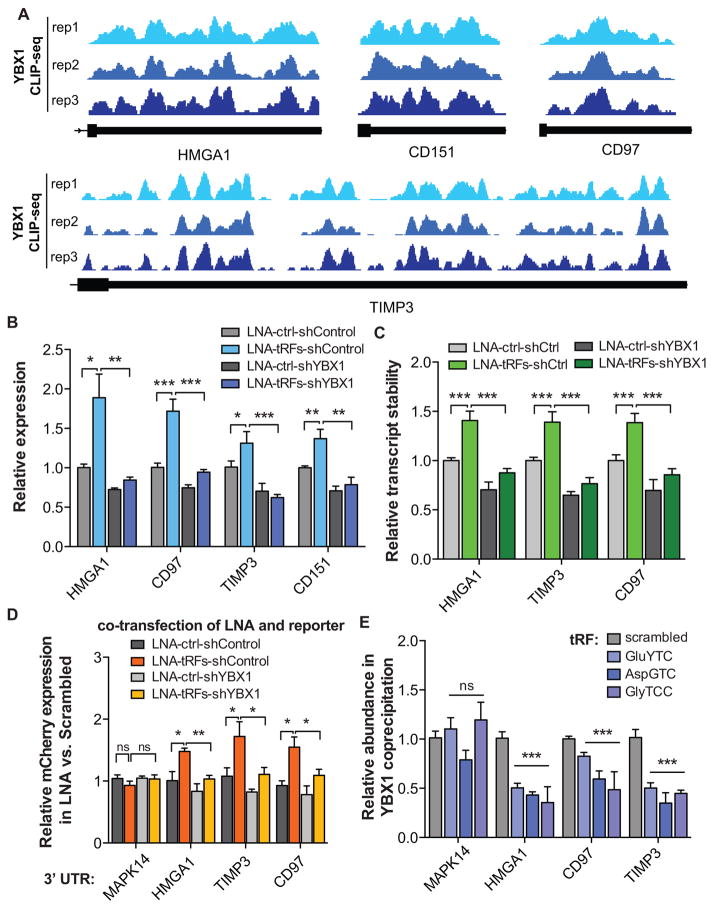
Whole-genome expression and stability measurements support a transcript displacement model for tRF-YBX1 interaction, wherein tRFs effectively compete with endogenous transcripts for YBX1 binding. In order to independently validate our observations for a specific set of targets, we chose HMGA1, CD151, CD97, and TIMP3 for target-specific follow-up experimental validation based on pervasive in vivo interactions with YBX1 along their 3′UTRs (Figure 5A). Transfection of tRF-specific antisense LNAs significantly up-regulated and stabilized these transcripts (Figures 5B–C). More importantly, their observed up-regulation was abrogated in YBX1-knockdown cells (Figures 5B–C).
Figure 5. YBX1 target transcripts and their response to changes in tRF levels.
(A) YBX1 interacts with the 3′UTRs of HMGA1, CD151, CD97, and TIMP3. The last exon of the indicated transcripts are shown with mapped reads from experimental replicates of YBX1 CLIP-seq. (B) Transfection of antisense LNAs against YBX1-binding tRFs resulted in the up-regulation of HMGA1, CD151, CD97, and TIMP3 transcripts, in a YBX1-dependent manner, as determined by qPCR measurements. (C) Similarly, transfecting antisense LNAs resulted in a significant stabilization of HMGA1, CD97, and TIMP3 transcripts in a YBX1-dependent manner. Whole-genome RNA stability measurements were perfomed using α-amanitin-mediated inhibition of RNA polymerase II (see Methods). (D) A GFP/mCherry dual-reporter assay was used to measure the effects of cloning HMGA1, CD97, and part of the TIMP3 3′ UTRs downstream of mCherry using qRT-PCR. The 3′ UTR of MAPK14, which is devoid of YBX1 tags, was included as a control. Consistent with our prior findings, LNA transfections resulted in a significant increase in relative mCherry expression in a YBX1-dependent manner. (E) Exogenously added tRF mimetics, while showing no effect on MAPK14 abundance, resulted in a significant depletion of HMGA1, CD97, and TIMP3 transcripts from the YBX1 co-immunoprecipitated RNA population. Statistical significance is measured using one-tailed Student’s t-test: *, p<0.05, **, p<0.01, and ***, p<0.001. Error bars in all panels indicate s.e.m. unless otherwise specified.
As mentioned earlier, under hypoxic conditions, in which the YBX1-bound tRFs were shown to be up-regulated in MDA-parental cells (Figure 2C), we observed a concomitant down-regulation of YBX1 target transcripts (Figure 2D). Importantly, this hypoxia-induced YBX1-dependent down-regulation, which was absent in highly metastatic MDA-LM2 cells, was diminished once hypoxic cells were transfected with antisense LNAs (Figure S5B). We also observed a similar expression pattern for HMGA1, CD97, and TIMP3 transcripts when tested under hypoxia and normoxia in both control and YBX1-knockdown cells in MDA-parental and MDA-LM2 backgrounds (Figure S5C). Consistently, we found that the 3′UTR sequences of these transcripts cloned downstream of a bidirectional reporter construct were sufficient to confer YBX1-dependent up-regulation of the reporter upon transfection of antisense LNAs targeting the four selected tRFs (Figure 5D). The MAPK14 3′ UTR, which was not bound by YBX1 as assessed by CLIP-seq, served as a comparative control. Importantly, the YBX1 dependence of this tRF-mediated response to LNA transfection further supports a role for YBX1 in stabilizing target transcripts through binding of 3′ UTR elements (Figure 5D).
Consistent with our proposed tRF-mediated transcript displacement model, we also observed that transfecting tRF mimetics into cells followed by YBX1 co-immunoprecipitation depleted HMGA1, TIMP3, and CD97 transcripts from the YBX1-bound RNA population (Figure 5E). This observation further supports direct competition between tRFs and endogenous transcripts for YBX1 binding in vivo.
tRF-mediated modulation of cancer progression and metastasis
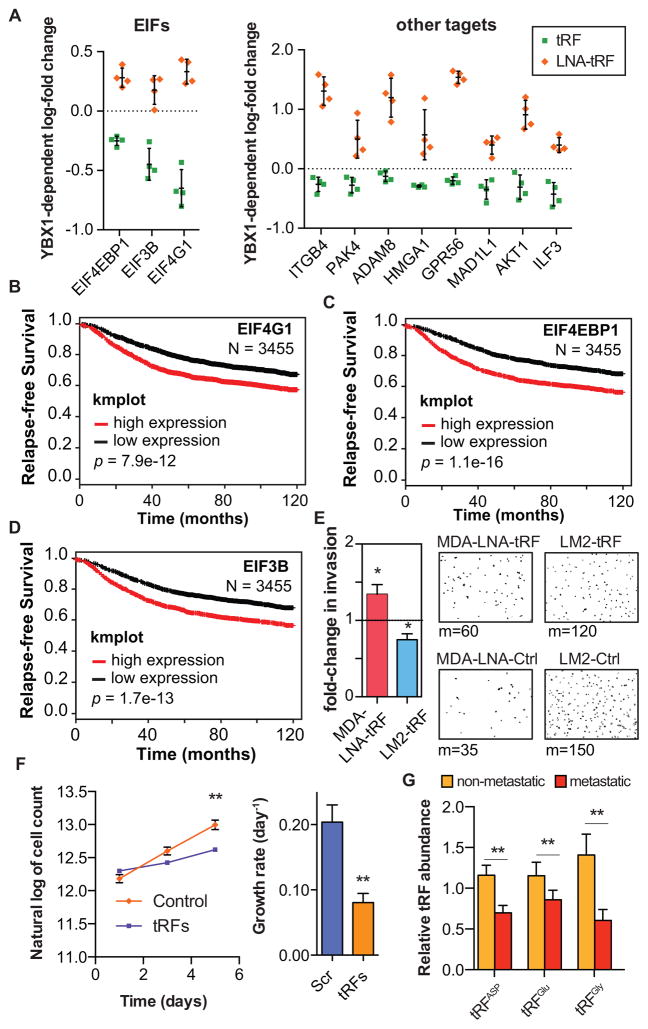
YBX1 has been implicated in cancer progression. YBX1 over-expression is correlated with tumorigenic phenotypes and has also been shown to promote cancer metastasis (Jurchott et al., 2010; Matsumoto and Bay, 2005; Uchiumi et al., 2006; Wu et al., 2012). Given the role of tRFs in suppressing the expression of YBX1 target genes, we hypothesized that this class of small RNAs may act to suppress cancer progression. For example, a tRF-YBX1 signature based on the average expression of roughly 70 YBX1-bound transcripts with robust modulations in response to antisense LNA or tRF mimetic transfections was found to be significantly associated with cancer stage as well as relapse-free survival of patients with breast cancer (Figure S6A–B). The entire YBX1-tRF regulon—defined as the subset of endogenous transcripts that are bound by YBX1 in vivo and whose expression is modulated by the transfection of tRF mimetics and antisense LNAs in a YBX1-dependent manner—contains hundreds of transcripts, including many known promoters of tumorigenesis and metastasis, such as AKT, EIF4G1, ITGB4, and HMGA1 (Figures 6A and S6C). We also noted highly significant associations between increased expression of multiple tRF-YBX1 targets that are key components of translation initiation (EIF4G1, EIF4EBP1, and EIF3B) and reduced relapse-free survival (p=8e-12, 1e-16, and 2e-13 respectively; N=3455; Figure 6B–D). The transcripts of these oncogenes, which play roles in various aspects of cellular function, including translation and cell signaling, were repressed by tRFs in breast cancer cells (Figure 6A). Transfection of antisense LNAs targeting the tRFs into MDA-parental and CN-parental cancer cells significantly enhanced cell invasion capacity in vitro (Figures 6E and S6D). Conversely, transfection of tRF mimetics into metastatic MDA-LM2 and CN-LM1a lines significantly reduced cancer cell invasion (Figures 6E and S6D). We also observed a substantial decrease in cell proliferation rate under serum-starved conditions in the presence of these tRFs, further highlighting their roles as components of a general stress response pathway (Figure 6F). These tRFs were ineffective at suppressing these phenotypes in cells depleted of YBX1—consistent with a required role for YBX1 in modulating these tRF-mediated responses (Figures S6E–F).
Figure 6. YBX1-binding tRFs play a significant role in modulating oncogenes.
(A) Competitive displacement of YBX1 from its target transcripts by tRNA-derived fragments resulted in the down-regulation of a large set of oncogenes and metastasis promoter genes. (B–D) Kaplan-Meier curves for three translation initiation factors that were modulated by tRFs via YBX1 binding (Gyorffy et al., 2012). (E) Exogenously added tRF mimetics or antisense LNAs resulted in a significant increase and decrease in cancer cell invasion, respectively. Shown are the fold-changes in cancer cell invasion for MDA-parental cells transfected with LNAs and MDA-LM2 cells transfected with tRFs. We have also included representative fields from the invasion inserts along with the median of cells observed in each cohort (n=7–8). (F) Growth rates (estimated based on an exponential model) under serum-starved conditions for MDA-LM2 cells transfected with tRF mimetics relative to mock-transfected cells (n=6). (G) qRT-PCR assays were used to quantify the levels of tRFAsp, tRFGlu, and tRFGly in metastatic (n=18) and non-metastatic (n=9) primary breast cancers. For comparing growth under serum-starved conditions, two-way ANOVA was used to measure statistical significance. For all other cases, statistical significance was measured using one-tailed Student’s t-test: *, p<0.05, **, p<0.01, and ***, p<0.001. Error bars in all panels indicate s.e.m. unless otherwise specified.
Importantly, we also detected tRFAsp, tRFGlu, and tRFGly in RNA samples from metastatic and non-metastatic primary tumors as well as normal breast tissue. Consistent with a tumor suppressive role for these tRFs, their levels were significantly lower in breast cancer tissue relative to normal breast tissue (Figure S6G). Moreover, if these specific tRFs suppress metastatic progression, we would expect that there would be a selection for reduced expression of these tRFs during this process. Indeed, we observed a significant trend towards reduced tRF levels in metastatic samples compared to primary tumors (p=0.003, N=27; Figure 6G).
To demonstrate the physiological relevance of this hypoxia-induced tRF-YBX1 pathway, we used a reporter driving the expression of luciferase under a hypoxia response promoter (Figure S7A). Consistent with MDA-parental breast cancer cells experiencing hypoxia early in the metastatic process in the lungs of xenografted mice, cells carrying this reporter exhibited induction of the hypoxia-induced pathway 24 hours post-injection (Figure S7B). To probe the in vivo expression of tRF-YBX1 targets, we constructed a lentiviral system with firefly luciferase reporter fused to 3′ UTRs of CD97 and TIMP3 (tRF-YBX1 targets) as well as MAPK14 (as a control). Consistent with YBX1 stabilizing these transcripts by binding to their 3′ UTRs, immediately after injection, we observed a significantly lower luciferase activity for CD97 and TIMP3 3′UTRs in YBX1-knockdown cells (Figure S7C). More importantly, the CD97 and TIMP3 reporters showed significantly lower day 3 to day 0 luciferase activity ratios in control cells compared to YBX1-knockdown cells (Figure S7D). These findings are consistent with the in vivo induction of tRFs under hypoxia and the YBX1-dependence of the associated response. It should be highlighted that this reduction was absent in the luciferase reporter carrying the MAPK14 3′UTR (Figure S7D).
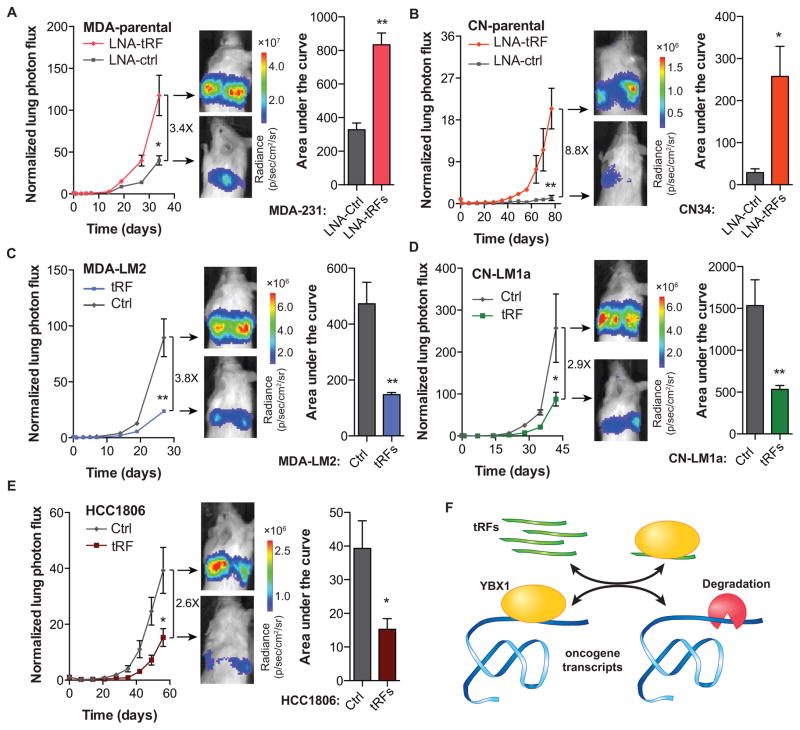
Consistent with the observed clinical associations and our in vitro as well as in vivo findings, transfection of tRF mimetics and antisense LNAs significantly impacted metastatic colonization in in vivo lung colonization assays by multiple independent cell lines. MDA-parental cells transfected with antisense LNAs exhibited significantly higher metastatic colonization activity relative to cells transfected with scrambled controls (Figures 7A and S7E). Similarly, transfection of inhibitory antisense LNAs in CN-parental cells also caused a marked and significant increase in metastatic colonization of the lungs (Figures 7B and S7E). In contrast, exogenous transfection of tRF mimetics into highly metastatic lines (MDA-LM2 and CN-LM1a cells) significantly reduced cancer metastasis to the lungs (Figures 7C–D and S7E). We also tested and validated the impact of tRF modulation on the metastatic activity of a third human breast cancer cell-line—HCC1806 (Figure 7E and S7E). It should be noted that in these in vivo lung colonization assays, a clear and significant difference in the normalized signal could be detected early in the in vivo experiments (Figure S7F). This early impact on metastasis is consistent with the role of tRFs in cancer cell invasion, which is an early determinant of metastatic progression. This early difference persists throughout the experiment despite the dilution of mimetics and anti-sense LNAs (Figure S7G) and the two cohorts fail to converge. Importantly, consistent with a YBX1-dependent mode of action, depleting YBX1 from cancer cells using RNAi made cells insensitive to tRF-mediated modulation of metastatic activity (Figure S7H).
Figure 7. YBX1-binding tRFs play a significant role as suppressors of tumor progression and metastasis.
(A–B) Bioluminescence imaging plot of metastatic lung colonization by MDA-parental and CN-parental cells transfected with synthetic antisense LNAs against all four YBX1-binding tRFs. Representative images along with quantification of the area-under-the-curve for each mouse are also included (n=3–5 in each cohort). (C–D) Bioluminescence imaging plot of lung metastasis by MDA-LM2 and CN-LM1a cells transfected with the four YBX1-binding tRFs. Representative images and area-under-the-curve quantifications are also included (n=4–5 in each cohort). (E) Bioluminescence imaging plot of lung metastasis by HCC1806 cells transfected with the four YBX1-binding tRFs. Representative images and area-under-the-curve quantifications are also included (n=5 in each cohort). (F) Schematic of tRF-mediated modulation of invasion and metastatic lung colonization through in vivo titration of YBX1 and the subsequent destabilization of its oncogenic and pro-metastatic targets. For comparing metastasis colonization assays, two-way ANOVA was used to measure statistical significance. For all other cases, statistical significance is measured using one-tailed Student’s t-test: *, p<0.05, **, p<0.01, and ***, p<0.001. Error bars in all panels indicate s.e.m. unless otherwise specified.
DISCUSSION
By integrating biochemical, molecular, computational, and phenotypic analyses, we have found that a specific set of tRNA-derived fragments functionally engages the oncogenic RNA-binding protein YBX1. These fragments, which contain a CU-box motif, post-transcriptionally suppress the expression of YBX1 transcripts by competitively displacing them from YBX1. We find that YBX1 binds and promotes the stability of a large set of transcripts, thus modulating a large regulon with broad consequences for cellular function. Our study reveals the first comprehensive and in vivo interaction map between YBX1, one of the most over-expressed oncogenes observed in human cancer (up-regulated in 10% of all cancer versus normal tissue datasets; Oncomine), and its post-transcriptional target transcripts (Lasham et al., 2012; Uchiumi et al., 2006; Wu et al., 2012). A number of these transcripts encode established drivers of oncogenesis, such as EIF4G1, ITGB4, AKT1, and ADAM8. YBX1 stabilization of oncogenic transcripts is mediated by its binding to the CU-box motif which is primarily located in the 3′UTR’s of transcripts. The displacement of oncogenic transcripts from YBX1 results in their destabilization and down-regulation. Consistent with a tumor-suppressive role for these YBX1-antagonistic small-RNAs, their introduction into breast cancer cells inhibited breast cancer growth under serum starvation, cell invasion, and metastasis. Conversely, inhibiting these fragments by antisense LNAs enhanced these phenotypes.
This tRF-mediated displacement mechanism of post-transcriptional silencing involving the binding of specific tRFs to YBX1 differs from RNAi-mediated silencing in that small tRNA-fragments do not serve as guides for transcript engagement by YBX1. Rather, they competitively displace and thus destabilize YBX1-bound transcripts. Our findings expand the repertoire of endogenous small-RNA mediated post-transcriptional modes of regulation that have been previously described (RNAi, microRNA, and CeRNA; Karaca et al., 2014; Lujambio and Lowe, 2012; Salmena et al., 2011).
Our findings reveal that a specific set of fragments contain tumor-suppressive and metastasis-suppressive activity. We propose that these fragments are generated as a result of oncogenic stress as an internal mechanism for tumor suppression. We speculate that during breast cancer evolution, two mechanisms counter this small-RNA mediated tumor suppressive mechanism: the first being the evasion of the hypoxia evoked induction of tumor suppressive tRFs and the second being YBX1 up-regulation. Consistent with these findings, tRNA fragments were detected at significantly lower levels in metastatic breast cancer relative to non-metastatic cancers, while YBX1 is known to be up-regulated as a function of cancer progression (Lasham et al., 2012).
We find that the repressive effects of these endogenous tRNA fragments on the abundance and stability of oncogenic transcripts is moderate in scale. Nonetheless, our loss-of-function and gain-of-function studies involving these fragments reveal robust in vitro and in vivo effects resulting from their modulation in breast cancer. We believe that these effects result from the coordinated post-transcriptional control of a large set of YBX1-dependent oncogenes whose concomitant suppression results in robust phenotypic effects. These observations parallel those seen with microRNAs implicated in cancer progression—moderate post-transcriptional suppressive effects on groups of transcripts involved in common oncogenic processes (Lujambio and Lowe, 2012).
An association between YBX1 and tRNA halves has been previously described (Emara et al., 2010). Transfection of tRNA halves arising from tRNAAla and tRNACys was found to globally inhibit translation. These tRNA halves were found to repress translation by ~30% and these effects seemed to result from the disengagement of translational initiation factor EIF4G1. Additionally, it was found that the effect of these tRNA-halves on translational repression was YBX1-dependent. These tRNA-halves were found to interact with YBX1 and both fragments contained a terminal oligoguanine motif. While molecular mechanisms for the global inhibition of translation by these tRNA-halves and their in vivo roles have yet to be delineated, the authors proposed that these fragments suppress translation by interfering with EIF4G protein in a YBX1-dependent manner. Our findings described here reveal a distinct mechanism of action by a different class of tRFs. The tRFs we have implicated belong to a distinct class of fragments and mediate post-transcriptional destabilization of a specific set of transcripts by directly engaging YBX1 in a sequence-specific manner. Importantly, our observations regarding the down-regulation of elongation initiation factors at the transcript level is in agreement with the broad inhibition of translation reported to be induced by a broader class of tRNA fragments. Moreover, the different mechanisms by which distinct classes of tRFs regulate YBX1-dependent gene expression highlight the significance of this stress-activated regulator in mammalian gene regulation.
Taken together, our findings support a role for endogenous tRFs in destabilizing oncogenic transcripts through their direct binding to YBX1. Transfer RNA derived fragment binding of YBX1 leads to displacement of endogenous oncogenic transcripts from YBX1—resulting in their destabilization (Figure 7F). Based on this model, specific tRFs mediate a unique post-transcriptional gene expression regulatory program through their engagement of YBX1. It should be noted however that the regulatory interactions mediated by tRFs are unlikely to be limited to YBX1 and they likely serve as a component of a larger regulatory network consisting of various RNA-binding proteins and small non-coding RNAs. We should also point out the possible role of RNA modifications in the functionality of tRNA fragments. Given that extensive base modifications in tRNAs are crucial for their function, future studies should address the potential role of these modifications in tRNA fragments as well. Two lines of evidence in our data suggest a substantial role for these modifications in modulating the regulatory effects of endogenous tRFs. Firstly, while the induction of endogenous tRFs under hypoxia in MDA-parental cells was substantially more modest than exogenous transfection of synthetic tRF mimetics, the ensuing YBX1-dependent down-regulation of the tRF-YBX1 regulon was higher in magnitude for endogenous fragments relative to unmodified transfected mimetics. Secondly, consistent with the possible importance of RNA modifications in tRFs, transfection of antisense LNAs that inhibit endogenous fragments was more potent than synthetic mimetics in eliciting a regulatory response in the cells. This higher potency of endogenous tRFs relative to synthetic mimetics could be explained by the presence of RNA modifications that are likely to affect the structure, stability, and binding affinity of these fragments.
While we have shown that the fragments described here modulate specific phenotypes through transcript displacement from YBX1, they may also modulate the activity of additional trans factors. From a broader perspective, we speculate that fragments arising from other classes of non-coding RNAs, such as ribosomal and sno-RNAs, might mediate similar effects by displacing distinct RNA-binding proteins (or other non-coding RNAs) from their endogenous downstream targets.
EXPERIMENTAL PROCEDURES
Tissue Culture
HEK293T, MDA-MB-231, CN34 cells and their derived sub-lines, CN-LM1a and MDA-LM2, were cultured in DMEM-based media supplemented with 10% FBS, glutamine, pyruvate, penicillin, streptomycin and fungizone was used. RNAi and DNA transfections were performed using Lipofectamine 2000 (Invitrogen) and TransIT-293 (Mirus), respectively.
Exogenous tRF and antisense LNA transfection
tRF antisense LNA oligonucleotides (Exiqon) or synthetic tRF miemtics (IDT) were transfected using Lipofectamine 2000 in Reduced Serum Media (Life Technologies) for a final concentration of 50nM consisting of equal parts of each tRF decoy or anti-tRF LNA. After 6 hours of incubation, transfection media was replaced with fresh media. Cells were subjected to in vitro and in vivo studies 48 hours post transfection. The sequences for the short and long tRF mimetics are provided in Figure S2B.
Animal Studies
All mouse studies were conducted according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the Rockefeller University.
Supplementary Material
Acknowledgments
We are grateful to Saeed Tavazoie, Claudio Alarcon, and Alexander Nguyen for their thoughtful comments on this manuscript. We thank Nora Pencheva for insightful discussions on YBX1. We thank C. Zhao, W. Zhang, C. Lai and S. Dewell of the Rockefeller Genomics Resource Center for assistance with next-generation RNA-sequencing and microarray profiling. We also thank H. Molina at Rockefeller Proteomics Resource Center. H.G. was previously supported by an Anderson Cancer Center Fellowship and is currently the recipient of a Ruth L. Kirschstein National Research Service Award from the NIH (T32CA009673-36A1). S.F.T. is a Department of Defense Era of Hope Scholar and a Department of Defense Breast Cancer Collaborative Scholars and Innovators Award recipient.
Footnotes
Information supplementing this article consists of seven figures and extended experimental procedures.
AUTHOR CONTRIBUTIONS
S.F.T. conceived the project and supervised all research. S.F.T., H.G., X.L., H.C.B.N, L.F., and S.Z. wrote the manuscript. H.G., X.L., H.C.B.N, S.Z. and L.F. designed, performed and analyzed the experiments.
ACCESSION NUMBERS
The data for high-throughput sequencing and microarray profiling experiments are deposited at GEO under the accession number GSE63605.
References
- Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue. Cancer research. 1977;37:3362–3366. [PubMed] [Google Scholar]
- Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nature reviews Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. Rna. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all Genomes and data types. Molecular cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived Stress-induced RNAs Promote Stress-induced Stress Granule Assembly. Journal of Biological Chemistry. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA biology. 2013;10 doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr-Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-Induced tRNA Fragments Inhibit Translation Initiation. Molecular cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurchott K, Kuban RJ, Krech T, Bluthgen N, Stein U, Walther W, Friese C, Kielbasa SM, Ungethum U, Lund P, et al. Identification of Y-Box Binding Protein 1 As a Core Regulator of MEK/ERK Pathway-Dependent Gene Signatures in Colorectal Cancer Cells. Plos Genet. 2010;6 doi: 10.1371/journal.pgen.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, et al. Human CLP1 Mutations Alter tRNA Biogenesis, Affecting Both Peripheral and Central Nervous System Function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lasham A, Samuel W, Cao H, Patel R, Mehta R, Stern JL, Reid G, Woolley AG, Miller LD, Black MA, et al. YB-1, the E2F pathway, and regulation of tumor cell growth. Journal of the National Cancer Institute. 2012;104:133–146. doi: 10.1093/jnci/djr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & Development. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Bay BH. Significance of the Y-box proteins in human cancers. Journal of molecular and genetic medicine : an international journal of biomedical research. 2005;1:11–17. doi: 10.4172/1747-0862.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer MW. Targeting hypoxia brings breath of fresh air to cancer therapy. Nature medicine. 2012;18:636–637. doi: 10.1038/nm0512-636b. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annual review of cell and developmental biology. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- Shen C, Kaelin WG., Jr The VHL/HIF axis in clear cell renal carcinoma. Seminars in cancer biology. 2013;23:18–25. doi: 10.1016/j.semcancer.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer. Cancer. 1979;44:2120–2123. doi: 10.1002/1097-0142(197912)44:6<2120::aid-cncr2820440623>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Fotovati A, Sasaguri T, Shibahara K, Shimada T, Fukuda T, Nakamura T, Izumi H, Tsuzuki T, Kuwano M, et al. YB-1 is important for an early stage embryonic development - Neural tube formation and cell proliferation. Journal of Biological Chemistry. 2006;281:40440–40449. doi: 10.1074/jbc.M605948200. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nature reviews Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- Wu Y, Yamada S, Izumi H, Li Z, Shimajiri S, Wang KY, Liu YP, Kohno K, Sasaguri Y. Strong YB-1 expression is associated with liver metastasis progression and predicts shorter disease-free survival in advanced gastric cancer. J Surg Oncol. 2012;105:724–730. doi: 10.1002/jso.23030. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nature Biotechnology. 2011;29:607–U686. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.