Abstract
Batrachochytrium dendrobatidis (Bd), a chytrid fungus, has increasingly been implicated as a major factor in the worldwide decline of amphibian populations. The fungus causes chytridiomycosis in susceptible species leading to massive die-offs of adult amphibians. Although Bd infects the keratinized mouthparts of tadpoles and negatively affects foraging behavior, these infections are non-lethal. An important morphogen controlling amphibian metamorphosis is thyroid hormone (T3). Tadpoles may be infected with Bd and the fungus may be exposed to T3 during metamorphosis. We hypothesize that exposure of Bd to T3 may induce the expression of factors associated with host colonization and pathogenicity. We utilized a proteomics approach to better understand the dynamics of the Bd-T3 interaction. Using liquid chromatography-mass spectrometry (LC-MS), we generated a data set of a large number of cytoplasmic and membrane proteins following exposure of Bd to T3. From these data, we identified a total of 263 proteins whose expression was significantly changed following T3 exposure. We provide evidence for expression of an array of proteins that may play key roles in both genomic and non-genomic actions of T3 in Bd. Additionally, our proteomics study shows an increase in several proteins including proteases and a class of uncommon crinkler and crinkler-like effector proteins suggesting their importance in Bd pathogenicity as well as those involved in metabolism and energy transfer, protein fate, transport and stress responses. This approach provides insights into the mechanistic basis of the Bd-amphibian interaction following T3 exposure.
Introduction
Batrachochytrium dendrobatidis (Bd), a chytrid fungus, has been implicated in widespread amphibian decline [1–4]. The fungus infects the keratin skin layer of metamorphosed amphibians causing the diseased animals to experience thickening of the epidermal layer and eventually sloughing of the skin [5]. The life cycle of Bd consists of substrate-independent motile zoospores and substrate-dependent sporangia [3]. However, little is known about the early events during the fungal-amphibian interaction leading to death of these animals.
The amphibian life cycle primarily consists of tadpole and adult animals, and the transition of tadpole to adult is known as metamorphosis [6]. Thyroid hormone plays an integral role in the metamorphosis of these animals. A strong association of metamorphosis with the rise in circulating plasma concentrations of thyroid hormone has been observed [7]. At the climax stage of metamorphosis, a surge of thyroid hormone occurs, however the level of the hormone is reduced at the end of this stage. Studies have shown that pre-metamorphic tadpoles that are lacking thyroid hormones had the ability to sense exogenous hormone. When pre-metamorphic tadpoles were exposed to thyroid hormone, they were capable of precocious metamorphosis [8], suggesting the importance of the thyroid hormone in the development of amphibians.
Although Bd infection negatively affects the feeding behavior of young tadpoles, the infection is not lethal [5], [9]. Various studies have reported massive die-offs of amphibians that have recently undergone metamorphosis due to Bd infections [5,10–12]. Additionally, Bd-infected amphibians exhibit a weakened immune response to the pathogen. Recent studies demonstrate that Bd may cause severe immune suppression in susceptible hosts [13–15]. We recently reported that a Bd subtilisin-like serine protease degrades frog anti-microbial peptides [16]. This may lead to increased susceptibility of the host to the fungus.
To better understand the early events during Bd-frog interactions, particularly, how Bd responds to host-derived thyroid hormone, a study of the proteome was carried out following in vitro exposure of Bd to thyroid hormone (T3). Proteomic analyses have been used to understand pathogenicity in fungi such as Botrytis cinerea [17] and Sclerotinia sclerotiorum [18]. Additionally, in the human pathogen, Candida albicans, a proteomics study demonstrated the importance of proteins involved in hyphal-yeast transitions [19]. Using proteomic and phenotypic profiling of Bd, a previous study showed that genotype is associated with virulence [20]. Here we present a global proteomics approach to document protein expression changes in Bd exposed to T3 and discuss its significance in understanding this fungal-amphibian interaction.
Methods
Cultivation of fungus
The VM1 isolate of Batrachochytrium dendrobatidis isolated from a diseased Western chorus frog (Pseudacris triseriata), was provided by Louise Rollins-Smith (Vanderbilt Univ). Bd was maintained on TGhL (1.6% tryptone, 0.2% gelatin hydrolysate, 0.4% lactose, 0.8% agar) plates and all experiments were conducted by inoculation in H-broth (1% tryptone, 0.32% glucose). The fungal culture was routinely maintained at room temperature (21°C) and incubated in the dark.
Exposure to thyroid hormone
Of the two forms of thyroid hormone, 3, 5, 3'- triiodothyronine (T3) and 3, 5, 3', 5'- tetraiodothyronine (thyroxine, T4), T3 is the biologically more active form in vertebrates [21] and was used in this study. T3 was used at the concentration at which it occurs physiologically in tadpoles [22]. A 10 mM stock solution of T3 was prepared by dissolving 3, 3′, 5-triiodo-L-thyronine sodium salt (Sigma-Aldrich) in methanol and stored at -20°C. The T3 stock was further diluted in water, which was then used for the experiments. The fungal cultures were exposed to a final concentration of 50 nM T3 for 3 hours at room temperature. Methanol was used as the solvent control. Three biological replicates of T3-treated and control cultures were used for the study.
Protein extraction
To pinpoint when Bd proteins are induced during exposure to T3 and at what level they are expressed, a time-course experiment was carried out. Following 7 days of growth in H-broth, Bd cells were exposed to a final concentration of 50 nM T3 for 1, 3, 6 or 12 hrs. After careful analysis, the 3-hr time point was chosen for this proteomics study. Proteins from Bd cells were extracted using a glass bead method. The cells were harvested at 10,000 rpm at 4°C for 10 minutes. The supernatant was discarded and the pellet was resuspended in breaking buffer (50 mM sodium monophosphate (pH 7.4), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM ethylenediaminetetraacetic acid (EDTA) and 5% glycerol). An equal volume of acid-washed glass beads (0.5 mm diameter) was added to the cell suspension. The pellet was then subjected to eight alternate cycles of vortexing and incubation on ice for 30 seconds. The mixture was then centrifuged at 14,000 x g for 10 minutes at 4°C. The resulting supernatant was transferred to a microfuge tube and subjected to 14,000 x g for 10 minutes to further remove any glass beads from the solution. The concentration of total soluble protein was estimated by the Bradford assay [23] and further used for LC-MS study.
One-dimensional (1-D) gel electrophoresis
Protein samples (100 μg) of 3 biological replicates from T3-treated and control were separated on a one-dimensional gel using SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The samples were mixed with protein loading buffer (2% SDS, 1% Tris-HCl, 10% v/v glycerol, 0.01% bromophenol blue, 5% beta-mercapto-ethanol, pH 6.8). The mixture was boiled for 5 minutes, centrifuged briefly and then loaded into a 12% resolving gel (BioRad). Following electrophoresis the gel was stained with 0.1% Coomassie Blue stain for 4 hours followed by destaining (10% methanol, 10% acetic acid, 2% glycerol and 78% water) overnight. The proteins were visualized using Alpha Innotech imaging system (Cell Science).
In-gel digestion and extraction of peptides
Each sample lane of the gel in which the proteins were separated was cut into 8 equal slices and each slice was kept in a 0.5 ml microfuge tube. In-gel digestion on each slice was carried out as described previously [24]. Briefly, these slices were washed in milli Q water for 5 minutes at 37°C at 600 rpm and then washed twice with acetonitrile (ACN)/100 mM NH4HCO3 (50/50) for 10 minutes to destain the gels. Subsequently, proteins in the gel pieces were subjected to reduction in 50 μl 10 mM dithiothreitol at 56°C at 600 rpm for one hour. These slices were then washed in water, alkylated in 50 μl of iodoacetamide solution (55 mM in 40 mM NH4HCO3), and incubated in the dark for 30 minutes. To remove any remaining dye, the gel slices were washed alternately in water and 50% ACN. After completely removing the dye, the gel pieces were incubated for one minute in 100 μl of 100% ACN and air-dried. The samples were digested using 30 μL of sequencing grade trypsin (Promega) solution (12.5 ng/μL in 25 mM NH4HCO3) and incubated overnight at 37°C. Peptide extraction was performed twice using 50% ACN containing 0.1% formic acid solution. The extracted peptide solutions were pooled and dried using a speed vacuum centrifuge and the peptides resuspended in 20 μl of 0.1% formic acid.
Nano LC-MS/MS
To analyze the peptides extracted from in-gel digestion, nano-flow liquid chromatography tandem mass spectrometry (nano-LC-MS/MS) was used on an LTQ-XL ion trap mass spectrometer (Thermo, CA, USA) at the Center for Biotechnology and Genomics, Texas Tech University. Chromatographic separation of the peptides was carried out using a Dionex nano-HPLC (Ultimate 3000) with a trapping column (C18, 3 μm, 100 Å, 75 μm × 2 cm) followed by a reverse phase column (C18, 2 μm, 100 Å, 75 μm × 15 cm, nanoViper). Peptides were first injected onto the trapping column, which was equilibrated with 1% ACN, 0.1% formic acid in mass spectrometric grade water. These peptides were trapped for 10 minutes using the loading pump at a flow rate of 5 μl/min. The trapped peptides were then loaded on the reverse-phase analytical column, and bound peptides were eluted using solvents A (2% ACN, 0.1% formic acid in water) and B (98% ACN, 2% water, 0.1% formic acid) at 300 nl/min. The gradient was maintained constant for the first 10 minutes at 4% solvent B followed by a gradual increase up to 30% solvent B in 20 minutes. Solvent B was further increased to 60% in 40 minutes followed by a rapid increase up to 90% over 5 minutes. The eluted peptides were directed into the nanospray ionization source of the LTQ-XL with a capillary voltage of ∼2 kV. The collected spectra were scanned over the mass range of 300–2000 atomic mass units. Data dependent scan settings were defined to choose the 6 most intense ions with dynamic exclusion list size of 100, exclusion duration of 30 seconds, repeat count of 2, and repeat duration of 15 seconds. To generate MS/MS spectra, collision-induced dissociation of the peptide ions at normalized collision energy of 35% was utilized.
Database search
To identify the proteins using the spectra acquired from the LTQ-XL mass spectrometer, Proteome Discoverer software (version 1.3, Thermo Scientific) was employed. For this purpose, SEQUEST cluster was used as the search engine (Thermo Electron Corp., San Jose CA) against a Bd database (www.broadinstitute.org). The following criteria were used by the search engine: precursor ion mass tolerance was set at 2.5 Da, and fragment ion mass tolerance at 0.8 Da. Additional parameters included fully tryptic enzyme specificity, two missed cleavages, and mass range 350–5000 Da and CID as the collision method. For all searches, carbamidomethylation of cysteines and oxidation of methionine were set as dynamic modifications. The false discovery rate (FDR), percentage of false positive identifications among all the tentative peptide identifications, was set at 1% using a decoy databases created from a reversed target database.
Quantitative proteomic analysis
ProteoIQ software (ProteoIQ 2.70, Premier Biosoft) was used for label-free comparative relative protein quantification using spectral counts. For protein quantification purposes, the following stringent filter criteria were employed: minimum number of spectra = 5, minimum percentage of replicates = 60 (2 out of 3 replicates) and maximum protein false discovery rate (FDR %) = 1. Additionally, probability filters including minimum peptide probability = 0.99 and minimum protein group probability = 0.95 were applied. Using the reversed target database as decoy, the protein FDR was calculated as protein FDR = (number of reverse proteins identified)/ (total protein identifications) x100. To calculate the peptide FDR, the formula, peptide FDR = 2*(number of reverse peptide identifications)/ (total peptide identifications) x100 was used. Protein abundance data were determined using the method described previously [25]. To calculate protein abundance data, normalized spectral abundance factors (NSAF) were employed. In this method, for each protein, k, in sample i, the number of spectral counts (SpC, the total number of MS/MS spectra) identifying the protein was divided by the apparent length of the protein. To calculate the protein length, molecular weight of the protein was divided by the molecular weight of an average amino acid. The NSAFi values for the sample i was determined as SpCk/Lengthk values normalized to the total by dividing by the sum (SpCk/Lengthk). The values in T3 normalized spectral count (T3 N-SC) Log2 relative expression are presented here. To calculate the absolute fold change, the conversion was applied as 2^ (T3 N-SC).
Statistical analysis
To determine the protein expression changes between T3 treatment and methanol control, a student t-test was performed. The t-test was performed using log transformed NSAF data, and those P-values less than 0.05 were measured to be statistically significant. These protein sets were further subjected to functional annotation.
Annotation and mapping
To gain evidence on functional annotation of identified Bd proteins, the nucleotide sequences of all proteins (Broad Institute) were matched to the NCBI non redundant (NR) protein database and the gene ontology (GO) database by Blast2GO software (Version 1.6) [26]. Using Mercator-Mapman annotation tool, metabolic pathways and other cellular processes in the fungus were mapped (S2 Table and S3 Table). This tool was used to map the differentially expressed proteins into various metabolic pathways [27].
Results
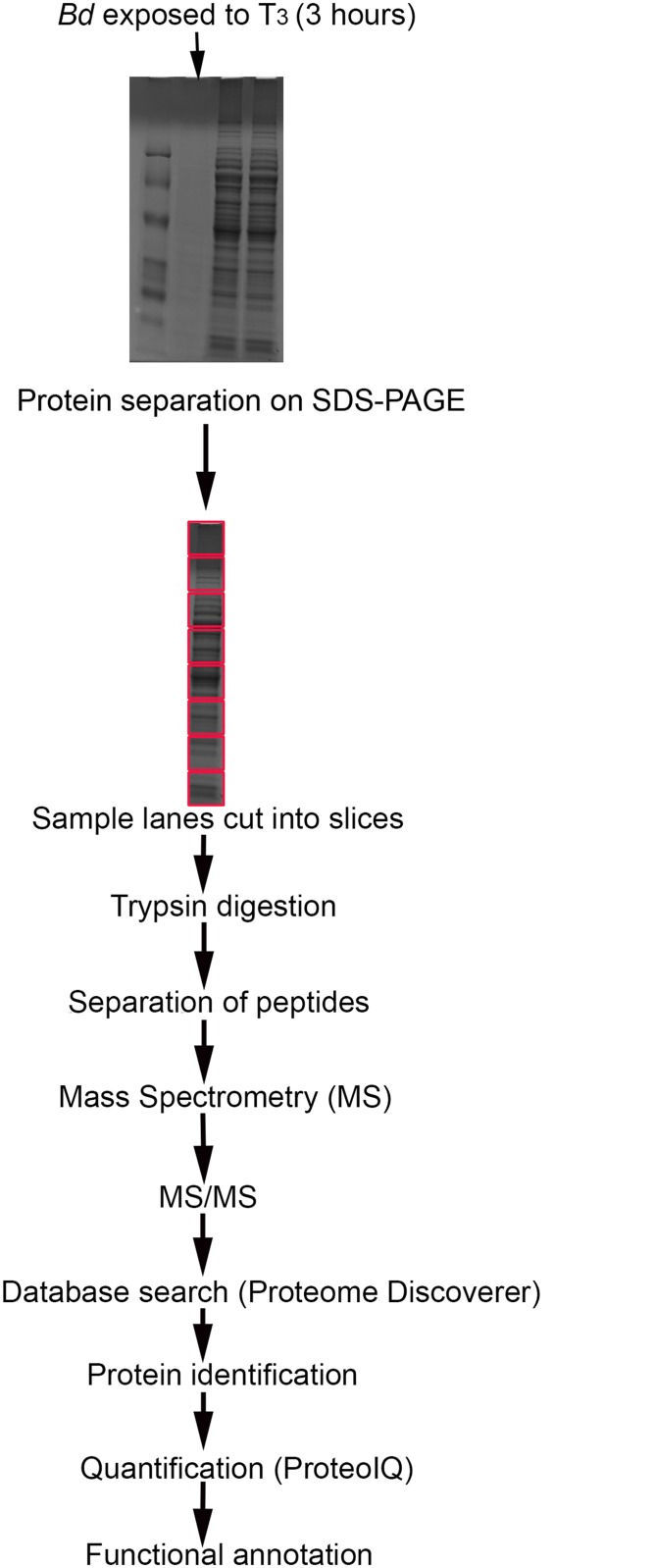
In an effort to understand the early events during Bd-T3 interaction, we used a quantitative proteomic approach and assessed the protein expression profiles of Bd exposed to T3. The strategy followed for protein preparation and profiling using LC-MS approach is shown in Fig 1. Using this approach, we identified and quantitatively analyzed the changes in relative abundance of Bd proteins (S1 Table). Among the identified proteins, we found expression differences of 263 proteins that were statistically significant (P value < 0.05). Of these, the expression of 104 proteins was found to have increased by more than 2-fold (Table 1) and 42 proteins were uniquely present (Table 2) in Bd cells following exposure to T3. We observed a more than 2-fold decrease of 29 proteins (Table 3) while 26 proteins were found to be undetectable (Table 4) in the T3-treated samples. Here we list those Bd proteins that were, (1) uniquely present or (2) that showed greater than a 2-fold change in abundance (either increase or decrease) following its exposure to T3.
Fig 1. Schematic illustration for the proteomics study to profile total proteins in Batrachochytrium dendrobatidis following exposure to T3.
Red boxes show regions where each sample lane of the gel was cut into slices for peptide extraction.
Table 1. List of proteins that showed more than 2-fold increase in Bd exposed to T3.
| Gene accession number a | Name/predicted name of protein | T3 (N-SC)Log2 relative expression |
|---|---|---|
| BDET_08575 | prolyl endopeptidase | 3.21 |
| BDET_05763 | ACTN1 protein | 3.1 |
| BDET_04253 | hypothetical protein similar to glutamate carboxypeptidase | 2.93 |
| BDET_01134 | hypothetical protein | 2.91 |
| BDET_07031 | conserved hypothetical protein | 2.9 |
| BDET_02940 | rab GDP dissociation inhibitor beta | 2.85 |
| BDET_03111 | conserved hypothetical protein | 2.61 |
| BDET_02571 | bifunctional purine biosynthesis protein ADE17 | 2.46 |
| BDET_04126 | bifunctional purine biosynthesis protein ADE17 | 2.46 |
| BDET_04144 | phosphoglucomutase | 2.46 |
| BDET_07811 | hypothetical protein | 2.41 |
| BDET_05116 | crinkler family protein | 2.4 |
| BDET_05132 | crinkler family protein | 2.4 |
| BDET_08307 | conserved hypothetical protein | 2.36 |
| BDET_08030 | glucose-6-phosphate isomerase | 2.31 |
| BDET_06818 | conserved hypothetical protein | 2.26 |
| BDET_08009 | conserved hypothetical protein | 2.24 |
| BDET_04007 | hypothetical protein similar to hydrolase | 2.2 |
| BDET_01275 | electron transfer flavoprotein subunit beta | 2.19 |
| BDET_03579 | conserved hypothetical protein | 2.18 |
| BDET_05121 | crinkler family protein | 2.16 |
| BDET_01745 | aminoacylase-1 | 2.15 |
| BDET_00627 | conserved hypothetical protein | 2.14 |
| BDET_07364 | conserved hypothetical protein | 2.14 |
| BDET_03035 | proliferating cell nuclear antigen | 2.08 |
| BDET_08526 | conserved hypothetical protein | 2.08 |
| BDET_05736 | triosephosphate isomerase | 2.07 |
| BDET_03419 | hypothetical protein similar to AhpC/TSA family protein | 2.07 |
| BDET_07373 | branched-chain-amino-acid aminotransferase, mitochondrial precursor | 2.03 |
| BDET_06577 | protein disulfide-isomerase erp38 precursor | 2.01 |
| BDET_07554 | monothiol glutaredoxin-4 | 2 |
| BDET_02020 | long-chain acyl CoA ligase | 1.95 |
| BDET_02521 | crinkler family missing secretion signal peptide | 1.95 |
| BDET_04065 | conserved hypothetical protein | 1.88 |
| BDET_03580 | ribosomal L-30 | 1.8 |
| BDET_02674 | crinkler family missing secretion signal peptide | 1.79 |
| BDET_02498 | enolase | 1.79 |
| BDET_01981 | conserved hypothetical protein | 1.73 |
| BDET_03190 | deoxyuridine 5'-triphosphate nucleotidohydrolase | 1.72 |
| BDET_05854 | isocitrate dehydrogenase subunit 1, mitochondrial precursor | 1.72 |
| BDET_05255 | conserved hypothetical protein | 1.72 |
| BDET_06670 | protein phosphatase PP2A regulatory subunit A | 1.71 |
| BDET_02031 | hypothetical protein similar to 5'-methylthioadenosine phosphorylase | 1.71 |
| BDET_01967 | conserved hypothetical protein | 1.68 |
| BDET_00885 | conserved hypothetical protein | 1.67 |
| BDET_00733 | 4-hydroxyphenylpyruvate dioxygenase | 1.64 |
| BDET_06414 | multidrug/metal resistance protein, ABC transporter | 1.6 |
| BDET_04177 | phosphoribosylaminoimidazole carboxylase | 1.59 |
| BDET_00864 | conserved hypothetical protein | 1.59 |
| BDET_01181 | hypothetical protein similar to aminopeptidase | 1.58 |
| BDET_05762 | ATP citrate synthase | 1.58 |
| BDET_06464 | conserved hypothetical protein | 1.54 |
| BDET_04424 | hypothetical protein similar to dipeptidyl peptidase III | 1.53 |
| BDET_00939 | conserved hypothetical protein | 1.53 |
| BDET_00944 | conserved hypothetical protein | 1.53 |
| BDET_06915 | conserved hypothetical protein | 1.51 |
| BDET_02099 | pre-mRNA-processing-splicing factor 8 | 1.51 |
| BDET_02010 | GPI anchor protein | 1.5 |
| BDET_06692 | NIF 3 like protein | 1.5 |
| BDET_06549 | tryptophanyl-tRNA synthetase | 1.47 |
| BDET_05202 | hypothetical protein | 1.46 |
| BDET_00751 | cell division control protein 3 | 1.44 |
| BDET_04460 | ubiquitin-activating enzyme E1 1 | 1.42 |
| BDET_08521 | peptidyl-prolyl cis-trans isomerase B precursor | 1.42 |
| BDET_05202 | dihydroorotase | 1.4 |
| BDET_06814 | cytochrome c peroxidase, mitochondrial precursor | 1.39 |
| BDET_02514 | predicted protein | 1.39 |
| BDET_02516 | predicted protein | 1.39 |
| BDET_03024 | protein phosphatase PP2A regulatory subunit B | 1.36 |
| BDET_01885 | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 1.35 |
| BDET_04151 | glucosamine-fructose-6-phosphate aminotransferase | 1.35 |
| BDET_06546 | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 1.33 |
| BDET_03560 | acetyl-CoA acetyltransferase | 1.33 |
| BDET_08315 | biofilm development protein YmgB/AriR | 1.3 |
| BDET_07061 | phosphoglucomutase | 1.3 |
| BDET_08261 | phosphoribosylglycinamide formyltransferase | 1.3 |
| BDET_01150 | conserved hypothetical protein | 1.29 |
| BDET_01159 | conserved hypothetical protein | 1.29 |
| BDET_03350 | organic hydroperoxide resistance protein | 1.29 |
| BDET_02250 | conserved hypothetical protein | 1.26 |
| BDET_04753 | acyl CoA oxidase | 1.25 |
| BDET_03317 | nucleoside diphosphate kinase 1 | 1.25 |
| BDET_03057 | isocitrate lyase | 1.22 |
| BDET_05399 | protein phosphatase regulatory subunit SDS22 | 1.2 |
| BDET_04703 | calmodulin | 1.2 |
| BDET_06960 | conserved hypothetical protein | 1.2 |
| BDET_02981 | hypothetical protein | 1.2 |
| BDET_00091 | conserved hypothetical protein | 1.18 |
| BDET_07377 | ATP synthase F1 gamma | 1.1 |
| BDET_05222 | heat shock protein 90 | 1.1 |
| BDET_07409 | nucleoside-triphosphatase/ nucleotide binding protein | 1.08 |
| BDET_03133 | aspartyl-tRNA synthetase | 1.08 |
| BDET_01550 | conserved hypothetical protein | 1.07 |
| BDET_05272 | inorganic pyrophosphatase | 1.07 |
| BDET_03724 | conserved hypothetical protein | 1.07 |
| BDET_00078 | tubulin alpha-6 chain | 1.06 |
Statistically significant expression at P <0.05.
The values are given as T3 normalized spectral count (N-SC) Log2 relative expression.
To calculate the absolute fold change, the conversion is applied as 2^ (T3 N-SC).
aAs given according to the www.broadinstitute.org.
Table 2. List of uniquely present proteins in Bd exposed to T3.
| aGene accession number | Name/predicted name of protein |
|---|---|
| BDET_00207 | conserved hypothetical protein |
| BDET_00091 | conserved hypothetical protein |
| BDET_00370 | hypothetical protein |
| BDET_00520 | hypothetical protein |
| BDET_00578 | cell division protein kinase 2 |
| BDET_00617 | diphosphomevalonate decarboxylase |
| BDET_01201 | succinate dehydrogenase iron-sulfur protein |
| BDET_01306 | hypothetical protein similar to glutathione transferase zeta 1 |
| BDET_01576 | cytochrome c1, mitochondrial precursor |
| BDET_01620 | hypothetical protein similar to alcohol dehydrogenase superfamily |
| BDET_01772 | leiomodin-1 |
| BDET_02033 | ran-specific gtpase-activating protein 1 |
| BDET_02201 | peptidyl-prolyl cis-trans isomerase pin1 |
| BDET_02231 | hypothetical protein similar to oxidoreductase |
| BDET_02645 | conserved hypothetical protein |
| BDET_02736 | leukotriene A-4 hydrolase |
| BDET_02989 | predicted protein |
| BDET_03180 | hypothetical protein |
| BDET_03405 | conserved hypothetical protein |
| BDET_03430 | ribonuclease p protein subunit p30 |
| BDET_03541 | conserved hypothetical protein |
| BDET_03674 | acyl-binding protein |
| BDET_03762 | valyl-tRNA synthetase |
| BDET_04022 | conserved hypothetical protein |
| BDET_04238 | phosphate induced protein |
| BDET_04676 | xanthine dehydrogenase/oxidase |
| BDET_05174 | novel protein containing Initiation factor 2 subunit family domain |
| BDET_05329 | conserved hypothetical protein |
| BDET_05479 | mt-GrpE |
| BDET_06040 | conserved hypothetical protein |
| BDET_06411 | phosphate induced protein |
| BDET_06369 | predicted protein |
| BDET_06502 | crinkler family protein |
| BDET_06621 | acetoacetyl-CoA synthetase |
| BDET_06834 | alanine aminotransferase 2 |
| BDET_07584 | Na/H exchanger |
| BDET_03192 | NADH dehydrogenase ubiquinone alpha |
| BDET_07703 | cytochrome oxidase |
| BDET_07599 | conserved hypothetical protein |
| BDET_08178 | cysteinyl-tRNA synthetase |
| BDET_08421 | 10 kDa heat shock protein, mitochondrial |
| BDET_08720 | crinkler family protein |
aAs given according to the www.broadinstitute.org.
Table 3. List of proteins that showed 2-fold decrease in Bd exposed to T3.
| aGene accession number | Name/predicted name of protein | T3 (N-SC)Log2 relative expression |
|---|---|---|
| BDET_00978 | long-chain acyl-CoA synthetase 7 | -2.66 |
| BDET_06102 | Rpb (RNA-polymerase) | -2.54 |
| BDET_07602 | rRNA 2'-O-methyltransferase fibrillarin | -2.24 |
| BDET_03865 | importin beta, transportin | -2.09 |
| BDET_00308 | conserved hypothetical protein | -1.97 |
| BDET_06700 | dual specificity protein kinase FUZ7 | -1.95 |
| BDET_01880 | calreticulin | -1.84 |
| BDET_00694 | conserved hypothetical protein | -1.83 |
| BDET_03549 | CTP synthase | -1.79 |
| BDET_06886 | vacuolar sorting protein | -1.59 |
| BDET_08238 | shwachman-bodian-diamond syndrome protein | -1.59 |
| BDET_03021 | conserved hypothetical protein | -1.50 |
| BDET_07660 | 26S proteasome subunit p45 | -1.39 |
| BDET_05219 | pyrD | -1.29 |
| BDET_04706 | proline dehydrogenase family protein | -1.29 |
| BDET_03460 | 60S ribosomal protein L2 | -1.24 |
| BDET_04352 | glutaminyl-tRNA synthetase | -1.23 |
| BDET_04880 | pullulanase | -1.20 |
| BDET_06734 | conserved hypothetical protein | -1.20 |
| BDET_08256 | G-protein beta | -1.16 |
| BDET_06205 | NAD-dependent epimerase/dehydratase family protein | -1.13 |
| BDET_00517 | conserved hypothetical protein | -1.16 |
| BDET_07664 | conserved hypothetical protein | -1.00 |
| BDET_02151 | conserved hypothetical protein | -1.00 |
| BDET_00579 | conserved hypothetical protein | -1.00 |
| BDET_01641 | conserved hypothetical protein | -1.00 |
| BDET_08055 | 26S protease regulatory subunit 8 | -1.00 |
| BDET_03232 | conserved hypothetical protein | -1.00 |
| BDET_00482 | polyadenylate-binding protein 1 | -1.00 |
Statistically significant expression at P <0.05.
The values are given as T3 normalized spectral count (N-SC) Log2 relative expression.
To calculate the absolute fold change, the conversion is applied as 2^ (T3 N-SC).
aAs given according to the www.broadinstitute.org.
Table 4. List of uniquely present proteins in Bd exposed to methanol control.
| aGene accession number | Name/ predicted name of protein |
|---|---|
| BDET_05257 | oligopeptide transporter opt family |
| BDET_08499 | extracellular elastinolytic metalloproteinase |
| BDET_04809 | pre-mRNA splicing factor |
| BDET_04728 | mrna turnover protein 4 homolog |
| BDET_03527 | hypothetical protein |
| BDET_02501 | hypothetical protein |
| BDET_06402 | hypothetical protein |
| BDET_02113 | conserved hypothetical protein |
| BDET_04582 | multiple coagulation factor deficiency isoform |
| BDET_00183 | TatD Dnase family Scn1 |
| BDET_00782 | retinoid dehydrogenase |
| BDET_01194 | zuotin |
| BDET_01995 | RNA polymerase |
| BDET_02205 | cyclophilin |
| BDET_02773 | deoxyhypusine hydroxylase |
| BDET_03497 | transporter SEC 24 |
| BDET_03858 | expressed protein |
| BDET_05019 | chromosome segregation protein SudA |
| BDET_05542 | septin |
| BDET_05655 | ARM repeat containing protein |
| BDET_06581 | SCF ubiquitin ligase complex subunit CulA |
| BDET_06582 | cullin-1 |
| BDET_06665 | transporter SEC 24 |
| BDET_07485 | actin-binding protein |
| BDET_08328 | U2 small nuclear ribonucleoprotein A |
| BDET_01389 | conserved hypothetical protein |
aAs given according to the www.broadinstitute.org.
To gain an integrated perspective of the Bd biological processes influenced following exposure to T3, the complete dataset of identified proteins was classified into Mapman functional categories. Mapman-Mercator analysis has been widely used in analyzing gene expression in higher plants [21] and green algae, Chlamydomonas reinhardtii [28]. Mapman pathway analysis in our study showed that Bd proteins that were identified are involved in metabolism and energy acquisition, cytoskeleton signaling, and ubiquitin and autophagy-dependent degradation. Such changes in protein expression levels belonging to diverse functional groups point to a broad fungal response when exposed to T3.
Discussion
Our experimental data support the hypothesis that exposure of Bd to T3 results in protein expression changes associated with various cellular roles in the fungus. This study shows the expression of a large number of proteins in Bd, which have been described in both genomic and non-genomic actions of T3 in vertebrates. Additionally, this study sheds light on the possible mechanism of how T3 may act in Bd.
Genomic action of T3 in Bd
In vertebrates, in addition to nuclear receptor regulators that control transcriptional activity in a hormone dependent manner, the action of thyroid hormone receptor can be controlled by other proteins [29]. Our study provides evidence for certain cellular proteins in Bd that may control the transcriptional activity of thyroid hormone receptor (TR). These TR-interacting proteins primarily consist of transcription modulators and cytoskeletal element regulators such as cyclin dependent kinases, 26 S proteasome subunit p45, ubiquitin-proteasome pathway components including SCF ubiquitin ligase complex subunit culA and cullin, cytoskeletal elements such as actin binding protein, tubulin and alpha-actinin. Additionally, a TR interacting protein- 13 (TRIP-13) (BDET_00690) has been identified in Bd. However, expression of this protein did not change following exposure to T3.
Non-genomic action of T3 in Bd
Membrane receptors play an important role in non-genomic actions of T3 [29]. These receptors could be proteins such as integrin or the G-protein coupled receptor (GPCR). Additionally, it has been shown that rapid response to T3 is moderated by the mitogen activated protein kinase (MAPK) signaling pathway [30]. Mitogen-activated protein kinases (MAPKs) belong to a family of serine-threonine protein kinases. These kinases are known to play important roles in the signal transduction of a large number of external stimuli and in development and differentiation processes [31]. Like other eukaryotic cells, fungi including yeast and human pathogens such as Candida albicans respond to several extracellular stimuli using highly conserved MAPK signaling cascades. Although not statistically significant, we observed a decrease in abundance of the protein Fuz7, a homolog of the yeast protein Ste7 which is found to be involved in the pheromone response pathway in the fungus [32]. In the case of the plant pathogen, Ustilago maydis, Fuz7 codes for a MEK/MAPKK homolog. Additionally, this protein has been implicated in a pathway that responds to plant signals [33]. The Fuz7 (BDET_06700) protein is known to be a key protein in the MAPK signaling pathway, which is mediated by G-proteins. It has been shown that GPCRs are involved in this signaling pathway, which results in a corresponding decrease in the receptor and subsequent hormone response. Thus reduced abundance of the Fuz7 protein in this study may be due to desensitization of a GPCR.
Action of T3 on plasma-membrane transport function
Na+/H+ transporter
The sodium-proton exchanger protein, which is known to play an important role in non-genomic action of T3 in vertebrates, was among uniquely present proteins in Bd exposed to T3. A previous study showed that a sodium-proton antiporter in humans was regulated by T3 [34]. Sodium-proton exchangers pump Na+ ions either out of cells or into cells in exchange for H+ [35]. A recent study in two pathogenic species of Candida elucidated the role of a membrane Na+/H+ exchanger in salt tolerance [36]. Since Bd lives in fresh water environments, the role of Na+/H+ exchanger in this fungus is not very clear. However, we have observed that a subtilisin-like protease (SSP), one of the possible pathogenicity factors in Bd, requires sodium ions for its optimal activity [12]. Taken together, our observation suggests that a Na+/H+ transporter (BDET_07584) may be important for Bd SSP function possibly during pathogenicity.
Ca2+ATPase, Calmodulin and Calreticulin
In eukaryotes, Ca2+ pumps and transporters have been shown to be important in maintaining the resting cytosolic free Ca2+ concentration [Ca2+] at very low levels. It has been shown that certain hormones and environmental signals cause a surge in Ca2+ concentration which further triggers several downstream signaling proteins such as protein kinase C (PKC) and Ca2+ /calmodulin (CAM)- binding kinases [37], [38]. Several studies have demonstrated the role of Ca2+ - modulated signal cascades in biological processes including, circadian rhythms, differentiation, cell cycle and stress responses in eukaryotic cells [39–42].
Our study showed changes in the abundance level of diverse calcium signaling proteins including Ca2+ATPase (BDET_ 06015), calmodulin (BDET_04703) and calreticulin (BDET_01880). We detected a 2-fold increase in abundance of Ca2+ATPase, a calcium pump- associated enzyme (Table 1). In eukaryotes, this protein is involved in maintaining intracellular calcium concentration at extremely low levels [43] and the activity of Ca2+ ATPase is moderated by thyroid hormone [44]. We also detected a 2-fold increase in calmodulin, a cytoplasmic intracellular Ca2+ binding protein, which is involved in the modulation of plasma membrane Ca2+ ATPase activity. Calmodulin is also important for the ability of thyroid hormone to enhance the activity of this ATPase [45].
As a Ca2+ receptor, calmodulin modulates several intracellular proteins in diverse signaling pathways [46]. Calmodulin has been reported in zoospores of the aquatic chytrid, Blastocladiella emersonii [47]. The Ca2+-calmodulin complex has been shown to play a key role during growth and sporulation in this fungus [48]. The high abundance of calmodulin following T3 treatment in this work suggests its role in maintaining low Ca+ levels in Bd. Additionally, in fungi; calmodulin has been implicated in stress responses, virulence, and morphogenesis. For instance, in fungal plant pathogens Magnaporthe grisea and Colletotrichium trifolli, calmodulin has been shown to be essential for the growth of specialized infection structures known as appressoria [49], [50]. Thus the increased abundance of calmodulin following T3 exposure supports a role of this hormone in pathogenicity of the fungus.
Calreticulin is an essential Ca2+ binding protein in the endoplasmic reticulum [51] and is involved in two major functions in the ER lumen including chaperoning and regulation of Ca2+ homoeostasis [52], [53]. Additionally, this protein has been involved in several cellular processes such as cell adhesion, migration and signal transduction [54], [55]. Our observation of a decrease in abundance of calreticulin in response to T3 implies that the hormone may cause a significantly reduced Ca2+ storage capacity in the ER in the fungus. Our observation also suggests a role for Ca2+ in physiological changes in Bd including reduced cell motility which may trigger chitin synthase actively favoring the transition from a wall less, motile zoospore to a walled sporangium.
Action of T3 on mitochondria
In the current study, we detected a higher abundance of enzymes involved in mitochondrial oxidative phosphorylation in the T3 treatment. For example, proteins that are implicated in oxidative phosphorylation including cytochrome c oxidase (BDET_07703) and NADH dehydrogenase subunit (BDET_03192) are uniquely present following fungal exposure to T3. We also observed an increase of 2-fold in the F1-ATPase subunit (BDET__07377) in Bd exposed to T3. This observation is consistent with that of mammalian systems where T3 stimulates mitochondrial respiration resulting in increased ATP production.
Fatty acid metabolism
Among the proteins that showed a dramatic increase in expression (3.9-fold change) was the long chain acyl CoA ligase (BDET_02020). We also found an increased (2.4-fold change) abundance of acyl CoA oxidase (BDET_04753). The role of fatty acid β-oxidation in fungal pathogenesis is highly suggested by the abundance level of lipid metabolism-associated genes when it infects its host. Successful fungal pathogens, such as C. albicans utilize proteins for respiratory catabolism such as long chain acyl CoA ligase and acyl CoA oxidase for efficient nutrient acquisition and energy production in vivo [56]. Abundance of these enzymes in Bd exposed to T3 suggests that the hormone treatment may influence fungal acquisition of nutrients or use fatty acids as an energy source.
Central carbohydrate metabolic processes
Exposure to T3 also stimulated expression of proteins involved in carbohydrate metabolism. A protein similar to phosphoglucomutase (BDET_07061) showed a 2.5-fold increase compared to the control. This protein plays a role in the reversible interconversion of glucose-1-phosphate to glucose-6-phosphate. In Aspergillus nidulans, phosphoglucomutase has been implicated in asexual development of the fungus [57]. Our observation of an increased expression of phosphoglucomutase suggests that in response to T3, this protein may aid the fungus in its developmental processes such as the transition from zoospores to walled reproductive thalli.
Pathogenicity-associated proteins
Exposure to T3 also showed an increase of seven proteins (BDET_06502, BDET_08720, BDET_05121, BDET_05116, BDET_05132, BDET_02674, and BDET_02521) that are highly similar to that of the crinkler family proteins or crinkler-like effectors (CRN) when compared to the control. Microbial effectors are implicated in the destruction of host defenses and thus are capable of changing the host cell metabolism [58]. These proteins were assumed to be present only in Oomycetes, a significant group of pathogens in fish and plants. In general, CRN effectors consist of a signal peptide, a translocator domain that helps the entry of CRN proteins into host cells, and a C-terminal domain that is found to be involved in host protein interaction. These effectors are denoted as crinkler proteins due to their involvement in leaf crinkling and cell death. Interestingly, Bd causes similar effects on amphibian skin [59]. The possibility that Bd might have acquired these genes from Oomycetes through horizontal gene transfer has been discussed [60]. Our observation suggests that crinkler proteins may play a key role in Bd virulence and may be regulated by T3. A recent genome-wide study in Bd showed an increased expression of Bd crinkler and CRN genes in the frog skin. Interestingly, this study also demonstrated a dramatic expression of CRN genes in the Bd zoospore as compared to reproductive thalli [61]. Our observation in the current study suggests a comprehensive analysis of CRN genes in Bd.
In addition to crinkler proteins, we identified proteins that may be implicated in proteolysis and thus fungal pathogenesis. For example, prolyl endopeptidase (peptidase S9) (BDET_08575) showed a 9.3-fold increase, aminopeptidase P (peptidase M24) (BDET_01181) had a 3-fold increase, and the dipeptidyl peptidase III (peptidase M49) (BDET_04424) had a 2.9-fold increase. Additionally, leukotriene A-4 hydrolase (M1) (BDET_02736), was uniquely present in Bd exposed to T3. Interestingly, these proteases were also identified in a recent study wherein anuran skin was exposed to supernatant of Bd zoospores which caused disruption of intercellular junctions in frog skin [62]. Like other successful pathogens, Bd may use proteases to modulate immune responses in amphibians. Genomic analysis of Bd has indicated an intense expansion of the fungalysin metallopeptidase and serine peptidase gene families in the fungus [63]. Serine proteases are important virulence factors in parasites and pathogenic microbes [64]. Bd-infected Silurana tropicalis showed an increase in expression of serine proteases [15] and a recent in vitro study showed a Bd subtilisin-like serine protease impairs frog antimicrobial peptides [16]. These observations suggest that these proteases are important in impairing vertebrate innate immunity. In this study, we identified a dramatic increase in both metalloprotease and serine peptidase proteins. Thus our study provides further evidence for proteins that may inhibit innate immune responses in the susceptible host species [13], [15], [65]. Previous studies with Bd-infected frogs showed a clear down-regulation of adaptive immunity in the animals [13], [15]. A recent study demonstrated Bd is capable of impairing host lymphocyte responses and inducing apoptosis [14]. In another study Ellison et al showed that Bd-infected frogs had lower expression of many genes involved in adaptive immunity including B-cell related genes, T-cell markers and many T-cell receptor components [66].
Mechanisms by which fungi cause immunosuppression involve the manipulation of host-immune receptors [67] and the discharge of toxins [68]. Bd may release toxic components [69] that impede immune responses in vitro and these components may be produced from the Bd cell wall [14]. In this study, we used whole-cell extracts and thus the proteins identified include mixed life-stages of Bd consisting of both cell walled sporangia and wall-less zoospores. A further proteomics study that separates developmental stages of Bd (zoospores and sporangia) may provide clearer evidence for gene expression changes associated with immunosuppression caused by Bd.
Heat shock proteins (Hsps)
In the current study, we observed a significant up-regulation of Bd proteins that are implicated during survival of stress conditions. These proteins include heat shock proteins Hsp10 (BDET_08421) Hsp 90 (BDET_05222) and Hsp101 (BDET_04470). Hsps are cellular chaperones that play a key role in protein folding homeostasis, revival and degradation of impaired proteins [70], and in thermo tolerance [71] [72]. Hsp90 is a vital and remarkably conserved chaperone in all eukaryotes and controls the role and stability of a variety of proteins including nuclear steroid receptors and protein kinases [73]. A recent study has shown that Hsp90 plays a key role in regulating morphogenetic switch from yeast to hypha and is temperature dependent [74]. In the case of aquatic chytrid fungus, B. emersonii, the expression of genes encoding cytoplasmic and endoplasmic reticulum Hsp90 proteins (Hsp90A and Hsp90 B) has been documented in response to thermal stress [75]. In addition to response to heat shock at 38°C, the levels of Hsp90A have been increased at physiological temperature (27°C) both during fungal germination and sporulation. The increase in expression of Hsp90 in our study suggests that Hsp90 might be involved in the morphological switch from zoospore to thallus. A recent study reported the up-regulation of several Bd Hsps including Hsp90 in frogs infected with Bd implying a role of Hsps under stress conditions [11].
Hsp10, a fungal equivalent of E. coli GroES has been known to function in association with Hsp60 to form a chaperonin that favors mitochondrial folding [76]. Additionally, Hsp10 has been shown to have a role in defending cells from various stresses due to infection and inflammation [77],[78]. In pathogenic fungi such as C. albicans, the role of Hsp10 remains unknown. The significance of the increase of Hsp10 in our study is not clear; perhaps this protein, like in other eukaryotes, might be involved in mitochondrial protein folding. Hsp101, a member of Hsp100/ClpB family of chaperones is vital for resistance to high temperature stress. The cytosolic Hsp101 in yeast and plants contributes to thermo tolerance [79–81]. It is important to note that Clp proteins are essential to cells that are not only exposed to heat stress, but also other forms of environmental stresses. It has been shown that Hsp101 in yeast, in addition to heat stress confers resistance to chemicals including ethanol and arsenite [82]. The increased abundance of Hsp101 in our study suggests that exposure to T3 may create chemical stress condition and that Hsp101 may allow Bd to tolerate the host immune response.
Mapman-Mercator pathway analysis
Mapman analysis revealed a high abundance of enzymes that are involved in amino acid biosynthesis, suggesting that when Bd is exposed to T3, the fungal cells may require increased protein synthesis for thallus formation. The transition from zoospore to thallus in Bd is important for fungal colonization of the host. However, this transition is an energy-expensive process, which requires a large supply of resources for repeated mitotic cell divisions and cell wall synthesis. Amino acids could conceivably be used as an energy source. During the infection process, Bd must obtain nutrients from its hosts and the fungus may primarily depend on amino acids generated by proteases. Recent studies have identified and characterized Bd proteases that may be involved during infection [16], [83]. In addition to serving as a source of carbon and protein building blocks, amino acids play other roles in fungi. For example, in the case of Allomyces, certain amino acids play an essential role in making sugars including fructose and mannose and thus may serve as sources for both carbon and nitrogen [84]. In mammalian systems, iodothyronines such as T3 are considered as a special class of amino acids from two tyrosine residues and that amino acid transporters play a key role in thyroid hormone uptake into several tissues [85], [86]. Our results indicate that increased amino acid synthesis in the fungus may further aid in thyroid hormone transport.
Analysis of central metabolic pathways showed a variation in protein abundance changes for those proteins involved in the tricarboxylic acid (TCA) cycle. For example, succinate dehydrogenase (SDH) was found in relative abundance, while malate dehydrogenase (MDH) was less abundant in Bd following its exposure to T3. The enzymes implicated in the glycolytic pathway were found to be up-regulated. It was also observed that the enzymes involved in the glyoxylate cycle including malate synthase (MS) and isocitrate lyase (ICL) were up-regulated in Bd following its exposure to T3. The glyoxylate cycle is involved in lipid metabolism and in fungi is a peroxisome-associated process. In Bd, as zoospores are released from the zoosporangium, they contain several lipid globules that are partly surrounded by the microbody, a key characteristic of Bd [3]. The lipids present in the zoospores might be broken down using the beta-oxidation pathway present on the peroxisome. These degradation products might be further processed through the glyoxylate cycle to maintain growth of new sporangia. A similar observation was also made in a nematode-trapping fungus, Arthrobotrys oligospora [87]. Using proteomics and genomics approaches, Yang and coworkers demonstrated the up-regulation of these enzymes in A. oligospora in response to nematode extract [87]. A previous study has demonstrated that glyoxylate cycle is essential for fungal virulence [88]. Additionally, our current study is consistent with that of C. albicans in that while the fungus infects macrophages, these key enzymes were found to be up- regulated [89].
In summary, the proteomics data described in our study help to understand cellular responses of Bd following its exposure to a host-derived morphogen. This response relies on the expression of a particular group of proteins or genes allowing the fungus to adapt to its environment. Our results implicate proteins involved in metabolism and energy, protein fate, transport, stress responses and pathogenesis in Bd that respond to exposure to T3. These observations provide a basis for further experimental exploration.
Supporting Information
(XLS)
(XLS)
The identified proteins were classified according to the previously defined Mapman functional categories by using the online Mapman-Mercator annotation tool.
(XLSX)
Acknowledgments
We thank the Department of Biological Sciences for support during this work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1. Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, Speare R. Emerging infectious diseases and amphibian population declines. Emerg Infect Dis. 1999;5: 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci. U S A. 2006;103: 3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Longcore JE, Pessier AP, Nichols DK. Batrachochytrium dendrobatidis gen et sp nov, a chytrid pathogenic to amphibians. Mycologia. 1999;91: 219–227. [Google Scholar]
- 4. Wake DB, Vredenburg VT. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Nat Acad Sci. U S A. 2008;105: 11466–11473. 10.1073/pnas.0801921105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci. U S A 1998;95: 9031–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi YB. Amphibian metamorphosis: From morphology to molecular biology. Ist ed New York: Wiley-Liss; 1999. [Google Scholar]
- 7. White B, Nicoll C. Hormonal control of amphibian metamorphosis In: Gilbert L, Frieden E, editors. Metamorphosis: A Problem in Developmental Biology. New York: Plenum Press; 1981. pp. 363–396. [Google Scholar]
- 8. Tata JR. Amphibian metamorphosis: An exquisite model for hormonal regulation of postembryonic development in vertebrates. Dev Growth Differ. 1996;38: 223–31. [DOI] [PubMed] [Google Scholar]
- 9. Venesky MD, Wassersug RJ, Parris MJ. Fungal pathogen changes the feeding kinematics of larval anurans. J Parasitol. 2010;96: 552–557. 10.1645/GE-2353.1 [DOI] [PubMed] [Google Scholar]
- 10. Bosch J, Martinez-Solano I, Garcia-Paris M. Evidence of a chytrid fungus infection involved of the decline of the common midwife toad (Alytes obstetricans) in protected areas in central Spain. Biol Conserv. 2001;97: 331–337. [Google Scholar]
- 11. Rachowicz LJ, Knapp RA, Morgan JAT, Stice MJ, Vredenburg VT, Parker JM, et al. Emerging infectious disease as a proximate cause of amphibian mass mortality. Ecology. 2006;87: 1671–1683. [DOI] [PubMed] [Google Scholar]
- 12. Walker SF, Bosch J, Gomez V, Garner TWJ, Cunningham AA, Schmeller DS, et al. Factors driving pathogenicity vs. prevalence of amphibian panzootic chytridiomycosis in Iberia. Ecol Letters. 2010;13: 372–382. 10.1111/j.1461-0248.2009.01434.x [DOI] [PubMed] [Google Scholar]
- 13. Rosenblum EB, Poorten TJ, Settles M, Murdoch GK, Robert J, Maddox N, et al. Genome-wide transcriptional response of Silurana (Xenopus) tropicalis to infection with the deadly chytrid fungus. PLoS ONE. 2009;4: e6494 10.1371/journal.pone.0006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fites JS, Ramsey JP, Holden WM, Collier SP, Sutherland DM, Reinert LK, et al. The invasive chytrid fungus of amphibians paralyzes lymphocyte responses. Science. 2013; 342: 366–369. 10.1126/science.1243316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ribas L, Li M-S, Doddington BJ, Robert J, Seidel JA, Kroll JS, et al. Expression profiling the temperature-dependent amphibian response to infection by Batrachochytrium dendrobatidis . PLoS ONE. 2009;4: e8408 10.1371/journal.pone.0008408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thekkiniath JC, Zabet-Moghaddam M, San Francisco SK, San Francisco MJ. A novel subtilisin-like serine protease of Batrachochytrium dendrobatidis is induced by thyroid hormone and degrades antimicrobial peptides. Fungal Biol. 2013;117: 451–461. 10.1016/j.funbio.2013.05.002 [DOI] [PubMed] [Google Scholar]
- 17. Fernández-Acero F, Jorge I, Calvo E, Vallejo I, Carbú M, Camafeita E, et al. Proteomic analysis of phytopathogenic fungus Botrytis cinerea as a potential tool for identifying pathogenicity factors, therapeutic targets and for basic research. Arch Microbiol. 2007; 187: 207–215. [DOI] [PubMed] [Google Scholar]
- 18. Yajima W, Kav NNV. The proteome of the phytopathogenic fungus Sclerotinia sclerotiorum . Proteomics. 2006;6: 5995–6007. [DOI] [PubMed] [Google Scholar]
- 19. Ebanks RO, Chisholm K, McKinnon S, Whiteway M, Pinto DM. Proteomic analysis of Candida albicans yeast and hyphal cell wall and associated proteins. Proteomics. 2006;6: 2147–2156. [DOI] [PubMed] [Google Scholar]
- 20. Fisher MC, Bosch J, Yin Z, Stead DA, Walker J, Selway L, et al. Proteomic and phenotypic profiling of the amphibian pathogen Batrachochytrium dendrobatidis shows that genotype is linked to virulence. Mol Ecol. 2009;18: 415–429. 10.1111/j.1365-294X.2008.04041.x [DOI] [PubMed] [Google Scholar]
- 21. Ingbar SH, Braverman LE. Active form of the thyroid hormone. Ann Rev Medicine. 1975;26: 443–449. [DOI] [PubMed] [Google Scholar]
- 22. Krain LP, Denver RJ. Developmental expression and hormonal regulation of glucocorticoid and thyroid hormone receptors during metamorphosis in Xenopus laevis . J Endocrinol. 2004;181: 91–104. [DOI] [PubMed] [Google Scholar]
- 23. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72: 248–254. [DOI] [PubMed] [Google Scholar]
- 24. Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2007;1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- 25. Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae . J Proteome Res. 2006;5: 2339–2347. [DOI] [PubMed] [Google Scholar]
- 26. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37: 914–939. [DOI] [PubMed] [Google Scholar]
- 28. May P, Wienkoop S, Kempa S, Usadel B, Christian N, Rupprecht J, et al. Metabolomics- and proteomics-assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii . Genetics. 2008;179: 157–166. 10.1534/genetics.108.088336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocrine Rev. 2010;31: 139–170. 10.1210/er.2009-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. D’Arezzo S, Incerpi S, Davis FB, Acconcia F, Marino M, Farias RN, et al. Rapid nongenomic effects of 3,5,3′-Triiodo-l-thyronine on the intracellular pH of L-6 myoblasts are mediated by intracellular calcium mobilization and kinase pathways. Endocrinology. 2004;145: 5694–5703. [DOI] [PubMed] [Google Scholar]
- 31. Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18: 128–131. [DOI] [PubMed] [Google Scholar]
- 32. Bardwell L, Cook JG, Inouye CJ, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae . Dev Biol. 1994;166: 363–379. [DOI] [PubMed] [Google Scholar]
- 33. Banuett F, Herskowitz I. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 1994;8: 1367–1378. [DOI] [PubMed] [Google Scholar]
- 34. Incerpi S, Luly P, De Vito P, Farias RN. Short-term effects of thyroid hormones on the Na/H antiport in L-6 myoblasts: High molecular specificity for 3, 3′, 5-triiodo-l-thyronine. Endocrinology. 1999;140: 683–689. [DOI] [PubMed] [Google Scholar]
- 35. Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta 2000;1465: 140–151. [DOI] [PubMed] [Google Scholar]
- 36. Krauke Y, Sychrova H. Functional comparison of plasma-membrane Na+/H+ antiporters from two pathogenic Candida species . BMC Microbiol. 2008;8: 1–9. 10.1186/1471-2180-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gehring CA, Irving HR, Parish RW. Effects of auxin and abscisic acid on cytosolic calcium and pH in plant cells. Proc Natl Acad Sci USA. 1990;87: 9645–9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bush DS. Regulation of cytosolic calcium in plants. Plant Physiol. 1993;103: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002;14: S401–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4: 517–529. [DOI] [PubMed] [Google Scholar]
- 41. Kraus PR, Heitman J. Coping with stress: calmodulin and calcineurin in model and pathogenic fungi. Biochem Biophy Res Comm. 2003;311: 1151–1157. [DOI] [PubMed] [Google Scholar]
- 42. Hepler PK. Calcium: a central regulator of plant growth and development. Plant Cell Online. 2005;17: 2142–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vincenzi FF, Larsen FL. The plasma membrane calcium pump: regulation by a soluble Ca2+ binding protein. Fed Proc. 1980;39: 2427–2431. [PubMed] [Google Scholar]
- 44. Galo MG, Unates LE, Farias RN. Effect of membrane fatty acid composition on the action of thyroid hormones on (Ca2+ + Mg2+)-adenosine triphosphatase from rat erythrocyte. J Biol Chem. 1981;256: 7113–7114. [PubMed] [Google Scholar]
- 45. Davis FB, Davis PJ, Blas SD. Role of calmodulin in thyroid hormone stimulation in vitro of human erythrocyte Ca2+-ATPase activity. J Clin Invest. 1983;71: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Williams RJP. Calcium and calmodulin. Cell Calcium. 1992;13: 355–362. [DOI] [PubMed] [Google Scholar]
- 47. Gomes SL, Mennucci L, Maia JC. A calcium-dependent protein activator of mammalian cyclic nucleotide phosphodiesterase from Blastocladiella emersonii . FEBS Lett 1979;99: 39–42. [DOI] [PubMed] [Google Scholar]
- 48. Vieira ALG, Gomes SL. Global gene expression analysis during sporulation of the aquatic fungus Blastocladiella emersonii . Eukaryotic Cell. 2010;9: 415–423. 10.1128/EC.00312-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee SC, Lee YH. Calcium/calmodulin-dependent signaling for appressorium formation in the plant pathogenic fungus Magnaporthe grisea . Mol cells. 1998;8: 698–704. [PubMed] [Google Scholar]
- 50. Warwar V, Dickman MB. Effects of calcium and calmodulin on spore germination and appressorium development in Colletotrichum trifolii . App Environ Microbiol. 1996;62: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Michalak M, Milner RE, Burns K, Opas M. Calreticulin. Biochem J 1992;285: 681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bleackley RC, Atkinson EA, Burns K, Michalak M. Calreticulin: a granule-protein by default or design? Pathways for Cytolysis. Curr Top Microbiol Immunol. 1995;198: 145–159. [DOI] [PubMed] [Google Scholar]
- 53. Krause KH, Michalak M. Calreticulin. Cell. 1997;88: 439–443. [DOI] [PubMed] [Google Scholar]
- 54. Nanney LB, Woodrell CD, Greives MR, Cardwell NL, Pollins AC, Bancroft TA, et al. Calreticulin enhances porcine wound repair by diverse biological effects. Am J Pathol. 2008;173: 610–630. 10.2353/ajpath.2008.071027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Coppolino M, Dedhar S. Ligand-specific, transient interaction between integrins and calreticulin during cell adhesion to extracellular matrix proteins is dependent upon phosphorylation/dephosphorylation events. Biochem J. 1999;340: 41–50. [PMC free article] [PubMed] [Google Scholar]
- 56. Gonzalez-Fernandez R, Jorrin-Novo JV. Contribution of proteomics to the study of plant pathogenic fungi. J Proteome Res. 2011;11: 3–16. 10.1021/pr200873p [DOI] [PubMed] [Google Scholar]
- 57. Hoffmann B, LaPaglia SK, Kübler E, Andermann M, Eckert SE, Braus GH. Developmental and metabolic regulation of the phosphoglucomutase-encoding gene, pgmB, of Aspergillus nidulans . Mol Gen Genet. 2000;262: 1001–1011. [DOI] [PubMed] [Google Scholar]
- 58. Kamoun S. A Catalogue of the effector secretome of plant pathogenic Oomycetes. Ann Rev Phytopathol. 2006;44: 41–60. [DOI] [PubMed] [Google Scholar]
- 59. Berger L, Speare R, Skerratt LF. Distribution of Batrachochytrium dendrobatidis and pathology in the skin of green tree frogs Litoria caerulea with severe chytridiomycosis. Dis Aquat Organ. 2005;68: 65–70. [DOI] [PubMed] [Google Scholar]
- 60. Sun G, Yang Z, Kosch T, Summers K, Huang J. Evidence for acquisition of virulence effectors in pathogenic chytrids. BMC Evol Biol. 2011;11: 195 10.1186/1471-2148-11-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosenblum EB, Poorten TJ, Joneson S, Settles M. Substrate-specific gene expression in Batrachochytrium dendrobatidis, the chytrid pathogen of amphibians. PLoS ONE. 2012; 7: e49924 10.1371/journal.pone.0049924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brutyn M, D’Herde K, Dhaenens M, Rooij PV, Verbrugghe E, Hyatt AD, et al. Batrachochytrium dendrobatidis zoospore secretions rapidly disturb intercellular junctions in frog skin. Fungal Genet Biol. 2012;49: 830–837. 10.1016/j.fgb.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 63. Rosenblum EB, Stajich JE, Maddox N, Eisen MB. Global gene expression profiles for life stages of the deadly amphibian pathogen Batrachochytrium dendrobatidis . Proc Natl Acad Sci. U S A 2008;105: 17034–17039. 10.1073/pnas.0804173105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McKerrow JH, Caffrey C, Kelly B, Loke PN, Sajid M. Proteases in parasitic diseases. Ann Rev Pathol: Mech Dis. 2006;1: 497–536. [DOI] [PubMed] [Google Scholar]
- 65. Rollins-Smith LA, Ramsey JP, Pask JD, Reinert LK, Woodhams DC. Amphibian immune defenses against chytridiomycosis: Impacts of changing environments. Int Comp Biol. 2011;51: 552–562. 10.1093/icb/icr095 [DOI] [PubMed] [Google Scholar]
- 66. Ellison AR, Savage AE, DiRenzo GV, Langhammer P, Lips KR, Zamudio KR. Fighting a losing battle: Vigorous immune response countered by pathogen suppression of host defenses in the chytridiomycosis-susceptible frog Atelopus zeteki . G3: Genes|Genomes|Genetics. 2014;4: 1275–1289 10.1534/g3.114.010744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brandhorst TT, Wüthrich M, Finkel-Jimenez B, Warner T, Klein BS. Exploiting Type 3 complement receptor for TNF-α suppression, immune evasion, and progressive pulmonary fungal infection. J Immunol. 2004;173: 7444–7453. [DOI] [PubMed] [Google Scholar]
- 68. Genevieve S. Bondy JJP Immunomodulation by fungal toxins. J Toxicol Environ Health, Part B. 2000;3: 109–143. [DOI] [PubMed] [Google Scholar]
- 69. McMahon TA, Brannelly LA, Chatfield MWH, Johnson PTJ, Joseph MB, McKenzie VJ et al. Chytrid fungus Batrachochytrium dendrobatidis has non-amphibian hosts and releases chemicals that cause pathology in the absence of infection. Proc Natl Acad Sci USA. 2013;110: 210–215. 10.1073/pnas.1200592110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Ann Rev Gen. 1993;27: 437–496. [DOI] [PubMed] [Google Scholar]
- 71. Sung D-Y, Kaplan F, Lee K-J, Guy CL. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003;8: 179–187. [DOI] [PubMed] [Google Scholar]
- 72. Montero-Barrientos M, Cardoza R, Gutiérrez S, Monte E, Hermosa R. The heterologous overexpression of hsp23, a small heat-shock protein gene from Trichoderma virens, confers thermotolerance to T. harzianum . Curr Gen. 2007;52: 45–53. [DOI] [PubMed] [Google Scholar]
- 73. Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11: 515–528. 10.1038/nrm2918 [DOI] [PubMed] [Google Scholar]
- 74. Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 2009;19: 621–629. 10.1016/j.cub.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pugliese L, Georg RC, Fietto LG, Gomes SL. Expression of genes encoding cytosolic and endoplasmic reticulum HSP90 proteins in the aquatic fungus Blastocladiella emersonii . Gene. 2008;411: 59–68. 10.1016/j.gene.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 76. Jia H, Halilou AI, Hu L, Cai W, Liu J, Huang B. Heat shock protein 10 (Hsp10) in immune-related diseases: one coin, two sides. Int J Biochem Mol Biol. 2011;2: 47–57. [PMC free article] [PubMed] [Google Scholar]
- 77. Johnson BJ, Le TT, Dobbin CA, Banovic T, Howard CB, Flores Fde M, et al. Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. J Biol Chem. 2005;280: 4037–4047. [DOI] [PubMed] [Google Scholar]
- 78. Corrao S, Campanella C, Anzalone R, Farina F, Zummo G, Conway de Macario E, et al. Human Hsp10 and Early Pregnancy Factor (EPF) and their relationship and involvement in cancer and immunity: current knowledge and perspectives. Life Sci. 2010;86: 145–152. 10.1016/j.lfs.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 79. Hong SW, Vierling E. Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J. 2001;27: 25–35. [DOI] [PubMed] [Google Scholar]
- 80. Keeler SJ, Boettger CM, Haynes JG, Kuches KA, Johnson MM, Thureen DL, et al. Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol. 2000;123: 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leonhardt S, Fearson K, Danese P, Mason T. HSP78 encodes a yeast mitochondrial heat shock protein in the Clp family of ATP-dependent proteases. Mol Cell Biol. 1993;13: 6304–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sanchez Y, Taulien J, Borkovich K, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO. 1992;11: 2357–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moss AS, Carty N, San Francisco MJ. Identification and partial characterization of an elastolytic protease in the amphibian pathogen Batrachochytrium dendrobatidis . Dis Aquat Organ. 2010;92: 149–158. 10.3354/dao02223 [DOI] [PubMed] [Google Scholar]
- 84. Machlis L. Effect of certain organic acids on the utilization of mannose and fructose by the filamentous watermold, Allomyces macrogynus . J Bacteriol. 1957;73: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lakshmanan M, Goncalves E, Lessly G, Foti D, Robbins J. The transport of thyroxine into mouse neuroblastoma cells, NB41A3: The effect of L-system amino acids. Endocrinology. 1990;126: 3245–3250. [DOI] [PubMed] [Google Scholar]
- 86. Friesema ECH, Jansen J, Milici C, Visser TJ. Thyroid hormone transporters. Vitam Horm. 2005;70: 137–167. [DOI] [PubMed] [Google Scholar]
- 87. Yang J, Wang L, Ji X, Feng Y, Li X, Zou C, et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011;7: e1002179 10.1371/journal.ppat.1002179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412: 83–86. [DOI] [PubMed] [Google Scholar]
- 89. Fernández-Arenas E, Cabezón V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, et al. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics. 2007;6: 460–478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
The identified proteins were classified according to the previously defined Mapman functional categories by using the online Mapman-Mercator annotation tool.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.