Abstract
The thyroid hormone receptor (TR) undergoes nucleocytoplasmic shuttling and regulates target genes involved in metabolism and development. Previously, we showed that TR follows a CRM1/calreticulin-mediated nuclear export pathway. However, two lines of evidence suggest TR also follows another pathway: export is only partially blocked by leptomycin B (LMB), a CRM1-specific inhibitor; and we identified nuclear export signals in TR that are LMB-resistant. To determine whether other exportins are involved in TR shuttling, we used RNA interference and fluorescence recovery after photobleaching shuttling assays in transfected cells. Knockdown of exportins 4, 5, and 7 altered TR shuttling dynamics, and when exportins 5 and 7 were overexpressed, TR distribution shifted towards the cytosol. To further assess the effects of exportin overexpression, we examined transactivation of a TR-responsive reporter gene. Our data indicate that multiple exportins influence TR localization, highlighting a fine balance of nuclear import, retention, and export that modulates TR function.
Keywords: thyroid hormone receptor, thyroid hormone, nuclear export, exportin
1. Introduction
Thyroid hormone (triiodothyronine, T3)1 is important in regulating genes responsible for metabolism, growth, and development. Encoded by two different genes, thyroid hormone receptors TRα1 and TRβ1 respond to T3 levels by activating or repressing target gene expression. Although primarily found in the nucleus at steady-state, TRα1 and TRβ1 can rapidly shuttle between the nucleus and cytoplasm (Baumann et al., 2001; Bunn et al., 2001; Grespin et al., 2008). Nuclear import and export of proteins occurs through the nuclear pore complexes, mediated by members of the karyopherin β family called importins and exportins, respectively (Kimura and Imamoto, 2014; Pemberton and Paschal, 2005). By coupling mutagenesis and localization studies, nuclear localization signal (NLS) and nuclear export signal (NES) motifs that interact with the transport machinery have been found in conserved domains of members of the nuclear receptor superfamily (Black et al., 2004; Holaska et al., 2002; Kanno et al., 2007; Kanno et al., 2005; Liu and DeFranco, 2000; Lombardi et al., 2008; Mavinakere et al., 2012; Nguyen et al., 2009; Pemberton and Paschal, 2005; Picard and Yamamoto, 1987; Saporita et al., 2003; Sorokin et al., 2007; Umemoto and Fujiki, 2012). This transport process provides a central regulatory point for coordinating cell signaling and gene expression.
From our prior studies, an increasingly complex picture has emerged of the intricate molecular mechanisms that regulate trafficking and function of TRα1 and TRβ1 (collectively referred to as TR hereinafter for simplicity). Previously, we showed that TR can exit the nucleus by a pathway mediated by the export factor CRM1 (chromosome region maintenance 1), also known as exportin 1, in cooperation with calreticulin (Grespin et al., 2008); however, the exact interaction and mechanism remain unclear. Two main lines of evidence suggested that TR might also follow a CRM1/calreticulin-independent nuclear export pathway. First, under conditions in which TR still shuttles, shuttling of p53 and the oncoprotein v-ErbA is completely blocked in the presence of the CRM1-specific inhibitor leptomycin B (LMB) (Bunn et al., 2001; DeLong et al., 2004). Also, during fluorescence recovery after photobleaching (FRAP) experiments, when one nucleus in a multinucleate HeLa cell was photobleached, recovery of fluorescence in the bleached nucleus in the presence of LMB was reduced by only 60%, relative to recovery in the absence of LMB (Grespin et al., 2008). Second, no CRM1-dependent NES in TR has yet been characterized (Mavinakere et al., 2012). Indeed, our studies showed that TR interacts directly with calreticulin but complex formation with CRM1 was not detectable in pull-down assays (Grespin et al., 2008). In an effort to identify and clarify the mode of TR nuclear export, we previously carried out a comprehensive analysis of TR to screen for NES motifs. We identified a region spanning helices 3 and 6 of the ligand-binding domain, that either houses two monopartite NESs or a single, bipartite NES, and we fully characterized another NES in helix 12 of the ligand-binding domain. These NES motifs were able to export a nucleus-localized fusion protein to the cytosol (Mavinakere et al., 2012). Intriguingly, these NES motifs were shown to be insensitive to LMB. In the presence of LMB, they were still able to direct the fusion protein to the cytosol, suggesting that they follow a CRM1-independent export pathway (Mavinakere et al., 2012).
In the present study, we sought to determine whether other exportins are involved in this alternative nuclear export pathway of TR, and to determine their relative contributions to TR export overall. To this end, we coupled RNA interference (RNAi) with FRAP experiments in live HeLa cells. Shuttling dynamics of TR were assessed upon knockdown of transportins 1 and 2, and exportins, 4, 5, 6, and 7. Additionally, we used overexpression assays and T3-responsive reporter gene assays to further assess the role of a panel of exportins in modulating TR function. Exportin-t and exportin 2 (CAS/CSE1L) were not included in our study, since they are specific for tRNA export (Arts et al., 1998) and importin α export (Kutay et al., 1997), respectively. We also excluded RanBP17; although a close homolog of exportin 7 (RanBP16), it is primarily expressed in the testis and pancreas, and a direct role for this protein in nuclear export has not been demonstrated (Koch et al., 2000; Kutay et al., 2000). Taken together, results presented here provide evidence that multiple exportins influence cellular localization of TRα1 and TRβ1 and, in this way, may play a role in modulating T3-mediated gene expression.
2. Methods
2.1. Plasmids
The plasmid pGFP-TRα1 encodes a functional green fluorescent protein (GFP)-tagged rat TRα1 fusion protein (Bunn et al., 2001). pGFP-TRβ1 encodes a functional GFP-tagged human TRβ1 (Mavinakere et al., 2012). Pre-designed SureSilencing short hairpin RNA (shRNA) plasmid sets consisting of four different shRNA expression plasmids for each target mRNA were purchased from SABioscience (Frederick, MD) for human transportin 1 (TNPO1), transportin 2 (TNPO2), exportin 4 (XPO4), exportin 5 (XPO5), exportin 6 (XPO6), exportin 7 (XPO7), and a scrambled sequence negative control. pk-Myc-exportin 5, pCMV-Myc, and pCMV-HA were obtained from Addgene (Cambridge, MA), Clontech Laboratories, Inc. (Mountain View, CA), and BD Biosciences (San Jose, CA), respectively. The HA-tagged exportin 7 expression plasmid (pMT2SM-RanBP16) was a gift from C. Smas (University of Toledo College of Medicine, Ohio). The mCherry-tagged exportin 4 expression plasmid (pmCherry-XPO4) was obtained from GenScript (Piscataway, NJ) and pmCherry-C1 was from Clontech. 2xDR4-SV40-Luc was a gift from J. L. Jameson, (Northwestern University) and consists of two copies of a positive, direct repeat TRE (DR+4) in the firefly luciferase vector pGL3. pGL4.74 encodes Renilla luciferase (Promega, Madison, WI).
2.2. Fluorescence recovery after photobleaching (FRAP)
HeLa cells (ATCC, #CCL-2) were cultured in Minimum Essential Medium (MEM) supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY), at 37°C under 5% CO2 and 98% humidity. Cells were seeded at 2.0–2.5 × 105 cells per 3-cm dish with a cover glass bottom (MatTek Corporation, Ashland, MA). Twenty four hours after seeding, cells were co-transfected with 1 μg GFP-TRα1 expression plasmid, and 1 μg of the appropriate target-specific or control shRNA expression plasmids, using the two shRNAs from each set of four (see Section 2.1) that showed the greatest knockdown efficacy as assessed by quantitative PCR (see Section 2.4). Transfection medium containing Lipofectamine 2000 (Life Technologies) was replaced with complete medium 9 h post-transfection. Twenty seven hours post-transfection, cells were prepared for live-cell imaging: cells were incubated in 2 mL of complete media containing 100 μg/mL cycloheximide (Sigma-Aldrich, St. Louis, MO), 100 units/mL penicillin, 100 μg/mL streptomycin, and 10 μg/mL wheat germ agglutinin conjugated to Alexa Fluor® 350 (Life Technologies). Cells were washed twice with Dulbecco’s phosphate-buffered saline and imaged. During the experiment, cells were incubated in MEM-α without phenol red, containing 50 μg/mL cycloheximide, 50 units/mL penicillin, and 50 μg/mL streptomycin. In preliminary studies, we tested a range of post-transfection incubation times (17 h, 24 h, 27 h, and 30 h), varied the amount of Lipofectamine 2000 and the time cells were exposed to the reagent, selected for knockdown cells with puromycin, and varied the shRNA plasmid amounts and combinations. The conditions described above were determined to have high transfection efficiency (70–80% of cells were transfected), effectively reduce the levels of exportins in cells (at least 50% knockdown), while still retaining cell viability. Altered conditions either decreased transfection efficiency, decreased knockdown efficiency, or led to increased cell mortality. Cell mortality was assessed by visual inspection of the number of adherent cells prior to transfection, compared with the number of cells remaining adhered post-transfection, with the standard set at >60% retention.
All FRAP experiments were performed in an OkoLab Incubation System (Warner Instruments, Inc., Hamden, CT) at 37°C under 5% CO2. Images were collected from an inverted Nikon A1Rsi confocal microscope Ti-E-PFS using a 40X water objective (Nikon Inc., Melville, NY). The 488-nm line of a krypton-argon laser with a band-pass 525/50 nm emission filter was used for GFP detection; the 405-nm line with a band-pass 450/50 emission filter was used for Alexa Fluor® 350 detection. Images were obtained using the stimulation/bleaching acquisition module of NIS-Elements AR (Nikon). An initial image was recorded from an area containing a GFP-expressing cell with two or more nuclei, using 1–4% laser power from the 488 nm line and 8–20% laser power from the 405 nm line. One nucleus within the multinucleated cell was exposed at 100% laser power for 10–12 sec using the 488 nm line. Post-bleach sequential images were then taken every 5 min for 24 cycles at the lower laser intensities noted above. For quantitative analysis of digitized images, fluorescent intensity values were generated using NIS-Elements AR (Nikon). Bleached and unbleached nuclei were each considered as independent regions of interest. In addition, these values took into account the background brightness levels during each experiment. Intensity values were subsequently normalized so that the total fluorescence within each multinucleated cell after bleaching was equal to 1.0 (arbitrary units). After normalization, convergence of the representative curves for bleached and unbleached nuclei toward one another represents the degree of fluorescence equilibration between these compartments. When one bleached and one unbleached nucleus are present, complete equilibration occurs at 0.5 fluorescence units (Grespin et al., 2008).
2.3. Fixation, immunofluorescence, and cell scoring
HeLa cells were seeded at 2.5–3.0 × 105 cells per well of a 6-well plate with glass coverslips (Fisher Scientific, Pittsburgh, PA). Twenty four hours after seeding, cells were transfected with 2 μg plasmid DNA, using Lipofectamine 2000 Reagent. Approximately 18 h post-transfection, cells were fixed in 3.7% formaldehyde and permeabilized with 0.2% Triton-X-100. The following antibodies were used at 1:500: anti-c-Myc (Clontech), Cy3-goat anti-mouse (Life Technologies/Zymed), and anti-HA tag (Abcam, Cambridge, MA). Texas Red anti-rabbit IgG (H+L) (Vector Laboratories, Burlingame, CA) antibody was used at 1:50. Coverslips were mounted in Fluoro-Gel II containing the DNA counter stain 4′, 6-diamidino-2′-phenylindole (DAPI, 0.5 μg/ml) (Electron Microscopy Sciences, Hatfield, PA).
Images were analyzed with an inverted Nikon ECLIPSE TE 2000-E fluorescence microscope. A Nikon Ultraviolet Excitation: UV-2E/C filter block for DAPI visualization, a Blue Excitation: B-2E/C filter block for GFP, and a Red Excitation: T-2E/C filter block for the Myc or HA tag were used with a Nikon Plan Apo 40X objective. NIS-Elements AR software was used for image acquisition and primary image processing. Cells were scored blind, without knowledge of the treatment conditions. The slides’ original labels were removed and replaced with random number labels by another lab member, who made a key and kept it secure until the scoring was completed and data were analyzed. All experiments consisted of a minimum of 3 replicates and at least 100–300 cells were scored per replicate. The state of the nuclei was assessed by visually examining the integrity and morphology of each DAPI-stained nucleus; only cells with intact nuclei were scored. Intracellular distribution patterns of TR in cells transfected with pGFP-TRα1 or pGFP-TRβ1 and pCMV-Myc or pMyc-Exp5 were scored into two categories: primarily nuclear (N), or distributions ranging from nuclear accumulation but a clearly visible cytosolic population to a whole cell distribution (N+C/WC). Intracellular distribution patterns of TR in cells transfected with pGFP-TRα1 or pGFP-TRβ1 and pmCherry-XPO4, pmCherry, pCMV-HA, or HA-exportin 7 (RanBP16) plasmids were scored into three categories: primarily nuclear (N), nuclear accumulation and a clearly visible cytosolic population (N+C); or whole cell, where the nucleus was not distinct (WC) (see supplementary data Fig. S1). Data were quantified as the percentage of cells in a given category (e.g., % of cells with a primarily nuclear distribution of TR).
2.4. Validation of RNA interference (RNAi) by quantitative PCR (qPCR)
HeLa cells seeded at 6–7 × 105 cells per 100-mm vented plate were transfected with 10 μg target-specific or control shRNA expression plasmids, using Lipofectamine 2000. Medium was replaced with complete medium 9 h post-transfection. Twenty seven hours post-transfection, RNA was extracted using the Aurum Total RNA Mini Kit (Bio-Rad, Hercules, CA) according to the manufacturer’s specifications, with the exception that the DNase I digestion was extended to 30 min. RNA quality was analyzed using an RNA 6000 Pico Total RNA Assay and Agilent 2100 BioAnalyzer’s Lab-on-a-Chip Technology (Santa Clara, CA). cDNA was synthesized using the SABioscience RT2 First Strand Kit, following the manufacturer’s specifications. Quantitative PCR (qPCR) was performed using the Real-Time RT2 qPCR Primer Assay (SABiosciences) with RT2 Real-Time SYBR Green/Fluorescein qPCR master mix and SABioscience validated primers specific for each exportin, or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. qPCR data were analyzed by the ΔΔCt (Livak) method (Livak and Schmittgen, 2001) using the Applied Biosystems StepOne Software Version 2.1 (Life Technologies).
2.5. Western blotting
Twenty seven hours post-transfection, HeLa cells were harvested in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 10 mM NaF, 10% glycerol, 1% Nonidet P-40 (Calbiochem, San Diego, CA), Complete Mini EDTA-free Protease Inhibitor Cocktail Tablet (1 tablet per 10 ml; Roche Diagnostics, Indianapolis, IN)). Lysates were centrifuged at 14,000 × g for 10 min at 4°C, and the supernatant protein concentration was determined using a NanoDrop® ND-1000 Full-spectrum UV/Vis Spectrophotometer. Lysates (40–60 μg of protein per lane) were separated by 8% SDS-PAGE and transferred to PVDF membrane using the iBlot Dry Blotting System (Life Technologies). The membranes were incubated overnight at 4ºC in blocking solution (Tris-buffered saline containing 0.1% Tween 20 [T-TBS], 1% bovine serum albumin). After 4–6 washes with T-TBS at room temperature, the membranes were incubated with primary antibodies for 1.5–2 h. All antibodies were incubated separately and used at the following concentrations: anti-β-tubulin (Santa Cruz Biotechnology Inc., Dallas, TX), 1:200; anti-exportin 5 (Abcam), 1:400; anti-transportin 1 (Abcam), 2.0 μg/ml; anti-transportin 2 (Santa Cruz), 1:1000; anti-exportin 4 (Santa Cruz), 1:500; anti-exportin 5 (Santa Cruz), 1:1000; anti-exportin 7 (Abcam), 0.5 μg/ml; and anti-GAPDH (Santa Cruz), 1:5000. Blots were then washed 4–6 times with T-TBS and incubated with the appropriate secondary antibody for 1.25 hours in blocking solution. Secondary antibodies were used at the following concentrations: horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG (GE Healthcare Life Sciences, Pittsburgh, PA), 1:25,000; HRP-sheep anti-mouse IgG (GE Healthcare Life Sciences), 1:25,000; and HRP-mouse anti-goat IgG (Santa Cruz), 1:10,000 or 1:25,000. Subsequently, blots were washed 6–8 times in T-TBS, followed by chemiluminescent detection using ECL Prime detection reagent (GE Healthcare Life Sciences). Protein size was monitored using Pre-Stained Kaleidoscope Protein Standards (Bio-Rad). X-ray films were quantified by scanning densitometry using ImageJ software (National Institutes of Health, Bethesda, MD).
2.6. Luciferase reporter gene assay
HeLa cells were seeded at 2.0 × 104 per well in a 96-well plate (PerkinElmer, Waltham, MA). Seventeen hours after seeding, cells were transiently transfected with 100 ng DNA, containing 25 ng each of expression plasmids for GFP-TRα1 or GFP-TRβ1, TRE (DR+4)-firefly luciferase reporter, Renilla luciferase internal control, and mCherry, mCherry-XPO4, Myc, Myc-XPO5, HA, or HA-XPO7. Transfection medium was replaced with complete medium 6 h post-transfection. Twelve hours post-transfection, complete medium was replaced with 100 μl MEM containing 10% charcoal-dextran stripped FBS (Life Technologies), supplemented or not with 100 nM T3. After an additional 12 h, a Dual-Glo® Luciferase Assay (Promega) was performed, according to the manufacturer’s protocol, using 100 μl of reagent per well.
2.7. Statistical analyses
Data represent the mean ±1 SEM of at least three independent experiments. Statistical differences between two groups were determined using an unpaired Student’s t test with the two-tailed P value. Results were considered significant at P<0.05.
3. Results
3.1. Knockdown of exportins 4, 5, and 7 slows nucleocytoplasmic shuttling of TRα1
Prior studies pointed to the possibility that, in addition to following a cooperative CRM1/calreticulin-dependent export pathway during nucleocytoplasmic shuttling, TR may also follow an alternative nuclear export pathway that does not rely on CRM1 (Bunn et al., 2001; Grespin et al., 2008; Mavinakere et al., 2012). To determine whether other exportins play a role in mediating TR nuclear export, we coupled in vivo approaches using FRAP and RNAi in transfected HeLa cells to examine how knockdown of individual exportins impacts the shuttling dynamics of TRα1. Since TRα1 is primarily nuclear at steady-state, but shuttles between the nucleus and the cytosol, knockdown of an essential export factor would be predicted to result in decreased nuclear export of TRα1. This effect would be visualized as greater retention of fluorescence in the unbleached nucleus, along with a significantly slower recovery of fluorescence to the bleached nucleus during a FRAP assay. It is important to note, however, that it was not expected that cells would ever show complete inhibition of TRα1 shuttling since RNAi leaves a portion of the target mRNA and protein in cells, and there are likely multiple pathways for export. In addition, if knockdown of a bidirectional transport factor affected nuclear import instead, this defect in import would be visualized as an accumulation of TRα1 in the cytosol.
To begin, shRNA-induced knockdown of target exportin mRNA and protein levels were validated by qPCR and western blotting, respectively. The levels of exportin mRNA in the presence of target shRNA were reduced by ≥75%, relative to the scrambled shRNA control (control mRNA expression was set at 100%) (Fig. 1A). Exportin protein levels were reduced relative to the scrambled control, on average, as follows: transportin 1, 31%; transportin 2, 56%; exportin 4, 41%; exportin 5, 30%; and exportin 7, 40% (Fig. 1B). We were unable to acquire a viable antibody for exportin 6, so in this case validation was restricted to qPCR. Taken together, these levels of knockdown confirm the efficacy of the RNAi system.
Fig. 1.
Validation of shRNA knockdown of target gene mRNA and protein levels. (A) HeLa cells were transiently transfected with a panel of exportin-specific shRNA expression plasmids, as indicated, or a scrambled shRNA plasmid as a control. qPCR was used to confirm knockdown of exportin mRNA levels. Bars indicate the relative expression level of exportin mRNA in cells treated with exportin-specific shRNA versus control cells (control mRNA expression set at 100%), normalized to the levels of the housekeeping mRNA, GAPDH. Error bars indicate ±1 SEM (n=3). TNPO1, transportin 1; TNPO2, transportin 2; XPO4, exportin 4; XPO5, exportin 5; XPO6, exportin 6; XPO7, exportin 7. (B) Western blot analysis was used to confirm knockdown of exportin protein levels relative to the control (control protein expression set at 100%), as indicated. Error bars indicate ±1 SEM (n=3, TNPO1, XPO4, XPO5, XPO7; n=5, TNPO2).
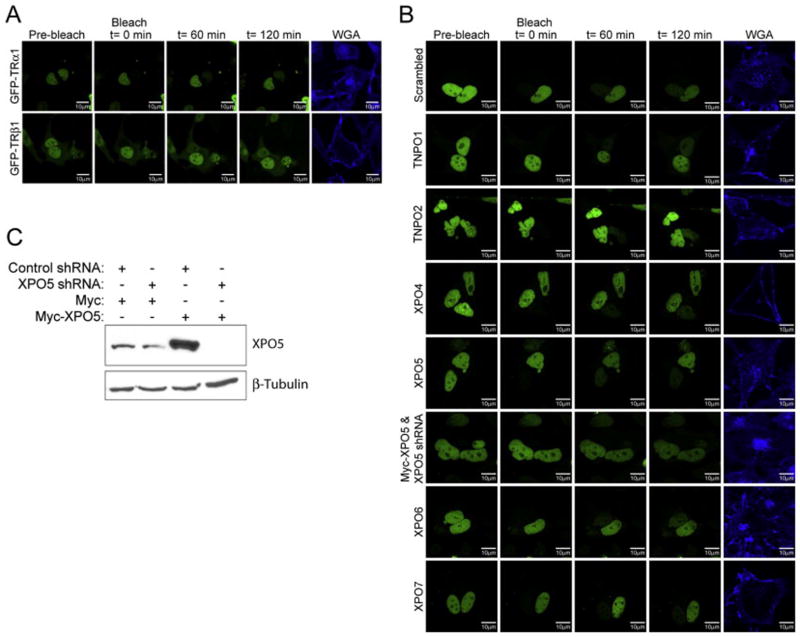
Next, we compared shuttling of GFP-TRα1 with GFP-TRβ1 under standard conditions, in the absence of shRNA expression (Fig. 2A, supplementary data Videos S1 and S2). To confirm that experiments were conducted in a single cell with multiple nuclei and not nuclei in adjacent, separate cells, transfected HeLa cells were incubated before visualization with fluorescent-tagged wheat germ agglutinin (WGA), an external plasma membrane marker for live-cell imaging (Fig. 2A). All FRAP experiments were also performed in the presence of cycloheximide to ensure that the fluorescence recovery of GFP in bleached nuclei was not due to de novo protein synthesis. As shown in Fig. 2A, both TRα1 and TRβ1 undergo shuttling; however, for TRβ1 a faint cytosolic population is visible at t=0, while TRα1 appears entirely nuclear. Thus, to avoid any ambiguity in interpreting FRAP results, we only performed knockdown assays with TRα1. To determine the effect of exportin knockdown on the dynamic shuttling of TRα1, HeLa cells were cotransfected with GFP-TRα1 and exportin-specific shRNA expression plasmids.
Fig. 2.
FRAP analysis of TR shuttling after knockdown of export factors. (A) HeLa cells were transiently transfected with expressions plasmids for GFP-TRα1 or GFP-TRβ1, as indicated. Two Quick-Time movie files are attached as supplemental materials (Videos S1 and S2). FRAP experiments were performed in multinucleated live cells to monitor the movement of GFP-tagged TRα1 or TRβ1 from unbleached to bleached nuclei. To identify cells with two or more nuclei, cells were treated with Alexa Fluor® 350-tagged wheat germ agglutinin (WGA), a plasma membrane marker. Representative images are shown pre-bleach and 0 min, 60 min, and 120 min post-bleach. (B) HeLa cells were transiently transfected with GFP-TRα1, scrambled shRNA control (n=7), or shRNA expression plasmids targeting transportin 1 (TNPO1, n=6), transportin 2 (TNPO2, n=6), exportin 4 (XPO4, n=7), exportin 5 (XPO5, n=9), exportin 6 (XPO6, n=7), or exportin 7 (XPO7, n=10). Alternatively, cells were cotransfected with GFP-TRα1 and Myc-exportin 5 (Myc-XPO5) expression plasmids, along with exportin 5 shRNA (n=4). Quick-Time movie files are attached as supplemental materials for the scrambled shRNA control and for exportins 4, 5, and 7 shRNA knockdown (Videos S3–S6). (C) HeLa cells were transiently transfected with GFP-TRα1 expression plasmid, Myc-tagged exportin 5 (Myc-XPO5) expression plasmid, Myc expression plasmid as a control, and shRNA expression plasmid targeting exportin 5 (XPO5 shRNA), or scrambled shRNA control. Cell lysates were subject to western blot analysis using antibodies specific for exportin 5 and β-tubulin as a loading control.
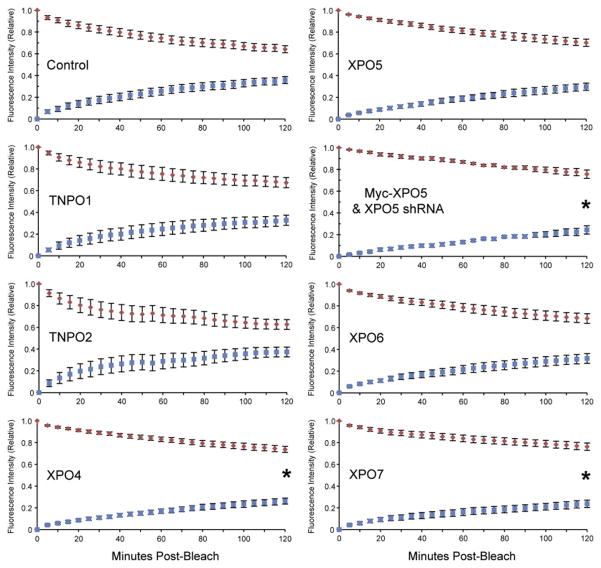
We first tested whether transportins 1 and 2 play a role in TRα1 nuclear export (Fig. 2B). Originally proposed to mediate bidirectional transport, more recent studies maintain that transportins 1 and 2 are restricted to mediating nuclear import of RNA-binding proteins that function as splicing regulators (Twyffels et al., 2014). Therefore, we predicted that the transportins would not be involved in TRα1 shuttling. For quantitation of FRAP (Fig. 3), bleached and unbleached nuclei were each considered as independent regions of interest. Intensity values were normalized so that the total fluorescence within each multinucleated cell after bleaching was equal to 1.0 (arbitrary units). After normalization, convergence of the representative curves for bleached and unbleached nuclei toward one another represents the degree of fluorescence equilibration between these compartments. When one bleached and one unbleached nucleus are present, complete equilibration occurs at 0.5 fluorescence units (Grespin et al., 2008). Recovery of fluorescence to bleached nuclei within live cells transfected with a scrambled control shRNA was measured at, on average, 51% fluorescence equilibration at 60 min and 72% at 120 min (Fig. 3). As expected, comparable shuttling dynamics were observed for cells transfected with transportin 1 shRNA, relative to the control (60 min, P=0.89; 120 min, P=0.59). At 60 min and 120 min, transportin 1-knockdown cells showed, on average, 49% and 65% fluorescence equilibration with unbleached nuclei, respectively. Similarly, there was no significant difference in TRα1 shuttling between the control and transportin 2 shRNA-transfected cells (60 min, P=0.68; 120 min, P=0.78). At 60 min and 120 min, transportin 2-knockdown cells showed, on average, 58% and 75% fluorescence equilibration with unbleached nuclei, respectively (Fig. 3). These data show that transportins 1 and 2 are not involved in nuclear retention or export of TRα1. Furthermore, no cytosolic accumulation of TRα1 was observed during the FRAP assay (Fig. 2B), indicating that knockdown of transportins 1 and 2 did not interfere with nuclear import of TRα1.
Fig. 3.
Nucleocytoplasmic shuttling of TRα1 is slowed by knockdown of exportins 4, 5, and exportin 7. Fluorescence recovery graphs summarize the data from the replicate FRAP experiments described in Fig. 2B. Blue squares are the intensity within bleached nuclei and red diamonds are the intensity within unbleached nuclei. Intensity values were normalized so that the total fluorescence within each multinucleated cell after bleaching was equal to 1.0 (arbitrary units). Convergence of the curves for bleached and unbleached nuclei toward one another represents the degree of fluorescence equilibration between the nuclei. When one bleached and one unbleached nucleus are present, complete equilibration occurs at 0.5 fluorescence units. Error bars indicate ±1 SEM. *P <0.05.
Exportin 4 is a bidirectional nuclear transport factor involved in nuclear export of translation initiation factor eIF-5A and transcriptional regulator Smad3 (Chook and Suel, 2011). Thus, we predicted this selective transporter would not play a role in promoting TRα1 nuclear export. At 60 min, there was no significant difference between TRα1 shuttling in control shRNA and exportin 4 shRNA-transfected cells (Figs. 2B and 3; supplementary data Videos S3 and S4) (P=0.06), with recovery in bleached nuclei in knockdown cells measured at 34% fluorescence equilibration, on average. However, unexpectedly, at 120 min there was a significant difference in the amount of recovery relative to the control (P=0.04); exportin 4 knockdown-cells only reached 52% fluorescence equilibration, on average (Fig. 3), suggesting that exportin 4 plays a role in TR nuclear export, either directly by facilitating exit of TRα1 from the nucleus or, indirectly by decreasing nuclear retention and thereby promoting interaction with other exportins. No cytosolic accumulation was observed (Fig. 2B), which if present would have been indicative of a defect in import, suggesting that exportin 4 does not play a role in mediating nuclear entry of TRα1.
The primary cargo of exportin 5 is microRNA (miRNA) precursors (Bohnsack et al., 2004; Lund et al., 2004); however, it also has been shown to mediate nuclear export of the androgen receptor, another member of the nuclear receptor superfamily (Shank et al., 2008). Thus, we predicted that knockdown of exportin 5 would alter TRα1 shuttling dynamics. Instead, although there appeared to be a modest decrease in TRα1 shuttling between cells transfected with control shRNA and exportin 5 shRNA knockdown, this effect was not significant at either 60 min (P=0.12) or 120 min (P=0.22) (Figs. 2B and 3). Fluorescence equilibration was measured, on average, at 38% and 60% for 60 min and 120 min, respectively (Fig. 3). Since exportin 5 mediates export of pre-miRNA and shRNA from the nucleus, there is a trade-off between down-regulating exportin 5, but still getting enough pre-shRNA out of the nucleus for sustained knockdown of exportin 5 mRNA levels. Previously, it was shown that overexpressing exportin 5 in the presence of shRNA expression plasmids increased the efficiency of RNAi (Yi et al., 2005). Thus, we tested whether overexpressing exportin 5 in the presence of Myc-tagged exportin 5 shRNA could, albeit counterintuitively, enhance knockdown. First, we validated the utility of this approach. In the presence of XPO5 shRNA, exportin 5 protein levels were decreased compared to the control shRNA, where no knockdown occurred (Fig. 2C; see also Fig. 1B). In contrast, when exportin 5 is overexpressed, there was even greater knockdown of exportin 5 protein levels with target shRNA. After determining that more efficient knockdown occurs when overexpressing exportin 5, we ran parallel FRAP experiments. In comparison to the shRNA scrambled control, we observed a significant reduction in shuttling of TRα1 (Figs. 2B and 3, supplementary data Video S5) at 60 min (P=0.03); however, after 120 min recovery was not significantly different from the control (P=0.05). The fluorescence equilbrations from unbleached nuclei to bleached nuclei were, on average, 26% at 60 min and 49% at 120 min (Fig. 3).
Next, we predicted that knockdown of exportins 6 and 7 would not alter TRα1 shuttling dynamics for the following reasons. Exportin 6 is a specific transporter of nuclear actin (Dopie et al., 2012; Stuven et al., 2003) and, although exportin 7 binds diverse cargo, it had not been shown to be involved in export of other members of the nuclear receptor superfamily (Mingot et al., 2004). As predicted, knockdown of exportin 6 resulted in similar TRα1 shuttling dynamics compared to the scrambled control, at both 60 min (P=0.44) and 120 min (P=0.46) (Figs. 2B and 3). Fluorescent equilibration of bleached nuclei was measured, on average, at 43% and 63% for 60 min and 120 min, respectively. In contrast, knockdown of exportin 7 resulted in altered shuttling of TRα1 (Figs. 2B and 3, supplementary data Video S6). Fluorescent equilibrations were measured, on average, at 32% and 48%, for 60 min and 120 min, respectively. Differences were not significant at 60 min (P=0.06); however, at 120 min there was a significant decrease in recovery (P=0.03), compared with the control, suggesting that exportin 7 plays a role in TRα1 cellular localization, either directly via mediating nuclear export, or indirectly via increasing intranuclear mobility and access to the export machinery.
Finally, we predicted that dual knockdown with combinations of shRNA against exportins 4, 5, and 7 would have a greater impact than single knockdowns. However, when we tested dual knockdown of exportins 4 and 5, exportins 5 and 6 (as a control), and exportins 5 and 7, these combinations did not result in further shifts in the shuttling pattern of TRα1 (data not shown), although this could well be due to increased cell mortality. These exportins are required for trafficking of proteins involved in many vital cell processes and, since cellular miRNAs potentially regulate the expression of hundreds of genes, saturation of the RNAi pathway with exogenous shRNA also can contribute to loss of cell viability (Scherr and Eder, 2007; Castonotto and Rossi, 2009). In addition, it is likely that the primarily nuclear location of TRα1 at steady-state limits how much the shuttling pattern can be altered over the time course of an experiment.
Taken together, these data provide evidence that in addition to CRM1 and calreticulin, exportins 4, 5, and 7 either directly or indirectly play a role in promoting nuclear export of TRα1. In contrast, transportins 1 and 2, and exportin 6 do not play a role, or at least not an essential role, in modulating TRα1 shuttling.
3.2. Overexpression of exportin 5 and exportin 7 promotes nuclear export of TRα1 and TRβ1
Having shown that nucleocytoplasmic shuttling of TRα1 is partially inhibited by knockdown of exportins 4, 5, or 7, we sought to ascertain whether their overexpression would alter the cellular localization of both TRα1 and TRβ1. We predicted that overexpression of exportins 4, 5, and 7 would cause a shift in the distribution of TRα1 and TRβ1, from the nucleus to the cytosol. To this end, HeLa cells were cotransfected with expression plasmids for GFP-tagged TRα1 or β1, and mCherry-tagged exportin 4, Myc-tagged exportin 5, or HA-tagged exportin 7, or mCherry, Myc tag, or HA tag, as controls. Distribution patterns were visualized by direct fluorescence or immunofluorescence microscopy (Figs. 4 and 5).
Fig. 4.
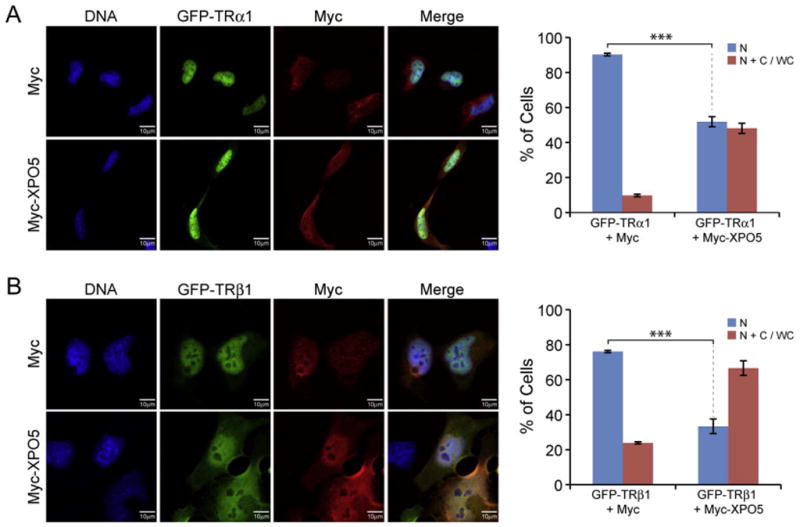
Overexpression of exportin 5 promotes nuclear export of TRα1 and TRβ1. (A) HeLa cells were transiently transfected with GFP-TRα1 expression plasmids, Myc or Myc-XPO5 (exportin 5), as indicated. Cells were immunostained with anti-Myc (red) and analyzed by fluorescence microscopy for the distribution of TRα1 (green). Nuclei were stained for DNA with DAPI (blue). Bar graph summarizes the effect of overexpressing exportin 5 on TRα1 distribution in two categories: primarily nuclear (N), or distribution patterns ranging from nuclear accumulation and a clearly visible cytosolic population to a whole cell distribution (N+C/WC). Error bars represent ±1 SEM (n=4 replicates each, with at least 300 cells scored per replicate). ***P<0.001. (B) Parallel experiments were performed with HeLa cells cotransfected with GFP-TRβ1 expression plasmid and Myc or Myc-XPO5 (n=4 replicates, 100–300 cells per replicate).
Fig. 5.

Overexpression of exportin 7 promotes nuclear export of TRα1 and TRβ1. (A) HeLa cells were transiently transfected with GFP-TRα1 or GFP-TRβ1, and mCherry or mCherry-tagged exportin 4 (XPO4) expression plasmids, as indicated. Bar graph summarizes the lack of effect of exportin 4 overexpression on TR distribution in three categories: primarily nuclear (N), nuclear accumulation and a clearly visible cytosolic population (N+C); or whole cell, where the nucleus was not distinct (WC). Error bars represent ±1 SEM (n=3 replicates, 100–300 cells scored per replicate). (B) Bar graph summarizing experiments performed with HeLa cells co-transfected with GFP-TRα1 or GFP-TRβ1, and HA tag or HA-tagged exportin 7 (XPO7) expression plasmids, as indicated. Error bars indicate ±1 SEM (n=5 replicates, 100–300 cells per replicate). ***P<0.001.
Consistent with our predictions, when exportin 5 was overexpressed, a significantly greater percentage of cells showed a shift towards a more cytosolic distribution for both TRα1 (P=0.00001) and TRβ1 (P=0.00006) (Fig. 4). In cells co-expressing GFP-TRα1 and the Myc tag (control), on average 90% of cells showed a primarily nuclear distribution of TRα1 (Fig. 4A). In contrast, in cells co-expressing GFP-TRα1 and Myc-tagged exportin 5, on average only 52% of cells showed a primarily nuclear distribution of TRα1 (Fig. 4A). In the cells expressing GFP-TRβ1 and the Myc tag, on average 76% of cells had a primarily nuclear distribution of TRβ1, compared with 34% of cells showing a primarily nuclear distribution of TRβ1 in the presence of Myc-tagged exportin 5 (Fig. 4B).
Interestingly, although knockdown of exportin 4 significantly slowed TR shuttling, overexpression of exportin 4 had no significant effect on the distribution patterns of either TRα1 (P=1.00) or TRβ1 (P=0.84) (Fig. 5A). In cells co-expressing GFP-TRα1 and mCherry (control) or mCherry-exportin 4, on average 77% of cells showed a nuclear localization of TRα1. In the cells co-expressing GFP-TRβ1 and mCherry or mCherry-exportin 4, on average 55% of cells showed a nuclear localization of TRβ1.
Comparable results to exportin 5 were obtained when exportin 7 was overexpressed; a significantly greater percentage of cells showed a shift towards a more cytosolic distribution for both TRα1 and TRβ1, relative to the control (P=0.00001) (Fig. 5B). In cells co-expressing GFP-TRα1 and the HA tag (control), on average 68% of cells showed a nuclear distribution of TRα1. In contrast, in cells co-expressing GFP-TRα1 and HA-tagged exportin 7, on average only 25% of cells showed a primarily nuclear distribution of TRα1. In the cells expressing GFP-TRβ1 and the HA tag, on average 59% of cells had a primarily nuclear distribution of TRβ1, compared with 22% of cells showing a primarily nuclear distribution of TRβ1 in the presence of HA-exportin 7.
3.3. Effect of enhanced nuclear export on TR-mediated ligand-independent and ligand-dependent gene expression
In the absence of T3, unliganded TRα1 and TRβ1 repress the expression of target genes that are under control of positive thyroid hormone response elements (TREs). In the presence of T3, the liganded receptors stimulate expression of these same genes. Thus, we sought to ascertain whether the cytosolic shift in the distribution of TRα1 and TRβ1 resulting from overexpression of exportins 5 and 7 would alter TR-mediated gene expression to a comparable extent. A firefly luciferase reporter gene under the control of a positive TRE (DR+4) was used to examine ligand-dependent transactivation by TRα1 (Fig. 6A) and TRβ1 (Fig. 6B), in the presence of exportin 4 (no cytosolic shift), exportin 5, or exportin 7. Exportin overexpression was confirmed by western blot analysis (data not shown). In the absence of T3, for cells overexpressing mCherry-exportin 4 or Myc-exportin 5, there was no significant difference in relative luciferase activity by unliganded TRα1 or TRβ1, compared with luciferase activity in cells overexpressing mCherry and Myc controls (TRα1 +XPO4, P=0.29; TRα1 +XPO5, P=0.07; TRβ1 +XPO4, P=0.36; TRβ1 +XPO5, P=0.54). Similarly, there was no significant difference in repression of luciferase activity by unliganded TRβ1 in cells overexpressing HA-exportin 7, compared to HA alone (P=0.31) In contrast, cells overexpressing HA-exportin 7 showed significantly greater luciferase reporter activity in the presence of unliganded TRα1, compared with HA alone (P=0.04). On average, luciferase activity levels were increased 3.0-fold in the presence of exogenous exportin 7, relative to levels in the absence of exogenous exportin 7, suggesting that exportin 7 has wider effects than on TRα1 export alone.
Fig. 6.
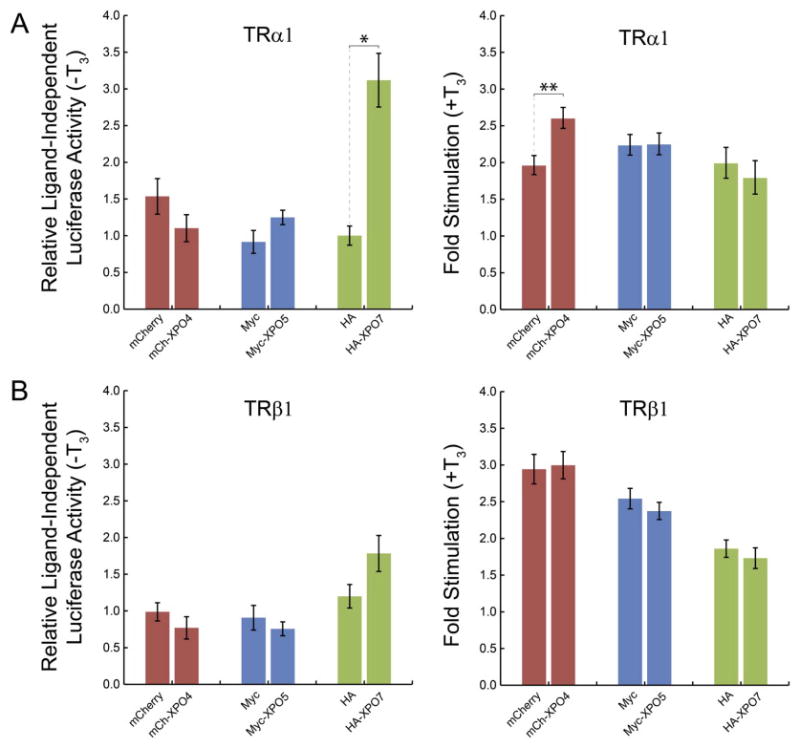
Overexpression of exportins 4, 5, and 7 has variable effects on TR-mediated ligand-independent and ligand-dependent gene regulation. (A) HeLa cells were transfected with GFP-TRα1, mCherry or mCherry-exportin 4 (mCh-XPO4), Myc or Myc-exportin 5 (Myc-XPO5), or with HA or HA-exportin 7 (HA-XPO7) expression plasmids, and TRE (DR+4)-firefly luciferase reporter and Renilla luciferase internal control. Data are presented as relative ligand-independent luciferase reporter activity (firefly/Renilla) (-T3), and fold stimulation in the presence of T3. Error bars indicate ±1 SEM (n=3 replicates of 8 wells per treatment); *P<0.05, **P<0.01. (B) Parallel experiments with GFP-TRβ1 (n=4 replicates).
We also examined the ability of liganded TR to stimulate TRE-luciferase reporter gene expression. In the presence of T3, fold stimulation of luciferase activity by liganded TRα1 and TRβ1 in cells overexpressing Myc-exportin 5 or HA-exportin 7 was not significantly different compared with fold stimulation in the presence of the Myc and HA tag controls (TRα1 +XPO5, P=0.94; TRα1 +XPO7, P=0.60; TRβ1 +XPO5, P=0.41; TRβ1 +XPO7, P=0.53), indicating that under these conditions a reduction in the percentage of cells with primarily nuclear TRα1 or TRβ1 of 38–42% does not have a measurable impact on reporter gene stimulation. Likewise, fold stimulation by liganded TRβ1 in the presence of overexpressed mCherry-exportin 4 also was not significantly different from the control (P=0.90). Interestingly, however, when TRα1 and mCherry-exportin 4 were co-expressed in the presence of T3, fold stimulation of luciferase was significantly increased, relative to mCherry alone (P=0.01). On average, luciferase activity levels were increased 1.3-fold, relative to levels in the absence of exogenous exportin 4, suggesting that exportin 4 may interact with TRα1 in ways other than simply mediating nuclear transport.
4. Discussion
Previously we showed that TRα1 participates in rapid nucleocytoplasmic shuttling. Further, our results pointed to the intriguing possibility that, in addition to following a cooperative CRM1/calreticulin-dependent export pathway during nucleocytoplasmic shuttling, TR also follows an alternative nuclear export pathway that does not rely on CRM1 (Bunn et al., 2001; Grespin et al., 2008; Mavinakere et al., 2012). Here, we propose that additional exportins play a role in modulating TR cellular localization. By coupling in vivo RNAi and FRAP experiments, we showed that knockdown of exportins 4, 5, or 7 altered nucleocytoplasmic shuttling of TRα1. In addition, overexpression of exportins 5 and 7 shifted the distribution pattern of TRα1 and TRβ1 toward a greater percentage of cells with a more cytosolic distribution. Interestingly, results differed for TRα1 and TRβ1-mediated transactivation in cells expressing exogenous exportins. When TRβ1 and exportins 4, 5, or 7 were co-expressed, there was no significant change in either repression or stimulation of T3-mediated gene expression. In contrast, when exportin 7 was co-expressed with unliganded TRα1, TRE-luciferase reporter gene activity was significantly greater compared with the control; that is, in the presence of exportin 7 unliganded TRα1 was less able to repress transcription. In addition, when exportin 4 was co-expressed with liganded TRα1, fold stimulation of the TRE-luciferase reporter gene was significantly increased compared with the control. TR subtype-specific regulation of target gene expression is not without precedent; distinct properties of the amino terminus of TRα1 have been shown to lead to 2-fold greater ligand-independent repression and ligand-dependent stimulation (Hollenberg et al., 1995). Taken together, our data provide evidence that in addition to the previously characterized role of CRM1 and calreticulin in TR export, exportins 4, 5, and 7 also influence TRα1 and TRβ1 cellular localization either directly by promoting nuclear export, or indirectly by decreasing nuclear retention. In this way, exportin levels may provide an additional level of control in modulation of the cellular response to T3.
Further investigation will be required to determine why overexpression of exportin 4 had no significant effect on TR distribution, while knockdown markedly inhibited shuttling. No cytosolic accumulation of TR was observed, suggesting that reduced recovery of fluorescence in the bleached nucleus is not due to inhibition of nuclear import. It may be that levels of endogenous exportin 4 are high enough already to be saturating for export, or exportin 4 may interact with TR in ways other than simply mediating nuclear transport. Additional roles in the cell for exportin 4 beyond transport activity are becoming better understood. A recent report implicates exportin 4 as a tumor suppressor that is down-regulated in hepatocellular carcinoma (Liang et al., 2011). In addition, exportin 4 plays a role in nuclear import of members of the Sox family of transcription factors (Gontan et al., 2009), and interacts directly with Sox 9, thereby blocking binding of Sox 9 to target genes (Tsuchiya et al., 2011). Here, we show that co-expressing exportin 4 with TRα1 increases ligand-dependent gene transactivation, but has no significant effect on ligand-independent repression. These data suggest that exportin 4 may exert its effects on TR localization indirectly, by altering nuclear retention and, thereby, promoting accessibility to the nuclear export machinery.
Recently, exportin 5 was shown to be important for mediating CRM1-independent nuclear export of the androgen receptor, through a NES located in the DNA binding and hinge domains of AR (Shank et al., 2008). Prior to this report, exportin 5 was only thought to be involved in regulating miRNA biogenesis by exporting the precursors of miRNAs out of the nucleus (Bohnsack et al., 2004; Lund et al., 2004; Yi et al., 2005). A role for exportin 5 in promoting nuclear export of TRα1 and TRβ1 further expands the potential cargo list for this versatile exportin. In addition to the cytosolic mislocalization of TR, since exportin 5 is critical for miRNA biogenesis, down-regulation of this exportin would be predicted to have widespread effects. More than 700 miRNAs have been identified in humans. With over two-thirds of protein-coding genes predicted as targets, these abundant small regulatory RNAs play important roles in modulating a broad range of cellular processes (Ebert and Sharp, 2012; Melo and Esteller, 2014). Indeed, inactivating mutations in exportin 5, and the concomitant trapping of pre-miRNA in the nucleus, are linked to human tumors with microsatellite instability, including colon, gastric, and endometrial tumors (Melo et al., 2010).
We conclude that exportin 7 is also a player in regulating shuttling of TRα1 and TRβ1. Exportin 7 (also known as RanBP16) mediates export of diverse proteins with variable NESs including eIF4A1, p50RhoGAP, 14-3-3σ (Mingot et al., 2004), and histones during erythroid maturation (Hattangadi et al., 2014). Intriguingly, despite promoting a significant cytosolic shift in the distribution pattern of TR, overexpression of exportin 7 had no significant effect on ligand-dependent stimulation; however, overexpression of exportin 7 did result in a significant increase in activity of a TRE-linked reporter gene by unliganded TRα1, suggesting less potent repressor activity. These findings suggest that exportin 7 overexpression has an impact on other factors that play a role in T3-independent gene regulation. TR does not act alone; the receptor requires association with coactivators to enhance transcription, and corepressors to silence transcription. In this way, TR activity can be modulated by the balance between coactivator and corepressor activity (Soriano et al., 2011). It may be that less potent TRα1-mediated reporter gene repression when exportin 7 is overexpressed in the absence of T3 reflects more rapid export of a corepressor. Interestingly, exportin 7 mediates nuclear export of liver kinase B1 (LKB1) (Liang et al., 2015). Cytosolic activated LKB1 is a “master kinase” that controls at least 13 downstream AMP-activated protein kinase (AMPK)-related kinases, and acts as a tumor suppressor (Dupuy et al., 2013; Gan and Li, 2014). Further, T3 has been shown to increase LKB1 expression in muscle, resulting in activation of LKB1/AMPK signalling pathway, and an increase in levels of peroxisome proliferator-activated receptorγ coactivator-1α (PGC-1α) (Branvold et al., 2008). PGC-1α enhances gene expression mediated by TR, while the silencing mediator of retinoic acid and thyroid hormone receptors (SMRT) represses gene expression. Although not linked to LKB1 levels, nuclear export of SMRT has been shown to be triggered by changes in cell activity (Guo et al., 2013; Soriano et al., 2011). It is tempting to speculate that exportin 7-promoted nuclear exit of LKB1 initiates a cascade of events, culminating in upregulation of TRα1 coactivators and downregulation of corepressors.
The results presented here highlight the complexity of TR shuttling, and provide further evidence of a role for multiple exportins in promoting nuclear export of this nuclear receptor, either through direct interactions or indirect mechanisms. Comprehensive analysis of how nuclear export integrates with other signaling pathways should help to identify the interacting partners in the fine balance of TR nuclear import, retention, and export. In addition, characterization of protein-protein interactions by in vitro and in vivo binding assays will clarify which of the exportins serve as a direct carrier for each of the multiple NES motifs in TRα1 and TRβ1, and will enhance understanding of how multiple pathways coordinate nuclear exit in response to cell-specific signals.
Supplementary Material
Highlights.
The thyroid hormone receptor (TR) undergoes nucleocytoplasmic shuttling.
We examine whether exportins in addition to CRM1 are involved in TR shuttling.
RNAi, FRAP, overexpression, and reporter gene assays were performed in cells.
We report that exportins 4, 5, and 7 influence TR localization and function.
Acknowledgments
This work was supported in part by National Institutes of Health grant 2R15DK058028-03 and National Science Foundation grant MCB 1120513 (to L.A.A.).
Footnotes
The abbreviations used are: T3, thyroid hormone (triiodothyronine); TRα1, thyroid hormone receptor α1; TRβ1, thyroid hormone receptor β1; NLS, nuclear localization signal; NES, nuclear export signal; CRM1, chromosome region maintenance 1; leptomycin B, LMB; FRAP, fluorescence recovery after photobleaching; RNAi, RNA interference; GFP, green fluorescent protein; shRNA, short hairpin RNA; qPCR, quantitative PCR; TRE, thyroid hormone response element; WGA, wheat germ agglutinin; microRNA (miRNA).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Baumann CT, Maruvada P, Hager GL, Yen PM. Nuclear cytoplasmic shuttling by thyroid hormone receptors - Multiple protein interactions are required for nuclear retention. J Biol Chem. 2001;276:11237–11245. doi: 10.1074/jbc.M011112200. [DOI] [PubMed] [Google Scholar]
- Black BE, Vitto MJ, Gioeli D, Spencer A, Afshar N, Conaway MR, Weber MJ, Paschal BM. Transient, ligand-dependent arrest of the androgen receptor in subnuclear foci alters phosphorylation and coactivator interactions. Mol Endocrinol. 2004;18:834–850. doi: 10.1210/me.2003-0145. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branvold DJ, Allred DR, Beckstead DJ, Kim HJ, Fillmore N, Condon BM, Brown JD, Sudweeks SN, Thomson DM, Winder WW. Thyroid hormone effects on LKB1, MO25, phospho-AMPK, phospho-CREB, and PGC-1α in rat muscle. J Appl Physiol (1985) 2008;105:1218–1227. doi: 10.1152/japplphysiol.00997.2007. [DOI] [PubMed] [Google Scholar]
- Bunn CF, Neidig JA, Freidinger KE, Stankiewicz TA, Weaver BS, McGrew J, Allison LA. Nucleocytoplasmic shuttling of the thyroid hormone receptor α. Mol Endocrinol. 2001;15:512–533. doi: 10.1210/mend.15.4.0619. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Suel KE. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong LJ, Bonamy GM, Fink EN, Allison LA. Nuclear export of the oncoprotein v-ErbA is mediated by acquisition of a viral nuclear export sequence. J Biol Chem. 2004;279:15356–15367. doi: 10.1074/jbc.M308214200. [DOI] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A. 2012;109:E544–552. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy F, Griss T, Blagih J, Bridon G, Avizonis D, Ling C, Dong Z, Siwak DR, Annis MG, Mills GB, Muller WJ, Siegel PM, Jones RG. LKB1 is a central regulator of tumor initiation and pro-growth metabolism in ErbB2-mediated breast cancer. Cancer Metab. 2013;1:18. doi: 10.1186/2049-3002-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan RY, Li HB. Recent progress on Liver Kinase B1 (LKB1): Expression, regulation, downstream signaling and cancer suppressive function. Int J Mol Sci. 2014;15:16698–16718. doi: 10.3390/ijms150916698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C, Guttler T, Engelen E, Demmers J, Fornerod M, Grosveld FG, Tibboel D, Gorlich D, Poot RA, Rottier RJ. Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J Cell Biol. 2009;185:27–34. doi: 10.1083/jcb.200810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grespin ME, Bonamy GMC, Roggero VR, Cameron NG, Adam LE, Atchison AP, Fratto VM, Allison LA. Thyroid hormone receptor α1 follows a cooperative CRM1/calreticulin-mediated nuclear export pathway. J Biol Chem. 2008;283:25576–25588. doi: 10.1074/jbc.M710482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Chen C, Liang Q, Karim MZ, Gorska MM, Alam R. Nuclear translocation of MEK1 triggers a complex T cell response through the corepressor silencing mediator of retinoid and thyroid hormone receptor. J Immunol. 2013;190:159–167. doi: 10.4049/jimmunol.1201657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattangadi SM, Martinez-Morilla S, Patterson HC, Shi J, Burke K, Avila-Figueroa A, Venkatesan S, Wang J, Paulsen K, Gorlich D, Murata-Hori M, Lodish HF. Histones to the cytosol: Exportin 7 is essential for normal terminal erythroid nuclear maturation. Blood. 2014 doi: 10.1182/blood-2013-11-537761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Black BE, Rastinejad F, Paschal BM. Ca2+-dependent nuclear export mediated by calreticulin. Mol Cell Biol. 2002;22:6286–6297. doi: 10.1128/MCB.22.17.6286-6297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg AN, Monden T, Wondisford FE. Ligand-independent and -dependent functions of thyroid hormone receptor isoforms depend upon their distinct amino termini. J Biol Chem. 1995;270:14274–14280. doi: 10.1074/jbc.270.24.14274. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Suzuki M, Miyazaki Y, Matsuzaki M, Nakahama T, Kurose K, Sawada J, Inouye Y. Difference in nucleocytoplasmic shuttling sequences of rat and human constitutive active/androstane receptor. Biochim Biophys Acta. 2007;1773:934–944. doi: 10.1016/j.bbamcr.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Suzuki M, Nakahama T, Inouye Y. Characterization of nuclear localization signals and cytoplasmic retention region in the nuclear receptor CAR. Biochim Biophys Acta. 2005;1745:215–222. doi: 10.1016/j.bbamcr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Kimura M, Imamoto N. Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic. 2014;15:727–748. doi: 10.1111/tra.12174. [DOI] [PubMed] [Google Scholar]
- Koch P, Bohlmann I, Schafer M, Hansen-Hagge TE, Kiyoi H, Wilda M, Hameister H, Bartram CR, Janssen JW. Identification of a novel putative Ran-binding protein and its close homologue. Biochem Biophys Res Commun. 2000;278:241–249. doi: 10.1006/bbrc.2000.3788. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Treichel N, Calado A, Carmo-Fonseca M, Prehn S, Kraft R, Gorlich D, Bischoff FR. Identification of two novel RanGTP-binding proteins belonging to the importin β superfamily. J Biol Chem. 2000;275:40163–40168. doi: 10.1074/jbc.M006242200. [DOI] [PubMed] [Google Scholar]
- Liang HJ, Chai RC, Li X, Kong JG, Jiang JH, Ma J, Vatcher G, Yu AC. Astrocytic exportin-7 responds to ischemia through mediating LKB1 translocation from the nucleus to the cytoplasm. J Neurosci Res. 2015;93:253–67. doi: 10.1002/jnr.23486. [DOI] [PubMed] [Google Scholar]
- Liang XT, Pan K, Chen MS, Li JJ, Wang H, Zhao JJ, Sun JC, Chen YB, Ma HQ, Wang QJ, Xia JC. Decreased expression of XPO4 is associated with poor prognosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26:544–549. doi: 10.1111/j.1440-1746.2010.06434.x. [DOI] [PubMed] [Google Scholar]
- Liu J, DeFranco DB. Protracted nuclear export of glucocorticoid receptor limits its turnover and does not require the exportin 1/CRM1-directed nuclear export pathway. Mol Endocrinol. 2000;14:40–51. doi: 10.1210/mend.14.1.0398. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lombardi M, Castoria G, Migliaccio A, Barone MV, Di Stasio R, Ciociola A, Bottero D, Yamaguchi H, Appella E, Auricchio F. Hormone-dependent nuclear export of estradiol receptor and DNA synthesis in breast cancer cells. J Cell Biol. 2008;182:327–340. doi: 10.1083/jcb.200712125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Mavinakere MS, Powers JM, Subramanian KS, Roggero VR, Allison LA. Multiple novel signals mediate thyroid hormone receptor nuclear import and export. J Biol Chem. 2012;287:31280–31297. doi: 10.1074/jbc.M112.397745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Esteller M. Disruption of microRNA nuclear transport in human cancer. Semin Cancer Biol. 2014;27:46–51. doi: 10.1016/j.semcancer.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, Yamamoto H, Schwartz S, Jr, Esteller M. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Mingot JM, Bohnsack MT, Jakle U, Gorlich D. Exportin 7 defines a novel general nuclear export pathway. EMBO J. 2004;23:3227–3236. doi: 10.1038/sj.emboj.7600338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MM, Dincer Z, Wade JR, Alur M, Michalak M, Defranco DB, Wang Z. Cytoplasmic localization of the androgen receptor is independent of calreticulin. Mol Cell Endocrinol. 2009;302:65–72. doi: 10.1016/j.mce.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X, Wang Z. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem. 2003;278:41998–42005. doi: 10.1074/jbc.M302460200. [DOI] [PubMed] [Google Scholar]
- Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444–449. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- Shank LC, Kelley JB, Gioeli D, Yang CS, Spencer A, Allison LA, Paschal BM. Activation of the DNA-dependent protein kinase stimulates nuclear export of the androgen receptor in vitro. J Biol Chem. 2008;283:10568–10580. doi: 10.1074/jbc.M800810200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FX, Leveille F, Papadia S, Bell KFS, Puddifoot C, Hardingham GE. Neuronal activity controls the antagonistic balance between peroxisome proliferator-activated receptor-γ coactivator-1α and silencing mediator of retinoic acid and thyroid hormone receptors in regulating antioxidant defenses. Antiox Redox Signal. 2011;14:1425–1436. doi: 10.1089/ars.2010.3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin A, Kim E, Ovchinnikov L. Nucleocytoplasmic transport of proteins. Biochem (Moscow) 2007;72:1439–1457. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- Stuven T, Hartmann E, Gorlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Ogawa H, Suzuki T, Sugiyama N, Haraguchi T, Hiraoka Y. Exportin 4 interacts with Sox9 through the HMG Box and inhibits the DNA binding of Sox9. PLoS One. 2011;6:e25694. doi: 10.1371/journal.pone.0025694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyffels L, Gueydan C, Kruys V. Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett. 2014;588:1857–1868. doi: 10.1016/j.febslet.2014.04.023. [DOI] [PubMed] [Google Scholar]
- Umemoto T, Fujiki Y. Ligand-dependent nucleo-cytoplasmic shuttling of peroxisome proliferator-activated receptors, PPARα and PPARγ. Genes Cells. 2012;17:576–596. doi: 10.1111/j.1365-2443.2012.01607.x. [DOI] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.