Abstract
Background
Immunoglobulin E (IgE) is a key mediator of allergic inflammation and is frequently elevated in allergic disorders.
Objective
To identify genetic variants associated with IgE levels in Latinos.
Methods
We performed a genome-wide association study (GWAS) and admixture mapping of total IgE levels in 3,334 Latinos from the Genes-environments & Admixture in Latino Americans (GALA II) study. Replication was evaluated in 454 Latinos, 1,564 European Americans, and 3,187 African Americans from independent studies.
Results
We confirmed associations of six genes identified by previous GWAS and identified a novel genome-wide significant association of a polymorphism in ZNF365 with total IgE (rs200076616, p=2.3x10−8). We next identified four admixture mapping peaks (6p21.32-p22.1, 13p22-31, 14q23.2, and 22q13.1) where local African, European, and/or Native American ancestry was significantly associated with IgE levels. The most significant peak was 6p21.32-p22.1, where Native American ancestry was associated with lower levels of IgE (p=4.95x10−8). All but 22q13.1 were replicated in an independent sample of Latinos, and two of the peaks were replicated in African Americans (6p21.32-p22.1 and 14q23.2). Fine mapping of 6p21.32-p22.1 identified six genome-wide significant single nucleotide polymorphisms in Latinos, two of which replicated in European Americans. Another SNP was peak-wide significant within 14q23.2 in African Americans (rs1741099, p=3.7x10−6), and replicated in non-African American samples (p=0.011).
Conclusion
We confirmed genetic associations at six genes, and identified novel associations within ZNF365, HLA-DQA1, and 14q23.2. Our results highlight the importance of studying diverse, multi-ethnic populations to uncover novel loci associated with total IgE levels.
Keywords: immunoglobulin E, genome-wide association study, admixture mapping, allergy, asthma, next-generation sequencing, Latinos, Hispanics, minority populations
INTRODUCTION
Immunoglobulin E (IgE) is a class of antibody involved in host defense mechanisms against parasite infections.1,2 Elevated levels of total IgE are strongly associated with allergic disorders1 and asthma severity.3 Monoclonal antibodies against IgE have proven to be effective in decreasing asthma exacerbations and allergic inflammation,4,5 highlighting the importance of IgE in the etiology of both asthma and allergic disease.
The high heritability of IgE levels (47–80%),6–8 its clinical and biological relationship with atopic diseases, and the ability to measure IgE serum levels with a high precision, make IgE a useful endophenotype for the study of atopic diseases.1,9 Consequently, many linkage analyses and candidate-gene association studies have identified genes associated with IgE levels.9 Recently, five genome wide association studies (GWAS) have revealed single nucleotide polymorphisms (SNPs) near 12 genes to be associated with IgE levels at a genome-wide significance level, including six genes within the major histocompatibility complex (MHC).10–13
Only 1% of individuals within prior GWAS were Latinos,10–14 and therefore studying diverse populations may uncover some of the additional missing heritability of IgE level.15 This is particularly relevant for studies of IgE, as it is known to vary by race-ethnicity in the United States, with higher levels reported in both Latinos and African Americans as compared to European Americans.16–19 Differences in IgE levels among ethnic groups may in part be due to variation in the frequencies of risk alleles for high IgE, resulting from different proportions of Native American, European, and African genetic ancestry at an individual locus (local ancestry). Consistent with this, African ancestry has been associated with higher IgE levels in African descent populations,20,21 suggesting that admixture mapping could be a powerful tool to identify novel loci associated with IgE levels as shown for other traits and diseases.22–24 Therefore, in this study we reasoned that novel genetic variants associated with IgE levels in Latinos could be identified through a combination of GWAS and admixture mapping.
METHODS
Study Populations
Genes-environments & Admixture in Latino Americans (GALA II) Study
A total of 3,334 participants were used for discovery (Table E1). GALA II is an ongoing multicenter case-control study of asthma in Latino children (Puerto Rican, Mexican and other Latino) recruited from mainland U.S. and Puerto Rico, and approved by all local institutional review boards. All participants/parents provided written assent/consent, respectively.
Total serum IgE levels were measured in duplicate on the ImmunoCAP™ 100 system (Phadia, Kalamazooo, MI). Subjects were genotyped on the Axiom® LAT1 array (World Array 4, Affymetrix, Santa Clara, CA),25 as described elsewhere.23
The Genetics of Asthma in Latino Americans (GALA I) study
Characteristics of the 454 asthma cases from GALA I used for replication are shown in Table E1. Individuals were recruited from U.S. and Mexico City. Total IgE levels were measured as described in GALA II, and genotyping was performed on the Affymetrix 6.0 GeneChip.22
Additional Replication Studies
Further replication was performed in European and African American studies from the EVE Consortium with measured IgE (Table E2).13,26 African American studies included the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE), the Genetic Research on Asthma in the African Diaspora in African American individuals (GRAAD-AA) and in Afro-Caribbean from Barbados (GRAAD-AC), and the Chicago Asthma Genetics study, Collaborative Studies on the Genetics of Asthma, and Severe Asthma Research Program (CAG/CSGA/SARP). European American studies with measured IgE included the Childhood Asthma Management Program (CAMP), the Childhood Asthma Research and Education Network (CARE), and CAG/CSGA/SARP.
Statistical analysis
GWAS
Quality control was performed by removing SNPs that had genotyping call rates below 95%, and/or deviated from Hardy-Weinberg equilibrium (p<10−6) within controls. Samples with discrepancy between genetic sex and reported gender, and with cryptic relatedness (proportion of identity by descent >0.3) were also removed. Allelic association testing between genotypes at individual SNPs with natural log transformed total serum IgE levels was performed assuming an additive model using linear regression in R 2.14,27 adjusting for age, sex, ethnicity, asthma status, and African and Native American global ancestries. We also evaluated a model adjusting for local African and Native American ancestry. Regions with more than one SNP associated with IgE levels at p≤5x10−6 underwent in silico fine mapping using genotype imputation. For that, genotyped SNPs were phased using SHAPE-IT28 followed by imputation using IMPUTE229 considering all populations from the 1000 Genomes Project Phase I v3 as a reference.30 In addition, we evaluated whether SNPs associated with IgE levels were associated with related phenotypes (atopic dermatitis, asthma, rhinitis, and allergic sensitization), or showed an interaction with asthma status.
Admixture mapping
Estimates of local ancestry were obtained using LAMP-LD under a 3-population model,31 using reference haplotypes from the HapMap phase II CEU (European) and YRI (African), and 71 Native American individuals genotyped on the Axiom LAT1 array.23 Inferred local ancestry was then used to perform admixture mapping by correlating levels of local African, European, and Native American ancestry across the genome with natural log transformed IgE levels using linear regression and adjusting for age, sex, ethnicity, global ancestry, and asthma status. To study if overlapping peaks found for IgE levels and asthma were due to pleiotropic effects (i.e. loci influencing IgE and asthma together) and not confounded by linkage disequilibrium, we used a Bayesian network method implemented in the package bnlearn for R.32 See the Online Repository Material for more details.
To account for multiple testing the effective number of ancestry blocks was estimated using an empirical autoregression framework with the package coda for R. Based on the number of ancestry blocks in GALA II (1041), we established a Bonferroni p-value threshold for significance as α=0.05/1041=4.8x10−5, similar to Shriner et al.33 Statistically significant admixture mapping peaks were brought forth for replication in GALA I and the African American EVE Consortium studies using linear regression models. A linear mixed effects model was employed for one study with extended pedigree structure (GRAAD-AC).34 To assess the overall effect size of local ancestry across studies, a meta-analysis was performed using METASOFT. 35
Fine mapping of significant admixture mapping peaks was performed using allelic association testing using both genotyped and imputed SNPs. A Bonferroni correction based on the number of genotyped SNPs within each peak was used to provide a peak-wide threshold of significance. Peak-wide significant SNPs identified in GALA II were brought for replication in the rest of studies. For Latinos and African Americans we adjusted for global ancestry estimates, and for European Americans we adjusted for two principal components from a principal component analysis.26
Functional annotation of associated SNPs
Functional annotation of associated SNPs and enhancer enrichment analysis was carried out with HaploReg.36 Expression quantitative trait loci (eQTLs) were identified querying the Geuvadis Data Browser.37
Exon Sequencing
We sequenced the coding and UTR exons of three genes, including one gene identified through GWAS (the zinc finger protein 365 [ZNF365]), and two genes encoding human leukocyte antigens (HLA) identified through fine mapping of admixture mapping peaks (HLA-DQB1 and HLA-DRB5 within 6p21–22). All Puerto Ricans from the GALA II study included in the study were sequenced (n=1,454). Exons were targeted using Molecular Inversion Probe (MIP) technology as described elsewhere.38,39 Targeted regions were sequenced on a MiSeq sequencer (Illumina Inc., San Diego, CA) using 150 bp reads. Output reads were aligned to the human genome (hg19) with BWA (Burrows-Wheeler Aligner),40 and variant calling was performed using GATK (Genome Analysis Toolkit).41,42 Variants were annotated using SeattleSeq (http://snp.gs.washington.edu).
Association testing was performed using linear regression models for individual variants using PLINK.43 Given that single-variant tests may have reduced statistical power to detect an association with rare variants we also analyzed the cumulative association of multiple rare variants with IgE levels using the Sequence Kernel Association Test (SKAT-O), adjusted by the same covariates as for individual allelic association testing.44 Sets of pooled nonsynonymous, synonymous, and non-coding variants were analyzed separately.
A flow chart with the study design is shown in Figure E1 and additional information on Methods can be found in the in the Online Repository Material.
RESULTS
GWAS
Manhattan plots of GWAS results for unadjusted models vs. models adjusted by local genetic ancestry are shown in Figures E2A and E2B, respectively. The quantile-quantile plot for an unadjusted model (Figure E2C) showed higher but still minimal genomic inflation (λGC=1.05) than the model adjusted for local ancestry (λGC=1.02, Figure E2D). Several SNPs within a 50 Kb window at 10p21.2 had p≤5x10−6 in the model adjusted by local ancestry (Table E3). Fine mapping of this region by imputing variants from the 1000 Genomes Project revealed a common insertion-deletion (indel) in ZNF365 that reached genome-wide significance (rs200076616, β=−0.23 for the insertion allele, p=2.3x10−8) (Figure 1). This indel was significantly associated with IgE levels in all three sub-groups of Latinos from GALA II: Mexicans (β=−0.25, p=9.2x10−5), Puerto Ricans (β=−0.21, p=8.4x10−4), and other Latinos (β=−0.21, p=0.020). However, the association was in the opposite direction in GALA I (β=0.22, p=0.038), and failed to replicate in African Americans and European Americans from the EVE consortium studies (β=−0.07, p=0.090 and β=−0.06, p=0.281, respectively). A summary of SNPs with p≤5x10−6 in models unadjusted by local ancestry is shown in Table E4.
Figure 1.
Fine mapping of ZNF365 in GALA II. Included in the plot are -log10(p-values) of association with total serum IgE by chromosome position. The most significant SNP is represented by a diamond and the results for the remaining SNPs are color coded to show their linkage disequilibrium (LD) with this SNP based on pairwise r2 values from the Latino populations of the 1000 Genomes Project. SNPs above the dashed line are genome-wide significant (p≤5x10−8).
We next tested for replication of 23 SNPs associated with IgE levels in prior GWAS.10–14 Fifteen of these SNPs were similarly associated with IgE in GALA II at nominal significance (p<0.05): 13 were associated in the same direction as the original study, one was associated in the opposite direction, and for one of them the direction could not be ascertained from the original study (Table E5). Nonetheless, only variants in RAD50, HLA-DRB1, and STAT6 were significantly associated with IgE levels following a Bonferroni correction (p≤2.2x10−3). However, when looking at the gene-level (±5 Kb) we observed a significant association after Bonferroni correction at a different SNP than initially reported for IL13 (min p=7.7x10−5, rs1295686), HLA-DRB1 (min p=1.1x10−7 for rs41284732), HLA-DQB1 (min p=3.5x10−6 for rs9273395), and IL4R/IL21R (min p=1.5x10−6,rs3024667) (Table E6). We did not find any associated SNPs within or nearby DARC, FCER1A, HLA-G, or HLA-G after correcting for multiple comparisons (Table E6).
Admixture Mapping
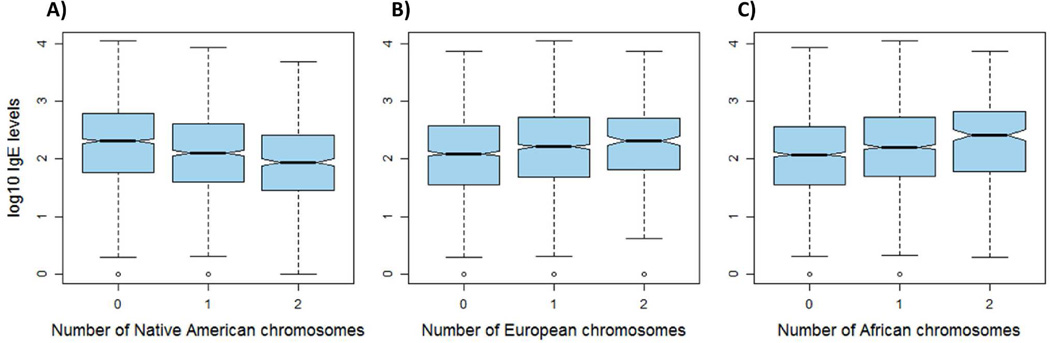
We identified 4 admixture mapping peaks whereby local genetic ancestry was significantly associated with IgE levels (p≤4.8x10−5) (Table 1). Additional peaks showing a suggestive association are listed in Table E7. Our most significant peak was located at 6p21.32-p22.1 within the MHC region (Figure E3A–C). The minimum p-value of the peak was located at rs3094691 in the HLA-B gene, where Native American ancestry was associated with lower levels of total IgE (β=−0.10, p=4.95x10−8), and European and African ancestries were associated with higher IgE levels (β=0.06, p=4.6x10−3 for European ancestry and β=0.07, p=1.6x10−4 for African ancestry) (Figure 2A–C). The top of the peak was located 300 Kb away from an admixture mapping peak for asthma previously identified in the same study population (GALA II).23 A stratified analysis of asthma cases and controls showed that the association of Native American ancestry with IgE at 6p21.32-p22.1 was independent of asthma status, as the effects were similar in both cases and controls (β=−0.23, p=4.8x10−5 for cases and β=−0.21, p=3.8x10−4 for controls). We further examined whether the association of Native American ancestry with IgE at 6p21.33 was mediating our prior association with asthma in the same region.23 Including IgE as a covariate in association testing for asthma yielded a result that was still significant (OR=0.83, 95%CI: 0.74–0.93, p=1.6x10−3 versus OR=0.78, 95%CI: 0.69–0.87, p=1.7x10−5, for the adjusted and unadjusted models, respectively). Furthermore, a Bayesian network analysis32 confirmed that the MHC region contains two independent associations with asthma and IgE, despite these associations are correlated through linkage disequilibrium (Figure E4). The three additional admixture mapping peaks detected included an association between local African ancestry and higher levels of IgE at 14q23.2 (β=0.25, p=3.0x10−5), and local African ancestry with lower levels of IgE at 13p22.1-q31.3 (β=−0.35, p=1.0x10−6), and 22q13.1 (β=−0.69, p=1.9x10−5) (Table 1).
Table 1.
Results of admixture mapping in GALA II and replication in GALA I. A total of four significant admixture mapping peaks were identified in GALA II, of which three were significantly replicated in the same direction and with the same ancestry in GALA I.
| Discovery GALA II | Replication GALA I | ||||||
|---|---|---|---|---|---|---|---|
| Band | Ancestry | Beta | 95% CI Beta | p-value | Beta | 95% CI Beta | p-value |
| 6p21.32- 22.1 | Native | −0.23 | −0.32-(−0.14) | 5.0x10−8 | −0.56 | −0.96-(−0.16) | 5.7x10−3 |
| African | 0.25 | 0.12–0.39 | 2.1x10−5 | 0.71 | 0.28–1.14 | 1.3x10−3 | |
| 13p22.1-q31.3 | African | −0.35 | −0.48-(−0.21) | 1.0x10−6 | −0.85 | −1.43-(−0.27) | 3.8x10−3 |
| European | 0.30 | 0.16–0.43 | 1.8x10−6 | 0.43 | 0.05–0.80 | 0.025 | |
| 14q23.2 | African | 0.25 | 0.12–0.39 | 3.0x10−5 | 0.80 | 0.20–1.40 | 9.1x10−3 |
| 22q13.1 | African | −0.69 | −1.01−(−0.37) | 1.9x10−5 | 0.35 | −0.25−0.94 | 0.252 |
Significant p-values following Bonferroni correction for multiple tests are in boldface.
CI: confidence interval.
Figure 2.
Boxplot showing the distribution of IgE levels for each copy of a chromosome of A) Native American, B) European, and C) African ancestry at 6p21.33.
Three of the four admixture mapping peaks were replicated in GALA I following a Bonferroni correction (4 peaks: p<0.013): 6p21.32-p22.1 (p=5.7x10−3 and p=1.3x10−3, for Native American and African ancestry, respectively), 13p22.1–q31.3 (p=3.8x10−3), and 14q23.2 (p=9.1x10−3) (Table 1). We then brought forward the three peaks showing positive replication in GALA I for replication in African Americans from the EVE studies and used a Bonferroni correction for the number of peaks tested (3 peaks: p<0.017). The peaks at 6p21.32-p22.1 and 14q23.2 were replicated in the combined analysis of 3,187 African American individuals from the EVE studies (p≤0.017, Table E8).
Fine mapping of admixture mapping peaks
We next performed fine mapping of three admixture mapping peaks that showed significant replication in GALA I (6p21.32-p22.1, 13p22.1-q31.3, and 14q23.2) via allelic association testing. A total of 29 SNPs within 6p21.32-p22.1 were significantly associated with IgE following Bonferroni correction for the number of genotyped SNPs within the admixture mapping peak (peak-wide significant, p≤8x10−6) (Table E9). Further fine mapping using imputed genotypes revealed 90 additional associated SNPs. A total of 39 of the 119 associated SNPs showed nominal replication in the same direction in GALA I (Table E10) and patterns of linkage disequilibrium were similar between GALA I and GALA II within this region (Figure E5). One of the imputed SNPs, located 21 kb 5' of HLA-DQA1, was genome-wide significant in GALA II (rs1846190, p=3.5x10−8, Figure 3) and replicated in the same direction in GALA I (p=0.003, meta-p=4.6x10−10). Among the 39 replicated SNPs, five additional SNPs reached genome-wide significance when the results from GALA II and GALA I were combined (Table 2). Two of these SNPs were further replicated in European Americans (rs113176001, p=0.010 and rs9270747, p=0.016), but not in African Americans (Table E10).
Figure 3.
Fine mapping results of 6p21.32-p22.1 in GALA II, focusing on the region showing genome-wide significant allelic associations. Included in the plot are -log10(p-values) of association with total serum IgE by chromosome position. The most significant SNP is represented by a diamond and the results for the remaining SNPs are color coded to show their linkage disequilibrium (LD) with this SNP based on pairwise r2 values from the Latino populations of the 1000 Genomes Project. SNPs above the dashed line are genome-wide significant (p≤5x10−8).
Table 2.
Summary of SNPs identified as genome-wide significantly associated with IgE through fine mapping of an admixture mapping peak at 6p21.32-p22.1.
| GALA II | GALA I | Meta-analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| rs # | Positiona | Gene / closest gene |
Allele 1/ Allele 2b |
Beta (95% CI) | p-value | Beta (95% CI) | p-value | Beta (95% CI) | p-value |
| rs41284732 | 32551427 | HLA-DRB1 | T/C | −0.27 (−0.37 – (−0.17)) | 1.06 x10−7 | −0.30 (−0.53 – (−0.07)) | 0.010 | −0.27 (−0.36 – (−0.18)) | 3.45x10−9 |
| rs186715542 | 32558702 | 3’ of HLA-DRB1 | C/T | 0.24 (0.15 – 0.33) | 4.96 x10−7 | 0.26 (0.02 – 0.50) | 0.034 | 0.25 (0.16 – 0.33) | 4.89x10−8 |
| rs9270747 | 32568292 | 3’ of HLA-DRB1 | A/G | −0.19 (−0.27 – (−0.11)) | 3.42 x10−6 | −0.33 (−0.53 – (−0.13)) | 0.002 | −0.21 (−0.29 – (−0.14)) | 3.56x10−8 |
| rs113176001 | 32571483 | 5’ of HLA-DRB1 | G/C | −0.29 (−0.41 – (−0.17)) | 1.03 x10−6 | −0.36 (−0.63 – (−0.09)) | 0.008 | −0.30 (−0.41 – (−0.20)) | 3.04x10−8 |
| rs1846190 | 32583813 | 5’ of HLA-DQA1 | G/A | 0.22 (0.14 – 0.30) | 3.51 x10−8 | 0.29 (0.10 – 0.48) | 0.003 | 0.23 (0.16 – 0.30) | 4.63x10−10 |
| rs557011 | 32587013 | 5’ of HLA-DQA1 | C/T | 0.21 (0.13 – 0.29) | 1.28 x10−7 | 0.27 (0.07 – 0.47) | 0.007 | 0.21 (0.14 – 0.29) | 3.55x10−9 |
p-values≤0.05 are in boldface.
According to the hg19 assembly
Allele 1 represents the allele for which effect size is reported. CI: confidence interval.
The SNP rs113176001 is located 14kb 5' of HLA-DRB1, within histone marks (H3k4me1, H3k9me3, H3k04me1, H3k27ac, and H3k79me2) in lymphoblastoid cells GM12878 identified by the ENCODE project, and it is in a region which acts as a strong enhancer based on the chromatin state segmentation.45 Furthermore, rs113176001 was previously identified as a strong eQTL for several proximal genes in Europeans from the 1000 Genomes Project (min p=3.2x10−17 for HLA-DQA1) (Table E11), but not in individuals of African descent. Rs9270747 is also a strong eQTL for several proximal genes (min p=1.8x10−60 for HLA-DRB5) (Table E11).
Within 13p22.1-q31.3 a total of 6 SNPs were peak-wide significantly associated with IgE levels in GALA II (min p=3.9x10−6, Table E9 and E12, Figure E6), but none of them were replicated in the rest of studies. However, these SNPs showed high variability in allele frequency across studies/populations (Figure E7). Furthermore, 4 of the SNPs had low quality imputation scores in one or more studies (info score <0.4).
Allelic association testing of SNPs within 14q23.2 in GALA II did not reveal any significantly associated genotyped SNPs following Bonferroni correction (Table E9). The most significant SNP was rs2012961 (p=1.4x10−4) (Figure E8), which is located in the 3'-UTR of the ras homolog family member J gene (RHOJ) and coincides with the top of the admixture mapping peak. This SNP was replicated in GALA I (p=0.019), but not in African Americans or European Americans in EVE (Table E12). Imputation in GALA II revealed an additional SNP that was slightly more significant than the strongest associated genotyped SNP (rs10129357, p=1.2x10−4 vs. p=1.4x10−4 for rs2012961). The imputed SNP was subsequently replicated in European Americans in EVE (p=0.024).
We then tested for allelic association within the admixture mapping peaks identified in GALA II in African Americans and European Americans, under the hypothesis that causal variants may be better tagged by different SNPs in other populations (Table E13). In African Americans, an intronic SNP within 14q23.2 was peak-wide significantly associated with IgE levels following Bonferroni correction for 6,511 tests (rs1741099, β=0.17, p=3.7x10−6). The association was replicated in all of the non-African American samples combined (β=0.08, p=0.011). The SNP rs1741099 is located in intron 1 within the gene encoding the glycoprotein hormone beta 5 (GPHB5) (Figure E9), and is 23 Kb away from rs2012961, the most associated SNP in Latinos. We did not find any significant associations in European Americans after adjusting for multiple tests (min p=1.1x10−4 for rs10483752 at the potassium voltage-gated channel, subfamily H [eag-related], member 5 [KCNH5]) (Table E13).
Association analysis with related phenotypes
SNPs from STAT6 and RAD50 were nominally associated with asthma, and SNPs from HLA-DQA1 were nominally associated with atopy and rhinitis (Table E14). The SNP in GPHB5 was nominally associated with asthma, eczema, and allergic rhinitis (Table E14). One SNP from IL4R/IL21R locus showed an interaction with asthma status at p<0.05 (Table E15).
Exon sequencing of Candidate Genes
Exon sequencing of HLA-DQB1, HLA-DRB5, and ZNF365 was performed in 1,395 Puerto Ricans from GALA II (856 asthma cases and 539 controls). Gene-based association testing revealed a significant contribution of multiple nonsynonymous variants to IgE levels in HLA-DQB1 (SKAT-O test, p=2.1x10−3, Figure E10), but not in HLA-DRB5 (p=1.0) or ZNF365 (p=0.245). No additional single variants were identified as being more strongly associated with IgE levels than those already identified through GWAS and imputation.
DISCUSSION
In this study we performed a GWAS of IgE levels in Latinos, and identified a novel genome-wide significant allelic association at ZNF365. We also found that the majority of prior associations identified from GWAS primarily in European populations, were applicable to Latino populations at the SNP and/or gene-based level. We further performed the first admixture mapping of IgE levels and identified a strong association between local ancestry at the MHC region and total serum IgE levels, which was replicated in Latinos from GALA I and in African Americans from the EVE consortium. Allelic association testing within the MHC peak in GALA II revealed several SNPs associated with IgE levels, which replicated in GALA I, and six of which were genome-wide significant in either GALA II alone or in a meta-analysis of the two studies. Two additional admixture mapping peaks identified in GALA II were replicated in GALA I (13p22.1-q31.3 and 14q23.2), and one of these peaks was further replicated in African Americans (14q23.2). Fine mapping of these two regions revealed allelic associations within the KCNH5-RHOJ-GPHB5 genes at 14q23.2 in a combination of Latinos, European and African Americans.
We performed admixture mapping in addition to a conventional GWAS, and identified additional associations with IgE. We predicted this would be a powerful approach because of the observed differences in IgE levels among Latinos, African Americans, and European Americans,16–19 and the association between genomic African ancestry and IgE levels.20,21 Our results demonstrate that local ancestry can capture additional, important genetic variation that is not attainable through conventional GWAS. As expected, individual alleles showing an association with IgE levels within the admixture mapping peaks had an enormous variation in minor allele frequency across populations (Figure E7). Exon sequencing further identified an association of pooled nonsynonymous variants with IgE levels in a gene implicated through admixture mapping (HLA-DQB1), but not in a gene implicated through conventional GWAS (ZNF365). Thus far our results suggest that the association detected with local ancestry at 6p21.32-p22.1 is driven by a combination of both common and rare variation.
ZNF365 encodes a protein involved in mitosis and genomic stability.46 Genetic variation in ZNF365 was associated with atopic dermatitis in Japanese and Chinese populations,47,48 but not in Europeans.49 Atopic dermatitis is an allergic disease modulated through IgE, and therefore our association with IgE reinforces the role of ZNF365 in atopic phenotypes. However, we note that the association of the genome-wide significant indel in ZNF365 was in the opposite direction in GALA I compared to GALA II. Allele frequencies were similar among studies, imputation performed well (info score ≥0.9), and we found no significant interaction with asthma. Therefore, our result could be attributed to differences in recruitment of patients or gene-environment interactions. The associated indel is located in the first intron of transcript variant D of ZNF365: an isoform whose differential expression has been previously associated with Crohn’s disease.50 The combined set of variants within the first intron of ZNF365 transcript variant D with p<10−6 showed a 2.6-fold enrichment in enhancers in the lymphoblastoid cell line GM12878 (p=0.027), and a 6.7-fold enrichment in DNase sensitivity sites in CD20+ B cells (p=0.036). Therefore, we speculate that variation associated with IgE in the first intron of ZNF365 affects the expression and/or splicing of ZNF365, and may ultimately affect recombination processes needed for the differentiation of B cells in IgE-producing cells as has been proposed for RAD50.51
We identified genome-wide significant associations through admixture mapping and fine mapping in the MHC region. Although this region was already known to contain genes that influence IgE levels,10, 11, 13 in this study we identified an important role of local genetic ancestry. In addition, our genome-wide significant associations are different from those previously described in GWAS involving European populations10,11,13 or Asian populations.14 Moreover, for the first time we implicated variation in HLA-DQA1 in affecting IgE levels. The most significantly associated SNP (rs113176001), is located within the intergenic region between HLA-DRB1 and HLA-DQA1, and is a strong eQTL for HLA-DQA1. Interestingly, this SNP was only an eQTL in lymphoblastoid cells in Europeans, but not in African individuals from the 1000 Genomes Project. Furthermore, it was not significantly associated with IgE levels in African Americans from the EVE consortium, although the direction of effect was the same. This could be due to variation in allele frequency between populations, as the minor allele of this SNP is more frequent in Latino and European populations from the 1000 Genomes Project (25% and 22% for the C allele, respectively) than in African populations (14%) (Figure E3).
Using admixture mapping we also uncovered an association of African ancestry at 13p22.1-q31.3 with lower levels of IgE. Fine mapping revealed significant allelic associations in GALA II that failed to replicate in GALA I, perhaps suggesting the presence of false positive associations, but may also be due to reduced statistical power from the smaller sample size in GALA I. African ancestry at 14q23.2 was associated with higher IgE levels both in Latinos and African Americans. Linkage studies previously identified 14q23 as a locus affecting IgE levels.52,53 However no associations of specific genes have been reported. Fine mapping of 14q23 in multiethnic groups narrowed the association to three candidate genes: KCNH5 and RHOJ through variants nominally associated in Latinos, and GPHB5 in African Americans. These three genes are expressed by immune cells such us T lymphocytes, B cells, and plasma cells, but they have not been associated before with IgE levels or allergic phenotypes. Therefore, the role of genetic variation in KCNH5, RHOJ, and GPHB5 on IgE levels requires additional study.
An important aspect of our study is that fine mapping of candidate regions using imputed genotypes from the 1000 Genomes project boosted our ability to detect novel associations. However, a GWAS using imputed data did not reveal any additional associations from those already identified in our fine mapping of regions identified by GWAS/admixture mapping (data not shown). One limitation of our study is that we did not account for possible interactions between genotype and early-life exposure to allergens. In addition, IgE levels were measured using different methods across the replication studies, which may account for the lack of the replication of some of the findings. Furthermore, we note that Latinos are a heterogeneous group and therefore findings from this study may not be generalizable to all Latino populations. Finally, future studies will be needed to disentangle the association at the MHC region by evaluating the association of classic alleles. However, better reference panels for HLA imputation need to be developed for Latino populations.
In summary, we confirmed associations at six loci previously associated with total serum IgE levels, and identified novel at ZNF365, HLA-DQA1, and 14q23.2, all of which are likely to play an important role in total serum IgE levels in Latinos. Sequencing of GWAS and admixture mapping peaks including non-coding regions followed by functional studies is required to identify the causal variation behind these associations.
Supplementary Material
Key Messages.
-
-
Although differences in IgE levels have been reported between racial/ethnic groups, genome-wide association studies have predominantly focused on populations of European descent.
-
-
We performed a genome-wide association study (GWAS) and admixture mapping in Latinos and found novel associations at three genes/loci including ZNF365, HLA-DQA1, and 14q23.2.
-
-
Our results demonstrate how GWAS and admixture mapping in diverse populations can identify novel genetic variation that is relevant to the health of global populations.
ACKNOWLEDGMENTS
The authors acknowledge the patients, families, numerous, recruiters, health care providers and community clinics for their participation. In particular, the authors thank Sandra Salazar for her support as GALA study coordinator and Choli Lee for his help with exonic sequencing.. Some computations were performed using the UCSF Biostatistics High Performance Computing System.
Sources of funding:
This work was supported by grants from National Institutes of Health: the National Heart, Lung and Blood Institute (HL088133, HL078885, HL004464, HL104608 and HL117004 to E.G.B; HL101651 to C.O.; HL093023 to R.K.; HL075419, HL65899, HL083069, HL066289, HL087680, HL101543, and HL101651 to S.T.W.; and HL111636 to J.M.G.); the National Institute of Environmental Health Sciences (ES015794 to E.G.B.); the National Institute on Minority Health and Health Disparities (MD006902 to E.G.B.); the National Institute of General Medical Sciences (GM007546 to E.G.B. and J.M.G; GM007175 to C.R.G.); the National Institute of Allergy and Infection Diseases (AI079139 and AI061774 to L.K.W); the National Cancer Institute (contract HHSN26120080001E by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research to C.A.W.); General Clinical Research Centers Program (M01-RR00188 to the Texas Children's Hospital General Clinical Research Center); the National Center for Advancing Translational Sciences (KL2TR000143 to J.M.G.). Other sources of funding included: Fundación Ramón Areces (M.P.Y.); American Asthma Foundation (E.E.E., L.K.W. and E.G.B.); RWJF Amos Medical Faculty Development Award (E.G.B.); the Sandler Foundation (E.G.B.); the Mary Beryl Patch Turnbull Scholar Program (K.C.B.); Ernest S. Bazley Grant (P.C.A); UCSF Chancellor’s Research and Dissertation Year Fellowships (C.R.G.); Hewett Fellowship (J.M.G); the Howard Hughes Medical Institute (E.E.E). The content of this publication is solely the responsibility of the authors and does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- CAG
Chicago Asthma Genetics Study
- CAMP
Childhood Asthma Management Program
- CARE
Childhood Asthma Research and Education
- CI
confidence interval
- CSGA
Collaborative Studies of the Genetics of Asthma
- eQTL
expression quantitative trait loci
- GALA II
Genes-environments &Admixture in Latino Americans
- GALA I
Genetics of Asthma in Latino Americans
- GPHB5
glycoprotein hormone beta 5
- GRAAD
Genetic Research on Asthma in the African Diaspora
- GWAS
genome wide association study
- HLA
human leukocyte antigen
- IgE
Immunoglobulin E
- KCNH5
potassium voltage-gated channel, subfamily H [eag-related], member 5 gene
- MHC
major histocompatibility complex
- MIP
Molecular Inversion Probe
- OR
odds ratio
- RHOJ
ras homolog family member J gene
- SAPPHIRE
Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity
- SARP
Severe Asthma Research Program
- SNP
Single nucleotide polymorphism
- SKAT
Sequence Kernel Association Test
- U.S.
United States
- ZNF365
zinc finger protein 365 gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Weidinger S, Baurecht H, Naumann A, Novak N. Genome-wide association studies on IgE regulation: are genetics of IgE also genetics of atopic disease? Curr Opin Allergy Clin Immunol. 2010;10:408–417. doi: 10.1097/ACI.0b013e32833d7d2d. [DOI] [PubMed] [Google Scholar]
- 2.Flohr C, Quinnell RJ, Britton J. Do helminth parasites protect against atopy and allergic disease? Clin Exp Allergy. 2009;39:20–32. doi: 10.1111/j.1365-2222.2008.03134.x. [DOI] [PubMed] [Google Scholar]
- 3.Naqvi M, Choudhry S, Tsai HJ, Thyne S, Navarro D, Nazario S, et al. Association between IgE levels and asthma severity among African American, Mexican, and Puerto Rican patients with asthma. J Allergy Clin Immunol. 2007;120:137–143. doi: 10.1016/j.jaci.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 4.Busse W, Corren J, Lanier BQ, McAlary M, Fowler-Taylor A, Cioppa GD, et al. Omalizumab, anti-IgE recombinant humanized monoclonal antibody, for the treatment of severe allergic asthma. J Allergy Clin Immunol. 2001;108:184–190. doi: 10.1067/mai.2001.117880. [DOI] [PubMed] [Google Scholar]
- 5.Lanier B, Bridges T, Kulus M, Taylor AF, Berhane I, Vidaurre CF. Omalizumab for the treatment of exacerbations in children with inadequately controlled allergic (IgE-mediated) asthma. J Allergy Clin Immunol. 2009;124:1210–1216. doi: 10.1016/j.jaci.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Palmer LJ, Burton PR, Faux JA, James AL, Musk AW, Cookson WO. Independent inheritance of serum immunoglobulin E concentrations and airway responsiveness. Am J Respir Crit Care Med. 2000;161:1836–1843. doi: 10.1164/ajrccm.161.6.9805104. [DOI] [PubMed] [Google Scholar]
- 7.Hopp RJ, Bewtra AK, Watt GD, Nair NM, Townley RG. Genetic analysis of allergic disease in twins. J Allergy Clin Immunol. 1984;73:265–270. doi: 10.1016/s0091-6749(84)80018-4. [DOI] [PubMed] [Google Scholar]
- 8.Hanson B, McGue M, Roitman-Johnson B, Segal NL, Bouchard TJ, Jr, Blumenthal MN. Atopic disease and immunoglobulin E in twins reared apart and together. Am J Hum Genet. 1991;48:873–879. [PMC free article] [PubMed] [Google Scholar]
- 9.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 10.Granada M, Wilk JB, Tuzova M, Strachan DP, Weidinger S, Albrecht E, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol. 2012;129:840–845. doi: 10.1016/j.jaci.2011.09.029. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin AM, Mathias RA, Huang L, Roth LA, Daley D, Myers RA, et al. A meta-analysis of genome-wide association studies for serum total IgE in diverse study populations. J Allergy Clin Immunol. 2012;131:1176–1184. doi: 10.1016/j.jaci.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yatagai Y, Sakamoto T, Masuko H, Kaneko Y, Yamada H, Iijima H, et al. Genome-Wide Association Study for Levels of Total Serum IgE Identifies HLA-C in a Japanese Population. PLoS One. 2013;8:e80941. doi: 10.1371/journal.pone.0080941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JJ, Burchard EG, Choudhry S, Johnson CC, Ownby DR, Favro D, et al. Differences in allergic sensitization by self-reported race and genetic ancestry. J Allergy Clin Immunol. 2008;122:820–827. doi: 10.1016/j.jaci.2008.07.044. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gergen PJ, Arbes SJ, Jr, Calatroni A, Mitchell HE, Zeldin DC. Total IgE levels and asthma prevalence in the US population: results from the National Health and Nutrition Examination Survey 2005–2006. J Allergy Clin Immunol. 2009;124:447–453. doi: 10.1016/j.jaci.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litonjua AA, Celedon JC, Hausmann J, Nikolov M, Sredl D, Ryan L, et al. Variation in total and specific IgE: effects of ethnicity and socioeconomic status. J Allergy Clin Immunol. 2005;115:751–757. doi: 10.1016/j.jaci.2004.12.1138. [DOI] [PubMed] [Google Scholar]
- 19.Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, et al. Ethnic differences in asthma and associated phenotypes: collaborative study on the genetics of asthma. J Allergy Clin Immunol. 2001;108:357–362. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- 20.Vergara C, Caraballo L, Mercado D, Jimenez S, Rojas W, Rafaels N, et al. African ancestry is associated with risk of asthma and high total serum IgE in a population from the Caribbean Coast of Colombia. Hum Genet. 2009;125:565–579. doi: 10.1007/s00439-009-0649-2. [DOI] [PubMed] [Google Scholar]
- 21.Vergara C, Murray T, Rafaels N, Lewis R, Campbell M, Foster C, et al. African ancestry is a risk factor for asthma and high total IgE levels in African admixed populations. Genet Epidemiol. 2013;37:393–401. doi: 10.1002/gepi.21702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torgerson DG, Gignoux CR, Galanter JM, Drake KA, Roth LA, Eng C, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012;130:76–82. doi: 10.1016/j.jaci.2012.02.040. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galanter JM, Gignoux CR, Torgerson DG, Roth LA, Eng C, Oh SS, et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: The Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol. 2014;134:295–305. doi: 10.1016/j.jaci.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiner AP, Beleza S, Franceschini N, Auer PL, Robinson JG, Kooperberg C, et al. Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. Am J Hum Genet. 2012;91:502–512. doi: 10.1016/j.ajhg.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann TJ, Zhan Y, Kvale MN, Hesselson SE, Gollub J, Iribarren C, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–430. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: A Language and Environment for Statistical Computing. Austria: Viena; 2008. [Google Scholar]
- 28.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baran Y, Pasaniuc B, Sankararaman S, Torgerson DG, Gignoux C, Eng C, et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics. 2012;28:1359–1367. doi: 10.1093/bioinformatics/bts144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagarajan R, Scutari M, Lèbre S. Bayesian Networks in R with Applications in Systems Biology. Springer eBooks; 2013. [Google Scholar]
- 33.Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS Comput Biol. 2011;7:e1002325. doi: 10.1371/journal.pcbi.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Park E, Kim CS, Paik JH. ZNF365 promotes stalled replication forks recovery to maintain genome stability. Cell Cycle. 2013;12:2817–2828. doi: 10.4161/cc.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 48.Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–694. doi: 10.1038/ng.851. [DOI] [PubMed] [Google Scholar]
- 49.Weidinger S, Willis-Owen SA, Kamatani Y, Baurecht H, Morar N, Liang L, et al. A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haritunians T, Jones MR, McGovern DP, Shih DQ, Barrett RJ, Derkowski C, et al. Variants in ZNF365 isoform D are associated with Crohn's disease. Gut. 2011;60:1060–1067. doi: 10.1136/gut.2010.227256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potaczek DP, Kabesch M. Current concepts of IgE regulation and impact of genetic determinants. Clin Exp Allergy. 2012;42:852–871. doi: 10.1111/j.1365-2222.2011.03953.x. [DOI] [PubMed] [Google Scholar]
- 52.Mansur AH, Bishop DT, Markham AF, Morton NE, Holgate ST, Morrison JF. Suggestive evidence for genetic linkage between IgE phenotypes and chromosome 14q markers. Am J Respir Crit Care Med. 1999;159:1796–1802. doi: 10.1164/ajrccm.159.6.9804036. [DOI] [PubMed] [Google Scholar]
- 53.Mansur AH, Bishop DT, Holgate ST, Markham AF, Morrison JF. Linkage/association study of a locus modulating total serum IgE on chromosome 14q13-24 in families with asthma. Thorax. 2004;59:876–882. doi: 10.1136/thx.2003.014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.