Abstract
Important viral and cellular gene products are regulated by stop codon readthrough and mRNA frameshifting, processes whereby the ribosome detours from the reading frame defined by three nucleotide codons after initiation of translation. In the last few years, rapid progress has been made in mechanistically characterizing both processes and also revealing that trans-acting factors play important regulatory roles in frameshifting. Here, we review recent biophysical studies that bring new molecular insights to stop codon readthrough and frameshifting. Lastly, we consider whether there may be common mechanistic themes in −1 and +1 frameshifting based on recent X-ray crystal structures of +1 frameshift-prone tRNAs bound to the ribosome.
Keywords: ribosome, protein synthesis, frameshifting, RNA structure, translocation
The ribosome is a macromolecular machine that performs protein synthesis, one of the most conserved cellular functions in all organisms. Translating the information encoded on the messenger RNA (mRNA) template into amino acids is a highly complex but coordinated process with four defined stages - initiation, elongation, termination and recycling (for review see reference (1)). Initiation is the most regulated step with the reading frame of the mRNA defined by the AUG start codon. The three letter mRNA template or codon is read by a complementary three nucleotide anticodon of transfer RNAs (tRNAs) during the elongation phase until protein factors recognize stop codons to terminate translation. Although much is known about protein synthesis, it remains unclear how the ribosome preserves or deviates in a controlled manner from a three nucleotide reading frame.
A feature that distinguishes the ribosome from DNA or RNA polymerases is that the polynucleotide template is transferred into an entirely different chemical, a polypeptide. DNA replication is tightly regulated for accurate inheritance of genetic information with proofreading repair mechanisms to minimize errors (2). RNA polymerase also has proofreading capability and in addition, mRNA degradation pathways that eliminate incorrectly synthesized mRNAs (3, 4). The ribosome is the most error-prone polymerase with error rates estimated at 10−3 – 10−4 per amino acid incorporated, at least for the most error-prone codon-anticodon pairs (5–7). The most common translational errors are missense or tRNA miscoding errors but non-initiation, stop codon readthrough and mRNA reading frame errors do occur, although at lower frequencies (8). All of these types of errors likely develop from noncanonical interactions between the mRNA codon and the tRNA anticodon, whose cognate interactions are required for accurate tRNA selection and movement of tRNAs through the ribosome during elongation.
The term “frameshift” or “recoding” refers to a shift in the three-nucleotide mRNA coding sequence, resulting in a movement in either the 5’ or the 3’ direction that produces an amino acid sequence distinct from the wild-type or 0 frame sequence (Fig. 1). Frameshift errors are predicted to occur rarely, ~1 out of 30,000 decoding events (9) while programmed frameshift events occur much more frequently, in some cases 50–80% of the expressed protein (10). Ribosomes that depart from the correct reading frame frequently encounter stop codons that abort translation and thereby, prevent the expression of the incorrect protein sequence (11). Because frameshifting usually produces a truncated, non-functional polypeptide, it is more deleterious than missense errors wherein a single incorrect amino acid is incorporated. The first evidence that ribosomes could decode a non three nucleotide codon was revealed over forty years ago where extragenic suppressors to insertions and deletions of the genetic code were found to be primarily located in tRNA genes, indicating that they functioned by altering protein synthesis (12–14). Suppressors-of-frameshift tRNAs do not cause a translational ‘error’, but rather they return the mRNA back to the 0 frame to allow wild-type protein expression. The ability of the ribosome to decode in alternate mRNA frames, that is, +1 or −1 frames (mRNA movement towards the 5’ or 3’ ends), provided evidence the genetic code was not immutable. Though frameshifting in viruses was demonstrated to be one way to regulate the expression of essential proteins in a defined ratio and in some retroviruses, to maximize limited genomes, this regulatory mechanism also occurs in bacteria and eukaryotes, demonstrating frameshifting as a universal gene expression mechanism (15–18).
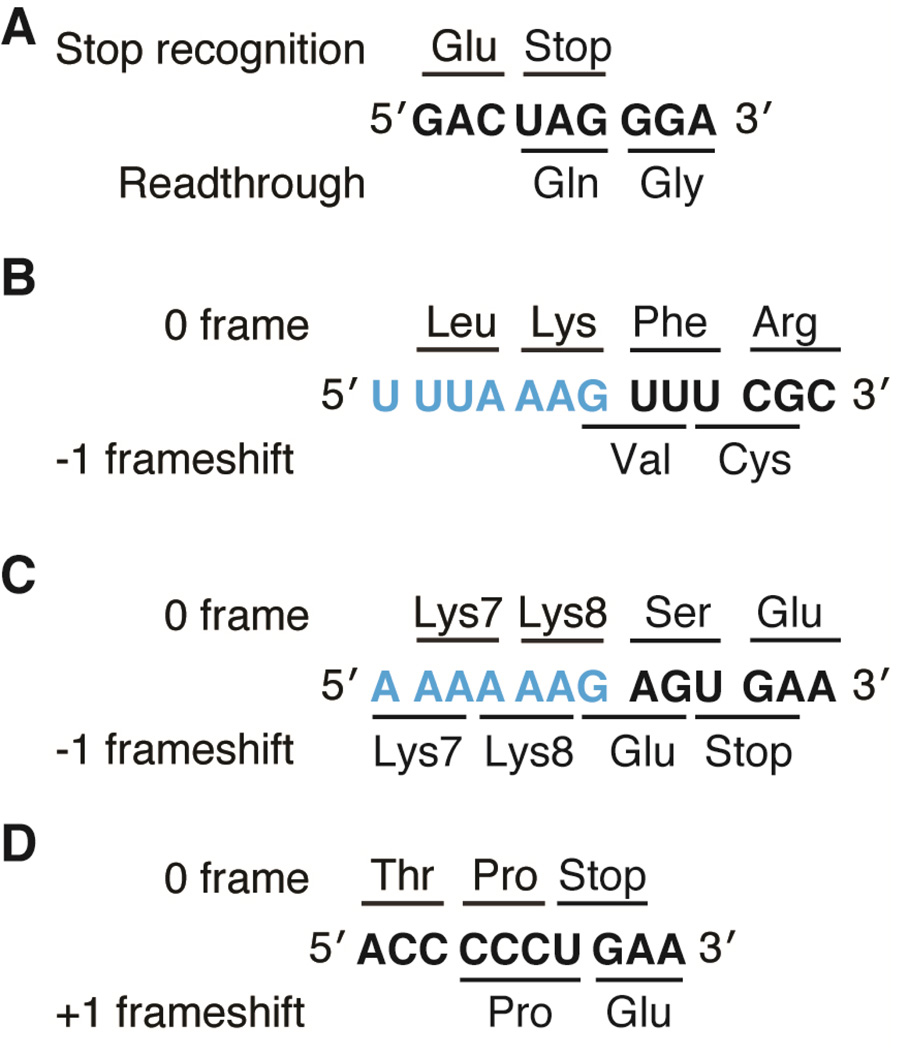
Figure 1. Examples of recoding events in viral and cellular genes.
A) MLV stop codon readthrough required for expression of the pol gene. B) An example of −1 frameshifting on the slippery sequence (blue) of the IBV 1a/1b gene (note wild-type sequence is 5’-U UUA AAC-3’) and C) on the slippery sequence (blue) of the dnaX gene. D) +1 frameshifting by frameshift suppressor tRNASufA6 on hisD3018 mRNA.
Genetic and molecular biology approaches have provided a wealth of data regarding the identification and causes of frameshifting but a defined molecular mechanism remains elusive. For example, where is the slippery sequence of the mRNA located on the ribosome when a frameshift occurs and what step of the elongation cycle does the frameshift affect? Are certain tRNAs prone to frameshifting and do tRNA modifications play a role in the stability of the mRNA-tRNA pair and thus canonical decoding properties? Can a concise model be developed to explain both viral and cellular frameshifts and can this model be extended to explain frameshifting in both directions on the mRNA? Here, we report on recent breakthroughs using biophysical and structural approaches to dissect the molecular mechanisms of +1 and −1 frameshifting. These studies explore the different origins of the shift and provide mechanistic insights that reveal potential commonalities between the different types of frameshifting.
mRNA conformational plasticity in the recoding of retrovirus messages
Programmed frameshifting was first identified in the Rous Sarcoma Virus (RSV) and explained how the gag structural gene followed by a pol enzymatic gene in a −1 reading frame produces a single Gag-Pol fusion polyprotein (19). Programmed frameshifts were subsequently discovered in diverse viruses where mRNA shifting was required to control the expression levels of Gag and Pol during viral replication (20–22). A typical −1 recoding mRNA signal consists of a ‘slippery’ nucleotide sequence usually defined as a run of the same nucleotides, followed by a base-paired mRNA, either a simple stem loop or a more complex pseudoknot structure proximal to the mRNA entrance tunnel (22–24). How the structured mRNA region influences the elongation process is unknown but it is thought to act synergistically with the slippery sequence to direct translation into a new frame (22).
In a subtle twist on this theme, the Murine Leukemia Virus (MLV) Pol protein is expressed by stop codon readthrough of the gag gene (25) (Fig. 1A). To determine the molecular basis of mRNA recoding required for Pol expression in MLV, the 3’ pseudoknot was characterized by NMR (26). Two conformational populations of the pseudoknot formed at physiological pH; their relative abundance could be influenced by either changing the pH or mutating the RNA to favor one state over the other. These two states were proposed to represent stop codon readthrough permissive or restrictive states that the ribosome encounters during translation. The relative abundance of each state also correlated with the extent of frameshifting in vivo. For example, the pseudoknot adopts the readthrough permissive state 6% of the time, which corresponds to the in vivo stop codon readthrough rate (26, 27). This study provides compelling evidence for how cis-acting RNA elements direct the frame, but several important issues remain. First, it is unclear if the structure solved in the absence of the ribosome recapitulates the conformations of the pseudoknot RNA when it is in the vicinity of the mRNA entrance channel (26). Second, since the proposed mechanism requires that the ribosome encounter one of the two conformational states during reading of the mRNA, this interconversion between the two states is required to be slower than the rate of translation. Future experiments to address how conformational switching affects frameshifting in the context of the ribosome are required to better understand this phenomenon.
Pre-steady state kinetic investigations of viral −1 frameshifting
Infectious Bronchitis Virus (IBV) undergoes high levels of −1 frameshifting at the 1a/1b gene containing a conserved slippery sequence of the type 5′-U UUA AAC-3′ (where the spacings separate three nucleotide codons in the 0 frame) (Fig. 1B). The IBV frameshift signal was the first recoding signal discovered containing a 3’ pseudoknot structure that promotes frameshifting (23). Frameshifting by the IBV signal can be recapitulated in the context of both bacterial and eukaryotic ribosomes implying functional conservation of ribosomal features required for this type of recoding (28). Recently, pre-steady state kinetic analysis of an E. coli in vitro translation system revealed the mechanism of the IBV frameshift signal using radiolabeled aminoacyl groups on tRNAs to monitor the nascent peptide chain elongation, along with fluorescent tRNAs or ribosomes to separately uncover the dynamics (29) (Figs. 2A & 2B). Unexpectedly, the amino acid incorporation rate during reading of the slippery sequence was found to be unaffected. However, both the rate of peptide bond formation for two overlapping downstream amino acids, corresponding to the 0 and −1 frame and proximal to the slippery sequence, were substantially slower. Surprisingly, the incorporation of the −1 frame Val (codon GUU; first G nucleotide is from the AAG codon of the slippery sequence) occurred at a faster rate than the 0 frame Phe (codon UUU; after the AAG slippery sequence codon), indicating movement into the −1 frame was the preferred elongation path (Fig. 1B). The overall slower rate of incorporation of either residue was coincidental with the IBV pseudoknot reaching the edge of mRNA entrance tunnel where unfolding would be required for the single stranded mRNA to enter. These studies demonstrated that the IBV frameshift occurs either during translocation of the codon following the slippery sequence or during accommodation of the downstream tRNA, again following the slippery sequence. The possibility that a shift into the −1 frame occurs during translocation was previously suggested from cryo-EM studies of the 80S bound to the IBV frameshift signal (not containing the slippery sequence) where eukaryotic elongation factor 2 (bacterial EF-G) was modeled bound (30).
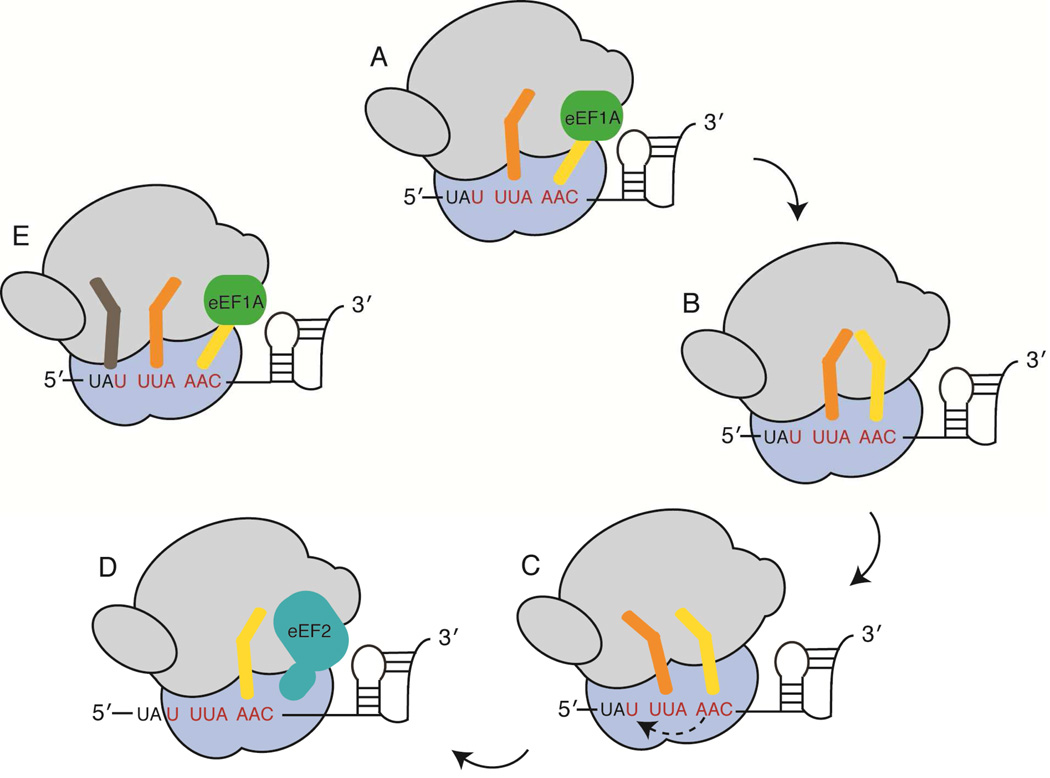
Figure 2. Selected probes used in recent biophysical studies of −1 programmed frameshifting.
A) Multiple conformational changes of ribosome components are necessary for mRNA-tRNA translocation during the elongation cycle. Opening and closing of the L1 arm is presumed to be associated with mRNA-tRNA translocation. The small ribosomal subunit body (b), head (h) and platform (pl) domains can move independently of one another while rotation of the small subunit (blue) relative to the large subunit (gray) and an orthogonal swiveling of the head domain are necessary for mRNA-tRNA translocation (30S shoulder domain closing is not shown). B) Radiolabeled methionine along with fluorescently labeled (purple star) tRNA (P-site tRNA is orange; A-site tRNA is yellow), EF-G and ribosomal proteins (S13, purple) were used to follow the mechanistic sub-steps of −1 programmed frameshifting on the Infectious Bronchitis Virus (IBV) pseudoknot frameshift signal (29). C) Fluorescent and fluorescence quencher oligonucleotides were hybridized to 16S rRNA helix 44 and 23S rRNA Helix 101 to follow small subunit rotation. tRNA or elongation factor arrival during translation of the dnaX frameshift signal was also followed by fluorescence (46). D) Qin et al. used multiple fluorescently labeled ribosomal proteins (S6 and L9 depicted in purple) to identify conformational states of the ribosome while programmed with the dnaX frameshift signal (45). E) A study by Kim et al. used smFRET between tRNA and ribosomal protein L1 to follow peptide bond formation and tRNA translocation on the dnaX frameshift signal (44).
Additional kinetic studies reveal no change in the A-site incorporation rate of fluorescent Lys-tRNALys suggesting the −1 shift follows this step. These data argue against the ‘9 Å’, ‘simultaneous slippage’ and the ‘three tRNA’ models which posit that viral −1 frameshifting occurs during tRNA accommodation at the second codon of the slippery sequence, i.e. with tRNALys decoding AAG in the IBV 1a/1b gene (31–33) (Figs. 1B and 3). Of the two remaining models, the ‘dynamic’ and ‘mechanical’ models (30, 34, 35) propose frameshifting occurs during translocation of the second and third codons (1a/1b gene codons UUA and AAG that encode for Leu and Lys residues, respectively) (Figs. 3C & 3D). An additional possibility is that the process of tRNA decoding downstream of the slippery sequence at either Val (−1 frame; GUU codon) or Phe (0 frame; UUU codon) plays a role in re-setting the reading frame. To test the latter, varying concentrations of Val-tRNAVal or Phe-tRNAPhe were added to programmed ribosome complexes which would be predicted to influence frameshift efficiencies. No effect on the frameshift efficiencies was observed, strongly suggesting that the reading frame was changed during movement of the preceding codon, that is, translocation of the slippery sequence UUA AAG from the P and A sites, to the E and P sites of the ribosome, respectively.
Figure 3. Models for −1 programmed frameshifting.
It has been proposed that frameshifting occurs A) during accommodation of aminoacyl tRNA on the third codon of the slippery sequence (‘9 Å’ model); B) after tRNA accommodation but before peptide bond formation (‘simultaneous slippage’ model); C) while tRNAs are in the hybrid state, which coincides with rotation of the small subunit (‘dynamic’ model); D) during eEF2 catalyzed formation of the post translocation state (‘mechanical’ model); E) during tRNA accommodation but with E- and P-site tRNAs in a noncanonical post translocation state. The organization of the figure is adapted from Brierley et al. (22) with the large subunit shown in grey, the small subunit in blue and the slippery mRNA sequence in red.
Translation is a dynamic process wherein the ribosome adopts multiple intermediate states in a stepwise manner to move the tRNAs through the three binding sites (36–39). Elongation involves rotation of the body and platform domains of the small ribosomal subunit (30S), swiveling of the 30S head domain, closing of the 30S shoulder and movement of the L1 protein on the large ribosomal subunit (50S) from an open to a closed state (37, 40) (Fig. 2A). These movements all contribute to at least four degrees of freedom that the ribosome samples, with each state being populated to varying degrees (41). Previously it was shown that when ribosomes encounter mRNAs containing stem loops or pseudoknots, ribosomal translocation or E-site tRNA ejection are compromised, depending upon the relative strength of the structured mRNA (42). Translation factor EF-G catalyzes translocation of mRNA-tRNA pairs through the ribosome, hydrolyzing GTP in the process. To dissect the sub-steps of the translocation process, fluorescently labeled EF-G, ribosomes or tRNAs were used by Caliskan et al. to demonstrate that the ribosome adopts two intermediate translocation states called POST1 and POST2 during movement of the slippery sequence, before adopting the canonical post-translocation state, POST3. These experiments indicate that the ribosomes commit to the frameshifting pathway early in the translocation process and that movement of the ribosome into the POST2 then POST3 state occurs more rapidly for ribosomes that switch to the −1 reading frame. These experiments further provide convincing evidence that frameshifting occurs during translocation and suggests that the ‘mechanical’ model best approximates how the IBV 1a/1b mRNA undergoes high levels of frameshifting (Fig. 3D) (30, 34).
Single molecule FRET analysis of the bacterial dnaX −1 frameshift signal
The E. coli dnaX gene exhibits high levels of −1 frameshifting required for expression of the γ subunit of DNA polymerase III while the 0 frame expresses the DNA polymerase III τ subunit (43). As with the IBV frameshift signal, the dnaX gene contains a slippery sequence located upstream of a base-paired RNA element. However, there are three differences between the mRNAs: the dnaX structured RNA is likely a stem loop rather than a pseudoknot; the dnaX mRNA contains an internal Shine-Dalgarno-like sequence upstream of the slippery sequence that enhances the frameshift efficiency (10); and the dnaX slippery sequence is 5’-A AAA AAG-3’ rather than the canonical −1 viral mRNA sequence of 5’-X XXY YYZ (where X, Y and Z represent different nucleotides) (Fig. 1C). Three separate groups have recently reported on how the dnaX mRNA promotes −1 frameshifting using single molecule Förster resonance energy transfer (smFRET) approaches (44–46).
Chen et al. used smFRET to address the timing of the frameshift event using fluorescent donor and quencher dyes attached to rRNA of both the 30S and the 50S subunits (Fig. 2C) (46). The ability to monitor the entire elongation cycle over multiple amino acid incorporations, including decoding of the slippery sequence, provided unprecedented mechanistic insights. The placement of the dyes allowed for monitoring of the relative subunit movements, which cycle from a non-rotated state (EF-Tu-aminoacyl-tRNA-GTP ternary complex delivery, accommodation and peptide bond formation), to a rotated state (EF-G binding and translocation and tRNA and 30S head domain movement towards the E site), and back to a non-rotated state (backward 30S body/platform rotation and 30S head domain swivel), approximating one round of elongation. These studies revealed that 75% of ribosomes undergo a −1 frameshift and have a 10-fold longer pause following decoding of the second codon of the slippery sequence (Lys7) as compared to the 25% of ribosomes that remain in the 0 frame (Fig. 2C). The longer pause is indicative of ribosomes in a rotated state waiting for EF-G to bind and translocate the mRNA-tRNA pairs. Analysis of the behavior of these two populations demonstrated that pausing in the rotated state at the Lys7 codon is concomitant with the 75% of ribosomes that underwent frameshifting. These data are interpreted to indicate the initial kinetic branch point determining the −1 or 0 reading frame is set before pausing, and is controlled by either formation of the upstream Shine-Dalgarno- antiShine-Dalgarno-like interaction or the conformation of the downstream RNA stem loop.
Analysis of the paused, rotated state revealed that EF-G binds and dissociates multiple times in competition with EF-Tu-Lys-tRNALys-GTP ternary complex sampling the AAG (Lys8) codon (Fig. 1C). Because the efficiency of frameshifting is dependent on the identity of the position 8 codon (46), the authors conclude that this continual A-site sampling by tRNALys defines the reading frame. In summary, these data support either the ‘9 Å’ or ‘three-tRNA’ models by which tRNA sampling promotes a shift in the mRNA frame at the third slippery codon. An extension of this model is that this sampling is a consequence of tRNALys being unable to recognize a rotated 30S stuck in a paused state, consistent with incomplete swiveling of the 30S head domain.
Using complementary smFRET analyses with a defined programmed ribosome, two additional studies investigated how the dnaX mRNA stem loop affects the ribosome’s conformational states (44, 45). Qin et al. used smFRET donor-acceptor pairs that tracked ribosomal protein S6 movement relative to protein L9 to define the amount of ribosomes in rotated or non-rotated states (Figs. 2A & 2D). When ribosomes are poised for mRNA-tRNA translocation, they can sample a rotated (hybrid) state and a non-rotated state. While it was established that multiple states of 30S subunit rotation are populated by the ribosome (36, 39, 47), Qin et al. found that the dnaX mRNA induces a previously unseen 30S conformation, termed the ‘hyper-rotated’ state, where the 30S is rotated by 22° from the non-rotated state (45). The hyper-rotated state is dependent on both the presence of the stem loop and the nucleotide spacing between it and the A-site codon; however the hyper-rotated state is independent of A-site tRNA occupancy. This study further demonstrated the hyper-rotated state reduces the dynamic transitions between the translocation sub-states possibly explaining the long pause previously seen in the rotated state observed by Chen et al. (46). Small angle x-ray scattering (SAXS), a technique that can define the low-resolution structure of a macromolecule, corroborates the formation of the hyper-rotated state deduced from FRET distances. Using FRET probes, the position of the L1 region of the ribosome was also followed (Fig. 2A). While L1 normally adopts a closed conformation on a rotated ribosome, FRET analysis of hyper-rotated ribosomes revealed L1 was in an open position. The L1 region was previously shown to be important in mRNA frame maintenance and an open L1 may explain the unexpected observation that E-site tRNA dissociates during the paused rotated state observed by Chen et al. (46, 48, 49) or upon the ribosome encountering a base-paired mRNA element at the mRNA entrance tunnel (42).
FRET probes positioned on tRNAs and L1 were used to follow the amount and inter-conversion rates of conformational states of the ribosome in the presence of the dnaX stem loop (44). When the ribosome was bound to the dnaX stem loop but in the absence of EF-G, destabilization of the rotated state occurred (defined as a high L1-tRNA FRET signal) while increasing the population of the non-rotated state (low L1-tRNA FRET signal). This interpretation is opposite to that of the previous two studies but may be reconciled if an open L1 conformation coexists with a rotated 30S subunit in the presence of the dnaX mRNA as seen by Qin et al. One possible reason for these discrepancies may be that L1 stalk dynamics are being monitored which may not directly correlate to subunit rotation. Upon addition of EF-G, similar ribosome behavior is observed as in the other smFRET study. Namely, dwell times in the hybrid state were increased with mRNA-tRNA translocation and adoption of a post-translocation state accumulates slowly. The authors speculate the dnaX stem loop promotes frameshifting by slowing translocation to allow more time to sample upstream conformations that allow slipping on the mRNA in the −1 direction. Additional experiments will be needed to understand precisely how ribosome conformations and dynamics control frameshifting but the studies described show smFRET will be a powerful tool in unraveling the mechanism of frameshifting.
Structural studies highlight the important roles tRNAs play in frameshifting
The best studied +1 frameshift examples are promoted by tRNAs that contain nucleotide insertions or deletions, lack nucleotide modifications or act under conditions where their cellular concentrations are altered (for review see reference (50)). The decoding of a three nucleotide codon by frameshift suppressor tRNAs is not a programmed gene expression event, but rather, this noncanonical decoding is a mechanism by which the wild-type protein amino acid sequence is preserved. Frameshift suppressor tRNAs have been invaluable tools to understand the general function of the ribosome despite the lack of clear molecular basis for their action. Recent structural studies shed light on the molecular basis of action of frameshift suppressor tRNAs that contain anticodon stem loop (ASL) insertions and also the roles of tRNA modifications in maintaining the reading frame (51, 52). Although models to explain −1 and +1 frameshifting have been restricted to each class (50, 53–56), single molecule and structural studies are beginning to provide molecular insights that indicate that each may not be as distinct as previously thought (29, 44–46, 51, 52).
Frameshift suppressor tRNASufA6, a derivative of tRNAProCGG, contains an insertion 3’ of the anticodon (after position 37). The absence of a N1-methyl (m1) base modification at G37 in tRNAProCGG is known to cause increased levels of +1 frameshifting (57, 58). tRNASufA6 stimulates frameshifting on sites with the codon signature CCC-N (where “N” denotes any nucleotide and is read as a proline CCC codon) (Fig. 1D), and multiple structures of the 70S ribosome were solved with the ASLs of tRNASufA6 decoding the following proline codons (CCC) containing an additional nucleotide decoded as part of the codon: CCC-U, CCC-G and CCC-A (51). In each case, two Watson-Crick base pairs form between the codon and anticodon, a non Watson-Crick C-C base pair forms at the third (or wobble) position, and no interaction occurs with the downstream nucleotide U/G/A that is part of the four nucleotide codon. These results establish a +1 slip of the reading frame likely occurs after completion of tRNA selection in the A site, either during mRNA-tRNA translocation to the P site, or within the P site itself. These structures also revealed that the U32-A38 base pair, which sits at the top of the anticodon loop, is disrupted in the context of structures containing a +1 frameshift-prone tRNA and corresponding four nucleotide codons (Fig. 4). Interestingly, the 32–38 interaction is restored when a +1 frameshift-prone tRNA interacts with a cognate codon that supports 0 frame decoding, signifying both the frameshift suppressor tRNA and a specific four nucleotide codon are required for the frameshift event (51). Interactions between this region of the ASL, specifically nucleotide 32, with either EF-G during translocation (59) or with the ribosome in the P site (60) suggest that tRNASufA6 may interact with EF-G in a noncanonical manner, resulting in incomplete or slowed translocation (51) (Fig. 4B). An additional possibility is that normal translocation occurs, but that the 32–38 pairing is not gripped tightly within the P site, which may facilitate movement between the anticodon and codon into a different frame (50, 56, 61).
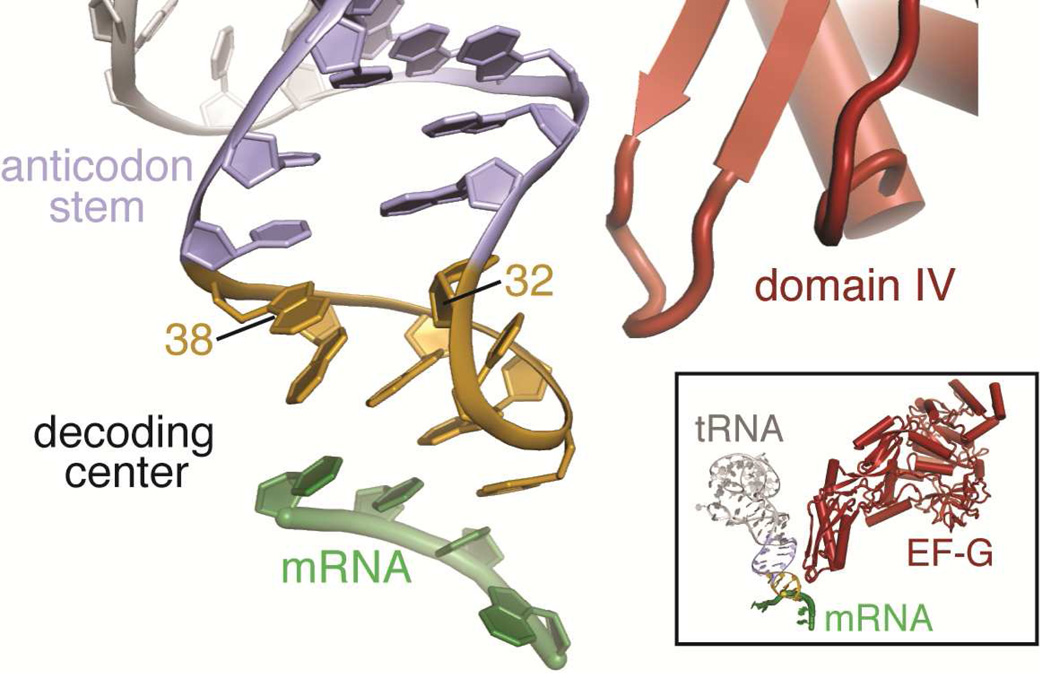
Figure 4. Base pairing at 32–38 of the ASL is important for frame maintenance.
Base pairing within the anticodon stem loop of tRNA (stem, purple; anticodon loop, yellow) is important for decoding of the mRNA codon (green). X-ray crystal structures of +1 frameshift-prone tRNAs bound to the 70S ribosome reveal disruption of the 32–38 pair. Cryo-electron microscopy particle reconstructions have shown domain IV of EF-G (red) contacts the ASL in the vicinity of the 32–38 base pair before translocation in the A site, suggesting how the 32–38 base pair may be linked to frameshifting (59).
Most tRNAs contain post-transcriptional modifications on nucleotide 37, adjacent to the anticodon, which influences frameshifting (62). Therefore 70S structures of ASLProCGG with and without the m1G37 modification bound to its cognate codon CCG were solved to determine the structural basis for this phenomenon. Surprisingly, the lack of the m1G37 modification in ASLProCGG also disrupts the 32–38 base pair (51). These data suggest a common mechanism underlying +1 frameshifting by frameshift-prone tRNAs and modification-deficient tRNAs.
Discovery of Programmed Frameshifting Mediated by Trans-acting Factors
Recent identification of protein and microRNA-induced frameshifting events indicate the possibility that there are many unknown factors that redirect the mRNA reading frame (63–65). This is not surprising given that −1 frameshifting sites were predicted to be widespread in the genomes of eukaryotes (66). However, validation of these studies reveals a new dimension in gene regulation.
The reasons viruses utilize frameshifting is clear: frameshifting allows for an important regulatory switch independent of the expression of additional factors, while also increasing viral genome density to encode multiple polypeptides per open reading frame. However, a high-density genome is not required for eukaryotic cells, therefore it is unclear why frameshifting would exist in eukaryotes. This point is further underscored by that fact that most computationally predicted −1 frameshift signals direct ribosomes to encounter stop codons that truncate the protein shortly after the shift (65). However, trans-activation of a −1 frameshifting signal would offer an additional layer of regulatory complexity of translational control. The recent identification of a microRNA (miRNA)-activated −1 frameshift site in the human CCR5 gene provides the first example of frameshifting by a trans-activating RNA molecule (65).
Putative miRNA sites within the CCR5 −1 frameshift signal suggested that the signal may be regulated in a RNA-mediated manner (65). The putative CCR5 −1 frameshift signal is similar to viral frameshift signals: a slippery sequence and a downstream pseudoknot located adjacent to the mRNA entrance tunnel. This study demonstrated that miR-1224 promoted −1 frameshifting by direct interaction with the CCR5 −1 frameshift signal. The authors also concluded that frameshifting is, in this case, not a mechanism to express a different protein, but rather, a mechanism by which the ribosome may be directed to an adjacent stop codon, stimulating the nonsense mediated decay of the CCR5 transcript. It is still unclear why mRNA degradation via −1 frameshifting would be initiated as a mechanism to control protein expression, but the authors propose a number of scenarios related to circumventing viral invasion as possibilities.
Another example of trans-activation of frameshifting was recently identified in which a viral protein redirects the mRNA frame. Again, prediction of a possible frameshifting site in the Porcine Reproductive and Respiratory Syndrome Virus (PRRV) was the starting point for these experiments (63). The first indication that this gene may undergo potentially novel regulation was the extremely high synonymous-site conservation in the 3′ region of the viral nsp2 coding region, and the absence of out-of-frame stop codons, typically representative of alternative, overlapping protein coding regions. The second indication was that the 3′ region of nsp2 revealed additional similarities to viral −1 frameshifting sites with a slippery site followed by a conserved sequence spaced 10 nucleotides downstream. However, other features were uncharacteristic of typical −1 viral frameshift signals; for example, ribosomes read a +1 mRNA reading frame and there was no predicted structured RNA region in the conserved downstream element (63).
Translation initiation provides three potential reading frames for any mRNA: the −1, 0 and +1 reading frames. In this study, an unusual −2 shift by the ribosome (which is macroscopically equivalent to a +1 shift) was confirmed by mass spectrometry analysis, the first demonstration of a −2 frameshift in a viral system (63). Interestingly, −1 frameshifting at the same site also occurred. This type of protein expression strategy results in three distinct C-terminal polypeptides appended to nsp2. The conserved nature of these events is underscored by the introduction of synonymous codon mutations within the frameshift signal that cause a 50 - 100 fold decrease in the efficiency of viral replication. Further experiments defined that deletion of an adjacent nsp1β gene controlled nsp2 frameshifting by direct binding of the nsp1β protein to the conserved downstream nsp2 element (64). These studies demonstrated for the first time a protein acting in trans to control a programmed frameshift event.
Converging Mechanistic Themes Across Diverse Frameshift Events
In a single year, tremendous mechanistic insights have advanced our understanding of what drives ribosomal frameshifting. While the ‘mechanical model’ best describes the behavior of the IBV 1a/1b frameshift signal (29, 30, 34), studies of the dnaX frameshift signal indicate aminoacyl-tRNA sampling of the A site plays a significant role in defining the mRNA frame (33, 46). Although both studies conclude that the frameshift signal causes noncanonical translocation behavior (29, 46), each supports a different model. Future experiments using different FRET probe sets, pre-steady state kinetics and structural biology approaches will help further define the models and provide additional important insights into how structured RNAs influence gene expression. Another important question will be in understanding the role of the hyper-rotated ribosomal state (45) and its impact on paused ribosomes.
Both +1 frameshift suppressor tRNAs and certain programmed −1 frameshift signals (e.g. IBV 1a/1b frameshift signal) function in the context of both prokaryotic and eukaryotic ribosomes, arguing the frameshift inducer is manipulating universally conserved features of the ribosome. Such conserved features include the ribosomal RNA core, adjacent to or forming direct interactions with mRNA, tRNAs and elongation factors. We reason that while the mechanisms of +1 frameshifting and −1 or −2 programmed frameshifting could be distinct, a common theme is beginning to emerge where frameshifting is promoted by altering conserved interactions of the substrate with the ribosome. Ribosome structure and dynamics studies conclude that the translocation step of elongation plays an important role in setting the mRNA frame (29, 44, 46, 51, 52). These studies indicate that conformational rearrangements of the ribosome required for mRNA-tRNA translocation are unable to adopt canonical positions in the context of a frameshifting element. Swiveling of the 30S head domain accomplishes the crucial hand-off of ASL-mRNA to the 30S P and E sites (38). Both +1 and −1 frameshifting may result from noncanonical interactions of ASL and mRNA with the 30S head domain during translocation.
Highlights.
We review +1 and −1 frameshifting promoted by viral and cellular mRNAs and tRNAs.
Frameshifting is an important regulatory mechanism for gene expression.
Single molecule and structural studies have provided insights into the mechanism of frameshifting.
One common theme is that frameshifting occurs during mRNA-tRNA translocation.
Data suggest similarities among different types of frameshifting may exist.
ACKNOWLEDGEMENTS
Research is supported by the National Institute of General Medical Sciences of the NIH under award number R01GM093278 and the Pew Scholar in the Biomedical Sciences Program (CMD). We thank Graeme L. Conn for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461(7268):1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19(6):732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136(4):763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Bouadloun F, Donner D, Kurland CG. Codon-specific missense errors in vivo. Embo J. 1983;2(8):1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelmann P, Gallant J. Mistranslation in E. coli. Cell. 1977;10(1):131–137. doi: 10.1016/0092-8674(77)90147-7. [DOI] [PubMed] [Google Scholar]
- 7.Manickam N, Nag N, Abbasi A, Patel K, Farabaugh PJ. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014;20(1):9–15. doi: 10.1261/rna.039792.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen F, Kurland CG. Processivity errors of gene expression in Escherichia coli. Journal of molecular biology. 1990;215(4):511–521. doi: 10.1016/S0022-2836(05)80164-0. [DOI] [PubMed] [Google Scholar]
- 10.Larsen B, Gesteland RF, Atkins JF. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50% Journal of molecular biology. 1997;271(1):47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manley JL, Gesteland RF. Suppression of amber mutants in vitro induced by low temperature. Journal of molecular biology. 1978;125(4):433–447. doi: 10.1016/0022-2836(78)90309-1. [DOI] [PubMed] [Google Scholar]
- 12.Riddle DL, Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol. 1973;242(121):230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- 13.Riddle DL, Roth JR. Frameshift suppressors. 3. Effects of suppressor mutations on transfer RNA. Journal of molecular biology. 1972;66(3):495–506. doi: 10.1016/0022-2836(72)90429-9. [DOI] [PubMed] [Google Scholar]
- 14.Riddle DL, Roth JR. Frameshift suppressors. II. Genetic mapping and dominance studies. Journal of molecular biology. 1972;66(3):483–493. doi: 10.1016/0022-2836(72)90428-7. [DOI] [PubMed] [Google Scholar]
- 15.Flower AM, McHenry CS. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990;87(10):3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 17.O'Connor M, Gesteland RF, Atkins JF. tRNA hopping: enhancement by an expanded anticodon. EMBO J. 1989;8(13):4315–4323. doi: 10.1002/j.1460-2075.1989.tb08618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsufuji S, Matsufuji T, Wills NM, Gesteland RF, Atkins JF. Reading two bases twice: mammalian antizyme frameshifting in yeast. EMBO J. 1996;15(6):1360–1370. [PMC free article] [PubMed] [Google Scholar]
- 19.Jacks T, Varmus HE. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein KM, Goff SP. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988;62(6):2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol. 2001;75(4):1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brierley I, Gilbert RJ, Pennell S. Pseudoknot-dependent Programmed-1 Ribosomal Frameshifting: Structures, Mechanisms and Models. In: Atkins JF, Gesteland RF, editors. Recoding: Expansoin of Decoding Rules Enriches Gene Expression, Nucleic Acids and Molecular Biology. New York; Dordrecht; Heidelberg; London: Springer; 2009. pp. 149–174. [Google Scholar]
- 23.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ten Dam EB, Pleij CW, Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus genes. 1990;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286(2):187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 26.Houck-Loomis B, et al. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature. doi: 10.1038/nature10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci U S A. 1985;82(6):1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brierley I, Meredith MR, Bloys AJ, Hagervall TG. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. Journal of molecular biology. 1997;270(3):360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. Programmed-1 Frameshifting by Kinetic Partitioning during Impeded Translocation. Cell. 2014;157(7):1619–1631. doi: 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441(7090):244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plant EP, et al. The 9-A solution: how mRNA pseudoknots promote efficient programmed-1 ribosomal frameshifting. RNA. 2003;9(2):168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacks T, et al. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 33.Leger M, Dulude D, Steinberg SV, Brakier-Gingras L. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed-1 ribosomal frameshift. Nucleic Acids Res. 2007;35(16):5581–5592. doi: 10.1093/nar/gkm578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss RB, Dunn DM, Shuh M, Atkins JF, Gesteland RF. E. coli ribosomes rephase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989;1(2):159–169. [PubMed] [Google Scholar]
- 35.Farabaugh PJ. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 36.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466(7304):329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 37.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325(5943):1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A. 2012;109(50):20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annual review of biophysics. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- 42.Chen C, et al. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat Struct Mol Biol. 2013;20(5):582–588. doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990;87(7):2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HK, et al. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci U S A. 2014;111(15):5538–5543. doi: 10.1073/pnas.1403457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin P, Yu D, Zuo X, Cornish PV. Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 2014;15(2):185–190. doi: 10.1002/embr.201337762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, et al. Dynamic pathways of-1 translational frameshifting. Nature. 2014 doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406(6793):318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 48.Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13(9):1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bekaert M, Rousset JP. An extended signal involved in eukaryotic-1 frameshifting operates through modification of the E site tRNA. Mol Cell. 2005;17(1):61–68. doi: 10.1016/j.molcel.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atkins JF, Bjork GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73(1):178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc Natl Acad Sci U S A. 2014;111(35):12740–12745. doi: 10.1073/pnas.1409436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fagan CE, Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA. 2014 doi: 10.1261/rna.046953.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley interdisciplinary reviews. RNA. 2012;3(5):661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tinoco I, Jr, Kim HK, Yan S. Frameshifting dynamics. Biopolymers. 2013;99(12):1147–1166. doi: 10.1002/bip.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus research. 2009;139(2):193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farabaugh PJ. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog Nucleic Acid Res Mol Biol. 2000;64:131–170. doi: 10.1016/s0079-6603(00)64004-7. [DOI] [PubMed] [Google Scholar]
- 57.Qian Q, et al. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol Cell. 1998;1(4):471–482. doi: 10.1016/s1097-2765(00)80048-9. [DOI] [PubMed] [Google Scholar]
- 58.Hagervall TG, Tuohy TM, Atkins JF, Bjork GR. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. Journal of molecular biology. 1993;232(3):756–765. doi: 10.1006/jmbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- 59.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A. 2013;110(52):20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 61.Nasvall SJ, Nilsson K, Bjork GR. The ribosomal grip of the peptidyl-tRNA is critical for reading frame maintenance. Journal of molecular biology. 2009;385(2):350–367. doi: 10.1016/j.jmb.2008.10.069. [DOI] [PubMed] [Google Scholar]
- 62.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. Journal of molecular biology. 2007;366(1):1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 63.Fang Y, et al. Efficient-2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci U S A. 2012;109(43):E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, et al. Transactivation of programmed ribosomal frameshifting by a viral protein. Proc Natl Acad Sci U S A. 2014;111(21):E2172–E2181. doi: 10.1073/pnas.1321930111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Belew AT, et al. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014;512(7514):265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belew AT, Hepler NL, Jacobs JL, Dinman JD. PRFdb: a database of computationally predicted eukaryotic programmed-1 ribosomal frameshift signals. BMC Genomics. 2008;9:339. doi: 10.1186/1471-2164-9-339. [DOI] [PMC free article] [PubMed] [Google Scholar]