Abstract
Isolation of monoclonal antibodies (MAbs) elicited by vaccination provides opportunities to define the development of effective immunity. Ab responses elicited by current HIV-1 envelope glycoprotein (Env) immunogens display narrow neutralizing activity with limited capacity to block infection by tier 2 viruses. Intense work in the field suggests that improved Env immunogens are forthcoming and it is therefore important to concurrently develop approaches to investigate the quality of vaccine-elicited responses at a higher level of resolution. Here, we cloned a representative set of MAbs elicited by a model Env immunogen in rhesus macaques and comprehensively characterized their genetic and functional properties. The MAbs were genetically diverse, even within groups of Abs targeting the same sub-region of Env, consistent with a highly polyclonal response. MAbs directed against two sub-determinants of Env, the CD4 binding site (CD4bs) and the V3 region, could in part account for the neutralizing activity observed in the plasma of the animal from which they were cloned, demonstrating the power of MAb isolation for a detailed understanding of the elicited response. Finally, through comparative analyses of MAb binding and neutralizing capacity of HIV-1 using matched Envs, we demonstrate complex relationships between epitope recognition and accessibility, highlighting the protective quaternary packing of the HIV-1 spike relative to vaccine-induced MAbs.
INTRODUCTION
The envelope glycoproteins (Env) of HIV-1 are large antigens, which despite their effective glycan and conformational shield, expose a number of immunogenic regions to the host immune system. Additional determinants may be exposed by Env immunogens that are imperfect mimics of the functional glycoprotein spike, as are most Env subunit vaccines tested pre-clinically or clinically to date. Generally, primate Abs elicited by Env immunization display narrow neutralizing profiles with limited capacity to block infection of tier 2 viruses. However, intense work in the field suggests that improved Env immunogens are forthcoming and, in anticipation of improved immune responses, it is important to concurrently develop approaches to interrogate the quality of vaccine-elicited responses at a high level of resolution. While serum binding and neutralization are measured in most Env immunogenicity studies, information is more limited regarding the diversity of antibody (Ab) sub-specificities elicited by Env immunization and their relative representation in the polyclonal B cell response.
Considerably more information is available from studies of chronically HIV-1-infected individuals, where neutralizing Ab responses elicited in several subjects are characterized in great detail. Several of these studies illustrate the extraordinarily complex evolutionary pathways required to develop broadly neutralizing Abs (bNAbs) during infection (1–5), emphasizing the challenge to elicit neutralizing breadth following vaccination. Efforts to mimic infection by stimulating vaccine-induced B cell responses to mature along defined pathways to promote the development of bNAbs have been proposed. These approaches are referred to as B cell lineage immunogen design (6) or antibody germline/maturation targeting strategies (7) and are undergoing current hypothesis-driven testing.
While bNAbs capable of neutralizing tier 2 viruses develop in some chronically infected individuals, this process almost invariably takes years to evolve. The development of infrequent broad neutralizing activity is usually preceded by neutralizing Ab responses that are restricted to sensitive tier 1 viruses and autologous tier 2 viruses (8, 9). Ab subspecificities responsible for mediating tier 1 neutralization during chronic HIV-1 replication include “F105-like” CD4 binding site (CD4bs)-directed Abs and variable region 3 (V3)-directed Abs, demonstrated over two decades ago by isolation of infection-induced monoclonal antibodies (10–12) (MAbs). The interest in cloning MAbs from chronically infected individuals has culminated in the recent isolation of several potent and broadly neutralizing MAbs that serve as templates for vaccine design (13–18). In addition, a subset of these bNAbs is capable of suppressing already established infection in experimental animal models (19, 20).
To date, bNAbs have not been elicited by Env immunization, but several studies demonstrate that Abs capable of neutralizing tier 1 viruses are readily induced in experimental systems (21–27) and, as well, in the VAX003 clinical trial (28). In a direct comparison, weaker and less sustained neutralizing Ab titers were detected in the RV144 trial (29) for reasons that are unclear and under investigation. Ab specificities elicited by Env immunization were not defined at the molecular level until relatively recently. Studies now demonstrate the isolation of CD4bs-directed neutralizing Abs from immunized rhesus macaques (30), V3-specific MAbs from Env-inoculated rabbits (31) and isolation of Env-specific MAbs from human subjects enrolled in either the RV144 trial (32, 33) or the GSK PRO HIV-002 trial (34). However, more comprehensive analyses of the genetic and functional properties of MAbs induced by HIV-1 Env immunization are still lacking and our current understanding of Env vaccine-elicited B cell responses are therefore largely based on analyses of polyclonal plasma or serum samples. The low resolution of these analyses provides limited molecular information about the composition of the induced B cell or antibody response.
To begin to understand primate immune responses to Env immunization, we recently investigated the contribution of individual Ab heavy chain V-gene segments in antigen-specific IgG-switched memory B cells from rhesus macaques immunized with soluble HIV-1 Env trimers in adjuvant (35). Using a highly specific flow cytometry-based strategy for single B cell sorting, we demonstrate that the pattern of immunoglobulin heavy chain variable (VH) gene segments usage among Env-specific memory B cells (based on VH sequences from over 500 single cells) is highly diverse, engaging a broad repertoire of VH gene segments similar to that identified in the expressed total IgG-switched memory B cell population. In our current study, we isolated and characterized the genetic properties of 52 cloned Env-specific MAbs, which mapped to distinct epitope regions of Env. We demonstrated that a subset of the MAbs, comprised of MAbs with specificity against the CD4bs or the V3 region, recapitulated the neutralizing activity in the plasma of the animal from which they were cloned. In some instances, these MAbs neutralized viruses beyond the activity measured in the corresponding plasma sample. We further demonstrated that HIV-1 neutralization depends both on epitope recognition on Env and epitope exposure on the matching virus, providing new and detailed information about the B cell response elicited by Env vaccination.
MATERIALS AND METHODS
Animals and ethics statement
Two rhesus macaques (Macaca Mulatta) of Chinese origin, designated F124 and F128, described elsewhere (22), were sampled in the present study. The animals were housed at the AAALAC accredited Astrid Fagraeus Laboratory at Karolinska Institutet. Housing and care procedures were in compliance with the guidelines of the Swedish Board of Agriculture. The facility has been assigned an Animal Welfare Assurance number by the Office of Laboratory Animal Welfare (OLAW) at the National Institute of Health. The Local Ethical Committee on Animal Experiments (Stockholms Norra Djurförsöksetiska Nämnd) (ethical permit number N85/09 and N32/12) approved all procedures. Before inclusion in the study all animals were tested and confirmed negative for simian immunodeficiency virus (SIV), simian T cell lymphotropic virus, and simian retrovirus type D.
Immunization and sampling
A detailed description of the immunization experiment was described previously (22). Briefly, all animals were inoculated intramuscularly five times at monthly intervals with soluble gp140 trimers derived from the HIV-1 YU2 isolate encoding a foldon (F) trimerization motif (hereafter referred to as gp140-F) (36) in an adjuvant formulation consisting of AbISCO-100 (Isconova AB, now Novavax) and CpG-C ODN2395 (Coley/Pfizer). Peripheral blood samples were collected before and after immunization and PBMCs were isolated by density-gradient centrifugation with Ficoll-Hypaque (GE Healthcare) followed by extensive washing in PBS. The PBMCs were then counted and frozen in 90% heat-inactivated FBS and 10% DMSO (Sigma-Aldrich). The MAbs described in the current study were isolated from IgG+ memory B cells after the 4th immunization from macaque F124, and following the 5th immunization from macaque F128.
Expression and purification of Env glycoproteins
The soluble YU2-derived gp140-F trimers (36) used as the immunogen were produced by transient transfection into Freestyle 293F suspension cells (Invitrogen) as previously described (37). The Env ligands used in the binding studies were the following trimeric proteins: gp140-F, gp120-F, gp120-F-ΔV3, gp120-F-ΔV1V2, gp140-F-D368R, and the following monomeric proteins: TriMut, TriMut-368/370, TriMut-368/370/474 and gp140-GCN4 were purified by lentil-lectin and gel filtration chromatography. The biotinylated gp140-F probe used for single cell sorting by flow cytometry was purified by lentil-lectin affinity chromatography and nickel-chelating chromatography (GE Healthcare, Uppsala, Sweden). All probes carried an Avi-tag for site-specific biotinylation at the C-termini of the proteins and biotinylation was performed with biotin ligase Bir A (Avidity, Denver, CO). All Env proteins were from the YU2 strain except the TriMut proteins, which were from HXBc2. The collagen-foldon protein was kindly received from the laboratory of Professor Rikard Holmdahl at Karolinska Institutet, recombinant Ovalbumin (Ova) protein was purchased from Sigma-Aldrich and recombinant influenza hemagglutinin 1 (HA1) was produced as previously described (38).
HIV-1 Env-specific single cell sorting by flow cytometry
The flow cytometric cell sorting details for Env-specific memory B cell isolation from the frozen PBMC samples from immunized macaques F124 and F128 are described elsewhere (35). In brief, Env-specific memory B cells were defined as CD3−, CD8−, Aqua Blue−, CD14−, CD20+, IgG+, CD27+, IgM−, gp140-F+, and were sorted at single-cell density into 96-well PCR plates containing 20 μl of cell lysis buffer (35) using a three-laser FACSAria cell sorter and stored at −80 ° C prior to RT-PCR. In sorts performed for F124, an additional 10 μg/ml carrier RNA (poly-A; Qiagen, CA) was included in the lysis buffer.
Single B cell RT-PCR and MAb cloning
Cell lysates from sorted HIV-1 Env-specific single memory B cells from animals F124 and F128 were used as a source of RNA for reverse transcription and V(D)J sequences were amplified as described previously (35, 39). Briefly, the 96-well plates, containing single B cell, were thawed at room temperature and reverse transcribed to cDNA by addition of random hexamers, dNTPs and SuperScript III reverse transcriptase (Invitrogen). The V(D)J sequences were amplified separately in 25 μl nested PCR reactions using 3 μl of cDNA in the 1st round PCR and 1.5 μl PCR product in the 2nd round PCR. The HotStar Taq Plus Kit (Qiagen, CA) and 5′ leader sequence–specific and 3′ IgG-specific primers were used. PCR products from the positive wells were purified, sequenced (GATC biotech) and analyzed. In frame unique sequences were submitted to GenBank. The productive heavy chain (HC) and light chain (LC) sequences were re-amplified in 25 μl cloning PCR reactions to add the cloning sites using 2 μl nested PCR product with Phusion Hot Start II High-Fidelity PCR enzyme (Thermo Scientific) and 5′ and 3′ custom cloning primers containing restriction sites previously described (39, 40).
Cloning PCR products were evaluated on 1% agarose gel for correct size (~450 bp for HC and ~350 bp for κ or λ LC), and then PCR purified. Cloning of the Ab sequences into expression vectors containing human Igγ1 H, Igκ1 L, or Igλ2 L constant regions (40) were performed with FastDigest restriction enzymes (Thermo Scientific) according to the manufacturer’s instructions. The digested PCR products were inserted into linearized, shrimp alkaline phosphatase-treated vectors using T4 DNA ligase (Thermo Scientific). XL10-Gold ultracompetent cells were then transformed by heat shock at 42 °C for 45 sec according to manufacturer’s protocol (Agilant Technologies). Positive colonies were verified for insert by PCR. Bacterial colonies containing plasmids with inserts of the correct size were then expanded followed by plasmid purification (Qiagen, CA) and Sanger sequencing (GATC biotech).
The Env-specific Ab heavy chain (HC) sequences used in the current study are available under accession numbers KF947536 – KF948098. The matching light chain (LC) sequences described here are available under GenBank accession numbers KP271293 – KP271344 at GenBank (http://www.ncbi.nlm.nih.gov/genbank/).
Expression of cloned MAbs
For Ab expression, 15 μg of each HC and LC vector DNA was transfected into FreeStyle 293-F cells, cultured in 30 ml of FreeStyle 293 expression medium (Life Technologies) at cell density 1 × 106 cells/ml and ≥ 90% viability, using 30 μl of FreeStyle Max reagent (Invitrogen) according to the manufacturer’s protocol. After 4–5 days cell culture supernatants were tested for total Ab production and binding to different HIV-1 Env ligands by ELISA. Cultures containing functional Env-specific Abs were then harvested and purified 7 days after transfection using Protein G Sepharose columns (GE Healthcare). All purified recombinant MAbs were further analyzed by SDS-PAGE under reducing condition using NuPAGE Novex 4–12% Bis-Tris polyacrylamide gels and NuPAGE reducing agent (Life Technologies) according to the manufacturer’s instructions.
Ig gene sequence analysis
Single cell RT-PCR-generated HC and LC sequences were analyzed by using the currently available database of rhesus macaque germline sequences (30) and the IgBLAST tool (41) to identify their V(D)J germline gene segments. CDR3 sequences of HC and LC were extracted with IMGT/V-QUEST (42).
Analysis of Ab sub-specificities
The purified recombinant MAbs were tested for their subspecificities in ELISA binding assays using different Env ligands. MaxiSorp 96-well plates (Nunc) were coated with 2 μg/ml gp140-F, gp120-F, gp140-F-D368R, gp120-F-ΔV3, gp120-F-ΔV1V2, gp140-GCN4, collagen-foldon, Ova or HA1, in PBS overnight at 4°C. Next, wells were washed six times with wash buffer (PBS + 0.05% Tween-20) and then blocked for 1.5 hour at 37 °C with blocking buffer (2% w/v nonfat dry milk, Sigma). The purified MAbs were 5-fold serially diluted in blocking buffer, starting with an initial concentration of 10 μg/ml and incubated for 1.5 hour at 37 °C. The secondary Ab, horseradish peroxidase (HRP)-conjugated goat-anti-rhesus (Nordic MUbio) or anti-human (Jackson ImmunoResearch) IgG Fcγ was added at 1:10,000 dilution in wash buffer followed by 1 hour incubation at room temperature. After washing the wells six times with washing buffer the Ab binding signal was developed for 5 min by adding 3,3,5,5 - tetramethylbenzidine (Life Technologies). The reaction was stopped by adding an equal volume of 1M H2SO4. The absorbance or optical density (OD) was measured at 450 nm. Binding curves were fit by non-linear regression using GraphPad prism version 6 software.
For MAbs that remained unmapped by these probes additional differential binding ELISA assays were performed as described previously (43). Briefly, ELISA plates were coated with 2 μg/ml of either TriMut core gp120 or TriMut core 368/370/474 overnight (ON) at 4°C. The TriMut core contains the I423M, N425K, G431E mutations that inhibit CD4 binding, while TriMut core 368/370/474 possesses three additional mutations in the CD4bs (D368R, E370F and D474A) that eliminate binding by all known CD4bs-directed antibodies. The glycan- specific bNAb, 2G12 or CD4bs-directed bNAb, VRC01, and the non-bNAb, b6, were used as controls in this assay. After blocking the plates with non-fat milk and fetal bovine serum (FBS), the plates were incubated with 5-fold serial dilutions of the NHP MAbs with an initial starting concentration of 10 μg/ml. After washing, HRP-conjugated anti-human IgG (1:5000) was added for detection. The plates were then developed by adding 3,3′,5,5′-tetramethylbenzidine chromogenic substrate (Life Technologies) and stopped by sulfuric acid. Absorbance was measured at 450 nm.
Fine-mapping of the V3-specific MAbs was performed by using twelve 15-mer peptides, overlapping by 11 residues, spanning the V3 region of strain YU2 (Genscript). A full-length YU2 V3 peptide was included as a positive control. All peptides were used at the concentration 2 μg/ml to coat the ELISA plates and the assays were performed as described above, with the exception that the purified V3-directed MAbs were used in 3-fold dilution series in blocking buffer, starting with an initial concentration 10 μg/ml.
Binding assays for DJ263.8, MW965.26, SS1196.01 and JRFL
To assess the binding of isolated MAbs to monomeric gp120 dissociated from pseudoviruses or Env transfected 293T cells, a sandwich ELISA was conducted as previously described (44). Briefly, ELISA plates were coated with the sheep polyclonal antibody D7324, which is specific for the gp120 C5 region. Supernatants containing gp120 shed from DJ263 or JRFL Env transiently expressed in transfected cells from plasmid DNA, or alternatively, pseudovirus lysates of MW965 and SS1196, were added to the ELISA wells and incubated at 37°C for 2 hours to accomplish gp120 capture. ELISA reactions were then developed as described above. The pseudovirus lysates were generated by solubilization in 0.5% Triton X-100 at 37°C for an hour.
HIV-1 neutralization assays
Neutralization assays were performed using a single round infectious HIV-1 Env pseudovirus assay with TZM-bl target cells (45). To determine the Ab concentration and the serum dilution that resulted in a 50% reduction in relative luciferace units (RLU), serial dilutions of the MAbs and the sera were performed and the neutralization dose-response curves were fit by non-linear regression using a 5 parameter hill slope equation using the R statistical software package. Neutralization capacities of MAbs were reported as the Ab concentration resulting in 50% virus neutralization (IC50) whereas the result for sera were reported as the serum neutralization ID50, which is the reciprocal of the serum dilution producing 50% virus neutralization. Diverse HIV-1 virus isolates were used in the neutralization assays. The sources of the Env-encoding plasmids were as follows: ADA, 89.6 and YU2 (Dana Gabuzda, Dana Farber Cancer Institute) SF162 (Leo Stamatatos, Seattle Biomedical Research Institute), JRFL and JRCSF (James Binley, Torrey Pines Institute). The BaL.26 (46) and SS1196.1 (45) Envs were previously described and the clade A DJ263.8 sequence was cloned from virus provided by Francine McCutchan and Vicky Polonis (U.S. Military HIV Research Program). The clade C MW965 Env plasmid was obtained from the AIDS Research and Reagent Repository.
RESULTS
Vaccine-elicited MAbs recognize distinct sub-determinants of Env
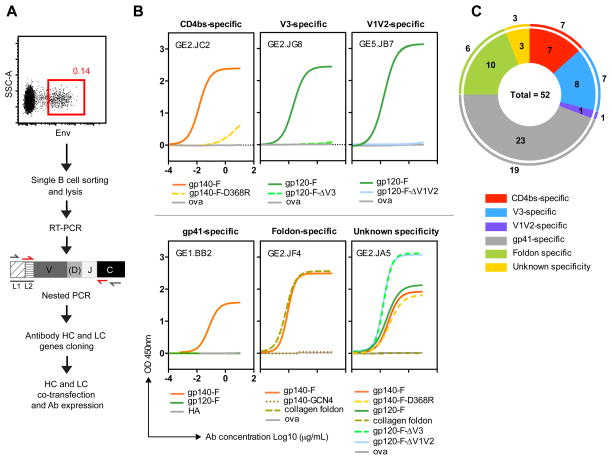
We used a flow cytometry-based strategy for single cell sorting antigen-specific IgG-switched memory B cells from rhesus macaques immunized with soluble HIV-1 gp140-F trimers (herein referred to as Env) administered in adjuvant (35). Individual Env-specific memory B cells were sorted based on expression of cell-surface CD20, CD27 and IgG, as well as by Ag-specific binding to a fluorochrome-conjugated Env trimer probe as previously described (30). As an example, we show a sorting experiment, in which the Env-specific population was found to be 0.14% of the total input cells corresponding to 4% of the IgG+ memory population (Figure 1A). Next, heavy chain V(D)J sequences from sorted Env-specific memory B cells were amplified with nested RT-PCR as previously described (39). Following processing to remove unproductive sequences, V(D)J gene segments were assigned using the currently available database of rhesus macaque germline sequences (30) and IgBLAST followed by CDR3 extraction using IMGT/V-Quest. Of these amplicons, 52 heavy chain VDJ and 52 matching light chain VJ sequences from two NHP donors were cloned and expressed as functional MAbs.
Figure 1. Isolation of Env-vaccine MAbs and epitope mapping.
(A) Schematic of Env-specific memory B cell sorting followed by amplification, cloning, and expression of antibody HC and LC. (B) Representative ELISA binding curves showing the epitope mapping using YU2 Env ligands as follows: gp140-F and gp140-F-D368R (CD4bs-specific), gp120-F and gp120-F-ΔV3 (V3-specific), gp120-F and gp120-F-ΔV1V2 (V1V2-specific), gp140-F and gp120-F (gp41-specific), gp140-F, gp140-GCN4 and collagen foldon (foldon-specific). MAbs that could not be mapped with this set of the probes used here were defined as unknown specificity. Ova or influenza hemagglutinin-1 (HA1) were used as a controls. Titration curves are shown as Log10 dilutions (μg/ml). (C) The pie chart demonstrates the distribution of Env-specific MAbs based on their epitope specificity with the total number of MAbs indicated in the center. The color of each slice represents a different Env specificity: red - CD4bs specific, blue – V3 specific, purple – V1V2 specific, grey – gp41specific, green – foldon specific, orange – unknown specificity, and the area of the slice is proportional to the total number of MAbs against a given specificity. The numbers outside each colored slice indicates the number of unique clones for each of the specificities.
The resulting MAbs were evaluated for binding to HIV-1 gp140-F by ELISA and were confirmed to be immunogen and probe-specific. We next mapped the sub-specificities of the 52 functional MAbs using a set of probes based on the same YU2 HIV-1 strain as the gp140-F trimers used for immunization, all described previously. The probes included the full-length gp140-F glycoprotein, a trimeric form of gp120 (gp120-F) containing a deletion of the gp41 ectodomain sequence from gp140-F and selectively truncated versions of gp120-F with deletions of variable regions 1 and 2 (gp120-F-ΔV1V2) or variable region 3 (gp120-F-ΔV3) and, as well, a gp140-F probe carrying the 368D/R mutation (gp140-F-D368R) that can be used to detect Ab reactivity directed to the gp120 CD4 binding site (CD4bs). All probes were previously described (47, 48). For MAbs that remained unmapped with this set of probes, we included three additional probes, TriMut, TriMut-368/370, TriMut-368/370/474 (21, 43), which allowed the identification of CD4bs-directed MAbs, which were not sensitive to the D368R mutation alone (Supplementary Figure S1). We also included a trimeric gp140 protein stabilized with a heterologous trimerization motif from the GCN4 transcription factor (gp140-GCN4) (49), a trimeric collagen probe stabilized by the foldon motif, collagen-F, and two negative control proteins, influenza hemaglutinin (HA) and ovalbumin (Ova). Collectively, these probes allowed us to map the gp140-F-elicited MAbs to the following possible sub-determinants of Env: the V1V2 region, the V3 region, gp41, the foldon motif and the CD4bs. All binding curves from the mapping studies are shown in Supplementary Figure S1 and representative binding curves are shown in Figure 1B. A summary of the specificities of the isolated MAbs is shown in Figure 1C. The numbers inside each “slice” of the pie chart indicate the total number of MAbs against a given specificity, while the numbers outside each slice indicates the number of unique clones for each of the specificities. From this analysis we mapped one MAb to the V1V2 region, 8 MAbs to the V3 region, 23 MAbs to gp41, 10 MAbs to the foldon motif, 7 MAbs to the CD4bs and 3 MAbs that could not be mapped with this set of probes.
Env-specific MAbs against distinct epitope regions are genetically diverse
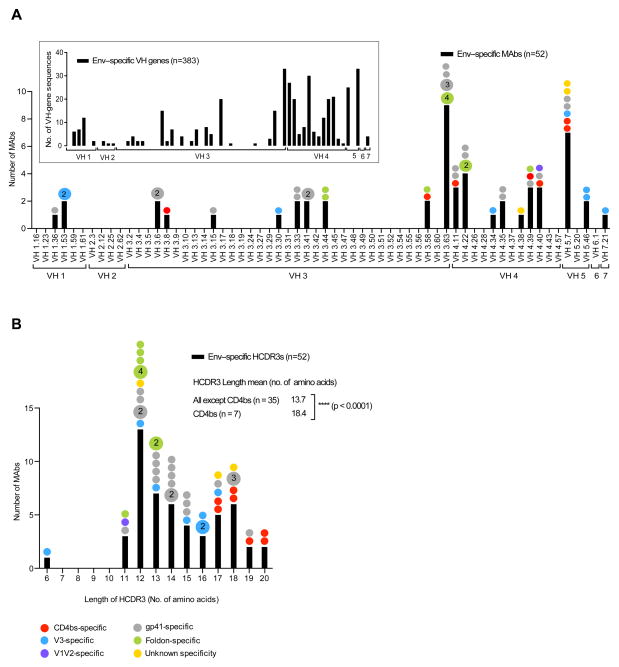
We next examined the genetic properties of the cloned MAbs. We first assigned each antibody heavy and light chain sequence to the closest germline sequence based on the currently available data base for rhesus macaque V(D)J gene segments (30). We then analyzed VH usage and found a marked diversity of gene segment usage among the different Env-specific MAbs, with 21 segments utilized out of a total 61 VH gene segments in the current rhesus macaque database (Figure 2A). The bars in Fig 2 indicate the number of cloned MAbs using a given VH gene segment and the dots above each bar indicate the epitope specificity of expressed MAbs using a given segment. In the cases where more than one somatic variant was isolated, a larger dot indicates the number of variants identified. The VH gene segments expressed by all Env-specific cells isolated from the two rhesus macaques (F124, n=194 and F128, n=189) are shown in the insert of Figure 2A (n=383), illustrating that the distribution of VH gene segments used among the cloned 52 MAbs was similar to that of the total number of HC transcripts sequenced from these animals (35). Most of the Abs expressed in the total B cell memory repertoire used segments from the VH3 and VH4 families and this was also the case for the MAbs isolated here. The most frequently used gene segment, VH3.63, was utilized by B cells expressing BCRs of two different Env subspecificities: gp41-directed and foldon-directed MAbs. In our previous study (35), we detect a significant overrepresentation of the VH5 family (p<0.01, Chi-square test) in the Env-specific B cells compared to the total IgG-switched B cells. In that study, VH5.46 is used with significantly increased frequently in the Env-specific memory B cell pool compared to the total IgG memory B cell pool. Here, we identified two MAbs that used the VH5.46 gene segment, both V3-directed. In addition, we identified another gene segment belonging to the VH5 family, VH5.7, which was frequently used. VH5.7 was represented by three different Env sub-specificities (CD4bs-directed, V3-directed, gp41-directed) and by two unmapped MAbs. Otherwise, we found a broad VH gene usage for all Env sub-specificities suggesting no overall bias toward a given gene segment by any of the epitopes. This is consistent with the notion that Ab HCDR3 region, not predominantly encoded by VH, dictates much of the specificity of typical vaccine-induced antibodies, as shown for previously isolated vaccine-elicited CD4bs-directed MAbs (50, 51).
Figure 2. Vaccine-induced Env-specific MAbs are genetically diverse.
The bars in both (A) and (B) indicate the number of cloned MAbs (Y-axis) using a given VH gene segment (A) or HCDR3 length (B). The colored dots above the bars indicate the epitope specificity of expressed MAbs for a given VH gene segment (A) or HCDR3 length (B). Larger dots indicate the number of clonally related MAbs identified. The VH gene segment expression profile by Env-specific IgG+ memory B cells isolated from the F124 and F128 together (n=383) is shown in the insert box in (A).
We also examined the length of the HCDR3 regions of the cloned MAbs and found that they varied between 11 and 20 amino acids (aa), except for one V3-directed MAb that had a HCDR3 of only 6 aa (Figure 2B). Interestingly, for the CD4bs-directed MAbs there was a bias toward longer HCDR3 regions, between 17–20 aa for the 7 MAbs included in the analysis. In contrast, the foldon-specific MAbs studied here possessed overall shorter HCDR3s, 11–13 aa. There was no bias toward longer or shorter HCDR3 regions for the other sub-specificities for the MAbs examined here. The bias toward longer HCDR3s for the CD4bs-directed MAbs compared to the other Ab sub-specificities was statistically significant (p<0.0001). We also analyzed the HCDR3 length of the MAbs together with the larger set of Env-specific (n=383) and total memory (n=259) HC sequences obtained from single cell isolation and RT-PCR of VDJ transcripts from animals F124 and F128 described in Sundling et al. (35). This analysis demonstrated that the distribution of HCDR3 lengths plotted for the cloned MAbs was representative also of the larger data set, with the majority of the HC sequences encoding HCDR3s of 11–20 aa (Supplementary Figure S2).
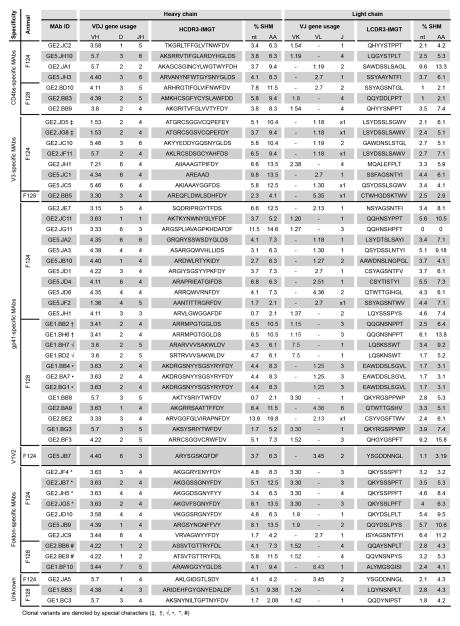
We next examined the light chain V gene usage and found that 25 of the MAbs used kappa chains while 27 MAbs used lambda chains. Several different kappa and lambda V gene segments from VK and VL gene families were used, again consistent with a diverse polyclonal response (Figure 3 and Supplementary Figure S2). The light chain V and J gene usage are described in Figure 3 as well as the heavy chain V, D and J gene usage and characteristics of the HCDR3 and LCDR3 regions. The length of the LCDR3 region was between 8–12 amino acids, which, as expected, is shorter than the HCDR3 regions due to the lack of a D segment. We also asked if any of the MAbs were clonally related based on a definition that clonal relatives have the same V(D)J gene usage and the same length and close identity of the HCDR3. We identified only a few clonally related antibodies in this set of 52 MAbs as indicated in Figure 3. Finally, we calculated the degree of somatic hypermutation (SHM) for each antibody at both the nucleotide (nt) and amino acid level. We found that the average SHM for the heavy chain variable region was 4.9% (nt) and 8.4% (aa) and the average SHM for the light chain variable region was 3.5% (nt) and 6.2 (aa), similar to the level of SHM observed in previous studies of vaccine-elicited antibody responses (30).
Figure 3. Summary of genetic properties of the vaccine-elicited Env-specific MAbs.
V(D)J gene annotations and CDR3 regions of HC and LC of Env-specific MAbs were extracted using IgBLAST and IMGT-VQUEST respectively. Antibody HC and LC sequences with the same V and J gene, identical CDR3 length, and≥ 80% CDR3 identity at the aa level were determined as clonal variants and are denoted by special characters: GE2.JD5‡ and GE2.JG8‡; GE1.BB2† and GE1.BH6†; GE1.BH7√ and GE1.BD2√; GE1.BB4•, GE2.BA7•, and GE2.BG1•; GE2.JF4*, GE2.JB7*, GE2.JH5*, and GE2.JG5*; GE2.BB6# and GE2.BE8#.
Collectively these data illustrate the genetic diversity and level of SHM of the Env vaccine-elicited Ab response, as well as the diversity among Abs recognizing specific Env epitope regions.
CD4bs- and V3-directed MAbs display distinct HIV-1 neutralization signatures
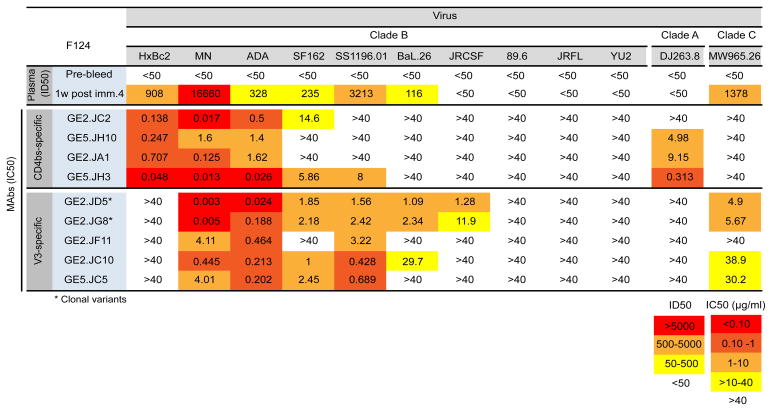
To examine the neutralizing capacity of the 52 vaccine-induced MAbs analyzed here, we chose a panel of commonly used HIV-1 Env pseudoviruses. The panel included “easy-to-neutralize” viruses represented by tier 1 viruses including HxBc2, MN, ADA and SF162 (all clade B), DJ263 (clade A) and MW965 (clade C); viruses that display an intermediate neutralization phenotype, classified as tier 1B viruses, including SS1196, BaL.26, JRCSF and 89.6 (clade B) and neutralization-resistant tier 2-like viruses, considered representative of primary circulating strains, such as JRFL and YU2 (clade B) (52).
The vaccine-elicited neutralizing MAbs isolated here were either CD4bs- or V3-specific, while no neutralizing activity was detected for the gp41-, foldon- or V1V2-specific MAbs. We therefore focused our subsequent analyses on animal F124, from which four CD4bs-directed MAbs were neutralizing and five of seven V3-directed MAbs were neutralizing. The neutralizing activities of these MAbs are quantified by the IC50 values shown in Figure 4. In brief, we observed distinct neutralization signatures depending on the sub-specificity of the Abs. All the CD4bs-directed MAbs neutralized HxBc2 and all but one neutralized the clade A virus, DJ263. These two viruses were not neutralized by any of the V3-directed MAbs. Five viruses in the panel; SF162, SS1196, BaL.26, JRCSF and MW965 were neutralized by all or most of the V3-directed MAbs, while these viruses were not neutralized by the CD4bs-directed MAbs, except in a few isolated cases. Two viruses in this panel, MN and ADA, were neutralized by both the CD4bs- and V3-directed MAbs. One of the CD4bs-directed MAbs, GE5.JH3, displayed superior potency reaching low (<0.05) IC50 values against HxBc2, MN and ADA. GE5.JH3 also neutralized two of the V3-sensitive viruses (SF162 and SS1196) not neutralized by the other CD4bs-directed MAbs, indicating a potentially unique mode of interaction with the viral spike for this MAb.
Figure 4. HIV-1 neutralizing activity of Env-specific MAbs.
The pre- and post-immunization plasma, and the CD4bs-directed and V3-specific MAbs isolated from animal F124 were tested for their capacities to neutralize a panel of Env-pseudoviruses. The plasma dilution and the MAb concentration inhibiting 50% of viral entry are shown as neutralization ID50 and IC50 values respectively. The ID50 values are color-coded where red indicates (>5000), orange (500–5000), yellow (50–500), and white (<50) whereas the IC50 values in red indicates (<0.1 μg/ml), dark orange (0.1–1 μg/ml), light orange (1–10 μg/ml), yellow (10–40 μg/ml) and white (>40 μg/ml).
We also measured the neutralizing activity of the unfractionated plasma from animal F124, shown as ID50 values. We included one pre-bleed sample and one sample collected one week after the 4th immunization, the same time point from which the MAbs were isolated. Six viruses (HxBc2, MN, ADA, SF162, SS1196 and MW965) were neutralized by the post-4 immunization plasma sample, while there was no detectable neutralizing activity in the pre-bleed control sample. These data demonstrate that all neutralizing activity measured in the plasma of F124 could in part be accounted for by the CD4bs- and V3-specific MAbs isolated from the same animal.
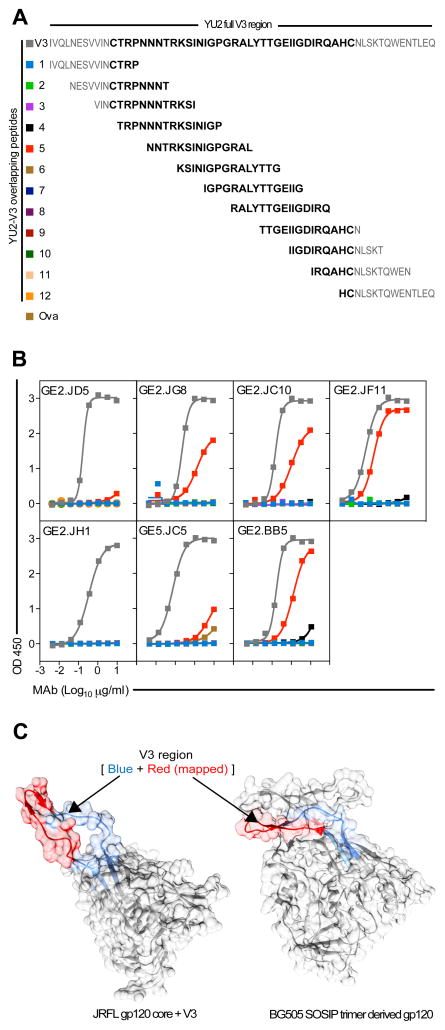
The V3-specific MAbs target the crown and flanking N-terminal region
To determine specifically within V3 the V3 region-directed MAbs bind, we performed binding studies using the full length V3 peptide and 15-mer peptides overlapping by 11 residues (Figure 5A). We included the six V3-specific MAbs from animal F124 (all except GE5.JC1) and one neutralizing V3-specific MAb from animal F128. All seven MAbs bound the full-length peptide, confirming their specificities for V3. Of the 15-mer peptides, only one peptide, peptide 5, was efficiently recognized by the vaccine-elicited MAbs, specifically by GE2.JC10, GE2.JF11, GE2.JG8, GE2.BB5 and GE5.JC5. GE2.JD5 bound peptide 5 only very weakly despite it being a clonal variant of GE2.JG8 suggesting a role for differences in somatic hypermutation between these MAbs (Figure 5B). These data suggest that a major part of the epitope is contained within the peptide 5 region. To examine the exposure of this epitope region in the context of the three-dimensional architecture of HIV-1 Env, we used the structure of the soluble CD4-complexed JRFL gp120 core containing the V3 region (53) compared to the core and V3 extracted from the structure of the cleaved, soluble BG505 SOSIP gp140 trimers (54) (PDB IDs- 2B4C and 4NCO). As shown in Figure 5C, the peptide 5 region is readily exposed on the post-CD4 JRFL core conformation, whereas, in contrast, the V3 region is occluded in the BG505 SOSIP structure, which is in the pre-CD4 triggered state. In this native-like conformation, the V1, V2 and V3 regions form a cap at the apex of the trimer, precluding recognition of this V3 epitope in this context and by inference, on the functional HIV-1 primary isolate spike.
Figure 5. Illustration of the epitope region targeted by the vaccine-elicited V3-directed MAbs.
(A) Amino acid sequences of the full-length YU2 V3 peptide (indicated in bold letters) and twelve 15-mer peptides overlapping by 11, where each peptide is indicated by a number and a color (B) Binding curves of the isolated V3-directed MAbs to the overlapping peptides. (C) A surface rendered crystal structures of two HIV-1 Envs - CD4-complexed JRFL gp120 core (PDB ID - 2B4C) and the cleaved, soluble BG505.SOSIP gp120 in native state (PDB ID – 4NCO) (gray) generated with Chimera (66) indicate structural rearrangement of the V3 loop (blue + red) in the post-CD4 binding (JRFL gp120 core) and pre-CD4 triggered (BG505.SOSIP) conformations. The peptide region to which the V3-specific MAbs bind is shown in red while the rest of the V3 region is shown in the blue.
HIV-1 neutralization depends on both epitope recognition and epitope exposure
To further define criteria for Ab neutralization, we focused on three strains, DJ263, MW965 and SS1196, for which we observed a clear difference in the ability of the CD4bs- and V3-directed MAbs to accomplish neutralization. We reasoned that the capacity of an Ab to neutralize depends on both 1) the presence of the cognate epitope on the target Env, assessed by measuring binding capacity and 2) the exposure of the epitope on the functional virus spike, assessed by measuring neutralizing capacity. To investigate the relative contribution of recognition and exposure, we used supernatants or lysates of cells transfected with the respective Envs to provide non-functional, monomeric forms of the gp120 ligands for use in binding studies. We selected three of the V3-specific MAbs and three of the CD4bs-specific MAbs for these analyses.
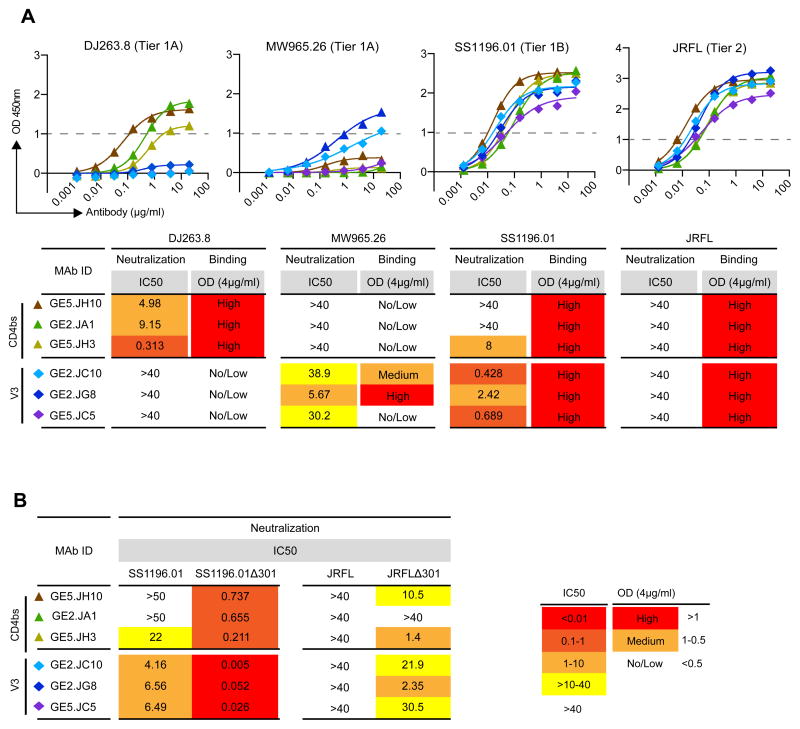
Starting with the DJ263 virus, we found that all three CD4bs-directed MAbs bound to monomeric DJ263-derived gp120, while the V3-specific MAbs exhibited no or very low binding affinity to this gp120 (Figure 6A, upper panels). These results indicate that the inability of the V3-specific MAbs to neutralize DJ263 is due to a lack of the cognate epitope on gp120. Interestingly, inspection of the primary amino acid sequence of DJ263 shows that peptide 5 is conserved in this Env (Supplementary Figure S3), thus it is possible that differences outside of the DJ263 peptide 5 contribute to recognition by these YU2-derived V3-directed MAbs. In contrast, the CD4bs-directed MAbs bind the monomeric Env and neutralize the functional virus spike demonstrating that the cognate CD4bs epitope is both present and available for Ab binding on this virus. For MW965, binding of monomeric gp120 and an association to neutralize virus was observed for 2 of the 3 V3-specific MAbs, while a lack of binding was associated with a similar lack of neutralizing capacity for the CD4bs-directed MAbs (Figure 6A, middle panels). Overall, for the “easy-to-neutralize” tier 1A viruses DJ263 and MW965, the antibody binding profile to monomeric gp120 correlated well with the neutralizing activity.
Figure 6. Comparison of MAb binding and neutralizing properties.
(A) Upper panels: ELISA binding curves of the vaccine-induced CD4bs- and V3-directed MAbs to monomeric gp120 from DJ263.8, MW965.26, SS1196.01, and JRFL are shown. Lower panels: A summary of gp120 binding activity and virus neutralizing activity for DJ263.8, MW965.26, SS1196.01, and JRFL are shown. Binding activity is indicated as OD values at Ab concentration, 4 μg/ml, where red indicates High (OD >1), light orange indicates Medium (OD 1–0.5), and white indicates No/Low (OD <0.5) binding. Virus neutralizing activity is indicated as IC50 values and is color-coded as described in Figure 4. (B) Comparison of neutralizing activity for SS1196.01 and SS1196.01Δ301 and for JRFL and JRFLΔ301.
For SS1196, we detected a different relationship between monomeric binding and virus neutralization. Efficient binding to gp120 by both the V3- and CD4bs-directed MAbs was detected by ELISA, which was associated with virus neutralization by all three V3-specific MAbs, but for only one of the CD4bs-directed MAbs, GE5.JH3 (Figure 6A, lower panels). From these results we interpret that SS1196 gp120 contains the epitope for both the V3- and the CD4bs-directed MAbs, but that the V3 region is exposed on the SS1196 functional spike while the CD4bs is less accessible. This is consistent with its classification as a tier 1B virus, which is more neutralization-resistant presumably due to steric hindrance of certain neutralizing Ab epitopes by quaternary spike packing. Furthermore, monomeric JRFL gp120 was also well recognized by both the CD4bs- and V3-directed MAbs, but none of these antibodies neutralized the JRFL virus, consistent with the tight packing of tier 2 virus Env spikes and their resistance to current vaccine-elicited V3- and CD4bs-directed MAbs.
To further investigate the question of recognition versus exposure, we generated a variant of SS1196 that lacked the site for N-linked glycosylation at position 301 and we compared the ability of the V3- and CD4bs-directed MAbs to neutralize the parental SS1196 virus compared to the SS1196Δ301 variant. We found that SS1196Δ301 was sensitive to the CD4bs-directed MAbs and displayed greatly increased sensitivity to the V3-directed MAbs. These data illustrate that glycan shielding at this position effectively prevents recognition of the functional spike by CD4bs-directed Abs and partly prevents recognition of V3-directed Abs. We performed a similar comparison of binding versus neutralization using the JRFL strain, which was not neutralized by any of the Env-elicted MAbs (Figure 4), but for which there already existed an engineered viral variant lacking a site for N-linked glycosylation at position 301 (JRFLΔ301). We previously showed that removal of the 301 glycan rendered JRFL sensitive to several non-bNAb CD4bs-directed MAbs (30, 55), illustrating effective glycan shielding at this position. Here, we found that all V3- and two of the CD4bs-directed MAbs neutralized the JRFLΔ301 virus (Figure 6B), suggesting a close relationship between exposure of the CD4bs and the V3 region on tier 2 viruses. In sum, these studies suggest that the capacity to achieve neutralization depends both on the presence of the cognate epitope on the specific Env gp120 and relative accessibility of a given epitope in the context of the native, functional virus spike.
DISCUSSION
Successful vaccination stimulates polyclonal B cell responses directed against multiple epitopes on target antigens of a given pathogen. The precise specificities of antibodies that mediate protection by anti-viral vaccines are usually not known as this level of resolution requires the isolation of monoclonal antibodies from vaccine recipients, an endeavor that is rarely undertaken if the vaccine is considered to be effective. In contrast, for vaccines that do not induce desired Ab responses, such as the HIV-1 Env immunogens studied to date, MAb isolation provides concrete information about the fine specificities of the response. This analysis both increases our understanding of biological events taking place following vaccination and reveals the fine specificities that are induced by a given immunogen (30, 50, 56). This information can guide the design of improved immunogens aimed to favorably alter the elicited B cell response to accelerate the development of HIV-1 vaccine candidates. Here, where we demonstrate engagement of a broad set of VH genes to multiple specificities, but such analysis is also ongoing for immunogens designed to target specific VH genes, such as the B cell germline engagement immunogen design approach (6).
In the current study, we isolated a set of MAbs elicited by immunization with a model Env trimer immunogen (22). We sorted single Env-specific memory B and sequenced their V(D)J transcripts, revealing a highly polyclonal, Env-specific VH repertoire usage. We cloned matching HC and LC pairs resulting in 52 Env-specific MAbs, of which 25% were neutralizing. The neutralizing MAbs, which were either CD4bs- or V3-specific, in part recapitulated the neutralizing activity observed in the plasma of the animal from which they were isolated. Interestingly, the CD4bs-directed MAbs neutralized DJ263, a virus that was not detectably neutralized by the plasma. Similarly, two of the V3-directed MAbs neutralized JRCSF, which was not neutralized by the plasma. These results demonstrate that enrichment of distinct specificities through MAb isolation can reveal additional neutralizing activities, providing considerably more information from a given pre-clinical or clinical vaccine trial. Furthermore, the higher resolution provided by MAb isolation confirms that Env-based vaccines can elicit CD4bs- and V3-directed neutralizing Abs and is consistent with studies using adsorption and competition assays on unfractionated plasma or serum samples (21, 48, 57–59). Among the cloned MAbs, additional Env sub-specificities identified were composed of gp41 (n=23) and the V1V2 region (n=1). The latter MAb may be worthy of further investigation, since studies of the RV144 clinical trial suggested a correlation between V1V2-directed antibodies and vaccine-induced protection (60, 61) stimulating a renewed interest in MAbs against this determinant (33).
The current study reveals that, remarkably, all V3-specific MAbs were directed against a linear determinant on the N-terminal flank of the V3 crown. Consistent with this specificity, this region is expected to be exposed on V3-sensitive tier 1 viruses such as MN, ADA and SF162. Interestingly SS1196, classified as a tier 1B virus, was also neutralized by the V3-specific MAbs, but not by most the CD4bs-directed MAbs, suggesting an intermediate Env conformation between tier 1A and tier 2 viruses. By comparing binding versus neutralization of SS1196 gp120 by selected MAbs, we demonstrated that the CD4bs-directed MAbs recognized their cognate epitope, but were unable to access it on the functional spike, consistent with epitope masking of conserved regions on more neutralization resistant viruses. Interestingly, a variant of SS1196 lacking the site for N-linked glycosylation at position 301, SS1196Δ301, was sensitive to the CD4bs-directed MAbs and displayed greatly increased sensitivity to the V3-directed MAbs, illustrating that glycan shielding at this position effectively prevents recognition of the functional spike by CD4bs-directed Abs and partly prevents recognition of V3-directed Abs. Similarly, the inability of the V3-specific MAbs to neutralize tier 2 viruses such as JRFL is likely due to steric hindrance, since removal of the N-linked glycan at position 301 of JRFL to generate the JRFLΔ301 virus, rendered this tier 2 virus sensitive to both V3- and CD4bs-directed MAbs. These data are consistent with the structural data of the soluble SOSIP trimers that indicate that these two epitope regions are proximal to one another. In contrast, the lack of HXBc2 neutralization by the V3-specific Abs can be explained by a mismatch of the V3 regions between these Envs as HxBc2 gp120 possesses an insertion of two residues near its V3 crown (Supplementary Figure S3). Collectively, these studies show that the availability of MAbs targeting different epitopes on Env and their use in functional assays can greatly facilitate our understanding of HIV-1 neutralizing determinants and relative exposure/accessibility.
Studies of infection-induced V3 MAbs in humans show an overrepresentation of antibodies using the human VH5-51 gene segment (62). Human VH5-51-using MAbs are non-broadly neutralizing and recognize their epitope on V3 in a manner referred to as the “cradle mode” (63). The cradle mode contrasts with the “ladle mode” used by the relatively broadly neutralizing, V3-directed MAb, 447-52D (64). The orthologue of the human VH5-51 gene segment in rhesus macaques is VH5.7, used by one of our V3-specific MAbs, GE2.JF11. Studies of V3-directed MAbs elicited in Env-immunized rabbits resulted in the isolation of the R56 MAb, which binds by the cradle mode via an epitope that overlaps with peptide 5 identified as the target epitope in our current study (31, 65). R56 was shown to neutralize a similar set of viruses as the V3-specific rhesus MAbs isolated here, suggesting that the V3-specific MAbs identified in our study bind by a similar mode of recognition. In contrast, PGT128, a human bNAb isolated from a chronically infected individual, binds further toward the C-terminal part of V3 to an epitope that also includes the basal N-linked glycans (16).
The CD4bs-specific MAbs isolated in this study display similar neutralizing properties to previously described vaccine-elicited CD4bs-specific MAbs, GE136, GE148 and GE356 (30, 50). Based on mutagenesis and binding studies to define the antibody-antigen interface combined with modelling studies these MAbs attempt to bind the Env spike by a vertical angle of approach that is not permitted on tier 2 viruses, thus limiting their neutralization capacities to tier 1 viruses only (50, 51). A similar mode of binding is likely for the CD4bs-directed MAbs isolated here. Similarly to GE136, GE148 and GE356, the CD4bs-directed MAbs described here possess long HCDR3 regions compared to the MAbs recognizing other epitope regions, suggesting that this property is required for access to the CD4bs. Interestingly, one of the CD4bs-directed MAbs, GE5.JH3, was more potent and displayed a modest increased in breadth of neutralization compared to the other vaccine-induced CD4bs-specific MAbs, suggesting a different mode of interaction with the functional spike. Whether Ab specificities displaying broader neutralizing activities exist at some low frequency in these animals is unknown but can be addressed with more selective strategies for B cell sorting.
In sum, the vaccine-induced MAbs described here reveal new information about how the primate B cell repertoire responds to Env immunization and demonstrate the utility of MAb isolation for defining the neutralizing Ab activity induced by vaccination. The availability of MAbs against defined epitope regions of HIV-1 Env provides broad utility to probe the antigenic and immunogenic surfaces of existing and forthcoming subunit Env-based vaccine candidates.
Supplementary Material
Acknowledgments
We thank Dr. Martin Corcoran for critically reading the manuscript.
We gratefully acknowledge financial support from the Swedish Research Council, Karolinska Institutet, the National Institutes of Health (NIH) grants P01 1P01AI104722-01A1, the NIH intramural research program and the International AIDS Vaccine Initiative (IAVI). IAVI’s funding is made possible by generous support from many donors with the full list of IAVI donors available at www.iavi.org.
References
- 1.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, Lynch RM, Alam SM, Moody MA, Ferrari G, Berrong M, Kelsoe G, Shaw GM, Hahn BH, Montefiori DC, Kamanga G, Cohen MS, Hraber P, Kwong PD, Korber BT, Mascola JR, Kepler TB, Haynes BF. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, Hahn BH, Shaw GM. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology. 2009;387:414–426. doi: 10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho DD, McKeating JA, Li XL, Moudgil T, Daar ES, Sun NC, Robinson JE. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J Virol. 1991;65:489–493. doi: 10.1128/jvi.65.1.489-493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsushita S, Robert-Guroff M, Rusche J, Koito A, Hattori T, Hoshino H, Javaherian K, Takatsuki K, Putney S. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J Virol. 1988;62:2107–2114. doi: 10.1128/jvi.62.6.2107-2114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posner MR, Hideshima T, Cannon T, Mukherjee M, Mayer KH, Byrn RA. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J Immunol. 1991;146:4325–4332. [PubMed] [Google Scholar]
- 13.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011 doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013 doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Lifson JD, Dimitrov D, Nussenzweig MC, Martin MA. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013 doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti B, Feng Y, Kumar S, McKee K, Karlsson GB, Hedestam LC, Montefiore MJR, DCWRT Robust Neutralizing Antibodies Elicited by HIV-1 JRFL Envelope Glycoprotein Trimers in Non-human Primates. 2013 doi: 10.1128/JVI.01247-13. Manuscript in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundling C, Forsell MN, O’Dell S, Feng Y, Chakrabarti B, Rao SS, Lore K, Mascola JR, Wyatt RT, Douagi I, Karlsson Hedestam GB. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacs JM, Nkolola JP, Peng H, Cheung A, Perry J, Miller CA, Seaman MS, Barouch DH, Chen B. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci U S A. 2012;109:12111–12116. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sellhorn G, Kraft Z, Caldwell Z, Ellingson K, Mineart C, Seaman MS, Montefiori DC, Lagerquist E, Stamatatos L. Engineering, expression, purification, and characterization of stable clade A/B recombinant soluble heterotrimeric gp140 proteins. J Virol. 2012;86:128–142. doi: 10.1128/JVI.06363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear WB, Schuman JT, Kraft Z, O’Malley J, Mori M, Srivastava I, Barnett S, Stamatatos L, Haigwood NL. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85:5262–5274. doi: 10.1128/JVI.02419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bontjer I, Melchers M, Tong T, van Montfort T, Eggink D, Montefiori D, Olson WC, Moore JP, Binley JM, Berkhout B, Sanders RW. Comparative Immunogenicity of Evolved V1V2-Deleted HIV-1 Envelope Glycoprotein Trimers. PLoS One. 2013;8:e67484. doi: 10.1371/journal.pone.0067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moody MA, Santra S, Vandergrift NA, Sutherland LL, Gurley TC, Drinker MS, Allen AA, Xia SM, Meyerhoff RR, Parks R, Lloyd KE, Easterhoff D, Alam SM, Liao HX, Ward BM, Ferrari G, Montefiori DC, Tomaras GD, Seder RA, Letvin NL, Haynes BF. Toll-like receptor 7/8 (TLR7/8) and TLR9 agonists cooperate to enhance HIV-1 envelope antibody responses in rhesus macaques. J Virol. 2014;88:3329–3339. doi: 10.1128/JVI.03309-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman PW, Huang W, Riddle L, Gray AM, Wrin T, Vennari J, Johnson A, Klaussen M, Prashad H, Kohne C, deWit C, Gregory TJ. Development of bivalent (B/E) vaccines able to neutralize CCR5-dependent viruses from the United States and Thailand. Virology. 1999;265:1–9. doi: 10.1006/viro.1999.0031. [DOI] [PubMed] [Google Scholar]
- 29.Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O’Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. High-Resolution Definition of Vaccine-Elicited B Cell Responses Against the HIV Primary Receptor Binding Site. Sci Transl Med. 2012;4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Vaine M, Wallace A, Han D, Wan S, Seaman MS, Montefiori D, Wang S, Lu S. A novel rabbit monoclonal antibody platform to dissect the diverse repertoire of antibody epitopes for HIV-1 Env immunogen design. J Virol. 2013;87:10232–10243. doi: 10.1128/JVI.00837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang KK, Gilbert PB, Huang Y, Gurley TC, Kozink DM, Marshall DJ, Whitesides JF, Tsao CY, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Kim JH, Michael NL, Tomaras GD, Montefiori DC, Lewis GK, DeVico A, Evans DT, Ferrari G, Liao HX, Haynes BF. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, Yu JS, Zhang R, Meyerhoff RR, Parks R, Scull JC, Wang L, Vandergrift NA, Pickeral J, Pollara J, Kelsoe G, Alam SM, Ferrari G, Montefiori DC, Voss G, Liao HX, Tomaras GD, Haynes BF. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. J Virol. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundling C, Zhang Z, Phad GE, Sheng Z, Wang Y, Mascola JR, Li Y, Wyatt RT, Shapiro L, Karlsson Hedestam GB. Single-Cell and Deep Sequencing of IgG-Switched Macaque B Cells Reveal a Diverse Ig Repertoire following Immunization. J Immunol. 2014 doi: 10.4049/jimmunol.1303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forsell MN, Dey B, Morner A, Svehla K, O’Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, Karlsson Hedestam GB, Wyatt RT. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 2008;4:e1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundling C, Martinez P, Soldemo M, Spangberg M, Bengtsson KL, Stertman L, Forsell MN, Karlsson Hedestam GB. Immunization of macaques with soluble HIV type 1 and influenza virus envelope glycoproteins results in a similarly rapid contraction of peripheral B-cell responses after boosting. J Infect Dis. 2013;207:426–431. doi: 10.1093/infdis/jis696. [DOI] [PubMed] [Google Scholar]
- 39.Sundling C, Phad G, Douagi I, Navis M, Karlsson Hedestam GB. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J Immunol Methods. 2012;386:85–93. doi: 10.1016/j.jim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Y, McKee K, Tran K, O’Dell S, Schmidt SD, Phogat A, Forsell MN, Karlsson Hedestam GB, Mascola JR, Wyatt RT. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site. J Biol Chem. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, O’Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. Mechanism of Neutralization by the Broadly Neutralizing HIV-1 Monoclonal Antibody VRC01. J Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, Barnett SW, Nabel GJ, Mascola JR. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine. 2007;25:1398–1408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dosenovic P, Chakrabarti B, Soldemo M, Douagi I, Forsell MN, Li Y, Phogat A, Paulie S, Hoxie J, Wyatt RT, Karlsson Hedestam GB. Selective expansion of HIV-1 envelope glycoprotein-specific B cell subsets recognizing distinct structural elements following immunization. J Immunol. 2009;183:3373–3382. doi: 10.4049/jimmunol.0900407. [DOI] [PubMed] [Google Scholar]
- 48.Douagi I, Forsell MN, Sundling C, O’Dell S, Feng Y, Dosenovic P, Li Y, Seder R, Lore K, Mascola JR, Wyatt RT, Karlsson Hedestam GB. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J Virol. 2010;84:1683–1695. doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol. 2000;74:4746–4754. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Navis M, Tran K, Bale S, Phad GE, Guenaga J, Wilson R, Soldemo M, McKee K, Sundling C, Mascola J, Li Y, Wyatt RT, Karlsson Hedestam GB. HIV-1 receptor binding site-directed antibodies using a VH1-2 gene segment orthologue are activated by Env trimer immunization. PLoS pathogens. 2014;10:e1004337. doi: 10.1371/journal.ppat.1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Karlsson Hedestam GB, Wyatt RT. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313:387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 56.Tran K, Poulsen C, Guenaga J, de Val Alda N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Karlsson Hedestam GB, Wyatt RT. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingale J, Tran K, Kong L, Dey B, McKee K, Schief W, Kwong PD, Mascola JR, Wyatt RT. Hyperglycosylated stable core immunogens designed to present the CD4 binding site are preferentially recognized by broadly neutralizing antibodies. J Virol. 2014;88:14002–14016. doi: 10.1128/JVI.02614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morner A, Douagi I, Forsell MN, Sundling C, Dosenovic P, O’Dell S, Dey B, Kwong PD, Voss G, Thorstensson R, Mascola JR, Wyatt RT, Karlsson Hedestam GB. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. J Virol. 2009;83:540–551. doi: 10.1128/JVI.01102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, Wenschuh H, Zerweck J, Greene K, Gao H, Berman PW, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Robb ML, Michael NL, Kim JH, Zolla-Pazner S, Haynes BF, Mascola JR, Self S, Gilbert P, Montefiori DC. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One. 2013;8:e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorny MK, Wang XH, Williams C, Volsky B, Revesz K, Witover B, Burda S, Urbanski M, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S, Nadas A. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol. 2009;46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorny MK, Sampson J, Li H, Jiang X, Totrov M, Wang XH, Williams C, O’Neal T, Volsky B, Li L, Cardozo T, Nyambi P, Zolla-Pazner S, Kong XP. Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure. PLoS One. 2011;6:e27780. doi: 10.1371/journal.pone.0027780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447–52D. Structure. 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Pan R, Sampson JM, Chen Y, Vaine M, Wang S, Lu S, Kong XP. Rabbit anti-HIV-1 monoclonal antibodies raised by immunization can mimic the antigen-binding modes of antibodies derived from HIV-1-infected humans. J Virol. 2013;87:10221–10231. doi: 10.1128/JVI.00843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE Principal Investigators PG. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.